Abstract
The growth and development of the vertebrate limb relies on homeobox genes of the Hox and Shox families, with their independent mutation often giving dose-dependent effects. Here we investigate whether Shox2 and Hox genes function together during mouse limb development by modulating their relative dosage and examining the limb for nonadditive effects on growth. Using double mRNA fluorescence in situ hybridization (FISH) in single embryos, we first show that Shox2 and Hox genes have associated spatial expression dynamics, with Shox2 expression restricted to the proximal limb along with Hoxd9 and Hoxa11 expression, juxtaposing the distal expression of Hoxa13 and Hoxd13. By generating mice with all possible dosage combinations of mutant Shox2 alleles and HoxA/D cluster deletions, we then show that their coordinated proximal limb expression is critical to generate normally proportioned limb segments. These epistatic interactions tune limb length, where Shox2 underexpression enhances, and Shox2 overexpression suppresses, Hox-mutant phenotypes. Disruption of either Shox2 or Hox genes leads to a similar reduction in Runx2 expression in the developing humerus, suggesting their concerted action drives cartilage maturation during normal development. While we furthermore provide evidence that Hox gene function influences Shox2 expression, this regulation is limited in extent and is unlikely on its own to be a major explanation for their genetic interaction. Given the similar effect of human SHOX mutations on regional limb growth, Shox and Hox genes may generally function as genetic interaction partners during the growth and development of the proximal vertebrate limb.
Keywords: epistasis, SHOX, chondrogenesis, perichondrium, fluorescence in situ hybridization
THE vertebrate limb is a valuable model for studying the genetic coordination of a complex developing structure. The proximodistal axis of the limb is composed of discrete segments, the growth and development of which are selectively perturbed when individual, or combinations of, homeobox genes are disrupted. In mice, mutations of the paralogous Hox9 and Hox10 genes result in shortened stylopodal elements (containing the humerus and femur) (Fromental-Ramain et al. 1996a; Wellik and Capecchi 2003), deletions of Hox11 genes result in truncated zeugopodal elements (radius/ulna and fibula/tibia) (Davis et al. 1995; Wellik and Capecchi 2003), and disruption of Hox13 genes results in agenesis of the autopod (metacarpals/metatarsals and the digits) (Fromental-Ramain et al. 1996b). Mutation of short stature homeobox (Shox) genes similarly gives rise to the disproportionate shortening of certain limb regions. In humans, loss of SHOX leads to the truncated zeugopod elements found in people with Leri–Weill, Turner, and Langer syndromes (Rao et al. 1997; Belin et al. 1998; Shears et al. 1998; Zinn et al. 2002). While rodents have uniquely lost the Shox gene among mammals (Gianfrancesco et al. 2001), disruption of the widely conserved Shox2 gene results in severely shortened stylopodal elements in mice (Cobb et al. 2006). Thus, Hox and Shox gene perturbations each give rise to regional phenotypes along the proximodistal axis, suggesting the possibility that these genes function together during limb development.
Limb chondrogenesis begins following the early stages of limb bud formation, where mesenchymal cells condense and differentiate into Col2a1-expressing chondrocytes. After a proliferative phase, the chondrocytes nearest the middle of the element stop dividing, undergo hypertrophy, and express Col10a1, a process that is associated with elongation of the skeletal element (Karsenty and Wagner 2002). Surrounding the chondrocytes is a layer of flattened and elongated cells, the perichondrium, that influences the developmental progression of the cartilage cells and is furthermore important for growth (Kronenberg 2007). Runx2, which is expressed both by chondrocytes and the perichondrium, is essential for proper chondrocyte hypertrophy and the formation of osteoblasts, thus being important for both the proper development of the cartilage template and its eventual replacement by bone (Otto et al. 1997; Yoshida et al. 2004). As mutation of Shox2 or Hox genes results in a strong reduction or loss of Runx2 expression, a lack of chondrocyte maturation likely underlies regional shortening in these animals (Boulet and Capecchi 2004; Cobb et al. 2006; Villavicencio-Lorini et al. 2010; Gross et al. 2012).
The regulation of individual Hox and Shox genes follows precise spatial and temporal patterns of gene expression in the limb. The regulation of Hox genes has been intensively studied, showing that their expression occurs in two main phases: an early phase of gene expression that is associated with the development of the more proximal limb (stylopod and zeugopod) and a later phase that is associated with the development of the distal limb (autopod) (Kmita et al. 2002; Tarchini and Duboule 2006; Andrey et al. 2013). This collinear strategy sets up partially overlapping domains of gene expression across the limb that underlie the discrete phenotypes that occur when paralagous Hox genes are mutated. The reported expression of human SHOX and SHOX2 is also regional, with SHOX2 expressed in the developing stylopod and SHOX expressed in the developing zeugopod (Clement-Jones et al. 2000). Mouse Shox2 expression, in contrast, occupies both the developing stylopodal and zeugopodal domains even though its mutation primarily disrupts the developing stylopod (Cobb et al. 2006). Within their broad segmental domains, Shox2 and at least some Hox genes are expressed in the proliferating chondrocytes and perichondrium of developing skeletal elements (Villavicencio-Lorini et al. 2010; Swinehart et al. 2013), being thus associated with their continual growth during development.
Gene expression levels can be critical during development, with variation in expression leading to dose-dependent responses. The effects of dosage variation can be assessed by modulating the expression of a single gene or varying the expression of multiple genes, where genetic interactions, or epistasis, become important. In the latter case, the effect of a given variant differs in the presence and absence of variation in another gene (Phillips 2008). For Hox genes, which harbor a large degree of functional redundancy, the quantitative nature of their function is manifest when a certain threshold of gene product is crossed. For instance, removing a single allele of Hoxa13 in an otherwise wild-type animal has far less effect on development than when it is removed in conjunction with Hoxd13 disruption (Fromental-Ramain et al. 1996b). The quantitative response to Hox gene function has been proposed to distinguish “short” bones from “long” bones (Gonzalez-Martin et al. 2014). SHOX function in humans is also dosage sensitive; a majority of individuals missing one allele of SHOX have moderately shortened zeugopodal segments, while individuals missing both SHOX alleles have a more penetrant and severe shortening (Rao et al. 1997; Belin et al. 1998; Shears et al. 1998; Zinn et al. 2002; Albuisson et al. 2012). SHOX duplications have furthermore been hypothesized to lead to tall stature (Ogata et al. 2000; Durand and Rappold 2013), highlighting the interest in SHOX dosage and its effect on limb development. Thus, examining the effects of dosage variation is key to understanding the role of Shox and Hox genes in development and disease.
