SUMMARY
Microbial genome sequencing platforms have produced a deluge of orphan biosynthetic pathways suspected of biosynthesizing new small molecules with pharmacological relevance. Genome synteny analysis provides an assessment of genomic island content, which is enriched in natural product gene clusters. Here we identified an atypical orphan carbohydrate-nonribosomal peptide synthetase (NRPS) genomic island in Photorhabdus luminescens using genome synteny analysis. Heterologous expression of the pathway led to the characterization of five new oligosaccharide metabolites with lysozyme inhibitory activities. The oligosaccharides harbor a 1,6-anhydro-β-D-N-acetyl-glucosamine (Glc-NAc) moiety, a rare structural feature for natural products. Gene deletion analysis and biochemical reconstruction of oligosaccharide production led to the discovery that a hypothetical protein in the pathway is a novel lytic transglycosylase responsible for bicyclic sugar formation. The example presented here supports a notion where targeting select genomic islands with a reduced reliance on known protein homologies could enhance the discovery of new metabolic chemistry and biology.
Introduction
Microbial secondary metabolism plays an important role in the discovery and development of new molecular probes and drugs, as their small molecule products have been molded within the constraints of evolutionary selection (Demain, 2014; Newman and Cragg, 2012). The biocatalysts that evolved to carry out the syntheses of these natural products can have value in the construction of building blocks with similarly privileged structural features. Next generation sequencing technologies continue to produce the sequences of microbial genomes at an exponential rate, illuminating countless orphan biosynthetic gene clusters responsible for the synthesis of currently unknown natural products (Winter et al., 2011; Bachmann et al., 2014). While many of the enzymes encoded in these gene clusters closely resemble proteins from classically studied biosynthetic systems, the number of hypothetical proteins with no known functions is similarly increasing. Undoubtedly, this growing reservoir of hypothetical proteins will harbor many new biocatalysts involved in novel bioactive small molecule syntheses.
Natural product gene clusters frequently reside on genomic islands and endow the producing hosts with chemical traits that can contribute to functional adaptations in their environmental niches (Ziemert et al., 2014). Genomic islands resulting from the horizontal transfer of sequences of chromosomal, plasmid, or phage origin can dramatically alter the chemical physiology of an organism. These evolutionary events can be observed using genome synteny analysis as lost or acquired sequences relative to phylogenetically-related organisms. Assessing microbial genomic island content provides a complementary vantage point for identifying atypical biosynthetic pathways that are not readily detected by homology (Vizcaino et al., 2014). Here we identified an unusual putative biosynthetic gene cluster in the entomopathogenic bacterium Photorhabdus luminescens TT01 using genome synteny analysis. P. luminescens participates in a multipartite symbiosis with Heterorhabditis nematodes and insect larvae in the soil, and its genome encodes a variety of known and currently uncharacterized natural products (Brachmann and Bode, 2013; Vizcaino et al., 2014). The Photorhabdus-Heterorhabditis bacterium-nematode complex collaboratively infect, kill, and consume the insect larvae with many of the natural products serving as virulence factors, mutualistic factors, antibiotics, and signaling molecules that aid in regulating the multipartite lifecycle.
The selected pathway was not recognized by homology-based pathway search programs, such as antiSMASH (Blin et al., 2013), indicating sequence divergence from previously studied biosynthetic systems. We reconstructed the pathway for heterologous expression in E. coli BAP1 (Pfeifer et al., 2001), which led to the structural characterization of five new metabolites containing a rare natural product feature, a 1,6-anhydro-β-D-Glc-NAc. Gene deletion and biochemical reconstitution studies revealed that a glycosyltransferase (GT) and a hypothetical protein encoded in the gene cluster were central to product formation. We demonstrate that the hypothetical protein is a novel lytic transglycosylase and shares parallel chemistry to enzymes of Gram-negative cell wall recycling pathways (Lee et al., 2013). Structurally diverse oligosaccharides can target a wide range of biological systems, underlying their potential pharmacological value (McCranie and Bachmann, 2014), rare sugars are important metabolic building blocks of natural products (Lin et al., 2013), and 1,6-anhydro sugars are commonly utilized for the laboratory synthesis of glycosylated molecules (Tanaka et al., 2009). This example illustrates that atypical pathways represent an avenue for the discovery of new biocatalytic chemistry.
