Significance
Excess androgens and abnormal follicle development, largely due to ovarian granulosa cell (GC) dysfunction, characterize polycystic ovary syndrome (PCOS), a common endocrinopathy of women predisposing to infertility. Thus, it is important to understand GC dysfunction. The androgen receptor (AR) is widely believed to be an essential regulator of GC biology. High expression of AR in GCs is primarily considered to associate with PCOS. However, we show that AR alternative splice variants in GCs disturb androgen metabolism and follicle growth, leading to PCOS because of impaired transcription factor function. These data considerably change our understanding of the role of AR in the etiology of PCOS, and inform the development of clinical diagnostic and classification tests as well as novel therapeutic interventions.
Keywords: AR, splicing, hyperandrogenism, folliculogenesis, PCOS
Abstract
Polycystic ovary syndrome (PCOS) is one of the most common female endocrine disorders and a leading cause of female subfertility. The mechanism underlying the pathophysiology of PCOS remains to be illustrated. Here, we identify two alternative splice variants (ASVs) of the androgen receptor (AR), insertion and deletion isoforms, in granulosa cells (GCs) in ∼62% of patients with PCOS. AR ASVs are strongly associated with remarkable hyperandrogenism and abnormalities in folliculogenesis, and are absent from all control subjects without PCOS. Alternative splicing dramatically alters genome-wide AR recruitment and androgen-induced expression of genes related to androgen metabolism and folliculogenesis in human GCs. These findings establish alternative splicing of AR in GCs as the major pathogenic mechanism for hyperandrogenism and abnormal folliculogenesis in PCOS.
In ovarian follicles, oocytes are surrounded by granulosa cells (GCs), which have crucial endocrine functions. Polycystic ovary syndrome (PCOS) is a heterogeneous hormonal disorder affecting one in 15 women, and is one of the most common causes of female infertility (1). Hyperandrogenism and abnormal follicle development, associated with excessively small follicles and ovulatory dysfunction largely due to GCs dysfunction, characterize the pathogenesis of PCOS. Although the underlying etiology remains unclear, androgens and the androgen receptor (AR) are considered important on account of their critical roles in the prevalence of hyperandrogenism and ovarian folliculogenesis in this disorder (1–3).
Androgens elicit their effects upon binding to AR, and AR functions primarily via genomic activities as a nuclear receptor. In the ovary, AR is predominantly expressed by GCs throughout most stages of follicular development. Nearly all reproductive phenotypes observed in global AR knockout mice have been attributed to a lack of AR expression in GCs (3). Haploinsufficiency for exon 3-deleted mutant AR is associated with a premature reduction in female fecundity (4), verifying the crucial role of classical genomic AR activity in normal ovarian function. Clinical studies have suggested that PCOS might be associated with AR (CAG)n repeats (5) and rs6152A (6) gene polymorphisms. These studies promoted interest in investigating the effects of AR in GC dysfunction in PCOS women. We therefore hypothesized that abnormally expressed and/or dysfunctional AR plays an important role in the pathogenesis of PCOS.
Results
Alternative Splice Variants of AR in GCs of PCOS Women.
Nested RT-PCR (Fig. S1A) was used to amplify AR cDNAs from GCs of Southeastern Han Chinese women undergoing in vitro fertilization and embryo transfer. We identified two alternative splice variants (ASVs) expressed exclusively in PCOS women. One ASV inserted 69 bp into intron 2 (ivs2) between exons 2 and 3 (insertion isoform, ins) whereas the other skipped exon 3 (deletion isoform, del), as demonstrated by agarose gel electrophoresis (Fig. 1A) and PCR product sequencing (Fig. 1B). AR ASVs were not detected in the GCs of controls; whereas ins and del ARs were coexpressed with wild-type (WT) in 36.8% (WT + ins) and 25.0% (WT + del) of PCOS patients, respectively. None of the PCOS patients expressed both ins and del variants (Fig. 1C). Consistent with previous work (7), total AR mRNA levels were up-regulated in the GCs of PCOS women compared with controls (Fig. 1D). However, surprisingly, the mRNA levels of WT AR were reduced (Fig. 1E), whereas those of AR ASVs were increased (Fig. 1 F and G). Notably, this pattern of expression was tissue-specific, as peripheral lymphocytes expressed only WT AR, which was up-regulated in PCOS women (Fig. S2).
Fig. 1.
Identification and genomic origin of ins and del AR ASVs in the GCs of women with PCOS. (A and B) Representative PCR products (A) and corresponding sequencing results (B) of AR ASVs in patient GCs. Exon boundaries are indicated with vertical double arrows and the sequence of the in-frame insertion from ivs2 of ins AR is shown in detail. (C) Percent distribution of AR splicing patterns in control (n = 120) and PCOS (n = 68) women. (D–G) Relative mRNA levels of total, WT, ins, and del AR in GCs of PCOS women compared with controls (*P < 0.05). (H) Optimal predicted 3D model of the protein structures of AR variants. α-Helices are purple or red (ivs2), and loops are light blue. Data are presented as mean ± SEM.
