Abstract
In response to fat intake, enteroendocrine K cells release the hormone glucose-dependent insulinotropic polypeptide (GIP). GIP acts on adipocytes to increase lipid uptake and enhance adipokine secretion, promoting weight gain and insulin resistance. Modulation of intestinal GIP release could therefore represent a therapeutic strategy for the treatment and prevention of obesity and diabetes. However, the prospects of using drugs to effectively target specific enteroendocrine cell types have been tempered by the realization that these cells share similar transcriptional programs and frequently employ common mechanisms of hormone secretion. To gain novel insights into the regulation of GIP release, we generated knock-in mice expressing green fluorescent protein (GFP) under the control of the endogenous GIP promoter that enable the isolation of a purified population of small intestine K cells. Using RNA sequencing, we comprehensively characterized the transcriptomes of GIPGFP cells as well as the entire enteroendocrine lineage derived from Neurogenin3-expressing progenitors. Among the genes differentially expressed in GIPGFP cells, we identified and validated fatty acid-binding protein 5 (FABP5) as a highly expressed marker of GIP-producing cells that is absent in other enteroendocrine cell types. FABP5 promotes intracellular transport and inactivation of endocannabinoids, including anandamide, which inhibits GIP release. Remarkably, we found that circulating levels of GIP were significantly decreased in FABP5-deficient mice in the fasting state and in response to acute, oral fat diet administration. Our findings highlight the power of RNA sequencing to uncover molecular signatures of specific enteroendocrine cell types that can potentially be exploited for therapeutic purposes in the treatment of metabolic disorders.
Hormones secreted by cells of the gastrointestinal tract play key roles in metabolic homeostasis and disease. The enteroendocrine system is recognized as an important player in the control of appetite and satiety (1) and is critical for intestinal nutrient absorption (2) and glucose and lipid metabolism (3). Due to their low density and dispersed nature, however, enteroendocrine cells have remained poorly defined and were initially classified on the basis of their secretion products using multilabel immunohistochemical techniques (4). More recently, transgenic reporter mice that allow the identification of genetically tagged hormone-expressing intestinal epithelial cell populations have been instrumental in providing new insights into their biology (5, 6). Interestingly, microarray analyses revealed a higher than anticipated degree of similarity among different types of enteroendocrine cells located within the same intestinal region (7, 8). Furthermore, specific enteroendocrine cell populations were recently found to express a diverse repertoire of gut hormones along with their major hormonal product (7, 8). These observations indicate the need for further investigation into the molecular mechanisms underlying the development and physiology of the gut endocrine lineage that can inform translational studies aimed to selectively modulate intestinal hormone secretion.
A subset of enteroendocrine cells known as K cells produces glucose-dependent insulinotropic polypeptide (GIP), a hormone that stimulates the postprandial insulin response. In addition to its incretin effect, GIP plays a role in lipid metabolism and has been implicated in the etiology of obesity and associated metabolic disturbances. GIP is released shortly after ingestion of carbohydrates and fat (9), and consumption of high-fat diets has been shown to induce K-cell hyperplasia and increase GIP expression and secretion (10–13). Humans and mouse models with obesity-diabetes exhibit elevated levels of circulating GIP and an exaggerated K-cell secretory response to nutrient ingestion (12, 14, 15). Moreover, GIP receptor activation in adipocytes promotes fatty acid synthesis and incorporation (13) and induces an inflammatory response (16), thereby contributing to the development and maintenance of obesity. These findings are supported by phenotypic analyses of mice deficient in either the hormone or its receptor (13, 17) and indicate that modulating the levels of circulating GIP by pharmacological blockade of GIP release may offer a novel therapeutic approach to the management of obesity.
To shed light on the mechanisms underlying GIP synthesis and secretion, we created a knock-in mouse line that allows the identification and isolation of K cells. We found that green fluorescent protein (GFP) expression was restricted to rare flask-shaped cells in both villi and crypts of the small intestine epithelium of GIP-GFP mice. Using fluorescence-activated cell sorting (FACS), we obtained a purified population of small intestine GIPGFP cells and performed next-generation RNA sequencing (RNA-Seq) analysis. By contrasting the gene expression profile of this specific enteroendocrine cell population to that of the entire enteroendocrine cell lineage, we uncovered known and novel transcriptional differences associated with the development and functioning of K cells. Among these, we found that fatty acid-binding protein 5 (FABP5) is exclusively expressed in GIP-producing cells, and we show here that FABP5 is required by K cells to maintain proper levels of circulating GIP, possibly by antagonizing the inhibitory effect of endocannabinoids on GIP secretion. Our findings demonstrate the existence of functionally relevant enteroendocrine cell type-specific modulators of intestinal hormone release that can be targeted for the prevention and treatment of metabolic diseases.
Materials and Methods
Animals
Animal care and procedures were approved by the Laboratory Animal Science Center at Boston University School of Medicine. GIP-GFP knock-in mice were generated as described in Supplemental Materials and Methods. Neurogenin3 (Ngn3)-Cre mice and LoxP-STOP-LoxP-Tomato mice were obtained from JAX (stocks 06333 and 07909, respectively), and FABP5 (Mal1) null mice (18) were generously provided by Gökhan S. Hotamisligil. Animals were housed in a barrier facility with a dark-light cycle of 10 and 14 hours, respectively, and given free access to food and water.
Preparation of RNA-Seq libraries and Illumina Sequencing
RNA extraction was performed using the RNeasy Micro kit (QIAGEN) with on column deoxyribonuclease digestion. RNA quality was assessed using a 2100 Bioanalyzer (Agilent Technologies), and only samples with RIN (RNA integrity Number) greater than 8 were included. For each of the 3 libraries, total RNA samples were pooled in equal amounts, and nonamplified polyadenylated mRNA was purified for the synthesis of cDNA. cDNA libraries were prepared using the Illumina TruSeq RNA Sample Preparation kit and sequenced with an Illumina Genome Analyzer II generating 40-base pair single-end reads. Sequenced reads were analyzed using FastQC and aligned with TopHat (version 1.5.0) to the Mus musculus UCSC mm10 reference genome. Differentially expressed transcripts were identified using the “Tuxedo Protocol” (19), and those showing low expression levels (fragments per kilobase of transcript per million fragments mapped [FPKM] < 0.1) were filtered out for subsequent analyses. RNA-Seq data were visualized and inspected using Integrative Genomics Viewer (20). Gene Ontology (GO) analysis was done using the DAVID gene functional classification tool (http://david.abcc.ncifcrf.gov).