Results and Discussion
Identification and heterologous expression of an orphan carbohydrate-NRPS genomic island
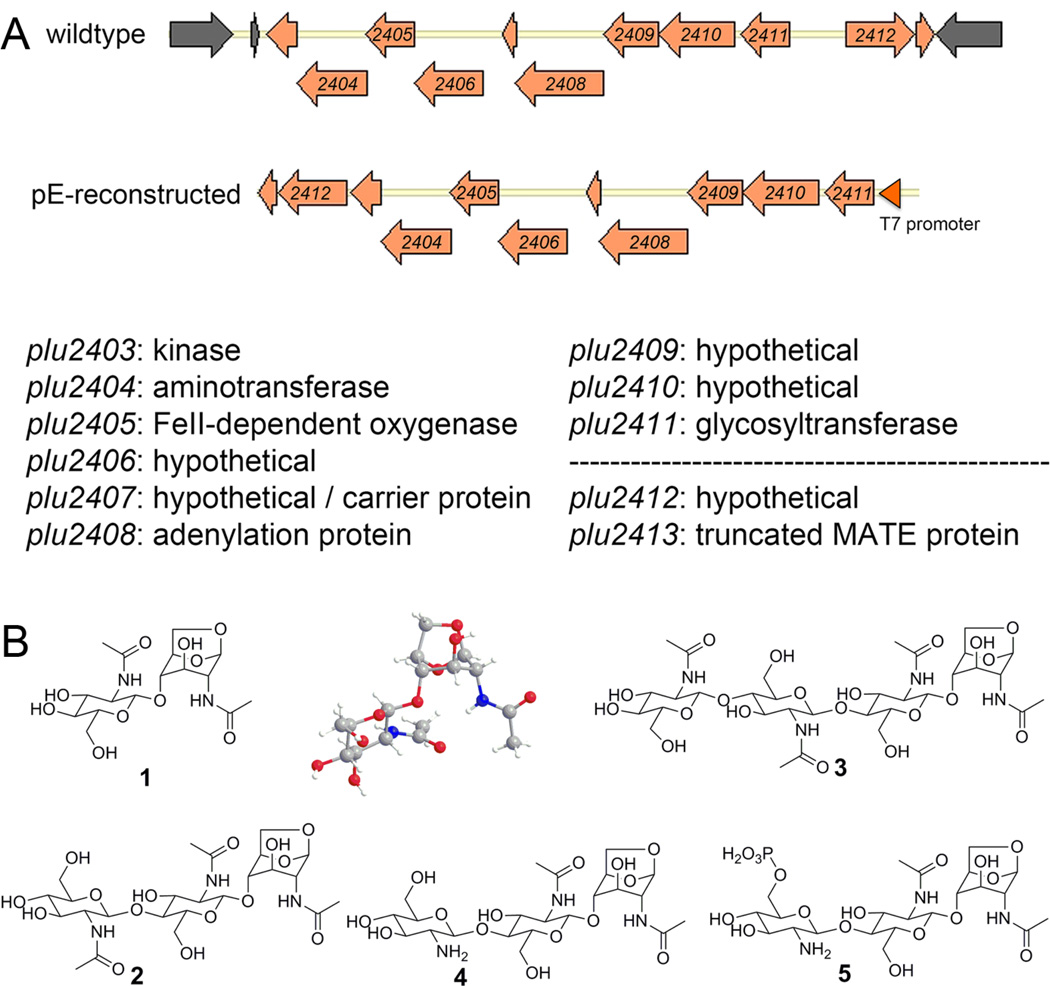
We searched the P. luminescens TT01 genome (Duchaud et al., 2003) using the MicroScope bioinformatics platform (Vallenet et al., 2009) for genomic island content that could potentially encode atypical secondary metabolites. We selected a unique genetic locus encoding an eclectic collection of 11 predicted proteins, including 5 hypothetical proteins (Figure 1A and Figure S1). Five of the proteins have divergent albeit recognizable sequence homologies to functionally described enzymes, including a nucleotide diphosphate kinase (Plu2403), an aminotransferase (Plu2404), an FeII-α-ketoglutarate-dependent oxygenase (Plu2405), a GT (Plu2411), and an adenylation protein (Plu2408). These types of proteins are frequently found in both primary and secondary metabolism, and barring adenylation sequences within NRPS biosynthetic systems, would not typically be used as search criteria for the discovery of orphan secondary metabolite pathways. Plu2413 at the edge of the gene cluster appears to be a truncated nonfunctional multidrug and toxic compound extrusion (MATE) protein. We reasoned that the remaining five hypotheticals could potentially encode new biocatalysts. Interestingly, hypothetical Plu2406 currently has no homologs in the NCBI database (E-value cutoff of 1×10−2). Hypothetical Plu2407 is distantly related to a few other hypothetical proteins (E-value 1×10−3) and Phyre2 analysis (Kelley and Sternberg, 2009) predicts related structural topology to carrier proteins like those found in fatty acid, polyketide, and nonribosomal peptide synthesis. This carrier protein prediction is consistent with the presence of adenylation protein Plu2408, which is predicted to activate Gly/Ala (Röttig et al., 2011). A portion of hypothetical Plu2410 shares distant homology to adenylation-domain containing proteins (E-value 1×10−3). Hypothetical Plu2412 exhibits some predicted structural homology to nucleotidyltransferases. Lastly, hypothetical Plu2409, which is a focus of this study, has only distant sequence homology to a variety of other hypothetical proteins and putative uncharacterized sugar binding proteins. The protein is unique to P. luminescens TT01 relative to closely related sequenced species in the Photorhabdus and Xenorhabdus genera, and its closest homologs exist in Actinobacteria Kutzneria sp. 744. (local alignment, id/sim/gap (%), 29.2/44.7/17.6) and Vibrio cholerae (31.0/46.5/10.1).
Figure 1. Biosynthetic gene cluster and characterized oligosaccharides 1–5.
(A) Atypical carbohydrate-NRPS genomic island. Wildtype genetic organization and reconstructed genetic organization used in heterologous expression studies are shown. Peripheral genes marked in grey are predicted to not be a part of the pathway. See also Figure S1. (B) Structures of oligosaccharides 1–5.