The AR protein consists of four functional domains: the N-terminal transactivation domain, the DNA binding domain (DBD) containing two zinc fingers translated from exons 2 and 3, hinge region, and ligand-binding domain (LBD) (Fig. S1B). Sequence alterations in both AR ASVs were in-frame and significantly altered the sequence of the DBD, especially that of the second zinc finger (Fig. S1C). Molecular modeling suggests that these changes dramatically altered the 3D structure of the DBD (Fig. 1H). These findings suggest that GC-specific AR ASVs, which are exclusively expressed in PCOS patients, may alter the DNA binding properties of AR.
Hyperandrogenism and Abnormal Folliculogenesis in PCOS Women with AR ASVs.
We examined whether AR ASV expression was associated with clinical features in PCOS women (Table 1). Hyperandrogenism is the most consistent and prominent diagnostic component of PCOS (1). Compared with control and PCOS women expressing WT alone, those with ins or del, especially the former, had notably higher serum total testosterone (TT), serum dehydroepiandrosterone (DHEA) and follicular fluid free androgen index (FAI). In addition, those with WT and ins had a longer cycle length as well as higher antral follicle count, compared with PCOS women with WT alone, whereas both those with ins and those with del were distinguished from controls. It is worth pointing out that only ins was associated with higher follicular fluid levels of anti-Müllerian hormone (AMH), a growth factor produced solely by small follicles (8). These results indicate that the GC AR splicing pattern is tightly linked to hyperandrogenism and abnormalities in folliculogenesis.
Table 1.
AR ASVs are associated with hyperandrogenism in women with PCOS
| Clinical parameter | Control | PCOS | |||
| WT | Total | WT | WT + ins | WT + del | |
| n | 120 | 68 | 26 | 25 | 17 |
| Age, year | 29.98 ± 0.29 | 29.09 ± 0.36 | 29.04 ± 0.56 | 28.84 ± 0.61 | 29.53 ± 0.74 |
| BMI, kg/m2 | 21.84 ± 0.29 | 22.57 ± 0.43 | 22.40 ± 0.70 | 21.84 ± 0.61 | 23.92 ± 0.96 |
| Cycle length, days | 29.37 ± 0.15 | 53.63 ± 2.98* | 51.31 ± 3.53* | 66.68 ± 8.21*† | 54.47 ± 4.02* |
| Antral follicle count | 9.28 ± 0.21 | 18.13 ± 0.49* | 17.76 ± 0.64* | 21.28 ± 1.06*† | 18.71 ± 1.07* |
| Day 3 serum LH/FSH | 0.70 ± 0.03 | 1.34 ± 0.09* | 1.43 ± 0.16* | 1.37 ± 0.16* | 1.16 ± 0.17* |
| Day 3 serum TT, nmol/L | 0.72 ± 0.04 | 1.38 ± 0.08* | 1.00 ± 0.08* | 1.79 ± 0.13*† | 1.43 ± 0.13*† |
| Day 3 serum DHEA, µmol/L | 6.62 ± 0.24 | 8.89 ± 0.51* | 6.86 ± 0.34 | 11.18 ± 1.14*† | 8.85 ± 0.62* |
| Day 3 serum E2, pmol/L | 122.20 ± 5.12 | 136.15 ± 7.85 | 120.86 ± 10.49 | 149.82 ± 15.33 | 140.21 ± 14.92 |
| r-FSH administered, IU | 2,289.9 ± 75.5 | 1,802.2 ± 63.3* | 1,817.0 ± 114.4* | 1,901.5 ± 98.3* | 1,793.5 ± 82.8 |
| FAI in folliclular fluid | 10.76 ± 0.66 | 20.42 ± 2.12* | 13.08 ± 1.62 | 24.59 ± 3.55*† | 25.56 ± 4.09*† |
| E2 in follicular fluid, pmol/L | 14,522 ± 446 | 14,148 ± 654 | 14,897 ± 719 | 13,128 ± 1030 | 14,739 ± 2024 |
| AMH in follicular fluid, µg/L | 4.22 ± 0.44 | 7.65 ± 0.73* | 5.82 ± 0.78 | 9.44 ± 1.30*† | 7.54 ± 1.71 |
| Insulin in follicular fluid, mU/L | 34.79 ± 3.23 | 42.73 ± 3.75 | 36.68 ± 4.08 | 43.29 ± 7.15 | 53.13 ± 8.65 |
| Leptin in follicular fluid, µg/L | 14.48 ± 1.16 | 18.31 ± 1.16* | 16.74 ± 1.36 | 17.74 ± 2.08 | 22.22 ± 2.71* |
AMH, anti-Müllerian hormone; BMI, body mass index; Day 3, third day of spontaneous menstrual cycle; E2, estradiol; FSH, follicle-stimulating hormone; LH, luteinizing hormone; r-FSH, recombinant FSH. Data are presented as mean ± SEM.
P < 0.05 versus control WT.
P < 0.05 versus PCOS WT.
The Origin of AR ASVs.