GIP secretion studies
The RAT/MOUSE GIP ELISA kit (Millipore) was used to determine total plasma GIP levels in overnight-fasted mice at 0 and 60 minutes after acute administration of olive oil (20 μL/g body weight). GIP levels were similarly determined in the supernatant of intestinal organoids cultured from crypts isolated from the proximal small intestine of FABP5 knockout and wild-type littermates. Cultures of intestinal organoids were prepared as previously described (21) with some modifications. Briefly, the upper small intestine was opened longitudinally and rinsed in cold PBS. The villi were then scraped off using a hemacytometer coverslip leaving the crypts attached. The tissue was washed extensively with cold PBS to remove remaining villi, chopped into 2- to 4-mm pieces, and incubated in PBS containing 2mM EDTA for 30 minutes on ice. After removal of EDTA, the tissue was vigorously suspended in cold PBS and centrifuged repeatedly to release the crypts. The crypt fraction was then passed through a 70-μm cell strainer to remove residual villous material. Isolated crypts were embedded in Matrigel and cultured in advanced DMEM/F12 supplemented with penicillin/streptomycin, 10mM HEPES, and 1× B27 (Life Technologies), containing 50-ng/mL Epidermal Growth Factor (EGF) 100-ng/mL Noggin, and 1-μg/mL R-spondin (all from R&D Systems, Inc). At day 6, organoids were collected in 1.5-mL Eppendorf tubes, washed with HBSS and incubated at room temperature in Hank's balanced salt solution (HBSS) for 2 hours. After centrifugation, the supernatant was used to measure GIP content and the cell fraction for genomic DNA isolation.
Statistical analysis
Student's t test was used to determine significance of the differences between groups. Data are presented as mean ± SE.
Results
A knock-in GIP-GFP reporter mouse for the study of enteroendocrine K cells
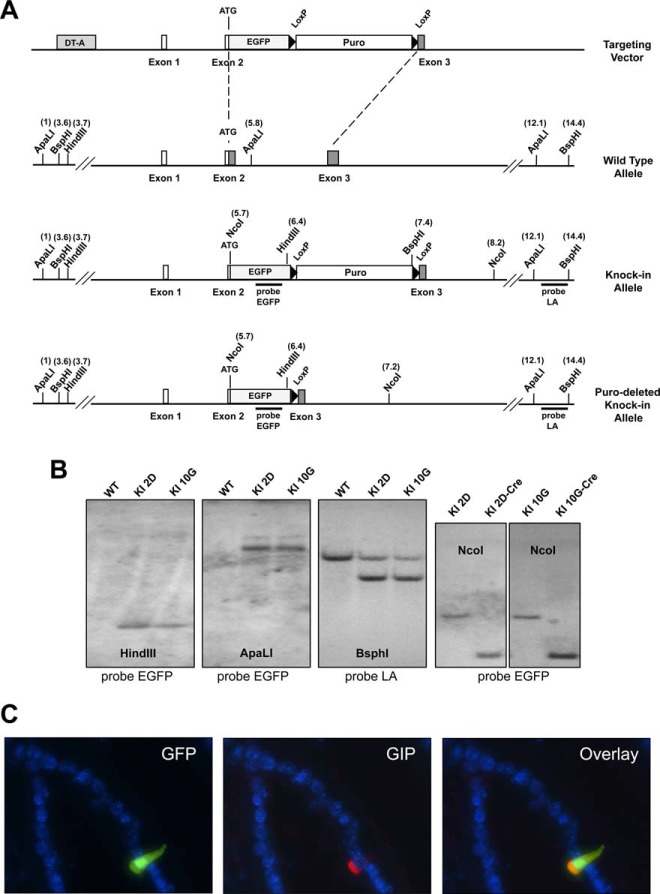
To enable the identification, isolation, and molecular characterization of enteroendocrine K cells, we generated knock-in mice that express the enhanced green fluorescent protein (EGFP) reporter gene from the endogenous GIP locus. To this end, we designed a vector targeting the ATG initiation codon of the GIP gene (Figure 1, A and B). Expression of EGFP was confined to scattered cells located predominantly in the duodenum and jejunum and, to a lesser extent, in the ileum of heterozygous GIP-GFP mice (Supplemental Figure 1A). These cells often displayed flask-shaped morphology and had a characteristic cytoplasmic process extending towards the gut lumen (Figure 1C). We were unable to detect expression of GIP or GFP transcripts by TaqMan real-time PCR in tissues other than the small intestine (Supplemental Figure 1B). In accordance with these results, we did not observe GFP+ cells in histological sections of extraintestinal tissues, and we confirmed these findings by immunohistochemistry using a GFP-specific antibody (Supplemental Figure 2). To validate that EGFP specifically labels GIP-expressing cells, we performed immunohistochemical analyses on small intestine tissue sections from GIP-GFP mice using a GIP-specific antibody. We found colocalization of EGFP and GIP proteins in nearly 90% of duodenal GFP+ cells (n = 12 slides). The GIP protein was frequently localized to the basolateral surface of enteroendocrine K cells, a pattern associated with GIP storage in mature secretory vesicles (Figure 1C). The morphology, distribution, and immunohistochemical features of GFP+ cells clearly indicate that EGFP knocked into the GIP locus faithfully parallels the normal expression pattern of the gene. In agreement with previous studies (22), we observed GFP+ cells (hereafter referred to as GIPGFP cells) not only in the intestinal villi but also in the crypts of GIP-GFP mice, frequently at or near position +4 relative to the crypt base, where GIP-expressing label-retaining intestinal stem cells were recently described (23) (Supplemental Figure 3). To characterize GIPGFP cells in more detail, we obtained single cell preparations of small intestine epithelial cells using collagenase digestion and employed FACS to isolate pure populations of cells based on GFP fluorescence (Figure 2A). Expression of GIP and GFP was approximately 17 000-fold and 13 500-fold higher, respectively, in FACS-sorted GIPGFP cells compared with nonfluorescent cells. In addition, GIPGFP cells contained higher levels of the enteroendocrine marker chromogranin A and the transcription factors paired box 6 (Pax6) and pancreatic and duodenal homeobox 1 (Pdx1) known to be important for GIP expression (Figure 2B). We conclude that GIPGFP cells in the small intestine of GIP-GFP knock-in mice are bona fide enteroendocrine K cells.