We reconstructed the unusual Gammaproteobacterial locus as a single polycistronic operon under the control of an upstream T7 promoter for heterologous expression in E. coli BAP1 (Figure 1A). Differential LC/MS analysis of cells carrying the reconstructed pathway compared to control cells carrying an empty vector led to the identification of five major small molecules dependent on a functional pathway (Figure 1B). These molecules were not detected in wildtype P. luminescens cultures, supporting a “silent” pathway in the native host under typical laboratory cultivation conditions. Genetic incorporation of additional rare tRNAs (pRARE2) in the heterologous expression strain further enhanced yields of these molecules by about 4- to 8-fold (Figure S2). HR Q-TOF MS revealed that the small molecules had ion signals of 407.1666, 608.2313, 813.3227, 568.2334, and 648.1976, consistent with the molecular formulas [C16H27N2O10]+, [C24H38N3O15]−, [C32H53N4O20]+, [C22H38N3O14]+, [C22H39N3O17P]+, respectively (Table S1). HR MS2 fragmentation analysis of this family supported structural modifications of a small molecule homologous series consistent with di-, tri-, and tetra-oligosaccharides. The five new metabolites were isolated and further characterized by detailed one- (1H) and two- (gCOSY, gHSQC, gHMBC, and ROESY) dimensional NMR. Structural elucidation efforts are detailed in the Supporting Information. Briefly, one- and two-dimensional NMR data indicated that small molecules 1, 2, and 3 consisted of β-Glc-NAc residues connected via 1,4-glycosidic linkages and terminating in a bicyclic 1,6-anhydro-β-Glc-NAc at the reducing end of the chains. Absolute configurations were determined by derivatization, chemical degradation, and GC/MS, which showed the presence of only D-Glc-NAc residues. Additionally, we crystallized 1 and confirmed its absolute configuration and conformation by X-Ray crystallography. Small molecules 1–3 are new natural products of metabolism, but they were previously reported as synthetic products of the thermal degradation of chitin (Koll et al., 1991). In contrast to neutral molecules 1–3, which are primarily secreted into the media, the two new small molecules 4 and 5 were mainly identified in the cell pellets. Metabolites 4 and 5 represented structurally related trisaccharides differing at the nonreducing end of the chain. 4 contained a glucosamine at this position, whereas 5 contained a glucosamine-6-phosphate.
Biosynthetic Gene Deletion Analysis
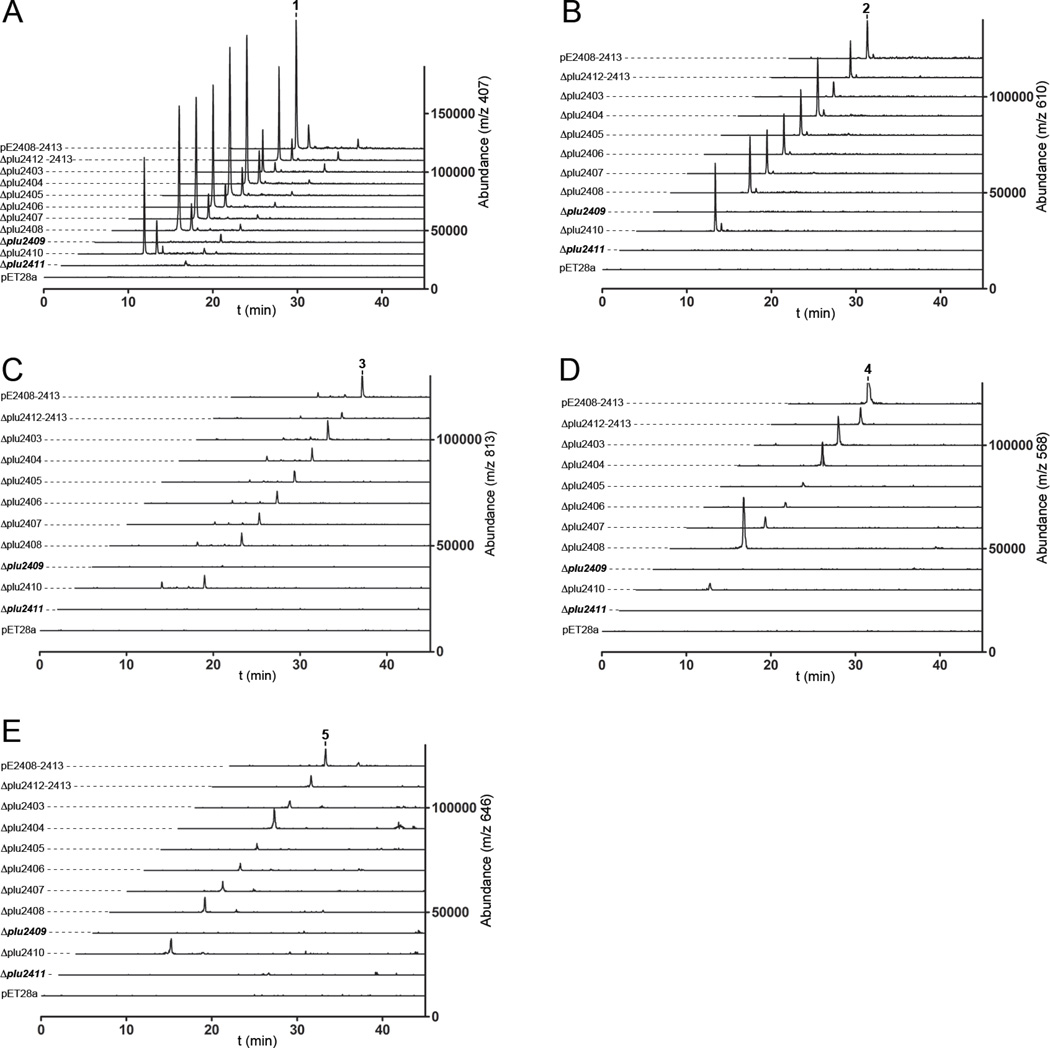
