Abstract
We previously reported the inhibitory role of thioredoxin-related protein of 14 kDa (TRP14), a novel disulfide reductase, in nuclear factor-κB (NF-κB) activation, but its biological function has remained to be explored. Here, we evaluated the role of TRP14 in the differentiation and function of osteoclasts (OCs), for which NF-κB and cellular redox regulation have been known to be crucial, using RAW 264.7 macrophage cells expressing wild-type TRP14 or a catalytically inactive mutant, as well as its small interfering RNA. TRP14 depletion enhanced OC differentiation, actin ring formation, and bone resorption, as well as the accumulation of reactive oxygen species (ROS). TRP14 depletion promoted the activation of NF-κB, c-Jun NH2-terminal kinase, and p38, the expression of c-Fos, and the consequent induction of nuclear factor of activated T cell, cytoplasmic 1 (NFATc1), a key determinant of osteoclastogenesis. However, pretreatment with N-acetylcysteine or diphenylene iodonium significantly reduced the OC differentiation, as well as the ROS accumulation and NF-κB activation, that were enhanced by TRP14 depletion. Furthermore, receptor activator of NF-κB ligand (RANKL)-induced ROS accumulation, NF-κB activation, and OC differentiation were inhibited by the ectopic expression of wild-type TRP14 but not by its catalytically inactive mutant. These results suggest that TRP14 regulates OC differentiation and bone resorption through its catalytic activity and that enhancing TRP14 may present a new strategy for preventing bone resorption diseases.
INTRODUCTION
A delicate balance between bone resorption by osteoclasts (OCs) and bone formation by osteoblasts is critical for bone homeostasis (1). Many bone diseases, such as osteoporosis and rheumatoid arthritis, are caused by excessive OC activity, which leads to increased bone resorption (2). An OC is a multinucleated giant cell derived from the hematopoietic lineage (3). Receptor activator of nuclear factor κB (NF-κB) ligand (RANKL), also known as tumor necrosis factor (TNF)-related activation-induced cytokine (4), osteoclast differentiation factor (5), and osteoprotegerin ligand (6), is an essential molecule for OC differentiation. RANKL stimulates morphological changes, cell survival, and osteolytic activity in OCs in cooperation with macrophage colony-stimulating factor (7).
Binding of RANKL to its receptor, RANK, induces the recruitment of TNF receptor-associated factor 6 (TRAF6), which in turn activates NF-κB and mitogen-activated protein kinase (MAPK) pathways, leading to the expression of c-Fos and nuclear factor of activated T cell, cytoplasmic 1 (NFATc1) (8–11). Previous studies have shown that c-Fos and NFATc1 play crucial roles in OC differentiation and in bone resorption and that NFATc1 is the downstream target of c-Fos during osteoclastogenesis (12–17).
Thioredoxin (Trx)-related protein of 14 kDa (TRP14) is a novel disulfide reductase with two active-site Cys residues in its WCPDC motif, which is comparable to the WCGPC motif of Trx (18). TRP14 was shown to inhibit NF-κB in TNF-α signaling, and dynein light chain LC8 was identified as its potential target by a catalytic mechanism-based substrate trapping method (19). LC8 was subsequently shown to mediate redox regulation of NF-κB by preventing IκBα phosphorylation by IκBα kinase through redox-dependent interaction with IκBα (20, 21). TRP14 was suggested to contribute to the regulation of NF-κB activation by facilitating the reduction of LC8 (20, 21).
Several genetic studies have shown that NF-κB and MAPKs are essential for RANKL-induced OC differentiation (22–25). Growing evidence suggests that RANKL induces transient production of reactive oxygen species (ROS) that mediate OC differentiation, and activation of NF-κB and MAPKs is regulated by cellular redox change (26–30). We recently showed that LC8 inhibits RANKL-induced OC differentiation by suppressing NF-κB and prevents inflammation-induced bone erosion (31). Thus, we sought to evaluate whether a novel cellular redox regulator, TRP14, regulates RANKL-induced activation of NF-κB and MAPKs and subsequent osteoclast differentiation by using RAW 264.7 macrophage cells with ectopic expression of wild-type TRP14 and a catalytically inactive mutant, as well as its small interfering RNA. Our results suggest that TRP14 inhibits OC differentiation and bone resorption through its catalytic activity and that the enhancement of TRP14 may be a new strategy for preventing the bone diseases caused by excessive OC activity.
MATERIALS AND METHODS
Reagents and antibodies.
Human recombinant RANKL was purchased from PeproTech, Inc. An antibody specific for TRP14 has been described previously (18). Rabbit polyclonal antibodies against phosphorylated IκBα (phospho-IκBα), phospho-c-Jun NH2-terminal kinase (phospho-JNK), phospho-extracellular-signal-regulated kinase (phospho-ERK), and phospho-p38 were purchased from Cell Signaling Technology. Rabbit polyclonal antibodies against IκBα, JNK1, p38, c-Fos, c-Jun, and TRAF6 and mouse monoclonal antibodies against α-tubulin, TATA-binding protein (TBP), ERK2, and NFATc1 were purchased from Santa Cruz Biotechnology, Inc. Rabbit polyclonal antibodies against β-actin and NF-κB p65 were purchased from Abcam and Millipore, respectively. N-acetylcysteine (NAC) and diphenylene iodonium (DPI) were purchased from Sigma-Aldrich.
Construction of TRP14-specific shRNA expression vectors.
Three small hairpin RNAs (shRNAs) that target mouse TRP14 mRNAs were based on nucleotides 105 to 123 (5′-GGA TAC CGA AGG GAA GAG C-3′), 145 to 163 (5′-GCT GAG CCA GTC ATT CGA G-3′), and 268 to 286 (5′-ATA ACC GCA GTG CCT ACA C-3′), respectively, relative to the translation start site. Each 64-bp duplex DNA, containing the TRP14 targeting sequence, which is the 19-bp sequence of the target transcript separated by a short spacer from the reverse complement of the same sequence plus five thymidines as the termination signal, was cloned into the BglII and HindIII sites of the pSUPER vector (32). Of the resulting plasmids, pSUPER-mTRP14-2 was digested with EcoRI and HindIII, and the remaining 309-bp DNA fragment was cloned into the corresponding sites of the pSuperior.puro vector that harbors the puromycin resistance gene. The resulting plasmid, pSuperiorpuro-mTRP14, was used for establishing the cells that stably express TRP14 shRNA.
Cloning and mutagenesis of mouse TRP14.
The DNA sequence encoding mouse TRP14 was amplified by PCR from RAW 264.7 cDNA with a forward primer (5′-CG GAATTC ATG GCC ACC TTC GAG GAG GTG-3′), containing both an EcoRI site (underlined) and the initiation codon (italicized), and a reverse primer (5′-GC CTCGAG TTA ATC TTC AGA GAA GAT CAT CTC C-3′), containing both an XhoI site (underlined) and the stop codon (italicized). The PCR product was purified, digested with EcoRI and XhoI, and cloned into the corresponding sites of the pCR3.1 vector. The resulting plasmid, pCR-mTRP14, was used as a template for mutagenesis. The mutant TRP14-C43S, in which Cys43 is replaced by serine, was generated by QuikChange II XL site-directed mutagenesis (Stratagene) with complementary primers containing a 1-bp mismatch which converts the codon for cysteine to one for serine.