In the present work, we investigate the functional relationship between Shox2 and Hox genes during mouse limb development. Modulating Shox2 transcript levels in a variety of Hox-mutant backgrounds produces nonadditive effects on limb growth, indicative of genetic interaction. This approach reveals that the functions of these genes are intimately associated, exhibiting aspects of both synergy and redundancy during limb development and functioning upstream of Runx2 during chondrogenesis.
Materials and Methods
Mice
Mouse lines used were Shox2fl/+ (Cobb et al. 2006), RosaCAG-STOP-Shox2 (Scott et al. 2011), HoxD+/− (Del9) (Spitz et al. 2001), HoxAfl/+ (Kmita et al. 2005), and Prrx1-Cre (Logan et al. 2002). The Life and Environmental Sciences Animal Care Committee approved all animal experiments.
In situ hybridization
Chromogenic or fluorescence whole-mount in situ hybridization was performed as previously outlined (Neufeld et al. 2013). For confocal microscopy, limb buds were imaged in 1% low-melt agarose. Single optical sections were taken at 40×, using a Zeiss LSM-510 META confocal microscope. Section ISH was performed on 10-μm cryosections as previously described (Alam et al. 2005) except that BMpurple (Roche) was used for signal development. Probes used were Shox2 (Cobb and Duboule 2005), Hoxd9 (Renucci et al. 1992), Hoxa11 (Cobb and Duboule 2005), Hoxd13 (Dolle et al. 1991), Hoxa13 (Warot et al. 1997), Col2a1 (Metsaranta et al. 1991), Runx2 (Cobb et al. 2006), and Col10a1 (Apte et al. 1992).
Quantitative real-time PCR
Embryos were dissected in PBS and the limb buds stored in RNAlater (QIAGEN, Valencia, CA) during genotyping. Limb bud tissue was disrupted using a motorized pestle (VWR), and RNA was isolated using the E.Z.N.A Total RNA Kit I (Omega Bio-Tek, Norcross, GA). RNA was reverse transcribed using qScript cDNA Super Mix (Quanta BioSciences, Gaithersburg, MD). Relative levels of selected cDNAs were measured using PerfeCTa SYBR Green Fastmix (Quanta BioSciences) and an iCycler IQ Real-Time PCR Detection System (Bio-Rad Laboratories, Hercules, CA) by comparison to a standard curve. Levels were normalized to Actb. Primer sequences used were Shox2 forward, TGGAACAACTCAACGAGCTGGAGA; Shox2 reverse, TTCAAACTGGCTAGCGGCTCCTAT; Actb forward, TTAATTTCTGAATGGCCCAGGTCT; and Actb reverse, ATTGGTCTCAAGTCAGTGTACAGG. Differences in transcript levels between groups were assessed with two-tailed unpaired t-tests, using between three and five animals per genotype.
Skeletal staining and length quantification
Skeletons of newborn mice were stained with Alizarin Red and Alcian Blue, using standard techniques. The lengths of the Alizarin-Red stained regions of the right stylopodal and zeugopodal elements were measured under a microscope, using a densely marked straightedge, giving measurements to the closest tenth of a millimeter. No differences in overall body length were observed between wild-type animals and mutants.
Graphing
Bar plots and interaction plots were made using GraphPad Prism software. Heat maps in Figure 2 were made using the gplots package in R (R Core Team 2013).
Figure 2.
Epistasis between Shox2 loss-of-function mutations and Hox genes during growth of the stylopodal and zeugopodal elements. (A) Deletion of a single copy of Shox2 has no effect on the newborn limb in an otherwise wild-type background, but leads to a shortened humerus in a HoxD−/− background (n = 4–6 for each genotype). Arrowhead points to humerus. (B) Deletion of both copies of Shox2 has a stronger effect on the growth of the ulna in a HoxAc/c background than in an otherwise wild-type background (n = 4–6 for each genotype). Arrowhead points to ulna. (A′ and B′) Interaction plots showing disproportionate effects, as shown by nonparallel lines, of the respective Shox2 mutation in each Hox background. Data are plotted as means ± SEM. Effect of Shox2 mutation is significantly different in each Hox background compared to an otherwise wild-type background. P < 0.001. (A′′ and B′′) Genotype–phenotype maps showing the effects of Shox2 mutations on skeletal length in multiple Hox backgrounds, arranged in decreasing dose, in both the fore- and hindlimbs.
Statistical analysis of genetic Interactions
To identify epistatic interactions between Shox2 and Hox genes during limb growth, the limbs from three to six animals were measured for each of the 36 genotypes analyzed. A factorial analysis of variance (ANOVA) was performed using the lm function in R, considering Shox2, HoxA, and HoxD as independent factors and the number of functional alleles as levels (or groups in each factor). The RosaShox2 transgene was considered as an additional Shox2 level. To minimize the number of limb measurements with zero length, which also had zero variance, the Shox2c/− genotype was excluded from the femur data set, and the HoxD−/− genotype was excluded from the radius and ulna data sets. Following these exclusions, the data sets for each element showed roughly uniform variance and nearly normally distributed residuals (as judged by residuals-vs.-fitted plots and quantile-quantile). Differential effects of a given Shox2 mutation between wild-type and mutant Hox backgrounds were assessed by performing contrasts, using the multcomp package (Hothorn et al. 2008) in R. The estimates from these contrasts can be calculated with the following equation (as seen in Jaccard 1998), substituting in the average lengths (designated μ) of the four following genotypes,
where ε is the difference between the expected and observed lengths, and Shox2(mut) and Hox(mut) are mutant genotypes for Shox2 and Hox genes. When there is no genetic interaction, the estimate is zero or close to zero. When a genetic interaction is present, the value is significantly different from zero. To account for multiple comparisons, a Bonferroni correction was used to generate a stringent significance threshold.