We conducted a full gene deletion analysis to identify biosynthetic genes required for 1,6-anhydro-oligosaccharide production. Independent deletion of every gene in the reconstructed pathway determined that GT plu2411 and hypothetical plu2409 were the only genes in the pathway required for synthesis of oligosaccharides 1–5 (Figure 2). The hypothetical gene requirement suggested the potential for a new biocatalyst involved in bicyclic sugar biosynthesis. These in vivo studies also support that either the iterative GT has substrate promiscuity during chain elongation to account for 4 versus 2, or 2 is a substrate of an endogenous E. coli hydrolase to generate 4. Additionally, the deletion studies demonstrate that the phosphorylation of 4 to 5 is not encoded within the pathway. 4 is likely either a substrate of an endogenous E. coli kinase or the kanamycin phosphotransferase used for pathway selection. Shunt product formation is common for overexpressed biosynthetic gene clusters due to intermediate accumulation.
Figure 2. LC/ESI-MS Extracted Ion Count (EIC) analysis of oligosaccharide 1–5 production in the deletion mutants.
(A) Oligosaccharide 1; (B) 2; (C) 3; (D) 4; and (E) 5. The reconstructed pathway (pE-reconstructed = pE2408–2413) was compared to gene deletions (plu2403–plu2413) and an empty control vector (pET28a) by LC/MS. Positive ion signals ([M+H]+) are shown for 1–4. Negative ion signal ([M−H]−) is shown for phosphorylated 5. See also Figure S2.
Biochemical Reconstitution of Oligosaccharide Synthesis
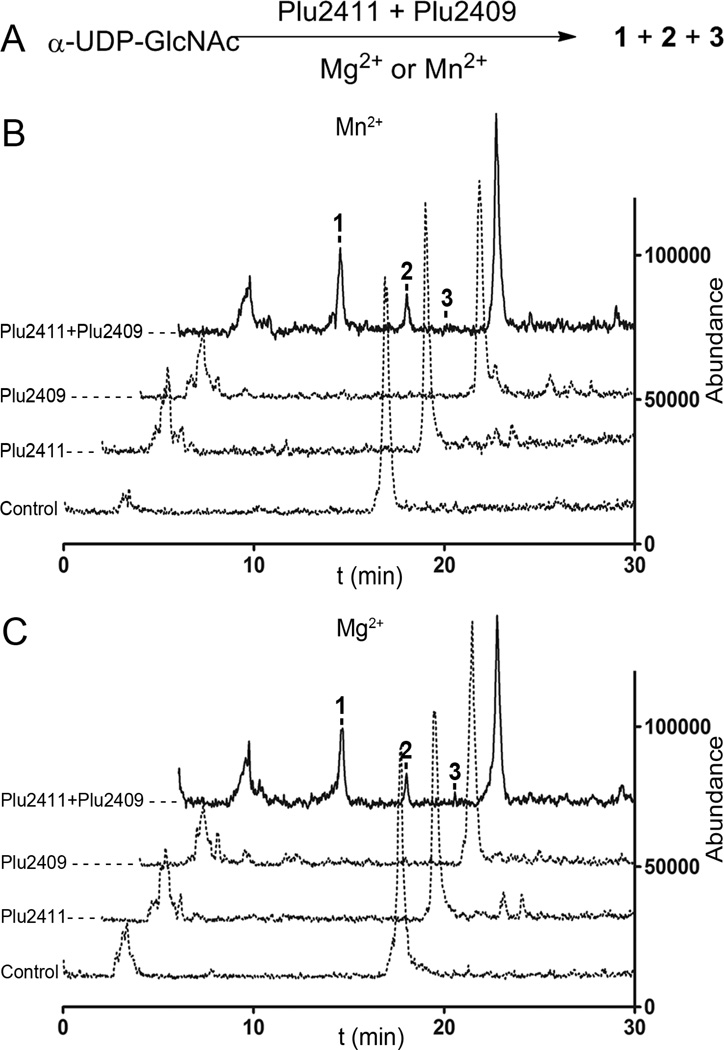
To determine the biosynthetic route for oligosaccharide production, we cloned plu2411 and plu2409, expressed their gene products in E. coli Rosetta2(DE3) as recombinant His6-tagged proteins, and purified them to homogeneity for in vitro biochemical analyses (Figure S3). The in vivo deletion studies above significantly focused our in vitro biosynthetic efforts. We hypothesized that the hypothetical protein either generated a 1,6-anhydro-β-D-Glc-NAc monosaccharide intermediate followed by iterative GT homologation or the GT first constructed oligosaccharides followed by hypothetical protein-mediated cleavage and concomitant cyclization. Within the known constraints of E. coli metabolism, we tested various possible monosaccharide substrates (α-UDP-Glc-NAc, Glc-NAc-6-phosphate, and Glc-NAc), metal cofactors (Mg+2, Mn+2, Zn+2, or Ca+2), and in vitro reaction buffer conditions (Tris and K/PO4). Ultimately, combination of both GT Plu2411 and hypothetical protein Plu2409 with α-UDP-Glc-NAc and Mg+2 (or Mn+2) in a Tris-buffer system led to the in vitro reconstruction of oligosaccharides 1–3 (Figure 3).
