Abstract
T-box genes often exhibit dynamic expression patterns, and their expression levels can be crucial for normal function. Despite the importance of these genes, there is little known about T-box gene regulation. We have focused on the Caenorhabditis elegans gene tbx-2 to understand how T-box gene expression is regulated, and here we demonstrate TBX-2 itself directly represses its own expression in a negative autoregulatory loop. tbx-2 is essential for normal pharyngeal muscle development, and a tbx-2 promoter gfp fusion (Ptbx-2::gfp) is transiently expressed in the pharynx during embryogenesis and in a small number of head neurons in larvae and adults. Reduced tbx-2 function resulted in ectopic Ptbx-2::gfp expression in the seam cells and gut in larvae and adults. Mutation of potential T-box binding sites within the tbx-2 promoter resulted in a similar pattern of ectopic Ptbx-2::gfp expression, and chromatin immunoprecipitation analyses show TBX-2 binds these sites in vivo. This pattern of ectopic Ptbx-2::gfp expression in tbx-2 mutants was very similar to that observed in mutants affecting the NF-Y complex, and our results comparing tbx-2 and nfyb-1 single- and double mutants suggest TBX-2 and NF-Y function in a single pathway to repress the tbx-2 promoter. The tbx-2 promoter is the first direct target identified for TBX-2, and we used it to ask whether SUMOylation is essential for TBX-2 repression. RNAi knockdown of SUMOylation pathway components led to ectopic Ptbx-2::gfp expression in the seam cells and gut. Ectopic Ptbx-2::gfp also was observed in the syncytial hypodermis, suggesting either the tbx-2 promoter is repressed by other SUMOylation dependent mechanisms, or that decreased SUMOylation leads to stable changes in seam cell nuclei as they fuse with the syncytial hypodermis. We suggest negative autoregulation is an important mechanism that allows precise control of tbx-2 expression levels and may allow rapid changes in gene expression during development.
Keywords: T-box, C. elegans, SUMOylation, negative autoregulation, NF-Y
T-box proteins are an ancient and highly conserved transcription factor family found in all animals, and members of this family play crucial roles in cell fate specification and organogenesis [reviewed in (Sebe-Pedros et al. 2013; Takashima and Suzuki 2013)]. T-box gene expression is highly regulated, and these genes often are expressed in dynamic patterns during development. For example in mouse, the closely related Tbx2 and Tbx3 genes are expressed at various developmental stages in tissues as different as the allantois, heart, limbs, eye, mammary gland, and brain [reviewed in (Abrahams et al. 2010)]. In many cases, appropriate levels of T-box gene expression are crucial for their function, and both under- and overexpression of T-box genes can lead to defects. In the mouse, progressively decreasing Tbx5 or Tbx20 activity results in distinct changes in target gene expression and phenotypic defects (Takeuchi et al. 2005; Mori et al. 2006). Likewise, in humans, haploinsufficiency for individual T-box genes underlies a variety of congenital diseases, including DiGeorge syndrome (TBX1), Ulnar-mammary syndrome (TBX3), Holt-Oram syndrome (TBX5), small patella syndrome (TBX4), and cleft palette (TBX22 and TBX10) [reviewed in (Takashima and Suzuki 2013)]. Conversely, overexpression of TBX2 and TBX3 is found in number of different cancers, where they are believed to inhibit cellular senescence by repressing expression of the cyclin-dependent kinase inhibitors p21WAF and p19ARF [reviewed in (Abrahams et al. 2010)].
Transcription factor gene expression frequently is controlled by autoregulatory mechanisms, and both positive and negative autoregulation has been observed in organisms as different as bacteria and humans (Alon 2007; Nevozhay et al. 2009). Positive autoregulation generally results in slower response times after initial gene activation with an eventual switch-like increase in gene expression, and it results in cell populations that exhibit broad or even bimodal levels of gene expression. In contrast, negative autoregulation results in rapid response times and reduces cell-to-cell variation in the level of gene expression. Both positive and negative autoregulation has been described for T-box genes in ascidians and humans, suggesting autoregulation may be a common feature of this gene family (Conlon et al. 1996; Takahashi et al. 1999; Mitani et al. 2001; Sun et al. 2004; Imai et al. 2006; Andreou et al. 2007).
We are functionally characterizing T-box factors in Caenorhabditis elegans, and here we focus on the Tbx2 subfamily factor TBX-2. TBX-2 is the sole C. elegans member of the Tbx2-subfamily, which in mammals includes Tbx2, Tbx3, Tbx4, and Tbx5, and it is most closely related to the transcriptional repressors Tbx2 and Tbx3 (Pocock et al. 2004). In C. elegans, tbx-2 is required during embryogenesis for formation of the subset of muscles in the pharynx derived from the ABa blastomere, and tbx-2(ok529)−null mutants arrest shortly after hatching due to an inability to feed (Roy Chowdhuri et al. 2006; Smith and Mango 2007). tbx-2 mutants also exhibit defects in cell fate specification and differentiation of the hermaphrodite specific neurons and phasmid type B neurons and in olfactory adaptations (Miyahara et al. 2004; Singhvi et al. 2008). The hypomorphic mutant tbx-2(bx59) contains a missense mutation affecting a conserved residue in the “dimerization domain” of the T-box, and these animals exhibit partially penetrant, temperature-sensitive larval lethality (Huber et al. 2013). However, many of these mutants grow to adulthood, which allows the effect of reduced TBX-2 activity to be examined in later larvae and adults.
We are interested in mechanisms regulating TBX-2 activity in C. elegans. We have previously shown that tbx-2 gene transcription is directly repressed by the NF-Y transcription factor, and strong genetic interactions between mutants affecting NF-Y and tbx-2(bx59) indicate this repression is crucial for tbx-2 function in vivo (Milton et al. 2013). We also have shown that TBX-2 protein interacts with enzymes that attach the small ubiquitin modifier (SUMO) to target proteins and that TBX-2 can be SUMOylated in mammalian cell assays (Roy Chowdhuri et al. 2006; Huber et al. 2013). Moreover, reducing SUMOylation phenocopies tbx-2−null mutants and enhances the lethality and the severity of pharyngeal defects of tbx-2(bx59) mutants (Huber et al. 2013), and we hypothesize that SUMOylation is essential for TBX-2 function in vivo. TBX-2 can repress gene expression in cotransfection assays in mammalian cells, and a microarray comparison of wild-type and tbx-2(bx59) C. elegans embryos identified approximately 1000 genes that are upregulated and approximately 200 genes down-regulated in tbx-2(bx59) mutants, suggesting that TBX-2 predominantly functions as a transcriptional repressor.
Here we show that TBX-2 directly represses tbx-2 gene expression in a negative autoregulatory loop. Expression of the endogenous tbx-2 gene and a tbx-2 promoter::gfp reporter is increased in tbx-2 mutants and tbx-2(RNAi) animals, and ectopic tbx-2::gfp expression was found in the gut and seam cells of the lateral hypodermis in these animals. This repression was mediated by consensus T-box binding sites within the tbx-2 promoter, and TBX-2 binds these sites in vivo. To test whether SUMOylation is required for TBX-2 repression, we examined the effect of inhibiting SUMOylation on tbx-2::gfp expression. We observed ectopic Ptbx-2::gfp expression in the seam cells and gut similar to that observed in tbx-2 mutants, but we also observed more widespread Ptbx-2::gfp expression throughout the hypodermis. These results indicate that TBX-2 targets its own promoter in a negative autoregulatory loop, and suggests that TBX-2 and perhaps other factors repress tbx-2 expression in the hypodermis and gut through SUMO-dependent mechanisms.