Results
To investigate the possibility that Shox2 and Hox genes function together during limb development, we examined their relative expression dynamics, using dual fluorescence in situ hybridization (FISH) in single embryos (Figure 1). Hoxd9, Hoxa11, and Hoxd13 were used as representative Hox genes that function in the individual segments along the proximodistal axis. At embryonic day 10.5 (E10.5), Shox2 transcripts occupied the majority of the limb bud, giving extensive overlap with each Hox gene (Figure 1A, inset shows expression of Shox2 and Hoxd13 in a single optical section). At E11.5, Shox2 expression was confined to the proximal limb and shared a similar distal expression border to Hoxd9 and Hoxa11 (Figure 1B), the latter of which selectively marks the developing zeugopod (Nelson et al. 1996). Hoxd13 was, in contrast, no longer expressed with Shox2, but expanded in the distal region of the limb bud in association with the developing autopod (Figure 1B). At E12.5, Shox2 expression is maintained proximally, along with Hoxd9 and Hoxa11, and Hoxd13 expression surrounds the developing digit condensations. As Hoxa13 is additionally expressed in the developing carpal region (Nelson et al. 1996), its domain closely juxtaposes that of Shox2 (Figure 1C). Thus Shox2 and Hox genes are coordinately expressed in broad domains that closely correspond to the discrete segments of the proximodistal axis. To investigate their expression in and around the proximal skeletal elements, we performed chromogenic ISH on sections at E12.5 and E13.5. Shox2, Hoxd9, and Hoxa11 were all expressed in the mesenchyme outside the elements, and each had at least some expression in the perichondrium (Figure 1D). Shox2 and Hoxa11 were additionally each expressed in the proliferating chondrocytes of the elements in their respective expression domains (Figure 1D).
Figure 1.
Characterization of Shox2 and Hox gene expression. (A–C) Fluorescence in situ hybridization of Shox2 and Hox genes in single embryos, with yellow signal in the merged image showing coexpression. (A) At E10.5, Hox genes are expressed in a nested pattern within the Shox2 expression domain, which occupies the majority of the limb bud. The inset for the Shox2/Hoxd13 series shows a single optical section through the limb bud, using confocal microscopy. (B) At E11.5, Shox2 expression is confined to the proximal limb and shares a similar distal expression border to that of the Hoxd9 and Hoxa11 expression domains. Hoxd13 is exclusively expressed in the distal limb bud. (C) At E12.5, Shox2 expression is maintained in the proximal limb. Hoxd9 and Hoxa11 are expressed in the zeugopod, with Hoxd9 additionally being expressed in the developing digits. Hoxd13 is expressed in the developing digits, and Hoxa13 is expressed in the developing digits and carpals. (D) Chromogenic in situ hybridization at E12.5 and E13.5 shows Shox2 and the Hox genes are expressed both outside and within the elements of the stylopod and zeugopod. Shox2 and Hoxa11 are each expressed in the proliferating chondroctyes within their respective expression domains. Expression outlining each element corresponds to the perichondrium. H, humerus; R, radius; U, ulna. Bar, 0.25 mm.
Given that the mutations of Shox2 or Hox genes similarly give rise to regional phenotypes and that they have closely associated expression dynamics, we sought to determine whether these genes might function together during limb development. Mice harboring Shox2 mutations (Cobb et al. 2006), a complete deficiency of the HoxD cluster (Spitz et al. 2001), and a Cre-dependent deletion of the entire HoxA cluster (Kmita et al. 2005) were crossed, and the limbs of resultant progeny were analyzed for evidence of epistasis as newborns (Figure 2). Intriguingly, while the removal of an individual Shox2 allele did not lead to limb shortening when the Hox genes were intact, this same mutation gave significantly truncated humeri when the HoxD cluster was disrupted (∼30% shorter than expected if HoxD genes did not affect the outcome of Shox2 mutation) [Figure 2, A and A′: nonparallel lines in interaction plots are characteristic of interaction (e.g., Zhu et al. 2014)]. Additionally, Prrx1-Cre; Shox2fl/− (or Shox2c/−) animals, which have both copies of Shox2 removed from the limb mesenchyme, have disproportionately shorter ulnae when the HoxA genes are concomitantly disrupted (∼25% shorter than expected if HoxA genes did not affect the outcome of Shox2 mutation) (Figure 2, B and B′). Thus, sensitized Hox backgrounds reveal dose-dependent roles for Shox2 in the growth of limb elements, including a previously unrecognized yet sizable role in the growth of the newborn zeugopod. To verify these results in a statistical framework and also extend the analysis to each element of the stylopod and zeugopod of both the fore- and hindlimbs, we used a linear model (ANOVA) to identify significant interactions between Shox2, HoxA, and HoxD alleles (see Materials and Methods) (Supporting Information, Table S1). This analysis revealed epistasis between Shox2 and Hox genes in all elements of the stylopod and zeugopod (with the exception of the developing fibula), where there were significant differences between the observed bone lengths and those expected if there were no interactions (shown as “estimates” in Table S2). These interactions were associated with entire series of genotypes and were often similar for corresponding skeletal elements of the fore- and hindlimbs (Figure 2, A′′ and B′′). Overall, there is pervasive epistasis between loss-of-function alleles of Shox2 and Hox genes, where modulating their relative gene dose produces graded changes in the lengths of individual skeletal elements.
To further analyze the role of Shox2 in limb growth, we also examined the effects of Shox2 gain-of-function on the developing limb. Does Shox2 overexpression lead to limb lengthening, as has been proposed for human SHOX? Or, as Shox2 and Hox genes function together, can Shox2 overexpression compensate for the loss of Hox gene function? To address these questions, a line of mice with a single copy of Shox2 targeted to the Rosa26 locus (Scott et al. 2011) was crossed to Prrx1-Cre mice, giving rise to embryos with ectopic Shox2 expression throughout the mesenchyme of the developing limb (Figure 3A) (referred to as “RosaShox2” animals). To define the relative level of Shox2 expression in these mice, entire E10.5 limb buds and proximal E12.5 limb buds were microdissected, and quantitative real-time polymerase chain reaction (RT-PCR) was performed. For each stage, there was an average increase of between 2 and 2.5 times compared to wild type (Figure 3B). Analysis of RosaShox2 hemizygotes failed to show any limb lengthening and actually showed a slightly reduced humeral length compared to wild type (Figure 3, C and C′). These animals furthermore lacked lateral ossifications in the autopod and were missing their scapular spine. The RosaShox2 allele could, importantly, functionally replace the native Shox2 gene, as RosaShox2; Shox2c/− animals had humeri of similar lengths to those of wild-type and RosaShox2 animals (Figure 3, C and C′). We crossed the RosaShox2 allele into the Hox cluster-deficient lines to generate animals that overexpress Shox2 in each Hox mutant background, measured the lengths of the limb segments in the progeny, and incorporated the data into our limb models. This analysis revealed that the RosaShox2 allele could extend the length of the humerus of the two most severe Hox mutants, although in a limited manner (Figure 3C′) (Table S2), indicating Shox2 overexpression can partially compensate for Hox gene loss. We further found that, in a HoxA/D mutant background, graded modulation of effective Shox2 levels through under- and overexpression gave stepwise changes in humerus length (Figure 3D). Thus, Shox2 is sufficient to drive humerus growth in the absence of the HoxA and HoxD clusters, but only in a limited and inefficient manner. These data suggest that Shox2 and Hox gene functions normally synergize to drive the robust growth of the humerus.