Figure 3. Reactions catalyzed by Plu2411 and Plu2409.
(A) Conversion of α-UDP-GlcNAc to 1–3 by Plu2411 and Plu2409 in the presence of Mn2+ or Mg2+; LC/ESI-MS(+) Total Ion Count (TIC) chromatogram of the reaction in the presence of Mn2+ (B) and Mg2+ (C).
Hypothetical Protein Utilizes Chitin as a Substrate
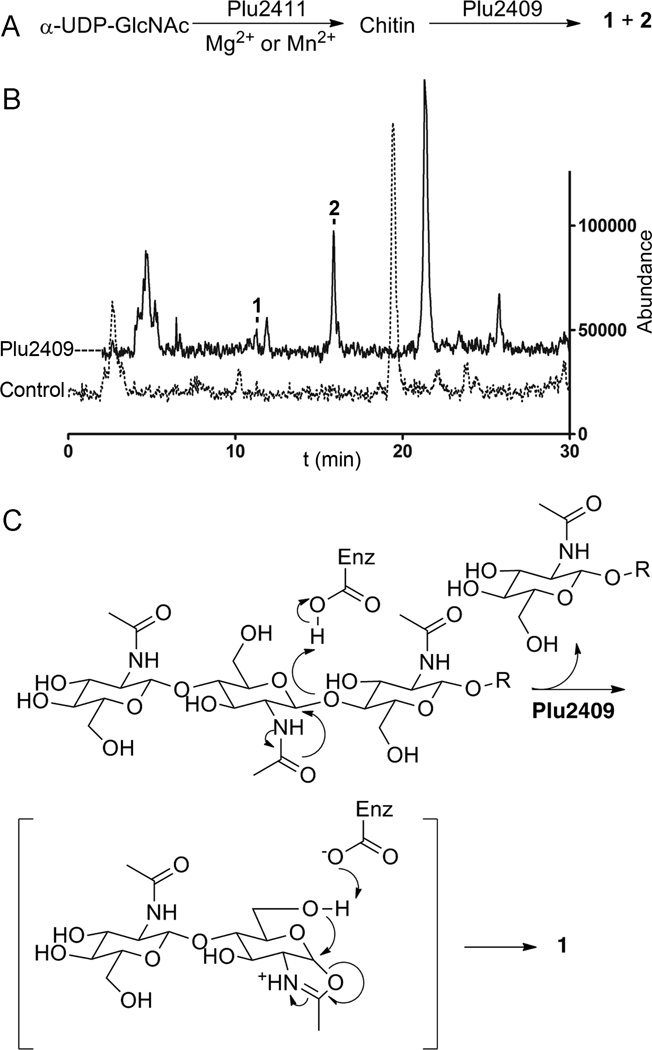
With the establishment of an in vitro dual-enzyme system, we then probed individual enzyme requirements for bicyclic sugar formation. Using α-UDP-Glc-NAc as a substrate, 1,6-anhydro-products could not be detected in single enzyme reactions. A molecule consistent with the 1,6-anhydro-Glc-NAc monosaccharide was identified by LC/MS as only a very minor constituent and only in the dual-enzyme reaction. In the GT-only reaction, we could detect UDP and free Glc-NAc products, but we were not able to detect short chain oligosaccharides by LC/MS. We postulated that GT Plu2411 could synthesize longer chitin chains, and consequently, we tested chitin as a substrate for the hypothetical protein. Chitin was determined to be an authentic substrate, leading to products 1–2 in vitro, albeit with an altered product output distribution (Figure 4AB). Metal supplements were not required for the Plu2409-mediated cleavage and cyclization reactions.
Figure 4. Reaction catalyzed by hypothetical Plu2409 with chitin substrate.
(A) Proposed transformations catalyzed by GT Plu2411 and hypothetical Plu2409; (B) LC/ESI-MS(+) TIC chromatogram for Plu2409 catalyzed reaction with chitin as a substrate; see also Figure S4. (C) Proposed biosynthesis of representative 1 catalyzed by Plu2409.