Establishment of stable RAW 264.7 cells.
RAW 264.7 murine macrophages were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and were incubated at 37°C in 5% CO2. To silence the expression of TRP14, the cells were transfected with pSuperior.puro-mTRP14 vector using a Nucleofector kit V (Amaxa) according to the manufacturer's protocol. For ectopic expression of TRP14, the cells were transfected with pCR-mTRP14 or pCR-mTRP14-C43S vectors. Forty-eight hours after transfection, the medium for cells transfected with pSuperior.puro-mTRP14 was replaced with fresh medium containing 1.5 μg/ml puromycin (Sigma) or 1 mg/ml G418 (A.G. Scientific, Inc.) for cells transfected with pCR-mTRP14 or pCR-mTRP14-C43S. The antibiotic-resistant clones stably transfected with the corresponding vectors were isolated and subjected to Western blot analysis to measure the abundance of TRP14.
OC differentiation and tartrate-resistant acid phosphatase staining.
RAW 264.7 cells were loaded onto a 48-well plate and were treated for 3 to 5 days with 100 ng/ml RANKL in α-minimal essential medium (α-MEM) supplemented with 10% fetal bovine serum. The cells were washed twice with 1× phosphate-buffered saline (PBS), fixed for 10 min with 4% paraformaldehyde, and stained for tartrate-resistant acid phosphatase (TRAP) using a leukocyte acid phosphatase cytochemistry kit (Sigma-Aldrich) according to the manufacturer's instructions. TRAP-positive multinucleated cells (TRAP+ MNCs) containing three or more nuclei were counted as OCs under a light microscope.
Actin ring staining.
After OC differentiation, the cells were fixed with 3.7% formaldehyde solution in PBS, permeabilized with 0.1% Triton X-100, and incubated with Alexa Fluor 488-phalloidin (Invitrogen) for 20 min. After washing with PBS, cells were incubated with 4′,6-diamidino-2-phenylindole (Roche) for 2 min and were photographed under a fluorescence microscope.
Bone resorption assay.
Cells were plated onto dentine discs (Immunodiagnostic Systems) and treated with 100 ng/ml RANKL for 9 days. The cells were completely removed from the dentine discs via abrasion with a cotton tip, and the dentine discs were stained with hematoxylin. Photographs of the resorption pits were obtained under a light microscope at ×40 magnification, and the areas were measured using Image-Pro Plus 4.5 software (Media Cybernetics).
Determination of intracellular ROS.
Cells were washed with α-MEM lacking phenol red and then incubated in the dark for 5 min with 5 μM 5-(and-6-)chloromethyl-2′,7′-dichlorodihydrofluorescein diacetate (Molecular Probes). The cells were collected by using scrapers and analyzed using a FACSCalibur flow cytometer (BD Biosciences).
Nuclear fractionation.
RAW 264.7 cells grown on 60-mm dishes were washed with ice-cold PBS, scraped into 500 μl of buffer A (10 mM HEPES, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM dithiothreitol [DTT], 0.05% NP-40, pH 7.9) containing a cocktail of protease inhibitors, and incubated on ice for 5 min. After centrifugation at 8,000 rpm for 5 min at 4°C, the supernatant was kept as the cytosolic fraction. The precipitate was resuspended in 50 μl of buffer B (5 mM HEPES, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM DTT, 26% glycerol, pH 7.9) containing a cocktail of protease inhibitors and incubated on ice for 30 min. After centrifugation at 14,000 rpm for 30 min at 4°C, the resulting supernatant was used as the nuclear fraction.
RNA isolation, reverse transcription, and real-time PCR analysis.
Total RNA was isolated using the TRIzol reagent (Invitrogen) according to the manufacturer's instructions and quantified by measurement of absorbance at 260 nm. Total RNA (2 μg) was incubated in a volume of 15 μl containing 0.5 μg of oligo(dT) primers at 70°C for 5 min and cooled immediately on ice. The first-strand cDNA was synthesized in a final volume of 25 μl, containing 200 units of Moloney murine leukemia virus (MMLV) reverse transcriptase (Promega), 24 units of RNase inhibitor, and 0.25 mM each deoxynucleoside triphosphate (dNTP) for 1 h at 42°C, followed by enzyme inactivation at 70°C for 10 min. A 0.8-μl aliquot of diluted (1:2.5) cDNA template was amplified in a final volume of 20 μl, containing 10 μl of 2× SYBR green PCR master mix and 1 μl of each gene-specific primer at 10 μM using an ABI 7300 real-time PCR system (Applied Biosystems). The primer sequences are shown in Table 1. The amplification protocol consisted of an initial step of 2 min at 50°C and 2 min at 95°C, followed by 40 cycles of 15 s at 95°C and 1 min at 60°C. Melting curve analysis and agarose gel electrophoresis were performed to ensure a single PCR product of the correct amplicon length. The level of each mRNA was normalized to the level of actin mRNA as an internal control gene.
TABLE 1.
Sequences of real-time PCR primers and amplicon sizes of PCR products
| Gene producta | Primers (sense, antisense) | Amplicon length (bp) |
|---|---|---|
| TRP14 | 5′-CCGCAGTGCCTACACTAC-3′, 5′-TCTCCACAAGGCTGGATTG-3′ | 81 |
| SOD2 | 5′-ATTAACGCGCAGATCATGCA-3′, 5′-TGTCCCCCACCATTGAACTT-3′ | 161 |
| COX2 | 5′-GATCATAAGCGAGGACCTG-3′, 5′-GTCTGTCCAGAGTTTCACC-3′ | 85 |
| c-Fos | 5′-GAGAAACGGAGAATCCGAAG-3′, 5′-GAGAAACGGAGAATCCGAAG-3′ | 97 |
| TRAF6 | 5′-GTGGAGTTTGACCCACCTCT-3′, 5′-TGCATCCCTTATGGATTTGA-3′ | 132 |
| NFATc1 | 5′-CTTCAGCTGGAGGACACC-3′, 5′-CCAATGAACAGCTGTAGCG-3′ | 79 |
| CK | 5′-ACCACTGCCTTCCAATACG-3′, 5′-CGTGGCGTTATACATACAAC-3′ | 99 |
| MMP9 | 5′-AGACGACATAGACGGCATC-3′, 5′-TGCTGTCGGCTGTGGTTC-3′ | 97 |
| β-Actin | 5′-ACCCTAAGGCCAACCGTG-3′, 5′-GCCTGGATGGCTACGTAC-3′ | 81 |
SOD2, superoxide dismutase 2; COX2, cyclooxygenase 2; CK, cathepsin K; MMP9, matrix metalloprotease 9.
TRAP activity assay.