To explore the genomic origin of AR ASVs in the GCs of PCOS women, we sequenced exon 2 and ∼100 bp of the flanking introns, as well as exon 3 and ∼2 kb of the flanking introns (Fig. S3A). No mutations were identified in conserved regions; however, one PCOS patient with ins was heterozygous (C/T) for a single nucleotide in ivs2, and the variant was produced from only the C allele (Fig. S3B), suggesting ins AR was not randomly generated. Furthermore, single nucleotide polymorphisms have been implicated in alternative RNA splicing (9). We examined the rs6152G/A polymorphism (6) in exon 1 of AR in the GCs of all patients; however, only the G/G genotype was observed. Epigenetic modifications, such as differential methylation at exon-intron boundaries, also have a role in splicing regulation (10). We analyzed the DNA methylation status of 11 cytosine phosphate guanine (CpG) sites of exon 3 and its flanking fragments in GCs by bisulfite genomic sequencing PCR (Fig. 2A). We observed altered methylation of two CpG sites, located at intron 2 and exon 3 junction, in PCOS individuals with ins (Fig. 2B), suggesting the possibility that the ins isoform may be the result of altered DNA methylation.
Fig. 2.
Altered DNA methylation in PCOS women with ins AR. (A) Bisulfite genomic sequencing PCR shows the methylation status of individual DNA strands of exon 3 and its flanking region of AR including 11 CpG sites for individuals with different AR splicing patterns, with ten clones (shown as lines) per sample; CpG sites are shown as blank (unmethylated) or filled (methylated) circles. (B) The above results are summarized as the average methylation ratio at each CpG site (n = 3, *P < 0.05) with a schematic representation of exon 3 and its flanking region of the AR gene.
Reduced Genome-Wide Recruitment Patterns of AR ASVs.
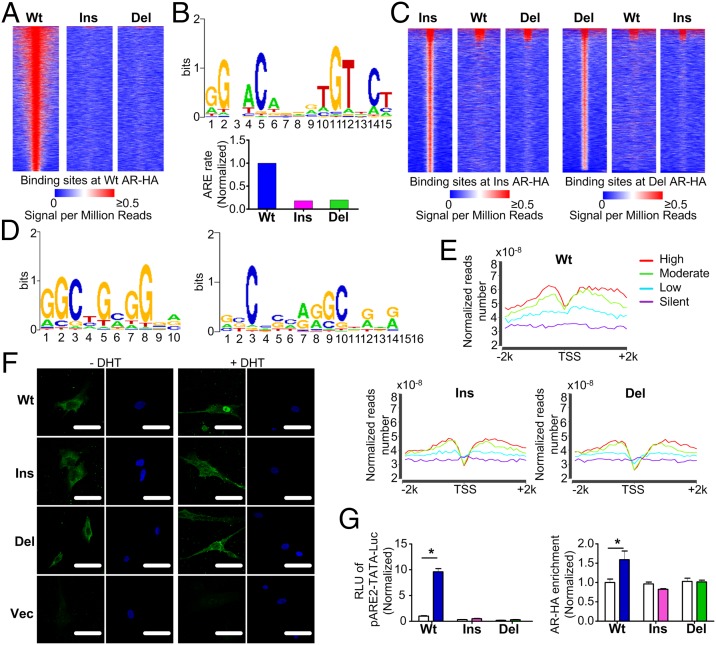
We used adenovirus-mediated expression of hemagglutinin (HA)-tagged WT, ins, and del ARs to map the genomic recruitment of AR variants in primary cultures of human GCs. Chromatin immunoprecipitation (ChIP) sequencing (ChIP-seq; GEO accession: GSE58628) showed that genome-wide AR-HA recruitment was dramatically reduced in GCs overexpressing ins or del (Fig. 3A). Binding sites of WT AR-HA were strongly enriched for androgen response element (ARE) motifs, whereas those of ins or del were not (Fig. 3B). Accordingly, ins and del ASVs had distinct genomic recruitment patterns and enriched motifs (Fig. 3 C and D and Table S1). Combined with RNA sequencing (RNA-seq) data, we found that GCs expressing ins or del displayed a reduced AR-HA binding signal in the regions flanking gene transcription start sites, especially for highly expressed genes, compared with WT (Fig. 3E).
Fig. 3.
Expression of AR ASVs in primary GCs alters genome-wide AR recruitment patterns. (A) Heat map of the ChIP sequencing HA binding signal from −1 kb to +1 kb surrounding the center of WT AR-HA binding sites in GCs transduced with AR variants. Each line represents a single AR-HA binding site and the color scale denotes the AR-HA signal in reads encompassing each locus per million total reads. (B) Top-scoring motif in GCs expressing WT AR (Upper) and its enrichment in GCs expressing AR ASVs (Lower). (C) ChIP sequencing heat maps of ins (Left) and del (Right) AR-HA binding sites. (D) Top-ranking binding motifs enriched in GCs expressing ins (Left) or del (Right) AR. (E) Combined RNA and ChIP sequencing shows reduced AR-HA signal in regions flanking (−2 kb to +2 kb) gene transcription start sites (TSSs) in GCs expressing AR variants. The different color lines represent the genes, the expression levels of which are high (red), moderate (green), low (blue) and silent (purple). (F) Primary GCs transduced with the control vector (Vec) or AR-HA variants were treated with DHT and fluorescently stained with anti-HA (green) or DAPI (blue, nuclei). (Scale bars, 50 μm.) (G) DHT-induced transcriptional activation of (left, luciferase assay), and recruitment of AR-HA variants to (right, ChIP assay), a classical ARE-luciferase reporter in HEK293FT cells (n = 4, *P < 0.05). White columns: DHT (-); colored columns: DHT (+). Data are presented as mean ± SEM.