In contrast to chitinases that are catabolic hydrolases that break down chitin, a polymer found in the insect exoskeleton, for example, hypothetical Plu2409 is not a hydrolase, but rather a 1,6-anhydropyranose synthase. We propose that the anabolic iterative inverting GT Plu2411 is capable of producing chitin molecules. These substrates are processed by the 1,6-anhydropyranose synthase Plu2409, liberating oligosaccharides 1–2 as the predominant products, which are then secreted into the extracellular environment. Based on the genes located in the genomic island, these molecules are likely isolable intermediates of a currently uncharacterized glycopeptide. The pathway identified here shares parallel features to cell wall recycling pathways in Gram-negative bacteria. A family of lytic transglycosylases act on the peptidoglycan ultimately releasing β-D-GlcNAc-(1→4)-1,6-anhydro-β-D-N-acetyl-muramic-acid-L-Ala-D-γ-Glu-meso-DAP-D-Ala, which is uptaken by the cell through a permease for recycling and subsequent de novo cell wall synthesis (Lee et al., 2013). Rather than acting on cell wall substrates for internalization, hypothetical protein Plu2409 in conjunction with GT Plu2411 catalyze the synthesis of the secreted 1,6-anhydro-di-, tri-, and tetra-oligosaccharides. Based on our results and biochemical studies of lytic transglycosylases (Fibriansah et al., 2012), a plausible catalytic route for the hypothetical protein is substrate-assisted oligosaccharide cleavage and concomitant cyclization with retention of β-configuration (Figure 4C).
Inhibitory Effects of Oligosaccharides 1–5 on Lysozyme
Oligosaccharide natural products contribute to a variety of functional biological roles (McCranie and Bachmann, 2014). Secondary metabolites 1–5 did not exhibit detectable antimicrobial activities against E. coli BL21(DE3), Bacillus subtilis NCIB 3610, or Saccharomyces cerevisiae in disk diffusion assays. In considering functional roles, we evaluated ecological contexts and Photorhabdus-host interactions. During infection, the invertebrate hosts launch coordinated innate immune responses but ultimately fail succumbing to septicemia (Eleftherianos et al., 2010). In the model invertebrate M. sexta, gene expression of lysozyme, an important antibacterial effector protein, is stimulated upon P. luminescens challenge. Given the partial structural similarities of 1–5 to the known lysozyme inhibitors chitobiose and chitotriose (Laible and Germaine, 1985), we examined their in vitro lysozyme inhibitory activities using the model chicken egg white lysozyme. Inhibition constants for oligosaccharides 1–5 and reference compound chitobiose against lysozyme were determined using Dixon plots (Table 1 and Figure S5). Trisaccharides 2 and 4 and tetrasaccharide 3 were notably more potent than chitobiose. All tested oligosaccharides showed characteristic competitive inhibitor behaviors while phosphorylated trisaccharide 5 behaved as a mixed type inhibitor (Figure S5). It is possible that the biosynthesized oligosaccharides, which intriguingly mimic breakdown products of the insect exoskeleton, and their currently uncharacterized downstream products may participate in immunomodulation during the bacterium’s multipartite lifecycle.
Table 1. Inhibition constants (Ki) of oligosaccharides for lysozyme.
See also Figure S5. Error bars represent standard deviation at 95% confidence.
| Compound | Ki (mM) | Type |
|---|---|---|
| chitobiose | 0.45±0.26 | competitive |
| 1 | 3.59±1.29 | competitive |
| 2 | 0.17±0.13 | competitive |
| 3 | 0.30±0.27 | competitive |
| 4 | 0.40±0.23 | competitive |
| 5 | 0.75±0.41 | mixed |
Significance
The functional elucidation of new enzymes has lagged far behind the discovery of new genes and proteins, leading to a stockpile of gene products with no known functions. Many of these gene products could represent new “parts” for natural host metabolic diversification or metabolic engineering (Hanson et al., 2010). Systematic approaches to illuminate the structure and function of unknown proteins have been implemented, such as in the Protein Structure Initiative (Gifford et al., 2012) and the Enzyme Function Initiative (Gerlt et al., 2011). Using protein structure as a guide combined with genome context, computational predictions, and experimental data provide powerful platforms for the discovery of new enzymes (Bastard et al., 2014; Zhao et al., 2013). Rather than using protein structures, we focused on a rare structural feature of a small family of carbohydrate natural products encoded by an atypical gene cluster as a guide to elucidate the function of a novel enzyme. Natural product gene clusters have traditionally provided a framework for elucidating enzyme functions, as enzymatic involvement in natural product synthesis can be queried, and ultimately, validated by in vitro biochemical analysis. Putative atypical biosynthetic pathways that are not readily recognized by homology-based searches can be spotted using genome synteny analysis and exist in a variety of microorganisms capable of producing bioactive small molecules. These atypical pathways often located in genomic islands, such as the example introduced here, provide significant discovery opportunities. Decoding these pathways will illuminate new biocatalysts involved in natural product synthesis and the privileged bioactive small molecules that they produce.