Figure 1. Generation of GIP-GFP reporter mice by homologous recombination in embryonic stem cells (ESCs).
A, Schematic representation of the targeting strategy. The protein-coding region of exon 2 and part of exon 3 of the GIP gene was replaced by a sequence encoding EGFP and the PGK1-Puro resistance cassette flanked by LoxP sites. In correctly targeted clones, the GIP promoter drives expression of EGFP. Gray boxes indicate coding regions; the numbers in parentheses indicate kb; DT-A, diphtheria toxin A fragment. B, Correctly targeted ESC clones (2D and 10G) were identified by Southern blot analysis and treated with Cre recombinase to remove the puro cassette. C, GFP expression is restricted to GIP-producing cells in the small intestine epithelium. Intestinal K cells expressing GFP stain positive with a GIP-specific antisera.
Figure 2. GIPGFP cells represent genuine intestinal K cells.
A, FACS of GFP+ cells of the small intestine enables enrichment of GIP-expressing enteroendocrine cells. B, TaqMan gene expression analysis of the GFP+ and GFP− cell populations confirms the identity of GIPGFP cells.
Transcriptome sequencing of enteroendocrine cells of the proximal small intestine
To gain novel insights into the development and physiology of intestinal K cells, we interrogated the whole transcriptome of FACS-isolated small intestine GIPGFP cells using high-throughput mRNA sequencing. For comparison, we first obtained the global gene expression patterns of the entire enteroendocrine cell lineage as well as the nonenteroendocrine cell population, comprising enterocytes, goblet cells, and Paneth cells. To achieve this, small intestine epithelial cells from male mice resulting from the breeding of Ngn3-Cre mice with ROSA26-LoxP-STOP-LoxP-TOMATO indicator mice were isolated based on TOMATO fluorescence and negative staining for cluster of differentiation 45 (CD45) (Supplemental Figure 4). This approach allows the investigation of cells that arise from Ngn3-expressing precursor cells, which are committed to the enteroendocrine lineage and do not contribute to nonenteroendocrine lineages (24). As expected from this model, scattered TOMATO+ cells were observed in histological sections prepared from proximal small intestine of Ngn3-Cre/ ROSA26-LSL-TOMATO mice (Supplemental Figure 4). Consistent with the low density of enteroendocrine cells, flow cytometry analysis of a preparation of dissociated duodenal cells revealed less than 1% GIPGFP cells and about 3% Ngn3TOMATO cells (Supplemental Figure 4). Due to the small cell numbers, we constructed each of the 3 RNA-Seq libraries (GIPGFP, Ngn3TOMATO, and Ngn3−) using a pool of equal amounts of individual RNA samples without RNA amplification. Nearly 130 million RNA-Seq reads were generated (all libraries combined). After quality and low complexity filtering, over 86% of all sequence reads were successfully mapped to the C57BL/6 reference genome. We employed Cufflinks to assemble aligned reads into transcripts and estimate their abundances in FPKM values, which are linearly proportional to original transcript levels (25). Ngn3TOMATO cells, comprising a heterogeneous population of hormone-producing cells all contributing to the final expression profile, exhibited abundant expression of GIP, somatostatin (Sst), secretin (Sct), cholecystokinin (Cck), ghrelin (Ghrl), neurotensin (Nts), and the proglucagon/Glucagon-like peptide-1 Glp-1 precursor (Gcg) (Supplemental Table 1). In contrast, transcripts for gut peptides were not found among the top 30 most abundant protein-coding transcripts in Ngn3− cells (Supplemental Table 2). We next run Cuffdiff to determine genes with differential expression in the enteroendocrine lineage. Compared with the nonenteroendocrine cell population, Ngn3TOMATO cells were highly enriched in transcripts encoding biologically active peptides with a role in the control of energy homeostasis: Cck, GIP, Ghrl, Gcg, Sst, Nts, peptide YY (Pyy), nerve growth factor inducible (VGF), cocaine- and amphetamine-regulated transcript (CART) prepropeptide, islet amyloid polypeptide, transthyretin, and urocortin 3 (Supplemental Table 3). To identify the most relevant biological processes associated with transcripts differentially up-regulated in Ngn3TOMATO cells, we selected those that showed Log2-transformed fold change greater than 3 and performed GO analysis using DAVID. Nearly all the significantly overrepresented GO terms (P < .01) were related to the synthesis, maturation, transport, storage, and secretion of hormone products (Supplemental Table 4). Genes classified into these functional categories included regulators of hormone release (eg, somatostatin receptor 5 (sstr5)), the prohormone convertase 1/3 and its inhibitor proprotein convertase subtilisin/kexin type 1 inhibitor (Pcsk1n), the prohormone-processing carboxypeptidase, chromogranin-secretogranin precursors (chromogranin A (Chga), chromogranin B (ChgB) and secretogranin V (Scg5)), cation channels (eg, transient receptor potential cation channel, subfamily A, member 1 (trpa1), calcium channel, voltage-dependent, alpha2/delta subunit 1 (Cacna2d1), and potassium channel, subfamily K, member 16 (kcnk16)), and amino acid transporters (eg, solute carrier family 38, member 1 (Slc38a1)). These results indicate that Ngn3TOMATO cells possess the distinct biological features of enteroendocrine cells and thereby provide a suitable control cell population for discovering genes that are differentially regulated in GIPGFP cells.