Materials and Methods
Nematode handling, transformation, and strain construction
C. elegans were grown under standard conditions (Lewis and Fleming 1995). Germline transformation was performed by microinjection with the plasmid pRF4 carrying rol-6(su1006) (100 ng/μL) and various gfp reporters (10 ng/μL) (Mello and Fire 1995). The following strains were used in these studies: N2, OK0592 cuIs23[Ptbx-2::gfp] III (Milton et al. 2013), OK0611 cuIs25[Ptbx-2::gfp] X, OK0969 rrf-3(pk1426) II; cuIs23 III (Simmer et al. 2002), OK0660 tbx-2(bx59) III (Huber et al. 2013), OK0460 tbx-2(ok529)/dpy-17(e164) unc-32(e189) (Roy Chowdhuri et al. 2006), OK0970 tbx-2(bx59); cuIs25 X, OK0856 cuEx685[Ptbx-2prox::gfp], OK0857 cuEx686[Ptb-2prox::gfp], OK0912 cuEx730[Ptbx-2dist::gfp], OK0914 cuEx732[Ptbx-2dist::gfp], OK0916 cuEx734[Ptbx-2prox+dist::gfp], OK0919 cuEx737[Ptbx-2prox+dist::gfp], OK0873 tbx-2(ok529); wgIs159, OK0814 nfyb-1(cu13) II (Milton et al. 2013), OK0661 nfyb-1(cu13) II; tbx-2(bx59) III, OK0898 nfyb-1(cu13) II; tbx-2(bx59) III; cuIs25 X, and OK1030 tbx-2(ok529)/dpy-17(e164) unc-32(e189) III; cuIs25 X. OK1029 nfyb-1(cu13) II; cuIs25 X. Newly constructed strains containing tbx-2(ok529), tbx-2(bx59), or nfyb-1(cu13) were molecularly genotyped as previously described (Roy Chowdhuri et al. 2006; Huber et al. 2013; Milton et al. 2013).
General methods for nucleic acid manipulations and plasmid construction
Standard methods were used to manipulate plasmid DNAs and oligonucleotides (Ausubel 1990), and all plasmid sequences are available from the authors. The proximal T-box site was mutated in the Ptbx-2::gfp plasmid pOK206.33 (AGGTGGCA to ATTGTGC) using the Stratagene QuikChange II kit to produce Ptbx-2prox::gfp plasmid pOK255.04. A 42-bp fragment (−1157 to −1116 upstream of the tbx-2 ATG) containing the pair of distal T-box sites was deleted by inverse polymerase chain reaction (PCR) of pOK206.33 or pOK255.04 with primers PO1256 [GACTCTAGAAGTGATAAGAAGCCGCGAG] and PO1257 [GACTCTAGATGCAGCACTGAATTGATGA], digested with XbaI, and recircularized by ligation, to produce Ptbx-2dist::gfp plasmid pOK278.02 and Ptbx-2prox+dist::gfp plasmid pOK278.03, respectively. All plasmids were sequenced to verify the presence of the mutations.
Identification of candidate T-box binding sites in the tbx-2 promoter
Candidate TBX-2 binding sites were identified by scanning the Ptbx-2 promoter with the WormBase function Annotate Sequence Motif using the GBrowse plugin MotifFinder (www.wormbase.org; gmod.org/wiki/MotifFinder.pm) with a T-box half-site position frequency matrix derived from JASPAR MA0009.1 (jaspar.genereg.net) at a threshold of 0.85.
>T-box_half_site:
A [40 0 0 0 0 0 1 40 31 ]
C [ 0 0 0 0 0 2 7 0 5 ]
G [ 0 40 40 0 40 0 28 0 0 ]
T [ 0 0 0 40 0 38 4 0 4 ]
This position frequency matrix is nearly identical to those experimentally derived for T-box factors from the mouse and two echinoderm species (Badis et al. 2009; Cheatle Jarvela et al. 2014). This analysis identified the proximal and one distal T-box site at −259 and −1156, respectively (both AGGTGGCA). Subsequent analysis with the Uniprobe database (the_brain.bwh.harvard.edu/uniprobe/) with reduced threshold identified the second distal T-box site at −1135 (ATGTGTGA).
RNA interference (RNAi) analyses
The feeding RNAi screen was performed with the use of Escherichia coli strains expressing dsRNA from transcription factor genes on OK0592 cuIs23[Ptbx-2::gfp] III or OK0969 rrf-3(pk1426) II; cuIs23 III at 20° or 16°, respectively, as previously described (Milton et al. 2013). RNAi by injection of double-stranded RNA was performed as previously described (Fire et al. 1998). dsRNA produced by in vitro transcription (Ambion) of cDNA from tbx-2 (yk112c4/pOK165.07) was injected into OK0592 cuIs23[Ptbx-2::gfp] strain at approximately 200−500 ng/μL (Roy Chowdhuri et al. 2006). Injected animals were transferred to fresh plates every 12 hr and F1 progeny laid 24−48 hr postinjection (20°) were scored for altered Ptbx-2::gfp expression.
Quantification of tbx-2 mRNA levels using real-time PCR
Synchronized N2 and tbx-2(bx59) L1 larvae were prepared by bleaching mixed-stage animals and allowing the isolated embryos to grow 12 hr on OP50 seeded NGM plates at 20° (Lewis and Fleming 1995). Total RNA isolation and real-time PCR was performed as described previously (Milton et al. 2013). Exonic primers spanning introns in tbx-2 [PO1048 (TCAAAACGAGAAGGTGACGG); PO1072 (ATGTGTGGGTAGTGAAGCGG)] and the control mRNA ama-1 [PO1061 (AGGCGAAGGATGTGTTGTG); PO1062 (TCACCGTGTTCTTTGGGTC)] in real-time PCR reactions. Three to four replicates were performed for each RNA sample, and three independently isolated RNA samples were assayed for each genotype. The tbx-2 mRNA level relative to that of ama-1 was compared in N2 and tbx-2(bx59) animals using the 2-ΔΔCt method (Livak and Schmittgen 2001).
Chromatin Immunoprecipitation (ChIP) and quantification
ChIP assays were performed on mixed-stage N2 and OK0873 tbx-2(ok529); wgIs159 animals as described (Mukhopadhyay et al. 2008). Animals were fed in liquid culture with E. coli HB101 at 20°, harvested by centrifugation, Dounce homogenized at room temperature in phosphate-buffered saline + 2% formaldehyde, incubated 20 min, and quenched by adding glycine to 120 mM. Chromatin was sheared to 200−500 bp by sonication (Branson Sonifier) at 30% amplitude for 8 cycles of 12-se pulses (0.9 sec on, 0.1 sec off), with cooling between cycles, and 10% of the lysate was saved for input. Preclearing of the lysate was performed twice, once with Dynabeads protein G (Invitrogen) bound to 2.5 μg of IgG preserum followed by preclearing with Dynabeads alone. Immunoprecipitation reactions containing approximately 2−3 mg of total protein were performed with 3 μg of anti-green fluorescent protein (anti-GFP; Roche 11814460001) or IgG preserum (Invitrogen 02-6502). No antibody controls were performed in parallel to experimental assays. Three independent assays were performed for each strain.
Crosslinking was reversed and the precipitated DNA was analyzed by semiquantitative PCR for 33 and 35 cycles to verify linear amplification. Primers flanking the proximal T-box site [PO1307 (CATCCATCCATGGACCATTC); PO1308 (CGTTTTGCCGCTCTATGACT)], the distal T-box sites [PO1311 (TCAGTGCTGCAAATGTGTGA); PO1312 (CATTCTCGCGGCTTCTTATC)], or sax-2 [PO1394 (TGGATCATCAGTGTGTGCCT); PO1395 (AAATTCACGTTCGATCCTCG)] were used.