After OC differentiation on a 96-well plate, the cells were lysed in 120 μl of 100 mM sodium acetate buffer (pH 5.2) containing 10 mM sodium tartrate, 1% Triton X-100, 10 μg/ml aprotinin, 10 μg/ml leupeptin, and 1 mg/ml para-nitrophenyl phosphate. The plate was incubated at 37°C for 30 min, and the reaction was stopped by adding 80 μl of 1 N NaOH. The amount of para-nitrophenol liberated during the reaction was determined by measuring the absorbance at 405 nm.
Statistical analysis.
Data are presented as the means ± standard deviations (SD). Statistical significance was calculated using the Student t test, and P values less than 0.05 were considered statistically significant.
RESULTS
TRP14 depletion enhances the RANKL-induced OC differentiation of RAW 264.7 cells.
In order to select the most effective target sequence for silencing mouse TRP14, we constructed three different shRNA plasmids, which were used to transiently transfect NIH 3T3 cells. The knockdown of TRP14 was most pronounced by transfection with pSUPER-mTRP14-2 (by ∼90% after 60 h) (Fig. 1A). RAW 264.7 cells were transfected with the TRP14 shRNA expression vector, and puromycin-resistant cell lines were isolated (Fig. 1B). Of the resulting cell lines, line 7 was chosen due to having the greatest depletion of TRP14. To assess the role of TRP14 in OC differentiation, we examined the effect of TRP14 depletion on the RANKL-induced OC differentiation of RAW 264.7 cells. The formation of TRAP-positive multinucleated cells containing more than three nuclei (TRAP+ MNCs) increased profoundly in TRP14-depleted cells (line 7) in comparison with their formation in control cells that were stably transfected with an empty vector (Fig. 1C). The augmentation of RANKL-induced OC differentiation by TRP14 depletion was also apparent in another stable cell line, line 10 (data not shown).
FIG 1.
Augmentation of RANKL-induced OC differentiation in TRP14-depleted cells. (A) NIH 3T3 cells were transfected for 60 h with empty vector (Mock) or one of three mouse TRP14 shRNA expression vectors (pSUPER-mTRP14-1, -2, and -3) by using FuGene6, and the cell lysates were subjected to immunoblot analysis using antibodies specific for TRP14 and β-actin. (B) RAW 264.7 cells were transfected with either an empty vector or pSuperior.puro-mTRP14 using an Amaxa transfection kit (reagent V) according to the manufacturer's manual. After 60 h, the cells were incubated in medium containing 1.5 μg/ml puromycin, and puromycin-resistant clones were isolated and subjected to immunoblot analysis using antibodies specific for TRP14 and β-actin. (C) RAW 264.7 cells stably transfected with either empty vector or pSuperior.puro-mTRP14 (line 7) were treated with 100 ng/ml of RANKL. At the indicated times, cells were stained with TRAP solution (left), and TRAP-positive multinucleated cells containing more than 3 nuclei (TRAP+ MNCs) were counted (right). Scale bars, 200 μm. Data represent means ± SD (n = 3). *, P < 0.05; **, P < 0.005.
TRP14 depletion promotes RANKL-induced actin ring formation and bone resorption.
Mature OCs have a sealing zone, which consists of a ring of filamentous actin that is required for bone resorption, and clear margins (33). Therefore, we decided to determine the effects of TRP14 depletion on actin ring formation and bone resorption. TRP14-depleted cells formed actin rings with clearer and denser margins than those formed in control cells (Fig. 2A). To determine whether the promotion of actin ring formation by TRP14 depletion affects the resorption ability of OCs, we induced OC differentiation in dentine discs. TRP14-depleted cells formed larger and deeper resorption pits than did control cells (Fig. 2B). These results suggest that TRP14 inhibits actin ring formation and bone resorption.
FIG 2.

Promotion of RANKL-induced actin ring formation and bone resorption in TRP14-depleted cells. (A) After incubation of RAW 264.7 cells with 100 ng/ml RANKL for 5 days, the cells were fixed with 3.7% formaldehyde solution in PBS, permeabilized with 0.1% Triton X-100, and incubated with Alexa Fluor 488-phalloidin for 20 min. After washing with PBS, cells were incubated with 4′,6-diamidino-2-phenylindole (DAPI) for 2 min and then photographed under a fluorescence microscope. Scale bars, 200 μm. (B) The RAW 264.7 cells on dentine discs were incubated with RANKL (100 ng/ml) for 9 days. (Left) The cells were removed from the dentine discs, and the resorption pits were visualized by staining with hematoxylin. Scale bars, 100 μm. (Right) The values for pit area represent means ± SD (n = 4). *, P < 0.005.
TRP14 depletion leads to an increase in the accumulation of ROS which may contribute to the promotion of OC differentiation.
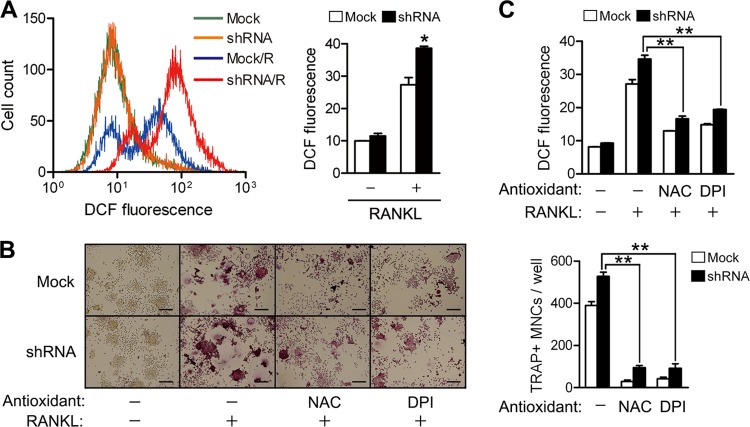
Since TRP14 is a disulfide reductase, its depletion may cause oxidative stress. The intracellular ROS level in TRP14-depleted cells is higher than that in control cells upon stimulation with RANKL (Fig. 3A). To assess whether the increased intracellular ROS contribute to the promotion of OC differentiation in the TRP14-depleted cells, cells were pretreated with NAC, a well-known antioxidant, or DPI, an NADPH oxidase (Nox) inhibitor, before RANKL stimulation. NAC and DPI substantially reduced OC differentiation, as well as intracellular ROS levels, in TRP14-depleted cells (Fig. 3B and C). These results suggest that TRP14 may inhibit OC differentiation by regulating the cellular redox status.
FIG 3.