Experimental Procedures
Cloning, Gene Deletion, and Protein Expression and Purification
LC-ESI-MS Metabolite Analysis
50 mL M9-glucose medium (10.5 g/L M9 minimal salts, 2 mM MgSO4, 0.1 mM CaCl2, 0.4% glucose, pH 7.5) supplemented with 0.5% casamino acids and 50 µg/mL kanamycin was inoculated with 1 mL of overnight seed culture (E. coli BAP1 transformed with pET-28a(+), pE-reconstructed, Δplu2403, Δplu2404, Δplu2405, Δplu2406, Δplu2407, Δplu2408, Δplu2409, Δplu2410, Δplu2411, Δplu2413 or Δplu2412–2413) and grown at 37 °C at 250 rpm until OD600 reached 0.6. Then the culture was cooled on ice and supplemented with 0.5 mM IPTG and incubated for another 48 hours at 18 °C. The cells were then centrifuged and the cell pellets were lyophilized, pulverized and extracted with 5 mL MeOH. The MeOH extracts were centrifuged, filtered, dried in vacuo and resuspended in 5 mL dH2O. The aqueous layer was extracted with 3×5 mL butanol and dried. The residues from aqueous phase were dissolved in dH2O while those from the butanol phase were dissolved in MeOH. The spent media from individual cultures were treated in the same way as the pellets. The extracts were analyzed by LC-ESI-MS with a 100×4.6 mm Hypercarb column.
Oligosaccharide Production Optimization, Isolation, and Purification
See Supplemental Experimental Procedures. Briefly, a 2 L scale up of M9-glucose cultivation conditions except with BAP1/pE-reconstructed+pRARE2 was prepared. Oligosaccharides were purified on a semi-prep 10×250 mm hypercarb column to afford 110.2 mg of 1, 9.7 mg of 2, 0.9 mg of 3, 1.1 mg of 4, and 4.7 mg of 5.
Plu2411 and Plu2409 Combined Activity Assay
100 µL assay buffer conditions (200 mM Tris-buffer, pH 8.0), Plu2411 (12 µM) and Plu2409 (12 µM) were incubated with α-UDP-GlcNAc (0.5 mM) in the presence of TCEP (1 mM) and 0.1 mM divalent metal ions (MnCl2, MgCl2, ZnSO4 or CaCl2) for 1 hour. Reaction mixtures were quenched with liquid nitrogen, lyophilized, extracted with 100 µL 1:1 MeOH:H2O, and analyzed by LC-ESI-MS on a 4.6 × 100 mm Hypercarb column.
Plu2409 Activity Assay Using Chitin as Substrate
100 µL assay buffer conditions (200 mM Tris-buffer, pH 8.0) with Plu2409 (12 µM) were incubated with chitin (~1 mg, washed 3 times with MeOH and then dH2O before use) in the presence of TCEP (1 mM) and 0.1 mM divalent metal ions (MnCl2 or MgCl2) for 3 hours. Reaction mixtures were quenched with liquid nitrogen, lyophilized, extracted with 100 µL 1:1 MeOH:dH2O, and analyzed by LC-ESI-MS on a 4.6 × 100 mm Hypercarb column.
Lysozyme Inhibition Activity Assay
Oligosaccharide Ki values were determined using EnzChek Lysozyme Assay kit using DQ™ lysozyme substrate (Micrococcus lysodeikticus labeled with fluorescein) according to the manufacturer’s instructions.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Institutes of Health (R00-GM097096) to JMC. Generous lab support was also provided by the Searle Scholars Program (13-SSP-210). We thank Dr. Brandon Mercado (Yale University) for X-ray crystallographic analysis. We also thank the Complex Carbohydrate Research Center (University of Georgia) for carbohydrate chemical degradation and GC/MS services.
Footnotes
Supplemental Information
Supplemental Information includes five figures, three tables, spectroscopic characterization of oligosaccharides 1–5, materials and Supplemental Experimental Procedures and can be found with this article online at xxx.
REFERENCES
- Bachmann BO, Van Lanen SG, Baltz RH. Microbial genome mining for accelerated natural products discovery: is a renaissance in the making? J. Ind. Microbiol. Biot. 2014;41:175–184. doi: 10.1007/s10295-013-1389-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastard K, Smith AAT, Vergne-Vaxelaire C, Perret A, Zaparucha A, De Melo-Minardi R, Mariage A, Boutard M, Debard A, Lechaplais C, et al. Revealing the hidden functional diversity of an enzyme family. Nat. Chem. Biol. 2014;10:42–49. doi: 10.1038/nchembio.1387. [DOI] [PubMed] [Google Scholar]
- Blin K, Medema MH, Kazempour D, Fischbach MA, Breitling R, Takano E, Weber T. antiSMASH 2.0-a versatile platform for genome mining of secondary metabolite producers. Nucleic Acids Res. 2013;41:W204–W212. doi: 10.1093/nar/gkt449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann AO, Bode HB. Identification and bioanalysis of natural products from insect symbionts and pathogens. Adv. Biochem. Eng. Biotechnol. 2013;135:123–155. doi: 10.1007/10_2013_192. [DOI] [PubMed] [Google Scholar]
- Demain AL. Importance of microbial natural products and the need to revitalize their discovery. J. Ind. Microbiol. Biot. 2014;41:185–201. doi: 10.1007/s10295-013-1325-z. [DOI] [PubMed] [Google Scholar]
- Duchaud E, Rusniok C, Frangeul L, Buchrieser C, Givaudan A, Taourit S, Bocs S, Boursaux-Eude C, Chandler M, Charles JF, et al. The genome sequence of the entomopathogenic bacterium Photorhabdus luminescens. Nat. Biotechnol. 2003;21:1307–1313. doi: 10.1038/nbt886. [DOI] [PubMed] [Google Scholar]
