Abstract
The animal gut serves as a primary location for the complex host-microbe interplay that is essential for homeostasis and may also reflect the types of ancient selective pressures that spawned the emergence of immunity in metazoans. In this review, we present a phylogenetic survey of gut host-microbe interactions and suggest that host defense systems arose not only to protect tissue directly from pathogenic attack but also to actively support growth of specific communities of mutualists. This functional dichotomy resulted in the evolution of immune systems much more tuned for harmonious existence with microbes than previously thought, existing as dynamic but primarily cooperative entities in the present day. We further present the protochordate Ciona intestinalis as a promising model for studying gut host-bacterial dialogue. The taxonomic position, gut physiology and experimental tractability of Ciona offer unique advantages in dissecting host-microbe interplay and can complement studies in other model systems.
Keywords: gut immune evolution, gut host-microbe interactions, Ciona intestinalis, gut immune effectors, mutualism, gut biofilms
Introduction
Complex communities of microbes colonize nearly every epithelial surface of animals, including mucosal epithelial surfaces that become highly valued locales for microbial attachment and growth immediately upon birth (Neish, 2014). The gastrointestinal (GI) system, in particular, has emerged as a prime example of an epithelium-lined organ that sustains an abundant and complex community of closely associated, generally species-specific microbiota, which can exert enormous physiological influence on animal hosts (Savage, 1977; Falk et al., 1998; Neish, 2009; Lozupone et al., 2012; Yatsunenko et al., 2012). The host-microbe interactions at the surface of the epithelium are benign or beneficial as a whole and generally are tolerated by the host, although pathogenic breach of the epithelium can result in strong host immune responses. Understanding the multiple types of microbe-host interactions in the gut may provide insight as to the types of selective pressures that spawned the emergence and elaboration of the immune systems of metazoan animals.
From an ecological standpoint, the alimentary canal (or GI system) is an external environment that passes through animals, generally via two openings (i.e., in and out). In the simplest terms, this canal evolved to deliver water and nutritional sustenance to an increasingly complex and generally sterile body. Although this function is essential for life in nearly all animals, such a direct juxtaposition of substances from the external environment inside the body nevertheless poses an enormous challenge for the host. Within the gut tube, the host encounters an essentially continuous microbiological load that can include pathogens as well as a myriad of either beneficial or innocuous species. This complexity is enhanced by the presence of distinctly differentiated compartments within the gut tube, each with potentially distinct and permanent commensal communities (Faust et al., 2012; Koren et al., 2013; Segata et al., 2012). One often studied example is the human colon, which contains several compartments, including the often controversial cecal appendix, and hosts up to 100 trillion (1014) bacterial cells at any given time, in effect generating orders of magnitude greater intraorganismal genetic complexity than that of the host genome itself (Andersson et al., 2008; Costello et al., 2009; Eckburg et al., 2005; Gill et al., 2006; Human Microbiome Project, 2012; Ley et al., 2006; Macfarlane and Macfarlane, 1997; Savage, 1977;). Reduced diversity and/or shifts in the composition of microbiota among different epithelial compartments can dramatically affect homeostasis and host health, in some cases promoting infections that endanger host wellbeing (Chan et al., 2013; Fallucca et al., 2014; Luckey et al., 2013; Mondot et al., 2013; Petrof et al., 2013; Power et al., 2014; Shim, 2013). Further, in mammals, the direct epithelial dialogue with bacteria is generally critical for immune maturation (Cebra, 1999; Cebra et al., 1998; Sommer and Backhed, 2013), and often can provide tolerogenic signals to the host (Lee, 2009; McFall-Ngai et al., 2013; Shin et al., 2011). It might seem at the outset that the monumental task of managing the complex microbial milieu of the alimentary canal would be performed by a multilayered system of immune attack that is effectively repelling an ever changing array of invaders intent on circumventing its defenses. Indeed, the gut of animals evolved so that its epithelial surface is fully immunocompetent, serving as a barrier to potential pathogens while simultaneously recognizing and communicating with microbes in both the lumen (inside) of the gut and the intra-organismal tissue that is generally sterile. [e.g., (Bosch et al., 2009; Duerkop et al., 2009; Duerr and Hornef, 2012; Pott and Hornef, 2012)]. This function is carried out in large part by immunocytes, which migrate to tissue spaces adjacent to the epithelial barrier early during development. In some species, these immunocytes form dense communities and produce an outward appearance of a defense system poised to attack, analogous to an army preparing to repel an invasion. In vertebrates, this tissue space is well recognized for the presence of gut-associated lymphoid tissue (GALT), one of the most prominent immune tissues (Pearson et al., 2012; Shields, 2000). Whereas certain aspects of host immunity certainly do involve these aggressive characteristics, an increasing amount of emerging data indicate that the system as a whole may be much more tuned for harmonious existence with the microbes it encounters and may have evolved such relationships in this cooperative manner (Eberl, 2010; Equileor and Ottaviani, 2011; Loker, 2012; Nyholm and Graf, 2012; Thomas and Parker, 2010).
In this review, we use an immune-phylogeny approach (Fig. 1) to argue that host defense systems have met the challenge of managing microbial communities by evolving strategies that not only directly protect the host's tissue from pathogenic infection but also actively support the growth of specific communities of microbes such that the maturation of these barriers becomes as much, if not more so, of a “welcome wagon” for mutualists in the lumen as opposed to a buildup for war against pathogens. This dichotomy serves to protect against pathogenic invasion, to ensure proper nutritional sustenance to the host and to train the host's immunological systems. The host-microbial dialogue, as part of the metaorganism or holobiont (Bosch and McFall-Ngai, 2011; McFall-Ngai et al., 2013; Rosenberg et al., 2010), has helped shape the evolution of gut immunity and exists as a continually dynamic but primarily cooperative system in the present day. Furthermore, we present the concept that protochordates, such as the tunicate Ciona intestinalis, can serve as unique and highly informative models for defining sets of chordate rules for host-bacterial dialogue in the gut. These organisms provide experimental advantages based on both their taxonomic position and specific gut physiology, and, when compared to vertebrates, can offer fascinating insights into the evolution of host-microbe relations in the gut.
Figure 1.
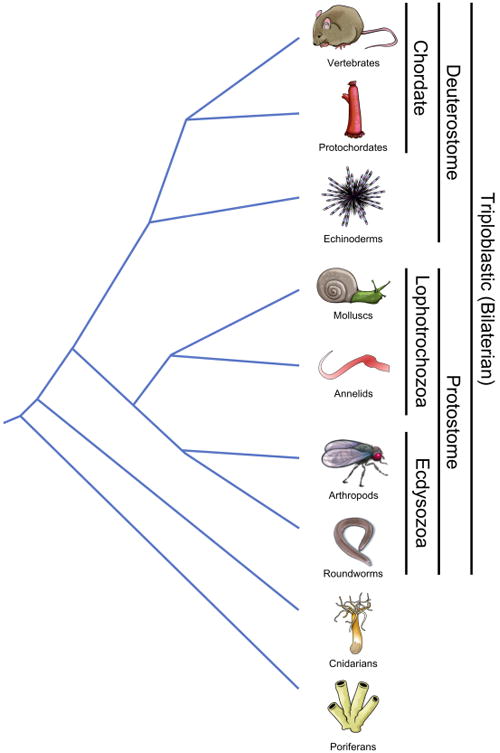
Phylogenetic relationships of the taxonomic groups discussed in this review. Relative times of divergence are not to scale. Groupings of particular phyla are indicated by text at right.
Simplest Epithelium
The most basic and phylogenetically conserved interactions between complex metazoan hosts and the microbial world occur across epithelial barriers (Fig. 2A). Hydra sp. are solitary freshwater Cnidarians with a simple body architecture consisting of a thin gelatinous mesoglea separating an internal endoderm and external ectoderm (Fig. 2B). They have become an indispensible model system for defining the origins of the most basic interactions across epithelial barriers (Augustin et al., 2012; Bosch, 2013). Distinct bacterial communities have been shown to colonize the epithelium of two Hydra species cultured under identical laboratory conditions; the microbial communities observed are representative of those identified in Hydra isolated from the wild (Fraune and Bosch, 2007). These observations constitute the first compelling evidence that even the simplest of animals maintain species-specific microbial communities and give rise to several questions: What regulates these bacterial communities? Could it be that ecological factors at the surface of the epithelium such as nutrient composition and pH create conditions that favor some microbial communities over others? Alternatively, does host immunity play a much more specific and active role in defining the communities, and if so, could genetic factors outweigh environmental influences? Further insight into the mechanisms affecting this unexpectedly high degree of selection can be gained by considering several key observations.
Figure 2.
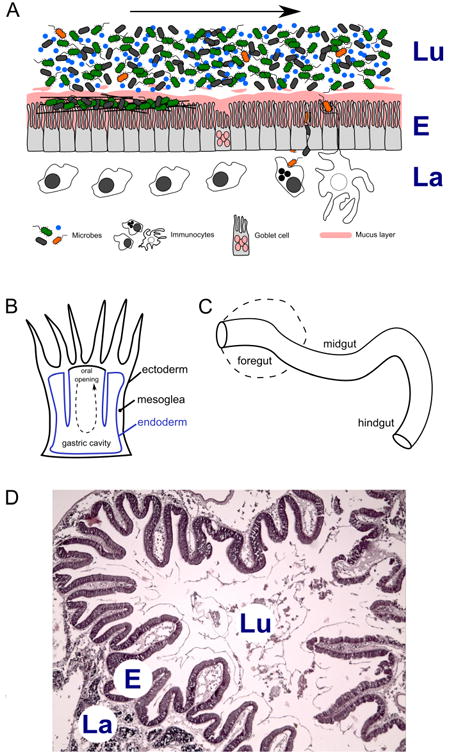
(A) Schematic diagram of a generic animal gut ecosystem. Dietary intake and microbes in the lumen (top) flow through the alimentary canal alongside the gut epithelium (middle), with immune and other host cell types occupying the lamina space interior to the gut wall (bottom). Bacteria in the lumen may be transient occupants (blue), commensal/biofilm residents (green), pathogenic invaders (orange) or opportunistic invaders (black). Flow of gut contents is indicated by arrow. (B) Schematic cross section through a Cnidarian polyp showing the simple gastric cavity; oral opening on top. Tissue layers and gut anatomical features are labeled. Flow of food into and out of the oral opening is indicated by arrow. (C) Segmented gut found in most bilaterian animals. Foregut may be a simple tube or elaborated into a stomach (dashed line). (D) Haematoxylin-eosin stained section of Ciona intestinalis stomach. Lu, lumen; E, gut epithelium; La, lamina.
First, in experiments lasting more than 15 weeks, colonization of the epithelial surface of Hydra was noted to occur in reproducible stages (Franzenburg et al., 2013a). Both experimental and mathematical concepts were applied to demonstrate that colonization is controlled first by competitive interactions among diverse bacteria for access to the host surface and resources. The initial colonization of the tissue appears to prime the response, which matures over some defined period of time. The result is reduced diversity of bacterial communities as well as a reduction in variation in the abundance of certain groups in a process that is influenced by host-derived factors. A key feature of the Hydra system (but unlike some other Cnidarians) is the apparent lack of mobile phagocytic cells. Microbial sensing in this species is mediated exclusively by the epithelium and involves the nuclear factor kappa B (NF-kB), Toll-like receptor (TLR), and Myd88 signaling cascades (Bosch et al., 2009). Preliminary sorting of bacteria apparently is carried out by diverse antimicrobial peptides (AMPs) that influence composition of the microbiota from the onset of development onwards (Franzenburg et al., 2013a; Franzenburg et al., 2013b; Fraune et al., 2010). The association of stable microbial communities is governed by epithelial homeostasis and is influenced by the physical process of cellular turnover (Boehm et al., 2012; Bosch, 2013; Fraune et al., 2009). In mammals, for comparison, stem-cell guided gut epithelial turnover helps to promote not only healing of injured surfaces but to re-establish the complex layering of mucin (Johansson, 2012; Johansson et al., 2011a; Johansson et al., 2011b), re-concentrate host-derived factors such as AMPs and prompt re-colonization of some adherent bacterial communities (Eri and Chieppa, 2013; Marchiando et al., 2010; Mukherjee et al., 2008; Sommer and Backhed, 2013). Thus, even in the most basic epithelial surface of the Hydra system, a vastly complex interplay between an array of factors, including local physiological properties, host-derived immune mediators, and microbiome-derived factors, governs the establishment, maintenance and displacement of adherent microbial communities.
The Cnidarian Gastric Cavity
The Cnidarian body plan consists of a pair of tissue layers. In actively feeding Cnidarians such as Hydra, as well as in some species of corals, anemones, and jellyfish, a single opening acts as both a mouth and anus (Fig. 2B). The oral opening empties into a sac that is lined with endoderm (a specialized digestive epithelium known as gastrodermis), which is analogous to the gut epithelium of higher animals. Food is ingested into a pouch-like area (i.e., the gastric cavity) where it is metabolized, after which fecal remains are released into the environment through the same oral opening. However, just as with the gut of higher animals, the gastric cavity remains an external environment within the internal confines of the host organism (in this case, the Cnidarian polyp). Recently, Agostini and colleagues (Agostini et al., 2012) demonstrated that this most primitive example of a gut-like structure is not as simple as it may seem. Previous work in corals (Sorokin, 1973) and anemones (Herndl and Velimirov, 1986) has demonstrated distinct groups of bacteria in the gastric cavity. It was concluded that either the animals were cultivating bacteria as a backup carbon source during times of starvation (Herndl and Velimirov, 1986) or that the bacteria contributed essential vitamins and nutrients for the host (Sorokin, 1973). The more recent analysis by Agostini and colleagues suggests that the corals manage communities of bacteria in distinct areas of the gastric cavity, forming a complex ecosystem of diet-, host-, and bacterial-derived factors that is associated with differences in pH, dissolved oxygen, essential nutrients, vitamin content and bacterial diversity within individual polyps (Agostini et al., 2012). The gastric cavity microbiota within the polyps were shown to differ from microbial populations of surrounding seawater, suggesting that this co-evolving ecosystem involves bacterial selection. Given what we know from the studies in Hydra and other Cnidarians, in which expression of immune factors has been shown in oral tissues (Kimura et al., 2009; Vidal-Dupiol et al., 2011), it is tempting to speculate that, host-derived immune mediators play a significant role in defining how the composition of the gastric cavity microbiome develops and is maintained in these ancient diploblastic organisms. Consistent with this view, strong immune gene expression is detected in endodermal tissues and/or oral ectoderm (Kimura et al., 2009; Vidal-Dupiol et al., 2011). Furthermore, complex expression patterns of immune molecules result when the animals are dosed and/or challenged with various bacteria. Such patterns, including the direct downregulation of a specific AMP (Vidal-Dupiol et al., 2011) by a known coral pathogen, point toward a complex dialogue between host and microbiota in the gastric cavity.
Ancient Alimentary Canals
The alimentary canal, which in the broadest sense is a feeding tube that on one end has a mouth and on the other an anus, is an ancient structure shared by all bilaterian animals (Fig. 2C). In triploblastic animals (three germ layers, a coelomic cavity, and bilateral symmetry; Fig. 1), the gut is derived from endoderm lining the yolk sac, which itself is enveloped by the developing coelom, a feature unique to animals with mesoderm (Hejnol and Martindale, 2008; Holland, 2000; Noah et al., 2011). Developmentally, some aspects of mesodermic formation of the coelom as well as the derivation of the gut from the earliest blastomere divide the bilaterians in two groups, the protostomes and the deuterostomes. The protostomes include the molting organisms (Ecdysozoans) such as arthropods and nematodes as well as the non-molting forms (Lophotrochozoans), which include annelids and molluscs. The deuterostomes include echinoderms, early chordates (the invertebrate protochordates) and the later chordates (the vertebrates) (Fig. 1). Three important aspects of the gut appear to be phylogenetically conserved: 1) compartmentalization so that along the alimentary canal, morphological, and in some cases biochemical, features separate the gut into distinct segments roughly within the confines of a foregut (mouth, esophagus, stomach), midgut (the middle intestines), and the hindgut (mostly the caecum, colon, anus), 2) the gut is a prominent immunological organ that exerts a strong influence on host physiology and 3) the gut has evolved in most taxa as an ecosystem to harbor microbial communities (in many cases including a fermentation chamber) that in most cases is of host benefit.
In general, many features of animal immunity have been conserved in phylogeny; however, there exists an astonishing diversity of specific immune mechanisms, consistent with the extensive variation among genomes and the enormous morphological complexity representative of the various phyla. In all animals, the expression of AMPs in the gut is a major effector response that is critical to the function and maintenance of homeostasis. Recent work has demonstrated a remarkable array of multi-functional potential for these diverse effector molecules (Diamond et al., 2009; Semple and Dorin, 2012; Semple et al., 2010). Functions include the “neutralization” of lipopolysaccharide (LPS), an important component of the cell wall of many bacteria. Such neutralization, which appears to facilitate the induction of intestinal epithelial cell (IEC) tolerance (Pulido et al., 2012), reduces the host's response to LPS, which might otherwise result in bacterial killing and/or unnecessary host inflammation. However, neutralization apparently does not eliminate the biological function of LPS in mutualistic bacteria which grow quite readily within environments rich in the AMPs (Gruenheid and Le Moual, 2012; Gutsmann et al., 2005). Thus, in addition to selective anti-bacterial properties, some AMPs demonstrate immune-modulating potential (Choi et al., 2012; Scott et al., 2007; Territo et al., 1989). This dual function may serve to protect the host against collateral damage (i.e., unnecessary or misdirected inflammation) and facilitate colonization by harmless or beneficial microbiota. It also is possible that some of these AMPs mobilize immunity to actively regulate the composition of adherent bacterial communities. Simpler model systems (e.g., those lacking adaptive immunity) can help dissect the diverse roles of AMPs in the gut.