Reduction of the increased intracellular ROS level and OC differentiation in TRP14-depleted cells by antioxidant. (A) RAW 264.7 cells were treated with RANKL (100 ng/ml) for 10 min, washed with α-MEM lacking phenol red, and then incubated with 5 μM 5-(and-6-)chloromethyl-2′,7′-dichlorodihydrofluorescein diacetate in the dark for 5 min. (Left) The cells were analyzed with a flow cytometer. R indicates RANKL treatment. (Right) The values represent means ± SD (n = 4). *, P < 0.005. (B) RAW 264.7 cells were incubated with RANKL (100 ng/ml) in the presence of 5 mM NAC or 0.1 mM DPI for 4 days. The cells were stained for TRAP (left), and the TRAP-positive MNCs containing more than 3 nuclei were counted (right) as described in the legend to Fig. 1C. Scale bar, 200 μm. The values are means ± SD (n = 3). Con, control; **, P < 0.0005. (C) After preincubation of RAW 264.7 cells with 5 mM NAC or 0.1 mM DPI for 60 min, the cells were treated with 100 ng/ml of RANKL for 10 min and the intracellular ROS levels were measured as described for panel A. The values represent means ± SD (n = 4). **, P < 0.0005.
TRP14 depletion promotes RANKL-induced NF-κB activation.
Because NF-κB is known to be redox sensitive, as well as one of the key transcription factors activated by RANKL during OC differentiation, we assessed the effects of TRP14 depletion on RANKL-induced NF-κB activation. In response to NF-κB activators, such as TNF-α, the inhibitory protein IκB, which binds to NF-κB and sequesters it in the cytoplasm under resting conditions, is phosphorylated and subsequently degraded by the ubiquitin-proteasome system, thereby allowing NF-κB to translocate to the nucleus and activate gene expression (34). Therefore, we examined the effect of TRP14 depletion on the time course of the phosphorylation and degradation of IκBα in RAW 264.7 cells stimulated by RANKL. Compared to the effect in control cells, TRP14 depletion increased the rate and extent of serine phosphorylation of IκBα, resulting in an elevated rate of IκBα degradation (Fig. 4A). We also determined the effect of TRP14 depletion on the nuclear translocation of NF-κB and the expression of its target genes, such as SOD2 and COX2. Consistent with the results described above, the amount of nuclear p65 and the mRNA levels of SOD and COX2 were significantly higher in TRP14-depleted cells than in control cells upon RANKL stimulation (Fig. 4B and C). Moreover, NAC and DPI considerably inhibited the increase of IκBα phosphorylation by TRP14 depletion (Fig. 4D). These results indicate that TRP14 inhibits RANKL-induced NF-κB activation by attenuating the phosphorylation of IκBα in a redox-dependent manner.
FIG 4.
Augmentation of RANKL-induced NF-κB activation by TRP14 depletion. (A) RAW 264.7 cells were exposed to RANKL (100 ng/ml) for the indicated times, and the cell lysates were subjected to immunoblot analysis using antibodies specific for phospho-IκBα (p-IκBα) and IκBα (top). The chemiluminescence signals for phospho-IκBα and IκBα were quantified and normalized based on the signals of β-actin (bottom). The values represent means ± SD (n = 3). *, P < 0.05. (B) RAW 264.7 cells were incubated with RANKL (100 ng/ml) for 10 min and subjected to nuclear fractionation as described in Materials and Methods. The proteins (10 μg) in the cytosolic and nuclear fraction were analyzed with antibodies specific to p65. Tubulin and TATA binding protein (TBP) were used as markers for the cytosol and nucleus, respectively. (C) RAW 264.7 cells were incubated with RANKL (100 ng/ml) for 4 h. Total RNA was extracted from the cells and used in real-time PCR to quantify the mRNA levels of SOD2 and COX2 genes. The relative levels of individual mRNAs were normalized to those of β-actin mRNA and are presented as fold induction values. The values represent means ± SD (n = 3). **, P < 0.001; ***, P < 0.0005. (D) After preincubation of RAW 264.7 cells with 5 mM NAC or 100 nM DPI for 60 min, the cells were treated with 100 ng/ml of RANKL for 6 min. The cell lysates were subjected to immunoblot analysis as described for panel A.
TRP14 depletion promotes RANKL-induced MAPK activation.
RANKL also induces the activation of MAPKs, including JNK, ERK, and p38, which are known to play crucial roles in OC differentiation (25) and to be redox sensitive. Therefore, we examined whether TRP14 depletion affects the RANKL-induced MAPK activation. MAPKs are activated by dual phosphorylation of threonine and tyrosine residues (Thr183 and Tyr185 for JNK, Thr180 and Tyr182 for p38, and Thr202 and Tyr204 for ERK) within a TXY motif. Immunoblot analysis with antibodies specific for dually phosphorylated proteins revealed that the rate and extent of RANKL-induced JNK and p38 phosphorylation increased significantly in TRP14-depleted cells compared with the results in control cells, whereas the time courses and extents of ERK phosphorylation were not significantly affected by TRP14 knockdown (Fig. 5A and B). In addition, the activation of JNK and p38 that was increased by TRP14 knockdown was inhibited by NAC and DPI (Fig. 5C).
FIG 5.
Enhancement of RANKL-induced MAPK activation by TRP14 depletion. (A) RAW 264.7 cells were exposed to 100 ng/ml of RANKL for the indicated times. The cell lysates were then subjected to immunoblot analysis using antibodies against phospho-JNK and JNK1, phospho-ERK and ERK2, or phospho-p38 and p38, and representative immunoblots are shown. (B) The chemiluminescence signals for phospho-JNK, phospho-ERK, and phospho-p38 were quantified and normalized based on the signals of JNK1, ERK2, and p38, respectively. The values represent means ± SD (n = 3). *, P < 0.05; **, P < 0.001. (C) After preincubation of RAW 264.7 cells with 5 mM NAC or 0.1 mM DPI for 60 min, the cells were treated with 100 ng/ml RANKL for 15 min. The cell lysates were subjected to immunoblot analysis as described for panel A.
TRP14 depletion promotes RANKL-induced c-Fos and NFATc1 expression.
Binding of RANKL to its receptor RANK induces the recruitment of TRAF6 to the cytoplasmic domain of the receptor, which triggers the activation of NF-κB and MAPK pathways, leading to the induction of genes, such as c-Fos and NFATc1, that are essential for OC differentiation (25, 35). Because NF-κB activation is known to be upstream from c-Fos/NFATc1 in RANKL-mediated OC differentiation (36) and TRP14 inhibits RANKL-induced NF-κB activation, we examined the effects of TRP14 depletion on the expression of c-Fos and NFATc1 in RANKL-stimulated RAW 264.7 cells. TRP14 depletion significantly augmented not only the mRNA expression of c-Fos and NFATc1 (Fig. 6A) but also their protein expression (Fig. 6B and C) upon stimulation with RANKL. TRP14 knockdown also enhanced the induction of NFATc1 target genes and TRAP activity (Fig. 6A and D). In addition, the TRAF6 expression under the basal condition was higher in TRP14-depleted cells than in control cells and was induced by RANKL (Fig. 6A and C). Moreover, NAC and DPI significantly reduced the expression of c-Fos and NFATc1 that was enhanced by TRP14 depletion (Fig. 6E and F).
FIG 6.