DNA was electrophoresed on 2–2.8% agarose gels and band intensities were measured using ImageJ (rsb.info.nih.gov/ij/). Gel images were inverted and background was subtracted with a rolling ball radius of 20. Enrichment of DNA was measured by averaging anti-GFP/Input or IgG/Input. Fold difference was calculated by difference of averaged GFP over IgG. Statistical significance was determined by two-tailed t-test with unequal variance comparing fold enrichment of the proximal or distal T-box sites to relative to sax-2 in N2 and tbx-2(ok529); wgIs159 using Microsoft Excel.
Microscopy
C. elegans were visualized using Zeiss Axioskop and AxioImager microscopes equipped for differential interference contrast and fluorescence microscopy. Images were captured using an Axiocam MRm camera and AxioVision software. Seam cells were identified by their position on the lateral surface of the animal and underlying the alae in L1 and adult animals (Altun and Hall 2009b). Hyp6 nuclei were identified by position in L1 animals (Altun and Hall 2009a).
Results
TBX-2 negatively regulates its own expression
We identified TBX-2 as a repressor of its own promoter in an RNAi-based screen where we knocked down expression of transcription factor genes in a strain containing a tbx-2 promoter gfp fusion (Ptbx-2::gfp) (Figure 1, A and B). In wild-type animals, Ptbx-2::gfp is expressed in the developing pharynx of embryos and becomes restricted to several neurons in the head near hatching, where expression persists through adulthood (Figure 1, C−F) (Roy Chowdhuri et al. 2006). We screened for changes in Ptbx-2::gfp expression in animals where 724 of the 924 predicted transcription factor genes were individually knocked down. Using this screen, we previously showed that knockdown of the NF-Y complex results in ectopic Ptbx-2::gfp expression in the gut and lateral hypodermal seam cells (Milton et al. 2013). We similarly found that tbx-2 knockdown also results in ectopic Ptbx-2::gfp expression in these tissues. Of note, none of the other 722 transcription factor genes examined in this RNAi screen produced ectopic Ptbx-2::gfp expression, indicating that loss of TBX-2 or NF-Y activity specifically derepresses expression of this reporter.
Figure 1.
Ptbx-2::gfp expression in embryos and late larvae. (A) Schematic diagram of the tbx-2 region contained in fosmid WRM063aG09 indicating the location of the 3.8-kb Ptbx-2 promoter fragment and the positions of consensus T-box binding sites within this fragment (red bars). (B) Diagram of a portion of the Ptbx-2 promoter indicating the positions of candidate T-box binding sites (red dots). For clarity the orientation of this fragment is reversed relative to (A) and only the region containing T-box sites is indicated. A promoter proximal site is located at −259 bp upstream of the tbx-2 ATG (AGGTGGCA, minus strand sequence), and a pair of closely spaced distal sites are located at -1135 bp (AGGTGGCA, plus strand) and −1156 bp (ATGTGTGA, plus strand) upstream of the tbx-2 ATG, respectively. (C, D) Green fluorescent protein (GFP; left) and differential interference contrast (DIC; right) images of Ptbx-2::gfp expression in the pharyngeal primordium in an embryo at the end of gastrulation (anterior to the left). (E, F) GFP (top) and DIC (bottom) images of an L4 animal expressing Ptbx-2::gfp in head neurons (arrows). Occasional GFP expression was also observed in tail neurons, but no expression was detected elsewhere. Gut cell cytoplasm contains autofluorescent gut granules (bracket).
We more carefully examined Ptbx-2::gfp expression in tbx-2(RNAi) animals and tbx-2 mutants. Although elimination of tbx-2 results in severe pharyngeal defects and L1 arrest, partially affected animals grow to adulthood (Roy Chowdhuri et al. 2006; Huber et al. 2013). We found that both tbx-2(RNAi) animals and mutants homozygous for the temperature-sensitive hypomorphic allele tbx-2(bx59) exhibited highly penetrant ectopic Ptbx-2::gfp expression in the seam cells and gut in late larval and adult stages (Table 1 and Figure 2). tbx-2(bx59) mutants exhibited this ectopic expression at both the permissive and nonpermissive temperatures (16° and 25°, respectively), suggesting that even a relatively small reduction in TBX-2 activity derepresses the tbx-2 promoter. This ectopic expression is specific for Ptbx-2::gfp, as we have examined expression of numerous other gfp reporters in tbx-2(bx59) mutants, including those regulated the D2096.6, T25E4.1, pqn-71, myo-5, nmgp-1, T12A7.6, cpn-4, and F41H10.5 promoters, but have never observed ectopic expression in the seam cells and gut [(Huber et al. 2013) and data not shown].
Table 1. Frequency of Ptbx-2::gfp expression in seam cells and gut.
| Genotype | % Animals Expressing GFP in Seam Cells | % Animals Expressing GFP in Gut | n |
|---|---|---|---|
| cuIs23[Ptbx-2::gfp]a | 0 | 6 | 148 |
| cuIs23[Ptbx-2::gfp]; tbx-2(RNAi)a,b | 97 | 100 | 228 |
| cuIs25[Ptbx-2::gfp]; tbx-2(bx59)a @16° | 44 | 98 | 59 |
| cuIs25[Ptbx-2::gfp]; tbx-2(bx59)a @25° | 100 | 100 | 100 |
| cuEx686[Ptbx-2prox::gfp]c | 32 | 60 | 77 |
| cuEx685Ptbx-2prox::gfp c | 33 | 53 | 164 |
| cuEx730[Ptbx-2dist::gfp]c | 7 | 98 | 111 |
| cuEx732[Ptbx-2dist::gfp]c | 2 | 98 | 178 |
| cuEx734Ptbx-2prox+dist::gfpc | 1 | 96 | 168 |
| cuEx737[Ptbx-2prox+dist::gfp]c | 0 | 99 | 148 |
GFP, green fluorescent protein.
GFP expression was scored in L4 to young adult hermaphrodites.
tbx-2(RNAi) was performed by feeding animals E. coli expressing tbx-2 dsRNA.
GFP expression was scored in L3 to young adult hermaphrodites.
Figure 2.
Ptbx-2::gfp is ectopically expressed in animals with reduced tbx-2 expression. Green fluorescence protein (GFP) and differential interference contrast (DIC) images of L4 and young adult animals of the indicated genotypes expressing Ptbx-2::gfp. Ectopic Ptbx-2::gfp expression detectable in seam cells (arrowheads) and gut nuclei (arrows) are indicated in tbx-2(RNAi) (C, D) and tbx-2(bx59) mutants (E, F).
In humans, T-box gene mutations often are haploinsufficient, with the loss of one functional allele underlying congenital diseases such as Holt-Oram and DiGeorge syndromes (Takashima and Suzuki 2013). Although tbx-2(ok529) null mutants are completely viable as heterozygotes (Roy Chowdhuri et al. 2006), we found that tbx-2(ok529)/+ heterozygotes exhibited ectopic Ptbx-2::gfp expression in the seam cells (Figure 3). No ectopic expression was observed in the gut of these animals. Thus, tbx-2(ok529) also exhibits a mild haploinsufficient phenotype.
Figure 3.

Ectopic Ptbx-2::gfp expression in tbx-2(ok529)/+ heterozygotes. Green fluorescence protein (left) and differential interference contrast (right) images of a young adult tbx-2(ok529)/+ heterozygote ectopically expressing Ptbx-2::gfp in seam cells (arrowheads). tbx-2(ok529) is a null allele containing a 1.1-kb deletion that removes sequences encoding the C-terminus of the T-box DNA-binding domain (Roy Chowdhuri et al. 2006).
tbx-2(ok529) homozygotes and approximately half of tbx-2(bx59) animals grown at the nonpermissive temperature arrest as L1 larvae, and we examined Ptbx-2::gfp expression in these L1 animals. At this stage, no ectopic Ptbx-2::gfp was detectable in the seam cells, and only occasional expression was observed in the gut. However, these animals exhibited a strong up-regulation of Ptbx-2::gfp expression in the head neurons where Ptbx-2::gfp is normally expressed (Figure 4, A−C).