Drosophila melanogaster (fruit fly) has been used widely and successfully as a model system to dissect host-microbe interactions. The genomes of various Drosophila species have been determined (Gilbert, 2007; Hahn et al., 2007; Rubin, 1996; Song et al., 2011), knockouts are available and knockdowns easily generated, and more aspects of their biology are understood at the genetic level than for any other species (e.g., see special Nature web focus: http://www.nature.com/nature/focus/drosophila/). Drosophila possess a long gut (several times their body length) that is compartmentalized into fore-, mid-, and hindgut. Intestinal stem cells that can facilitate the renewal of gut barriers are present (Buchon et al., 2013b; Lemaitre and Miguel-Aliaga, 2013; Micchelli, 2012; Micchelli et al., 2011). The Drosophila immune system has been studied extensively, frequently using systems biology approaches (Bodenmiller et al., 2007; Cuomo and Bonaldi, 2010; de Morais Guedes et al., 2005; Engstrom et al., 2004; Lilley and Griffiths, 2003; Stuart et al., 2007; Vierstraete et al., 2005), and has been thoroughly described elsewhere (Charroux and Royet, 2010; Kingsolver et al., 2013; Kuraishi et al., 2013; Lemaitre and Hoffmann, 2007; Wang et al., 2014). Briefly, Drosophila relies on NF-kB modulation of antimicrobial defenses in response to recognition and activation of either immune deficiency (IMD) or TLR signaling pathways. In gut epithelial cross-talk with bacteria, Drosophila also utilizes (like many other animals) the dual oxidase (DUOX) system to modulate inflammation and discriminate among potential pathogens (Kim and Lee, 2014; Lee et al., 2013; Ryu et al., 2010). Immune homeostasis of gut microbial communities, in Drosophila, largely is governed largely by the activity of AMPs and the modulation thereof (Kuraishi et al., 2013).
Complex communities of bacteria, strongly affected by diet and environment, can be sampled from the Drosophila gut, particularly in wild-type flies (Broderick and Lemaitre, 2012; Buchon et al., 2013a). The gut hosts a much lower compositional diversity than is found in surrounding environments, suggesting that Drosophila modulate the communities of bacteria that persist in their gut (Chandler et al., 2011), likely by creating either an environment that is not hospitable to most microbes or one that supports the growth of some species over others. However, the inability to consistently recover identical bacterial operational taxonomic units (OTUs, typically equivalent to bacteria matched to a sequence identity of >97%) across individual flies and populations calls into question the possibility that flies select for or are dependent on a limited core set of microbiota (Wong et al., 2013). Intriguingly, Drosophila, like bumblebees and honeybees (Martinson et al., 2011) as well as fireflies (Sudakaran et al., 2012), does maintain particular species of bacteria in the gut. These gut microbiota have now been shown to modulate various aspects of gut biology, including host innate immunity (Buchon et al., 2013a; Glittenberg et al., 2011; Lee et al., 2013) as well as intestinal epithelia cell turnover (Buchon et al., 2009a; Buchon et al., 2009b; Lee, 2009). Modulation of host responses is not limited to the gut, and likely can exert enormous influences on host behavior and physiology (Engel et al., 2012; Koropatnick et al., 2004; Salem et al., 2013; Sharon et al., 2011; Storelli et al., 2011). This phenomenon demonstrates a co-evolving partnership between host and gut microbiota, despite the observation that many of the microbial species in the gut of Drosophila do not colonize the animal permanently, but rather need to be replenished periodically to preserve the composition of the gut flora (Blum et al., 2013). Some microbes may find it difficult to establish permanent communities, possibly as a consequence of epithelial turnover and peritrophic matrix renewal in the midgut (Kuraishi et al., 2011). Additional difficulty could arise via the activity of a broad spectrum of AMPs expressed in each compartment of the gut.
In the simple nematode, Caenorhabditis elegans, the entire gut is descended from a single embryonic cell (Deppe et al., 1978) and results in a simple intestine (20 somatic cells) lacking stem cells, and subsequently, renewal potential, which in turn influences lifespan. This trait is likely evolutionarily derived in C. elegans, and other organisms (Rost-Roszkowska and Undrul, 2008), as renewal of gut epithelium is a common feature among most animals. Because C. elegans feeds primarily on microorganisms, host-microbe interactions are initiated in the intestines and colonization of the gut can occur there; pathogens can also attempt to colonize and reduce lifespan of the worm either by adhering to the outer cuticle and causing damage or by secreting toxins that cause intestinal damage and/or systemic injury (Zhang and Hou, 2013). When confronted with pathogens, C. elegans can utilize avoidance behaviors that involve signaling through its single TLR, Tol-1 (Pradel et al., 2007). However, protection against pathogenic invasion of the intestines also is dependent on Tol-1 (Tenor and Aballay, 2008) and can involve a variety of complex protective defense strategies, including detoxification of bacterial products (Marsh and May, 2012; Zhang and Hou, 2013). These responses rely heavily on the expression of various immune mediators such as: lectins and lysozyme (Mallo et al., 2002), AMPs (Roeder et al., 2010), orthologs of Toll-interleukin-1 receptor (TIR) domain protein sterile alpha and armadillo-motif containing protein (SARM) and mitogen activated protein kinases (Marsh and May, 2012; Shivers et al., 2009). Olfactory-driven avoidance behaviors are also present (Zhang et al., 2005). Studies in C. elegans were among the first to suggest that gut-bacterial interactions could influence longevity in the host (Garigan et al., 2002; Gusarov et al., 2013; Ottaviani et al., 2011; Portal-Celhay et al., 2012; So et al., 2011). Whereas persistence of gut microbiota likely is completely dependent on diet, host fitness and outcome depend on relationships developed with certain bacteria (Cabreiro and Gems, 2013).
Among the non-molting protostomes and much like other animals, the gut of the earthworm (a segmented worm, or annelids) is highly compartmentalized and relies on bacteria as part of its diet and/or to: help metabolize food, process calories, establish bioavailable vitamins and nutrients, and protect against pathogens (Drake and Horn, 2007; Idowu et al., 2008; Parthasarathi et al., 2007; Thakuria et al., 2010). However, the bacterial groups seen most consistently in surveying earthworm gut microbiota often do not represent those found in the diet or experimental bedding, suggesting that some bacteria likely are retained for some time in the gut and then proliferate in response to certain diets (Rudi et al., 2009). It seems that some bacteria are resistant to or can tolerate the antimicrobial activity of digestive fluids derived from the earthworm gut (Khomiakov et al., 2007) and, not surprisingly, the immune system of this species demonstrates both specificity and memory components (Kvell et al., 2007). The earthworm gut likely has the capacity to discriminate among beneficial and harmful microbiota.
At least one species of polychaete worm (also an annelid) has been shown to maintain specific communities of bacteria within distinct regions of the gut (consistent with anterior, middle, and posterior sections, as referred to by the authors)(Li et al., 2009). Surprisingly, the deep-sea annelid tubeworms lack a gut but instead have an anterior “plum” organ that is highly vascularized and used for exchanging dissolved molecules and capturing material from seawater. Within the coelomic cavity is a specialized organ, the trophosome, which houses symbiotic bacteria responsible for nearly all of the dietary metabolic activity sustaining the animals (Cavanaugh et al., 1981). A detailed transcriptomics study under in situ conditions in Ridgeia piscasae (the deep sea tubeworm) (Nyholm et al., 2012) identified innate immune pattern recognition receptors (PRRs) as major players in helping to establish and maintain homeostasis through a carefully orchestrated dialogue between host and bacterial symbionts of the trophosome. Another marine invertebrate filter-feeder, the oyster Crassostrea virginica, also has been shown to also maintain distinct bacteria in distinct compartments of the gut (King et al., 2012). Here, as in other examples noted above, the gut likely is functioning as a co-evolved system in which successful partnerships emerge from a continuous dialogue involving local ecological properties of the gut compartments and an active innate defense system utilizing PRRs (including TLRs), AMPs, and the specific recognition and effector responses mediated by epithelial cells as well as immunocytes (Bachere et al., 2004; Buchon et al., 2013a; Nyholm and Graf, 2012; Rosa et al., 2012; Schmitt et al., 2012; Zhang et al., 2014).
The simplest deuterostomes include diverse groups of highly derived echinoderms, some of which demonstrate exceptionally complex immune systems (Smith et al., 2010). One of the most dramatic findings from the complete genome of the purple sea urchin (Strongylocentrotus purpuratus) (Davidson, 2006; Sea Urchin Genome Sequencing et al., 2006) was the presence of massively expanded repertoires of innate immune receptors and effectors (Hibino et al., 2006; Rast et al., 2006). The sea urchin immune system has undergone extensive expansion and diversification of genes primarily related to pattern recognition of microbial products, including TLRs, nucleotide-binding oligomerization domain receptors (NLRs, cytoplasmic receptors) and the scavenger receptor cysteine-rich (SRCR). Other key elements of innate immunity, including some components of the complement system, peptidoglycan recognition proteins (PGRPs) and Gram-negative binding proteins (GNBPs), are present in the sea urchin genome in equivalent numbers to their homologs in protostomes or other deuterostomes (Hibino et al., 2006). The sea urchin also expresses, in immunocytes and other tissues, an unusual gene family that is potentially unique to these animals [Sp185/333, (Ghosh et al., 2010)]. Sp185/333 gene products exhibit exceptional diversity in immunocytes (Majeske et al., 2013) and it is likely that Sp185/333 proteins play an integral role in echinoderm immunity, possibly through coupling interactions among other PRRs.
Not surprisingly, most of the expanded and diverged genes are expressed in the gut of the sea urchin (Hibino et al., 2006) and, like many of these PRRs in mammals (Abreu, 2010; Cario et al., 2002; Rakoff-Nahoum et al., 2004), may be involved in helping to define physiologically distinct regions of the gut. As in numerous other animals, the gut of the adult echinoderm is highly compartmentalized and includes anaerobic chambers. Diverse communities of bacteria, many of which are not common in the animal's surroundings, reside within these compartments in sea urchins (Meziti et al., 2007; Thorsen, 1998; Thorsen et al., 2003), as well as in sea cucumbers (Zhang et al., 2012; Zhang et al., 2013) and a deep sea holothurian (Amaro et al., 2009); some of these bacteria may represent members of the permanent gut flora. Holothurians present an interesting case and a unique opportunity to study bacterial (re)colonization of guts from adult animals as they can undergo autotomy (i.e., self amputation), in most cases resulting in a nearly complete loss of internal organs (Byrne, 2001; Mashanov and Garcia-Arraras, 2011; Wilkie, 2001); under the proper conditions, these species can regenerate their internal organs (e.g., reconstruct the entire intestinal tract). It is likely that immunity plays vital roles in the regenerative process as development leads to re-exposure to microbes. Regeneration is also associated with a unique immune profile in some tunicates (an invertebrate chordate group, see below) (Rinkevich et al., 2007). Because of the complexity of interactions that involve innate immunity and frequently occur across mucosal barriers, invertebrate defense systems are far from simple (Loker et al., 2004). It is now apparent that the host immune dialogue with the external environment and associated microbiota is essential in maintaining tissue homeostasis, influencing renewal processes and ultimately driving immune evolution and innovation (Loker, 2012). Although it remains to be determined, the unusual expansion of innate immune receptors among some deuterostome invertebrates suggests the existence of a complex dialogue with microbiota, particularly in the gut where most of them are expressed. Complex mutualisms, as well as pathogen discrimination, within the gut lumen could drive the diversification of these PRRs (Dishaw et al., 2012b; Hibino et al., 2006; Rast et al., 2006).
Tunicates: representatives of the early chordate gut
Protochordates are invertebrate chordates such as sea squirts (Urochordata or Tunicata) and the lancelet, amphioxus (Cephalochordata). These species diverged prior to the vertebrates (Fig. 1) and as such lack classical adaptive immunity; nevertheless, they retain and share key, chordate-specific, developmental features that include a notochord, hollow dorsal nerve cord, pharyngeal slits, endostyle and post-anal tail (Brown et al., 2008; Cameron et al., 2000; Cañestro et al., 2003b; Katz, 1983; Kubo et al., 2009; Meinertzhagen and Okamura, 2001; Paris and Laudet, 2008; Shi et al., 2005; Swalla et al., 2000; Swalla and Smith, 2008). Ciona intestinalis, a tunicate with a compartmentalized gut (Burighel and Cloney, 1997; Hirano and Nishida, 2000; Nakazawa et al., 2013) that undergoes epithelial turnover and renewal (Ermak, 1981), is being adapted by our laboratory as a model to help define the evolution of bacterial-immune dialogue in the chordate gut (Dishaw et al., 2012a; Dishaw et al., 2011). Ciona is a suspension (filter) feeder that siphons water continuously, a process that results in the exposure to and consumption of ample amounts of dietary microorganisms. This basic process exposes the gut to potential colonizers and/or pathogens. A variety of cell types can be discerned within the stomach epithelium of tunicates (Fig. 2D), including absorptive, zymogenic, endocrine, ciliated mucous and undifferentiated cells (Koyama et al., 2012) as well as various lineages of blood-derived hemocytes. The gut also produces copious amounts of mucus (Goddard and Hoggett, 1982). Whereas the amphioxus genome experienced a considerable expansion of innate immune receptors almost as extensive as that described in the sea urchin (Dishaw et al., 2012b; Holland et al., 2008; Huang et al., 2008) and, through domain duplication, may have even resulted in novel signaling pathways (Zhang et al., 2008), the same is not true of Ciona intestinalis. Rather, Ciona appears to have undergone a considerable genome reduction and likely represents a core immune gene set (Cañestro et al., 2003; Dehal et al., 2002; Kubo et al., 2009). At a minimum, Ciona could represent the basic functional requirements for homeostasis in the gut of chordates. Ciona possesses a competent innate immune system (Azumi et al., 2003; Shida et al., 2003) and includes AMPs (Di Bella et al., 2011; Fedders and Leippe, 2008), two TLRs with multi-ligand binding potential (Sasaki et al., 2009), GNBPs, lipopolysaccharide binding protein (LBP), a variety of C-type lectins and other PRRs, tumor necrosis factor (TNF) (Azumi et al., 2003), mannose binding lectin (MBL) (Skjoedt et al., 2010), complement protein C3 (Marino et al., 2002), as well as the variable region-containing chitin-binding proteins (VCBPs), a relatively small family of immunoglobulin domain-containing proteins (Dishaw et al., 2011).
Like most other animals, Ciona likely uses AMPs as a first line of defense (Di Bella et al., 2011; Fedders and Leippe, 2008; Lu et al., 2014); however, response to specific bacterial products also can be viewed as being consistent with the patterns of expression of TLRs and other PRRs in gut tissues, resulting in the induction of pro-inflammatory molecules, e.g., TNF (Sasaki et al., 2009). Although the Ciona genome encodes only two TLRs, both are expressed, along with VCBPs, in distinct locations along the gut. Having only two TLRs, each interacting with multiple ligands, could be interpreted to be disadvantageous. However, multiple PRRs may be coupled to downstream pathways, which may facilitate more discriminating ligand recognition.
Current laboratory investigations (Dishaw et al., unpublished) suggest that Ciona likely maintains a state of balance (i.e., homeostasis) between tolerance and protection through host epithelial–microbe interactions that promote the maintenance of stable, adherent, microbial communities (Dishaw et al., 2014). Ciona TLRs (Sasaki et al., 2009), as well as additional PRRs [e.g., MBL (Skjoedt et al., 2010)], are differentially expressed in unique sections of the gut. The innate immune repertoire of Ciona likely is augmented by VCBPs. At least one of these proteins (VCBP-C) is secreted into the gut lumen by specialized epithelial cells, can bind bacteria, and acts as an opsonin (Dishaw et al., 2011). It is likely that these innate immune molecules interact with the gut flora and modulate adherence patterns. VCBPs are expressed in distinct regions along the gut at the onset of developmental maturation and as well as at the onset of feeding, and are highly responsive to microbial products (Liberti et al., 2014). Ciona intestinalis from distinct populations have now been shown to harbor a core set of microbiota in the gut (Dishaw et al., 2014) in which adherent communities likely play key functional roles. How the Ciona immune system helps to discriminate among colonizing bacteria and regulate the population of gut symbionts is unclear, given the reduced complexity of its innate immune repertoire. Mariculture of Ciona is well established; animals can be grown under microbe-free conditions by the hundreds on standard tissue culture dishes that facilitate the production of large clonal replicates (Fig. 3) (Dishaw et al., 2012a). Current efforts are focused on characterizing the biology of the compartmentalized gut in Ciona and in determining the roles of VCBPs in modulating the composition of bacteria in the gut and facilitating homeostasis. More global studies of the mechanisms of temporal and developmental regulation of microbial populations in this species are in progress.
Figure 3.