Figure 3.
Epistasis between Shox2 gain-of-function and Hox genes during growth of the stylopod. (A) Shox2 ISH on wild-type and RosaShox2 limbs at E12.5. (B) Real-time PCR determining the relative Shox2 levels in wild-type and RosaShox2 animals in whole E10.5 forelimb buds and proximal E12.5 forelimb buds. (C) Newborn forelimb skeletons showing the effects of Shox2 overexpression in wild-type and Shox2c/- backgrounds. Arrowheads point to humerus. (C′) Interaction plot displaying the effects of overexpressing Shox2 in wild-type, Shox2c/−, HoxAc/+; HoxD−/−, and HoxAc/c; HoxD−/− backgrounds (n = 3–6 for each genotype). Data are plotted as means ± SEM. Effect of RosaShox2 is significantly different in each mutant background compared to an otherwise wild-type background. P < 0.001. (D) Newborn forelimb skeletons and quantification of the effect of modulating Shox2 dose in a HoxAc/c; HoxD−/− background (n = 3–6 for each genotype). Arrowheads point to humerus. P < 0.05.
To investigate the cellular basis of the limb shortening in Shox2 and Hox mutants, we went on to examine the expression of markers and regulators of chondrogenesis. Previous studies have demonstrated a delay in humeral chondrogenesis, associated with a loss or reduction in Runx2 levels, when Shox2 is deleted in the limb mesenchyme (Cobb et al. 2006; Yu et al. 2007). To ascertain whether Shox2 and Hox mutants have a similar delay in chondrogenesis, we therefore examined Col2a1, Runx2, and Col10a1 expression, in wild-type, Shox2c/−, and HoxAc/c; HoxD−/− animals (Figure 4A). Sections at E14.5, when chondrogenesis is well underway, showed that both wild-type and mutant humeri contained Col2a1-positive cells, with wild-type animals additionally showing down regulation in the middle of the element, corresponding to the zone of maturation. Runx2 was strongly expressed in the chondrocytes and the perichondrium of wild-type animals, but this expression was depressed in the humeri of both Shox2 and Hox mutants. Furthermore, wild-type animals showed strong Col10a1 expression in the middle of the humerus, which was also absent in both Shox2c/− and HoxAc/c; HoxD−/− animals. These data suggest that Shox2 and Hox mutants fail to undergo timely chondrocyte maturation. In support of this view, hematoxylin and eosin staining showed that wild-type humeri contained hypertrophic chondrocytes in the middle of the element, while Shox2 and Hox mutants displayed only immature chondrocytes (Figure 4B). Thus the coordinated action of Shox2 and Hox genes likely drives cartilage maturation upstream of Runx2 during development.
Figure 4.
Independent mutation of Shox2 and Hox genes results in a similar delay in humeral chondrocyte maturation. Shown are in situ hybridization and histological staining of E14.5 forelimb sections. (A) Wild-type, Shox2c/−, and HoxAc/c; HoxD−/− humeri all show Col2a1 expression, with wild-type limbs additionally showing downregulation of expression in the middle of the element. Wild-type humeri have prominent Runx2 and Col10a1 expression, while Shox2c/− and HoxAc/c; HoxD−/− animals show reduced Runx2 levels and a lack of Col10a1 expression. Arrowheads point to the middle of the humerus. sc, scapula. (B) Hematoxylin and eosin staining showing that wild-type limbs contain hypertrophic chondrocytes, while Shox2c/− and HoxAc/c; HoxD−/− animals lack these cells.
Given that Shox2 and Hox genes functionally interact and have associated expression dynamics, we considered that they might influence one another’s transcription. In agreement with earlier studies (Cobb et al. 2006; Yu et al. 2007), no changes in Hox gene expression were found in Shox2c/− animals (data not shown), suggesting Shox2 does not function upstream of Hox transcription. Considering the converse relationship, we examined whether Hox genes may activate Shox2 expression. In situ hybridization at E10.5, when there is maximal overlap between Shox2 and Hox expression (Figure 1), revealed a reproducible dampening of Shox2 transcript levels in HoxAc/c; HoxD−/− animals, compared to wild type, throughout the limb bud (n = 3/3) (Figure 5). This regulation could reflect direct trans-activation by HOX proteins and/or indirect regulation through intermediary molecules. Supporting a contribution from the latter scenario, it was reported that Shh, which is activated by Hox function (Kmita et al. 2005), is required for normal Shox2 transcript levels at E10.5 (Probst et al. 2011). This downregulation of Shox2 expression is transient, however, as HoxAc/c; HoxD−/− animals show similar Shox2 expression at E11 to wild type, despite having a drastic difference in limb bud morphology (Figure 5). A recent report demonstrated that Hox11 genes are required for Shox2 expression in the proliferating chondrocytes of the zeugopodal elements (Gross et al. 2012), suggesting Hox genes could generally function upstream of Shox2 expression in these cells. To assess whether Hox genes are necessary for Shox2 activation in the developing humerus, ISH was performed on sections of wild-type and HoxA/D mutant limbs at E14.5. However, both wild-type and HoxAc/c; HoxD−/− animals had similar Shox2 expression in the humeral chondrocytes (n = 3/3) (Figure 5), suggesting any Hox-mediated transcriptional control of Shox2 in this cell population is limited. There was, however, a lack of Shox2 expression in the perichondrium of the humerus in Hox mutants (n = 3/3) (Figure 5). As the strongest perichondrial expression of Shox2 in wild-type animals is adjacent to the zone of maturation (Figure 5), which HoxAc/c; HoxD−/− animals lack, this absence of Shox2 expression is likely, at least in part, a secondary consequence of a failure to undergo chondrocyte maturation. However, we also recently showed that chondrocyte-specific deletion of Shox2 leads to a considerably less severe phenotype than its removal from the entire limb (Bobick and Cobb 2012), implicating the presence of additional Shox2-expressing cells during growth. Given the intimate association of the perichondrium with the underlying chondrocytes and their critical functional interactions (Kronenberg 2007), perichondrial Shox2 expression represents a potential candidate for making contributions to limb growth. Thus, the absence of Shox2 expression in the perichondrium of Hox mutants may indeed contribute to a lack of growth in these animals.