Figure 4.
Ectopic Ptbx-2::gfp expression detectable in the gut in tbx-2(bx59) and nfyb-1(cu13); tbx-2(bx59) L1 animals. Green fluorescence protein (GFP) and differential interference contrast (DIC) images of L1 animals of the indicated genotypes expressing Ptbx-2::gfp. GFP expressing head neurons (arrowheads), gut cells (arrows), hyp6 (lines), and tail neurons (t) are marked. The microscopy setup was identical for each of these images, and the exposure time for each fluorescence image is indicated in milliseconds (ms). Note that Ptbx-2::gfp is very highly expressed in head neurons in tbx-2 and nfyb-1 single and double mutants, and, when compared to wild-type animals, the exposure times are much shorter.
We next asked whether TBX-2 negatively regulates the endogenous tbx-2 gene by using quantitative real-time PCR to compare tbx-2 mRNA levels in wild-type and tbx-2(bx59) L1 animals grown at the nonpermissive temperature. We found tbx-2 mRNA was overexpressed fourfold in tbx-2(bx59) compared with the wild type (Table 2). These results are consistent with those of a genome-wide microarray showing tbx-2 mRNA levels are increased in tbx-2(bx59) mutants (Huber et al. 2013). Although these results using whole worm RNA cannot determine whether endogenous tbx-2 is up-regulated in the same tissues where we observe ectopic Ptbx-2::gfp expression, they do demonstrate the endogenous gene is subject to negative autoregulation.
Table 2. Endogenous tbx-2 expression levels.
| Genotype | Relative tbx-2 mRNA expression ± SD |
|---|---|
| +/+ | 1.00 ± 0.06 |
| tbx-2(bx59) | 4.00 ± 0.01 |
Collectively, these results indicate TBX-2 is a dose-dependent regulator of its own promoter. We suggest this negative autoregulation of the tbx-2 promoter functions to precisely control tbx-2 gene expression.
T-box binding sites mediate tbx-2 repression
Most T-box factors bind similar sites related to AGGTGTGA (Wilson and Conlon 2002), and we identified three potential T-box binding sites in the Ptbx-2 promoter fragment (Figure 1A). We asked whether these sites are involved in repression of Ptbx-2::gfp and found mutation of the promoter proximal site (Ptbx-2::gfpprox) or deletion of a pair of closely linked distal sites (Ptbx-2::gfpdist) led to ectopic expression in seam cells and gut in a pattern similar to that observed when wild-type Ptbx-2::gfp expression was examined in tbx-2 mutants and tbx-2(RNAi) animals (Table 1). Although these results show that both the proximal and distal sites repress tbx-2 promoter activity, we also found that the proximal and distal sites have distinct effects on expression. Two independent transgenic lines expressed Ptbx-2prox::gfp at moderate frequency in both the seam cells and gut. In contrast, nearly every transgenic animal from independent lines expressed Ptbx-2dist::gfp in the gut, whereas relatively few of these animals expressed this construct in the seam cells. We expected that a double mutant containing both a disrupted proximal site and deleted distal sites (Ptbx-2prox+dist) would exhibit an additive expression pattern, but Ptbx-2prox+dist was expressed in a pattern very similar to the Ptbx-2::gfpdist single mutant, with very frequent expression in the gut but very little expression in the seam cells (Table 1). Together, these results indicate that both the proximal and distal T-box sites repress Ptbx-2::gfp expression, although their roles differ in seam cells and gut.
TBX-2 directly represses its own promoter in vivo
The modENCODE Consortium provided a tbx-2 transgene wgIs159 containing wild-type tbx-2 in the fosmid WRM063aG09 tagged at the 3′-end with gfp (Sarov et al. 2012). wgIs159 expresses a full-length TBX::2GFP fusion protein, and we have found that it rescues the null mutant tbx-2(ok529) (P. Huber and P. Okkema, unpublished data). The modENCODE Consortium has used similar transgenes to map transcription factor binding sites throughout the genome by ChIP using anti-GFP antibodies (Niu et al. 2011).
We performed ChIP on mixed-stage tbx-2(ok529); wgIs159 hermaphrodites and found TBX-2 specifically binds both the proximal and distal sites in the tbx-2 promoter (Figure 5). Primers flanking the proximal and distal sites were used to amplify DNA isolated from chromatin immunoprecipitated with an anti-GFP antibody or with nonspecific IgG, or in mock precipitations with no antibody. Because wgIs159 contains multiple copies of the tbx-2 gene that increase the amount of DNA immunoprecipitated in ChIPs (Niu et al. 2011), we also examined binding to a region of the sax-2 gene, which is also located in WRM063aG09 and does not contain any predicted T-box binding sites. The proximal and distal sites were enriched 7.1-fold and 2.7-fold when precipitated with anti-GFP, respectively, compared with precipitations with IgG, and this enrichment was significantly more than that observed for the sax-2 site (Figure 5, A and C). In comparison, neither of these sites was enriched in wild-type animals lacking TBX-2::GFP protein (Figure 5, B and C). We conclude that TBX-2 specifically binds the proximal and distal T-box sites in vivo and that TBX-2 binding at these sites directly represses tbx-2 expression in a negative autoregulatory loop.
Figure 5.
TBX-2 binds T-box sites in the tbx-2 promoter in vivo. Semiquantitative polymerase chain reaction analyses of representative chromatin immunoprecipitation from tbx-2(ok529); wgIs159 animals expressing a TBX-2::GFP fusion protein (A) and wild-type N2 animals lacking any GFP (B) immunoprecipitated with anti-GFP, nonspecific IgG or no antibody as indicated. 0.25% of the total protein lysate used in the immunoprecipitation reactions is shown as input. (C) Bar graph indicating the amount of product precipitated with anti-GFP relative to the amount precipitated with IgG. Data represents the average for 3 experiments with error bars indicating standard deviation. P < 0.03 (*) or P < 0.01 (**). GFP, green fluorescent protein.
Repression tbx-2 expression requires SUMOylation
Our previous biochemical and genetic evidence led us to hypothesize that TBX-2 function is affected by SUMOylation (Roy Chowdhuri et al. 2006; Huber et al. 2013), and this hypothesis predicts that reducing SUMOylation levels will affect Ptbx-2::gfp expression similarly to reducing TBX-2 activity. We performed RNAi to knockdown expression of the E2 SUMO conjugating enzyme UBC-9, the E3 SUMO ligase GEI-17, and the SUMO SMO-1 in a transgenic strain expressing Ptbx-2::gfp. RNAi performed by injection of ubc-9 or smo-1 dsRNA these genes produces highly penetrant embryonic lethality (Jones et al. 2002; Roy Chowdhuri et al. 2006) and would prevent us from examining Ptbx-2::gfp expression in larvae. Therefore, RNAi was performed by feeding animals E. coli expressing dsRNA to reduce the lethality of ubc-9(RNAi) and smo-1(RNAi) (Kamath and Ahringer 2003). We observed significant embryonic lethality in these experiments, indicating that the RNAi treatment was effective, but enough animals progressed through larval development to allow GFP expression to be examined in L4s and young adults.