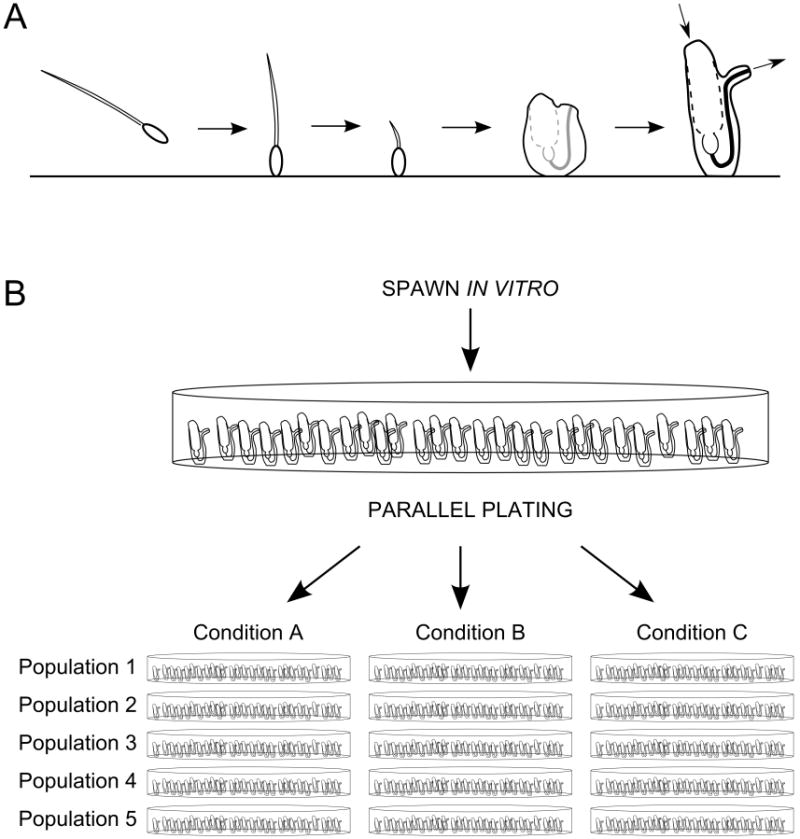
(A) Life cycle of Ciona intestinalis, including from left to right: swimming tadpole larva stage, attached larva, tailbud retraction and onset of metamorphosis, early juvenile (not feeding; unopened siphons) and adult (with siphon flow indicated by arrows). (B) Illustration of high-throughput, parallel experiments using the Ciona system to investigate the response of five parallel populations of idividually attached Ciona to three experimental conditions (standard tissue culture dishes shown). “Populations” are interchangeable with “replicates;” additional replicates allow the production of more genetic material for study. Pooled replicates from different genetic backgrounds can also be performed to strengthen observations.
Vertebrates: immune management of complex flora
The epithelial barriers and underlying mucosal tissues of the intestine are integral components of the highly complex and integrated vertebrate immune system (Bates et al., 2006; Buchon et al., 2009a; Duerr and Hornef, 2012; Mazmanian et al., 2005; Pott and Hornef, 2012; Round and Mazmanian, 2009). The gut in general, and the distal gut (i.e., the lower intestines) in particular, possess a number of distinct compartments each housing complex communities of microbes. Each compartment within the gut is defined by a complex set of microbial, physiologic and immunologic parameters, and these compartments act together as a major site of immune detection and response (Abreu, 2010; Clarke et al., 2010; Hooper et al., 2001; Lentle and Janssen, 2008; Lentle et al., 2013; Ley et al., 2006; Macdonald and Monteleone, 2005; Ortega-Cava et al., 2003; Savage, 1977;), where host survival requires achieving a balance between resistance and tolerance (Fujimura et al., 2010; hill and Artis, 2010; Medzhitov, 2010). This balance involves a continuous crosstalk, vital to immune development and gut homeostasis, between the gut microbial community and the immune system (Cebra, 1999; Edelman and Kasper, 2008; Hooper and Gordon, 2001; Kanther and Rawls, 2010; Mazmanian et al., 2005). This crosstalk varies considerably along the length of the alimentary canal (e.g., distinct compartments) where physical properties such as flow rate (Pacha, 2000), which itself is affected by diet (Hammer et al., 1998), as well as localized mucus production can vary significantly (Johansson et al., 2011a; Johansson et al., 2008; Ley et al., 2006). Shifts in the population structure of the microbiota can have significant downstream consequences (Bates et al., 2006; Rawls et al., 2006; Chow and Mazmanian, 2009; Round and Mazmanian, 2009; Kanther and Rawls, 2010; Vijay-Kumar et al., 2010) e.g., the breakdown of immune tolerance, resulting in chronic inflammatory conditions (Cario and Podolsky, 2005; Schreiber et al., 2005; Round and Mazmanian, 2009; Round et al., 2010a).
Despite the enormous complexity of microbiota encountered by and maintained within the gut of mammals, a single cell layer of IECs provides the interface for communication with this immense array of microbial and food components (Duerkop et al., 2009; Vaishnava et al., 2008). Innate immune receptors (e.g., TLRs and NLRs), which are expressed by IECs (Abreu, 2010; Artis, 2008) along with a diverse set of mucosal-derived factors (Hooper and Macpherson, 2010; Macfarlane and Macfarlane, 1997), provide the front line of communications between the innate immune system and the resident gut bacteria. While recognition complexity of adaptive immune receptors involves somatic rearrangement of immunoglobulin genes, it now seems clear that this essential dialogue with resident microbiota is not to be re-learned each generation and thus is governed by heritable, germline-encoded, innate immune receptors. In vertebrates, homeostasis in the gut is likely maintained further via complex interactions with the adaptive immune system across the basal lamina propria (Round and Mazmanian, 2009). Further details of the rules that govern symbiotic interactions and sustain homeostasis in the intestines remain largely unknown (Hooper, 2009; Medzhitov, 2010). It is well established that the proper developmental ontogeny of the mammalian gut is dependent on the proper timing and exposure to appropriate microbiota. In the absence of proper colonization, appropriate maturation of the gut immune system, and in particular, gut-associated lymphoid tissue (GALT) is lacking (Atarashi et al., 2011; Cebra, 1999; Chung et al., 2012; Deplancke and Gaskins, 2001; Ivanov et al., 2009; Tlaskalova-Hogenova et al., 2002; Umesaki and Setoyama, 2000; Williams et al., 2006). Studies in various model organisms demonstrate a significant influence on host epithelial behavior and gut immune responses by the colonizing flora, via microbe-specific metabolic products or small molecules.
Bacterial recruitment and colonization is also influenced significantly by physical properties of the gut, which includes mucus type and availability (Johansson et al., 2011a), which together with host diet, can help attract bacteria to the epithelial surface (Derrien et al., 2010; Falk et al., 1998; Fischbach and Sonnenburg, 2011; Juge, 2012; Kashyap et al., 2013a; Kashyap et al., 2013b; Marcobal et al., 2013; Martens et al., 2008; Sonnenburg et al., 2005). This co-evolved mutualism, consequently, involves host physiological responses to bacterial-derived capsular polysaccharides that influence microbial colonization and host immune tolerance (Macfarlane and Macfarlane, 1997; Mazmanian, 2008; Mazmanian and Kasper, 2006; Mazmanian et al., 2005; Mazmanian et al., 2008; Round and Mazmanian, 2009, 2010; Round et al., 2010b). These examples demonstrate a co-evolved mutualism with gut flora that is hundreds of millions of years old and consistently, across diverse taxonomic lineages, modulates host tolerance and influences the process of homeostasis (Dalmasso et al., 2011; Hooper et al., 1998; Ley et al., 2008; McFall-Ngai et al., 2013).
Our understanding regarding the overall function of the vertebrate GALT has undergone a revolution within the past 10 years. The idea that the immune system evolved, in part, to support symbiotic bacteria has been most fully developed in mammals, particularly humans and laboratory rodents. However, developments in the field of immunology have always been strongly driven by the need to prevent infectious disease, leading to a misdirected early view of the immune system as being antagonistic toward all microbes, even those that normally colonize the epithelial surfaces of the host. As will be discussed below, the idea that the immune system is antagonistic toward all non-host species is still influential, with unfortunate consequences for human health.
The realization that the immune system evolved, in part, to actually support the growth of mutualistic bacteria in mammals originally derived from studies in xenotransplantation (Parker and Lesher, 2005). Researchers investigating the possibility of implementing clinical xenotransplants found that “natural” antibodies, or those antibodies present without a known history of infection or immunization, were responsible for xenograft rejection. Subsequent investigation revealed that: (a) the natural antibody repertoire is very stable in the population, not changing over the course of time (Galili et al., 1999; Parker et al., 1999), (b) mutualistic bacteria express antigens recognized by those antibodies (Casali and Schettino, 1996; MacDonald and Pettersson, 2000; Shimoda et al., 1999; Sonnenwirth, 1979; Tagliabue et al., 1983), and (c) the expression of those antigens by mutualistic bacteria does not appear to be obligatory (Stowell et al., 2010); i.e., the bacterial antigens recognized by xenoreactive antibodies are not constitutive in nature, and tended to vary depending on the bacterial isolate. These three observations, when combined, suggested that the immune system is not antagonistic toward the entire microbiome, as was thought at the time. If such antagonism existed, expression of bacterial antigens recognized by natural antibodies, particularly non-constitutive antigens, should decrease over time. Indeed, in the face of a potent immune response, natural selection should drive the microbiome to mimic the host in terms of antigen expression. With this in mind, the stable expression by gut bacteria of the antigens recognized by natural antibodies suggested that the traditional view in immunology was misguided: that immune recognition is beneficial rather than detrimental to survival of some bacteria (Everett et al., 2004). These observations, combined with earlier work demonstrating that intestinal bacteria produce their own receptors for host immune molecules (Friman et al., 1996; Schalen, 1993; Wold and Adlerberth, 2000; Wold et al., 1990), led to a series of studies indicating that the immune system supports the growth of mutualistic bacteria in the gut of mammals (Bollinger et al., 2003) (Fig 4).
Figure 4.
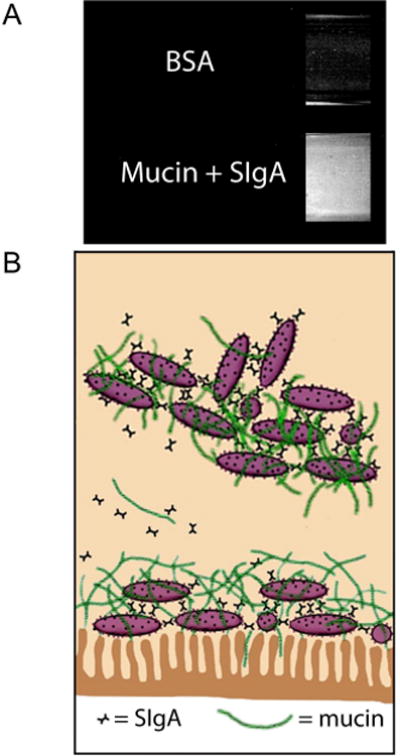
Biofilm formation mediated by the human immune system. (A) side views of two test tubes (top, bottom) coated with human epithelial cells are shown. In each tube, Escherichia coli bacteria were incubated with nutrient rich media that was replenished on a regular basis. In the top tube, 0.5 mg/ml bovine serum albumin (BSA; a serum protein unrelated to immune function) was added to the media, and in the bottom tube, porcine gastric mucin (50 mg/ml) and human secretory IgA (SIgA; 0.5 mg/ml) were added to the media. Biofilm growth was evident in the tube containing immune-related molecules as shown by increased staining (bottom), but not in the tube without immune related molecules. (B) Schematic diagram demonstrating host-mediated biofilm growth (bottom, attached to epithelium, i.e., immune inclusion), as well as shedding of biofilm fragments (top, unattached to epithelium, i.e., immune exclusion).
The current view regarding molecular mechanisms underpinning this host-microbe association is that both host and symbiont produce an array of recognition molecules that involve both the innate and adaptive immune system of the host in a mutually beneficial way. Most of the work directed at understanding the complex relationships between gut microbes and immunity has focused on specific antigens recognized by the antigen binding domain of particular antibodies and on the expression of the type I pilus, a mannose dependent receptor found in enterobacteria such as E. coli. Studies in mammalian hosts have focused on immunoglobulin, particularly IgA, and on mucin. The model resulting from these studies (Fig. 4B) is based in part on the findings that a substantial amount of host-derived immunoglobulin is adherent to mucus (in distinct gut compartments) and assists in the formation of biofilms among select bacterial communities (Bollinger et al., 2003; Bollinger et al., 2006; Lee et al., 2011; Thomas and Parker, 2010). In a direct physical sense, the successful maintenance of biofilms protects the host against aggressive penetrating pathogens.
It seems clear that the view of the immune system as actively supporting the growth of symbiotic microbiota should no longer be a concept at the fringes of mainstream immunology. In fact, mainstream immunology may currently be witnessing a paradigm shift in recognizing that host immune protection was born as a consequence of a system that evolved initially to facilitate and manage symbiotic interactions. The immune systems of modern animals have evolved to establish harmonious interfaces with the associated microbiota, if not to be the host's primary ambassador in the complex dialogue between host and associated microbiota. The gut remains the most pivotal organ from which to study how these diverse immune systems evolved.
Application of the Ciona system to future studies
As defined above, epithelial barriers and associated innate immune functions are phylogenetically ancient and have evolved diverse languages to help sustain a stable dialogue with adherent microbiota. As a result of this long collaboration, normal animal development and many aspects of tissue physiology (i.e., gut homeostasis) are highly dependent on microbial interactions (McFall-Ngai, 2002; Sommer and Backhed, 2013); the animal gut serves as a primary location of this interplay (Hooper and Gordon, 2001; O'Hara and Shanahan, 2006). For example, some microbial capsular polysaccharides and/or metabolic by-products readily interact with or traverse the epithelium and are sensed by innate immunocytes in vertebrates. The resulting co-evolved dialogue can produce a cascade of interactions that shape not only gut development and maturation of distinct lymphocyte lineages (Cebra et al., 1999; Koropatnick et al., 2004; Edelman and Kasper, 2008; Sommer and Backhed, 2013) but also immunological tolerance and gut homeostasis (Peterson et al., 2007; Mazmanian et al., 2008; Chow and Mazmanian, 2010; Aychek and Jung, 2014; Mortha et al., 2014).
While it is now recognized that microbes colonize the gut of most animals almost immediately after birth producing richly diverse and stable microbial communities (Savage, 1977;), the details of this carefully orchestrated dialogue are only now being revealed. In vertebrates, the distal gut possesses the most complex communities of microbes, which can vary dramatically among individuals (Hooper and Gordon, 2001; Qin et al., 2010). The respective roles of interindividual variation and effects of diet, environment and lifestyle in defining the host-microbe interactions in the gut of mammals are furthermore difficult to dissect owing to the coordinate influences of both innate and adaptive immunity along the length of the gut. Several mechanisms underlying the homeostasis of the gut have been defined (Duerkop et al., 2009) and include (a) a compartmentalized gut with epithelial renewal properties and a mucin-glycocalyx barrier (Johansson, 2012; Johansson et al., 2011a; Johansson et al., 2011b; Johansson et al., 2008), (b) complex epithelial interactions with the lumenal microbial communities (Artis, 2008; Saenz et al., 2008) including the polarized expression patterns of PRRs (Gewirtz et al., 2001; Vijay-Kumar et al., 2008), and (c) multifunctional innate effectors (Diamond et al., 2009; Scott et al., 2007; Semple and Dorin, 2012). Although much is known about how the host protects itself from pathogenic invasion, much less is understood about how symbiotic communities are tolerated and cultivated by the immune system. With this in mind, model systems such as the invertebrate chordate, Ciona intestinalis, hold great promise for future investigations targeted at the mechanism(s) involving host support of mutualistic microbial growth in the gut.
Research in host-microbial relations has intensified in the face of dramatically increasing rates of occurrences of gastrointestinal-associated diseases, including inflammatory bowel disease and a wide range of food allergies in westernized human cultures. The gut ecosystem, the major site of immune detection and response to a potentially overwhelming antigenic load, is failing in its ability to maintain a balance of resistance and tolerance (Rook and Brunet, 2005). The continuous crosstalk that has evolved between the immune system and the microbiome is breaking down, as is the closely associated development of host immunity (Bickler and DeMaio, 2008). To a significant extent, the root cause of these immune problems can be traced to a protracted history of concern about infectious agents, which has reinforced the traditional (and overly simplistic) view that the immune system is more of a defense system than an interface with the environment. As a result, a profound depletion of the diversity of the “human biome,” the life associated with the ecosystem of the human body, has impacted human health dramatically (Bilbo et al., 2011; Parker and Ollerton, 2013). Relevant animal models, particularly less complex models such as Ciona, will serve as invaluable means to address not only the ramifications of this “biome depletion” for host-microbe interactions, but also the anticipated biome enrichment procedures (Parker et al., 2012) that are expected to help resolve the problem. And since accumulation of immunocytes and lamina thickening (i.e., immune maturation) in the gut of Ciona appears to be intimately linked to exposure to microbiota (Dishaw et al, unpublished), the Ciona system offers an unprecedented opportunity to dissect the role of microbiota in directing immune maturation which may inform some processes that shape GALT formation in vertebrates.
In recent years, efficient isolator systems, which allow the researcher to completely control the timing of microbial colonization as well as the microbial community structure, have become increasingly popular (Gordon, 1960; Wagner, 2008). Mouse and zebrafish are the primary models for studying the interplay between gut microbiota and the host immune system using these isolator systems (Rawls et al., 2006). Since gut microbiota often reflect flora that are most readily acquired from the environment, critical differences likely exist between the microbiota of aquatic and terrestrial animals; this disparity may become important when comparing experimental findings between the systems. Marine environments, for example, possess abundant taxa (Pedros-Alio 2006, 2012; Munn 2011; Gibbons et al, 2013) that may influence symbiotic relationships. However, comparative studies hold enormous potential to reveal both complex and conserved features that sustain stable host-microbial partnerships (Rawls et al, 2004). For example, reciprocal microbial transplants between germ-free zebrafish and mice indicated the existence of selective local environments within each recipient that could shape the composition of related taxa, resulting in predictable assemblages of microbiota (Rawls et al, 2006). Furthermore, utilization of the zebrafish model has resulted in the finding that specific bacterial phyla within the zebrafish gut microbial community either can directly increase or decrease the absorption of dietary lipids by the gut, with clear implications for translational studies of probiotics in obesity (Semova et al., 2012). While these studies have been highly informative, a simpler chordate system may be necessary to further elucidate some of the basic underlying mechanisms conserved in the phylum to which vertebrates belong. Although most marine invertebrates do not thrive and/or develop properly in germ-free environments, Ciona adapts to these conditions for sufficient time to explore the biology of colonization and thus may serve as an excellent complementary model system.
