Rab11a is a key modulator of vesicular trafficking processes, but there is limited information about the GEFs and GAPs that regulate its GTP-GDP cycle. TBC1D9B is identified as a Rab11a GAP in MDCK cells, where it regulates the Rab11a-dependent basolateral-to-apical transcytotic pathway.
Abstract
Rab11a is a key modulator of vesicular trafficking processes, but there is limited information about the guanine nucleotide-exchange factors and GTPase-activating proteins (GAPs) that regulate its GTP-GDP cycle. We observed that in the presence of Mg2+ (2.5 mM), TBC1D9B interacted via its Tre2-Bub2-Cdc16 (TBC) domain with Rab11a, Rab11b, and Rab4a in a nucleotide-dependent manner. However, only Rab11a was a substrate for TBC1D9B-stimulated GTP hydrolysis. At limiting Mg2+ concentrations (<0.5 mM), Rab8a was an additional substrate for this GAP. In polarized Madin–Darby canine kidney cells, endogenous TBC1D9B colocalized with Rab11a-positive recycling endosomes but less so with EEA1-positive early endosomes, transferrin-positive recycling endosomes, or late endosomes. Overexpression of TBC1D9B, but not an inactive mutant, decreased the rate of basolateral-to-apical IgA transcytosis—a Rab11a-dependent pathway—and shRNA-mediated depletion of TBC1D9B increased the rate of this process. In contrast, TBC1D9B had no effect on two Rab11a-independent pathways—basolateral recycling of the transferrin receptor or degradation of the epidermal growth factor receptor. Finally, expression of TBC1D9B decreased the amount of active Rab11a in the cell and concomitantly disrupted the interaction between Rab11a and its effector, Sec15A. We conclude that TBC1D9B is a Rab11a GAP that regulates basolateral-to-apical transcytosis in polarized MDCK cells.
INTRODUCTION
Collectively Rab GTPases form a large family of evolutionarily conserved proteins (42 subfamilies in humans) that coordinate vesicle fission, transport, tethering, and fusion (Stenmark, 2009). Their function is regulated by specific guanine nucleotide-exchange factors (GEFs), GTPase-activating proteins (GAPs), and GDP dissociation inhibitors. Whereas GEFs activate their cognate Rabs by promoting the exchange of GDP for GTP, GAPs facilitate the hydrolysis of GTP to GDP, returning Rabs to their inactive, GDP-bound state. Studies in the past few years have identified a family of Tre2-Bub2-Cdc16 (TBC) domain–containing proteins, several members of which have Rab GAP activity. This function depends on two conserved motifs in their TBC domain: an arginine or R finger within an IxxDxxR motif, and a glutamine or Q finger within an YxQ motif. Whereas the conserved Y residue in the Q finger interacts with the conserved switch II glutamate residue of the Rab GTPase, the R and Q residues of TBC proteins coordinate the Rab-bound GTP, promoting its hydrolysis (Pan et al., 2006; Gavriljuk et al., 2012). The Rab specificity and GAP activity have been defined for some TBC-domain proteins (Fukuda, 2011; Frasa et al., 2012), but there is limited information for the other family members, including TBC1D9B.
Structurally, TBC1D9B is composed of two N-terminal GRAM domains, a central TBC domain (which includes R and Q fingers), and a C-terminal EF hand motif (Doerks et al., 2000; Frasa et al., 2012). The closest nonvertebrate orthologue to TBC1D9B is the yeast protein Gyp2p, which has a similar domain architecture (but has only one GRAM domain) and apparently acts as a GAP for Ypt31p, a homologue of Rab11a (Sciorra et al., 2005). No studies have defined whether vertebrate TBC1D9B has GAP activity or have determined its Rab specificity.
Rab11a is a well-studied Rab GTPase that regulates exocytosis and is critical for a variety of cellular processes, including membrane protein recycling (Ullrich et al., 1996; Wang et al., 2000b), transcytosis (Wang et al., 2000b; Su et al., 2010; Xu et al., 2011), regulated exocytosis (Khandelwal et al., 2008, 2013), cytokinesis (Fielding et al., 2005; Wilson et al., 2005; Yu et al., 2007), ciliogenesis (Knodler et al., 2010; Westlake et al., 2011), lumenogenesis (Desclozeaux et al., 2008; Bryant et al., 2010), and autophagosome formation (Fader et al., 2008; Longatti et al., 2012). Whereas many studies have focused on identifying Rab11a effectors (e.g., FIP1-5 and Sec15A; Hales et al., 2001; Lindsay et al., 2002; Wallace et al., 2002; Zhang et al., 2004; Wu et al., 2005; Oztan et al., 2007; Horgan and McCaffrey, 2009; Baetz and Goldenring, 2013), much less is known about the specific GEFs and GAPs that regulate vertebrate Rab11a activity, and the majority of insights have come from studies in model organisms. In yeast, the multisubunit TRAPII complex is reported to act as a GEF for Ypt31p and Ypt32p (Jones et al., 2000; Morozova et al., 2006), which is an additional orthologue of Rab11a. However, the purified TRAPII complexes may lack GEF activity in mammalian cells (Yamasaki et al., 2009). In addition, the Drosophila protein Crag acts as a GEF for Rab11 in photoreceptor cells (Xiong et al., 2012) and for Rab10 in in vitro studies (Yoshimura et al., 2010). In the case of GAPs, the TBC-domain-containing protein Evi5 regulates border cell migration during Drosophila oocyte development by acting as a Rab11a GAP (Laflamme et al., 2012). However, whether Evi5 acts as a GAP for vertebrate Rab11a is controversial. Some reports indicate that it lacks Rab11a GAP activity, whereas others point to the opposite conclusion (Dabbeekeh et al., 2007; Fuchs et al., 2007; Westlake et al., 2007; Hehnly et al., 2012). Another TBC-containing protein, TBC1D14, appears to bind to Rab11a and Rab11b and regulate autophagosome formation but apparently does not function as a GAP but does function as a Rab11a/b effector (Longatti et al., 2012). Thus, the regulatory proteins that regulate the vertebrate Rab11a GTPase cycle remain to be defined and characterized.
Using a polarized Madin–Darby canine kidney (MDCK) cell culture model, coupled with in vitro assays, we observed the following. Although able to interact with Rab11a, Rab11b, and Rab4a in a nucleotide dependent manner, TBC1D9B only stimulated Rab11a GTP hydrolysis in vitro. Furthermore, endogenous TBC1D9B colocalized with Rab11a-dependent endosomes, as well as with cargoes that transit these endosomes, including immunoglobulin A (IgA) bound to the polymeric immunoglobulin receptor (pIgR), the archetypal basolateral-to-apical transcytotic protein. We further observed that overexpression of TBC1D9B, but not an inactive mutant, slowed the rate of basolateral-to-apical IgA transcytosis, where short hairpin RNA (shRNA)–mediated depletion of TBC1D9B increased the rate of this process. Furthermore, expression of TBC1D9B decreased the amount of active Rab11a in the cell and concomitantly disrupted the interaction between Rab11a and its effector, Sec15A. We discuss the likelihood that TBC1D9B acts as a Rab11a GAP in polarized MDCK cells, where it regulates the Rab11a-dependent IgA-pIgR transcytotic pathway. We also discuss the possibility that TBC1D9B may act as an effector of Rab11b and/or Rab4a.
RESULTS
TBC1D9B interacts with specific Rab GTPases
TBC1D9B is an ∼140-kDa TBC domain–containing protein with a comparable domain architecture to Gyp2p (Figure 1A). Furthermore, the overall similarity of these proteins is reasonably high at 49% and increases to 64% when the TBC domains are assessed individually. The TBC domain of TBC1D9B contains conserved R, Y, and Q residues, which form the “R” and “Q” fingers that are critical for GTP hydrolysis in other TBC proteins (Figure 1B; Pan et al., 2006; Gavriljuk et al., 2012).
FIGURE 1:
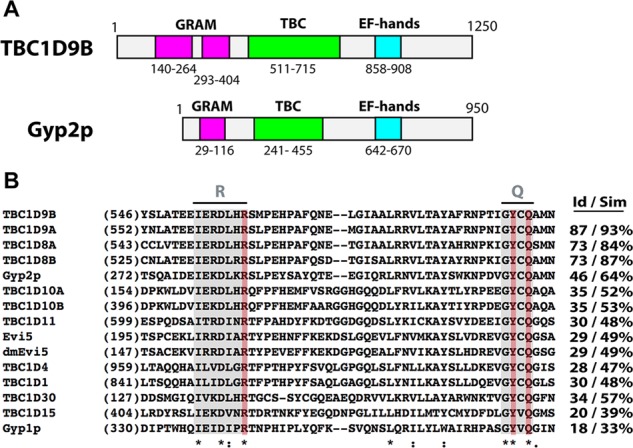
Domain structure of TBC1D9B and sequence comparisons of its TBC domain with other TBC domain–containing proteins. (A) Schematic of human TBC1D9B and Saccharomyces cerevisiae Gyp2p domain structure. Predicted GRAM domains are colored pink, TBC domains are colored green, and EF hands are colored light blue. (B) Alignment of the indicated region of the TBC domains using MAFFT software (Larkin et al., 2007; Katoh and Standley, 2013). Sequences including the arginine (R) and glutamine (Q) fingers are shaded gray; key amino acids in the R and Q fingers are shaded red. Identical residues are indicated with an asterisk, conserved amino acid substitutions by a colon, and semiconserved substitutions by a period. The percentage of identity (Id) or similarity (Sim) between the TBC domain of TBC1D9B and those of the indicated proteins is given in the right-hand columns.
To identify possible substrates for TBC1D9B, we first tested whether TBC1D9B could bind to Rab GTPases—Rab11a in particular—using a yeast two-hybrid assay with TBC1D9B as bait and different Rabs as prey (Supplemental Figure S1). A positive interaction was found between TBC1D9B and Rab11aQ70L (abbreviated Rab11a-QL), a mutant of Rab11a that was proposed to be defective in GTP hydrolysis (Adari et al., 1988; Stenmark et al., 1994; Ullrich et al., 1996). Notwithstanding a previous report that this mutant is able to hydrolyze GTP and therefore may not be constitutively active (Casanova et al., 1999), a more recent study demonstrates that Rabs with Q-to-L mutations retain the ability to hydrolyze GTP, albeit at a reduced level to the wild-type protein (Gavriljuk et al., 2012). No interaction was observed when a GDP-locked mutant of Rab11a was used (Rab11aS25N; abbreviated Rab11a-SN) or when other Rab GTPases (Rab5, Rab6, Rab7, Rab33, and Rab4), nominally in their GTP-locked or GDP-locked configuration, were tested. Haas et al. (2005) previously described that an inactivating mutation in the R finger of the TBC domain can enhance the interaction between a TBC domain–containing protein and its substrate. When we mutated Arg-559 in TBC1D9B to an Ala residue (R559A; abbreviated TBC1D9B-RA), we observed that the interaction with Rab11a-QL was stronger (Supplemental Figure S1). However, under these conditions, we also observed interactions with Rab11a-SN and to a lesser extent with Rab4 and the empty pB42AD prey vector.
As an additional method to screen for TBC1D9B interactions, we determined whether flag-TBC1D9B could coimmunoprecipitate a broader array of GFP-tagged, GTP-locked mutants of Rab proteins previously implicated in endocytic and exocytic membrane traffic. Of the Rabs tested, we observed that Rab11a-QL, Rab11b-QL, and Rab4a-QL formed significant interactions with flag-TBC1D9B (Figure 2). In contrast, little interaction was observed with nominally GTP-locked mutants of Rab3a, Rab5a, Rab6a, Rab7, Rab8a, Rab10, Rab13, Rab25, Rab27a, or Rab33a (Figure 2). Taken together, these results indicate that TBC1D9B forms interactions with a selective number of Rab GTPases, including Rab11a, Rab11b, and Rab4a.
FIGURE 2:
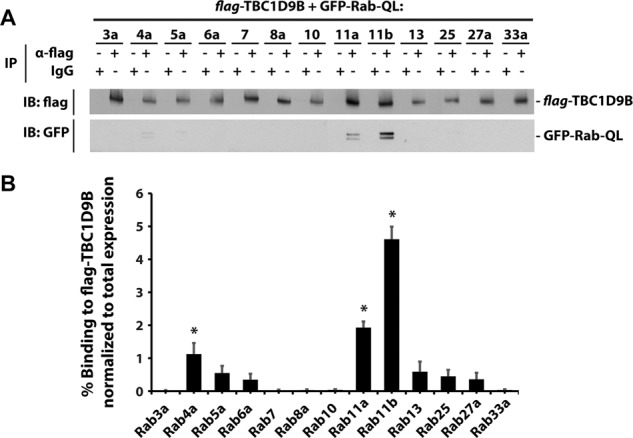
TBC1D9B interacts with Rab GTPases. (A) Flag-tagged TBC1D9B, along with the indicated GFP-Rabs containing an activating Q-to-L mutation, were coexpressed in HEK cells. Flag-TBC1D9B was recovered using an anti-flag tag antibody and coimmunopreciptated GFP-tagged Rab-QL detected using an anti-GFP antibody. IgG was used as a nonspecific control. (B) Quantification of the percentage GFP-Rab-QL coimmunoprecipitated with TBC1D9B normalized to the total amount of GFP-Rab-QL in the lysate. Data were obtained from three independent experiments, and the mean ± SEM is shown. Values significantly different from the group means, assessed by ANOVA, are indicated (*p < 0.05).
TBC1D9B stimulates Rab11a GTP hydrolysis
We next sought to determine whether TBC1D9B had GAP activity against any of its binding partners. We first performed comparative protein structure modeling using the known three-dimensional (3D) structure of the TBC1D4 TBC domain as a template (Protein Data Bank [PDB] file 3QYB; Park et al., 2011). Figure 3A shows the predicted structure of the TBC domain of TBC1D9B superimposed on the known structure of the TBC1D4 TBC domain. The structural overlap parameter obtained was 64%, confirming that the two structures were structurally related (Geourjon et al., 2001). In addition, the putative catalytic residues (R559, Y592, and Q594) of TBC1D9B were exposed similarly to those of TBC1D4 and other TBC proteins such as Gyp1p (unpublished data). Thus, the sequence homologies and predicted structure are consistent with the possibility that TBC1D9B was a functional GAP.