Increase of RANKL-induced expression of c-Fos and NFATc1 in TRP14-depleted cells. (A) RAW 264.7 cells were incubated with RANKL (100 ng/ml) for 8 h (c-Fos), 1 day (TRAF6), or 2 days (NFATc1, MMP9, and CK). Total RNA was extracted from the cells and used in real-time PCR to quantify the mRNA levels. The relative levels of individual mRNAs were normalized to those of β-actin mRNA and are presented as fold induction values. The values represent means ± SD (n = 4). *, P < 0.05; **, P < 0.01; ***, P < 0.0005. (B and C) RAW 264.7 cells were treated with 100 ng/ml of RANKL for the indicated times. Immunoblot analysis of c-Fos (B) and NFATc1 (C) was performed. (D) RAW 264.7 cells were incubated with RANKL (100 ng/ml) for 3 days. TRAP activity was determined as described in Materials and Methods. The values represent means ± SD (n = 3). *, P < 0.05. (E and F) After preincubation of RAW 264.7 cells with 5 mM NAC or 50 nM DPI for 60 min, the cells were treated with 100 ng/ml of RANKL for 8 h (E) or 2 days (F). The cell lysates were subjected to immunoblot analysis of c-Fos and NFATc1.
RANKL-induced OC differentiation is inhibited by wild-type TRP14 but not by its catalytically inactive mutant.
To further confirm the function of TRP14 in OC differentiation, we also established RAW 264.7 cell lines that stably express either wild-type TRP14 or its catalytically inactive mutant TRP14-C43S, in which the active-site Cys43 was mutated to serine. After transfection of RAW 264.7 cells with expression vectors for wild-type or mutant TRP14, we isolated several cell lines, of which lines 9 and 40 were selected for further study of wild-type and mutant TRP14, respectively, because of their similar levels of expression of TRP14 (Fig. 7A). When the cells were treated with RANKL, ectopic expression of wild-type TRP14 significantly inhibited both the formation of TRAP-positive multinucleated cells and TRAP activity, whereas the expression of the catalytically inactive mutant TRP14-C43S did not affect OC formation and slightly promoted TRAP activity (Fig. 7B and C). To clarify whether the catalytic activity of TRP14 is involved in the suppression of ROS levels which leads to NF-κB inhibition, we examined intracellular ROS levels and the extent of IκBα phosphorylation when TRP14 or its catalytically inactive mutant is overexpressed. Upon stimulation with RANKL, the expression of wild-type TRP14 significantly decreased both the levels of ROS and IκBα phosphorylation, whereas the expression of the catalytically null mutant slightly increased ROS levels and could not inhibit IκBα phosphorylation (Fig. 7D and E).
FIG 7.
Effects of ectopic expression of wild-type TRP14 or its catalytically inactive mutant on RANKL-induced OC differentiation of RAW 264.7 cells. (A) RAW 264.7 cells were transfected with pCR3.1 vector (Mock), pCR-mTRP14 (WT), or pCR-mTRP14-C43S (C43S), and G418-resistant cell lines were isolated. The abundance of TRP14 was assessed by immunoblot analysis. (B and C) Cells stably transfected with pCR3.1 vector (Mock), pCR-mTRP14 (line 9), or pCR-mTRP14-C43S (line 40) were treated with 100 ng/ml of RANKL. At the indicated times, cells were stained with TRAP solution (B, left), TRAP-positive multinucleated cells containing more than 3 nuclei (TRAP+ MNCs) were counted (B, right), and TRAP activity was measured (C). The values represent means ± SD (n = 3). *, P < 0.05; **, P < 0.01; ***, P < 0.005. (D) RAW 264.7 cells were treated with RANKL (100 ng/ml) for 10 min, washed with α-MEM lacking phenol red, and incubated with 5 μM 5-(and-6-)chloromethyl-2′,7′-dichlorodihydrofluorescein diacetate in the dark for 5 min. The cells were analyzed with a flow cytometer. The values represent means ± SD (n = 3). *, P < 0.01; **, P < 0.0005. (E) RAW 264.7 cells were treated with 100 ng/ml of RANKL for 6 min, and the cell lysates were subjected to immunoblot analysis using antibodies specific for phospho-IκBα and IκBα.
DISCUSSION
For the first time, the biological role of TRP14 was demonstrated to be to inhibit the formation and function of OC by regulating cellular redox balance. We also showed that the inhibitory role of TRP14 in OC differentiation depends on its catalytic activity.
TRP14 depletion promoted the activation of NF-κB and MAPKs, including JNK and p38, and the induction of c-Fos and NFATc1 upon stimulation with RANKL. Several genetic studies have shown that NF-κB plays a crucial role in OC differentiation: NF-κB p50 and p52 double-knockout mice or IKKβ-deficient mice have severe osteopetrosis caused by the failure of OC formation (22, 23). Studies using either mice lacking JNK or the p38 inhibitor SB203580 (37–39) suggest that JNK and p38 play important roles in OC differentiation. Therefore, TRP14 may attenuate OC differentiation by inhibiting NF-κB, JNK, and p38.
NF-κB induces the initial expression of NFATc1, a master regulator of osteoclastogenesis (40). c-Fos is also involved in the robust induction of NFATc1 expression as a component of the activator protein-1 (AP-1) transcription factor (16). Previous studies have shown that c-Fos plays critical roles in OC differentiation and that NFATc1 is the downstream target of c-Fos: c-Fos knockout mice were deficient in OCs and exhibited an osteopetrotic phenotype (13), but the ectopic expression of NFATc1 in c-Fos-deficient cells restored the transcription of osteoclastogenesis-associated genes and resumed OC formation (14). In addition, c-Fos and NFATc1 were not induced by RANKL in NF-κB p50/p52 double-knockout OC precursor cells (36), suggesting that c-Fos/NFATc1 is downstream from NF-κB. JNK can directly phosphorylate c-Jun, which is one of the major components of transcription factor AP-1, indicating that JNK may contribute to osteoclastogenesis by modulating AP-1 activity. Thus, the enhanced activation of NF-κB and JNK by TRP14 depletion may lead to the increased expression of c-Fos and NFATc1, resulting in the promotion of OC differentiation.
However, TRP14 depletion also increased the basal level of TRAF6. It has been observed that TRAF6 mediates RANKL-induced NF-κB activation and ROS generation (41), that its interaction with RANK is required for the actin ring formation and bone resorption activity of mature OCs (42), and that mice deficient in TRAF6 exhibit osteopetrosis (43). The promoter region of the mouse TRAF6 gene is predicted to contain four putative AP-1 binding sites, which are located within ∼1.2 kb upstream from tentative transcriptional start sites. The human promoter region also has three putative AP-1 binding sites that are properly aligned with the positions of the corresponding sites in the mouse promoter. Given that TRP14 depletion promotes the expression of c-Fos, as well as the activation of JNK, which mediates direct phosphorylation of c-Jun, the level of TRP14 is expected to affect TRAF6 expression by modulating AP-1 activity. Hence, the inhibitory effects of TRP14 on RANKL-induced signal transduction and OC differentiation may stem from the regulation of TRAF6 expression.