Ciona intestinalis has been shown previously to be a relevant model system for studies of development (Baghdiguian et al., 2007; Cañestro et al., 2003; Davidson, 2007; Katz, 1983; Meinertzhagen and Okamura, 2001; Sasakura et al., 2009; Shi et al., 2005) and immune defense (Fujita et al., 2004; Parrinello et al., 2008; Sasaki et al., 2009; Zucchetti et al., 2009) and presently is being developed for studies of gut host-bacterial interactions (Dishaw et al., 2012a; Dishaw et al., 2011). By permitting the experimental dissection of the conversation between host and microbiota at the onset of colonization, the Ciona model promises to reveal chordate rules that govern the biology of colonization. Experimentally, cultured Ciona is adaptable to address questions such as:
What signals do bacteria send during colonization of previously sterile tissue?
Do most stable bacterial communities exist as biofilms on or within epithelial-associated mucus?
How are the first biofilms formed and what roles do they play?
What role does the immune system play in that dialogue, in supporting biofilm formation and stability?
What are the relative contributions between physiologic versus immunologic factors that define distinct gut compartments?
How have factors such as flow rate, compartment diameter, and physiological criteria such as pH, coevolved with the expression of various immunologic factors to produce distinct GI compartments?
Addressing these questions across a broad spectrum of animal models will help identify chordate-specific details of the host-microbial dialogue that achieve and maintain lifelong homeostasis. As discussed previously, Ciona has undergone considerable reductions (gene loss) in many of the innate immune gene families (Dehal et al., 2002; Hughes and Friedman, 2005) that have expanded in other deuterostome invertebrates, implicating a streamlined version of phenomena that modulate host-bacterial interactions in the gut of of this chordate. This gene reduction can be highly advantageous in defining the first steps in the dialogue by reducing the set of candidate effectors in the system. For example, while Ciona only appears to possess two TLRs (Sasaki et al., 2009), recent experimental observations suggest that the gut is likely discriminating among microbes in a process coupled to immune mediators possessing immunoglobulin domains (Dishaw et al, 2011; Liberti et al, 2014) to selectively maintain a core microbiota (Dishaw et al, 2014) quite possibly in a fashion reminiscent of vertebrate immune inclusion-exclusion (Everett et al, 2004).
With a completed genome, well-developed mariculture methods and exceptional experimental tractability, development of Ciona as a host-microbe interaction model not only will complement the studies being performed in mouse and zebrafish but also will provide a system that: (1) can produce very high numbers of offspring with a very short generation time (i.e., sample and experimental replicates), (2) can be conveniently manipulated, unlike other marine invertebrates, into a specific pathogen-free or gnotobiotic (bacteria free) system, (3) allows extremely rapid shifting of microbial exposure due to its filter-feeding, (4) performs its immunological discrimination of microbes outside of the influence of adaptive immunity and (5) may include a single molecular species (the VCBP family) that is unique to the protochordate system but may function in a manner analogous to other classes of immune effectors found in the vertebrates. In summary, the Ciona model can help address some of the fundamental questions that remain in this field such as how do we define homeostasis in the gut, as governed by innate immunity, and what role(s) do stably associated microbiota serve in that process and in shaping the maturation of host immune responses?
Highlights.
We review the evolution of host-microbe dialog in the gut
We review the role of host immunity in recognition of gut microbes
We provide evidence that host immunity supports the growth of gut bacteria
We argue that Ciona intestinalis is a viable model in studies of gut homeostasis
Acknowledgments
LJD is supported by the All Children's Hospital Foundation Research Award and a USF College of Medicine Internal Award and GWL by the National Institute of Health (NIH AI23338).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abreu MT. Toll-like receptor signalling in the intestinal epithelium: how bacterial recognition shapes intestinal function. Nat Rev Immunol. 2010;10:131–144. doi: 10.1038/nri2707. [DOI] [PubMed] [Google Scholar]
- Agostini S, Suzuki Y, Higuchi T, Casareto BE, Yoshinaga K, Nakano Y, Fujimura H. Biological and chemical characteristics of the coral gastric cavity. Coral Reefs. 2012;31:147–156. [Google Scholar]
- Amaro T, Witte H, Herndl GJ, Cunha MR, Billett DSM. Deep-sea bacterial communities in sediments and guts of deposit-feeding holothurians in Portuguese canyons (NE Atlantic) Deep-Sea Research I. 2009;56:1834–1843. [Google Scholar]
- Andersson AF, Lindberg M, Jakobsson H, Backhed F, Nyren P, Engstrand L. Comparative analysis of human gut microbiota by barcoded pyrosequencing. PLoS One. 2008;3:e2836. doi: 10.1371/journal.pone.0002836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artis D. Epithelial-cell recognition of commensal bacteria and maintenance of immune homeostasis in the gut. Nat Rev Immunol. 2008;8:411–420. doi: 10.1038/nri2316. [DOI] [PubMed] [Google Scholar]
- Atarashi K, Umesaki Y, Honda K. Microbiotal influence on T cell subset development. Semin Immunol. 2011;23:146–153. doi: 10.1016/j.smim.2011.01.010. [DOI] [PubMed] [Google Scholar]
- Augustin R, Fraune S, Franzenburg S, Bosch TC. Where simplicity meets complexity: hydra, a model for host-microbe interactions. Adv Exp Med Biol. 2012;710:71–81. doi: 10.1007/978-1-4419-5638-5_8. [DOI] [PubMed] [Google Scholar]
- Aychek T, Jung S. Immunology. The axis of tolerance. Science. 2014;343:1439–1440. doi: 10.1126/science.1252785. [DOI] [PubMed] [Google Scholar]
- Azumi K, De Santis R, De Tomaso A, Rigoutsos I, Yoshizaki F, Pinto MR, Marino R, Shida K, Ikeda M, Arai M, Inoue Y, Shimizu T, Satoh N, Rokhsar DS, Du Pasquier L, Kasahara M, Satake M, Nonaka M. Genomic analysis of immunity in a Urochordate and the emergence of the vertebrate immune system: “waiting for Godot”. Immunogenetics. 2003;55:570–581. doi: 10.1007/s00251-003-0606-5. [DOI] [PubMed] [Google Scholar]
- Bachere E, Gueguen Y, Gonzalez M, de Lorgeril J, Garnier J, Romestand B. Insights into the anti-microbial defense of marine invertebrates: the penaeid shrimps and the oyster Crassostrea gigas. Immunol Rev. 2004;198:149–168. doi: 10.1111/j.0105-2896.2004.00115.x. [DOI] [PubMed] [Google Scholar]
- Baghdiguian S, Martinand-Mari C, Mangeat P. Using Ciona to study developmental programmed cell death. Semin Cancer Biol. 2007;17:147–153. doi: 10.1016/j.semcancer.2006.11.005. [DOI] [PubMed] [Google Scholar]
- Bates J, Mittge E, Kuhlman J, Baden K, Cheesman S, Guillemin K. Distinct signals from the microbiota promote different aspects of zebrafish gut differentiation. Dev Biol. 2006;297:374–386. doi: 10.1016/j.ydbio.2006.05.006. [DOI] [PubMed] [Google Scholar]
- Bickler SW, DeMaio A. Western diseases: current concepts and implications for pediatric surgery research and practice. Pediatr Surg Int. 2008;24:251–255. doi: 10.1007/s00383-007-2095-3. [DOI] [PubMed] [Google Scholar]
- Bilbo SD, Wray GA, Perkins SE, Parker W. Reconstitution of the human biome as the most reasonable solution for epidemics of allergic and autoimmune diseases. Med Hypotheses. 2011;77:494–504. doi: 10.1016/j.mehy.2011.06.019. [DOI] [PubMed] [Google Scholar]
- Blum JE, Fischer CN, Miles J, Handelsman J. Frequent replenishment sustains the beneficial microbiome of Drosophila melanogaster. MBio. 2013;4:e00860–00813. doi: 10.1128/mBio.00860-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodenmiller B, Malmstrom J, Gerrits B, Campbell D, Lam H, Schmidt A, Rinner O, Mueller LN, Shannon PT, Pedrioli PG, Panse C, Lee HK, Schlapbach R, Aebersold R. PhosphoPep--a phosphoproteome resource for systems biology research in Drosophila Kc167 cells. Mol Sys Biol. 2007;3:139. doi: 10.1038/msb4100182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm AM, Khalturin K, Anton-Erxleben F, Hemmrich G, Klostermeier UC, Lopez-Quintero JA, Oberg HH, Puchert M, Rosenstiel P, Wittlieb J, Bosch TC. FoxO is a critical regulator of stem cell maintenance in immortal Hydra. Proc Natl Acad Sci U S A. 2012;109:19697–19702. doi: 10.1073/pnas.1209714109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollinger RR, Everett ML, Palestrant D, Love SD, Lin SS, Parker W. Human secretory immunoglobulin A may contribute to biofilm formation in the gut. Immunology. 2003;109:580–587. doi: 10.1046/j.1365-2567.2003.01700.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollinger RR, Everett ML, Wahl SD, Lee YH, Orndorff PE, Parker W. Secretory IgA and mucin-mediated biofilm formation by environmental strains of Escherichia coli: role of type 1 pili. Mol Immunol. 2006;43:378–387. doi: 10.1016/j.molimm.2005.02.013. [DOI] [PubMed] [Google Scholar]
- Bosch TC. Cnidarian-microbe interactions and the origin of innate immunity in metazoans. Annu Rev Microbiol. 2013;67:499–518. doi: 10.1146/annurev-micro-092412-155626. [DOI] [PubMed] [Google Scholar]
- Bosch TC, Augustin R, Anton-Erxleben F, Fraune S, Hemmrich G, Zill H, Rosenstiel P, Jacobs G, Schreiber S, Leippe M, Stanisak M, Grotzinger J, Jung S, Podschun R, Bartels J, Harder J, Schroder JM. Uncovering the evolutionary history of innate immunity: the simple metazoan Hydra uses epithelial cells for host defence. Dev Comp Immunol. 2009;33:559–569. doi: 10.1016/j.dci.2008.10.004. [DOI] [PubMed] [Google Scholar]
- Bosch TC, McFall-Ngai MJ. Metaorganisms as the new frontier. Zoology. 2011;114:185–190. doi: 10.1016/j.zool.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broderick NA, Lemaitre B. Gut-associated microbes of Drosophila melanogaster. Gut Microbes. 2012;3:307–321. doi: 10.4161/gmic.19896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown FD, Prendergast A, Swalla BJ. Man is but a worm: chordate origins. Genesis. 2008;46:605–613. doi: 10.1002/dvg.20471. [DOI] [PubMed] [Google Scholar]
- Buchon N, Broderick N, Poidevin M, Pradervand S, Lemaitre B. Drosophila intestinal response to bacterial infection: activation of host defense and stem cell proliferation. Cell Host & Microbe. 2009a;5:200–211. doi: 10.1016/j.chom.2009.01.003. [DOI] [PubMed] [Google Scholar]
- Buchon N, Broderick NA, Chakrabarti S, Lemaitre B. Invasive and indigenous microbiota impact intestinal stem cell activity through multiple pathways in Drosophila. Genes Dev. 2009b;23:2333–2344. doi: 10.1101/gad.1827009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchon N, Broderick NA, Lemaitre B. Gut homeostasis in a microbial world: insights from Drosophila melanogaster. Nat Rev Microbiol. 2013a;11:615–626. doi: 10.1038/nrmicro3074. [DOI] [PubMed] [Google Scholar]
- Buchon N, Osman D, David FP, Fang HY, Boquete JP, Deplancke B, Lemaitre B. Morphological and molecular characterization of adult midgut compartmentalization in Drosophila. Cell Reports. 2013b;3:1725–1738. doi: 10.1016/j.celrep.2013.04.001. [DOI] [PubMed] [Google Scholar]
- Burighel P, Cloney RA. Microscopic Anatomy of Invertebrates, Vol 15: Hemichordates, Chaetognatha, and the Invertebrate Chordates. Wiley-Liss; 1997. Urochordata Ascidiacea; pp. 221–347. [Google Scholar]
- Byrne M. The morphology of autotomy structures in the sea cucumber Eupentacta quinquesemita before and during evisceration. J Exp Biol. 2001;204:849–863. doi: 10.1242/jeb.204.5.849. [DOI] [PubMed] [Google Scholar]
- Cabreiro F, Gems D. Worms need microbes too: microbiota, health and aging in Caenorhabditis elegans. EMBO Mol Med. 2013;5:1300–1310. doi: 10.1002/emmm.201100972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron CB, Garey JR, Swalla BJ. Evolution of the chordate body plan: new insights from phylogenetic analyses of deuterostome phyla. Proc Natl Acad Sci U S A. 2000;97:4469–4474. doi: 10.1073/pnas.97.9.4469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cañestro C, Bassham S, Postlethwait J. Seeing chordate evolution through the Ciona genome sequence. Genome Biol. 2003;4:208. doi: 10.1186/gb-2003-4-3-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cario E, Brown D, McKee M, Lynch-Devaney K, Gerken G, Podolsky DK. Commensal-associated molecular patterns induce selective toll-like receptor-trafficking from apical membrane to cytoplasmic compartments in polarized intestinal epithelium. Am J Pathol. 2002;160:165–173. doi: 10.1016/S0002-9440(10)64360-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cario E, Podolsky D. Intestinal epithelial TOLLerance versus inTOLLerance of commensals. Mol Immunol. 2005;42:887–893. doi: 10.1016/j.molimm.2004.12.002. [DOI] [PubMed] [Google Scholar]
- Casali P, Schettino EW. Structure and function of natural antibodies. Curr Top Micro Immol. 1996;210:167–179. doi: 10.1007/978-3-642-85226-8_17. [DOI] [PubMed] [Google Scholar]
- Cavanaugh CM, Gardiner SL, Jones ML, Jannasch HW, Waterbury JB. Prokaryotic Cells in the Hydrothermal Vent Tube Worm Riftia pachyptila Jones: Possible Chemoautotrophic Symbionts. Science. 1981;213:340–342. doi: 10.1126/science.213.4505.340. [DOI] [PubMed] [Google Scholar]
- Cebra JJ. Influences of microbiota on intestinal immune system development. Am J Clin Nutr. 1999;69:1046S–1051S. doi: 10.1093/ajcn/69.5.1046s. [DOI] [PubMed] [Google Scholar]
- Cebra JJ, Periwal SB, Lee G, Lee F, Shroff KE. Development and maintenance of the gut-associated lymphoid tissue (GALT): the roles of enteric bacteria and viruses. Dev Immunol. 1998;6:13–18. doi: 10.1155/1998/68382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan YK, Estaki M, Gibson DL. Clinical consequences of diet-induced dysbiosis. Ann Nutr Metab. 2013;63(Suppl 2):28–40. doi: 10.1159/000354902. [DOI] [PubMed] [Google Scholar]
- Chandler JA, Lang JM, Bhatnagar S, Eisen JA, Kopp A. Bacterial communities of diverse Drosophila species: ecological context of a host-microbe model system. PLoS Genet. 2011;7:e1002272. doi: 10.1371/journal.pgen.1002272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charroux B, Royet J. Drosophila immune response: From systemic antimicrobial peptide production in fat body cells to local defense in the intestinal tract. Fly. 2010;4:40–47. doi: 10.4161/fly.4.1.10810. [DOI] [PubMed] [Google Scholar]
- Choi KY, Chow LN, Mookherjee N. Cationic host defence peptides: multifaceted role in immune modulation and inflammation. J Innate Immun. 2012;4:361–370. doi: 10.1159/000336630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow J, Mazmanian S. Getting the bugs out of the immune system: do bacterial microbiota “fix” intestinal T cell responses? Cell Host & Microbe. 2009;5:8–12. doi: 10.1016/j.chom.2008.12.006. [DOI] [PubMed] [Google Scholar]