FIGURE 3:
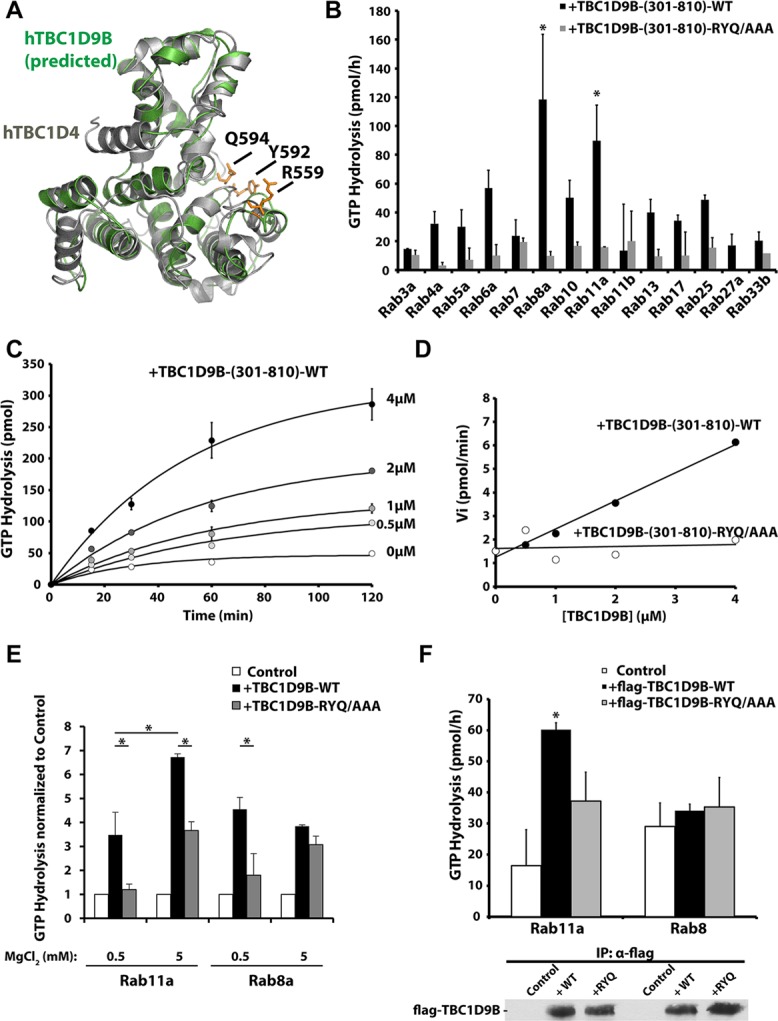
TBC1D9B in vitro GAP activity. (A) TBC1D4 (PDB: 3QYB) was used as a template (gray) to model the 3D structure of the TBC1D9B TBC domain (green). (B) GTP hydrolysis by the indicated wild-type, GST-tagged Rab loaded with [γ-32P]GTP and incubated with 2 μM of either wild-type or mutant TBC1D9B-(301-810) for 60 min at 30°C. Data are corrected for reactions lacking the TBC1D9B fragment. Those values significantly different from the group means, assessed by ANOVA, are indicated (*p < 0.05). (C) Kinetics of Rab11a GTP hydrolysis loaded with [γ-32P]GTP and incubated with 0, 0.5, 1, 2, or 4 μM recombinant TBC1D9B-(301-810). (D) Initial rates of Rab11a GTP hydrolysis plotted against the concentration of wild-type or mutant TBC1D9B-(301-810). (E) In vitro GAP assays performed in the presence of Rab11a or Rab8a. In these reactions, Mg2+ mixed at a 1:1 M ratio with GTP, was added at a final concentration of 0.5 mM. Alternatively, the reaction was supplemented with 5 mM MgCl2. Reactions were incubated for 30 min at 30°C. Data were normalized to control reactions in which no TBC1D9B-(301-810) was added. Values for TBC1D9B-(301-810) that were significantly different (p < 0.05) from matched incubations performed in the presence of TBC1D9B-RYQ/AAA (301-810) or between the indicated reactions are identified with an asterisk. (F) Top, full-length flag-TBC1D9B-WT or flag-TBC1D9B-RYQ/AAA was immunoprecipitated from HeLa cell lysates and a GAP assay performed using Rab11a or Rab8a loaded with [γ-32P]GTP and incubated for 60 min at 30°C. Lysates from nontransfected cells were used as controls. Bottom, Western blot of a 5-μl aliquot of the immunoprecipitates used in the in vitro assay was detected using an anti-flag antibody. (B–F) Data were obtained from three or more independent experiments performed in duplicate, and the mean ± SEM is shown. In E and F, values significantly different between TBC1D9B-WT and TBC1D9B-RYQ/AAA are indicated with an asterisk (p < 0.05).
Next, we evaluated whether TBC1D9B had measurable GAP activity. Because we were unable to express sufficient quantities of the full-length protein in Escherichia coli (TBC1D9B has a molecular weight of ∼140 kDa), we synthesized fragments of TBC1D9B and tested these for GAP activity (Supplemental Figure S2A). The fragment 301–810, which we refer to as TBC1D9B-(301-810), retained maximal in vitro GAP activity against Rab11a and was used in our subsequent studies (Supplemental Figure S2B). Of the 14 Rabs tested, a significant increase in mean GAP activity was observed only in the case of Rab8a and Rab11a (Figure 3B). However, no GAP activity was detected for the other Rabs, including Rab4a, or the other Rab11 family members Rab11b and Rab25. Further analysis revealed a linear dependence between the TBC1D9B-(301-810) concentration and the initial rate of Rab11a-GTP hydrolysis (Figure 3, C and D). At 4 μM, the extent of GTP hydrolysis was 9.7 times greater than GTP hydrolysis in the absence of TBC1D9B-(301-810) (Figure 3C), and the calculated rate of GTP hydrolysis was 4.8 times that of the intrinsic rate of Rab11a GTPase activity in the absence of TBC1D9B-(301-810) (Figure 3D). The catalytic efficiency of wild-type TBC domain was calculated as 158.3 M−1 s−1.
As a control for these studies, we generated variants of TBC1D9B that contained mutations in key amino acids exposed in the active site of the protein (Figures 1B and 3A) and were therefore likely to lack activity. To screen for inactive TBC1D9B mutants, we used nonpolarized HEK cells. In these cells, we observed a significant redistribution of Rab11a from recycling endosomes (which appear as a juxtanuclear cluster in these images) to the cytosol when TBC1D9B was overexpressed (Supplemental Figure S3A). However, this redistribution was not observed in cells expressing TBC1D9B-Y592A/Q594A (abbreviated YQ/AA) or TBC1D9B-R559A/Y592A/Q594A (abbreviated RYQ/AAA) (Supplemental Figure S3, A and B). In contrast, expression of TBC1D9B-RA did not differ in effect from the wild-type TBC1D9B. This may indicate that this mutant retains some or all of its GAP activity, as opposed to the previously described loss of activity caused by equivalent mutations in the Gyp1p and Gyp7p yeast GAPs (Albert et al., 1999). As a control for these experiments, we observed that wild-type or mutant variants of TBC1D9B had no effect on the distribution of Rab33b (Supplemental Figure S3, A and C), which we showed is not an apparent substrate for TBC1D9B (Figure 3B). As further confirmation that TBC1D9B-RYQ/AAA is an inactive mutant, we showed that it lacked significant GAP activity in the in vitro assays described earlier (Figure 3, B–D).
We were initially surprised that Rab8a-QL served as a substrate for TBC1D9B in the GAP assays (Figure 3B) but did not coimmunoprecipitate with flag-TBC1D9B (Figure 2). Additional experimentation revealed that these differences were likely related to the amount of Mg2+ used in these assays. Whereas the in vitro GAP assays were initially performed in minimal concentrations of Mg2+ (GTP bound with Mg2+ was used in the assays at a final concentration of 0.5 mM), similar to previous reports (Krugmann et al., 2004; Haas et al., 2005; Fuchs et al., 2007; Yoshimura et al., 2008), the coimmunoprecipitation studies in Figure 2 were performed in the presence of an additional 2.5 mM Mg2+ (Oztan et al., 2007; Tsun et al., 2013). Of interest, when we removed Mg2+ from the coimmunoprecipitation studies, we observed interactions between GFP-Rab8a-QL and flag-TBC1D9B (see later discussion of Figure 5C). In contrast, supplementing the concentration of Mg2+ in the GAP assays by 5 mM MgCl2 (Du et al., 1998; Albert et al., 1999; Casanova et al., 1999) evoked a significant increase in Rab11a GTPase activity (Figure 3E) but not for Rab8a (Figure 3E) or any of the other Rab GTPases tested (Supplemental Figure S2C). Finally, we expressed full-length flag-TBC1D9B in HeLa cells and then recovered the protein by immunoprecipitation. Like TBC1D9B-(301-810), the full-length protein showed GTPase activity only against Rab11a and not Rab8a (Figure 3F).
FIGURE 5:
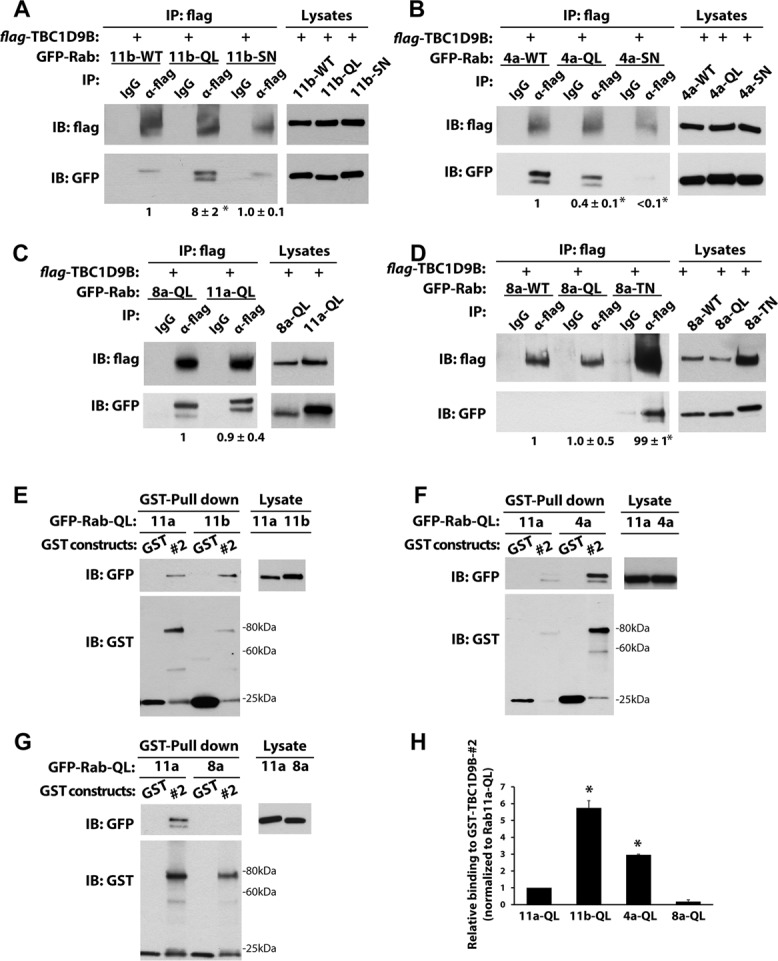
TBC1D9B interacts with Rab4a and Rab11b. (A) Left, coimmunoprecipitation of flag-tagged TBC1D9B with GFP-Rab11b-WT (11b-WT), GFP-Rab11b-Q70L (11b-QL), or GDP-locked GFP-Rab11bS25N (11b-SN) coexpressed in HEK cells. Anti-flag antibody was used to immunoprecipitate flag-TBC1D9B, and anti-GFP antibody was used to detect the coimmunoprecipitated GFP-tagged Rab protein. IgG was used as a nonspecific control. Right, 2% of each lysate was resolved by SDS–PAGE and overexpressed proteins detected by Western blot. Values were normalized to the total expression of each protein and then normalized to the amount of Rab11b-WT lysate. (B) Same as in A, but HEK cells coexpressed flag-TBC1D9B and GFP-Rab4a-WT (4a-WT), GFP-Rab4aQ72L (4a-QL), or GFP-Rab4aS27N (4a-SN). (C) Same as A, but the cells expressed Rab8aQ67L (8a-QL) or Rab11aQ70L (11a-QL), and after lysis, the incubations were performed in the absence of MgCl2. (D) Same as in A, but HEK cells coexpressed flag-TBC1D9B with GFP-Rab8a-WT (8a-WT), GFP-Rab8a-Q67L (8a-QL), or GFP-Rab8a-T22N (8a-TN). (E–G) Top left, GST alone or GST-TBC1D9B-(301-810) (#2 fragment) was used to affinity capture GFP-Rab11a-QL and GFP-Rab11b-QL (E), GFP-Rab4a-QL (F). or GFP-Rab8a-QL (G). Right, 2% of each lysate was resolved by SDS–PAGE and GFP-Rab-QL detected by Western blot using anti-GFP antibody. Bottom left, GST constructs were resolved by SDS–PAGE and proteins detected on Western blots using an anti-GST antibody. (H) Quantification of data from E–G. Values are normalized to the total expression of the protein first and then to the values obtained for the GFP-Rab11a-WT pull down. Data were obtained from at least three independent experiments, and the mean ± SEM is shown. Values significantly different from the group means, as assessed by ANOVA, are indicated (*p < 0.05).
In summary, these results demonstrate that when the assay reactions are supplemented with 5.0 mM Mg2+, TBC1D9B shows greatest activity toward Rab11a.
Rab11a binds to the TBC domain of TBC1D9B
Next, we explored the nature of the interaction between TBC1D9B and Rab11a. In general, GAPs are believed to interact most robustly to Rabs in their GTP-bound state and less so with Rabs in their GDP-bound state (Will and Gallwitz, 2001; Haas et al., 2005; Fuchs et al., 2007; Hannemann et al., 2012). Consistent with this finding, we observed that immunoisolated flag-tagged TBC1D9B coimmunoprecipitated approximately five times more avidly with Rab11a-QL than with Rab11a-WT or Rab11a-SN (Figure 4A). Mutations of the R and/or Q fingers in the TBC domain are reported to alter TBC–protein interaction with Rab-GTP, either enhancing it (Will and Gallwitz, 2001; Haas et al., 2005; Itoh et al., 2006) or impairing it (Itoh et al., 2006). In the case of TBC1D9B, we observed that full-length flag-TBC1D9B-RYQ/AAA coimmunoprecipitated Rab11a-WT in a manner that was greater than for the wild-type flag-TCB1D9B protein (Figure 4B). Thus, our results were similar to those reported for RUTBC-3/RabGAP-5, USP6NL/RN-Tre, and Gyp6p, all of which show increased binding to variants of their cognate TBC GAPs that contain mutations in their active-site residues (Will and Gallwitz, 2001; Haas et al., 2005).