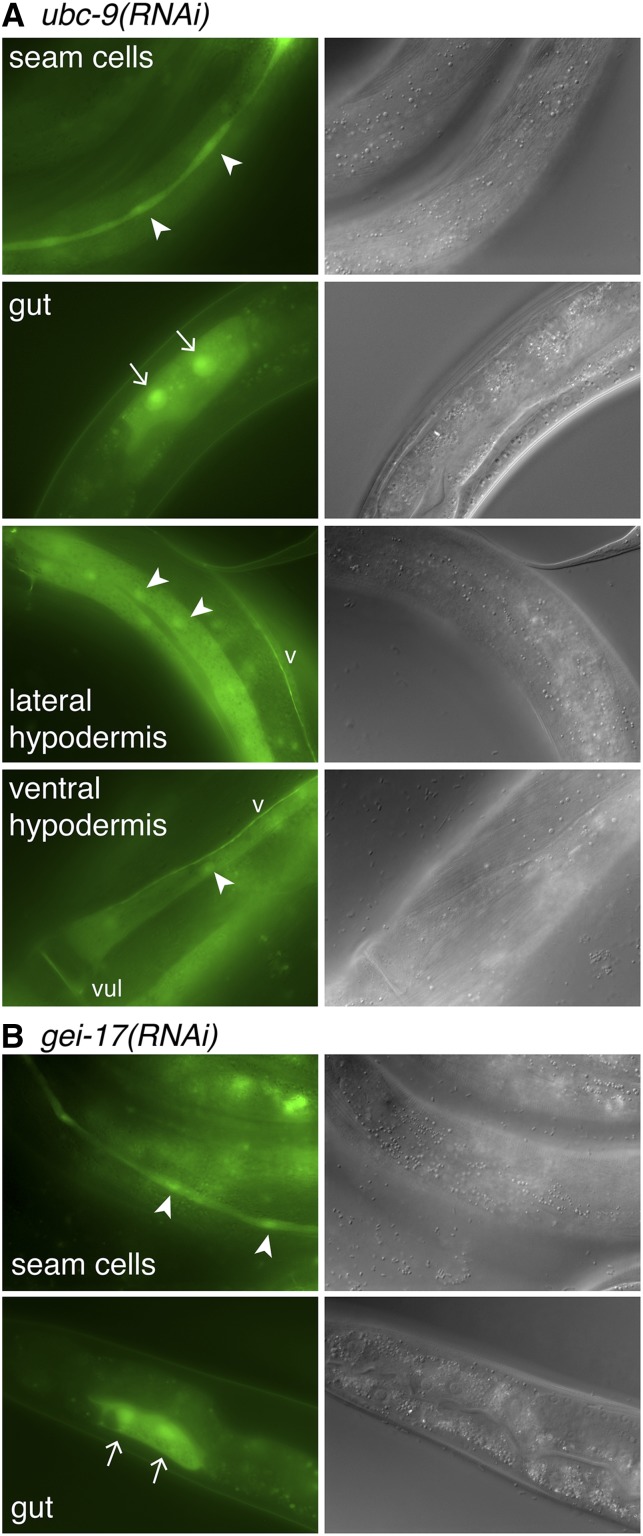
We predicted that reducing SUMOylation would lead to ectopic Ptbx-2::gfp expression in seam cells and the gut in a pattern similar to what we observed in tbx-2(RNAi) and tbx-2 mutants, and we did observe Ptbx-2::gfp expression in both of these tissues in ubc-9(RNAi) and gei-17(RNAi) animals (Table 3 and Figure 6). In comparison, ectopic Ptbx-2::gfp expression was found in the seam cells of smo-1(RNAi) animals but was not observed in the gut. Surprisingly, we also observed frequent ectopic Ptbx-2::gfp expression in the nonseam, syncytial hypodermis in ubc-9(RNAi) and smo-1(RNAi) animals, and at a low frequency in gei-17(RNAi) animals (Figure 6 and Table 3). In addition, occasional Ptbx-2::gfp expression also was observed in the ventral nerve cord. Hypodermal expression typically was found in the lateral and ventral regions of the hyp7 syncytium flanking the seam cells. This pattern of ectopic Ptbx-2::gfp in the nonseam hypodermis and ventral nerve cord was only very rarely observed in tbx-2 mutants, suggesting the tbx-2 promoter may be repressed by additional SUMO-dependent mechanisms in these tissues. Alternatively, because the majority of hyp7 nuclei are derived from fusion with seam cell descendants (Sulston and Horvitz 1977), this ectopic expression could result from stable changes in tbx-2 promoter activity that are maintained as seam cell nuclei join the hyp7 syncytium.
Table 3. Ectopic Ptbx-2::gfp expression in animals with reduced SUMOylation.
| Genotypea | % Animals Expressing GFP in Seam Cells | % Animals Expressing GFP in Gut | % Animals Expressing GFP in Syncytial Hypodermis | n |
|---|---|---|---|---|
| Ptbx-2::gfp | 0 | 6 | 0 | 148 |
| Ptbx-2::gfp; tbx-2(RNAi) | 97 | 100 | 0 | 228 |
| Ptbx-2::gfp; ubc-9(RNAi) | 24 | 12 | 67 | 101 |
| Ptbx-2::gfp; gei-17(RNAi) | 40 | 87 | 6 | 100 |
| Ptbx-2::gfp; smo-1(RNAi) | 24 | 0 | 81 | 143 |
SUMO, small ubiquitin modifier; GFP, green fluorescent protein.
Expression was scored in L4 to young adult hermaphrodites.
Figure 6.
Disruption of SUMOylation causes misexpression of Ptbx-2::gfp. Green fluorescence protein (GFP; left) and differential interference contrast (right) images of young adult ubc-9(RNAi) (A) and gei-17(RNAi) (B) animals ectopically expressing Ptbx-2::gfp ectopically in the indicated tissues. Nuclei of GFP-expressing seam cells, lateral hypodermis, and ventral hypodermis are marked with arrowheads, whereas nuclei of GFP expressing gut cells are marked with arrows. ‘v’ marks the ventral nerve cord, and ‘vul’ marks the vulva on the ventral surface of the animal.
Ptbx-2::gfp expression is not expanded in nfyb-1(cu13); tbx-2(bx59) double mutants
We have previously shown that the NF-Y complex also represses Ptbx-2::gfp expression in the seam cells and gut similarly to TBX-2 (Milton et al. 2013). To ask whether NF-Y and TBX-2 function in parallel to repress the tbx-2 promoter, we compared Ptbx-2::gfp expression in nfyb-1(cu13); tbx-2(bx59) double mutants with that of wild-type animals and tbx-2(bx59) and nfyb-1(cu13) single mutants. nfyb-1(cu13); tbx-2(bx59) double mutants exhibit temperature-sensitive, synthetic lethality at the L1 stage, so we examined expression in L1 animals grown at the nonpermissive temperature (25°).
In wild-type L1s, Ptbx-2::gfp was expressed most frequently in head neurons with less frequent expression in the anterior-most seam cell H0 and the syncytial head hypodermal cell hyp6 (Figure 4A and Table 4). Both tbx-2(bx59) and nfyb-1(cu13) single mutants exhibited a strong increase in the level of Ptbx-2::gfp expression in head neurons (Figure 4, C and D). In addition these mutants exhibited a moderately increased frequency of Ptbx-2::gfp expression in hyp6, and a small but significant increase in the number of neurons in the head and tail that expressed this construct (Table 4 and Table 5). Notably all nfyb-1(cu13) mutants exhibited Ptbx-2::gfp expression in H0 and the gut. The nfyb-1(cu13); tbx-2(bx59) double mutant expressed Ptbx-2::gfp in a pattern that was very similar to the nfyb-1(cu13) single mutant, with very strong expression in the head neurons and frequent expression in the gut (Figure 4E and Table 4). There was also a small but significant increase in the number of Ptbx-2::gfp expressing neurons in the head and tail expression that might result from additive effects of these mutants. No additional expression was observed in other tissues in the double mutants. These results strongly suggest that NF-Y and TBX-2 do not function in independent parallel pathways to repress the tbx-2 promoter. Furthermore, they suggest that synthetic lethality of the nfyb-1(cu13); tbx-2(bx59) double mutant does not result from synergistic increase in tbx-2 promoter activity.