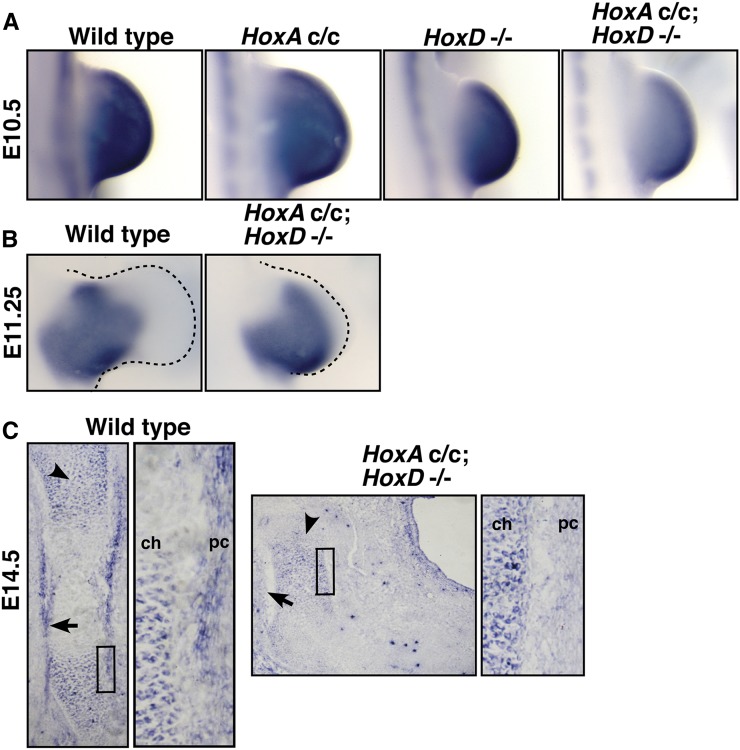
Figure 5.
Disruption of Hox genes gives a transient decrease in Shox2 expression in the early limb bud and a lack of Shox2 expression in the perichondrium, as seen through in situ hybridization. (A) At E10.5, HoxAc/c; HoxD−/− animals have a decrease in Shox2 expression, compared to wild-type, HoxAc/c, and HoxD−/− animals (n = 3/3). (B) At E11, HoxAc/c; HoxD−/− animals have similar Shox2 levels to those of wild-type controls (n = 2/2). (C) At E14.5, Shox2 is expressed in the proliferating chondrocytes (arrowhead) and the perichondrium (arrow) of wild-type animals, but expression is selectively absent in the perichondrium of HoxAc/c; HoxD−/− animals (n = 3/3). ch, chondrocytes; pc, perichondrium.
Discussion
This work investigates the genetic relationship between Shox2 and Hox genes during the growth and development of the limb segments. We analyzed their relative expression dynamics during normal development, using representative Hox genes, showing they have associated expression changes that occur in broad domains along the proximodistal axis. We then manipulated the relative expression levels of Shox2 and Hox genes, demonstrating these adjustments tune the length of individual skeletal elements through epistatic interactions. Finally, we show that both Shox2 and Hox genes function upstream of Runx2 expression and are required for normal chondrocyte maturation.
Characterization of the relative gene expression patterns of Shox2 and Hox genes showed that they are dynamic and occupy broad regions along the proximodistal axis. While Shox2 is initially coexpressed with multiple Hox genes, it is restricted to the stylopodal and zeugopodal domains at later stages, along with Hoxd9 and Hoxa11, and juxtaposes the distal expression of Hoxd13 and Hoxa13. Thus, Shox2 expression closely resembles the “early” phase of Hox expression (Tarchini and Duboule 2006; Andrey et al. 2013). Interestingly, it was recently demonstrated that a Shox2-Cre knock-in allele also drives reporter activity in a proximally restricted manner (Sun et al. 2013), suggesting the lineage of early Shox2-expressing cells does not contribute to digit development. Thus, while Shox2 expression appears throughout the early limb bud (Figure 1A), it is likely not expressed in the presumptive digit cells. This view is consistent with a recent study showing fish Hox enhancers have activity in the “distal” part of the early mouse limb bud and that this domain later corresponds to the proximal limb (Woltering et al. 2014). The coincidence of Shox2 and the early phase of Hox expression may reflect an ancestral functional relationship in the fins of fish, as both Hox genes and Shox2 are expressed in the developing zebrafish fins (Sordino et al. 1995; Thisse and Thisse 2004).
It is currently unknown how the rodent evolutionary lineage tolerated the loss of Shox. However, given that Shox is otherwise conserved in vertebrates, expressed in the developing zeugopods of chicks and humans, and required for normal human limb development (Rao et al. 1997; Belin et al. 1998; Shears et al. 1998), its loss likely required compensatory changes. We suggest that multiple factors were involved, and one of them could have been expression of Shox2 in the developing zeugopod. This proposal is based on multiple lines of evidence. First, mouse Shox2 is expressed in both the developing stylopodal and zeugopodal regions (Cobb et al. 2006; Bobick and Cobb 2012), and since Shox2 and Shox are homologous genes, this could have helped buffer the effects of SHOX loss. It has indeed been previously shown that a human SHOX knock-in allele is able to compensate for the loss of mouse Shox2 in the developing forelimb (Liu et al. 2011). Interestingly, the reported SHOX2 expression domains in chicks and humans are restricted to the stylopod (Clement-Jones et al. 2000; Tiecke et al. 2006), suggesting that the distally extended expression of mouse Shox2 may be a derived feature in rodents. And second, although the individual mutation of mouse Shox2 has little effect on the newborn zeugopod, its mutation in sensitized Hox backgrounds demonstrates its prominent function in this region. Based on this model, identifying additional factors that helped compensate for Shox loss could be an active area for future research.
Shox2 and Hox genes play critical roles in the formation of the limb skeleton, as evidenced by the strong phenotypes observed following their genetic ablation. Emerging evidence indicates that the activation of Runx2 is a critical point of regulation by these genes (Cobb et al. 2006; Villavicencio-Lorini et al. 2010; Gross et al. 2012). The present work supports this general model and extends it by revealing that Shox2 and Hox genes genetically interact during the development of both the stylopod and zeugopod elements. Given the similar regional shortening in the limbs of humans with SHOX mutations, we furthermore propose that Shox and Hox genes may generally coordinate Runx2 expression in the stylopodal and zeugopodal elements of vertebrate limbs.