FIGURE 4:
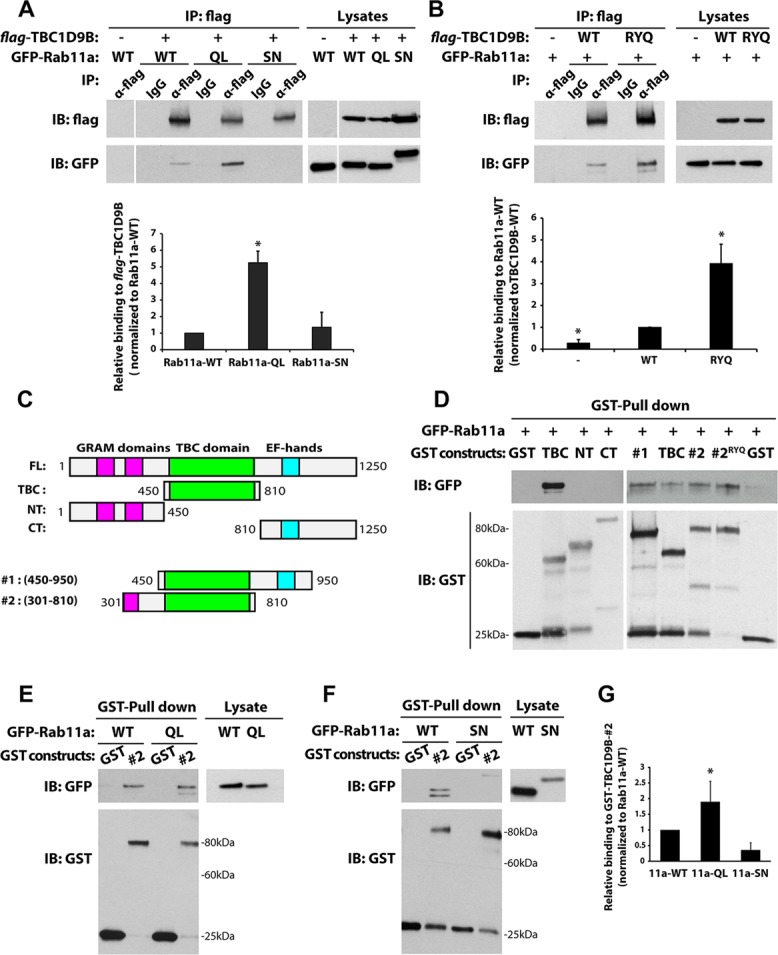
Rab11a binds to the TBC domain of TBC1D9B. (A) Left, coimmunoprecipitation of flag-tagged TBC1D9B with GFP-Rab11a-wild-type (WT), GTP-locked GFP-Rab11aQ70L (QL), or GDP-locked GFP-Rab11aS25N (SN) coexpressed in HEK cells. Anti-flag antibody was used to recover flag-TBC1D9B, and coimmunopreciptated GFP-Rab11a was detected using an anti-GFP antibody. Controls included use of IgG instead of the flag-tag antibody and expression of GFP-Rab11a alone (first lane). Right, 2% of each lysate was resolved by SDS–PAGE and the indicated proteins detected by Western blot. Bottom, quantification of coimmunoprecipitations. Values were normalized to the total expression of each protein first and then to the values obtained in experiments using the Rab11a-WT lysate. (B) HEK cells were cotransfected with GFP-Rab11a and either flag-TBC1D9B-wild-type (WT) or the flag-TBC1D9B-RYQ/AAA mutant (RYQ). Flag-tagged TBC1D9B was recovered by immunoprecipitation, and the amount of GFP-Rab11a was quantified. Control reactions were performed for cells that only expressed GFP-Rab11a, or in some cases IgG was substituted for the anti-flag antibody. Right, 2% of each lysate was resolved by SDS–PAGE and the indicated proteins detected by Western blot. Bottom, amount of coimmunoprecipitated GFP-Rab11a normalized to the amount recovered from the TBC1D9B-WT lysate. (C) Fragments of TBC1D9B used in pull-down studies. (D) Top, GST-TBC1D9B fragments, labeled according to C, were used to affinity capture GFP-Rab11a in HEK cell lysates. Here #2RYQ denotes the use of fragment #2 with the RYQ/AAA mutations. GST alone was used as a control. Bottom, GST constructs were resolved by SDS–PAGE and proteins blotted with anti-GST antibody. (E, F) Top left, GST-TBC1D9B-#2 fragment was used to affinity capture GFP-Rab11aQ70L (QL) or GFP-Rab11aS25N (SN) from HEK cell lysates. Right, 2% of each lysate was resolved by SDS–PAGE and GFP-Rab11a detected by Western blot using anti-GFP antibody. Bottom left, GST constructs were resolved by SDS–PAGE and proteins detected using an anti-GST antibody. (G) Quantification of data from E and F. Values were normalized to total expression of the protein of interest and then to the values obtained for the GFP-Rab11a-WT pull down. For A, B, and G, data are from at least three independent experiments, and the mean ± SEM is shown. Values significantly different from the group means, as assessed by ANOVA, are indicated (*p < 0.05).
The foregoing results indicate that R559, Y592, and Q594 are unlikely to mediate the binding between TBC1D9B and Rab11a. To assess which domain(s) of TBC1D9B were responsible for binding Rab11a, we generated glutathione S-transferase (GST)–tagged fragments of TBC1D9B containing or lacking the TBC domain (Figure 4C). These were used to pull down GFP-Rab11a-WT produced in HEK cells (Figure 4D). Fragments lacking the TBC domain did not interact with GFP-Rab11a (Figure 4D, left), whereas all fragments containing the TBC domain, including those with flanking N-and C-terminal amino acids, interacted with Rab11a (Figure 4D, right). As expected, the TBC1D9B-(301-810)-RYQ/AAA mutant retained the ability to interact with Rab11a (Figure 4D, lane #2RYQ). Finally, we observed that, like the full-length protein, flag-TBC1D9B-(301-810) showed the greatest interaction with Rab11a-QL and less with Rab11a-WT or Rab11a-SN (Figure 4, E–G).
The foregoing results demonstrate that Rab11a interacts with TBC1D9B via its TBC domain and that this interaction is dependent on the nucleotide state of Rab11a. We further observe that mutants that alter the TBC active site show increased binding to Rab11a.
TBC1D9B interacts with Rab11b and Rab4a in a nucleotide-dependent manner
Next, we further explored the interaction of TBC1D9B with Rab11b and Rab4a. Although not substrates for TBC1D9B (Figure 3B), both Rabs were found to coimmunoprecipitate with flag-TBC1D9B expressed in HEK cells (Figure 2). Of interest, when wild-type (WT), GTP-locked (QL), and GDP-locked (SN) mutants of Rab11b were compared in an immunoprecipitation assay (Figure 5A), we observed that flag-TBC1D9B interacted most efficiently with Rab11b-QL. In a somewhat similar manner, both the WT and QL forms, but not the SN mutant, of Rab4a also interacted with TBC1D9B (Figure 5B). However, we did note that TBC1D9B interacted more strongly with Rab4a-WT than Rab4a-QL. Whereas full-length flag-TBC1D9B showed the rank efficiency for interactions with its cognate Rabs as Rab11b > Rab11a > Rab4a (Figure 2), the efficiency of interaction for the GST-TBC1D9B-(301-810) fusion protein was Rab11b > Rab4a > Rab11a (Figure 5, E, F, and H). Thus, there were some differences in the interactions between the full-length protein and the 301–810 protein fragment. We also studied the interaction between TBC1D9B and Rab8a, which was observed when the Mg2+ in the buffer was removed. Under these conditions, there was little difference in the binding efficiency between flag-TBC1D9B and Rab8a-QL and Rab11a-QL (Figure 5C). However, when the assay was supplemented with 2.5 mM Mg2+, neither Rab8a-WT nor Rab8a-QL interacted with flag-TBC1D9B (Figure 5D). Oddly, in the presence of Mg2+, Rab8a-SN showed a strong interaction with flag-TBC1D9B.
These results demonstrate that in the presence of 2.5 mM Mg2+, TBC1D9B selectively interacts with GTP-locked mutants of Rab4a and Rab11b but not with GDP-locked variants or with Rab8a-WT/-QL.
TBC1D9B localizes to Rab11a-positive endosomes
To better understand the site of TBC1D9B function in the cell, we studied its subcellular localization in polarized epithelial MDCK cells. In these cells, endogenous TBC1D9B was vesicular in appearance and accumulated in the apical cytoplasm of the cell up to the level of the apical membrane (defined by GP-135 or actin staining in Figure 6A). TBC1D9B did not localize to EEA1-positive early endosomes or to LAMP2-labeled late endosomes/lysosomes (Figure 6, B–D; Manders coefficient of colocalization of 0.031 ± 0.005 and 0.035 ± 0.005, respectively). TBC1D9B also partially localized with the giantin-labeled Golgi (coefficient of colocalization of 0.31 ± 0.04; Figure 6, B and E), which is consistent with previous observations that Rab11a is localized in part to this organelle (Ullrich et al., 1996; Chen et al., 1998). Small amounts of TBC1D9B colocalized with the transferrin receptor (TfR; coefficient of colocalization of 0.20 ± 0.05; Figure 6, B and F), which shows only modest colocalization with Rab11a in MDCK cells (Brown et al., 2000; Leung et al., 2000). However, as expected for a protein that interacts with Rab11a, a relatively large fraction of TBC1D9B was localized to Rab11a-positive vesicles (coefficient of colocalization of 0.48 ± 0.1; Figure 6, B and G.
FIGURE 6:
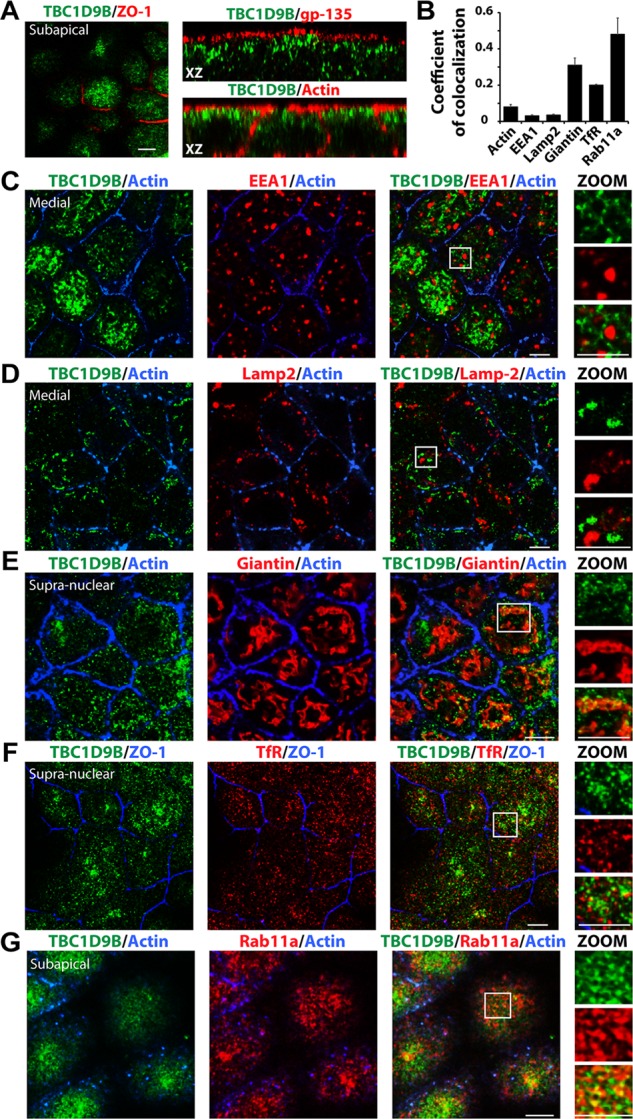
Subcellular localization of TBC1D9B in MDCK cells. Filter-grown MDCK cells were immunolabeled to show the distribution of the following endogenous proteins. (A) TBC1D9B and the tight junction–associated protein ZO-1 (xy-section, left), the apical membrane protein gp-135 (xz-section, top right), or actin (xz-section, bottom right). (B) Coefficient of colocalizations for endogenous TBC1D9B and the following markers: (C) TBC1D9B, actin, and EEA1 (an early endosome marker); (D) TBC1D9B, actin, and Lamp2 (a late endosome/lysosome marker); (E) TBC1D9B, actin, and giantin (a Golgi marker); (F) TBC1D9B, ZO-1, and TfR (a marker of the common recycling endosome and basolateral early endosomes); and (G) TBC1D9B, actin, and Rab11a (which is associated with the apical recycling endosomes in these cells). In A and C–G, single optical sections from the region just under the apical membrane (subapical), just above the nucleus (supranuclear), or at the level of the nucleus along the lateral membranes (medial) are shown. Boxed regions are magnified in the images below the word ZOOM. Scale bar, 5 μm. In B, data are mean ± SEM from three experiments (n > 100 cells).
We also measured the amount of colocalization between endogenous TBC1D9B and GFP-Rab11a, GFP-Rab11b, GFP-Rab25, or endogenous Rab4a and Rab8a (Supplemental Figure S4). Colocalization was observed between TBC1D9B and most of the Rabs tested (Supplemental Figure S4D), but the amount of colocalization of TBC1D1B with Rab25 was low (coefficient of colocalization of ∼0.1). It is prudent to note that Rab11b and Rab25 exhibit a moderate to large degree of colocalization with Rab11a (Casanova et al., 1999; Wang et al., 2000b; Lapierre et al., 2003; Butterworth et al., 2012), and we observed that there was significant overlap between endogenous Rab11a with either Rab4a or Rab8a (coefficient of colocalization of 0.49 ± 0.05 and 0.60 ± 0.05, respectively; unpublished data). Unfortunately, we were unable to perform triple-label studies, as antibodies from additional species did not work in our hands or were not available.
Next, we determined whether TBC1D9B was localized to vesicle populations where Rab11a was likely to be in its active, GTP-bound state. One such vesicle pool is the apical recycling endosome, which is accessed by the pIgR late in its transit from the basolateral-to-apical poles of the cell (Apodaca et al., 1994; Casanova et al., 1999; Wang et al., 2000a). As predicted, endogenous TBC1D9B colocalized with Rab11a and the basolaterally internalized pIgR ligand, IgA, in subapical vesicles (coefficient of colocalization of 0.59 ± 0.02; Figure 7, A and D). In addition, TBC1D9B colocalized with vesicles positive for the pIgR (coefficient of colocalization of 0.54 ± 0.07; Figure 7, B and D) or the Rab11a (and Rab8a) effector, Sec15A, one subunit of the octameric exocyst complex (coefficient of colocalization of 0.33 ± 0.04; Figure 7, C and D; Guo et al., 1999; Zhang et al., 2004; Wu et al., 2005; Novick et al., 2006; Oztan et al., 2007).
FIGURE 7:
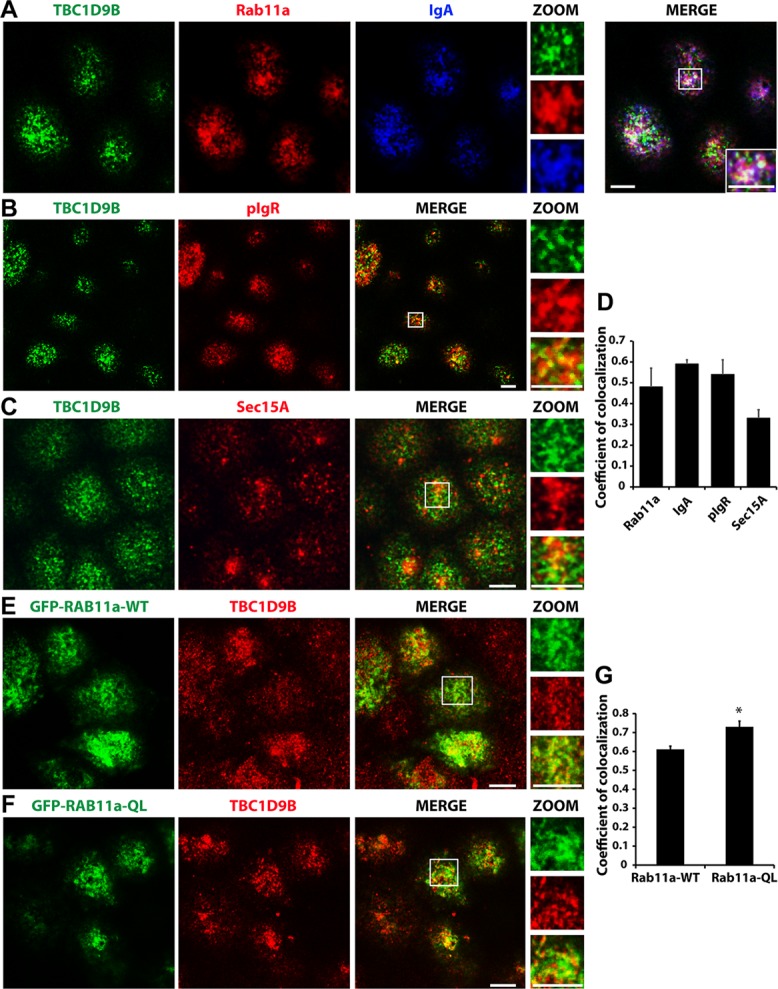
Localization of TBC1D9B at sites of active Rab11a function. (A) Filter-grown MDCK cells were pulse labeled with 200 μg/ml IgA at the basolateral surface for 20 min at 18°C and chased for 20 min at 37°C in the presence of apical Cy5-labeled anti-IgA antibodies. The cells were fixed and then immunolabeled to detect endogenous TBC1D9B (green), endogenous Rab11a (red), and transcytosed IgA (blue). In the merged panel, the boxed region is magnified in the bottom right inset. Magnified views of the individual channels are found in the insets under ZOOM. (B, C) MDCK cells were stained for endogenous TBC1D9B and either the (B) pIgR or (C) endogenous Sec15A. (D) Coefficient of colocalization for endogenous TBC1D9B and the indicated proteins. (E, F) MDCK cells were transfected with cDNA encoding GFP-Rab11a-WT (E) or GFP-Rab11a-QL (F) and after 72 h fixed and then stained for endogenous TBC1D9B. (G) Coefficient of colocalization for TBC1D9B and the indicated Rab11a construct. In (B, C, E, and F), the boxed region is magnified in the panels under the label ZOOM. Scale bar, 5 μm. In D and G, data were obtained from at least three independent experiments, and the means ± SEM are shown (n ≥ 150 cells). In G, a Student's t test was used to show that difference between these two sample groups was significantly different (*p < 0.05).