Table 4. Ptbx-2::gfp ectopic expression in tbx-2(bx59) and nfyb-1(cu13) mutants at the L1 stage.
| Genotype | Percent L1 Animals Expressing Ptbx-2::gfp in: | n | ||||
|---|---|---|---|---|---|---|
| Head Neurons | H0 Seam Cells | Gut | Tail Neurons | Hyp6 | ||
| cuIs23[Ptbx-2::gfp] | 100 | 14 | 0 | 0 | 5 | 56 |
| tbx-2(bx59); cuIs25[Ptbx-2::gfp] | 100 | 38 | 2 | 73 | 13 | 60 |
| nfyb-1(cu13); cuIs25[Ptbx-2::gfp] | 100 | 100 | 100 | 55 | 21 | 61 |
| nfyb-1(cu13); tbx-2(bx59); cuIs25[Ptbx-2::gfp] | 100 | 100 | 100 | 100 | 52 | 50 |
Table 5. Number of Ptbx-2::gfp-expressing neurons.
| Genotype | Head Neurons | Tail Neurons | n |
|---|---|---|---|
| cuIs23[Ptbx-2::gfp] | 5.40 ± 0.7 | 0 ± 0 | 59 |
| tbx-2(bx59); cuIs25[Ptbx-2::gfp] | 6.69 ± 1.7 | 1.23 ± 0.9 | 59 |
| nfyb-1(cu13); cuIs25[Ptbx-2::gfp] | 7.17 ± 1.4 | 0.85 ± 0.9 | 60 |
| nfyb-1(cu13); tbx-2(bx59); cuIs25[Ptbx-2::gfp] | 7.76 ± 1.2 | 2.65 ± 1.2 | 47 |
Discussion
Here we show that TBX-2 binds sites within its own promoter and negatively autoregulates its own expression. Knockdown of TBX-2 activity or mutation of T-box binding sites within the promoter leads to ectopic Ptbx-2::gfp expression in the hypodermal seam cells and the gut of larvae and adults and increased expression in head neurons in the L1 stage. TBX-2 protein binds these sites in the tbx-2 promoter in vivo, indicating this negative autoregulation is direct. This regulation also affects endogenous tbx-2 expression as tbx-2 mRNA is increased in tbx-2 mutants, although genome editing experiments could be done to verify this regulation occurs through the sites we have identified using Ptbx-2::gfp . SUMOylation is required for tbx-2 repression, as RNAi knockdown of the UBC-9 E2 SUMO-conjugating enzyme, the GEI-17 E3 SUMO-ligase, or the SMO-1 SUMO peptide also resulted in ectopic Ptbx-2::gfp expression. These studies identify tbx-2 as the first target regulated by TBX-2, and they demonstrate directly that TBX-2 functions as a transcriptional repressor whose function in vivo depends on SUMOylation. We note that using the Ptbx-2::gfp reporter as a molecular readout for TBX-2 activity facilitated our ability to detect defects in negative autoregulation, because it uncouples increased promoter activity from increased TBX-2 repressor activity.
TBX-2 negatively autoregulates its own promoter
Negative autoregulation allows rapid transcriptional response time and reduces cell-to-cell variability in gene expression, and it can provide a linear response of target genes to the dose of key transcription factors (Alon 2007; Nevozhay et al. 2009). Negative autoregulation is observed for over 50% of the transcription factors in E. coli (Alon 2007), and it plays important roles in animal development (Crews and Pearson 2009). For example in the Drosophila embryo, the Hox factor Ubx maintains its expression or even completely represses its expression in different tissues through negative autoregulation (Irvine et al. 1993). Likewise, in the mouse and zebrafish, time delayed, negative autoregulation of basic helix-loop-helix (bHLH) transcription factors leads to oscillating gene expression levels during somitogenesis (Hirata et al. 2002; Lewis 2003). Negative autoregulation of transcription factors and signaling molecules is frequent in Ciona and is believed to be a key developmental feature of gene regulation (Imai et al. 2006).
T-box genes frequently are expressed in highly dynamic patterns, and their expression levels are critical for normal function (Naiche et al. 2005). Negative autoregulation may be one mechanism to precisely control T-box gene expression, and it has been observed for human TBX22 and the Ciona T-box genes Brachyury, Tbx2/3, and Tbx6b/c/d (Imai et al. 2006; Andreou et al. 2007). Our studies indicate that TBX-2 negatively regulates its promoter in the head neurons, where Ptbx-2::gfp is normally expressed, and in the seam cells and gut, where Ptbx-2::gfp expression is not detected. We suggest that negative autoregulation in the neurons maintains tbx-2 expression at levels to appropriately regulate downstream target genes. Low level expression of endogenous tbx-2 also has been detected in the hypodermis, which includes the seam cells, and the gut (Spencer et al. 2011), and we suggest that negative autoregulation reversibly represses tbx-2 expression perhaps to allow rapid tbx-2 induction under specific developmental or environmental conditions. Although the seam and gut cells are not lineally related (Sulston et al. 1983), they do undergo DNA replication at each larval stage (Sulston and Horvitz 1977; Hedgecock and White 1985), and we have speculated previously that tbx-2 may be primed for expression in these tissues to promote DNA replication (Milton et al. 2013).
Although knockdown of tbx-2 results in strong derepression of Ptbx-2::gfp in both seam cells and gut, mutation of TBX-2 binding sites in the tbx-2 promoter differently affects Ptbx-2::gfp expression in these tissues. Mutation of the proximal T-box binding site leads to ectopic expression in both the seam cells and gut, whereas deletion of the distal sites leads to frequent ectopic expression only in the gut. The double mutant containing both the proximal site mutation and deletion of the distal sites did not exhibit an additive phenotype; rather, it behaved identically to the single mutant containing the distal site deletion. These results could be explained if deletion of the distal sites not only affects TBX-2 repression but also removes a site activating expression in seam cells. We have not identified potential binding sites in this region for factors known to promote seam cell proliferation or differentiation, including GATA- and Runt-family factors (Koh and Rothman 2001; Nimmo et al. 2005; Smith et al. 2005), implying another seam cell regulatory factor might bind this deleted region.
tbx-2 gene repression requires SUMOylation
Our previous results led us to hypothesize that TBX-2 functions as a SUMOylation-dependent transcriptional repressor (Roy Chowdhuri et al. 2006; Huber et al. 2013). TBX-2 binds the E2 SUMO-conjugating enzyme UBC-9 and the E3 SUMO ligase GEI-17 in yeast 2-hybrid assays, and TBX-2 can be SUMOylated in mammalian cells. In C. elegans, RNAi knockdown of ubc-9 phenocopies tbx-2 null mutants, whereas gei-17 knockdown phenocopies weak tbx-2 mutants. Moreover, partially reducing ubc-9 or the smo-1 SUMO enhances the lethality and pharyngeal defects in the tbx-2(bx59) hypomorphic mutant, indicating SUMOylation is crucial for TBX-2 function.