To elucidate the molecular basis for the diverse effects of AR ASVs, we examined their nuclear shuttling dynamics as well as their ability to bind the AREs of AR target genes. Treatment of transduced primary GCs with dihydrotestosterone (DHT) induced the nuclear translocation of WT AR, but only partially for ins or del (Fig. 3F). Next, we overexpressed AR variants in HEK293FT cells and performed luciferase assays using the pARE2-TATA-Luc reporter (11). DHT induced remarkable activation of the reporter in cells transfected with WT, but not in cells expressing ins or del (Fig. 3G), although coexpression of WT partially restored reporter activity in cells expressing ins or del, especially in the latter case (Fig. S4). ChIP assays further demonstrated reduced DHT-stimulated recruitment of ins and del AR to the classical ARE in pARE2-TATA-Luc (Fig. 3G). Thus, attenuated nuclear shuttling and ARE binding result in distinct genome-wide recruitment patterns of AR ASV.
Altered Gene Expression Related to Follicular Development in GCs with ASVs.
We used RNA-seq to detect and quantitative RT-PCR (RT-qPCR) to further confirm the transcription effects of AR ASVs in primary cultures of human GCs. A cluster of genes were found dysregulated in GCs expressing AR ASVs (Fig. S5 and Dataset S1). DHT-mediated activation of WT AR significantly altered the expression of genes related to folliculogenesis including DHRS13 and DHCR24 (Fig. S6A), steroidogenesis including PRSS23, PGK1, C1GALT, IGFBP5, IGFBP7, and TMSB10 (Fig. S6B) and ovulation including HSPG2, FBN1, SPARC, and PLOD2 (Fig. S6C). DHT-induced gene expression profiles in GCs expressing ins or del AR differed considerably from those with WT AR in both variant- and gene-specific manners. Therefore, DHT-regulated expression of genes related to follicular growth, steroidogenesis and ovulation are significantly altered in GCs expressing ins or del ASVs.
Effects of AR ASVs in GCs on Hyperandrogenism.
We further investigated how altered GC steroidogenesis contributes to ins and del AR ASV-associated hyperandrogenism. RT-qPCR showed that, unlike GCs transduced with WT AR, those transduced with ins or del did not exhibit DHT-induced up-regulation of CYP19A1 encoding aromatase, a rate-limiting enzyme for the conversion of androgens to estrogens (12) (Fig. 4A). Accordingly, GCs expressing ins or del failed to up-regulate the ratios of E2 to TT and estrone (E1) to androstenedione (A) (Fig. 4B), which are indicators of aromatase activity (13). In addition, DHT-induced expression of CYP17A1 encoding 17α-hydroxylase, which catalyzes A biosynthesis (14), was increased only in GCs transduced with ins (Fig. 4C), and caused elevated A levels in the conditioned medium (Fig. 4D). To verify the effects of AR ASVs on androgen metabolism in vivo, we performed similar assays on GCs and follicular fluids from control and PCOS women, and obtained consistent results (Fig. 4 E–H). These data suggest crucial roles for GC AR ASVs in excess androgen accumulation, subsequently leading to hyperandrogenism.
Fig. 4.
Roles of AR ASVs in GCs in ovarian hyperandrogenism. (A–D) In vitro effects of overexpressed AR variants in primary GC as indicated by CYP19A1 (aromatase) mRNA levels (A), estrogen to androgen ratios (B), CYP17A1 (17α-hydroxylase) mRNA levels (C), and A production (D) (n = 6, *P < 0.05). (E–H) In vivo effects of AR variants on androgen metabolism in control and PCOS women as indicated by GC CYP19A1 expression (E), follicular fluid estrogen to androgen ratios (F), CYP17A1 expression (G), and follicular fluid A levels (H) (n = 28, 12, 17 and 5 in the control WT, PCOS WT, ins, and del groups, respectively, *P < 0.05). (I) ChIP assay measuring the recruitment of HA-tagged AR to the U1 ARE site in the CYP19A1 promoter of primary GCs transduced with AR variants (n = 9, *P < 0.05). (J) Schematic diagram of the roles of AR ASVs in GCs in the context of hyperandrogenism and abnormal follicular development. Green (upward) and red (downward) triangles denote increases and decreases, respectively. Data are presented as mean ± SEM.
Next, we tested the ability of AR ASVs to bind potential AREs in the CYP19A1 promoter. In silico analysis of the genomic DNA 5 kb upstream and downstream of the CYP19A1 transcription start site (TSS) identified four high-scoring putative AREs (Fig. S7A). ChIP assays in primary GCs demonstrated DHT-induced recruitment of WT AR-HA to the U1 ARE (Fig. S7B). The transcriptional activity of WT AR at this ARE was confirmed in HEK293FT using an U1-TATA-Luc reporter (Fig. S7C). Then, we used ChIP assays to demonstrate reduced DHT-stimulated recruitment of ins and del AR ASVs to the CYP19A1 U1 ARE in GCs (Fig. 4I). Our findings strongly suggest that the suppression of aromatase expression by ins or del AR results from diminished binding to ARE in the CYP19A1 promoter.