We also determined whether TBC1D9B colocalized to a greater extent with the GFP-tagged Rab11aQ70L mutant. Indeed, we observed higher degree of colocalization between endogenous TBC1D9B and GFP-tagged Rab11a-QL than with GFP-tagged Rab11a-WT (coefficient of colocalization of 0.73 ± 0.03 and 0.61 ± 0.02, respectively; Figure 7, E–G). We also determined whether the localization of Rab11a in MDCK cells was altered by expressing CFP-TBC1D9B-WT or CFP-TBC1D9B-RYQ/AAA. However, we found no difference in the Rab11a distribution (Supplemental Figure S5, A and B; see Figure 9A later in this article), which is consistent with previous reports that the Rab11a-SN mutant remains membrane bound in MDCK cells (Wang et al., 2000b). There was also no change in Rab11a colocalization with these TBC1D9B variants (coefficient of colocalization of 0.72 ± 0.06 and 0.70 ± 0.06 for WT and RYQ/AAA, respectively; Supplemental Figure S5C). In contrast, in HEK cells, there was a significant redistribution of Rab11a from recycling endosomes to the cytosol when TBC1D9B-WT, but not when the inactive mutant TBC1D9B-RYQ/AAA, was expressed (Supplemental Figure S2, A and B). This is the expected result for HEK cells if the GDP-bound form of Rab11a increased as a result of expressing TBC1D9B-WT. Finally, when Rab11a expression was depleted using a specific shRNA, endogenous TBC1D9B's normal accumulation in the apical half of the cell was altered, and its distribution became more dispersed throughout the cell (Supplemental Figure S5D).
FIGURE 9:
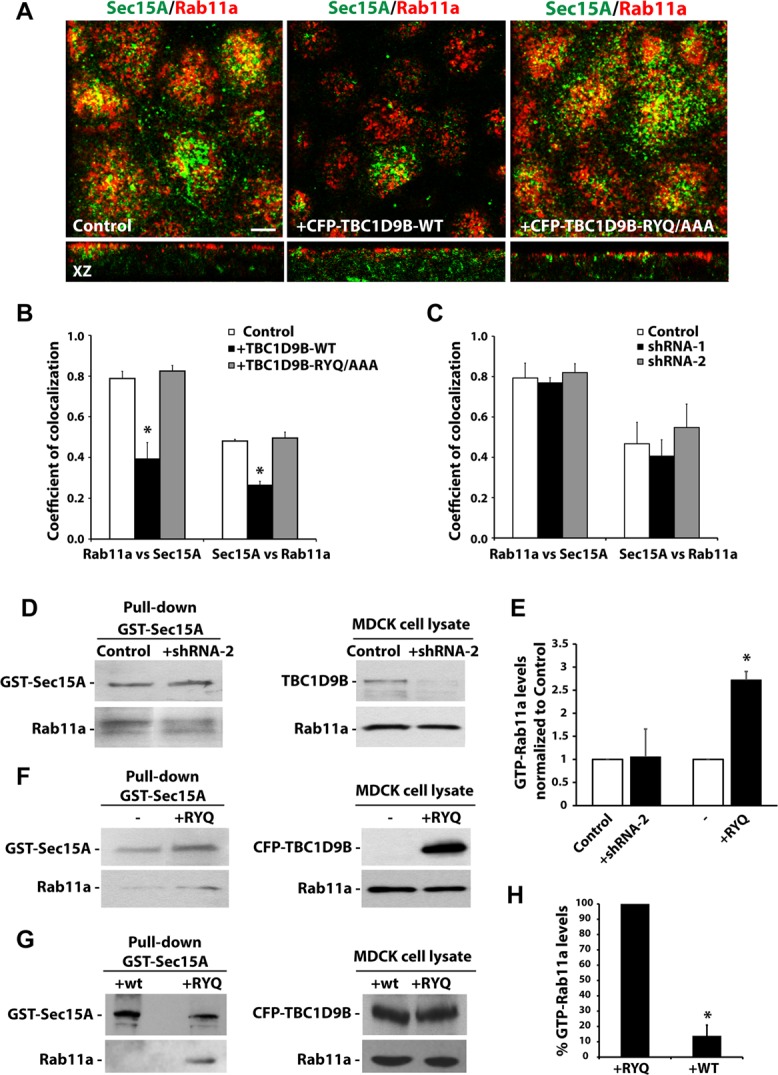
TBC1D9B has GAP activity in cellula. (A) Localization of endogenous Rab11a and endogenous Sec15A in cells transduced with adenovirus encoding CFP-TBC1D9B-WT or the inactive mutant (-RYQ/AAA). Top, from the subapical region of the cell; bottom, xz-sections. (B, C) Coefficient of colocalization for the fraction of Rab11a that colocalizes with Sec15A (or vice versa) in MDCK cells transduced with adenovirus encoding CFP-TBC1D9B-WT or -RYQ/AAA (B) or in cells expressing shRNA-1 or shRNA-2 against canine TBC1D9B (C). In B, GFP expression was used as control, and in C, scrambled shRNA was used as control. (D) GST-Sec15A–mediated pull down of activated Rab11a in MDCK cells expressing scrambled shRNA (Control) or shRNA-2 specific for canine TBC1D9B. Left, immunoprecipitate from the pull down was analyzed by Western blot, using anti-GST and anti-Rab11a antibodies. Right, 2% of each lysate was analyzed by Western blotting, using anti-TBC1D9B and anti-Rab11a antibodies. (E) Data from D and F are quantified and normalized to control incubations. (F) GST-Sec15A pull down of activated Rab11a in MDCK cells expressing CFP-TBC1D9B-RYQ/AAA (+RYQ) compared with control cells expressing endogenous TBC1D9B (–). Left, the precipitate from the pull down was analyzed by Western blot, using anti-GST and anti-Rab11a antibodies. Right, 2% of each lysate was analyzed by Western blot, using anti-GFP and anti-Rab11a antibodies. (G) GST-Sec15A pull down of activated Rab11a in MDCK cells expressing CFP-TBC1D9B-WT (+WT) or -RYQ/AAA (+RYQ). CFP-TBC1D9B-WT and -RYQ/AAA were detected using anti-GFP antibody, endogenous Rab11a was detected using anti-Rab11a antibody, and GST-Sec15A was detected using GST antibody. (H) Data from G are quantified. In B, C, E, and H, data were obtained from at least three independent experiments, and mean ± SEM is shown. Values different from the control, assessed by ANOVA, are indicated (*p < 0.05).
Taken together, these results indicate that TBC1D9B is associated with endocytic vesicles, including those likely to contain active Rab11a. Moreover, the loss of Rab11a expression results in an altered distribution of TBC1D9B, further pointing to an interaction between these proteins.
TBC1D9B regulates basolateral-to-apical transcytosis but not transferrin recycling or epidermal growth factor degradation
If TBC1D9B acts as a Rab11a-GAP, we reasoned that increasing its expression should down-regulate Rab11a activity and in doing so alter Rab11a-dependent trafficking events. We first tested the effects of TBC1D9B expression on the efficiency of basolateral-to-apical IgA transcytosis, a known Rab11a-dependent trafficking event (Casanova et al., 1999; Wang et al., 2000b). As expected, expression of TBC1D9B-WT, but not TBC1D9B-RYQ/AAA, significantly slowed the rate of IgA transcytosis, particularly at early time points (Figure 8A). We also tested the effects of TBC1D9B-WT expression on the fraction of IgA that recycles apically, a trafficking event that shows partial dependence on Rab11a (Wang et al., 2000b). Although the effect was not pronounced, there was a small but significant inhibition in the amount of IgA that recycled apically (Figure 8B). We also tested two pathways known to be Rab11a independent, including basolateral recycling of transferrin (Tf) and degradation of basolaterally internalized epidermal growth factor (EGF; Barbieri et al., 2000; Wang et al., 2000b). In both cases, expression of TBC1D9B-WT (or TBC1D9B-RYQ/AAA) had no significant effect (Figure 8, C and D).
FIGURE 8:
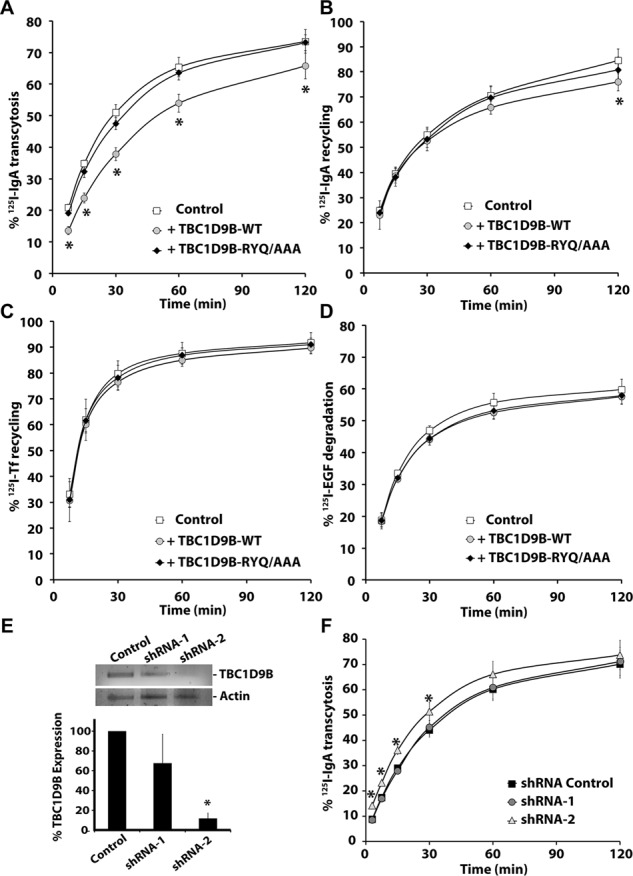
TBC1D9B regulates basolateral-to-apical transcytosis of IgA. The fraction of basolaterally internalized [125I]IgA that was transcytosed (A), the fraction of apically internalized [125I]IgA that was recycled (B), the fraction of basolaterally internalized [125I]Tf that was recycled (C), or the fraction of basolaterally internalized [125I]EGF that was degraded (D) was evaluated in filter-grown MDCK cells expressing CFP-TBC1D9B-WT or CFP-TBC1D9B-RYQ/AAA. (E) Top, RT-PCR analysis of MDCK cells expressing scrambled shRNA or specific shRNAs (shRNA1/2) that targeted canine TBC1D9B expression. Bottom, data are quantified. (F) Basolateral-to-apical transcytosis of [125I]IgA was measured in MDCK cells expressing either scrambled shRNA (shRNA Control) or shRNA-1/shRNA-2. In A–F, data were obtained from three independent experiments performed in triplicate, and the means ± SD are shown. Values different from the control, assessed by ANOVA, are indicated (*p < 0.05).
Finally, we rationalized that if TBC1D9B negatively regulates IgA transcytosis by its action on Rab11a, then depletion of TBC1D9B should increase the rate or extent of IgA transcytosis. TBC1D9B expression was knocked down in MDCK cells using specific shRNAs (shRNA-2), which decreased TBC1D9B expression by 88% (Figure 8E). We observed a modest but significant increase in the kinetics of IgA transcytosis (Figure 8F). However, no significant effect was observed if an inefficient shRNA (shRNA-1) was used, acting as a second negative control. As expected, shRNA-mediated depletion of TBC1D9B had no effect on the Rab11a-independent pathways, basolateral recycling of Tf, or degradation of basolaterally internalized EGF (Supplemental Figure S6, A and B). These results confirm a role of TBC1D9B in basolateral-to-apical transcytosis of IgA, a Rab11a-dependent pathway.
Expression of TBC1D9B-WT reduces the amount of active Rab11a
The foregoing results are consistent with the possibility that TBC1D9B acts as a Rab11a GAP but do not directly address whether TBC1D9B affects the amount of active Rab11a in the cell. To examine this possibility more directly, we determined whether expression of TBC1D9B-WT altered the amount of active Rab11a in the cell and thereby affected its association with its effector Sec15A. As predicted, expression of TBC1D9B-WT decreased the amount of colocalization between Rab11a and Sec15A, whereas expression of TBC1D9B-RYQ/AAA was without effect (Figure 9, A and B). We also observed that the normally apically distributed Sec15A became more random in distribution when cells expressed TBC1D9B-WT but not when TBC1D9B-RYQ/AAA was expressed (Figure 9A, middle). The total amount of Sec15A protein expressed in the cell was not affected by expression of TBC1D9B-WT (Supplemental Figure S7). However, a twofold increase in the expression of Sec15A was observed in cells expressing TBC1D9B-RYQ/AAA (Supplemental Figure S7B).
We also sought to determine whether shRNA-mediated depletion of TBC1D9B would increase the amount of colocalization between Rab11a and its effector, Sec15A, or whether it would affect the amount of Rab11a-GTP in the cell. However, depletion of TBC1D9B has no significant effect on the amount of colocalization between Rab11a and Sec15A (Figure 9C). To measure the amount of Rab11-GTP in the cells, we used an effector-based pull-down assay, using purified GST-tagged Sec15A to recover active Rab11a-GTP from cell extracts. We found no effect of decreasing TBC1D9B expression on the amount of Rab11a-GTP (Figure 9, D and E). However, when TBC1D9B-RYQ/AAA was expressed, we observed a 2.5-fold increase in the nominal amount of Rab11a-GTP, as indicated by increased interaction between Rab11a and Sec15A (Figure 9, E and F). In contrast, expression of TBC1D9B-WT reduced the amount of interaction between Rab11a and Sec15, likely by decreasing the amount of Rab11a-GTP in the cell (Figure 9, G and H).
In summary, overexpression of TBC1D9B decreases the amount of Rab11a-GTP in the cell and, in doing so, alters the interaction between Rab11a and Sec15A. In contrast, expression of TBC1D9B-RYQ/AAA has the opposite effects.