RANKL stimulation produces ROS through a signaling cascade involving TRAF6, Rac1, and NADPH oxidase 1 (Nox1) (28, 30). RANKL-induced ROS accumulation was enhanced by TRP14 depletion, reduced by ectopic expression of TRP14, and unaffected by the catalytically inactive TRP14 mutant. However, the basal ROS level was hardly affected by the manipulation of TRP14 expression. In addition, the increased intracellular ROS and OC differentiation in TRP14-depleted cells were reduced by pretreatment with NAC and DPI. These data suggest that TRP14 inhibits OC differentiation by regulating cellular redox status through its catalytic activity. In addition to having a disulfide reductase activity, TRP14 has the ability to reduce H2O2. However, TRP14 exhibits a high Km value for H2O2 (18), which may be the reason why the basal ROS level was not significantly affected by the manipulation of TRP14 expression. Many cellular proteins contain redox-sensitive cysteine residues that are readily oxidized by ROS to form intra- or intermolecular disulfide bonds. Some of these disulfides would be reduced by the action of TRP14, as exemplified by LC8 and phosphotyrosine phosphatase 1B (20, 21, 44). TRP14 was recently reported to be an efficient l-cystine reductase (45). The resulting oxidized TRP14 is reduced by the NADPH-dependent enzyme thioredoxin reductase, resulting in the overall reaction of the NADPH-dependent reduction of ROS. Therefore, TRP14-mediated cyclic interconversion of proteinaceous or free cysteine residues between thiol and disulfide forms can lead to the scavenging of ROS. Direct scavenging of ROS by TRP14, though not efficient, is also a possibility.
NAC and DPI significantly suppressed the activation of NF-κB and MAPK and the ROS accumulation promoted by TRP14 depletion. In response to various stimuli, ROS have been shown to regulate NF-κB and MAPKs through the oxidative modification of a conserved cysteine residue of redox-sensitive target molecules. We previously unraveled the mechanism underlying the redox-dependent regulation of TNF-α-induced NF-κB activation, in which ROS oxidize LC8 to a homodimer linked by a reversible intermolecular disulfide bond, which promotes its dissociation from IκBα, allowing IκBα to be phosphorylated and degraded and resulting in NF-κB activation (20, 21). Recently, we also showed that LC8 inhibits RANKL-induced osteoclastogenesis by regulating NF-κB and protects against inflammation-induced or ovariectomy-induced bone loss in mice (31). Thus, it is likely that TRP14 attenuates OC differentiation by inhibiting NF-κB through controlling the redox status of LC8.
MAPKs are activated through the dual phosphorylation of tyrosine and threonine by MAPK kinases and are inactivated by dephosphorylation by MAPK phosphatases (MKPs) (46). Therefore, the activity of MAPKs, such as JNK and p38, could be defined by the balance of their upstream kinase and MKP activities. Many protein tyrosine phosphatases have been shown to be inactivated through reversible oxidation of their catalytic cysteine in cells exposed to growth factors (47, 48). In addition, ROS promote sustained JNK activation by inhibiting MKP-1, MKP-3, MKP-5, and MKP-7, all of which act as JNK phosphatases (49). Thus, it is of interest to explore the changes of redox status of these MKPs and the relevance of TRP14 to those during RANKL-induced OC differentiation.
Although TRP14 was not induced during RANKL-induced OC differentiation, the ectopic expression of TRP14 inhibited OC differentiation. But the expression of the catalytically inactive TRP14 mutant did not affect OC formation and slightly inhibited OC function, indicating that the catalytic activity of TRP14 is essential for inhibiting OC differentiation. Therefore, the enhancement of TRP14 may present a new strategy for preventing the bone diseases caused by excessive OC activity. Finding bone diseases or pathophysiological conditions where TRP14 is downregulated allows greater opportunity for research on the regulation of TRP14 expression and for the development of a method to induce TRP14.
ACKNOWLEDGMENTS
This work was supported by the National Research Foundation of Korea (NRF) (grants M10642040002-07N4204-00210, 2014R1A2A2A01003983, 2012M3A9C5048708, and 2012R1A5A1048236), funded by the Ministry of Science, ICT, and future Planning (MSIP).
Footnotes
Published ahead of print 7 July 2014
REFERENCES
- 1.Takayanagi H, Kim S, Taniguchi T. 2002. Signaling crosstalk between RANKL and interferons in osteoclast differentiation. Arthritis Res. 4(Suppl 3):S227–S232. 10.1186/ar581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rodan GA, Martin TJ. 2000. Therapeutic approaches to bone diseases. Science 289:1508–1514. 10.1126/science.289.5484.1508 [DOI] [PubMed] [Google Scholar]
- 3.Bar-Shavit Z. 2007. The osteoclast: a multinucleated, hematopoietic-origin, bone-resorbing osteoimmune cell. J. Cell. Biochem. 102:1130–1139. 10.1002/jcb.21553 [DOI] [PubMed] [Google Scholar]
- 4.Wong BR, Rho J, Arron J, Robinson E, Orlinick J, Chao M, Kalachikov S, Cayani E, Bartlett FS, III, Frankel WN, Lee SY, Choi Y. 1997. TRANCE is a novel ligand of the tumor necrosis factor receptor family that activates c-Jun N-terminal kinase in T cells. J. Biol. Chem. 272:25190–25194. 10.1074/jbc.272.40.25190 [DOI] [PubMed] [Google Scholar]
- 5.Yasuda H, Shima N, Nakagawa N, Yamaguchi K, Kinosaki M, Mochizuki S, Tomoyasu A, Yano K, Goto M, Murakami A, Tsuda E, Morinaga T, Higashio K, Udagawa N, Takahashi N, Suda T. 1998. Osteoclast differentiation factor is a ligand for osteoprotegerin/osteoclastogenesis-inhibitory factor and is identical to TRANCE/RANKL. Proc. Natl. Acad. Sci. U. S. A. 95:3597–3602. 10.1073/pnas.95.7.3597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lacey DL, Timms E, Tan HL, Kelley MJ, Dunstan CR, Burgess T, Elliott R, Colombero A, Elliott G, Scully S, Hsu H, Sullivan J, Hawkins N, Davy E, Capparelli C, Eli A, Qian YX, Kaufman S, Sarosi I, Shalhoub V, Senaldi G, Guo J, Delaney J, Boyle WJ. 1998. Osteoprotegerin ligand is a cytokine that regulates osteoclast differentiation and activation. Cell 93:165–176. 10.1016/S0092-8674(00)81569-X [DOI] [PubMed] [Google Scholar]