Discussion
Studies conducted in recent decades have mainly focused on the expression, quantification, and genetic polymorphisms of AR, and have built a convincing argument that an abnormal AR is associated with PCOS; however, few studies provided direct evidence for this. We have uniquely documented the roles of AR ASVs in GCs in the pathogenesis of PCOS, although abnormal splicing of AR has been considered an important mechanism accounting for androgen-related diseases in men, including prostate cancer and androgen insensitivity syndrome (15). The findings that two AR ASVs including ins and del are expressed exclusively in the GCs of PCOS women, and not any of women without PCOS, and that these ASVs are notably associated with the clinical features of PCOS, suggest that the abnormal AR splicing is a cause of PCOS in these patients. However, it is worth further verification in other ethnicities. Our results provide a basis for pursuing studies on the functional consequences of these variants in term of GC dysfunction in PCOS. Enhanced expression of AR ASVs in normal GCs alters genome-wide AR recruitment patterns and androgen-induced gene expression, via attenuated nuclear transport and ARE binding, ultimately disrupting androgen metabolism and folliculogenesis. These observations suggest that abnormal splicing of AR is involved in a signaling cascade that augments the transcription of related genes, and subsequently results in hyperandrogenism and abnormalities in folliculogenesis in PCOS.
Our inspection of the clinical characteristics of subjects revealed that both ins and del AR were strongly associated with higher androgen levels in serum and follicles, particularly ins, indicating their roles in peripheral and ovarian hyperandrogenism in PCOS. Of interest, ins was linked to a longer cycle and a greater number of antral follicles, implying its role in defective ovulation and unregulated follicular growth in women with PCOS. Consistently, ins was also connected with elevated follicular levels of AMH, due to GC dysfunction (16) and related to the polycystic ovary phenotype independently of hyperandrogenism (17). These results might explain, in part, not only the differences in clinical appearance between PCOS women and controls, but also the heterogeneity among PCOS individuals. This finding might facilitate the development of clinical diagnostic and classification tests.
With regard to the regulation of AR splicing, the fact that ins or del AR always coexisted with WT, but not overlapping with each other, and the GC-specific expression of AR ASVs, together with the case of a PCOS individual with a heterozygous intron 2, which demonstrates preferential production in a parental specific manner, made it worth exploring the rule-based generation system of AR ASVs. Both ins and del variants were previously reported in men of a family with receptor-positive partial androgen insensitivity (18), resulting from a T/A mutation 11 bp upstream of exon 3. Unfortunately, we did not find any change in cis-acting factors of splicing regulation by DNA sequencing. Nevertheless, it is still promising that changes in transacting factors in the splicing machinery could result in AR ASVs generation in PCOS women, for example epigenetic modifications (19). As these effects are reversible, this may open up novel therapeutic approaches to treat PCOS.
What are the potential mechanisms of the distinct transcription effects of AR ASVs? As a classical nuclear receptor, AR is first activated by binding with androgen; it then translocates into the nucleus and finally binds to tissue-specific AREs to enable the recruitment of an array of coregulators and the general transcription machinery, thereby triggering the transcription of androgen-dependent genes. Firstly, because exons 5–8 are responsible for the coding hinge and LBD, it is expected that the ligand binding ability of ins and del is intact, as also evidenced by previous work (18). Secondly, efficient nuclear shuttling of AR depends on its nuclear localization signal, which is located in the junction between the second zinc finger domain of the DBD and the hinge region (20). The structural changes in the α-helix of the second zinc finger domain of ins and del are likely to interfere with the nuclear localization signal, which in turn would result in impaired nuclear shuttling of ins and del ARs, supported by our findings using immunofluorescent labeling of HA-tagged AR. Moreover, it could be possible that AR ASVs change the binding affinity to nuclear import factors (21) or scaffold proteins (22), which subsequently retards nuclear translocation. Our related study to investigate this question is under the way. Thirdly, in AR DBD, the first zinc finger, encoded by exon 2, defines DNA binding specificity, whereas the second one, encoded by exon 3, facilitates receptor dimerization and stabilization of the DNA–receptor complex (23). Our predictive data revealed that these two ASVs changed their secondary and spatial structures in the α-helix of the second zinc finger in DBD, implying at least partial loss of stability of the DNA–AR complex for both ins and del. This finding was further confirmed by our ChIP results. In particular, a dramatic reduction in binding efficiency with CYP19 ARE of ins and del AR led to deficient aromatase expression and impaired androgen conversion to estrogens, thus contributing to hyperandrogenism. However, our results showed that these ASVs could up-regulate the expression of some genes including CYP17A1 and IGFBP7, which may be not ARE-mediated, possibly due to additional transcriptional activities. The GCs used in the present study were collected from patients undergoing IVF (in vitro fertilization). On the other hand, much of the production of testosterone occurs in the theca cells (24) that surround the follicle and the GCs. Therefore, it is of significance to further confirm our findings in unstimulated and nonluteinized GCs and theca cells.
In summary, these findings demonstrate a previously unsuspected etiology of PCOS whereby alternative splicing of AR in GCs can alter the expression of genes related to androgen metabolism and folliculogenesis, thereby resulting in hyperandrogenism and abnormalities in folliculogenesis (Fig. 4J).
Materials and Methods
Patient Selection and Sample Collection.