DISCUSSION
Rab11a is a critical regulator of exocytosis, but there is a limited understanding of the proteins that regulate its GTPase cycle. Our studies focused on TBC1D9B, a protein that has been included in several biological screens (Itoh et al., 2006; Fuchs et al., 2007; Yoshimura et al., 2007; Ishibashi et al., 2009; Popovic et al., 2012), but there are no reports on whether it has GAP activity, nor is there information about its Rab substrates. In this work, we identified TBC1D9B as Rab GAP for vertebrate Rab11a. This conclusion derives from the following observations: 1) TBC1D9B is the closest homologue of the yeast protein Gyp2p, which is reported to be a GAP for the Rab11a homologue Ypt31p (Sciorra et al., 2005); 2) the TBC domain of TBC1D9B interacts with Rab11a and accelerates Rab11a GTPase activity in vitro, whereas the TBC1D9B-RYQ/AAA mutant is without effect; 3) endogenous TBC1D9B is found associated with Rab11a-positive compartments; 4) in HEK cells, overexpression of TBC1D9B causes Rab11a to dissociate from recycling endosomes, apparently resulting in its redistribution to the cytosol; 5) in MDCK cells, overexpression of TBC1D9B impairs IgA transcytosis, a biological activity dependent on Rab11a, whereas down-regulation of TBC1D9B causes a modest but significant increase in IgA transcytosis; and 6) overexpression of TBC1D9B decreases the apparent levels of the GTP-bound form of Rab11a in MDCK cells.
We calculated the catalytic efficiency of TBC1D9B toward Rab11a to be 158 M−1 s−1, which is modest compared with the catalytic efficiencies reported for TBC1D1 (5.3 × 103 M−1 s−1), TBC1D4 (2.8 × 103 M−1 s−1), and TBC1D20 (2.7 × 103 M−1 s−1; Sklan et al., 2007; Park et al., 2011) but comparable to the activity of other TBC-domain containing proteins such as the Caenorhabditis elegans homologue of TBC1D2, TBC-2 (∼50 M−1 s−1; Chotard et al., 2010), and full-length TBC1D20 (360 M−1 s−1; Sklan et al., 2007). However, the value we calculated for TBC1D9B is likely to be an underestimate, as we did not determine the catalytic efficiency of the full-length protein or under different concentrations of Mg2+, which we showed affected TBC1D9B activity.
Although some GAPs are reported to have multiple Rab substrates (Du et al., 1998; Nottingham et al., 2011; Frasa et al., 2012; Lachmann et al., 2012), our experiments indicate that TBC1D9B may be selective for Rab11a. Indeed, we observed that TBC1D9B has only low levels of activity against a number of Rab GTPases, including Rab11b and Rab4a, both of which interacted with TBC1D9B in their GTP-locked states. We also observed that the TBC domain of TBC1D9B has activity against Rab8a; however, this was noted only when low concentrations of Mg2+ were present in the assays. Thus, it is possible that under Mg2+-limiting conditions, Rab8a may serve as an additional substrate for TBC1D9B. We also observed that TBC1D9B was localized to Rab11a-containing endosomes and much less to EEA1-positive (and Rab5-positive) early endosomes or LAMP2-positive late endosomes/lysosomes. Moreover, only Rab11a-dependent endocytic pathways—for example, basolateral-to-apical transcytosis—were affected when TBC1D9B was overexpressed or when its endogenous protein levels were decreased by shRNA. Our results indicate that TBC1D9B is Rab11a specific at concentrations of Mg2+ in the 2.5–5.0 mM range. However, additional experiments may reveal that this protein targets other Rabs in other pathways, other cell types, or other physiological and developmental contexts.
In addition to TBC1D9B, there may be additional Rab11a GAPs, as is the case for other Rab GTPases (Fukuda, 2011; Frasa et al., 2012; Chaineau et al., 2013). This could explain why depleting endogenous TBC1D9B had only a modest effect on the rate of IgA transcytosis. In addition, the transcytotic pathway is also regulated by other Rabs (including Rab3b/d, Rab17, and Rab25; Hunziker and Peters, 1998; Zacchi et al., 1998; Larkin et al., 2000; Wang et al., 2000b; van IJzendoorn et al., 2002), whose activities are unlikely to be affected by the loss of TBC1D9B expression. The presence of additional Rab11a GAPs could also explain why TBDC1D9B depletion had no effect on the nominal levels of GTP-Rab11a in the cell (as assessed by binding to its Sec15A effector). Examples of potential Rab11a GAPs include Evi5, which has been described as a Rab11GAP in Drosophila melanogaster (Laflamme et al., 2012). However, whether Evi5 acts as a Rab11aGAP in mammalian cells is still controversial. For example, mammalian Evi5 interacts with Rab11a and can regulate Rab11a interaction with its effectors but apparently in a manner that is independent of its GAP activity (Westlake et al., 2007; Hehnly et al., 2012). Of interest, TBC1D14 lacks GAP activity toward Rab11a but does interact with Rab11a in its GTP-bound state, indicating that it acts as an effector rather than a GAP for this GTPase (Longatti et al., 2012). Furthermore, TBC1D11/GAPCen, a Rab4 and Rab6 GAP, and TBC1D15, a Rab7 GAP, are reported to act as GAPs for Rab11a in vitro (Zhang et al., 2005; Fuchs et al., 2007). However, their activity has not been assessed in cellula. Finally, we note that TBC1D9B is a member of a subgroup named TBC-A (Gabernet-Castello et al., 2013), which includes TBC1D8, TBC1D8B, TBC1D9, TBC1D9B, and Gyp2p. With the exception of TBC1D9B, which was studied here, the other mammalian TBC proteins in this subgroup are poorly characterized, and no substrates have been reported. However, it is possible that because of their high similarity, they could share similar substrates. In a similar manner, TBC1D10A and TBC1D10B were both identified as GAPs for Rab27A and, in a different study, as GAPs for Rab35 (Itoh and Fukuda, 2006; Hsu et al., 2010).
An interesting but perplexing finding is that TBC1D9B-RYQ/AAA, like RA mutants in other TBC-containing proteins (Will and Gallwitz, 2001; Haas et al., 2005), shows increased interaction with its cognate Rab. Moreover, we observed that expression of TBC1D9B-RYQ/AAA increases the nominal amount of Rab11a-GTP in the cell (measured by showing increased association between Rab11a and one of its effector proteins, Sec15A). These results are consistent with the possibility that this mutant acts in a dominant manner. However, this does not square with the lack of an obvious dominant effect on IgA transcytosis. One possibility is that TBC1D9B-RYQ/AAA stabilizes the Rab11a/Sec15A interaction, but any gains achieved in forming this interaction (likely at the level of tethering transcytotic vesicles) are offset by slowing the dissociation of Rab11a from Sec15A before vesicle fusion. Thus, it is possible that this mutant does have a dominant effect, but it was not revealed by our experiments. One difference between TBC1D9B and other TBC GAPs studied is that a single R-to-A mutation commonly used in other studies (Will and Gallwitz, 2001; Haas et al., 2005; Itoh et al., 2006; Hannemann et al., 2012) is not enough to produce an inactive protein. This may indicate that other residues in or near the catalytic pocket of TBC1D9B can substitute or that the interaction between TBC1D9B and Rab11a is not adequately represented by the current structural models.
Although we identified TBC1D9B as a Rab11a GAP, there remains much to understand about its physiological functions in the cell. Of great interest are previous reports that the pIgR is transcytosed more efficiently when it binds IgA (Song et al., 1994), which stimulates activation of protein kinase C, release of inositol 1,4,5-trisphosphate, and increase in intracellular calcium (Cardone et al., 1996). Of importance, TBC1D9B contains an EF hand, a well-described Ca2+-binding motif in other proteins. Thus, one possibility is that Ca2+ binding to the EF hand may allosterically impair the function of TBC1D9B and in doing so increase the amount of active Rab11a and therefore extent of pIgR-IgA transcytosis. One possible way that Ca2+ may exert this influence would be by altering the function of the N-terminal GRAM domains, which by analogy to other proteins are likely to regulate the association of TBC1D9B with phospholipids (Doerks et al., 2000; Tsujita et al., 2004).
Finally, it has been proposed in the yeast secretory system that Gyp2p may be recruited by a Rab that acts downstream of Ypt31/32, possibly as an effector of Sec4p (or a similar GTPase), to regulate secretory vesicle maturation. By engaging Gyp2p, Sec4p would ensure compartmental identity by promoting the inactivation and dissociation of Ypt31/32 from maturing vesicles (Sciorra et al., 2005). By analogy, we hypothesize that Rab4a and/or Rab11b could recruit TBC1D9B and in doing so generate endosomal microdomains/compartments that are depleted of active Rab11a. In the case of Rab4, this would be a mechanism to sequester Rab4a cargoes, which undergo rapid recycling (Sonnichsen et al., 2000; Grant and Donaldson, 2009), from the relatively slow recycling pathways usually associated with Rab11a. Similarly, it could explain how the cell generates Rab11b-positive microdomains, which are critical for the recycling of cargoes distinct from those regulated by Rab11a (Lapierre et al., 2003; Silvis et al., 2009; Butterworth et al., 2012).
MATERIALS AND METHODS
Antibodies and reagents
All reagents were obtained from Sigma-Aldrich (St. Louis, MO) unless otherwise stated. Rhodamine-phalloidin and TO-PRO-3 were obtained from Molecular Probes-Invitrogen/Life Sciences (Grand Island, NY). Antibodies used were as follows: mouse anti-flag M2 and rabbit anti-GFP (Abcam, Cambridge, MA), mouse anti-Rab11a (8H10) and rabbit anti-Rab11a (Goldenring et al., 1996; Lapierre et al., 2003), goat anti-GST (GE Healthcare, Pittsburgh, PA), rat anti-ZO1 hybridoma R40.76 culture supernatant (D. A. Goodenough, Harvard University, Cambridge, MA), mouse anti-gp135 hybridoma 3F2/D8 (G. Ojakian, State University of New York, New York, NY), mouse anti-EEA1 (BD Biosciences, San Jose, CA), mouse anti–LAMP-2 antibody AC17 (E. Rodriguez-Boulan, Cornell University, New York, NY), mouse anti-giantin (Adam Linstedt, Carnegie Mellon University, Pittsburgh, PA), mouse anti-Tf receptor antibody H68.4 (Invitrogen, Carlsbad, CA), goat anti-Rab4a (Santa Cruz Biotechnology, Dallas, TX), mouse anti-Rab8a (BD Biosciences), human polymeric IgA (purchased from J. P. Vaerman, Catholic University of Louvain, Louvain, Belgium), mouse anti-pIgR antibody SC166 (Solari et al., 1985), goat anti-Sec15A (Santa Cruz Biotechnology), and goat anti–glyceraldehyde-3-phosphate dehydrogenase (Proteintech, Chicago). Horseradish peroxidase–, CY5-, and Dylight 488–, 549–, and 649–conjugated, affinity-purified, and minimal cross-reacting secondary antibodies were from Jackson ImmunoResearch Laboratories (West Grove, PA).
Cloning of TBC1D9B and sequence analysis
For mammalian expression, full-length human TBC1D9B (Accession Number NP_942568.2) was obtained from Open Biosystems (Thermo Fisher Scientific, Waltham, MA). The cDNA was amplified by PCR and cloned into pEGFP-C1 vector for GFP tagging or pCDN3.1+ for flag tagging. The inactive mutant was produced by mutagenesis using the QuikChange Multi Site-Directed Mutagenesis Kit (Agilent Technologies, Santa Clara, CA), and the following residues were mutated: R559 to A, Y592 to A, and Q594 to A. Sequence alignments were performed using National Center for Biotechnology Information's BLASTp, and the Clustal format was produced with MAFFT software (Larkin et al., 2007; Katoh and Standley, 2013). Structure prediction and comparison was performed using Geneo3D (http://geno3d-pbil.ibcp.fr), with TBC1D4 (PDB 3QYB) as a template. Images were produced and structures were aligned with PyMol (DeLano, 2002).
Cell culture and transfection
HeLa and HEK 293FT (HEK; Invitrogen) cells were grown in DMEM (Cellgro, Herndon, VA) supplemented with 10% fetal bovine serum (FBS; Hyclone, Logan, UT) and 1% penicillin/streptomycin in a 37°C incubator gassed with 5% CO2. For transient transfection, these cells were plated on coverslips or on 10-cm tissue culture plates according to the experiment requirements, and after 24 h they were transfected with DNA using Lipofectamine 2000 according to the manufacturer's instructions (Invitrogen).
MDCK type II cells expressing the wild-type rabbit pIgR (pWe) have been described (Breitfeld et al., 1989). Cells were grown in MEM (Cellgro, Herndon, VA) containing 10% (vol/vol) FBS (Hyclone) and 1% penicillin/streptomycin (pen/strep) in a 37°C incubator gassed with 5% CO2. Cells were cultured on 0.4-μm and 12-mm Transwells (Costar, Cambridge, MA) and used 3–4 d after culture as described (Breitfeld et al., 1989). Cells were transfected in suspension as described previously (Mo et al., 2010), using the appropriate vector and Lipofectamine 2000 reagent.
Immunoprecipitation
Immunoprecipitations were performed as described previously with minor modifications (Oztan et al., 2007). Briefly, HEK cells were cotransfected, as described, with flag-tagged TBC1D9B and GFP-tagged Rab constructs. The GFP-tagged Rab constructs with Q-to-L mutation in their switch II active region, used for the Rab screen (Figure 2A), were described previously (Azouz et al., 2012). After 24–48 h transfection, HEK cells were washed with ice-cold phosphate-buffered saline (PBS), gently scraped into cold PBS using a flexible cell scraper, and then recovered by centrifugation at 1500 × g. The cells were resuspended in IP buffer (10 mM Tris, pH 7.4, 150 mM NaCl, 2.5 mM MgCl2, 5 mM EDTA, 10% glycerol, 1% NP-40, 1 mM phenylmethylsulfonyl fluoride [PMSF], and 5 μg/ml each of pepstatin, leupeptin, and antipain), and after incubation for 30 min on ice, the cells were passed 20 times through a 25-gauge syringe and the lysate centrifuged 10 min at maximum speed in a 5424R microcentrifuge at 4°C (Eppendorf, Westbury, NY). The supernatants were transferred into a new tube and then incubated overnight at 4°C on a rotator with 1 μg of anti-flag M2 antibody or 1 μg of nonspecific mouse IgG as a control. Next, 30 μl of a 50% (vol/vol) slurry of Protein G Sepharose 4FastFlow beads (GE Healthcare) was added to each tube, and the reaction was incubated at 4°C for 1 h on a rotator. Tubes were centrifuged for 1 min at 2000 × g, and the beads were washed five times with IP buffer and then resuspended in 25 μl of 2× Laemmli sample buffer. When the Rab screen was performed (Figure 2A), the washes were made using the IP buffer supplemented with 300 mM NaCl. When stated in the text, MgCl2 was omitted from the IP buffer. The samples were heated at 70°C for 10 min, the cell lysate was resolved by SDS–PAGE, and Western blots, performed as described previously (Oztan et al., 2007), were probed with the indicated primary antibodies. To quantify the binding efficiency of the coimmunoprecipitating proteins, the bands detected in the immunoblots were quantified using ImageJ software (National Institutes of Health, Bethesda, MD). Values were normalized to total expression of each protein.