- Eleftherianos I, ffrench-Constant RH, Clarke DJ, Dowling AJ, Reynolds SE. Dissecting the immune response to the entomopathogen Photorhabdus. Trends Microbiol. 2010;18:552–560. doi: 10.1016/j.tim.2010.09.006. [DOI] [PubMed] [Google Scholar]
- Fibriansah G, Gliubich FI, Thunnissen AMWH. On the Mechanism of Peptidoglycan Binding and Cleavage by the endo-Specific Lytic Transglycosylase MltE from Escherichia coli. Biochemistry. 2012;51:9164–9177. doi: 10.1021/bi300900t. [DOI] [PubMed] [Google Scholar]
- Gerlt JA, Allen KN, Almo SC, Armstrong RN, Babbitt PC, Cronan JE, Dunaway-Mariano D, Imker HJ, Jacobson MP, Minor W, et al. The Enzyme Function Initiative. Biochemistry. 2011;50:9950–9962. doi: 10.1021/bi201312u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gifford LK, Carter LG, Gabanyi MJ, Berman HM, Adams PD. The Protein Structure Initiative Structural Biology Knowledgebase Technology Portal: a structural biology web resource. J. Struct. Funct. Genomics. 2012;13:57–62. doi: 10.1007/s10969-012-9133-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson AD, Pribat A, Waller JC, de Crecy-Lagard V. 'Unknown' proteins and 'orphan' enzymes: the missing half of the engineering parts list - and how to find it. Biochem. J. 2010;425:1–11. doi: 10.1042/BJ20091328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley LA, Sternberg MJE. Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- Koll P, Borchers G, Metzger JO. Preparative isolation of oligomers with a terminal anhydrosuger unit by thermal degradation of chitin and cellulose. Journal of Analytical and Applied Pyrolysis. 1991;17:319–327. [Google Scholar]
- Laible NJ, Germaine GR. Bactericidal activity of human lysozyme, muramidase-inactive lysozyme, and cationic polypeptides against Streptococcus sanguis and Streptococcus faecalis: inhibition by chitin oligosaccharides. Infect. Immun. 1985;48:720–728. doi: 10.1128/iai.48.3.720-728.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M, Hesek D, Llarrull LI, Lastochkin E, Pi HL, Boggess B, Mobashery S. Reactions of All Escherichia coli Lytic Transglycosylases with Bacterial Cell Wall. J. Am. Chem. Soc. 2013;135:3311–3314. doi: 10.1021/ja309036q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CI, McCarty RM, Liu HW. The biosynthesis of nitrogen-, sulfur-, and high-carbon chain-containing sugars. Chem. Soc. Rev. 2013;42:4377–4407. doi: 10.1039/c2cs35438a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCranie EK, Bachmann BO. Bioactive oligosaccharide natural products. Nat. Prod. Rep. 2014;31:1026–1042. doi: 10.1039/c3np70128j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman DJ, Cragg GM. Natural Products As Sources of New Drugs over the 30 Years from 1981 to 2010. J. Nat. Prod. 2012;75:311–335. doi: 10.1021/np200906s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer BA, Admiraal SJ, Gramajo H, Cane DE, Khosla C. Biosynthesis of complex polyketides in a metabolically engineered strain of E. coli. Science. 2001;291:1790–1792. doi: 10.1126/science.1058092. [DOI] [PubMed] [Google Scholar]
- Röttig M, Medema MH, Blin K, Weber T, Rausch C, Kohlbacher O. NRPSpredictor2--a web server for predicting NRPS adenylation domain specificity. Nucleic Acids Res. 2011;39:W362–W367. doi: 10.1093/nar/gkr323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Huang WC, Noguchi M, Kobayashi A, Shoda S. Direct synthesis of 1,6-anhydro sugars from unprotected glycopyranoses by using 2-chloro-1,3-dimethylimidazolinium chloride. Tetrahedron Lett. 2009;50:2154–2157. [Google Scholar]
- Vallenet D, Engelen S, Mornico D, Cruveiller S, Fleury L, Lajus A, Rouy Z, Roche D, Salvignol G, Scarpelli C, et al. MicroScope: a platform for microbial genome annotation and comparative genomics. Database-Oxford. 2009 doi: 10.1093/database/bap021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizcaino MI, Guo X, Crawford JM. Merging chemical ecology with bacterial genome mining for secondary metabolite discovery. J. Ind. Microbiol. Biot. 2014;41:285–299. doi: 10.1007/s10295-013-1356-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter JM, Behnken S, Hertweck C. Genomics-inspired discovery of natural products. Curr. Opin. Chem. Biol. 2011;15:22–31. doi: 10.1016/j.cbpa.2010.10.020. [DOI] [PubMed] [Google Scholar]
- Zhao SW, Kumar R, Sakai A, Vetting MW, Wood BM, Brown S, Bonanno JB, Hillerich BS, Seidel RD, Babbitt PC, et al. Discovery of new enzymes and metabolic pathways by using structure and genome context. Nature. 2013;502:698–702. doi: 10.1038/nature12576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziemert N, Lechner A, Wietz M, Millan-Aguinaga N, Chavarria KL, Jensen PR. Diversity and evolution of secondary metabolism in the marine actinomycete genus Salinispora. Proc. Natl. Acad. Sci. USA. 2014;111:E1130–E1139. doi: 10.1073/pnas.1324161111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.