PCOS patients, diagnosed according to the Rotterdam Consensus (European Society for Human Reproduction and Embryology/American Society for Reproductive Medicine criteria) and women with tubal factor infertility (serving as controls) seeking IVF treatment at the Women's Hospital of the School of Medicine of Zhejiang University were recruited and gave their written informed consent. The ovarian morphology assessed by ultrasound in the controls was normal for every patient. The long agonist protocol for controlled ovarian hyperstimulation (COH) and the collection of follicular fluids and GCs, obtained by follicular aspiration from women undergoing oocyte retrieval for IVF, were performed as described (25). All patient studies were approved by the Ethics Committee of the Women's Hospital, School of Medicine, Zhejiang University.
Primary Culture.
Human GCs of control subjects were isolated and cultured as previously described (25).
Reverse Transcription Nested PCR and Quantitative Real-Time PCR.
Total RNA was isolated using the RNAiso Reagent and cDNA was prepared using the PrimeScript RT kit. Nested PCR was carried out using the Premix Ex Taq Hot Start Version, and quantitative real-time PCR was performed with the Premix Ex Taq (Probe qPCR) system or the SYBR Premix Ex Taq (Tli RNaseH Plus) system (TAKARA). Glyceraldehyde-3-phosphate dehydrogenase served as the internal control.
Protein Structure Prediction.
The 3D structure of WT, ins, and del AR was predicted using I-TASSER computer modeling and visualized using VMD1.9.1.
Measurement of Hormones.
The levels of day 3 serum hormones were measured in the clinical laboratory of Women's Hospital, School of Medicine, Zhejiang University by chemiluminescence immunoassay (CLIA). TT levels in serum were measured after exaction (26) using CLIA (Roche). TT, SHBG, E2 levels in follicular fluids were detected as described (25). FAI was calculated as TT divided by SHBG × 100 (27). Follicular levels of AMH, insulin, and leptin were measured by ELISA (R&D).
Cell Transfections and Luciferase Assay.
Expression vectors for WT, ins, and del AR, as well as the pARE2-TATA-Luc (28) and U1-TATA-Luc reporter vectors, were purchased from GeneCopoeia. Constructs were transfected into HEK293FT cells using the FuGENE HD transfection reagent (Roche). The luciferase assay was performed using the Luciferase Assay System (Promega).
Adenovirus Infections.
Adenoviral expression systems for HA-tagged WT, ins, and del AR were purchased from SinoGenoMax, Beijing, China. Transduction of primary cultured human GCs was performed as described (29).
Immunofluorescence.
GCs were immunostained with primary antibodies against HA (Cell Signaling, catalog #2367) using a standard protocol as described (25). Cell nuclei were stained with DAPI and staining was visualized using a laser-scanning confocal microscope fitted with a 10 × 60 objective lens (Zeiss).
ARE Screening and Chromatin Immunoprecipitation (ChIP) Analysis.
Putative androgen-response elements (ARE) were identified in the CYP19A1 promoter sequence using the Regulatory Sequence Analysis Tools (RSAT) program (30). ChIP was performed as described (31) with primary antibodies against HA (Abcam, ab9110). Precipitated DNA was amplified by quantitative real-time PCR.
RNA-seq.
Total RNA was extracted using the RNeasy Mini kit according to the manufacturer’s protocol (Qiagen). Total RNA was used to generate an Illumina RNA-seq library using the mRNA-seq 8 Sample Preparation kit (Illumina). RNA-seq libraries were sequenced and quantified on a HiSeq 2000 apparatus (Illumina). Raw reads were mapped to the hg19 using the TopHat version (v2.0.13) (32). We assigned RPKM (Reads per kilobase per million) as an expression value for each gene using Cufflinks version 2.2.1 (33). Heat maps were generated by using the Cluster 3.0 and Treeview. The Cuffdiff was used to identify differentially expressed genes (34) in GCs from DHT (+) versus the DHT (−). Functional enrichment analysis was performed on the differential expressed genes using DAVID Bioinformatics Resources 6.7 (david.abcc.ncifcrf.gov) (35).
ChIP-Seq and Data Processing.
Library preparation and Illumina sequencing were performed according to the manufacturer’s protocol. AR–HA samples were sequenced on a HiSeq 2000 apparatus (Illumina). Sequenced reads were mapped to the human genome (hg19) by SOAP2 (36) and peaks were identified using MACS (37). WT, ins, and del AR heatmaps were ordered by strength of HA binding signals in cells overexpressing HA-tagged WT AR. Enriched motifs from TRANSFAC (38) were found among ChIP regions by the SeqPos motif tool of CISTROME (39) through scanning the sequences of the AR–HA-binding regions using position weight matrices (PWMs) for known transcription factors from the TRANSAC databases, with the entire human genome as a background model. The occurrence of known motifs within the strongest 2,000 AR–HA-binding sites was calculated with MAST (40). Enrichment of AR–HA signals within gene bodies was divided into four categories according to the level of gene expression (high, moderate, low, and silent). Two-kilobase regions upstream and downstream of the gene TSS were divided into 20 intervals, and the tag density of each interval was shown in tag density plots.
Statistics.