- Chow J, Mazmanian SK. A pathobiont of the microbiota balances host colonization and intestinal inflammation. Cell Host Microbe. 2010;7:265–276. doi: 10.1016/j.chom.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung H, Pamp SJ, Hill JA, Surana NK, Edelman SM, Troy EB, Reading NC, Villablanca EJ, Wang S, Mora JR, Umesaki Y, Mathis D, Benoist C, Relman DA, Kasper DL. Gut immune maturation depends on colonization with a host-specific microbiota. Cell. 2012;149:1578–1593. doi: 10.1016/j.cell.2012.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke T, Davis K, Lysenko E, Zhou A, Yu Y, Weiser J. Recognition of peptidoglycan from the microbiota by Nod1 enhances systemic innate immunity. Nat Med. 2010;16:228–231. doi: 10.1038/nm.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. Bacterial community variation in human body habitats across space and time. Science. 2009;326:1694–1697. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuomo A, Bonaldi T. Systems biology “on-the-fly”: SILAC-based quantitative proteomics and RNAi approach in Drosophila melanogaster. Methods Mol Biol. 2010;662:59–78. doi: 10.1007/978-1-60761-800-3_3. [DOI] [PubMed] [Google Scholar]
- Dalmasso G, Nguyen HT, Yan Y, Laroui H, Charania MA, Ayyadurai S, Sitaraman SV, Merlin D. Microbiota modulate host gene expression via microRNAs. PLoS One. 2011;6:e19293. doi: 10.1371/journal.pone.0019293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson B. Ciona intestinalis as a model for cardiac development. Semin Cell Dev Biol. 2007;18:16–26. doi: 10.1016/j.semcdb.2006.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson EH. The sea urchin genome: where will it lead us? Science. 2006;314:939–940. doi: 10.1126/science.1136252. [DOI] [PubMed] [Google Scholar]
- de Morais Guedes S, Vitorino R, Domingues R, Tomer K, Correia AJ, Amado F, Domingues P. Proteomics of immune-challenged Drosophila melanogaster larvae hemolymph. Biochem Biophys Res Commun. 2005;328:106–115. doi: 10.1016/j.bbrc.2004.12.135. [DOI] [PubMed] [Google Scholar]
- Dehal P, Satou Y, Campbell R, Chapman J, Degnan B, De Tomaso A, Davidson B, Di Gregorio A, Gelpke M, Goodstein D, Harafuji N, Hastings K, Ho I, Hotta K, Huang W, Kawashima T, Lemaire P, Martinez D, Meinertzhagen I, Necula S, Nonaka M, Putnam N, Rash S, Saiga H, Satake M, Terry A, Yamada L, Wang H, Awazu S, Azumi K, Boore J, Branno M, Chin-Bow S, DeSantis R, Doyle S, Francino P, Keys D, Haga S, Hayashi H, Hino K, Imai K, Inaba K, Kano S, Kobayashi K, Kobayashi M, Lee B, Makabe K, Manohar C, Matassi G, Medina M, Mochizuki Y, Mount S, Morishita T, Miura S, Nakayama A, Nishizaka S, Nomoto H, Ohta F, Oishi K, Rigoutsos I, Sano M, Sasaki A, Sasakura Y, Shoguchi E, Shin-i T, Spagnuolo A, Stainier D, Suzuki M, Tassy O, Takatori N, Tokuoka M, Yagi K, Yoshizaki F, Wada S, Zhang C, Hyatt P, Larimer F, Detter C, Doggett N, Glavina T, Hawkins T, Richardson P, Lucas S, Kohara Y, Levine M, Satoh N, Rokhsar D. The draft genome of Ciona intestinalis: insights into chordate and vertebrate origins. Science. 2002;298:2157–2167. doi: 10.1126/science.1080049. [DOI] [PubMed] [Google Scholar]
- Deplancke B, Gaskins HR. Microbial modulation of innate defense: goblet cells and the intestinal mucus layer. Am J Clin Nutr. 2001;73:1131S–1141S. doi: 10.1093/ajcn/73.6.1131S. [DOI] [PubMed] [Google Scholar]
- Deppe U, Schierenberg E, Cole T, Krieg C, Schmitt D, Yoder B, von Ehrenstein G. Cell lineages of the embryo of the nematode Caenorhabditis elegans. Proc Natl Acad Sci U S A. 1978;75:376–380. doi: 10.1073/pnas.75.1.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien M, van Passel MW, van de Bovenkamp JH, Schipper RG, de Vos WM, Dekker J. Mucin-bacterial interactions in the human oral cavity and digestive tract. Gut Microbes. 2010;1:254–268. doi: 10.4161/gmic.1.4.12778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Bella MA, Fedders H, De Leo G, Leippe M. Localization of antimicrobial peptides in the tunic of Ciona intestinalis (Ascidiacea, Tunicata) and their involvement in local inflammatory-like reactions. Results in Immunol. 2011;1:70–75. doi: 10.1016/j.rinim.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond G, Beckloff N, Weinberg A, Kisich KO. The roles of antimicrobial peptides in innate host defense. Curr Pharm Des. 2009;15:2377–2392. doi: 10.2174/138161209788682325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dishaw LJ, Flores-Torres J, Lax S, Gemayel K, Leigh B, Melillo D, Mueller MG, Natale L, Zucchetti I, De Santis R, Pinto MR, Litman GW, Gilbert JA. The gut of geographically disparate Ciona intestinalis harbors a core microbiota. PLoS One. 2014;9:e93386. doi: 10.1371/journal.pone.0093386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dishaw LJ, Flores-Torres JA, Mueller MG, Karrer CR, Skapura DP, Melillo D, Zucchetti I, De Santis R, Pinto MR, Litman GW. A basal chordate model for studies of gut microbial immune interactions. Front Immunol. 2012a;3:96. doi: 10.3389/fimmu.2012.00096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dishaw LJ, Giacomelli S, Melillo D, Zucchetti I, Haire RN, Natale L, Russo NA, De Santis R, Litman GW, Pinto MR. A role for variable region-containing chitin-binding proteins (VCBPs) in host gut-bacteria interactions. Proc Natl Acad Sci U S A. 2011;108:16747–16752. doi: 10.1073/pnas.1109687108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dishaw LJ, Haire RN, Litman GW. The amphioxus genome provides unique insight into the evolution of immunity. Brief Funct Gen. 2012b;11:167–176. doi: 10.1093/bfgp/els007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake HL, Horn MA. As the worm turns: the earthworm gut as a transient habitat for soil microbial biomes. Annu Rev Microbiol. 2007;61:169–189. doi: 10.1146/annurev.micro.61.080706.093139. [DOI] [PubMed] [Google Scholar]
- Duerkop BA, Vaishnava S, Hooper LV. Immune responses to the microbiota at the intestinal mucosal surface. Immunity. 2009;31:368–376. doi: 10.1016/j.immuni.2009.08.009. [DOI] [PubMed] [Google Scholar]
- Duerr CU, Hornef MW. The mammalian intestinal epithelium as integral player in the establishment and maintenance of host-microbial homeostasis. Semin Immunol. 2012;24:25–35. doi: 10.1016/j.smim.2011.11.002. [DOI] [PubMed] [Google Scholar]
- Eberl G. A new vision of immunity: homeostasis of the superorganism. Mucosal Immunol. 2010;3:450–460. doi: 10.1038/mi.2010.20. [DOI] [PubMed] [Google Scholar]
- Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelman S, Kasper D. Symbiotic commensal bacteria direct maturation of the host immune system. Curr Opin Gastroenterol. 2008;24:720–724. doi: 10.1097/MOG.0b013e32830c4355. [DOI] [PubMed] [Google Scholar]
- Engel P, Martinson VG, Moran NA. Functional diversity within the simple gut microbiota of the honey bee. Proc Natl Acad Sci U S A. 2012;109:11002–11007. doi: 10.1073/pnas.1202970109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engstrom Y, Loseva O, Theopold U. Proteomics of the Drosophila immune response. Trends Biotechnol. 2004;22:600–605. doi: 10.1016/j.tibtech.2004.09.002. [DOI] [PubMed] [Google Scholar]
- Equileor Md, Ottaviani E. The central role of immunity in the symbiotic event referred as parasitism. Invert Surv J. 2011;8:227–230. [Google Scholar]
- Eri R, Chieppa M. Messages from the Inside. The Dynamic Environment that Favors Intestinal Homeostasis. Front Immunol. 2013;4:323. doi: 10.3389/fimmu.2013.00323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ermak TH. A comparison of cell proliferation patterns in the digestive tract of ascidians. J Exp Zool. 1981;217:325–339. [Google Scholar]
- Everett ML, Palestrant D, Miller SE, Bollinger RB, Parker W. Immune exclusion and immune inclusion: a new model of host-bacterial interactions in the gut. Clin Appl Imm Rev. 2004;5:321–332. [Google Scholar]
- Falk PG, Hooper LV, Midtvedt T, Gordon JI. Creating and maintaining the gastrointestinal ecosystem: what we know and need to know from gnotobiology. Microbiol Mol Biol Rev. 1998;62:1157–1170. doi: 10.1128/mmbr.62.4.1157-1170.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallucca F, Porrata C, Fallucca S, Pianesi M. Influence of diet on gut microbiota, inflammation and type 2 diabetes mellitus. First experience with macrobiotic Ma-Pi 2 diet Diabetes. Metab Res Rev. 2014;30:48–54. doi: 10.1002/dmrr.2518. [DOI] [PubMed] [Google Scholar]
- Faust K, Sathirapongsasuti JF, Izard J, Segata N, Gevers D, Raes J, Huttenhower C. Microbial co-occurrence relationships in the human microbiome. PLoS Comp Bio. 2012;8:e1002606. doi: 10.1371/journal.pcbi.1002606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedders H, Leippe M. A reverse search for antimicrobial peptides in Ciona intestinalis: identification of a gene family expressed in hemocytes and evaluation of activity. Dev Comp Immunol. 2008;32:286–298. doi: 10.1016/j.dci.2007.06.003. [DOI] [PubMed] [Google Scholar]
- Fischbach MA, Sonnenburg JL. Eating for two: how metabolism establishes interspecies interactions in the gut. Cell Host & Microbe. 2011;10:336–347. doi: 10.1016/j.chom.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzenburg S, Fraune S, Altrock PM, Kunzel S, Baines JF, Traulsen A, Bosch TC. Bacterial colonization of Hydra hatchlings follows a robust temporal pattern. ISME J. 2013a;7:781–790. doi: 10.1038/ismej.2012.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzenburg S, Walter J, Kunzel S, Wang J, Baines JF, Bosch TC, Fraune S. Distinct antimicrobial peptide expression determines host species-specific bacterial associations. Proc Natl Acad Sci U S A. 2013b;110:E3730–3738. doi: 10.1073/pnas.1304960110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraune S, Abe Y, Bosch TC. Disturbing epithelial homeostasis in the metazoan Hydra leads to drastic changes in associated microbiota. Environ Microbiol. 2009;11:2361–2369. doi: 10.1111/j.1462-2920.2009.01963.x. [DOI] [PubMed] [Google Scholar]
- Fraune S, Augustin R, Anton-Erxleben F, Wittlieb J, Gelhaus C, Klimovich VB, Samoilovich MP, Bosch TC. In an early branching metazoan, bacterial colonization of the embryo is controlled by maternal antimicrobial peptides. Proc Natl Acad Sci U S A. 2010;107:18067–18072. doi: 10.1073/pnas.1008573107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraune S, Bosch TC. Long-term maintenance of species-specific bacterial microbiota in the basal metazoan Hydra. Proc Natl Acad Sci U S A. 2007;104:13146–13151. doi: 10.1073/pnas.0703375104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friman V, Adlerberth I, Connell H, Svanborg C, Hanson LA, Wold AE. Decreased expression of mannose-specific adhesins by Escherichia coli in the colonic microflora of immunoglobulin A-deficient individuals. Infect Immun. 1996;64:2794–2798. doi: 10.1128/iai.64.7.2794-2798.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimura K, Slusher N, Cabana M, Lynch S. Role of the gut microbiota in defining human health. Expert Rev Anti Infect Ther. 2010;8:435–454. doi: 10.1586/eri.10.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita T, Endo Y, Nonaka M. Primitive complement system--recognition and activation. Mol Immunol. 2004;41:103–111. doi: 10.1016/j.molimm.2004.03.026. [DOI] [PubMed] [Google Scholar]
- Galili U, Wang L, LaTemple DC, Radic MZ. The natural anti-Gal antibody. Sub-Cell Biochem. 1999;32:79–106. doi: 10.1007/978-1-4615-4771-6_4. [DOI] [PubMed] [Google Scholar]
- Garigan D, Hsu AL, Fraser AG, Kamath RS, Ahringer J, Kenyon C. Genetic analysis of tissue aging in Caenorhabditis elegans: a role for heat-shock factor and bacterial proliferation. Genetics. 2002;161:1101–1112. doi: 10.1093/genetics/161.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gewirtz AT, Navas TA, Lyons S, Godowski PJ, Madara JL. Cutting edge: bacterial flagellin activates basolaterally expressed TLR5 to induce epithelial proinflammatory gene expression. J Immunol. 2001;167:1882–1885. doi: 10.4049/jimmunol.167.4.1882. [DOI] [PubMed] [Google Scholar]
- Ghosh J, Buckley KM, Nair SV, Raftos DA, Miller C, Majeske AJ, Hibino T, Rast JP, Roth M, Smith LC. Sp185/333: a novel family of genes and proteins involved in the purple sea urchin immune response. Dev Comp Immunol. 2010;34:235–245. doi: 10.1016/j.dci.2009.10.008. [DOI] [PubMed] [Google Scholar]
- Gibbons SM, Caporaso JG, Pirrung M, Field D, Knight R, Gilbert JA. Evidence for a persistent microbial seed bank throughout the global ocean. Proc Natl Acad Sci U S A. 2013;110:4651–4655. doi: 10.1073/pnas.1217767110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert DG. DroSpeGe: rapid access database for new Drosophila species genomes. Nucleic Acids Res. 2007;35:D480–485. doi: 10.1093/nar/gkl997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill SR, Pop M, Deboy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, Gordon JI, Relman DA, Fraser-Liggett CM, Nelson KE. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glittenberg MT, Kounatidis I, Christensen D, Kostov M, Kimber S, Roberts I, Ligoxygakis P. Pathogen and host factors are needed to provoke a systemic host response to gastrointestinal infection of Drosophila larvae by Candida albicans. Dis Model Mech. 2011;4:515–525. doi: 10.1242/dmm.006627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard CK, Hoggett AK. Gut contents of the ascidian Pyura praeputialis: endogenous and exogenous components. J Zool. 1982;196:489–497. [Google Scholar]
- Gordon HA. The germ-free animal. Its use in the study of “physiologic” effects of the normal microbial flora on the animal host. Am J Dig Dis. 1960;5:841–867. doi: 10.1007/BF02232187. [DOI] [PubMed] [Google Scholar]
- Gruenheid S, Le Moual H. Resistance to antimicrobial peptides in Gram-negative bacteria. FEMS Microbiol Lett. 2012;330:81–89. doi: 10.1111/j.1574-6968.2012.02528.x. [DOI] [PubMed] [Google Scholar]
- Gusarov I, Gautier L, Smolentseva O, Shamovsky I, Eremina S, Mironov A, Nudler E. Bacterial nitric oxide extends the lifespan of C. elegans. Cell. 2013;152:818–830. doi: 10.1016/j.cell.2012.12.043. [DOI] [PubMed] [Google Scholar]
- Gutsmann T, Hagge SO, David A, Roes S, Bohling A, Hammer MU, Seydel U. Lipid-mediated resistance of Gram-negative bacteria against various pore-forming antimicrobial peptides. J Endotoxin Res. 2005;11:167–173. doi: 10.1179/096805105X37330. [DOI] [PubMed] [Google Scholar]
- Hahn MW, Han MV, Han SG. Gene family evolution across 12 Drosophila genomes. PLoS Genet. 2007;3:e197. doi: 10.1371/journal.pgen.0030197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer J, Hammer K, Kletter K. Lipids infused into the jejunum accelerate small intestinal transit but delay ileocolonic transit of solids and liquids. Gut. 1998;43:111–116. doi: 10.1136/gut.43.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hejnol A, Martindale MQ. Acoel development indicates the independent evolution of the bilaterian mouth and anus. Nature. 2008;456:382–386. doi: 10.1038/nature07309. [DOI] [PubMed] [Google Scholar]
- Herndl G, Velimirov B. Deuxieme colloque international de bacteriologie marine. IFREMER; Brest, France: 1986. Bacteria in the coelenteron of Anthozoa: control of coelenteric bacterial density by the coelenteric fluid; pp. 407–414. [Google Scholar]
- Hibino T, Loza-Coll M, Messier C, Majeske AJ, Cohen AH, Terwilliger DP, Buckley KM, Brockton V, Nair SV, Berney K, Fugmann SD, Anderson MK, Pancer Z, Cameron RA, Smith LC, Rast JP. The immune gene repertoire encoded in the purple sea urchin genome. Dev Biol. 2006;300:349–365. doi: 10.1016/j.ydbio.2006.08.065. [DOI] [PubMed] [Google Scholar]
- Hill D, Artis D. Intestinal bacteria and the regulation of immune cell homeostasis. Ann Rev Immunol. 2010;28:623–667. doi: 10.1146/annurev-immunol-030409-101330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T, Nishida H. Developmental fates of larval tissues after metamorphosis in the ascidian, Halocynthia roretzi. II Origin of endodermal tissues of the juvenile. Dev Genes Evol. 2000;210:55–63. doi: 10.1007/s004270050011. [DOI] [PubMed] [Google Scholar]