Our analysis extensively investigated the genetic relationship of Shox and Hox genes, revealing multiple interesting features. When these genes are independently and fully deleted, they give prominent phenotypes, although they have some ability to tolerate intermediate changes in dose. These relatively “steep” phenotypic responses are in contrast to the changes that occur from the concomitant disruption of Shox2 and Hox genes. In these cases, the limb is much more sensitive to Shox2 and Hox disruption, giving less drastic, yet more widespread, changes in phenotype. These genetic interactions thus normally confer the ability to buffer the effects of some variation in gene product levels. Such perspectives provide insight into how the level of robustness of a system influences its response to mutation and is relevant to the establishment of genetic disease. In this regard, it is interesting to note that both human SHOX and human HOXA11 are considered haplo-insufficient loci (Rao et al. 1997; Thompson and Nguyen 2000), suggesting the zeugopod genetic network they function in may be somewhat sensitive to mutation in general. While the sources of the dosage sensitivity in Hox–Shox networks are unknown, it may furthermore extend to Runx2 regulation, as disruption of a single Runx2 allele causes limb defects (Otto et al. 1997). We also extensively explored the relationship between under- and overexpression of Shox2. In both wild-type and many Hox mutant backgrounds, Shox2 overexpression did not extend limb length, suggesting Shox2 levels were already present in excess or “saturating”. This was in contrast to the most severe Hox backgrounds, where the humerus responded nearly linearly to deletions of Shox2 and, conversely, Shox2 overexpression gave significant gains in length. Thus, only in those backgrounds where the limb was most sensitive to Shox2 reductions was the humerus also responsive to elevated Shox2 levels. In addition to providing insight into the constancy of developmental phenotypes, these data may be furthermore relevant to the effects of human SHOX dosage. It is hypothesized that SHOX overexpression leads to tall stature in humans, which is based on the observation that SHOX deletions are dosage sensitive and that people with three or more sex chromosomes (and therefore supernumerary copies of SHOX, as SHOX resides on the pseudoautosomal region of the sex chromosomes) are significantly taller than expected (Durand and Rappold 2013). However, associated with chromosome aneuploidies are changes in hormone levels, which influence the maturation of the growth plate during longitudinal bone growth (Smith et al. 1994; Morishima et al. 1995), thus confounding interpretations of the cause of tall stature (Ottesen et al. 2010). As SHOX deletions are dosage sensitive, SHOX overexpression alone may very well lead to longer limbs, but we also suggest there are likely limitations to its effects. Available data suggest that the response to SHOX deletions is quite nonlinear, where individuals with two deleted copies have a much more severe phenotype than those with a deletion in a single copy (Belin et al. 1998; Shears et al. 1998; Schiller et al. 2000; Zinn et al. 2002; Albuisson et al. 2012), suggesting the amount of functional SHOX levels may be approaching saturation in people with two intact copies. Thus, any substantial role SHOX overexpression has in overgrowth may be in conjunction with associated changes of having additional sex chromosomes, in line with the hypothesis that SHOX overexpression interacts with hormonal changes (Ogata et al. 2000).
The possibility that Hox genes regulate Shox2 expression was examined in considerable detail. We found that the HoxA/D clusters are required for full activation of Shox2 early on, but this regulation on its own may be of limited consequence as it is transient. However, given that Hox genes likely have multiple roles during limb development, even a transient decrease in Shox2 expression could make contributions to quantitative Hox phenotypes. Mutation of Hox genes additionally resulted in the loss of Shox2 expression in the perichondrium of the developing humerus, which was associated with a failure to undergo chondrocyte maturation. Future experiments are required to address any causal role of perichondrial Shox2 expression in driving the growth of the humerus or whether this expression is involved in later steps of skeletogenesis, such as the generation of bone. As Hox genes are also hypothesized to exert a large part of their effect on the developing skeleton from their expression in the perichondrium (Villavicencio-Lorini et al. 2010; Swinehart et al. 2013), their cell autonomous role in this structure should also be investigated. Of note, although Hox genes are required for normal Shox2 expression, such regulation cannot completely account for the role of Hox genes in growth, as Shox2 overexpression failed to fully rescue the Hox mutant phenotype. Thus, interactions between Shox2 and Hox genes during mouse limb development likely involve diverse mechanisms, which could also be the case for any interactions between human SHOX and HOX genes. Elucidation of the biochemical mechanism underlying SHOX and HOX cooperativity will be of considerable interest. SHOX and HOX proteins could, for example, coordinately bind common enhancers, bind distinct enhancers that synergistically activate transcription, or regulate different target genes that themselves act in a cooperative manner. Thus, identifying genomic regions that are bound by SHOX2 and HOX proteins during limb development could help illuminate the basis of their genetic interaction.
Supplementary Material
Acknowledgments
We are grateful to D. Duboule and M. Kmita for mice, L. Harder and C. Olito for helpful discussions on the statistical analysis, and members of the J. Cobb laboratory for comments on the manuscript. This work was supported by Natural Sciences and Engineering Research Council of Canada grant RGPIN/355731-2008.
Footnotes
Supporting information is available online at http://www.genetics.org/lookup/suppl/doi:10.1534/genetics.114.167460/-/DC1.