Transcriptional signatures of enteroendocrine K cells revealed by RNA-Seq
Not surprisingly, we found that GIP was the most highly expressed transcript in GIPGFP cells (Supplemental Table 5). Remarkably, transcripts encoding hormones traditionally associated with other enteroendocrine cell types, including Sct, Sst, Cck, and Pyy, were highly abundant in the GIPGFP library (Supplemental Table 5). The finding that the absolute levels of hormone-encoding transcripts were among the highest in both GIPGFP and Ngn3TOMATO libraries and the substantial similarity in terms of their gene expression profiles (r = 0.887, Pearson correlation coefficient) (Figure 3, A and D) are in line with recent studies, indicating a strong overlap between enteroendocrine cell types (8) and coexpression of peptide hormone precursors (7, 8). Notwithstanding these observations, the results of Cuffdiff differential expression testing suggest that K cells have reached a certain level of specification. Compared with Ngn3TOMATO cells, GIPGFP cells exhibited approximately 7.3-fold and approximately 2-fold higher expression of GIP and Pyy, respectively. Transcripts encoding other intestinal hormones were instead down-regulated in GIPGFP cells (Figure 3, A and B). Consistent with previous reports, we found enrichment of the transcription factors Pdx1, Pax6, ISL1 transcription factor, LIM/homeodomain (Isl1) and regulatory factor X, 6 (Rfx6) in the GIPGFP library (Table 1) (8, 26). In addition, we detected increased levels of the cell surface receptors cannabinoid receptor type 1 (Cnr1) and G protein-coupled receptor 119 (Gpr119), which have been implicated in the secretory response of K cells (27, 28). Furthermore, we identified novel transcripts that were differentially expressed in GIPGFP cells and may be important for maintaining the identity of K cells (Table 1). Importantly, these gene expression signatures were confirmed on individual RNA samples (n = 3) using TaqMan real-time PCR analysis (Figure 3, B and C), supporting the reliability of our RNA-Seq data and the validity of the pooling strategy. As a further indication of a higher degree of cell differentiation, genes that characterize other enteroendocrine cell types were depleted in GIPGFP cells (Table 1). These included the extracellular calcium-sensing receptor that allows cholecystokinin (CCK)-secreting I cells to respond to amino acids (6, 29) and the transient receptor potential cation channel Trpa1 and the vesicular monoamine transporters Slc18a1 and Slc18a2 that mediate serotonin storage and release from enterochromaffin cells (30, 31). Taken together, our results indicate that K cells normally coexpress transcripts for additional gut hormones and do so at relatively high levels. However, K cells can be distinguished among enteroendocrine cells by unique gene expression signatures that reflect molecular pathways important for their physiology.
Figure 3. RNA-Seq analysis of enteroendocrine cell populations isolated by FACS.
A, Scatter plots of normalized FPKM expression values comparing the transcriptomes of the enteroendocrine lineage (Ngn3TOMATO cells) with either the GIP-producing enteroendocrine cell subset (GIPGFP cells) or the nonenteroendocrine lineage (Ngn3− cells). Transcripts for gut peptides showing up- and down-regulation in Ngn3TOMATO cells are indicated in red and green, respectively. B and C, Validation of the RNA-Seq data in individual RNA samples using TaqMan assays specific for gut hormones and enteroendocrine cell markers. A strong correlation was found between the RNA-Seq data and the qPCR data (r = 0.942, P < .01), confirming the utility of pooling RNA samples for the construction of the RNA-Seq libraries. D, Frequency distribution of transcript abundances in the 3 libraries. r, Pearson correlation coefficient.
Table 1.
Selected Differentially Expressed Transcripts in GIPGFP Cells vs Ngn3TOMATO Cells
| Up-Regulated in K Cells | FPKM GIPGFP Cells | FPKM Ngn3TOMATO Cells | Log2 (fold change) | Down-Regulated in K Cells | FPKM GIPGFP Cells | FPKM Ngn3TOMATO Cells | Log2 (fold Change) |
|---|---|---|---|---|---|---|---|
| Transcription factors | Transcription factors | ||||||
| Fbxo15 | 28 | 2.51 | 3.48 | Lmx1a | 0.25 | 15.22 | −5.9 |
| Rfx6 | 26.22 | 4.57 | 2.52 | Atoh1 | 1.06 | 18.85 | −4.15 |
| Etv5 | 45.16 | 10.57 | 2.09 | Nr4a1 | 9.01 | 49.63 | −2.46 |
| Isl1 | 41.93 | 10.09 | 2.05 | Fosb | 24.21 | 108.27 | −2.16 |
| Pax6 | 14.32 | 3.52 | 2.02 | Atf3 | 12.81 | 55.77 | −2.12 |
| Pdx1 | 10.6 | 2.67 | 1.99 | Pax4 | 19.09 | 71.26 | −1.9 |
| Channels/receptors | Pitx2 | 3.62 | 13.45 | −1.89 | |||