Here we use the tbx-2 promoter as a direct readout of TBX-2 repressor activity to provide additional genetic evidence that TBX-2 requires SUMOylation in vivo. We found that knocking down expression of ubc-9, smo-1, or gei-17 derepresses Ptbx-2::gfp expression similarly to what we observed in tbx-2 mutants and tbx-2(RNAi). Although these results are consistent with our hypothesis, we cannot rule out the possibility that other SUMO-dependent mechanisms repress tbx-2. We did observe tissue specific differences in this derepression in these experiments. For example, while tbx-2(RNAi), ubc-9(RNAi) and gei-17(RNAi) resulted in ectopic Ptbx-2::gfp expression in both seam cells and the gut, smo-1(RNAi) resulted in expression in seam cells but not in gut. This difference might result from less efficient RNAi knockdown of smo-1 in gut than in seam cells. More interestingly, we also observed ectopic expression of Ptbx-2::gfp in the syncytial hypodermis when SUMOylation was knocked down, particularly in ubc-9(RNAi) or smo-1(RNAi) animals, but this expression was only very rarely observed in tbx-2 mutants. This pattern of ectopic Ptbx-2::gfp expression may indicate that additional SUMO-dependent mechanisms repress expression in the syncytial hypodermis. Such mechanisms could involve transcription factors that directly repress Ptbx-2::gfp expression in the syncytial hypodermis, or mechanisms that indirectly affect Ptbx-2::gfp by affecting normal seam cell or hypodermal differentiation. These mechanisms could affect factors such as the PcG component SOP-2 or the nuclear hormone receptor NHR-25, which are SUMOylated and play important roles in seam cell and hypodermal differentiation (Zhang et al. 2004; Silhankova et al. 2005; Cai et al. 2008; Ward et al. 2013). Alternatively, as the majority of the nuclei in the syncytial hypodermis are derived from fusion with seam cell descendants (Sulston and Horvitz 1977), this ectopic expression could result from stable changes in tbx-2 promoter activity in seam cell nuclei.
Two mechanisms repressing tbx-2
We have identified the NF-Y complex and TBX-2 itself as negative regulators of tbx-2 expression (this work; (Milton et al. 2013)). In both cases, these factors directly target the tbx-2 promoter and repress Ptbx-2::gfp expression in the seam cells and gut. We did not observe increased expression of Ptbx-2::gfp expression in nfyb-1(cu13); tbx-2(bx59) double mutants compared to each of the single mutants, strongly suggesting NF-Y and TBX-2 function together to repress Ptbx-2::gfp expression rather than through independent, parallel pathways. NF-Y has been suggested to function as a “pioneer” factor that binds its sites in chromatin, regardless of its modification state, and recruits histone modifiers to facilitate binding of other transcription factors (Fleming et al. 2013). In the tbx-2 promoter, the proximal T-box site is located ∼65 bp from a functional NF-Y binding site (Milton et al. 2013), and we suggest NF-Y binding may facilitate TBX-2 binding to the proximal site and repressing tbx-2 promoter activity.
nfyb-1(cu13); tbx-2(bx59) double mutants exhibit a highly penetrant synthetic lethal phenotype indicating these factors have common functions in vivo, but we do not yet know the cause of this lethality (Milton et al. 2013). One possibility is that these factors have partially overlapping function regulating additional downstream genes, and loss of both TBX-2 and NF-Y regulation leads to double mutant lethality. This and other hypotheses can be tested by identifying and characterizing addition genes regulated by these factors.
Acknowlegments
The authors are indebted to Paul Huber, Alena Kozlova, Mihail Sarov, and Valerie Reinke for reagents, strains, and advice, and to Paul Huber and two anonymous reviewers for critical comments on the manuscript. This project was supported by National Institutes of Health (NIH) grant R01 GM82865 (P.G.O.), an NIH Kirschstein NRSA F31 GM090675 (A.C.M.), and a UIC LAS Award for Faculty in the Natural Sciences. Some strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). We also thank the Research Open Access Publishing (ROAAP) Fund of the University of Illinois at Chicago for financial support towards the open access publishing fee for this article.
Footnotes
Communicating editor: B. J. Andrews
Literature Cited
- Abrahams A., Parker M. I., Prince S., 2010. The T-box transcription factor Tbx2: its role in development and possible implication in cancer. IUBMB Life 62: 92–102. [DOI] [PubMed] [Google Scholar]
- Alon U., 2007. Network motifs: theory and experimental approaches. Nat. Rev. Genet. 8: 450–461. [DOI] [PubMed] [Google Scholar]
- Altun, Z. F., and D. H. Hall, 2009a Epithelial system, hypodermis, in WormAtlas. 10.3908/wormatlas.1.13. Available at http://www.wormatlas.org/hermaphrodite/hypodermis/mainframe.htm. Accessed: April 20, 2015.
- Altun, Z. F., and D. H. Hall, 2009b Epithelial system, seam cells, in WormAtlas. 10.3908/wormatlas.1.14. Available at http://www.wormatlas.org/hermaphrodite/seam%20cells/mainframe.htm. Accessed: April 20, 2015.
- Andreou A. M., Pauws E., Jones M. C., Singh M. K., Bussen M., et al. , 2007. TBX22 missense mutations found in patients with X-linked cleft palate affect DNA binding, sumoylation, and transcriptional repression. Am. J. Hum. Genet. 81: 700–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel F. M., 1990. Current Protocols in Molecular Biology. Wiley, New York. [Google Scholar]
- Badis G., Berger M. F., Philippakis A. A., Talukder S., Gehrke A. R., et al. , 2009. Diversity and complexity in DNA recognition by transcription factors. Science 324: 1720–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Q., Sun Y., Huang X., Guo C., Zhang Y., et al. , 2008. The Caenorhabditis elegans PcG-like gene sop-2 regulates the temporal and sexual specificities of cell fates. Genetics 178: 1445–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheatle Jarvela A. M., Brubaker L., Vedenko A., Gupta A., Armitage B. A., et al. , 2014. Modular evolution of DNA-binding preference of a Tbrain transcription factor provides a mechanism for modifying gene regulatory networks. Mol. Biol. Evol. 31: 2672–2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conlon F. L., Sedgwick S. G., Weston K. M., Smith J. C., 1996. Inhibition of Xbra transcription activation causes defects in mesodermal patterning and reveals autoregulation of Xbra in dorsal mesoderm. Development 122: 2427–2435. [DOI] [PubMed] [Google Scholar]
- Crews S. T., Pearson J. C., 2009. Transcriptional autoregulation in development. Curr. Biol. 19: R241–R246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A., Xu S., Montgomery M. K., Kostas S. A., Driver S. E., et al. , 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391: 806–811. [DOI] [PubMed] [Google Scholar]
- Fleming J. D., Pavesi G., Benatti P., Imbriano C., Mantovani R., et al. , 2013. NF-Y coassociates with FOS at promoters, enhancers, repetitive elements, and inactive chromatin regions, and is stereo-positioned with growth-controlling transcription factors. Genome Res. 23: 1195–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedgecock E. M., White J. G., 1985. Polyploid tissues in the nematode Caenorhabditis elegans. Dev. Biol. 107: 128–133. [DOI] [PubMed] [Google Scholar]
- Hirata H., Yoshiura S., Ohtsuka T., Bessho Y., Harada T., et al. , 2002. Oscillatory expression of the bHLH factor Hes1 regulated by a negative feedback loop. Science 298: 840–843. [DOI] [PubMed] [Google Scholar]
- Huber P., Crum T., Clary L. M., Ronan T., Packard A. V., et al. , 2013. Function of the C. elegans T-box factor TBX-2 depends on SUMOylation. Cell. Mol. Life Sci. 70: 4157–4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai K. S., Levine M., Satoh N., Satou Y., 2006. Regulatory blueprint for a chordate embryo. Science 312: 1183–1187. [DOI] [PubMed] [Google Scholar]