- Holland LZ. Body-plan evolution in the Bilateria: early antero-posterior patterning and the deuterostome-protostome dichotomy. Curr Opin Genet Dev. 2000;10:434–442. doi: 10.1016/s0959-437x(00)00109-x. [DOI] [PubMed] [Google Scholar]
- Holland LZ, Albalat R, Azumi K, Benito-Gutierrez E, Blow MJ, Bronner-Fraser M, Brunet F, Butts T, Candiani S, Dishaw LJ, Ferrier DE, Garcia-Fernandez J, Gibson-Brown JJ, Gissi C, Godzik A, Hallbook F, Hirose D, Hosomichi K, Ikuta T, Inoko H, Kasahara M, Kasamatsu J, Kawashima T, Kimura A, Kobayashi M, Kozmik Z, Kubokawa K, Laudet V, Litman GW, McHardy AC, Meulemans D, Nonaka M, Olinski RP, Pancer Z, Pennacchio LA, Pestarino M, Rast JP, Rigoutsos I, Robinson-Rechavi M, Roch G, Saiga H, Sasakura Y, Satake M, Satou Y, Schubert M, Sherwood N, Shiina T, Takatori N, Tello J, Vopalensky P, Wada S, Xu A, Ye Y, Yoshida K, Yoshizaki F, Yu JK, Zhang Q, Zmasek CM, de Jong PJ, Osoegawa K, Putnam NH, Rokhsar DS, Satoh N, Holland PW. The amphioxus genome illuminates vertebrate origins and cephalochordate biology. Genome Res. 2008;18:1100–1111. doi: 10.1101/gr.073676.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper L. Do symbiotic bacteria subvert host immunity? Nat Rev Micro. 2009;7:367–374. doi: 10.1038/nrmicro2114. [DOI] [PubMed] [Google Scholar]
- Hooper L, Gordon J. Commensal host-bacterial relationships in the gut. Science. 2001;292:1115–1118. doi: 10.1126/science.1058709. [DOI] [PubMed] [Google Scholar]
- Hooper L, Macpherson A. Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat Rev Immunol. 2010;10:159–169. doi: 10.1038/nri2710. [DOI] [PubMed] [Google Scholar]
- Hooper L, Wong M, Thelin A, Hansson L, Falk P, Gordon J. Molecular analysis of commensal host-microbial relationships in the intestine. Science. 2001;291:881–884. doi: 10.1126/science.291.5505.881. [DOI] [PubMed] [Google Scholar]
- Hooper LV, Bry L, Falk PG, Gordon JI. Host-microbial symbiosis in the mammalian intestine: exploring an internal ecosystem. Bioessays. 1998;20:336–343. doi: 10.1002/(SICI)1521-1878(199804)20:4<336::AID-BIES10>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Huang S, Yuan S, Guo L, Yu Y, Li J, Wu T, Liu T, Yang M, Wu K, Liu H, Ge J, Yu Y, Huang H, Dong M, Yu C, Chen S, Xu A. Genomic analysis of the immune gene repertoire of amphioxus reveals extraordinary innate complexity and diversity. Genome Res. 2008;18:1112–1126. doi: 10.1101/gr.069674.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes A, Friedman R. Loss of ancestral genes in the genomic evolution of Ciona intestinalis. Evol Dev. 2005;7:196–200. doi: 10.1111/j.1525-142X.2005.05022.x. [DOI] [PubMed] [Google Scholar]
- Human Microbiome Project, C. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idowu AB, Edema MO, Adeyi AO. Gut Microflora and Microfauna of Earthworm Species in the Soilds of the Research Farms of the University of Agriculture, Abeokuta, Nigeria. Biol Agri Hort. 2008;25:185–200. [Google Scholar]
- Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, Tanoue T, Imaoka A, Itoh K, Takeda K, Umesaki Y, Honda K, Littman DR. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson ME. Fast renewal of the distal colonic mucus layers by the surface goblet cells as measured by in vivo labeling of mucin glycoproteins. PLoS One. 2012;7:e41009. doi: 10.1371/journal.pone.0041009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson ME, Ambort D, Pelaseyed T, Schutte A, Gustafsson JK, Ermund A, Subramani DB, Holmen-Larsson JM, Thomsson KA, Bergstrom JH, van der Post S, Rodriguez-Pineiro AM, Sjovall H, Backstrom M, Hansson GC. Composition and functional role of the mucus layers in the intestine. Cell Mol Life Sci. 2011a;68:3635–3641. doi: 10.1007/s00018-011-0822-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson ME, Larsson JM, Hansson GC. The two mucus layers of colon are organized by the MUC2 mucin, whereas the outer layer is a legislator of host-microbial interactions. Proc Natl Acad Sci U S A. 2011b;108(Suppl 1):4659–4665. doi: 10.1073/pnas.1006451107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson ME, Phillipson M, Petersson J, Velcich A, Holm L, Hansson GC. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc Natl Acad Sci U S A. 2008;105:15064–15069. doi: 10.1073/pnas.0803124105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juge N. Microbial adhesins to gastrointestinal mucus. Trends Microbiol. 2012;20:30–39. doi: 10.1016/j.tim.2011.10.001. [DOI] [PubMed] [Google Scholar]
- Kanther M, Rawls J. Host-microbe interactions in the developing zebrafish. Curr Opin Immunol. 2010;22:10–19. doi: 10.1016/j.coi.2010.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashyap PC, Marcobal A, Ursell LK, Larauche M, Duboc H, Earle KA, Sonnenburg ED, Ferreyra JA, Higginbottom SK, Million M, Tache Y, Pasricha PJ, Knight R, Farrugia G, Sonnenburg JL. Complex interactions among diet, gastrointestinal transit, and gut microbiota in humanized mice. Gastroenterology. 2013a;144:967–977. doi: 10.1053/j.gastro.2013.01.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashyap PC, Marcobal A, Ursell LK, Smits SA, Sonnenburg ED, Costello EK, Higginbottom SK, Domino SE, Holmes SP, Relman DA, Knight R, Gordon JI, Sonnenburg JL. Genetically dictated change in host mucus carbohydrate landscape exerts a diet-dependent effect on the gut microbiota. Proc Natl Acad Sci U S A. 2013b;110:17059–17064. doi: 10.1073/pnas.1306070110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz MJ. Comparative anatomy of the tunicate tadpole, Ciona intestinalis. Biol Bull. 1983;164:1–27. [Google Scholar]
- Khomiakov NV, Kharin SA, Nechitailo T, Golyshin PN, Kurakov AV, Byzov BA, Zviagintsev DG. Reaction of microorganisms to the digestive fluid of the earthworms. Mikrobiologiia. 2007;76:55–65. [PubMed] [Google Scholar]
- Kim SH, Lee WJ. Role of DUOX in gut inflammation: lessons from model of gut-microbiota interactions. Front Cell Infec Microbiol. 2014;3:116. doi: 10.3389/fcimb.2013.00116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura A, Sakaguchi E, Nonaka M. Multi-component complement system of Cnidaria: C3, Bf, and MASP genes expressed in the endodermal tissues of a sea anemone, Nematostella vectensis. Immunobiology. 2009;214:165–178. doi: 10.1016/j.imbio.2009.01.003. [DOI] [PubMed] [Google Scholar]
- King GM, Judd C, Kuske CR, Smith C. Analysis of stomach and gut microbiomes of the eastern oyster (Crassostrea virginica) from coastal Louisiana, USA. PLoS One. 2012;7:e51475. doi: 10.1371/journal.pone.0051475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsolver MB, Huang Z, Hardy RW. Insect antiviral innate immunity: pathways, effectors, and connections. J Mol Biol. 2013;425:4921–4936. doi: 10.1016/j.jmb.2013.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren O, Knights D, Gonzalez A, Waldron L, Segata N, Knight R, Huttenhower C, Ley RE. A guide to enterotypes across the human body: meta-analysis of microbial community structures in human microbiome datasets. PLoS Comp Biol. 2013;9:e1002863. doi: 10.1371/journal.pcbi.1002863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koropatnick TA, Engle JT, Apicella MA, Stabb EV, Goldman WE, McFall-Ngai MJ. Microbial factor-mediated development in a host-bacterial mutualism. Science. 2004;306:1186–1188. doi: 10.1126/science.1102218. [DOI] [PubMed] [Google Scholar]
- Koyama H, Taneda Y, Ishii T. The postbranchial digestive tract of the ascidian, Polyandrocarpa misakiensis (Tunicata: Ascidiacea). 2. Stomach. Zoolog Sci. 2012;29:97–110. doi: 10.2108/zsj.29.97. [DOI] [PubMed] [Google Scholar]
- Kubo A, Imai KS, Satou Y. Gene-regulatory networks in the Ciona embryos. Brief Funct Gen Prot. 2009;8:250–255. doi: 10.1093/bfgp/elp018. [DOI] [PubMed] [Google Scholar]
- Kuraishi T, Binggeli O, Opota O, Buchon N, Lemaitre B. Genetic evidence for a protective role of the peritrophic matrix against intestinal bacterial infection in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2011;108:15966–15971. doi: 10.1073/pnas.1105994108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuraishi T, Hori A, Kurata S. Host-microbe interactions in the gut of Drosophila melanogaster. Front Physiol. 2013;4:375. doi: 10.3389/fphys.2013.00375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvell K, Cooper EL, Engelmann P, Bovari J, Nemeth P. Blurring borders: innate immunity with adaptive features. Clin Dev Immunol. 2007;2007:83671. doi: 10.1155/2007/83671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KA, Kim SH, Kim EK, Ha EM, You H, Kim B, Kim MJ, Kwon Y, Ryu JH, Lee WJ. Bacterial-derived uracil as a modulator of mucosal immunity and gut-microbe homeostasis in Drosophila. Cell. 2013;153:797–811. doi: 10.1016/j.cell.2013.04.009. [DOI] [PubMed] [Google Scholar]
- Lee WJ. Bacterial-modulated host immunity and stem cell activation for gut homeostasis. Genes Dev. 2009;23:2260–2265. doi: 10.1101/gad.1858709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YH, Su KY, Wyse A, Barbas A, Palestrandt D, Shieh K, Everett ML, Devalapalli A, Orndorff PE, Bollinger RR, Parker W. Incorporation of secretory immunoglobulin A into biofilms can decrease their resistance to ciprofloxacin. Microbiol Immunol. 2011;55:174–183. doi: 10.1111/j.1348-0421.2010.00297.x. [DOI] [PubMed] [Google Scholar]
- Lemaitre B, Hoffmann J. The host defense of Drosophila melanogaster. Annu Rev Immunol. 2007;25:697–743. doi: 10.1146/annurev.immunol.25.022106.141615. [DOI] [PubMed] [Google Scholar]
- Lemaitre B, Miguel-Aliaga I. The digestive tract of Drosophila melanogaster. Annu Rev Genet. 2013;47:377–404. doi: 10.1146/annurev-genet-111212-133343. [DOI] [PubMed] [Google Scholar]
- Lentle RG, Janssen PW. Physical characteristics of digesta and their influence on flow and mixing in the mammalian intestine: a review. J Comp Physiol B. 2008;178:673–690. doi: 10.1007/s00360-008-0264-x. [DOI] [PubMed] [Google Scholar]
- Lentle RG, Janssen PW, Deloubens C, Lim YF, Hulls C, Chambers P. Mucosal microfolds augment mixing at the wall of the distal ileum of the brushtail possum. Neurogastroenterol Motil. 2013;25:881–e700. doi: 10.1111/nmo.12203. [DOI] [PubMed] [Google Scholar]
- Ley RE, Lozupone CA, Hamady M, Knight R, Gordon JI. Worlds within worlds: evolution of the vertebrate gut microbiota. Nat Rev Microbiol. 2008;6:776–788. doi: 10.1038/nrmicro1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124:837–848. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- Li M, Yang H, Gu JD. Phylogenetic diversity and axial distribution of microbes in the intestinal tract of the polychaete Neanthes glandicincta. Microb Ecol. 2009;58:892–902. doi: 10.1007/s00248-009-9550-8. [DOI] [PubMed] [Google Scholar]
- Liberti A, Melillo D, Zucchetti I, Natale L, Dishaw LJ, Litman GW, De Santis R, Pinto MR. Expression of Ciona intestinalis Variable Region-Containing Chitin- Binding Proteins during Development of the Gastrointestinal Tract and Their Role in Host-Microbe Interactions. PLoS One. 2014;9:e94984. doi: 10.1371/journal.pone.0094984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley KS, Griffiths DR. Proteomics in Drosophila melanogaster. Brief Funct Gen Prot. 2003;2:106–113. doi: 10.1093/bfgp/2.2.106. [DOI] [PubMed] [Google Scholar]
- Loker ES. Macroevolutionary Immunology: A Role for Immunity in the Diversification of Animal life. Front Immunol. 2012;3:25. doi: 10.3389/fimmu.2012.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loker ES, Adema CM, Zhang SM, Kepler TB. Invertebrate immune systems--not homogeneous, not simple, not well understood. Immunol Rev. 2004;198:10–24. doi: 10.1111/j.0105-2896.2004.0117.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489:220–230. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Zhuang Y, Liu J. Mining antimicrobial peptides from small open reading frames in Ciona intestinalis. J Pept Sci. 2014;20:25–29. doi: 10.1002/psc.2584. [DOI] [PubMed] [Google Scholar]
- Luckey D, Gomez A, Murray J, White B, Taneja V. Bugs & us: The role of the gut in autoimmunity. Indian J Med Res. 2013;138:732–743. [PMC free article] [PubMed] [Google Scholar]
- Macdonald T, Monteleone G. Immunity, inflammation, and allergy in the gut. Science. 2005;307:1920–1925. doi: 10.1126/science.1106442. [DOI] [PubMed] [Google Scholar]
- MacDonald TT, Pettersson S. Bacterial regulation of intestinal immune responses. Inflamm Bowel Diseases. 2000;6:116–122. doi: 10.1097/00054725-200005000-00008. [DOI] [PubMed] [Google Scholar]
- Macfarlane GT, Macfarlane S. Human colonic microbiota: ecology, physiology and metabolic potential of intestinal bacteria. Scand J Gastroenterol Suppl. 1997;222:3–9. doi: 10.1080/00365521.1997.11720708. [DOI] [PubMed] [Google Scholar]
- Majeske AJ, Oleksyk TK, Smith LC. The Sp185/333 immune response genes and proteins are expressed in cells dispersed within all major organs of the adult purple sea urchin. Innate Immun. 2013;19:569–587. doi: 10.1177/1753425912473850. [DOI] [PubMed] [Google Scholar]
- Mallo GV, Kurz CL, Couillault C, Pujol N, Granjeaud S, Kohara Y, Ewbank JJ. Inducible antibacterial defense system in C. elegans. Curr Biol. 2002;12:1209–1214. doi: 10.1016/s0960-9822(02)00928-4. [DOI] [PubMed] [Google Scholar]
- Marchiando AM, Graham WV, Turner JR. Epithelial barriers in homeostasis and disease. Ann Rev Pathol. 2010;5:119–144. doi: 10.1146/annurev.pathol.4.110807.092135. [DOI] [PubMed] [Google Scholar]
- Marcobal A, Southwick AM, Earle KA, Sonnenburg JL. A refined palate: bacterial consumption of host glycans in the gut. Glycobiology. 2013;23:1038–1046. doi: 10.1093/glycob/cwt040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino R, Kimura Y, De Santis R, Lambris JD, Pinto MR. Complement in urochordates: cloning and characterization of two C3-like genes in the ascidian Ciona intestinalis. Immunogenetics. 2002;53:1055–1064. doi: 10.1007/s00251-001-0421-9. [DOI] [PubMed] [Google Scholar]
- Marsh EK, May RC. Caenorhabditis elegans, a model organism for investigating immunity. Appl Environ Microbiol. 2012;78:2075–2081. doi: 10.1128/AEM.07486-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens EC, Chiang HC, Gordon JI. Mucosal glycan foraging enhances fitness and transmission of a saccharolytic human gut bacterial symbiont. Cell Host Microbe. 2008;4:447–457. doi: 10.1016/j.chom.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinson VG, Danforth BN, Minckley RL, Rueppell O, Tingek S, Moran NA. A simple and distinctive microbiota associated with honey bees and bumble bees. Mol Ecol. 2011;20:619–628. doi: 10.1111/j.1365-294X.2010.04959.x. [DOI] [PubMed] [Google Scholar]
- Mashanov VS, Garcia-Arraras JE. Gut regeneration in holothurians: a snapshot of recent developments. Biol Bull. 2011;221:93–109. doi: 10.1086/BBLv221n1p93. [DOI] [PubMed] [Google Scholar]
- Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122:107–118. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- Mazmanian SK. Capsular polysaccharides of symbiotic bacteria modulate immune responses during experimental colitis. J Pediatr Gastroenterol Nutr. 2008;46(Suppl 1):E11–12. doi: 10.1097/01.mpg.0000313824.70971.a7. [DOI] [PubMed] [Google Scholar]
- Mazmanian SK, Kasper DL. The love-hate relationship between bacterial polysaccharides and the host immune system. Nat Rev Immunol. 2006;6:849–858. doi: 10.1038/nri1956. [DOI] [PubMed] [Google Scholar]
- Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453:620–625. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- McFall-Ngai M. Unseen forces: the influence of bacteria on animal development. Dev Biol. 2002;242:1–14. doi: 10.1006/dbio.2001.0522. [DOI] [PubMed] [Google Scholar]