| Kcnj3 | 114.59 | 3.31 | 5.11 | Fos | 83.86 | 270.54 | −1.69 |
| Cnr1 | 49.16 | 1.66 | 4.89 | Foxa3 | 6.26 | 18.26 | −1.54 |
| Cacna2d1 | 25.54 | 1.15 | 4.47 | Channels/receptors | |||
| Chrm4 | 100.52 | 6.26 | 4 | Trpa1 | 0.21 | 20.99 | −6.6 |
| Kcnj5 | 10.17 | 0.69 | 3.89 | Slc38a11 | 0.99 | 74.64 | −6.23 |
| Kcnj4 | 16.94 | 1.38 | 3.62 | Kcnk16 | 0.52 | 18.46 | −5.16 |
| Kcnc3 | 15.61 | 2.53 | 2.93 | Clca3 | 6.69 | 168.26 | −4.65 |
| Gprc6a | 10.74 | 1.42 | 2.92 | Slc18a2 | 8.3 | 195.2 | −4.55 |
| Gpr75 | 12.88 | 1.89 | 2.76 | Trpm2 | 1.29 | 26.62 | −4.36 |
| Gpr142 | 10.69 | 1.76 | 2.6 | Grm4 | 0.77 | 12.3 | −4 |
| Cacnb2 | 33.98 | 5.71 | 2.57 | Scn3a | 21.73 | 327.4 | −3.91 |
| Gpr22 | 35.74 | 6.06 | 2.56 | Kcnk6 | 1.03 | 13.78 | −3.73 |
| Gpr119 | 36.08 | 7.03 | 2.36 | Vipr1 | 1.83 | 19.59 | −3.42 |
| Kcnj11 | 23.26 | 4.69 | 2.3 | Slc25a35 | 3.27 | 33.01 | −3.34 |
| Kcnb2 | 15.71 | 3.63 | 2.11 | Slc5a9 | 1.68 | 15.71 | −3.22 |
| Scarb1 | 67.01 | 16.98 | 1.98 | Slc18a1 | 6.98 | 63.02 | −3.17 |
| Grik3 | 13.89 | 3.63 | 1.94 | Casr | 1.32 | 10.79 | −3.03 |
| Slc26a4 | 43.51 | 12.56 | 1.79 | Cacnb3 | 11.41 | 87.71 | −2.94 |
| Trpc1 | 19.95 | 6.48 | 1.62 | Trpm5 | 269.46 | 1763.31 | −2.71 |
| Slc16a11 | 18.35 | 6.16 | 1.58 | Slc4a8 | 2.62 | 12.72 | −2.28 |
| Others | Glp1r | 3.28 | 13.28 | −2.01 | |||
| Lipk | 428.95 | 10.71 | 5.32 | Slc1a5 | 12.71 | 47.71 | −1.91 |
| Fabp4 | 17.59 | 0.62 | 4.81 | Erbb2 | 21.19 | 68.74 | −1.7 |
| Glb1l2 | 33.53 | 2.08 | 4.01 | Others | |||
| Alpl | 53.38 | 3.38 | 3.98 | Wif1 | 0.23 | 10.52 | −5.53 |
| Fabp5 | 186.38 | 13.74 | 3.76 | Chga | 197.17 | 7203.13 | −5.19 |
| Alox15 | 10.81 | 1.05 | 3.36 | Ern2 | 0.72 | 17.34 | −4.59 |
| Fstl4 | 10.55 | 1.09 | 3.27 | Rgs2 | 18.42 | 388.65 | −4.4 |
| Rgs16 | 149.7 | 17.29 | 3.16 | Ccl6 | 11.53 | 241.47 | −4.38 |
| Rgs4 | 63.56 | 7.92 | 3 | Dusp2 | 0.72 | 10.84 | −3.91 |
| Cd44 | 1524.22 | 193.6 | 2.98 | Fgf14 | 6.49 | 86.62 | −3.74 |
| Ddc | 23.57 | 3.01 | 2.97 | Tgfb1 | 0.83 | 10.78 | −3.69 |
| Pik3c2g | 181.13 | 26.51 | 2.77 | Guca2a | 48.52 | 516.79 | −3.42 |
| Itprip | 14.59 | 2.17 | 2.74 | Rgs13 | 2.29 | 21.97 | −3.26 |
| Rgs11 | 58.55 | 9.06 | 2.69 | Apobec1 | 1.16 | 10.81 | −3.22 |
| Fstl1 | 26.46 | 5.6 | 2.24 | Dusp1 | 10.61 | 88.86 | −3.06 |
| Sfrp5 | 34.55 | 7.32 | 2.24 | Vgf | 3.66 | 26.97 | −2.88 |
| Bmp1 | 45.29 | 9.82 | 2.2 | Mgll | 75.65 | 486.96 | −2.69 |
| Nxn | 22.83 | 5.26 | 2.16 | Itpr3 | 5.52 | 34.54 | −2.65 |
| Irs1 | 13.69 | 3.69 | 1.89 | Hbegf | 2.37 | 13.45 | −2.5 |
| Notch3 | 36.03 | 9.72 | 1.89 | Ppic | 3.31 | 14.04 | −2.09 |
| Fzd7 | 10.78 | 3.09 | 1.8 | Chgb | 35.79 | 104.56 | −1.54 |
FABP5 is a marker of K cells
Among the transcriptional differences between GIPGFP cells and Ngn3TOMATO cells, the gene encoding FABP5 showed more than 13.5-fold up-regulation in the GIP library (Table 1 and Figure 4A). Notably, not only was FABP5 differentially up-regulated in GIPGFP cells, it was also among the top 30 most abundant transcripts in this library (Supplemental Table 5). We confirmed the enrichment of FABP5 in GIPGFP cells relative to Ngn3TOMATO cells by TaqMan qPCR analysis and were unable to detect the transcript in Ngn3− cells (data not shown). To validate these findings, we performed immunohistochemistry on small intestine tissue sections from GIP-GFP mice. In agreement with the RNA-Seq data, less than 1% of intestinal epithelial cells stained positive for FABP5 (Figure 4B), and virtually every GIPGFP cell was also FABP5+. We observed a low percentage of FABP5+/GFP− cells, which we speculate are enteroendocrine K cells that have not activated the knocked in allele. Indeed, immunohistochemistry analysis of duodenal sections from GIP-GFP homozygous mice showed a higher percentage of GIPGFP cells and almost 100% double stained cells with very rare FABP5+/GFP− cells. Also consistent with these findings, the FABP5 protein was detected in about 10% of Ngn3TOMATO duodenal cells and was absent in Ngn3− cells (Figure 4B). Collectively, these results demonstrate that in the intestinal epithelium, FABP5 is highly and exclusively expressed in enteroendocrine K cells.
Figure 4. Identification of FABP5 as a marker of GIP-producing enteroendocrine cells.
A, Normalized RNA-Seq coverage (ie, number of reads matching an annotated gene) in the 3 libraries is displayed above the FABP5 gene. B, Virtually every GIPGFP cell in the small intestine of GIP-GFP mice but only a subset of enteroendocrine cells in the small intestine of Ngn3− TOMATO mice expresses FABP5 (arrows). Arrowheads indicate Ngn3TOMATO enteroendocrine cells that do not produce detectable levels of the FABP5 protein.