Communicating editor: T. R. Magnuson
Literature Cited
- Alam S., Zinyk D., Ma L., Schuurmans C., 2005. Members of the Plag gene family are expressed in complementary and overlapping regions in the developing murine nervous system. Dev. Dyn. 234: 772–782. [DOI] [PubMed] [Google Scholar]
- Albuisson J., Schmitt S., Baron S., Bezieau S., Benito-Sanz S., et al. , 2012. Clinical utility gene card for: Leri-Weill dyschondrosteosis (LWD) and Langer mesomelic dysplasia (LMD). Eur. J. Hum. Genet. 20: PMC3400739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrey G., Montavon T., Mascrez B., Gonzalez F., Noordermeer D., et al. , 2013. A switch between topological domains underlies HoxD genes collinearity in mouse limbs. Science 340: 1234167. [DOI] [PubMed] [Google Scholar]
- Apte S. S., Seldin M. F., Hayashi M., Olsen B. R., 1992. Cloning of the human and mouse type X collagen genes and mapping of the mouse type X collagen gene to chromosome 10. Eur. J. Biochem. 206: 217–224. [DOI] [PubMed] [Google Scholar]
- Belin V., Cusin V., Viot G., Girlich D., Toutain A., et al. , 1998. SHOX mutations in dyschondrosteosis (Leri-Weill syndrome). Nat. Genet. 19: 67–69. [DOI] [PubMed] [Google Scholar]
- Bobick B. E., Cobb J., 2012. Shox2 regulates progression through chondrogenesis in the mouse proximal limb. J. Cell Sci. 125: 6071–6083. [DOI] [PubMed] [Google Scholar]
- Boulet A. M., Capecchi M. R., 2004. Multiple roles of Hoxa11 and Hoxd11 in the formation of the mammalian forelimb zeugopod. Development 131: 299–309. [DOI] [PubMed] [Google Scholar]
- Clement-Jones M., Schiller S., Rao E., Blaschke R. J., Zuniga A., et al. , 2000. The short stature homeobox gene SHOX is involved in skeletal abnormalities in Turner syndrome. Hum. Mol. Genet. 9: 695–702. [DOI] [PubMed] [Google Scholar]
- Cobb J., Duboule D., 2005. Comparative analysis of genes downstream of the Hoxd cluster in developing digits and external genitalia. Development 132: 3055–3067. [DOI] [PubMed] [Google Scholar]
- Cobb J., Dierich A., Huss-Garcia Y., Duboule D., 2006. A mouse model for human short-stature syndromes identifies Shox2 as an upstream regulator of Runx2 during long-bone development. Proc. Natl. Acad. Sci. USA 103: 4511–4515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis A. P., Witte D. P., Hsieh-Li H. M., Potter S. S., Capecchi M. R., 1995. Absence of radius and ulna in mice lacking hoxa-11 and hoxd-11. Nature 375: 791–795. [DOI] [PubMed] [Google Scholar]
- Dolle P., Izpisua-Belmonte J. C., Boncinelli E., Duboule D., 1991. The Hox-4.8 gene is localized at the 5′ extremity of the Hox-4 complex and is expressed in the most posterior parts of the body during development. Mech. Dev. 36: 3–13. [DOI] [PubMed] [Google Scholar]
- Durand C., Rappold G. A., 2013. Height matters—from monogenic disorders to normal variation. Nat. Rev. Endocrinol. 9: 171–177. [DOI] [PubMed] [Google Scholar]
- Fromental-Ramain C., Warot X., Lakkaraju S., Favier B., Haack H., et al. , 1996a Specific and redundant functions of the paralogous Hoxa-9 and Hoxd-9 genes in forelimb and axial skeleton patterning. Development 122: 461–472. [DOI] [PubMed] [Google Scholar]
- Fromental-Ramain C., Warot X., Messadecq N., Lemeur M., Dolle P., et al. , 1996b Hoxa-13 and Hoxd-13 play a crucial role in the patterning of the limb autopod. Development 122: 2997–3011. [DOI] [PubMed] [Google Scholar]
- Gianfrancesco F., Sanges R., Esposito T., Tempesta S., Rao E., et al. , 2001. Differential divergence of three human pseudoautosomal genes and their mouse homologs: implications for sex chromosome evolution. Genome Res. 11: 2095–2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Martin M. C., Mallo M., Ros M. A., 2014. Long bone development requires a threshold of Hox function. Dev. Biol. 392: 454–465. [DOI] [PubMed] [Google Scholar]
- Gross S., Krause Y., Wuelling M., Vortkamp A., 2012. Hoxa11 and Hoxd11 regulate chondrocyte differentiation upstream of Runx2 and Shox2 in mice. PLoS ONE 7: e43553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hothorn T., Bretz F., Westfall P., 2008. Simultaneous inference in general parametric models. Biom. J. 50: 346–363. [DOI] [PubMed] [Google Scholar]
- Jaccard, J., 1998 Interaction Effects in Factorial Analysis of Variance. SAGE Publications, Thousand Oaks, California. [Google Scholar]
- Karsenty G., Wagner E. F., 2002. Reaching a genetic and molecular understanding of skeletal development. Dev. Cell 2: 389–406. [DOI] [PubMed] [Google Scholar]
- Kmita M., Fraudeau N., Herault Y., Duboule D., 2002. Serial deletions and duplications suggest a mechanism for the collinearity of Hoxd genes in limbs. Nature 420: 145–150. [DOI] [PubMed] [Google Scholar]
- Kmita M., Tarchini B., Zakany J., Logan M., Tabin C. J., et al. , 2005. Early developmental arrest of mammalian limbs lacking HoxA/HoxD gene function. Nature 435: 1113–1116. [DOI] [PubMed] [Google Scholar]
- Kronenberg H. M., 2007. The role of the perichondrium in fetal bone development. Ann. N. Y. Acad. Sci. 1116: 59–64. [DOI] [PubMed] [Google Scholar]
- Liu H., Chen C. H., Espinoza-Lewis R. A., Jiao Z., Sheu I., et al. , 2011. Functional redundancy between human SHOX and mouse Shox2 genes in the regulation of sinoatrial node formation and pacemaking function. J. Biol. Chem. 286: 17029–17038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan M., Martin J. F., Nagy A., Lobe C., Olson E. N., et al. , 2002. Expression of Cre Recombinase in the developing mouse limb bud driven by a Prxl enhancer. Genesis 33: 77–80. [DOI] [PubMed] [Google Scholar]
- Metsaranta M., Toman D., De Crombrugghe B., Vuorio E., 1991. Specific hybridization probes for mouse type I, II, III and IX collagen mRNAs. Biochim. Biophys. Acta 1089: 241–243. [DOI] [PubMed] [Google Scholar]
- Morishima A., Grumbach M. M., Simpson E. R., Fisher C., Qin K., 1995. Aromatase deficiency in male and female siblings caused by a novel mutation and the physiological role of estrogens. J. Clin. Endocrinol. Metab. 80: 3689–3698. [DOI] [PubMed] [Google Scholar]
- Nelson C. E., Morgan B. A., Burke A. C., Laufer E., Dimambro E., et al. , 1996. Analysis of Hox gene expression in the chick limb bud. Development 122: 1449–1466. [DOI] [PubMed] [Google Scholar]
- Neufeld S. J., Zhou X., Vize P. D., Cobb J., 2013. mRNA fluorescence in situ hybridization to determine overlapping gene expression in whole-mount mouse embryos. Dev. Dyn. 242: 1094–1100. [DOI] [PubMed] [Google Scholar]
- Ogata T., Kosho T., Wakui K., Fukushima Y., Yoshimoto M., et al. , 2000. Short stature homeobox-containing gene duplication on the der(X) chromosome in a female with 45,X/46,X, der(X), gonadal dysgenesis, and tall stature. J. Clin. Endocrinol. Metab. 85: 2927–2930. [DOI] [PubMed] [Google Scholar]
- Ottesen A. M., Aksglaede L., Garn I., Tartaglia N., Tassone F., et al. , 2010. Increased number of sex chromosomes affects height in a nonlinear fashion: a study of 305 patients with sex chromosome aneuploidy. Am. J. Med. Genet. A. 152A: 1206–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto F., Thornell A. P., Crompton T., Denzel A., Gilmour K. C., et al. , 1997. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell 89: 765–771. [DOI] [PubMed] [Google Scholar]
- Phillips P. C., 2008. Epistasis—the essential role of gene interactions in the structure and evolution of genetic systems. Nat. Rev. Genet. 9: 855–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Probst S., Kraemer C., Demougin P., Sheth R., Martin G. R., et al. , 2011. SHH propagates distal limb bud development by enhancing CYP26B1-mediated retinoic acid clearance via AER-FGF signalling. Development 138: 1913–1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team, 2013 A Language and Environment for Statistical Computing R Foundation for Statistical Computing, Vienna. Available at: http://www.R-project.org/.