- McFall-Ngai M, Hadfield MG, Bosch TC, Carey HV, Domazet-Loso T, Douglas AE, Dubilier N, Eberl G, Fukami T, Gilbert SF, Hentschel U, King N, Kjelleberg S, Knoll AH, Kremer N, Mazmanian SK, Metcalf JL, Nealson K, Pierce NE, Rawls JF, Reid A, Ruby EG, Rumpho M, Sanders JG, Tautz D, Wernegreen JJ. Animals in a bacterial world, a new imperative for the life sciences. Proc Natl Acad Sci U S A. 2013;110:3229–3236. doi: 10.1073/pnas.1218525110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medzhitov R. Innate immunity: quo vadis? Nat Immunol. 2010;11:551–553. doi: 10.1038/ni0710-551. [DOI] [PubMed] [Google Scholar]
- Meinertzhagen IA, Okamura Y. The larval ascidian nervous system: the chordate brain from its small beginnings. Trends Neurosci. 2001;24:401–410. doi: 10.1016/s0166-2236(00)01851-8. [DOI] [PubMed] [Google Scholar]
- Meziti A, Kormas KA, Pancucci-Papadopoulou MA, Thessalou-Legaki M. Bacterial phylotypes associated with the digestive tract of the sea urchin Paracentrotus lividus and the ascidian Microcosmus sp. Russian J Mar Biol. 2007;33:84–91. [Google Scholar]
- Micchelli CA. The origin of intestinal stem cells in Drosophila. Dev Dyn. 2012;241:85–91. doi: 10.1002/dvdy.22759. [DOI] [PubMed] [Google Scholar]
- Micchelli CA, Sudmeier L, Perrimon N, Tang S, Beehler-Evans R. Identification of adult midgut precursors in Drosophila. Gene Expr Patterns. 2011;11:12–21. doi: 10.1016/j.gep.2010.08.005. [DOI] [PubMed] [Google Scholar]
- Mondot S, de Wouters T, Dore J, Lepage P. The human gut microbiome and its dysfunctions. Dig Dis. 2013;31:278–285. doi: 10.1159/000354678. [DOI] [PubMed] [Google Scholar]
- Mortha A, Chudnovskiy A, Hashimoto D, Bogunovic M, Spencer SP, Belkaid Y, Merad M. Microbiota-dependent crosstalk between macrophages and ILC3 promotes intestinal homeostasis. Science. 2014;343:1249288. doi: 10.1126/science.1249288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S, Vaishnava S, Hooper LV. Multi-layered regulation of intestinal antimicrobial defense. Cell Mol Life Sci. 2008;65:3019–3027. doi: 10.1007/s00018-008-8182-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munn C. Marine Microbiology, Ecology and Applications. 2nd. Garland Science; New York, NY: 2011. [Google Scholar]
- Nakazawa K, Yamazawa T, Moriyama Y, Ogura Y, Kawai N, Sasakura Y, Saiga H. Formation of the digestive tract in Ciona intestinalis includes two distinct morphogenic processes between its anterior and posterior parts. Dev Dyn. 2013;242:1172–1183. doi: 10.1002/dvdy.24009. [DOI] [PubMed] [Google Scholar]
- Neish AS. Microbes in gastrointestinal health and disease. Gastroenterology. 2009;136:65–80. doi: 10.1053/j.gastro.2008.10.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neish AS. Mucosal immunity and the microbiome. Ann Am Thor Soc. 2014;11(Suppl 1):S28–32. doi: 10.1513/AnnalsATS.201306-161MG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noah TK, Donahue B, Shroyer NF. Intestinal development and differentiation. Exp Cell Res. 2011;317:2702–2710. doi: 10.1016/j.yexcr.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyholm SV, Graf J. Knowing your friends: invertebrate innate immunity fosters beneficial bacterial symbioses. Nat Rev Microbiol. 2012;10:815–827. doi: 10.1038/nrmicro2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyholm SV, Song P, Dang J, Bunce C, Girguis PR. Expression and putative function of innate immunity genes under in situ conditions in the symbiotic hydrothermal vent tubeworm Ridgeia piscesae. PLoS One. 2012;7:e38267. doi: 10.1371/journal.pone.0038267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Hara A, Shanahan F. The gut flora as a forgotten organ. EMBO reports. 2006;7:688–693. doi: 10.1038/sj.embor.7400731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega-Cava CF, Ishihara S, Rumi MA, Kawashima K, Ishimura N, Kazumori H, Udagawa J, Kadowaki Y, Kinoshita Y. Strategic compartmentalization of Toll-like receptor 4 in the mouse gut. J Immunol. 2003;170:3977–3985. doi: 10.4049/jimmunol.170.8.3977. [DOI] [PubMed] [Google Scholar]
- Ottaviani E, Ventura N, Mandrioli M, Candela M, Franchini A, Franceschi C. Gut microbiota as a candidate for lifespan extension: an ecological/evolutionary perspective targeted on living organisms as metaorganisms. Biogerontology. 2011;12:599–609. doi: 10.1007/s10522-011-9352-5. [DOI] [PubMed] [Google Scholar]
- Pacha J. Development of intestinal transport function in mammals. Physiol Rev. 2000;80:1633–1667. doi: 10.1152/physrev.2000.80.4.1633. [DOI] [PubMed] [Google Scholar]
- Paris M, Laudet V. The history of a developmental stage: metamorphosis in chordates. Genesis. 2008;46:657–672. doi: 10.1002/dvg.20443. [DOI] [PubMed] [Google Scholar]
- Parker W, Lesher AP. In pursuit of xenoreactive antibodies: where has it gotten us? Immunol Cell Biol. 2005;83:413–417. doi: 10.1111/j.1440-1711.2005.01349.x. [DOI] [PubMed] [Google Scholar]
- Parker W, Lin SS, Yu PB, Sood A, Nakamura YC, Song A, Everett ML, Platt JL. Naturally occurring anti-alpha-galactosyl antibodies: relationship to xenoreactive anti-alpha-galactosyl antibodies. Glycobiology. 1999;9:865–873. doi: 10.1093/glycob/9.9.865. [DOI] [PubMed] [Google Scholar]
- Parker W, Ollerton J. Evolutionary biology and anthropology suggest biome reconstitution as a necessary approach toward dealing with immune disorders. Evol Med Publ Health. 2013;2013:89–103. doi: 10.1093/emph/eot008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker W, Perkins SE, Harker M, Muehlenbein MP. A prescription for clinical immunology: the pills are available and ready for testing. Curr Med Res Opin. 2012;28:1193–1202. doi: 10.1185/03007995.2012.695731. [DOI] [PubMed] [Google Scholar]
- Parrinello N, Vizzini A, Arizza V, Salerno G, Parrinello D, Cammarata M, Giaramita F, Vazzana M. Enhanced expression of a cloned and sequenced Ciona intestinalis TNFalpha-like (CiTNF alpha) gene during the LPS-induced inflammatory response. Cell Tissue Res. 2008;334:305–317. doi: 10.1007/s00441-008-0695-4. [DOI] [PubMed] [Google Scholar]
- Parthasarathi K, Ranganathan LS, Anandi V, Zeyer J. Diversity of microflora in the gut and casts of tropical composting earthworms reared on different substrates. J Env Biol. 2007;28:87–97. [PubMed] [Google Scholar]
- Pearson C, Uhlig HH, Powrie F. Lymphoid microenvironments and innate lymphoid cells in the gut. Trends Immunol. 2012;33:289–296. doi: 10.1016/j.it.2012.04.004. [DOI] [PubMed] [Google Scholar]
- Pedros-Alio C. Marine microbial diversity: can it be determined? Trends Microbiol. 2006;14:257–263. doi: 10.1016/j.tim.2006.04.007. [DOI] [PubMed] [Google Scholar]
- Pedros-Alio C. The rare bacterial biosphere. Ann Rev Mar Sci. 2012;4:449–466. doi: 10.1146/annurev-marine-120710-100948. [DOI] [PubMed] [Google Scholar]
- Peterson DA, McNulty NP, Guruge JL, Gordon JI. IgA response to symbiotic bacteria as a mediator of gut homeostasis. Cell Host Microbe. 2007;2:328–339. doi: 10.1016/j.chom.2007.09.013. [DOI] [PubMed] [Google Scholar]
- Petrof EO, Claud EC, Gloor GB, Allen-Vercoe E. Microbial ecosystems therapeutics: a new paradigm in medicine? Ben Micro. 2013;4:53–65. doi: 10.3920/BM2012.0039. [DOI] [PubMed] [Google Scholar]
- Portal-Celhay C, Bradley ER, Blaser MJ. Control of intestinal bacterial proliferation in regulation of lifespan in Caenorhabditis elegans. BMC Microbiol. 2012;12:49. doi: 10.1186/1471-2180-12-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pott J, Hornef M. Innate immune signalling at the intestinal epithelium in homeostasis and disease. EMBO Rep. 2012;13:684–698. doi: 10.1038/embor.2012.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power SE, O'Toole PW, Stanton C, Ross RP, Fitzgerald GF. Intestinal microbiota, diet and health. Br J Nutr. 2014;111:387–402. doi: 10.1017/S0007114513002560. [DOI] [PubMed] [Google Scholar]
- Pradel E, Zhang Y, Pujol N, Matsuyama T, Bargmann CI, Ewbank JJ. Detection and avoidance of a natural product from the pathogenic bacterium Serratia marcescens by Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2007;104:2295–2300. doi: 10.1073/pnas.0610281104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulido D, Nogues MV, Boix E, Torrent M. Lipopolysaccharide neutralization by antimicrobial peptides: a gambit in the innate host defense strategy. J Innate Immunol. 2012;4:327–336. doi: 10.1159/000336713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Li R, Raes J, Arumugam M, Burgdorf K, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, Mende D, Li J, Xu J, Li S, Li D, Cao J, Wang B, Liang H, Zheng H, Xie Y, Tap J, Lepage P, Bertalan M, Batto J, Hansen T, Le Paslier D, Linneberg A, Nielsen H, Pelletier E, Renault P, Sicheritz-Ponten T, Turner K, Zhu H, Yu C, Jian M, Zhou Y, Li Y, Zhang X, Qin N, Yang H, Wang J, Brunak S, Doré J, Guarner F, Kristiansen K, Pedersen O, Parkhill J, Weissenbach J, Bork P, Ehrlich S, Consortium M. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–241. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- Rast JP, Smith LC, Loza-Coll M, Hibino T, Litman GW. Genomic insights into the immune system of the sea urchin. Science. 2006;314:952–956. doi: 10.1126/science.1134301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawls JF, Samuel BS, Gordon JI. Gnotobiotic zebrafish reveal evolutionarily conserved responses to the gut microbiota. Proc Natl Acad Sci USA. 2004;101:4596–4601. doi: 10.1073/pnas.0400706101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawls J, Mahowald M, Ley R, Gordon J. Reciprocal gut microbiota transplants from zebrafish and mice to germ-free recipients reveal host habitat selection. Cell. 2006;127:423–433. doi: 10.1016/j.cell.2006.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinkevich Y, Douek J, Haber O, Rinkevich B, Reshef R. Urochordate whole body regeneration inaugurates a diverse innate immune signaling profile. Dev Biol. 2007;312:131–146. doi: 10.1016/j.ydbio.2007.09.005. [DOI] [PubMed] [Google Scholar]
- Roeder T, Stanisak M, Gelhaus C, Bruchhaus I, Grotzinger J, Leippe M. Caenopores are antimicrobial peptides in the nematode Caenorhabditis elegans instrumental in nutrition and immunity. Dev Comp Immunol. 2010;34:203–209. doi: 10.1016/j.dci.2009.09.010. [DOI] [PubMed] [Google Scholar]
- Rook GA, Brunet LR. Microbes, Immunoregulation, and the Gut. Gut. 2005;54:317–320. doi: 10.1136/gut.2004.053785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa RD, de Lorgeril J, Tailliez P, Bruno R, Piquemal D, Bachere E. A hemocyte gene expression signature correlated with predictive capacity of oysters to survive Vibrio infections. BMC Genomics. 2012;13:252. doi: 10.1186/1471-2164-13-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg E, Sharon G, Atad I, Zilber-Rosenberg I. The evolution of animals and plants via symbiosis with microorganisms. Environ Microbiol Reports. 2010;2:500–506. doi: 10.1111/j.1758-2229.2010.00177.x. [DOI] [PubMed] [Google Scholar]
- Rost-Roszkowska MM, Undrul A. Fine structure and differentiation of the midgut epithelium of Allacma fusca (Insecta: Collembola: Symphypleona) Zool Studies. 2008;47:200–206. doi: 10.2108/zsj.25.753. [DOI] [PubMed] [Google Scholar]
- Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. 2009;9:313–323. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Round J, O'Connell R, Mazmanian S. Coordination of tolerogenic immune responses by the commensal microbiota. J Autoimmun. 2010a;34:J220–225. doi: 10.1016/j.jaut.2009.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci U S A. 2010;107:12204–12209. doi: 10.1073/pnas.0909122107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Round JL, O'Connell RM, Mazmanian SK. Coordination of tolerogenic immune responses by the commensal microbiota. J Autoimmun. 2010b;34:J220–225. doi: 10.1016/j.jaut.2009.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin GM. Around the genomes: the Drosophila genome project. Genome Res. 1996;6:71–79. doi: 10.1101/gr.6.2.71. [DOI] [PubMed] [Google Scholar]
- Rudi K, Odegard K, Lokken TT, Wilson R. A feeding induced switch from a variable to a homogenous state of the earthworm gut microbiota within a host population. PLoS One. 2009;4:e7528. doi: 10.1371/journal.pone.0007528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu JH, Ha EM, Lee WJ. Innate immunity and gut-microbe mutualism in Drosophila. Dev Comp Immunol. 2010;34:369–376. doi: 10.1016/j.dci.2009.11.010. [DOI] [PubMed] [Google Scholar]
- Saenz SA, Taylor BC, Artis D. Welcome to the neighborhood: epithelial cell-derived cytokines license innate and adaptive immune responses at mucosal sites. Immunol Rev. 2008;226:172–190. doi: 10.1111/j.1600-065X.2008.00713.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salem H, Kreutzer E, Sudakaran S, Kaltenpoth M. Actinobacteria as essential symbionts in firebugs and cotton stainers (Hemiptera, Pyrrhocoridae) Environ Microbiol. 2013;15:1956–1968. doi: 10.1111/1462-2920.12001. [DOI] [PubMed] [Google Scholar]
- Sasaki N, Ogasawara M, Sekiguchi T, Kusumoto S, Satake H. Toll-like receptors of the ascidian Ciona intestinalis: prototypes with hybrid functionalities of vertebrate Toll-like receptors. J Biol Chem. 2009;284:27336–27343. doi: 10.1074/jbc.M109.032433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasakura Y, Inaba K, Satoh N, Kondo M, Akasaka K. Ciona intestinalis and Oxycomanthus japonicus, representatives of marine invertebrates. Exp Anim. 2009;58:459–469. doi: 10.1538/expanim.58.459. [DOI] [PubMed] [Google Scholar]
- Savage DC. Microbial ecology of the gastrointestinal tract. Annu Rev Microbiol. 1977;31:107–133. doi: 10.1146/annurev.mi.31.100177.000543. [DOI] [PubMed] [Google Scholar]
- Schalen C. Prevalence of IgA receptors in clinical isolates of Streptococcus pyogenes and Streptococcus agalactiae: serologic distinction between the receptors by blocking antibodies. FEMS Immunol Med Microbiol. 1993;7:39–45. doi: 10.1111/j.1574-695X.1993.tb00379.x. [DOI] [PubMed] [Google Scholar]
- Schmitt P, Rosa RD, Duperthuy M, de Lorgeril J, Bachere E, Destoumieux-Garzon D. The Antimicrobial Defense of the Pacific Oyster, Crassostrea gigas. How Diversity may Compensate for Scarcity in the Regulation of Resident/Pathogenic Microflora. Front Microbiol. 2012;3:160. doi: 10.3389/fmicb.2012.00160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber S, Rosenstiel P, Albrecht M, Hampe J, Krawczak M. Genetics of Crohn disease, an archetypal inflammatory barrier disease. Nat Rev Genet. 2005;6:376–388. doi: 10.1038/nrg1607. [DOI] [PubMed] [Google Scholar]
- Scott MG, Dullaghan E, Mookherjee N, Galvas N, Waldbrook M, Thompson A, Wang A, Lee K, Doria S, Hamill P, Yu JJ, Li Y, Donini O, Guarna MM, Finlay BB, North JR, Hancock RE. An anti-infective peptide that selectively modulates the innate immune response. Nat Biotechnol. 2007;25:465–472. doi: 10.1038/nbt1288. [DOI] [PubMed] [Google Scholar]