- Irvine K. D., Botas J., Jha S., Mann R. S., Hogness D. S., 1993. Negative autoregulation by Ultrabithorax controls the level and pattern of its expression. Development 117: 387–399. [DOI] [PubMed] [Google Scholar]
- Jones D., Crowe E., Stevens T. A., Candido E. P., 2002. Functional and phylogenetic analysis of the ubiquitylation system in Caenorhabditis elegans: ubiquitin-conjugating enzymes, ubiquitin-activating enzymes, and ubiquitin-like proteins. Genome Biol. 3: RESEARCH0002.0001–0002.0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath R. S., Ahringer J., 2003. Genome-wide RNAi screening in Caenorhabditis elegans. Methods 30: 313–321. [DOI] [PubMed] [Google Scholar]
- Koh K., Rothman J. H., 2001. ELT-5 and ELT-6 are required continuously to regulate epidermal seam cell differentiation and cell fusion in C. elegans. Development 128: 2867–2880. [DOI] [PubMed] [Google Scholar]
- Lewis J., 2003. Autoinhibition with transcriptional delay: a simple mechanism for the zebrafish somitogenesis oscillator. Curr. Biol. 13: 1398–1408. [DOI] [PubMed] [Google Scholar]
- Lewis J. A., Fleming J. T., 1995. Basic culture methods, pp. 4–30 in Caenorhabditis elegans: Modern Biological Analysis of an Organism, edited by Epstein H. F., Shakes D. C. Academic Press, San Diego, CA. [Google Scholar]
- Livak K. J., Schmittgen T. D., 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)). Method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Mello C., Fire A., 1995. DNA transformation, pp. 451–482 in Caenorhabditis elegans: Modern Biological Analysis of an Organism, edited by Epstein H. F., Shakes D. C.. Academic Press, San Diego, CA. [Google Scholar]
- Milton A. C., Packard A. V., Clary L., Okkema P. G., 2013. The NF-Y complex negatively regulates Caenorhabditis elegans tbx-2 expression. Dev. Biol. 382: 38–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitani Y., Takahashi H., Satoh N., 2001. Regulation of the muscle-specific expression and function of an ascidian T-box gene, As-T2. Development 128: 3717–3728. [DOI] [PubMed] [Google Scholar]
- Miyahara K., Suzuki N., Ishihara T., Tsuchiya E., Katsura I., 2004. TBX2/TBX3 transcriptional factor homologue controls olfactory adaptation in Caenorhabditis elegans. J. Neurobiol. 58: 392–402. [DOI] [PubMed] [Google Scholar]
- Mori A. D., Zhu Y., Vahora I., Nieman B., Koshiba-Takeuchi K., et al. , 2006. Tbx5-dependent rheostatic control of cardiac gene expression and morphogenesis. Dev. Biol. 297: 566–586. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay A., Deplancke B., Walhout A. J., Tissenbaum H. A., 2008. Chromatin immunoprecipitation (ChIP) coupled to detection by quantitative real-time PCR to study transcription factor binding to DNA in Caenorhabditis elegans. Nat. Protoc. 3: 698–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naiche L. A., Harrelson Z., Kelly R. G., Papaioannou V. E., 2005. T-box genes in vertebrate development. Annu. Rev. Genet. 39: 219–239. [DOI] [PubMed] [Google Scholar]
- Nevozhay D., Adams R. M., Murphy K. F., Josic K., Balazsi G., 2009. Negative autoregulation linearizes the dose-response and suppresses the heterogeneity of gene expression. Proc. Natl. Acad. Sci. USA 106: 5123–5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimmo R., Antebi A., Woollard A., 2005. mab-2 encodes RNT-1, a C. elegans Runx homologue essential for controlling cell proliferation in a stem cell-like developmental lineage. Development 132: 5043–5054. [DOI] [PubMed] [Google Scholar]
- Niu W., Lu Z. J., Zhong M., Sarov M., Murray J. I., et al. , 2011. Diverse transcription factor binding features revealed by genome-wide ChIP-seq in C. elegans. Genome Res. 21: 245–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pocock R., Ahringer J., Mitsch M., Maxwell S., Woollard A., 2004. A regulatory network of T-box genes and the even-skipped homologue vab-7 controls patterning and morphogenesis in C. elegans. Development 131: 2373–2385. [DOI] [PubMed] [Google Scholar]
- Roy Chowdhuri S., Crum T., Woollard A., Aslam S., Okkema P. G., 2006. The T-box factor TBX-2 and the SUMO conjugating enzyme UBC-9 are required for ABa-derived pharyngeal muscle in C. elegans. Dev. Biol. 295: 664–677. [DOI] [PubMed] [Google Scholar]
- Sarov M., Murray J. I., Schanze K., Pozniakovski A., Niu W., et al. , 2012. A genome-scale resource for in vivo tag-based protein function exploration in C. elegans. Cell 150: 855–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebe-Pedros A., Ariza-Cosano A., Weirauch M. T., Leininger S., Yang A., et al. , 2013. Early evolution of the T-box transcription factor family. Proc. Natl. Acad. Sci. USA 110: 16050–16055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silhankova M., Jindra M., Asahina M., 2005. Nuclear receptor NHR-25 is required for cell-shape dynamics during epidermal differentiation in Caenorhabditis elegans. J. Cell Sci. 118: 223–232. [DOI] [PubMed] [Google Scholar]
- Simmer F., Tijsterman M., Parrish S., Koushika S. P., Nonet M. L., et al. , 2002. Loss of the putative RNA-directed RNA polymerase RRF-3 makes C. elegans hypersensitive to RNAi. Curr. Biol. 12: 1317–1319. [DOI] [PubMed] [Google Scholar]
- Singhvi A., Frank C. A., Garriga G., 2008. The T-box gene tbx-2, the homeobox gene egl-5 and the asymmetric cell division gene ham-1 specify neural fate in the HSN/PHB lineage. Genetics 179: 887–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J. A., McGarr P., Gilleard J. S., 2005. The Caenorhabditis elegans GATA factor elt-1 is essential for differentiation and maintenance of hypodermal seam cells and for normal locomotion. J. Cell Sci. 118: 5709–5719. [DOI] [PubMed] [Google Scholar]
- Smith P. A., Mango S. E., 2007. Role of T-box gene tbx-2 for anterior foregut muscle development in C. elegans. Dev. Biol. 302: 25–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer W. C., Zeller G., Watson J. D., Henz S. R., Watkins K. L., et al. , 2011. A spatial and temporal map of C. elegans gene expression. Genome Res. 21: 325–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston J. E., Horvitz H. R., 1977. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev. Biol. 56: 110–156. [DOI] [PubMed] [Google Scholar]
- Sulston J. E., Schierenberg E., White J. G., Thomson J. N., 1983. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev. Biol. 100: 64–119. [DOI] [PubMed] [Google Scholar]
- Sun G., Lewis L. E., Huang X., Nguyen Q., Price C., et al. , 2004. TBX5, a gene mutated in Holt-Oram syndrome, is regulated through a GC box and T-box binding elements (TBEs). J. Cell. Biochem. 92: 189–199. [DOI] [PubMed] [Google Scholar]
- Takahashi H., Mitani Y., Satoh G., Satoh N., 1999. Evolutionary alterations of the minimal promoter for notochord-specific Brachyury expression in ascidian embryos. Development 126: 3725–3734. [DOI] [PubMed] [Google Scholar]
- Takashima Y., Suzuki A., 2013. Regulation of organogenesis and stem cell properties by T-box transcription factors. Cell. Mol. Life Sci. 70: 3929–3945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi J. K., Mileikovskaia M., Koshiba-Takeuchi K., Heidt A. B., Mori A. D., et al. , 2005. Tbx20 dose-dependently regulates transcription factor networks required for mouse heart and motoneuron development. Development 132: 2463–2474. [DOI] [PubMed] [Google Scholar]
- Ward J. D., Bojanala N., Bernal T., Ashrafi K., Asahina M., et al. , 2013. Sumoylated NHR-25/NR5A regulates cell fate during C. elegans vulval development. PLoS Genet. 9: e1003992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson V., Conlon F. L., 2002. The T-box family. Genome Biol. 3: REVIEWS3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Smolen G. A., Palmer R., Christoforou A., van den Heuvel S., et al. , 2004. SUMO modification is required for in vivo Hox gene regulation by the Caenorhabditis elegans Polycomb group protein SOP-2. Nat. Genet. 36: 507–511. [DOI] [PubMed] [Google Scholar]