- 7.Teitelbaum SL. 2000. Bone resorption by osteoclasts. Science 289:1504–1508. 10.1126/science.289.5484.1504 [DOI] [PubMed] [Google Scholar]
- 8.Boyle WJ, Simonet WS, Lacey DL. 2003. Osteoclast differentiation and activation. Nature 423:337–342. 10.1038/nature01658 [DOI] [PubMed] [Google Scholar]
- 9.Bai S, Zha J, Zhao H, Ross FP, Teitelbaum SL. 2008. Tumor necrosis factor receptor-associated factor 6 is an intranuclear transcriptional coactivator in osteoclasts. J. Biol. Chem. 283:30861–30867. 10.1074/jbc.M802525200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walsh MC, Kim N, Kadono Y, Rho J, Lee SY, Lorenzo J, Choi Y. 2006. Osteoimmunology: interplay between the immune system and bone metabolism. Annu. Rev. Immunol. 24:33–63. 10.1146/annurev.immunol.24.021605.090646 [DOI] [PubMed] [Google Scholar]
- 11.Takayanagi H. 2007. Osteoimmunology: shared mechanisms and crosstalk between the immune and bone systems. Nat. Rev. Immunol. 7:292–304. 10.1038/nri2062 [DOI] [PubMed] [Google Scholar]
- 12.Ishida N, Hayashi K, Hoshijima M, Ogawa T, Koga S, Miyatake Y, Kumegawa M, Kimura T, Takeya T. 2002. Large scale gene expression analysis of osteoclastogenesis in vitro and elucidation of NFAT2 as a key regulator. J. Biol. Chem. 277:41147–41156. 10.1074/jbc.M205063200 [DOI] [PubMed] [Google Scholar]
- 13.Grigoriadis AE, Wang ZQ, Cecchini MG, Hofstetter W, Felix R, Fleisch HA, Wagner EF. 1994. c-Fos: a key regulator of osteoclast-macrophage lineage determination and bone remodeling. Science 266:443–448. 10.1126/science.7939685 [DOI] [PubMed] [Google Scholar]
- 14.Matsuo K, Galson DL, Zhao C, Peng L, Laplace C, Wang KZ, Bachler MA, Amano H, Aburatani H, Ishikawa H, Wagner EF. 2004. Nuclear factor of activated T-cells (NFAT) rescues osteoclastogenesis in precursors lacking c-Fos. J. Biol. Chem. 279:26475–26480. 10.1074/jbc.M313973200 [DOI] [PubMed] [Google Scholar]
- 15.Asagiri M, Sato K, Usami T, Ochi S, Nishina H, Yoshida H, Morita I, Wagner EF, Mak TW, Serfling E, Takayanagi H. 2005. Autoamplification of NFATc1 expression determines its essential role in bone homeostasis. J. Exp. Med. 202:1261–1269. 10.1084/jem.20051150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wagner EF, Eferl R. 2005. Fos/AP-1 proteins in bone and the immune system. Immunol. Rev. 208:126–140. 10.1111/j.0105-2896.2005.00332.x [DOI] [PubMed] [Google Scholar]
- 17.Takayanagi H, Kim S, Koga T, Nishina H, Isshiki M, Yoshida H, Saiura A, Isobe M, Yokochi T, Inoue J, Wagner EF, Mak TW, Kodama T, Taniguchi T. 2002. Induction and activation of the transcription factor NFATc1 (NFAT2) integrate RANKL signaling in terminal differentiation of osteoclasts. Dev. Cell 3:889–901. 10.1016/S1534-5807(02)00369-6 [DOI] [PubMed] [Google Scholar]
- 18.Jeong W, Yoon HW, Lee SR, Rhee SG. 2004. Identification and characterization of TRP14, a thioredoxin-related protein of 14 kDa. New insights into the specificity of thioredoxin function. J. Biol. Chem. 279:3142–3150. 10.1074/jbc.M307932200 [DOI] [PubMed] [Google Scholar]
- 19.Jeong W, Chang TS, Boja ES, Fales HM, Rhee SG. 2004. Roles of TRP14, a thioredoxin-related protein in tumor necrosis factor-alpha signaling pathways. J. Biol. Chem. 279:3151–3159. 10.1074/jbc.M307959200 [DOI] [PubMed] [Google Scholar]
- 20.Jung Y, Kim H, Min SH, Rhee SG, Jeong W. 2008. Dynein light chain LC8 negatively regulates NF-kappaB through the redox-dependent interaction with IkappaBalpha. J. Biol. Chem. 283:23863–23871. 10.1074/jbc.M803072200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jeong W, Jung Y, Kim H, Park SJ, Rhee SG. 2009. Thioredoxin-related protein 14, a new member of the thioredoxin family with disulfide reductase activity: implication in the redox regulation of TNF-alpha signaling. Free Radic. Biol. Med. 47:1294–1303. 10.1016/j.freeradbiomed.2009.07.021 [DOI] [PubMed] [Google Scholar]
- 22.Iotsova V, Caamano J, Loy J, Yang Y, Lewin A, Bravo R. 1997. Osteopetrosis in mice lacking NF-kappaB1 and NF-kappaB2. Nat. Med. 3:1285–1289. 10.1038/nm1197-1285 [DOI] [PubMed] [Google Scholar]
- 23.Ruocco MG, Maeda S, Park JM, Lawrence T, Hsu LC, Cao Y, Schett G, Wagner EF, Karin M. 2005. IκκB kinase (IKK)β, but not IKKα, is a critical mediator of osteoclast survival and is required for inflammation-induced bone loss. J. Exp. Med. 201:1677–1687. 10.1084/jem.20042081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alles N, Soysa NS, Hayashi J, Khan M, Shimoda A, Shimokawa H, Ritzeler O, Akiyoshi K, Aoki K, Ohya K. 2010. Suppression of NF-kappaB increases bone formation and ameliorates osteopenia in ovariectomized mice. Endocrinology 151:4626–4634. 10.1210/en.2010-0399 [DOI] [PubMed] [Google Scholar]
- 25.Lee ZH, Kim HH. 2003. Signal transduction by receptor activator of nuclear factor kappa B in osteoclasts. Biochem. Biophys. Res. Commun. 305:211–214. 10.1016/S0006-291X(03)00695-8 [DOI] [PubMed] [Google Scholar]
- 26.Bax BE, Alam AS, Banerji B, Bax CM, Bevis PJ, Stevens CR, Moonga BS, Blake DR, Zaidi M. 1992. Stimulation of osteoclastic bone resorption by hydrogen peroxide. Biochem. Biophys. Res. Commun. 183:1153–1158. 10.1016/S0006-291X(05)80311-0 [DOI] [PubMed] [Google Scholar]
- 27.Lean JM, Davies JT, Fuller K, Jagger CJ, Kirstein B, Partington GA, Urry ZL, Chambers TJ. 2003. A crucial role for thiol antioxidants in estrogen-deficiency bone loss. J. Clin. Invest. 112:915–923. 10.1172/JCI18859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee NK, Choi YG, Baik JY, Han SY, Jeong DW, Bae YS, Kim N, Lee SY. 2005. A crucial role for reactive oxygen species in RANKL-induced osteoclast differentiation. Blood 106:852–859. 10.1182/blood-2004-09-3662 [DOI] [PubMed] [Google Scholar]