Yeast two-hybrid assay
For yeast two-hybrid experiments the MATCHMAKER GAL4 Two-Hybrid System (Clontech Laboratories, Mountain View, CA) was used. TBC1D9B-WT or TBC1D9B-RA were cloned into pLexA vector. Human Rab4, Rab5, Rab6, Rab7, Rab11a, and Rab33 were cloned into pB42AD vector. Experiments were performed according to the directions supplied by the manufacturer.
Purification of GST-fusion proteins
cDNAs encoding full-length mammalian Rab GTPases fused to GST were described previously (Pan et al., 2006) and kindly provided by D. G. Lambright (University of Massachusetts, Worcester, MA). GST-Rab11b was subcloned from GFP-hRab11b (kindly provided by Robert Edinger, University of Pittsburgh, Pittsburgh, PA) into the pGEX4T1 vector. TBC1D9B fragments (residues 450–950, 450–810, 301–810, 1–450, and 810–1250) were subcloned into the pGEX4T1 vector. Plasmids encoding the Rabs or TCB1D9B fragments were introduced into BL21-CodonPlus (DE3)-RIPL E. coli cells (Agilent Technologies, Santa Clara, CA) by transformation and expression induced with 0.5 mM isopropyl-β-d-thiogalactoside for 16 h at 30°C. Cells were collected by centrifugation at 8000 × g and the cell pellet dissolved in lysis buffer (20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid [HEPES], pH 8, 500 mM NaCl, 0.5% [vol/vol] Triton X-100, 5 mM MgCl2, 1 mM dithiothreitol [DTT], 1 mM PMSF, and 5 μg/ml each of pepstatin, leupeptin, and antipain). When purifying the Rabs, 200 μM GDP was included in the lysis buffer. DNA was sheared by treating the lysates with 10 sonic pulses (15-s duration each) using a Model 100 Sonic Dismembrator (Fisher Scientific). After centrifugation for 30 min at 4°C at 15,000 × g, the supernatants were recovered and subsequently incubated with reduced glutathione (GSH) Sepharose-4B beads (GE Healthcare) for 3 h at 4°C with rotation. The proteins bound to the beads were then recovered by centrifugation at 5000 × g, washed four times with lysis buffer, and GST-fusion proteins released from the beads by incubating them with elution buffer (10 mM GSH in 100 mM Tris, pH 8, containing 1 mM DTT). Proteins were concentrated fourfold and equilibrated in dialysis buffer (10 mM Tris, pH 7.5, 0.1 mM DTT, 2 mM MgCl2, 150 mM NaCl) using Amicon-Ultra-4 centrifugal filters (EMD Millipore, Billerica, MA). Quantification of proteins was performed using the bicinchoninic acid assay (BCA) Kit (Promega, Madison, WI). Glycerol was added to a final concentration of 10% (vol/vol) and the purified proteins were divided into aliquots, snap-frozen using liquid nitrogen, and then stored at −80°C.
In vitro GAP assay
The efficiency of GTP binding to purified Rabs and GAP assays were performed as described previously (Haas et al., 2005; Fuchs et al., 2007; Yoshimura et al., 2008). To estimate the former, 100 pmol of GST-Rab protein was mixed on ice with 100 μl of assay buffer (50 mM HEPES-NaOH, pH 6.8, 1 mM DTT, 0.2 mg/ml bovine serum albumin, 1 mM EDTA, pH 8.0, and 0.5 mM of an equal mixture of GTP and MgCl2) containing 2 μl of [γ-32P]GTP (10 mCi/ml; 4500 Ci/mmol; MP Biomedicals, Solon, OH), and the reaction was then incubated for 15 min at 30°C. The efficiency of binding was evaluated by nitrocellulose-filter assay (Kabcenell et al., 1990). For GAP assays, the indicated concentration and fragment of TBC1D9B was added to this reaction mixture and then incubated at 30°C for up to 60 min. In some experiments, 5 mM MgCl2 was added in the GAP reaction as described previously (Du et al., 1998; Albert et al., 1999; Nottingham et al., 2011; Lachmann et al., 2012). A 2-μl aliquot of the reaction was counted directly to measure the specific activity in cpm/picomole GTP. The 32Pi release was quantified by taking duplicate 2-μl samples from the GAP reaction and mixing them with 795 ml of ice-cold activated charcoal (5% vol/vol in 50 mM NaH2PO4), which specifically binds to Pi. The charcoal was recovered by centrifugation at 10,000 × g, and the radioactivity within a 400-μl aliquot of the cleared supernatant was measured by Cerenkov counting in a Wallac 1409 liquid scintillation counter (PerkinElmer, Waltham, MA). The amount of hydrolyzed GTP (32Pi release) was calculated from the specific activity of the reaction mixture (Yoshimura et al., 2008) and normalized to the fraction of GTP-bound Rab, which was measured as described using the nitrocellulose-binding assay (Kabcenell et al., 1990). Where indicated, data were fitted to a pseudo-first-order Michaelis–Menten model in which y(t) = Amax[1 − exp(−Kobst)], where Amax corresponds to the maximum amount of hydrolyzed GTP and Kobs is the observed rate constant for the GTP hydrolysis. For each tested concentration of TBC1D9B-(301-810), a Kobs value was obtained, and the catalytic efficiency (Kcat/Km) relative to the intrinsic rate constant of GTP hydrolysis (Kintr) was calculated from the linear dependence between Kobs and the evaluated GAP concentrations, Kobs = Kintr + (Kcat/Km)[GAP] (Pan et al., 2006). Initial velocities (Vi) were calculated as Vi = AmaxKobs, where Amax and Kobs were obtained from the described fitted pseudo-first-order curves.
A previously described procedure was used to measure the GAP activity of full-length TBC1D9B (Nottingham et al., 2011). Briefly, HeLa cells were transfected with either flag-TBC1D9B-WT or flag-TBC1D9B-RYQ/AAA, and 24 h later, the cells were solubilized in lysis buffer as described and the total amount of protein was quantified using the BCA Kit (Promega, Madison, MI). One milligram of total protein was incubated with 0.5 μg of anti-flag antibody prebound to Protein G Sepharose 4 FastFlow beads for 2 h at 4°C with end-to-end rotation. The flag-TBC1D9B-beads were washed three times with lysis buffer, four times with PBS, and two times with assay buffer. An aliquot was kept to confirm the efficiency of the immunoprecipitation, and the remaining beads containing bound GAP were directly added to the described GAP assay. Untransfected HeLa cells were lysed, treated as described, and used as a control in the GAP assay.
Detection of interactions between TBC1D9B fusion proteins and Rab GTPases
HEK cells were transfected with cDNA encoding GFP-Rab constructs (either wild-type or mutant forms) and lysed 24–48 h posttransduction in IP buffer containing 1 mM GTP. The cell lysate was centrifuged and the supernatant “precleared” by incubating the lysate with GSH Sepharose-4B beads (GE Healthcare) for 30 min at 4°C. After a brief centrifugation at 2000 × g, an aliquot of lysate containing 250 μg of total protein was mixed with 1 μg of purified TBC1D9B fusion protein and interacting complexes recovered by the addition of 25 μl of GSH beads and incubation for 3 h at 4°C on a rotator. The reactions were centrifuged for 1 min at 2000 × g, and the beads were washed five times with IP lysis buffer. The beads were resuspended in 25 μl of 2× Laemmli sample buffer, heated at 70°C for 10 min, the proteins resolved by SDS–PAGE, and Western blots probed with the indicated primary antibodies. When stated in the text, no MgCl2 was added to the IP buffer. To quantify the binding efficiency in the pull-down experiments, we quantified the bands detected in the immunoblots using ImageJ software. Values were normalized to total expression of each protein in the lysate and the corresponding amount of GST protein used in each experiment.
Immunofluorescence and colocalization analysis
For immunofluorescence studies, MDCK cells were washed in PIPES-KOH buffer (80 mM 1,4-piperazinediethanesulfonic acid [PIPES]–KOH, pH 6.8, containing 2 mM MgCl2 and 5 mM ethylene glycol tetraacetic acid) and then fixed for 5 min with 4% (wt/vol) paraformaldehyde (Electron Microscopy Sciences, Hatfield, PA) dissolved in PIPES-KOH buffer (adjusted to pH 6.5 by the addition of HCl), followed by 10 min in 4% (wt/vol) paraformaldehyde dissolved in 100 mM sodium borate buffer (pH 11.0; Bacallao and Stelzer, 1989; Apodaca et al., 1994). After fixation, unreacted paraformaldehyde was quenched for 10 min at room temperature with PBS containing 20 mM glycine, pH 8.0, and 75 mM NH4Cl, with the addition of 0.1% Triton X-100. Fixed cells were incubated with block buffer (0.025% [wt/vol] saponin and 8.5 mg/ml fish-skin gelatin dissolved in PBS) for 60 min at room temperature. Cells were incubated with primary antibody, diluted in block buffer for 1 h at room temperature or 16 h at 4°C, washed three times with block buffer for 5 min, and then incubated with fluorescence-labeled secondary antibodies (diluted in block buffer) for 1 h at room temperature. After three additional 5-min washes with block buffer, the cells were rinsed with PBS, fixed with 4% (wt/vol) paraformaldehyde in 100 mM sodium cacodylate, pH 7.4, for 10 min at room temperature, and then mounted as described previously (Apodaca et al., 1994).
To label transcytosing IgA, filter-grown MDCK cells were washed with MEM/bovine serum albumin (BSA; MEM containing 0.45 g/ml NaHCO3, 0.6% [wt/vol] BSA, 10 mM HEPES, pH 7.4, and pen/strep/Fungizone) and incubated at 18°C for 15 min. IgA, 200 μg/ml, was internalized from the basolateral surface of the cells for 20 min at 18°C. The cells were then incubated for 20 min at 37°C, during which 25 μg/ml Cy5-labeled anti–human IgA was added to the apical media. The cells were then washed with ice-cold MEM/BSA and then PBS and finally fixed with 4% (wt/vol) paraformaldehyde in 100 mM sodium cacodylate, pH 7.4, for 10 min at room temperature (Oztan et al., 2007). Unreacted paraformaldehyde was quenched, and the cells were labeled as described.
Images were captured using an HCX PL APO 100×/numerical aperture (NA) 1.4 oil objective (Leica, Wetzlar, Germany) and the appropriate laser lines of a Leica TCS SP5 CW-STED confocal microscope (in normal confocal mode). The photomultipliers were set at 800–1000 V and zoom at 4×, and 8-bit images were collected using four to eight line averages combined with four to eight frame averages. Serial 0.25-μm z-sections were acquired. The images (512 × 512 pixels) were imported into Volocity 3D software (PerkinElmer, Waltham, MA) and, after image reconstruction, exported as TIFF files. The contrast of the latter was corrected in Photoshop CS5 (Adobe, San Jose, CA), and the composite images were prepared in Adobe Illustrator CS5.
Measures of colocalization were performed as described previously (Khandelwal et al., 2008; Khandelwal et al., 2013). Briefly, stacks of dual-labeled confocal sections were imported into Volocity software, background noise was removed using the fine (3 × 3) median noise reduction filter, and a scatter plot of voxel intensities for each of the markers was generated using the colocalization function. The images were thresholded using a fixed value of 40, and Mx, the Manders colocalization coefficient, was calculated for the entire 3D volume. An Mx value of 1.0 indicates that the ratio for all x intensity values that have a corresponding y intensity (i.e., are colocalized) divided by the sum of all x intensity values is 100%. In contrast, a value of 0.0 indicates that there is no colocalization.
Antibody production for anti-TBC1D9B in rabbits
The anti-TBC1D9B antibody is a rabbit polyclonal antibody made by Cocalico Biologicals (Reamstown, PA), using a KLH-conjugated peptide EEDEPPAPELHQDAARELQ (amino acids 1082–1100) from the human TBC1D9B isoform (NP_942568.2). The antibody production was approved by the University of Pittsburgh Institutional Animal Care and Use Committee, Protocol 0809099-1. This antibody was used in immunoblots and immunofluorescence studies.
Adenovirus production and transduction
Full-length TBC1D9B-WT and -RYQ-AAA were cloned into the pECFP vector (Clontech Laboratories ) and subcloned into pAdLOX vector (Hardy et al., 1997). These constructs were transfected into Cre8 cells, and the adenovirus was produced and amplified as previously described (Henkel et al., 1998; Khandelwal et al., 2008). Other constructs used were previously described: GFP/HA-Rab11a-WT, GFP/HA-Rab11a-S25N, GFP-Rab25 (Khandelwal et al., 2008), and GFP-Rab8a-T22N (Khandelwal et al., 2013). Adenovirus was transduced into MDCK cells as described previously (Henkel et al., 1998; Oztan et al., 2007). Adenoviral-mediated expression of CFP-TBC1D9B was detected by immunofluorescence using anti-GFP staining.
Lentivirus shRNA production and transduction
To decrease the expression of canine TBC1D9B, shRNA sequences were generated using an (AA)N19 algorithm and iRNAi software (www.mekentosj.com). The target sequences were (5′–3′): shRNA-1, GACGACATGTCCATGTCCT; shRNA-2, GGTGGAGAGACAGTTCAGC; and scrambled, CCGCAGGTATGCACGCGT. The sequences were cloned into the pLKO.1 vector (Addgene) and lentiviruses produced in 293FT cells using the ViraPower packaging kit (Invitrogen) according to the manufacturer's instructions. Infection of the MDCK cells was performed as described previously (Bryant et al., 2010), and cells expressing the shRNA were enriched by incubating the transduced cells in medium containing 5 μg/ml puromycin. The expression of endogenous TBC1D9B was detected by reverse transcription (RT)-PCR using the RETROscript First Strand Synthesis Kit (Life Technologies), followed by PCR using Taq polymerase (Life Technologies). PCR primers, designed using the predicted sequences of canine TBC1D9B (XM_538581.4), were as follows (5′–3′): forward primer, GCAGGACGCATGTTCCGGCT, and reverse primer, CACCGTGGCGATGGCATGGT. Actin was used as control. Sequencing was used to confirm the identity of the PCR products. To quantify the effectiveness of the shRNAs, PCR reaction products were separated on agarose gels, photographed using a Gel Doc XR+ Imaging System (Bio-Rad, Hercules, CA), and then quantified using ImageJ software. Values were normalized to actin expression. To deplete cells of Rab11a, shRNA specific for canine Rab11a was described previously (Bryant et al., 2010) and kindly provided by D. M. Bryant (University of California, San Francisco, San Francisco, CA).
125I-IgA transcytosis, 125I-IgA apical recycling, 125I-Tf recycling, and 125I-EGFR degradation
The postendocytotic fates of 125I-IgA, 125I-Tf and 125I-EGF were determined as described previously (Apodaca et al., 1994; Maples et al., 1997; Leung et al., 1999; Oztan et al., 2007).
Effector-based, pull-down assay to measure activated Rab11a
For this assay, a GST-ratSec15A full-length construct was used to produce recombinant protein as described previously (Oztan et al., 2007). MDCK cells were transduced with CFP-TBC1D9B-WT or -RYQ/AAA adenovirus as described. After 48 h, the cells were solubilized in IP buffer as described, and the lysate was incubated with 1 μg GST-Sec15A for 2 h at room temperature with rotation. GSH Sepharose-4B beads were used to pull down GST-Sec15A and the endogenous GTP-bound Rab11a associated with Sec15A. The beads were washed four times with lysis buffer and resuspended in 2× Laemmli sample buffer. The samples were heated at 95°C for 2 min and resolved by SDS–PAGE, and Rab11a was detected by Western blot. A similar protocol was used with MDCK cells transduced with lentivirus expressing scrambled shRNA or shRNA-2 targeting canine TBC1D9B.