FABP5 is required to maintain normal levels of circulating GIP
FABPs are lipid chaperones frequently expressed at high levels and regulating fundamental cellular processes (32). Recently, FABP5 was implicated in the proliferation and survival of neural stem/progenitor cells during neurogenesis (33). Because the development of both enteroendocrine cells and the neuronal lineage is regulated by similar proteins and signaling pathways (34), we first investigated whether K-cell differentiation was affected in mice that are deficient in FABP5. TaqMan qPCR analysis using RNA isolated from the proximal small intestine of FABP5−/− (Mal1−/−) mice revealed that levels of GIP, Cck, Ghrl, Gcg, Sst, Nts, Pyy, Sct, or the enteroendocrine cell markers chromogranin A and B were similar to wild type (data not shown). In addition, we observed comparable numbers of K cells in histological sections of small intestine from wild-type and FABP5−/− mice (9.6 vs 10.1 cells/section; n = 3). These results indicate that FABP5 is not required for GIP expression in the proximal small intestine. Additionally, the ability of FABP5 to mediate lipid signaling and its restricted expression to K cells in the intestinal epithelium suggested the possibility that this protein might be required for the release of GIP, which is frequently enhanced in the obese state (12, 14, 15). We reasoned that FABP5 could function in fully differentiated K cells to fine-tune the levels of circulating GIP triggered by changes in nutrient composition or overall metabolic state. A possible mechanism would rely on the ability of FABP5 to transport and mediate the intracellular hydrolysis of anandamide (AEA) (35, 36), a major endocannabinoid produced in the proximal small intestine that modulates intestinal physiology (37). Importantly, binding of AEA to the Cnr1, which we found elevated in GIPGFP cells compared with Ngn3TOMATO cells (Log2 fold change = 4.89) (Table 1), inhibits GIP secretion during fasting (27). Therefore, FABP5 could play a pivotal role in K-cell physiology by promoting rapid AEA breakdown, thereby attenuating the inhibitory effect of Cnr1 activation on GIP secretion. Because AEA levels increase 7-fold during starvation (38), we determined whether the lack of FABP5 leads to a more pronounced inhibition of GIP release in overnight-fasted mice. Remarkably, circulating levels of GIP were significantly decreased in FABP5−/− mice both in the fasting state and after administration of an oral bolus of olive oil (Figure 5A). To further delineate the role of FABP5 in the control of GIP secretion, we established ex vivo cultures of small intestine epithelium and measured GIP levels in the supernatant. Consistent with the in vivo results, we found decreased levels of GIP in the supernatant of FABP5−/− organoids compared with wild-type organoids (Figure 5B). Finally, although addition of exogenous AEA resulted in a further 30% decrease in GIP levels compared with the control (data not shown), this difference was not statistically significant (P < .18, Student's t test). We speculate that endogenously generated AEA and possibly other endocannabinoids and lipid molecules that inhibit GIP secretion and whose inactivation is mediated by FABP5 may contribute to decreased GIP secretion in the small intestine of FABP5−/− mice. Overall, our findings indicate that FABP5 has a role in the control of GIP release from K cells and is required to maintain physiological levels of circulating GIP.
Figure 5. FABP5 is required for GIP secretion.
A, Plasma GIP levels in FABP5 null mice and wild-type littermates after an overnight fast and at 1 hour after oral gavage with olive oil (n = 4–6). B, Isolated crypts efficiently form intestinal organoids after ex vivo expansion in Matrigel, regardless of genotype, but basal GIP secretion is attenuated in FABP5-deficient organoids. Data are mean ± SE. *, P < .05 and **, P < .01 by Student's t test.
Discussion
GIP-GFP reporter mice faithfully recapitulate the expression of GIP and offer a reliable animal model to study enteroendocrine cell differentiation and the physiology of K cells in normal conditions and under metabolic stress. In contrast to a previous study (39) and in agreement with a recent report (8), we were not able to detect GFP signal or its transcript in the adult pancreas. Importantly, homozygous GIP-GFP mice are deficient in GIP production and may provide a valuable tool for investigating the effects of GIP not only on pancreas and adipose tissue but also on bone and endothelial cells (40).
To the best of our knowledge, this is the first study reporting the use of high-throughput transcriptome sequencing to comprehensively characterize the expression profile of a purified population of enteroendocrine cells of the small intestine. Due to the relative scarcity of enteroendocrine cells, our analysis was performed using RNA pooled from a number of mice. However, the major conclusions were validated in individual samples. Although the nonfluorescent intestinal epithelial cell fraction has been typically used as a comparator in most previous reports (7, 26, 41), here we employed a lineage-tagging strategy to profile the progeny of Ngn3-expressing progenitor cells. This approach yielded a cell population that morphologically and molecularly resembles the enteroendocrine lineage and as such represents an improved control for gene expression analyses. Importantly, the digital nature of the RNA-Seq data will allow accurate comparisons to be made between particular enteroendocrine cell populations as the data generated within and between different labs become available. Finally, it is anticipated that methodologies that allow whole-transcriptome profiling of single enteroendocrine cells (Single-cell RNA-Seq) will provide a better understanding of the molecular mechanisms behind gut peptide expression and secretion in the gastrointestinal tract.
The notion that terminal differentiation of enteroendocrine progenitors produces cells specialized in the expression of a particular hormone has been recently challenged (42). In these studies, small intestine cells expressing a fluorescent tag from the promoter region of a gut peptide hormone precursor gene were shown to contain transcripts for additional hormonal products (7, 8). Here, we confirm and extend these findings by showing that GIP-producing cells express high levels of Sct, Sst, Cck, and Pyy. The endocrine hormone repertoire of purified GIPGFP cells revealed in this study by RNA-Seq analysis is highly similar to that reported by Habib et al (8) using microarray profiling of duodenal K cells. In light of a recent conclusion that Cck-producing cells coexpress a number of other hormones but not Sst (7), our findings point to the existence of at least 2 major populations of K cells characterized by abundant coexpression of either Sst or Cck. It is possible that in addition to its well-known paracrine effects on hormone release, Sst acts in an autocrine fashion to cell-autonomously block GIP secretion after activation of the Sst receptors present on the surface of K cells (27). Such an autoregulatory loop would prevent further release of GIP from the Sst-producing K-cell subpopulation.
The identification of transcripts that are enriched in K cells provides a foundation for hypothesis-driven basic and translational studies of GIP expression and release. However, because enteroendocrine cells differentiate from a common progenitor, it is possible that the expression of “K cell-specific” functionally relevant genes (eg, transcriptional regulators or receptors) is not entirely restricted to this enteroendocrine cell type. For example, the endocannabinoid receptors GPR119 and Cnr1 that we found up-regulated in GIPGFP cells (Table 1), have been described as being highly enriched in other enteroendocrine cell types (5, 41). On the other hand, we found down-regulation of calcium-sensing receptor CasR in K cells (Table 1), and this is consistent with the poor response of these cells to amino acids that is instead characteristic of Cck-secreting I cells, which express high levels of the transporter (6, 29). Thus, although enteroendocrine cells are increasingly being recognized as having more similarities than differences (42), our findings suggest that careful examination of digital gene expression datasets generated from specific populations of enteroendocrine cells may uncover genes and pathways associated with the synthesis and release of a particular hormone. The validity of this approach is illustrated in this study by the identification of a lipid chaperone that is abundantly and exclusively expressed in GIP-producing cells but not in other intestinal epithelial cells.