- 29.Kim HJ, Chang EJ, Kim HM, Lee SB, Kim HD, Su Kim G, Kim HH. 2006. Antioxidant alpha-lipoic acid inhibits osteoclast differentiation by reducing nuclear factor-kappaB DNA binding and prevents in vivo bone resorption induced by receptor activator of nuclear factor-kappaB ligand and tumor necrosis factor-alpha. Free Radic. Biol. Med. 40:1483–1493. 10.1016/j.freeradbiomed.2005.10.066 [DOI] [PubMed] [Google Scholar]
- 30.Wang Y, Lebowitz D, Sun C, Thang H, Grynpas MD, Glogauer M. 2008. Identifying the relative contributions of Rac1 and Rac2 to osteoclastogenesis. J. Bone Miner. Res. 23:260–270. 10.1359/jbmr.071013 [DOI] [PubMed] [Google Scholar]
- 31.Kim H, Hyeon S, Yang Y, Huh JY, Park DR, Lee H, Seo DH, Kim HS, Lee SY, Jeong W. 2013. Dynein light chain LC8 inhibits osteoclast differentiation and prevents bone loss in mice. J. Immunol. 190:1312–1318. 10.4049/jimmunol.1202525 [DOI] [PubMed] [Google Scholar]
- 32.Brummelkamp TR, Bernards R, Agami R. 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science 296:550–553. 10.1126/science.1068999 [DOI] [PubMed] [Google Scholar]
- 33.Teitelbaum SL, Ross FP. 2003. Genetic regulation of osteoclast development and function. Nat. Rev. Genet. 4:638–649. 10.1038/nrg1122 [DOI] [PubMed] [Google Scholar]
- 34.Ghosh S, Karin M. 2002. Missing pieces in the NF-kappaB puzzle. Cell 109(Suppl):S81–S96. 10.1016/S0092-8674(02)00703-1 [DOI] [PubMed] [Google Scholar]
- 35.Kim HH, Lee DE, Shin JN, Lee YS, Jeon YM, Chung CH, Ni J, Kwon BS, Lee ZH. 1999. Receptor activator of NF-kappaB recruits multiple TRAF family adaptors and activates c-Jun N-terminal kinase. FEBS Lett. 443:297–302. 10.1016/S0014-5793(98)01731-1 [DOI] [PubMed] [Google Scholar]
- 36.Yamashita T, Yao Z, Li F, Zhang Q, Badell IR, Schwarz EM, Takeshita S, Wagner EF, Noda M, Matsuo K, Xing L, Boyce BF. 2007. NF-kappaB p50 and p52 regulate receptor activator of NF-kappaB ligand (RANKL) and tumor necrosis factor-induced osteoclast precursor differentiation by activating c-Fos and NFATc1. J. Biol. Chem. 282:18245–18253. 10.1074/jbc.M610701200 [DOI] [PubMed] [Google Scholar]
- 37.David JP, Sabapathy K, Hoffmann O, Idarraga MH, Wagner EF. 2002. JNK1 modulates osteoclastogenesis through both c-Jun phosphorylation-dependent and -independent mechanisms. J. Cell Sci. 115:4317–4325. 10.1242/jcs.00082 [DOI] [PubMed] [Google Scholar]
- 38.Matsumoto M, Sudo T, Saito T, Osada H, Tsujimoto M. 2000. Involvement of p38 mitogen-activated protein kinase signaling pathway in osteoclastogenesis mediated by receptor activator of NF-kappa B ligand (RANKL). J. Biol. Chem. 275:31155–31161. 10.1074/jbc.M001229200 [DOI] [PubMed] [Google Scholar]
- 39.Lee SE, Woo KM, Kim SY, Kim HM, Kwack K, Lee ZH, Kim HH. 2002. The phosphatidylinositol 3-kinase, p38, and extracellular signal-regulated kinase pathways are involved in osteoclast differentiation. Bone 30:71–77. 10.1016/S8756-3282(01)00657-3 [DOI] [PubMed] [Google Scholar]
- 40.Asagiri M, Takayanagi H. 2007. The molecular understanding of osteoclast differentiation. Bone 40:251–264. 10.1016/j.bone.2006.09.023 [DOI] [PubMed] [Google Scholar]
- 41.Wong BR, Josien R, Lee SY, Vologodskaia M, Steinman RM, Choi Y. 1998. The TRAF family of signal transducers mediates NF-kappaB activation by the TRANCE receptor. J. Biol. Chem. 273:28355–28359. 10.1074/jbc.273.43.28355 [DOI] [PubMed] [Google Scholar]
- 42.Armstrong AP, Tometsko ME, Glaccum M, Sutherland CL, Cosman D, Dougall WC. 2002. A RANK/TRAF6-dependent signal transduction pathway is essential for osteoclast cytoskeletal organization and resorptive function. J. Biol. Chem. 277:44347–44356. 10.1074/jbc.M202009200 [DOI] [PubMed] [Google Scholar]
- 43.Lomaga MA, Yeh WC, Sarosi I, Duncan GS, Furlonger C, Ho A, Morony S, Capparelli C, Van G, Kaufman S, van der Heiden A, Itie A, Wakeham A, Khoo W, Sasaki T, Cao Z, Penninger JM, Paige CJ, Lacey DL, Dunstan CR, Boyle WJ, Goeddel DV, Mak TW. 1999. TRAF6 deficiency results in osteopetrosis and defective interleukin-1, CD40, and LPS signaling. Genes Dev. 13:1015–1024. 10.1101/gad.13.8.1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dagnell M, Frijhoff J, Pader I, Augsten M, Boivin B, Xu J, Mandal PK, Tonks NK, Hellberg C, Conrad M, Arner ES, Ostman A. 2013. Selective activation of oxidized PTP1B by the thioredoxin system modulates PDGF-beta receptor tyrosine kinase signaling. Proc. Natl. Acad. Sci. U. S. A. 110:13398–13403. 10.1073/pnas.1302891110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pader I, Sengupta R, Cebula M, Xu J, Lundberg JO, Holmgren A, Johansson K, Arner ES. 2014. Thioredoxin-related protein of 14 kDa is an efficient L-cystine reductase and S-denitrosylase. Proc. Natl. Acad. Sci. U. S. A. 111:6964–6969. 10.1073/pnas.1317320111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Davis RJ. 2000. Signal transduction by the JNK group of MAP kinases. Cell 103:239–252. 10.1016/S0092-8674(00)00116-1 [DOI] [PubMed] [Google Scholar]
- 47.Lee SR, Kwon KS, Kim SR, Rhee SG. 1998. Reversible inactivation of protein-tyrosine phosphatase 1B in A431 cells stimulated with epidermal growth factor. J. Biol. Chem. 273:15366–15372. 10.1074/jbc.273.25.15366 [DOI] [PubMed] [Google Scholar]
- 48.Meng TC, Fukada T, Tonks NK. 2002. Reversible oxidation and inactivation of protein tyrosine phosphatases in vivo. Mol. Cell 9:387–399. 10.1016/S1097-2765(02)00445-8 [DOI] [PubMed] [Google Scholar]
- 49.Kamata H, Honda S, Maeda S, Chang L, Hirata H, Karin M. 2005. Reactive oxygen species promote TNFalpha-induced death and sustained JNK activation by inhibiting MAP kinase phosphatases. Cell 120:649–661. 10.1016/j.cell.2004.12.041 [DOI] [PubMed] [Google Scholar]