Data are presented as mean ± SEM (n is the number of tissue preparations, cells or separate experiments, as indicated in the figure legends). Statistical analysis was performed by unpaired two-tailed Student’s t tests, one-way analysis of variance with post hoc tests, or their equivalent nonparametric tests (version 16.0; SPSS). P < 0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
This study was supported by National Natural Science Foundation–Canadian Institutes of Health Research (NSFC-CIHR) Joint Health Research Programs 81361128007 (to J.S.); Canadian Institutes for Health Research China–Canada Joint Health Research Initiative Grant CCI-132570 (to P.C.K.L.); National Basic Research Program of China 2012CB944900 (to H.H.) and 2011CB944502 (to J.S.); National Natural Science Foundation of China 81450038 and 81490742 (to H.H.) and 81270708 (to J.S.); National Science and Technology Support Program 2012BAI32B01 (to H.H.); Youth Science Fund Project of China 81401167 (to F.W.); Fundamental Research Funds for the Central Universities 2014FZA7001 (to F.W.); the Key Projects in the National Science Technology Pillar Program in the Twelve Five–year Plan Period, China (2012BAI32B04; to F.Q.); and Youth Science Fund Project of China 81200485 (to G.-L.D.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE58628).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1418216112/-/DCSupplemental.
References
- 1.Norman RJ, Dewailly D, Legro RS, Hickey TE. Polycystic ovary syndrome. Lancet. 2007;370(9588):685–697. doi: 10.1016/S0140-6736(07)61345-2. [DOI] [PubMed] [Google Scholar]
- 2.Hillier SG, Tetsuka M. Role of androgens in follicle maturation and atresia. Baillieres Clin Obstet Gynaecol. 1997;11(2):249–260. doi: 10.1016/s0950-3552(97)80036-3. [DOI] [PubMed] [Google Scholar]
- 3.Sen A, Hammes SR. Granulosa cell-specific androgen receptors are critical regulators of ovarian development and function. Mol Endocrinol. 2010;24(7):1393–1403. doi: 10.1210/me.2010-0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walters KA, et al. Female mice haploinsufficient for an inactivated androgen receptor (AR) exhibit age-dependent defects that resemble the AR null phenotype of dysfunctional late follicle development, ovulation, and fertility. Endocrinology. 2007;148(8):3674–3684. doi: 10.1210/en.2007-0248. [DOI] [PubMed] [Google Scholar]
- 5.Shah NA, et al. Association of androgen receptor CAG repeat polymorphism and polycystic ovary syndrome. J Clin Endocrinol Metab. 2008;93(5):1939–1945. doi: 10.1210/jc.2008-0038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peng CY, Long XY, Lu GX. Association of AR rs6152G/A gene polymorphism with susceptibility to polycystic ovary syndrome in Chinese women. Reprod Fertil Dev. 2010;22(5):881–885. doi: 10.1071/RD09190. [DOI] [PubMed] [Google Scholar]
- 7.Catteau-Jonard S, et al. Anti-Mullerian hormone, its receptor, FSH receptor, and androgen receptor genes are overexpressed by granulosa cells from stimulated follicles in women with polycystic ovary syndrome. J Clin Endocrinol Metab. 2008;93(11):4456–4461. doi: 10.1210/jc.2008-1231. [DOI] [PubMed] [Google Scholar]
- 8.Visser JA, Schipper I, Laven JS, Themmen AP. Anti-Müllerian hormone: An ovarian reserve marker in primary ovarian insufficiency. Nat Rev Endocrinol. 2012;8(6):331–341. doi: 10.1038/nrendo.2011.224. [DOI] [PubMed] [Google Scholar]
- 9.Jensen CJ, Stankovich J, Butzkueven H, Oldfield BJ, Rubio JP. Common variation in the MOG gene influences transcript splicing in humans. J Neuroimmunol. 2010;229(1-2):225–231. doi: 10.1016/j.jneuroim.2010.07.027. [DOI] [PubMed] [Google Scholar]
- 10.Laurent L, et al. Dynamic changes in the human methylome during differentiation. Genome Res. 2010;20(3):320–331. doi: 10.1101/gr.101907.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaku N, Matsuda K, Tsujimura A, Kawata M. Characterization of nuclear import of the domain-specific androgen receptor in association with the importin alpha/beta and Ran-guanosine 5′-triphosphate systems. Endocrinology. 2008;149(8):3960–3969. doi: 10.1210/en.2008-0137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Payne AH, Hales DB. Overview of steroidogenic enzymes in the pathway from cholesterol to active steroid hormones. Endocr Rev. 2004;25(6):947–970. doi: 10.1210/er.2003-0030. [DOI] [PubMed] [Google Scholar]
- 13.la Marca A, et al. Insulin-lowering treatment reduces aromatase activity in response to follicle-stimulating hormone in women with polycystic ovary syndrome. Fertil Steril. 2002;78(6):1234–1239. doi: 10.1016/s0015-0282(02)04346-7. [DOI] [PubMed] [Google Scholar]
- 14.Patel SS, Beshay VE, Escobar JC, Suzuki T, Carr BR. Molecular mechanism for repression of 17alpha-hydroxylase expression and androstenedione production in granulosa cells. J Clin Endocrinol Metab. 2009;94(12):5163–5168. doi: 10.1210/jc.2009-1341. [DOI] [PubMed] [Google Scholar]
- 15.Dehm SM, Tindall DJ. Alternatively spliced androgen receptor variants. Endocr Relat Cancer. 2011;18(5):R183–R196. doi: 10.1530/ERC-11-0141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Webber LJ, et al. Formation and early development of follicles in the polycystic ovary. Lancet. 2003;362(9389):1017–1021. doi: 10.1016/s0140-6736(03)14410-8. [DOI] [PubMed] [Google Scholar]