FABP5 has been shown to regulate developmental gene expression programs induced by lipid messengers such as retinoic acid (43). Therefore, the finding that FABP5 marks GIP-expressing intestinal epithelial cells suggested a possible role in enteroendocrine cell differentiation. Analysis of FABP5−/− mice nevertheless indicated that the protein is dispensable for enteroendocrine cell development and has no apparent effect on hormone expression in the small intestine. Instead, we found that genetic depletion of FABP5 is associated with a significant decrease in plasma GIP concentration both in fasting conditions and after acute lipid intake. Because deficiency of FABP5 has negative effects on neurogenesis (33, 43), the abnormal GIP levels could stem from impaired neural regulation of GIP secretion. However, intestinal organoids made up entirely of epithelial cells recapitulate this defect ex vivo, indicating that FABP5 controls GIP release in a cell autonomous manner. Existing evidence and our findings suggest that FABP5 may compete for and/or promote the inactivation of lipid molecules that normally inhibit GIP release. However, we cannot rule out the possibility that FABP5 is involved in the budding of transport vesicles containing secreted proteins from the K-cell endoplasmic reticulum, as has been shown previously in enterocytes for other members of the FABP family (44). Our results demonstrate that FABP5 is a key determinant of the identity and physiology of K cells that could potentially be targeted for therapeutic purposes.
Acknowledgments
We thank Dr Gökhan S. Hotamisligil for providing the FABP5 null mice, Shi-Ying Ding for excellent technical assistance, and Vipin T. Sreedharan (Rätsch lab) for help with RNA-Seq data analysis. We also thank the assistance of the Flow Cytometry Core, the Microarray Core, and the Illumina Sequencing Core Facility at Boston University School of Medicine. C.A.S. is an Evans Center Fellow. RNA-Seq data are deposited at NCBI GEO database.
This work was supported in part by a seed grant from the Boston University Genome Science Institute.
Disclosure Summary: The authors have nothing to disclose.
Funding Statement
This work was supported in part by a seed grant from the Boston University Genome Science Institute.
Footnotes
- AEA
- anandamide
- Cck
- cholecystokinin
- Cnr1
- cannabinoid receptor type 1
- EGFP
- enhanced green fluorescent protein
- FABP
- fatty acid-binding protein
- FACS
- fluorescence-activated cell sorting
- FPKM
- fragments per kilobase of transcript per million fragments mapped
- Gcg
- proglucagon/Glp-1 precursor
- GFP
- green fluorescent protein
- Ghrl
- ghrelin
- GIP
- glucose- dependent insulinotropic polypeptide
- GO
- Gene Ontology
- Gpr119
- G protein-coupled receptor 119
- Ngn3
- Neurogenin3
- Nts
- neurotensin
- Pax6
- paired box 6
- Pdx1
- pancreatic and duodenal homeobox 1
- Pyy
- peptide YY
- qPCR
- quantitative PCR
- RNA-Seq
- RNA sequencing
- Sct
- secretin
- Sst
- somatostatin
- Trpa1
- transient receptor potential cation channel, subfamily A, member 1.
References
- 1. Murphy KG, Bloom SR. Gut hormones and the regulation of energy homeostasis. Nature. 2006;444:854–859. [DOI] [PubMed] [Google Scholar]
- 2. Wang J, Cortina G, Wu SV, et al. Mutant neurogenin-3 in congenital malabsorptive diarrhea. N Engl J Med. 2006;355:270–280. [DOI] [PubMed] [Google Scholar]
- 3. Mellitzer G, Beucher A, Lobstein V, et al. Loss of enteroendocrine cells in mice alters lipid absorption and glucose homeostasis and impairs postnatal survival. J Clin Invest. 2010;120:1708–1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Roth KA, Kim S, Gordon JI. Immunocytochemical studies suggest two pathways for enteroendocrine cell differentiation in the colon. Am J Physiol. 1992;263:G174–G180. [DOI] [PubMed] [Google Scholar]
- 5. Reimann F, Habib AM, Tolhurst G, Parker HE, Rogers GJ, Gribble FM. Glucose sensing in L cells: a primary cell study. Cell Metab. 2008;8:532–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Liou AP, Sei Y, Zhao X, et al. The extracellular calcium-sensing receptor is required for cholecystokinin secretion in response to L-phenylalanine in acutely isolated intestinal I cells. Am J Physiol Gastrointest Liver Physiol. 2011;300:G538–G546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Egerod KL, Engelstoft MS, Grunddal KV, et al. A major lineage of enteroendocrine cells coexpress CCK, secretin, GIP, GLP-1, PYY, and neurotensin but not somatostatin. Endocrinology. 2012;153:5782–5795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Habib AM, Richards P, Cairns LS, et al. Overlap of endocrine hormone expression in the mouse intestine revealed by transcriptional profiling and flow cytometry. Endocrinology. 2012;153:3054–3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Schirra J, Katschinski M, Weidmann C, et al. Gastric emptying and release of incretin hormones after glucose ingestion in humans. J Clin Invest. 1996;97:92–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tseng CC, Jarboe LA, Wolfe MM. Regulation of glucose-dependent insulinotropic peptide gene expression by a glucose meal. Am J Physiol. 1994;266:G887–G891. [DOI] [PubMed] [Google Scholar]
- 11. Ross SA, Dupre J. Effects of ingestion of triglyceride or galactose on secretion of gastric inhibitory polypeptide and on responses to intravenous glucose in normal and diabetic subjects. Diabetes. 1978;27:327–333. [DOI] [PubMed] [Google Scholar]
- 12. Flatt PR, Bailey CJ, Kwasowski P, Swanston-Flatt SK, Marks V. Abnormalities of GIP in spontaneous syndromes of obesity and diabetes in mice. Diabetes. 1983;32:433–435. [DOI] [PubMed] [Google Scholar]
- 13. Miyawaki K, Yamada Y, Ban N, et al. Inhibition of gastric inhibitory polypeptide signaling prevents obesity. Nat Med. 2002;8:738–742. [DOI] [PubMed] [Google Scholar]
- 14. Creutzfeldt W, Ebert R, Willms B, Frerichs H, Brown JC. Gastric inhibitory polypeptide (GIP) and insulin in obesity: increased response to stimulation and defective feedback control of serum levels. Diabetologia. 1978;14:15–24. [DOI] [PubMed] [Google Scholar]
- 15. Salera M, Giacomoni P, Pironi L, et al. Gastric inhibitory polypeptide release after oral glucose: relationship to glucose intolerance, diabetes mellitus, and obesity. J Clin Endocrinol Metab. 1982;55:329–336. [DOI] [PubMed] [Google Scholar]