- Rao E., Weiss B., Fukami M., Rump A., Niesler B., et al. , 1997. Pseudoautosomal deletions encompassing a novel homeobox gene cause growth failure in idiopathic short stature and Turner syndrome. Nat. Genet. 16: 54–63. [DOI] [PubMed] [Google Scholar]
- Renucci A., Zappavigna V., Zakany J., Izpisua-Belmonte J. C., Burki K., et al. , 1992. Comparison of mouse and human HOX-4 complexes defines conserved sequences involved in the regulation of Hox-4.4. EMBO J. 11: 1459–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiller S., Spranger S., Schechinger B., Fukami M., Merker S., et al. , 2000. Phenotypic variation and genetic heterogeneity in Leri-Weill syndrome. Eur. J. Hum. Genet. 8: 54–62. [DOI] [PubMed] [Google Scholar]
- Scott A., Hasegawa H., Sakurai K., Yaron A., Cobb J., et al. , 2011. Transcription factor short stature homeobox 2 is required for proper development of tropomyosin-related kinase B-expressing mechanosensory neurons. J. Neurosci. 31: 6741–6749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shears D. J., Vassal H. J., Goodman F. R., Palmer R. W., Reardon W., et al. , 1998. Mutation and deletion of the pseudoautosomal gene SHOX cause Leri-Weill dyschondrosteosis. Nat. Genet. 19: 70–73. [DOI] [PubMed] [Google Scholar]
- Smith E. P., Boyd J., Frank G. R., Takahashi H., Cohen R. M., et al. , 1994. Estrogen resistance caused by a mutation in the estrogen-receptor gene in a man. N. Engl. J. Med. 331: 1056–1061. [DOI] [PubMed] [Google Scholar]
- Sordino P., Van Der Hoeven F., Duboule D., 1995. Hox gene expression in teleost fins and the origin of vertebrate digits. Nature 375: 678–681. [DOI] [PubMed] [Google Scholar]
- Spitz F., Gonzalez F., Peichel C., Vogt T. F., Duboule D., et al. , 2001. Large scale transgenic and cluster deletion analysis of the HoxD complex separate an ancestral regulatory module from evolutionary innovations. Genes Dev. 15: 2209–2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C., Zhang T., Liu C., Gu S., Chen Y., 2013. Generation of Shox2-Cre allele for tissue specific manipulation of genes in the developing heart, palate, and limb. Genesis 51: 515–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swinehart I. T., Schlientz A. J., Quintanilla C. A., Mortlock D. P., Wellik D. M., 2013. Hox11 genes are required for regional patterning and integration of muscle, tendon and bone. Development 140: 4574–4582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarchini B., Duboule D., 2006. Control of Hoxd genes’ collinearity during early limb development. Dev. Cell 10: 93–103. [DOI] [PubMed] [Google Scholar]
- Thisse, B., and C. Thisse, 2004 Fast Release Clones: A High Throughput Expression Analysis ZFIN Direct Data Submission. Available at: http://zfin.org.
- Thompson A. A., Nguyen L. T., 2000. A megakaryocytic thrombocytopenia and radio-ulnar synostosis are associated with HOXA11 mutation. Nat. Genet. 26: 397–398. [DOI] [PubMed] [Google Scholar]
- Tiecke E., Bangs F., Blaschke R., Farrell E. R., Rappold G., et al. , 2006. Expression of the short stature homeobox gene Shox is restricted by proximal and distal signals in chick limb buds and affects the length of skeletal elements. Dev. Biol. 298: 585–596. [DOI] [PubMed] [Google Scholar]
- Villavicencio-Lorini P., Kuss P., Friedrich J., Haupt J., Farooq M., et al. , 2010. Homeobox genes d11-d13 and a13 control mouse autopod cortical bone and joint formation. J. Clin. Invest. 120: 1994–2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warot X., Fromental-Ramain C., Fraulob V., Chambon P., Dolle P., 1997. Gene dosage-dependent effects of the Hoxa-13 and Hoxd-13 mutations on morphogenesis of the terminal parts of the digestive and urogenital tracts. Development 124: 4781–4791. [DOI] [PubMed] [Google Scholar]
- Wellik D. M., Capecchi M. R., 2003. Hox10 and Hox11 genes are required to globally pattern the mammalian skeleton. Science 301: 363–367. [DOI] [PubMed] [Google Scholar]
- Woltering J. M., Noordermeer D., Leleu M., Duboule D., 2014. Conservation and divergence of regulatory strategies at Hox loci and the origin of tetrapod digits. PLoS Biol. 12: e1001773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida C. A., Yamamoto H., Fujita T., Furuichi T., Ito K., et al. , 2004. Runx2 and Runx3 are essential for chondrocyte maturation, and Runx2 regulates limb growth through induction of Indian hedgehog. Genes Dev. 18: 952–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L., Liu H., Yan M., Yang J., Long F., et al. , 2007. Shox2 is required for chondrocyte proliferation and maturation in proximal limb skeleton. Dev. Biol. 306: 549–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C. T., Ingelmo P., Rand D. M., 2014. GxGxE for lifespan in Drosophila: mitochondrial, nuclear, and dietary interactions that modify longevity. PLoS Genet. 10: e1004354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinn A. R., Wei F., Zhang L., Elder F. F., Scott C. I., Jr, et al. , 2002. Complete SHOX deficiency causes Langer mesomelic dysplasia. Am. J. Med. Genet. 110: 158–163. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.