Statistical analysis
Data are reported as mean ± SEM. Statistically significant differences between means were determined using a two-tailed Student's t test; p < 0.05 was considered statistically significant. One-way analysis of variance (ANOVA), with Bonferonni's correction, was used when making multiple comparisons.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grants R37-DK54425 and P30-DK079307 (to G.A.) and R01-CA83817 and R01-CA111456 (to X.-M.Y.) and by the Kidney Imaging Core of the Pittsburgh Center for Kidney Research (P30-DK079307).
Abbreviations used:
- EGF
epidermal growth factor
- GAP
GTPase-activating protein
- GDP
guanosine diphosphate
- GEF
guanine nucleotide-exchange factor
- GTP
guanosine triphosphate
- IgA
immunoglobulin A
- MDCK
Madin-Darby canine kidney
- pIgR
polymeric immunoglobulin receptor
- TBC
Tre2-Bub2-Cdc16
- Tf
transferrin
- TfR
transferrin receptor
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-10-0604) on September 17, 2014.
REFERENCES
- Adari H, Lowy DR, Willumsen BM, Der CJ, McCormick F. Guanosine triphosphatase activating protein (GAP) interacts with the p21 ras effector binding domain. Science. 1988;240:518–521. doi: 10.1126/science.2833817. [DOI] [PubMed] [Google Scholar]
- Albert S, Will E, Gallwitz D. Identification of the catalytic domains and their functionally critical arginine residues of two yeast GTPase-activating proteins specific for Ypt/Rab transport GTPases. EMBO J. 1999;18:5216–5225. doi: 10.1093/emboj/18.19.5216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apodaca G, Katz LA, Mostov KE. Receptor-mediated transcytosis of IgA in MDCK cells is via apical recycling endosomes. J Cell Biol. 1994;125:67–86. doi: 10.1083/jcb.125.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azouz NP, Matsui T, Fukuda M, Sagi-Eisenberg R. Decoding the regulation of mast cell exocytosis by networks of Rab GTPases. J Immunol. 2012;189:2169–2180. doi: 10.4049/jimmunol.1200542. [DOI] [PubMed] [Google Scholar]
- Bacallao R, Stelzer EH. Preservation of biological specimens for observation in a confocal fluorescence microscope and operational principles of confocal fluorescence microscopy. Methods Cell Biol. 1989;31:437–452. doi: 10.1016/s0091-679x(08)61621-0. [DOI] [PubMed] [Google Scholar]
- Baetz NW, Goldenring JR. Rab11-family interacting proteins define spatially and temporally distinct regions within the dynamic Rab11a-dependent recycling system. Mol Biol Cell. 2013;24:643–658. doi: 10.1091/mbc.E12-09-0659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbieri MA, Roberts RL, Gumusboga A, Highfield H, Alvarez-Dominguez C, Wells A, Stahl PD. Epidermal growth factor and membrane trafficking. EGF receptor activation of endocytosis requires Rab5a. J Cell Biol. 2000;151:539–550. doi: 10.1083/jcb.151.3.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitfeld PP, Harris JM, Mostov KE. Postendocytotic sorting of the ligand for the polymeric immunoglobulin receptor in Madin-Darby canine kidney cells. J Cell Biol. 1989;109:475–486. doi: 10.1083/jcb.109.2.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown PS, Wang E, Aroeti B, Chapin SJ, Mostov KE, Dunn KW. Definition of distinct compartments in polarized Madin-Darby canine kidney (MDCK) cells for membrane-volume sorting, polarized sorting and apical recycling. Traffic. 2000;1:124–140. doi: 10.1034/j.1600-0854.2000.010205.x. [DOI] [PubMed] [Google Scholar]
- Bryant DM, Datta A, Rodriguez-Fraticelli AE, Peranen J, Martin-Belmonte F, Mostov KE. A molecular network for de novo generation of the apical surface and lumen. Nat Cell Biol. 2010;12:1035–1045. doi: 10.1038/ncb2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butterworth MB, Edinger RS, Silvis MR, Gallo LI, Liang X, Apodaca G, Frizzell RA, Johnson JP. Rab11b regulates the trafficking and recycling of the epithelial sodium channel (ENaC). Am J Physiol. Renal Physiol. 2012;302:F581–F590. doi: 10.1152/ajprenal.00304.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardone MH, Smith BL, Mennitt PA, Mochly-Rosen D, Silver RB, Mostov KE. Signal transduction by the polymeric immunoglobulin receptor suggests a role in regulation of receptor transcytosis. J Cell Biol. 1996;133:997–1005. doi: 10.1083/jcb.133.5.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova JE, Wang X, Kumar R, Bhartur SG, Navarre J, Woodrum JE, Altschuler Y, Ray GS, Goldenring JR. Association of Rab25 and Rab11a with the apical recycling system of polarized Madin-Darby canine kidney cells. Mol Biol Cell. 1999;10:47–61. doi: 10.1091/mbc.10.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaineau M, Ioannou MS, McPherson PS. Rab35: GEFs, GAPs and effectors. Traffic. 2013;14:1109–1117. doi: 10.1111/tra.12096. [DOI] [PubMed] [Google Scholar]
- Chen W, Feng Y, Chen D, Wandinger-Ness A. Rab11 is required for trans-Golgi network-to-plasma membrane transport and a preferential target for GDP dissociation inhibitor. Mol Biol Cell. 1998;9:3241–3257. doi: 10.1091/mbc.9.11.3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chotard L, Mishra AK, Sylvain MA, Tuck S, Lambright DG, Rocheleau CE. TBC-2 regulates RAB-5/RAB-7-mediated endosomal trafficking in Caenorhabditis elegans. Mol Biol Cell. 2010;21:2285–2296. doi: 10.1091/mbc.E09-11-0947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabbeekeh JT, Faitar SL, Dufresne CP, Cowell JK. The EVI5 TBC domain provides the GTPase-activating protein motif for RAB11. Oncogene. 2007;26:2804–2808. doi: 10.1038/sj.onc.1210081. [DOI] [PubMed] [Google Scholar]
- DeLano WL. Unraveling hot spots in binding interfaces: progress and challenges. Curr Opin Struct Biol. 2002;12:14–20. doi: 10.1016/s0959-440x(02)00283-x. [DOI] [PubMed] [Google Scholar]
- Desclozeaux M, Venturato J, Wylie FG, Kay JG, Joseph SR, Le HT, Stow JL. Active Rab11 and functional recycling endosome are required for E-cadherin trafficking and lumen formation during epithelial morphogenesis. Am J Physiol. Cell Physiol. 2008;295:C545–C556. doi: 10.1152/ajpcell.00097.2008. [DOI] [PubMed] [Google Scholar]
- Doerks T, Strauss M, Brendel M, Bork P. GRAM, a novel domain in glucosyltransferases, myotubularins and other putative membrane-associated proteins. Trends Biochem Sci. 2000;25:483–485. doi: 10.1016/s0968-0004(00)01664-9. [DOI] [PubMed] [Google Scholar]
- Du LL, Collins RN, Novick PJ. Identification of a Sec4p GTPase-activating protein (GAP) as a novel member of a Rab GAP family. J Biol Chem. 1998;273:3253–3256. doi: 10.1074/jbc.273.6.3253. [DOI] [PubMed] [Google Scholar]
- Fader CM, Sanchez D, Furlan M, Colombo MI. Induction of autophagy promotes fusion of multivesicular bodies with autophagic vacuoles in k562 cells. Traffic. 2008;9:230–250. doi: 10.1111/j.1600-0854.2007.00677.x. [DOI] [PubMed] [Google Scholar]
- Fielding AB, Schonteich E, Matheson J, Wilson G, Yu X, Hickson GR, Srivastava S, Baldwin SA, Prekeris R, Gould GW. Rab11-FIP3 and FIP4 interact with Arf6 and the exocyst to control membrane traffic in cytokinesis. EMBO J. 2005;24:3389–3399. doi: 10.1038/sj.emboj.7600803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frasa MA, Koessmeier KT, Ahmadian MR, Braga VM. Illuminating the functional and structural repertoire of human TBC/RABGAPs. Nat Rev. Mol Cell Biol. 2012;13:67–73. doi: 10.1038/nrm3267. [DOI] [PubMed] [Google Scholar]
- Fuchs E, Haas AK, Spooner RA, Yoshimura S, Lord JM, Barr FA. Specific Rab GTPase-activating proteins define the Shiga toxin and epidermal growth factor uptake pathways. J Cell Biol. 2007;177:1133–1143. doi: 10.1083/jcb.200612068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda M. TBC proteins: GAPs for mammalian small GTPase Rab? Biosci Rep. 2011;31:159–168. doi: 10.1042/BSR20100112. [DOI] [PubMed] [Google Scholar]
- Gabernet-Castello C, O’Reilly AJ, Dacks JB, Field MC. Evolution of Tre-2/Bub2/Cdc16 (TBC) Rab GTPase-activating proteins. Mol Biol Cell. 2013;24:1574–1583. doi: 10.1091/mbc.E12-07-0557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavriljuk K, Gazdag EM, Itzen A, Kotting C, Goody RS, Gerwert K. Catalytic mechanism of a mammalian Rab.RabGAP complex in atomic detail. Proc Natl Acad Sci USA. 2012;109:21348–21353. doi: 10.1073/pnas.1214431110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geourjon C, Combet C, Blanchet C, Deleage G. Identification of related proteins with weak sequence identity using secondary structure information. Protein Sci. 2001;10:788–797. doi: 10.1110/ps.30001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldenring JR, Smith J, Vaughan HD, Cameron P, Hawkins W, Navarre J. Rab11 is an apically located small GTP-binding protein in epithelial tissues. Am J Physiol. 1996;270:G515–G525. doi: 10.1152/ajpgi.1996.270.3.G515. [DOI] [PubMed] [Google Scholar]
- Grant BD, Donaldson JG. Pathways and mechanisms of endocytic recycling. Nat Rev. Mol Cell Biol. 2009;10:597–608. doi: 10.1038/nrm2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, Roth D, Walch-Solimena C, Novick P. The exocyst is an effector for Sec4p, targeting secretory vesicles to sites of exocytosis. EMBO J. 1999;18:1071–1080. doi: 10.1093/emboj/18.4.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas AK, Fuchs E, Kopajtich R, Barr FA. A GTPase-activating protein controls Rab5 function in endocytic trafficking. Nat Cell Biol. 2005;7:887–893. doi: 10.1038/ncb1290. [DOI] [PubMed] [Google Scholar]
- Hales CM, Griner R, Hobdy-Henderson KC, Dorn MC, Hardy D, Kumar R, Navarre J, Chan EK, Lapierre LA, Goldenring JR. Identification and characterization of a family of Rab11-interacting proteins. J Biol Chem. 2001;276:39067–39075. doi: 10.1074/jbc.M104831200. [DOI] [PubMed] [Google Scholar]
- Hannemann M, Sasidharan N, Hegermann J, Kutscher LM, Koenig S, Eimer S. TBC-8, a putative RAB-2 GAP, regulates dense core vesicle maturation in Caenorhabditis elegans. PLoS Genet. 2012;8:e1002722. doi: 10.1371/journal.pgen.1002722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy S, Kitamura M, Harris-Stansil T, Dai Y, Phipps ML. Construction of adenovirus vectors through Cre-lox recombination. J Virol. 1997;71:1842–1849. doi: 10.1128/jvi.71.3.1842-1849.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hehnly H, Chen CT, Powers CM, Liu HL, Doxsey S. The centrosome regulates the Rab11-dependent recycling endosome pathway at appendages of the mother centriole. Curr Biol. 2012;22:1944–1950. doi: 10.1016/j.cub.2012.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henkel JR, Apodaca G, Altschuler Y, Hardy S, Weisz OA. Selective perturbation of apical membrane traffic by expression of influenza M2, an acid-activated ion channel, in polarized madin-darby canine kidney cells. Mol Biol Cell. 1998;9:2477–2490. doi: 10.1091/mbc.9.9.2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horgan CP, McCaffrey MW. The dynamic Rab11-FIPs. Biochem Soc Trans. 2009;37:1032–1036. doi: 10.1042/BST0371032. [DOI] [PubMed] [Google Scholar]
- Hsu C, Morohashi Y, Yoshimura S, Manrique-Hoyos N, Jung S, Lauterbach MA, Bakhti M, Gronborg M, Möbius W, Rhee J, et al. Regulation of exosome secretion by Rab35 and its GTPase-activating proteins TBC1D10A-C. J Cell Biol. 2010;189:223–232. doi: 10.1083/jcb.200911018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunziker W, Peters PJ. Rab17 localizes to recycling endosomes and regulates receptor-mediated transcytosis in epithelial cells. J Biol Chem. 1998;273:15734–15741. doi: 10.1074/jbc.273.25.15734. [DOI] [PubMed] [Google Scholar]
- Ishibashi K, Kanno E, Itoh T, Fukuda M. Identification and characterization of a novel Tre-2/Bub2/Cdc16 (TBC) protein that possesses Rab3A-GAP activity. Genes Gells. 2009;14:41–52. doi: 10.1111/j.1365-2443.2008.01251.x. [DOI] [PubMed] [Google Scholar]
- Itoh T, Fukuda M. Identification of EPI64 as a GTPase-activating protein specific for Rab27A. J Biol Chem. 2006;281:31823–31831. doi: 10.1074/jbc.M603808200. [DOI] [PubMed] [Google Scholar]
- Itoh T, Satoh M, Kanno E, Fukuda M. Screening for target Rabs of TBC (Tre-2/Bub2/Cdc16) domain-containing proteins based on their Rab-binding activity. Genes Cells. 2006;11:1023–1037. doi: 10.1111/j.1365-2443.2006.00997.x. [DOI] [PubMed] [Google Scholar]
- Jones S, Newman C, Liu F, Segev N. The TRAPP complex is a nucleotide exchanger for Ypt1 and Ypt31/32. Mol Biol Cell. 2000;11:4403–4411. doi: 10.1091/mbc.11.12.4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabcenell AK, Goud B, Northup JK, Novick PJ. Binding and hydrolysis of guanine nucleotides by Sec4p, a yeast protein involved in the regulation of vesicular traffic. J Biol Chem. 1990;265:9366–9372. [PubMed] [Google Scholar]
- Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khandelwal P, Prakasam HS, Clayton DR, Ruiz WG, Gallo LI, van Roekel D, Lukianov S, Peranen J, Goldenring JR, Apodaca G. A Rab11a-Rab8a-Myo5B network promotes stretch-regulated exocytosis in bladder umbrella cells. Mol Biol Cell. 2013;24:1007–1019. doi: 10.1091/mbc.E12-08-0568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khandelwal P, Ruiz WG, Balestreire-Hawryluk E, Weisz OA, Goldenring JR, Apodaca G. Rab11a-dependent exocytosis of discoidal/fusiform vesicles in bladder umbrella cells. Proc Natl Acad Sci USA. 2008;105:15773–15778. doi: 10.1073/pnas.0805636105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knodler A, Feng S, Zhang J, Zhang X, Das A, Peranen J, Guo W. Coordination of Rab8 and Rab11 in primary ciliogenesis. Proc Natl Acad Sci USA. 2010;107:6346–6351. doi: 10.1073/pnas.1002401107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krugmann S, Williams R, Stephens L, Hawkins PT. ARAP3 is a PI3K- and rap-regulated GAP for RhoA. Curr Biol. 2004;14:1380–1384. doi: 10.1016/j.cub.2004.07.058. [DOI] [PubMed] [Google Scholar]