- 17.Stubbs SA, et al. Anti-müllerian hormone protein expression is reduced during the initial stages of follicle development in human polycystic ovaries. J Clin Endocrinol Metab. 2005;90(10):5536–5543. doi: 10.1210/jc.2005-0907. [DOI] [PubMed] [Google Scholar]
- 18.Brüggenwirth HT, et al. Molecular analysis of the androgen-receptor gene in a family with receptor-positive partial androgen insensitivity: An unusual type of intronic mutation. Am J Hum Genet. 1997;61(5):1067–1077. doi: 10.1086/301605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maunakea AK, et al. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature. 2010;466(7303):253–257. doi: 10.1038/nature09165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chan SC, Li Y, Dehm SM. Androgen receptor splice variants activate androgen receptor target genes and support aberrant prostate cancer cell growth independent of canonical androgen receptor nuclear localization signal. J Biol Chem. 2012;287(23):19736–19749. doi: 10.1074/jbc.M112.352930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cutress ML, Whitaker HC, Mills IG, Stewart M, Neal DE. Structural basis for the nuclear import of the human androgen receptor. J Cell Sci. 2008;121(Pt 7):957–968. doi: 10.1242/jcs.022103. [DOI] [PubMed] [Google Scholar]
- 22.Sen A, et al. Androgens regulate ovarian follicular development by increasing follicle stimulating hormone receptor and microRNA-125b expression. Proc Natl Acad Sci USA. 2014;111(8):3008–3013. doi: 10.1073/pnas.1318978111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ferraldeschi R, Welti J, Luo J, Attard G, de Bono JS. Targeting the androgen receptor pathway in castration-resistant prostate cancer: Progresses and prospects. Oncogene. 2014 doi: 10.1038/onc.2014.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McAllister JM, et al. Overexpression of a DENND1A isoform produces a polycystic ovary syndrome theca phenotype. Proc Natl Acad Sci USA. 2014;111(15):E1519–E1527. doi: 10.1073/pnas.1400574111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qu F, et al. A molecular mechanism underlying ovarian dysfunction of polycystic ovary syndrome: Hyperandrogenism induces epigenetic alterations in the granulosa cells. J Mol Med (Berl) 2012;90(8):911–923. doi: 10.1007/s00109-012-0881-4. [DOI] [PubMed] [Google Scholar]
- 26.Janse F, et al. Assessment of androgen concentration in women: Liquid chromatography-tandem mass spectrometry and extraction RIA show comparable results. Eur J Endocrinol. 2011;165(6):925–933. doi: 10.1530/EJE-11-0482. [DOI] [PubMed] [Google Scholar]
- 27.Doi SA, Al-Zaid M, Towers PA, Scott CJ, Al-Shoumer KA. Steroidogenic alterations and adrenal androgen excess in PCOS. Steroids. 2006;71(9):751–759. doi: 10.1016/j.steroids.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 28.Moilanen AM, et al. Identification of a novel RING finger protein as a coregulator in steroid receptor-mediated gene transcription. Mol Cell Biol. 1998;18(9):5128–5139. doi: 10.1128/mcb.18.9.5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bondestam J, et al. Engagement of activin and bone morphogenetic protein signaling pathway Smad proteins in the induction of inhibin B production in ovarian granulosa cells. Mol Cell Endocrinol. 2002;195(1-2):79–88. doi: 10.1016/s0303-7207(02)00219-8. [DOI] [PubMed] [Google Scholar]
- 30.Thomas-Chollier M, et al. RSAT 2011: Regulatory sequence analysis tools. Nucleic Acids Res. 2011;39(web server Issue):W86–W91. doi: 10.1093/nar/gkr377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Feng D, et al. A circadian rhythm orchestrated by histone deacetylase 3 controls hepatic lipid metabolism. Science. 2011;331(6022):1315–1319. doi: 10.1126/science.1198125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trapnell C, Pachter L, Salzberg SL. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25(9):1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28(5):511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trapnell C, et al. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol. 2013;31(1):46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 36.Li R, et al. SOAP2: An improved ultrafast tool for short read alignment. Bioinformatics. 2009;25(15):1966–1967. doi: 10.1093/bioinformatics/btp336. [DOI] [PubMed] [Google Scholar]
- 37.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9(9):R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matys V, et al. TRANSFAC: Transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 2003;31(1):374–378. doi: 10.1093/nar/gkg108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu T, et al. Cistrome: An integrative platform for transcriptional regulation studies. Genome Biol. 2011;12(8):R83. doi: 10.1186/gb-2011-12-8-r83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bailey TL, Gribskov M. Combining evidence using p-values: application to sequence homology searches. Bioinformatics. 1998;14(1):48–54. doi: 10.1093/bioinformatics/14.1.48. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.