- 16. Timper K, Grisouard J, Sauter NS, et al. Glucose-dependent insulinotropic polypeptide induces cytokine expression, lipolysis, and insulin resistance in human adipocytes. Am J Physiol Endocrinol Metab. 2013;304:E1–E13. [DOI] [PubMed] [Google Scholar]
- 17. Althage MC, Ford EL, Wang S, Tso P, Polonsky KS, Wice BM. Targeted ablation of glucose-dependent insulinotropic polypeptide-producing cells in transgenic mice reduces obesity and insulin resistance induced by a high fat diet. J Biol Chem. 2008;283:18365–18376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Maeda K, Uysal KT, Makowski L, et al. Role of the fatty acid binding protein mal1 in obesity and insulin resistance. Diabetes. 2003;52:300–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Trapnell C, Roberts A, Goff L, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Thorvaldsdóttir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14:178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sato T, Vries RG, Snippert HJ, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. [DOI] [PubMed] [Google Scholar]
- 22. Sei Y, Lu X, Liou A, Zhao X, Wank SA. A stem cell marker-expressing subset of enteroendocrine cells resides at the crypt base in the small intestine. Am J Physiol Gastrointest Liver Physiol. 2011;300:G345–G356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Buczacki SJ, Zecchini HI, Nicholson AM, et al. Intestinal label-retaining cells are secretory precursors expressing Lgr5. Nature. 2013;495:65–69. [DOI] [PubMed] [Google Scholar]
- 24. Bjerknes M, Cheng H. Neurogenin 3 and the enteroendocrine cell lineage in the adult mouse small intestinal epithelium. Dev Biol. 2006;300:722–735. [DOI] [PubMed] [Google Scholar]
- 25. Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. [DOI] [PubMed] [Google Scholar]
- 26. Suzuki K, Harada N, Yamane S, et al. Transcriptional regulatory factor X6 (Rfx6) increases gastric inhibitory polypeptide (GIP) expression in enteroendocrine K-cells and is involved in GIP hypersecretion in high fat diet-induced obesity. J Biol Chem. 2013;288:1929–1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Moss CE, Marsh WJ, Parker HE, et al. Somatostatin receptor 5 and cannabinoid receptor 1 activation inhibit secretion of glucose-dependent insulinotropic polypeptide from intestinal K cells in rodents. Diabetologia. 2012;55:3094–3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chu ZL, Carroll C, Alfonso J, et al. A role for intestinal endocrine cell-expressed g protein-coupled receptor 119 in glycemic control by enhancing glucagon-like peptide-1 and glucose-dependent insulinotropic peptide release. Endocrinology. 2008;149:2038–2047. [DOI] [PubMed] [Google Scholar]
- 29. Wang Y, Chandra R, Samsa LA, et al. Amino acids stimulate cholecystokinin release through the Ca2+-sensing receptor. Am J Physiol Gastrointest Liver Physiol. 2011;300:G528–G537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nozawa K, Kawabata-Shoda, Doihara H, et al. TRPA1 regulates gastrointestinal motility through serotonin release from enterochromaffin cells. Proc Natl Acad Sci USA. 2009;106:3408–3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Essand M, Vikman S, Grawé J, et al. Identification and characterization of a novel splicing variant of vesicular monoamine transporter 1. J Mol Endocrinol. 2005;35:489–501. [DOI] [PubMed] [Google Scholar]
- 32. Furuhashi M, Hotamisligil GS. Fatty acid-binding proteins: role in metabolic diseases and potential as drug targets. Nat Rev Drug Discov. 2008;7:489–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Matsumata M, Sakayori N, Maekawa M, Owada Y, Yoshikawa T, Osumi N. The effects of Fabp7 and Fabp5 on postnatal hippocampal neurogenesis in the mouse. Stem Cells. 2012;30:1532–1543. [DOI] [PubMed] [Google Scholar]
- 34. Schonhoff SE, Giel-Moloney M, Leiter AB. Minireview: development and differentiation of gut endocrine cells. Endocrinology. 2004;145:2639–2644. [DOI] [PubMed] [Google Scholar]
- 35. Kaczocha M, Glaser ST, Deutsch DG. Identification of intracellular carriers for the endocannabinoid anandamide. Proc Natl Acad Sci USA. 2009;106:6375–6380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Berger WT, Ralph BP, Kaczocha M, et al. Targeting fatty acid binding protein (FABP) anandamide transporters - a novel strategy for development of anti-inflammatory and anti-nociceptive drugs. PLoS One. 2012;7:e50968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. DiPatrizio NV, Astarita G, Schwartz G, Li X, Piomelli D. Endocannabinoid signal in the gut controls dietary fat intake. Proc Natl Acad Sci USA. 2011;108:12904–12908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gómez R, Navarro M, Ferrer B, et al. A peripheral mechanism for CB1 cannabinoid receptor-dependent modulation of feeding. J Neurosci. 2002;22:9612–9617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fujita Y, Wideman RD, Asadi A, et al. Glucose-dependent insulinotropic polypeptide is expressed in pancreatic islet α-cells and promotes insulin secretion. Gastroenterology. 2010;138:1966–1975. [DOI] [PubMed] [Google Scholar]
- 40. Campbell JE, Drucker DJ. Pharmacology, physiology, and mechanisms of incretin hormone action. Cell Metab. 2013;17:819–837. [DOI] [PubMed] [Google Scholar]
- 41. Sykaras AG, Demenis C, Case RM, McLaughlin JT, Smith CP. Duodenal enteroendocrine I-cells contain mRNA transcripts encoding key endocannabinoid and fatty acid receptors. PLoS One. 2012;7:e42373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Brubaker PL. A beautiful cell (or two or three?). Endocrinology. 2012;153:2945–2948. [DOI] [PubMed] [Google Scholar]
- 43. Yu S, Levi L, Siegel R, Noy N. Retinoic acid induces neurogenesis by activating both retinoic acid receptors (RARs) and peroxisome proliferator-activated receptor β/δ (PPARβ/δ). J Biol Chem. 2012;287:42195–42205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Neeli I, Siddiqi SA, Siddiqi S, et al. Liver fatty acid-binding protein initiates budding of pre-chylomicron transport vesicles from intestinal endoplasmic reticulum. J Biol Chem. 2007;282:17974–17984. [DOI] [PubMed] [Google Scholar]