- Lachmann J, Barr FA, Ungermann C. The Msb3/Gyp3 GAP controls the activity of the Rab GTPases Vps21 and Ypt7 at endosomes and vacuoles. Mol Biol Cell. 2012;23:2516–2526. doi: 10.1091/mbc.E11-12-1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laflamme C, Assaker G, Ramel D, Dorn JF, She D, Maddox PS, Emery G. Evi5 promotes collective cell migration through its Rab-GAP activity. J Cell Biol. 2012;198:57–67. doi: 10.1083/jcb.201112114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapierre LA, Dorn MC, Zimmerman CF, Navarre J, Burnette JO, Goldenring JR. Rab11b resides in a vesicular compartment distinct from Rab11a in parietal cells and other epithelial cells. Exp Cell Res. 2003;290:322–331. doi: 10.1016/s0014-4827(03)00340-9. [DOI] [PubMed] [Google Scholar]
- Larkin JM, Woo B, Balan V, Marks DL, Oswald BJ, LaRusso NF, McNiven MA. Rab3D, a small GTP-binding protein implicated in regulated secretion, is associated with the transcytotic pathway in rat hepatocytes. Hepatology. 2000;32:348–356. doi: 10.1053/jhep.2000.9110. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Leung SM, Rojas R, Maples C, Flynn C, Ruiz WG, Jou TS, Apodaca G. Modulation of endocytic traffic in polarized Madin-Darby canine kidney cells by the small GTPase RhoA. Mol Biol Cell. 1999;10:4369–4384. doi: 10.1091/mbc.10.12.4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung SM, Ruiz WG, Apodaca G. Sorting of membrane and fluid at the apical pole of polarized Madin-Darby canine kidney cells. Mol Biol Cell. 2000;11:2131–2150. doi: 10.1091/mbc.11.6.2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay AJ, Hendrick AG, Cantalupo G, Senic-Matuglia F, Goud B, Bucci C, McCaffrey MW. Rab coupling protein (RCP), a novel Rab4 and Rab11 effector protein. J Biol Chem. 2002;277:12190–12199. doi: 10.1074/jbc.M108665200. [DOI] [PubMed] [Google Scholar]
- Longatti A, Lamb CA, Razi M, Yoshimura S, Barr FA, Tooze SA. TBC1D14 regulates autophagosome formation via Rab11- and ULK1-positive recycling endosomes. J Cell Biol. 2012;197:659–675. doi: 10.1083/jcb.201111079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maples CJ, Ruiz WG, Apodaca G. Both microtubules and actin filaments are required for efficient postendocytotic traffic of the polymeric immunoglobulin receptor in polarized Madin-Darby canine kidney cells. J Biol Chem. 1997;272:6741–6751. doi: 10.1074/jbc.272.10.6741. [DOI] [PubMed] [Google Scholar]
- Mo D, Potter BA, Bertrand CA, Hildebrand JD, Bruns JR, Weisz OA. Nucleofection disrupts tight junction fence function to alter membrane polarity of renal epithelial cells. Am J Physiol. Renal Physiol. 2010;299:F1178–F1184. doi: 10.1152/ajprenal.00152.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morozova N, Liang Y, Tokarev AA, Chen SH, Cox R, Andrejic J, Lipatova Z, Sciorra VA, Emr SD, Segev N. TRAPPII subunits are required for the specificity switch of a Ypt-Rab GEF. Nat Cell Biol. 2006;8:1263–1269. doi: 10.1038/ncb1489. [DOI] [PubMed] [Google Scholar]
- Nottingham RM, Ganley IG, Barr FA, Lambright DG, Pfeffer SR. RUTBC1 protein, a Rab9A effector that activates GTP hydrolysis by Rab32 and Rab33B proteins. J Biol Chem. 2011;286:33213–33222. doi: 10.1074/jbc.M111.261115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick P, Medkova M, Dong G, Hutagalung A, Reinisch K, Grosshans B. Interactions between Rabs, tethers, SNAREs and their regulators in exocytosis. Biochem Soc Trans. 2006;34:683–686. doi: 10.1042/BST0340683. [DOI] [PubMed] [Google Scholar]
- Oztan A, Silvis M, Weisz OA, Bradbury NA, Hsu SC, Goldenring JR, Yeaman C, Apodaca G. Exocyst requirement for endocytic traffic directed toward the apical and basolateral poles of polarized MDCK cells. Mol Biol Cell. 2007;18:3978–3992. doi: 10.1091/mbc.E07-02-0097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X, Eathiraj S, Munson M, Lambright DG. TBC-domain GAPs for Rab GTPases accelerate GTP hydrolysis by a dual-finger mechanism. Nature. 2006;442:303–306. doi: 10.1038/nature04847. [DOI] [PubMed] [Google Scholar]
- Park SY, Jin W, Woo JR, Shoelson SE. Crystal structures of human TBC1D1 and TBC1D4 (AS160) RabGTPase-activating protein (RabGAP) domains reveal critical elements for GLUT4 translocation. J Biol Chem. 2011;286:18130–18138. doi: 10.1074/jbc.M110.217323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popovic D, Akutsu M, Novak I, Harper JW, Behrends C, Dikic I. Rab GTPase-activating proteins in autophagy: regulation of endocytic and autophagy pathways by direct binding to human ATG8 modifiers. Mol Cell Biol. 2012;32:1733–1744. doi: 10.1128/MCB.06717-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sciorra VA, Audhya A, Parsons AB, Segev N, Boone C, Emr SD. Synthetic genetic array analysis of the PtdIns 4-kinase Pik1p identifies components in a Golgi-specific Ypt31/rab-GTPase signaling pathway. Mol Biol Cell. 2005;16:776–793. doi: 10.1091/mbc.E04-08-0700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvis MR, Bertrand CA, Ameen N, Golin-Bisello F, Butterworth MB, Frizzell RA, Bradbury NA. Rab11b regulates the apical recycling of the cystic fibrosis transmembrane conductance regulator in polarized intestinal epithelial cells. Mol Biol Cell. 2009;20:2337–2350. doi: 10.1091/mbc.E08-01-0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sklan EH, Serrano RL, Einav S, Pfeffer SR, Lambright DG, Glenn JS. TBC1D20 is a Rab1 GTPase-activating protein that mediates hepatitis C virus replication. J Biol Chem. 2007;282:36354–36361. doi: 10.1074/jbc.M705221200. [DOI] [PubMed] [Google Scholar]
- Solari R, Kuhn L, Kraehenbuhl JP. Antibodies recognizing different domains of the polymeric immunoglobulin receptor. J Biol Chem. 1985;260:1141–1145. [PubMed] [Google Scholar]
- Song W, Apodaca G, Mostov K. Transcytosis of the polymeric immunoglobulin receptor is regulated in multiple intracellular compartments. J Biol Chem. 1994;269:29474–29480. [PubMed] [Google Scholar]
- Sonnichsen B, De Renzis S, Nielsen E, Rietdorf J, Zerial M. Distinct membrane domains on endosomes in the recycling pathway visualized by multicolor imaging of Rab4, Rab5, and Rab11. J Cell Biol. 2000;149:901–914. doi: 10.1083/jcb.149.4.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenmark H. Rab GTPases as coordinators of vesicle traffic. Nat Rev. Mol Cell Biol. 2009;10:513–525. doi: 10.1038/nrm2728. [DOI] [PubMed] [Google Scholar]
- Stenmark H, Parton RG, Steele-Mortimer O, Lutcke A, Gruenberg J, Zerial M. Inhibition of rab5 GTPase activity stimulates membrane fusion in endocytosis. EMBO J. 1994;13:1287–1296. doi: 10.1002/j.1460-2075.1994.tb06381.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su T, Bryant DM, Luton F, Verges M, Ulrich SM, Hansen KC, Datta A, Eastburn DJ, Burlingame AL, Shokat KM, et al. A kinase cascade leading to Rab11-FIP5 controls transcytosis of the polymeric immunoglobulin receptor. Nat Cell Biol. 2010;12:1143–1153. doi: 10.1038/ncb2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsujita K, Itoh T, Ijuin T, Yamamoto A, Shisheva A, Laporte J, Takenawa T. Myotubularin regulates the function of the late endosome through the gram domain-phosphatidylinositol 3,5-bisphosphate interaction. J Biol Chem. 2004;279:13817–13824. doi: 10.1074/jbc.M312294200. [DOI] [PubMed] [Google Scholar]
- Tsun ZY, Bar-Peled L, Chantranupong L, Zoncu R, Wang T, Kim C, Spooner E, Sabatini DM. The folliculin tumor suppressor is a GAP for the RagC/D GTPases that signal amino acid levels to mTORC1. Mol Cell. 2013;52:495–505. doi: 10.1016/j.molcel.2013.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullrich O, Reinsch S, Urbe S, Zerial M, Parton RG. Rab11 regulates recycling through the pericentriolar recycling endosome. J Cell Biol. 1996;135:913–924. doi: 10.1083/jcb.135.4.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van IJzendoorn SC, Tuvim MJ, Weimbs T, Dickey BF, Mostov KE. Direct interaction between Rab3b and the polymeric immunoglobulin receptor controls ligand-stimulated transcytosis in epithelial cells. Dev Cell. 2002;2:219–228. doi: 10.1016/s1534-5807(02)00115-6. [DOI] [PubMed] [Google Scholar]
- Wallace DM, Lindsay AJ, Hendrick AG, McCaffrey MW. Rab11-FIP4 interacts with Rab11 in a GTP-dependent manner and its overexpression condenses the Rab11 positive compartment in HeLa cells. Biochem Biophys Res Commun. 2002;299:770–779. doi: 10.1016/s0006-291x(02)02720-1. [DOI] [PubMed] [Google Scholar]
- Wang E, Brown PS, Aroeti B, Chapin SJ, Mostov KE, Dunn KW. Apical and basolateral endocytic pathways of MDCK cells meet in acidic common endosomes distinct from a nearly-neutral apical recycling endosome. Traffic. 2000a;1:480–493. doi: 10.1034/j.1600-0854.2000.010606.x. [DOI] [PubMed] [Google Scholar]
- Wang X, Kumar R, Navarre J, Casanova JE, Goldenring JR. Regulation of vesicle trafficking in madin-darby canine kidney cells by Rab11a and Rab25. J Biol Chem. 2000b;275:29138–29146. doi: 10.1074/jbc.M004410200. [DOI] [PubMed] [Google Scholar]
- Westlake CJ, Baye LM, Nachury MV, Wright KJ, Ervin KE, Phu L, Chalouni C, Beck JS, Kirkpatrick DS, Slusarski DC, et al. Primary cilia membrane assembly is initiated by Rab11 and transport protein particle II (TRAPPII) complex-dependent trafficking of Rabin8 to the centrosome. Proc Natl Acad Sci USA. 2011;108:2759–2764. doi: 10.1073/pnas.1018823108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westlake CJ, Junutula JR, Simon GC, Pilli M, Prekeris R, Scheller RH, Jackson PK, Eldridge AG. Identification of Rab11 as a small GTPase binding protein for the Evi5 oncogene. Proc Natl Acad Sci USA. 2007;104:1236–1241. doi: 10.1073/pnas.0610500104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will E, Gallwitz D. Biochemical characterization of Gyp6p, a Ypt/Rab-specific GTPase-activating protein from yeast. J Biol Chem. 2001;276:12135–12139. doi: 10.1074/jbc.M011451200. [DOI] [PubMed] [Google Scholar]
- Wilson GM, Fielding AB, Simon GC, Yu X, Andrews PD, Hames RS, Frey AM, Peden AA, Gould GW, Prekeris R. The FIP3-Rab11 protein complex regulates recycling endosome targeting to the cleavage furrow during late cytokinesis. Mol Biol Cell. 2005;16:849–860. doi: 10.1091/mbc.E04-10-0927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Mehta SQ, Pichaud F, Bellen HJ, Quiocho FA. Sec15 interacts with Rab11 via a novel domain and affects Rab11 localization in vivo. Nat Struct Mol Biol. 2005;12:879–885. doi: 10.1038/nsmb987. [DOI] [PubMed] [Google Scholar]
- Xiong B, Bayat V, Jaiswal M, Zhang K, Sandoval H, Charng WL, Li T, David G, Duraine L, Lin YQ, et al. Crag is a GEF for Rab11 required for rhodopsin trafficking and maintenance of adult photoreceptor cells. PLoS Biol. 2012;10:e1001438. doi: 10.1371/journal.pbio.1001438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S, Edman M, Kothawala MS, Sun G, Chiang L, Mircheff A, Zhu L, Okamoto C, Hamm-Alvarez S. A Rab11a-enriched subapical membrane compartment regulates a cytoskeleton-dependent transcytotic pathway in secretory epithelial cells of the lacrimal gland. J Cell Sci. 2011;124:3503–3514. doi: 10.1242/jcs.088906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki A, Menon S, Yu S, Barrowman J, Meerloo T, Oorschot V, Klumperman J, Satoh A, Ferro-Novick S. mTrs130 is a component of a mammalian TRAPPII complex, a Rab1 GEF that binds to COPI-coated vesicles. Mol Biol Cell. 2009;20:4205–4215. doi: 10.1091/mbc.E09-05-0387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura S, Egerer J, Fuchs E, Haas AK, Barr FA. Functional dissection of Rab GTPases involved in primary cilium formation. J Cell Biol. 2007;178:363–369. doi: 10.1083/jcb.200703047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura S, Gerondopoulos A, Linford A, Rigden DJ, Barr FA. Family-wide characterization of the DENN domain Rab GDP-GTP exchange factors. J Cell Biol. 2010;191:367–381. doi: 10.1083/jcb.201008051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura S, Haas AK, Barr FA. Analysis of Rab GTPase and GTPase-activating protein function at primary cilia. Methods Enzymol. 2008;439:353–364. doi: 10.1016/S0076-6879(07)00426-0. [DOI] [PubMed] [Google Scholar]
- Yu X, Prekeris R, Gould GW. Role of endosomal Rab GTPases in cytokinesis. Eur J Cell Biol. 2007;86:25–35. doi: 10.1016/j.ejcb.2006.10.002. [DOI] [PubMed] [Google Scholar]
- Zacchi P, Stenmark H, Parton RG, Orioli D, Lim F, Giner A, Mellman I, Zerial M, Murphy C. Rab17 regulates membrane trafficking through apical recycling endosomes in polarized epithelial cells. J Cell Biol. 1998;140:1039–1053. doi: 10.1083/jcb.140.5.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XM, Ellis S, Sriratana A, Mitchell CA, Rowe T. Sec15 is an effector for the Rab11 GTPase in mammalian cells. J Biol Chem. 2004;279:43027–43034. doi: 10.1074/jbc.M402264200. [DOI] [PubMed] [Google Scholar]
- Zhang XM, Walsh B, Mitchell CA, Rowe T. TBC domain family, member 15 is a novel mammalian Rab GTPase-activating protein with substrate preference for Rab7. Biochem Biophys Res Commun. 2005;335:154–161. doi: 10.1016/j.bbrc.2005.07.070. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