- Sea Urchin Genome Sequencing, C. Sodergren E, Weinstock GM, Davidson EH, Cameron RA, Gibbs RA, Angerer RC, Angerer LM, Arnone MI, Burgess DR, Burke RD, Coffman JA, Dean M, Elphick MR, Ettensohn CA, Foltz KR, Hamdoun A, Hynes RO, Klein WH, Marzluff W, McClay DR, Morris RL, Mushegian A, Rast JP, Smith LC, Thorndyke MC, Vacquier VD, Wessel GM, Wray G, Zhang L, Elsik CG, Ermolaeva O, Hlavina W, Hofmann G, Kitts P, Landrum MJ, Mackey AJ, Maglott D, Panopoulou G, Poustka AJ, Pruitt K, Sapojnikov V, Song X, Souvorov A, Solovyev V, Wei Z, Whittaker CA, Worley K, Durbin KJ, Shen Y, Fedrigo O, Garfield D, Haygood R, Primus A, Satija R, Severson T, Gonzalez-Garay ML, Jackson AR, Milosavljevic A, Tong M, Killian CE, Livingston BT, Wilt FH, Adams N, Belle R, Carbonneau S, Cheung R, Cormier P, Cosson B, Croce J, Fernandez-Guerra A, Geneviere AM, Goel M, Kelkar H, Morales J, Mulner-Lorillon O, Robertson AJ, Goldstone JV, Cole B, Epel D, Gold B, Hahn ME, Howard-Ashby M, Scally M, Stegeman JJ, Allgood EL, Cool J, Judkins KM, McCafferty SS, Musante AM, Obar RA, Rawson AP, Rossetti BJ, Gibbons IR, Hoffman MP, Leone A, Istrail S, Materna SC, Samanta MP, Stolc V, Tongprasit W, Tu Q, Bergeron KF, Brandhorst BP, Whittle J, Berney K, Bottjer DJ, Calestani C, Peterson K, Chow E, Yuan QA, Elhaik E, Graur D, Reese JT, Bosdet I, Heesun S, Marra MA, Schein J, Anderson MK, Brockton V, Buckley KM, Cohen AH, Fugmann SD, Hibino T, Loza-Coll M, Majeske AJ, Messier C, Nair SV, Pancer Z, Terwilliger DP, Agca C, Arboleda E, Chen N, Churcher AM, Hallbook F, Humphrey GW, Idris MM, Kiyama T, Liang S, Mellott D, Mu X, Murray G, Olinski RP, Raible F, Rowe M, Taylor JS, Tessmar-Raible K, Wang D, Wilson KH, Yaguchi S, Gaasterland T, Galindo BE, Gunaratne HJ, Juliano C, Kinukawa M, Moy GW, Neill AT, Nomura M, Raisch M, Reade A, Roux MM, Song JL, Su YH, Townley IK, Voronina E, Wong JL, Amore G, Branno M, Brown ER, Cavalieri V, Duboc V, Duloquin L, Flytzanis C, Gache C, Lapraz F, Lepage T, Locascio A, Martinez P, Matassi G, Matranga V, Range R, Rizzo F, Rottinger E, Beane W, Bradham C, Byrum C, Glenn T, Hussain S, Manning G, Miranda E, Thomason R, Walton K, Wikramanayke A, Wu SY, Xu R, Brown CT, Chen L, Gray RF, Lee PY, Nam J, Oliveri P, Smith J, Muzny D, Bell S, Chacko J, Cree A, Curry S, Davis C, Dinh H, Dugan-Rocha S, Fowler J, Gill R, Hamilton C, Hernandez J, Hines S, Hume J, Jackson L, Jolivet A, Kovar C, Lee S, Lewis L, Miner G, Morgan M, Nazareth LV, Okwuonu G, Parker D, Pu LL, Thorn R, Wright R. The genome of the sea urchin Strongylocentrotus purpuratus. Science. 2006;314:941–952. doi: 10.1126/science.1133609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segata N, Haake SK, Mannon P, Lemon KP, Waldron L, Gevers D, Huttenhower C, Izard J. Composition of the adult digestive tract bacterial microbiome based on seven mouth surfaces, tonsils, throat and stool samples. Genome Biol. 2012;13:R42. doi: 10.1186/gb-2012-13-6-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semova I, Carten JD, Stombaugh J, Mackey LC, Knight R, Farber SA, Rawls JF. Microbiota regulate intestinal absorption and metabolism of fatty acids in the zebrafish. Cell Host Microbe. 2012;12:277–288. doi: 10.1016/j.chom.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semple F, Dorin JR. beta-Defensins: multifunctional modulators of infection, inflammation and more? J Innate Immunol. 2012;4:337–348. doi: 10.1159/000336619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semple F, Webb S, Li HN, Patel HB, Perretti M, Jackson IJ, Gray M, Davidson DJ, Dorin JR. Human beta-defensin 3 has immunosuppressive activity in vitro and in vivo. Eur J Immunol. 2010;40:1073–1078. doi: 10.1002/eji.200940041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon G, Segal D, Zilber-Rosenberg I, Rosenberg E. Symbiotic bacteria are responsible for diet-induced mating preference in Drosophila melanogaster, providing support for the hologenome concept of evolution. Gut Microbes. 2011;2:190–192. doi: 10.4161/gmic.2.3.16103. [DOI] [PubMed] [Google Scholar]
- Shi W, Levine M, Davidson B. Unraveling genomic regulatory networks in the simple chordate, Ciona intestinalis. Genome Res. 2005;15:1668–1674. doi: 10.1101/gr.3768905. [DOI] [PubMed] [Google Scholar]
- Shida K, Terajima D, Uchino R, Ikawa S, Ikeda M, Asano K, Watanabe T, Azumi K, Nonaka M, Satou Y, Satoh N, Satake M, Kawazoe Y, Kasuya A. Hemocytes of Ciona intestinalis express multiple genes involved in innate immune host defense. Biochem Biophys Res Commun. 2003;302:207–218. doi: 10.1016/s0006-291x(03)00113-x. [DOI] [PubMed] [Google Scholar]
- Shields JW. The functional evolution of GALT: a review. Lymphology. 2000;33:47–57. [PubMed] [Google Scholar]
- Shim JO. Gut Microbiota in Inflammatory Bowel Disease. Ped Gastro Hepatol Nutr. 2013;16:17–21. doi: 10.5223/pghn.2013.16.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimoda M, Inoue Y, Azuma N, Kanno C. Natural polyreactive immunoglobulin A antibodies produced in mouse Peyer's patches. Immunology. 1999;97:9–17. doi: 10.1046/j.1365-2567.1999.00755.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin SC, Kim SH, You H, Kim B, Kim AC, Lee KA, Yoon JH, Ryu JH, Lee WJ. Drosophila microbiome modulates host developmental and metabolic homeostasis via insulin signaling. Science. 2011;334:670–674. doi: 10.1126/science.1212782. [DOI] [PubMed] [Google Scholar]
- Shivers RP, Kooistra T, Chu SW, Pagano DJ, Kim DH. Tissue-specific activities of an immune signaling module regulate physiological responses to pathogenic and nutritional bacteria in C. elegans. Cell Host Microbe. 2009;6:321–330. doi: 10.1016/j.chom.2009.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skjoedt MO, Palarasah Y, Rasmussen K, Vitved L, Salomonsen J, Kliem A, Hansen S, Koch C, Skjodt K. Two mannose-binding lectin homologues and an MBL-associated serine protease are expressed in the gut epithelia of the urochordate species Ciona intestinalis. Dev Comp Immunol. 2010;34:59–68. doi: 10.1016/j.dci.2009.08.004. [DOI] [PubMed] [Google Scholar]
- Smith LC, Ghosh J, Buckley KM, Clow LA, Dheilly NM, Haug T, Henson JH, Li C, Lun CM, Majeske AJ, Matranga V, Nair SV, Rast JP, Raftos DA, Roth M, Sacchi S, Schrankel CS, Stensvag K. Echinoderm immunity. Adv Exp Med Biol. 2010;708:260–301. doi: 10.1007/978-1-4419-8059-5_14. [DOI] [PubMed] [Google Scholar]
- So S, Tokumaru T, Miyahara K, Ohshima Y. Control of lifespan by food bacteria, nutrient limitation and pathogenicity of food in C. elegans. Mech Ageing Dev. 2011;132:210–212. doi: 10.1016/j.mad.2011.02.005. [DOI] [PubMed] [Google Scholar]
- Sommer F, Backhed F. The gut microbiota--masters of host development and physiology. Nat Rev Microbiol. 2013;11:227–238. doi: 10.1038/nrmicro2974. [DOI] [PubMed] [Google Scholar]
- Song X, Goicoechea JL, Ammiraju JS, Luo M, He R, Lin J, Lee SJ, Sisneros N, Watts T, Kudrna DA, Golser W, Ashley E, Collura K, Braidotti M, Yu Y, Matzkin LM, McAllister BF, Markow TA, Wing RA. The 19 genomes of Drosophila: a BAC library resource for genus-wide and genome-scale comparative evolutionary research. Genetics. 2011;187:1023–1030. doi: 10.1534/genetics.111.126540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenburg JL, Xu J, Leip DD, Chen CH, Westover BP, Weatherford J, Buhler JD, Gordon JI. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science. 2005;307:1955–1959. doi: 10.1126/science.1109051. [DOI] [PubMed] [Google Scholar]
- Sonnenwirth AC. Antibody response to anaerobic bacteria. Rev Infect Dis. 1979;1:337–341. doi: 10.1093/clinids/1.2.337. [DOI] [PubMed] [Google Scholar]
- Sorokin YI. On the feeding of some scleractinian corals with bacteria and dissolved organic matter. Limonol Oceano. 1973;18:380–385. [Google Scholar]
- Storelli G, Defaye A, Erkosar B, Hols P, Royet J, Leulier F. Lactobacillus plantarum promotes Drosophila systemic growth by modulating hormonal signals through TOR-dependent nutrient sensing. Cell Metabol. 2011;14:403–414. doi: 10.1016/j.cmet.2011.07.012. [DOI] [PubMed] [Google Scholar]
- Stowell SR, Arthur CM, Dias-Baruffi M, Rodrigues LC, Gourdine JP, Heimburg-Molinaro J, Ju T, Molinaro RJ, Rivera-Marrero C, Xia B, Smith DF, Cummings RD. Innate immune lectins kill bacteria expressing blood group antigen. Nat Med. 2010;16:295–301. doi: 10.1038/nm.2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart LM, Boulais J, Charriere GM, Hennessy EJ, Brunet S, Jutras I, Goyette G, Rondeau C, Letarte S, Huang H, Ye P, Morales F, Kocks C, Bader JS, Desjardins M, Ezekowitz RA. A systems biology analysis of the Drosophila phagosome. Nature. 2007;445:95–101. doi: 10.1038/nature05380. [DOI] [PubMed] [Google Scholar]
- Sudakaran S, Salem H, Kost C, Kaltenpoth M. Geographical and ecological stability of the symbiotic mid-gut microbiota in European firebugs, Pyrrhocoris apterus (Hemiptera, Pyrrhocoridae) Mol Ecol. 2012;21:6134–6151. doi: 10.1111/mec.12027. [DOI] [PubMed] [Google Scholar]
- Swalla BJ, Cameron CB, Corley LS, Garey JR. Urochordates are monophyletic within the deuterostomes. Syst Biol. 2000;49:52–64. doi: 10.1080/10635150050207384. [DOI] [PubMed] [Google Scholar]
- Swalla BJ, Smith AB. Deciphering deuterostome phylogeny: molecular, morphological and palaeontological perspectives. Philos Trans R Soc Lond B Biol Sci. 2008;363:1557–1568. doi: 10.1098/rstb.2007.2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagliabue A, Nencioni L, Villa L, Keren DF, Lowell GH, Boraschi D. Antibody-dependent cell-mediated antibacterial activity of intestinal lymphocytes with secretory IgA. Nature. 1983;306:184–186. doi: 10.1038/306184a0. [DOI] [PubMed] [Google Scholar]
- Tenor JL, Aballay A. A conserved Toll-like receptor is required for Caenorhabditis elegans innate immunity. EMBO Rep. 2008;9:103–109. doi: 10.1038/sj.embor.7401104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Territo MC, Ganz T, Selsted ME, Lehrer R. Monocyte-chemotactic activity of defensins from human neutrophils. J Clin Invest. 1989;84:2017–2020. doi: 10.1172/JCI114394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakuria D, Schmidt O, Finan D, Egan D, Doohan FM. Gut wall bacteria of earthworms: a natural selection process. ISME J. 2010;4:357–366. doi: 10.1038/ismej.2009.124. [DOI] [PubMed] [Google Scholar]
- Thomas AD, Parker W. Cultivation of epithelial-associated microbiota by the immune system. Fut Microbiol. 2010;5:1483–1492. doi: 10.2217/fmb.10.108. [DOI] [PubMed] [Google Scholar]
- Thorsen MS. Microbial activity, oxygen status and fermentation in the gut of the irregular sea urchin Echinocardium cordatum (Spatangoida: Echinodermata) Mar Biol. 1998;132:423–433. [Google Scholar]
- Thorsen MS, Wieland A, Ploug P, Kragelund C, Nielsen PH. Distribution and activity of endosymbiotic sulfur bacteria in anoxic aggregates from the hindgut of the sea urchin Echinocardium cordatum. Mar Biol Res. 2003;57:1–12. [Google Scholar]
- Tlaskalova-Hogenova H, Tuckova L, Lodinova-Zadnikova R, Stepankova R, Cukrowska B, Funda DP, Striz I, Kozakova H, Trebichavsky I, Sokol D, Rehakova Z, Sinkora J, Fundova P, Horakova D, Jelinkova L, Sanchez D. Mucosal immunity: its role in defense and allergy. Int Arch Allergy Immunol. 2002;128:77–89. doi: 10.1159/000059397. [DOI] [PubMed] [Google Scholar]
- Umesaki Y, Setoyama H. Structure of the intestinal flora responsible for development of the gut immune system in a rodent model. Microbes Infect. 2000;2:1343–1351. doi: 10.1016/s1286-4579(00)01288-0. [DOI] [PubMed] [Google Scholar]
- Vaishnava S, Behrendt C, Ismail A, Eckmann L, Hooper L. Paneth cells directly sense gut commensals and maintain homeostasis at the intestinal host-microbial interface. Proc Nat Acad Sci U S A. 2008;105:20858–20863. doi: 10.1073/pnas.0808723105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal-Dupiol J, Ladriere O, Destoumieux-Garzon D, Sautiere PE, Meistertzheim AL, Tambutte E, Tambutte S, Duval D, Foure L, Adjeroud M, Mitta G. Innate immune responses of a scleractinian coral to vibriosis. J Biol Chem. 2011;286:22688–22698. doi: 10.1074/jbc.M110.216358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierstraete E, Verleyen P, De Loof A, Schoofs L. Differential proteomics for studying Drosophila immunity. Ann N Y Acad Sci. 2005;1040:504–507. doi: 10.1196/annals.1327.104. [DOI] [PubMed] [Google Scholar]
- Vijay-Kumar M, Aitken J, Carvalho F, Cullender T, Mwangi S, Srinivasan S, Sitaraman S, Knight R, Ley R, Gewirtz A. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science. 2010;328:228–231. doi: 10.1126/science.1179721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijay-Kumar M, Aitken JD, Gewirtz AT. Toll like receptor-5: protecting the gut from enteric microbes. Seminars in Immunopathology. 2008;30:11–21. doi: 10.1007/s00281-007-0100-5. [DOI] [PubMed] [Google Scholar]
- Wagner R. Effects of microbiota on GI health: gnotobiotic research. Adv Exp Med Biol. 2008;635:41–56. doi: 10.1007/978-0-387-09550-9_4. [DOI] [PubMed] [Google Scholar]
- Wang L, Kounatidis I, Ligoxygakis P. as a model to study the role of blood cells in inflammation, innate immunity and cancer. Front Cell Infect Microbiol. 2014;3:113. doi: 10.3389/fcimb.2013.00113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkie IC. Autotomy as a prelude to regeneration in echinoderms. Micro Res Tech. 2001;55:369–396. doi: 10.1002/jemt.1185. [DOI] [PubMed] [Google Scholar]
- Williams AM, Probert CS, Stepankova R, Tlaskalova-Hogenova H, Phillips A, Bland PW. Effects of microflora on the neonatal development of gut mucosal T cells and myeloid cells in the mouse. Immunology. 2006;119:470–478. doi: 10.1111/j.1365-2567.2006.02458.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wold AE, Adlerberth I. Breast feeding and the intestinal microflora of the infant--implications for protection against infectious diseases. Adv Exp Med Biol. 2000;478:77–93. doi: 10.1007/0-306-46830-1_7. [DOI] [PubMed] [Google Scholar]
- Wold AE, Mestecky J, Tomana M, Kobata A, Ohbayashi H, Endo T, Eden CS. Secretory immunoglobulin A carries oligosaccharide receptors for Escherichia coli type 1 fimbrial lectin. Infect Immun. 1990;58:3073–3077. doi: 10.1128/iai.58.9.3073-3077.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong AC, Chaston JM, Douglas AE. The inconstant gut microbiota of Drosophila species revealed by 16S rRNA gene analysis. ISME J. 2013;7:1922–1932. doi: 10.1038/ismej.2013.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, Heath AC, Warner B, Reeder J, Kuczynski J, Caporaso JG, Lozupone CA, Lauber C, Clemente JC, Knights D, Knight R, Gordon JI. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Li L, Zhu Y, Zhang G, Guo X. Transcriptome Analysis Reveals a Rich Gene Set Related to Innate Immunity in the Eastern Oyster (Crassostrea virginica) Mar Biotechnol (NY) 2014;16:17–33. doi: 10.1007/s10126-013-9526-z. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Zmasek CM, Dishaw LJ, Mueller MG, Ye Y, Litman GW, Godzik A. Novel genes dramatically alter regulatory network topology in amphioxus. Genome Biol. 2008;9:R123. doi: 10.1186/gb-2008-9-8-r123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Hou A. Host-Microbe Interactions in Caenorhabditis elegans. ISRN Microbiol. 2013;2013:356451. doi: 10.1155/2013/356451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Nakahara T, Miyazaki M, Nogi Y, Taniyama S, Arakawa O, Inoue T, Kudo T. Diversity and function of aerobic culturable bacteria in the intestine of the sea cucumber Holothuria leucospilota. J Gen App Microbiol. 2012;58:447–456. doi: 10.2323/jgam.58.447. [DOI] [PubMed] [Google Scholar]
- Zhang X, Nakahara T, Murase S, Nakata H, Inoue T, Kudo T. Physiological characterization of aerobic culturable bacteria in the intestine of the sea cucumber Apostichopus japonicus. J Gen App Microbiol. 2013;59:1–10. doi: 10.2323/jgam.59.1. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Lu H, Bargmann CI. Pathogenic bacteria induce aversive olfactory learning in Caenorhabditis elegans. Nature. 2005;438:179–184. doi: 10.1038/nature04216. [DOI] [PubMed] [Google Scholar]
- Zucchetti I, De Santis R, Grusea S, Pontarotti P, Du Pasquier L. Origin and evolution of the vertebrate leukocyte receptors: the lesson from tunicates. Immunogenetics. 2009;61:463–481. doi: 10.1007/s00251-009-0373-z. [DOI] [PubMed] [Google Scholar]


