Abstract
Background
Despite an aggressive therapeutic approach, the prognosis for most patients with glioblastoma (GBM) remains poor. The aim of this study was to determine the significance of preoperative MRI variables, both quantitative and qualitative, with regard to overall and progression-free survival in GBM.
Methods
We retrospectively identified 94 untreated GBM patients from the Cancer Imaging Archive who had pretreatment MRI and corresponding patient outcomes and clinical information in The Cancer Genome Atlas. Qualitative imaging assessments were based on the Visually Accessible Rembrandt Images feature-set criteria. Volumetric parameters were obtained of the specific tumor components: contrast enhancement, necrosis, and edema/invasion. Cox regression was used to assess prognostic and survival significance of each image.
Results
Univariable Cox regression analysis demonstrated 10 imaging features and 2 clinical variables to be significantly associated with overall survival. Multivariable Cox regression analysis showed that tumor-enhancing volume (P = .03) and eloquent brain involvement (P < .001) were independent prognostic indicators of overall survival. In the multivariable Cox analysis of the volumetric features, the edema/invasion volume of more than 85 000 mm3 and the proportion of enhancing tumor were significantly correlated with higher mortality (Ps = .004 and .003, respectively).
Conclusions
Preoperative MRI parameters have a significant prognostic role in predicting survival in patients with GBM, thus making them useful for patient stratification and endpoint biomarkers in clinical trials.
Keywords: glioblastoma, imaging, overall survival, progression free survival, TCGA
Glioblastoma (GBM) is the most common primary malignant brain tumor in adults, accounting for 53.9% of gliomas.1 The current standard of care for newly diagnosed GBM involves surgery and concomitant regional radiotherapy with temozolomide (TMZ), followed by adjuvant TMZ.2 Despite an aggressive therapeutic approach, the prognosis for most patients with GBM remains poor, with a median survival of ∼14.6 months and a 5-year survival rate of <5%.1,2
A number of variables have been found to influence patient prognosis—age, preoperative performance score, and extent of tumor resection are among the well-known variables.3–12 Currently, the extent of resection of enhancing tumor is one of the most important independent factors predicting survival in patients with GBM.11 With advances in whole genome sequencing, molecular biomarkers have also been identified as impacting survival in GBM patients.13–17 Silencing of methylguanine-DNA methyltransferase is associated with longer survival in patients undergoing treatment with alkylating agents, such as TMZ.13 Yan et al14 showed that patients with tumors harboring isocitrate dehydrogenase (IDH) 1 or 2 mutations had a better outcome than those with IDH wild type. Multiple studies focusing on imaging correlates of survival in GBM patients are also available.18–30 Pope et al21 evaluated the relationship between 15 descriptive MRI variables and survival in patients with anaplastic astrocytoma and GBM. Nonenhancing tumor, edema, multifocality, and satellite lesions were found to be indicators of poor outcome. Gutman et al26 investigated a limited number of qualitative imaging variables; using 4 qualitative imaging features and a single measure of lesion size, the visually estimated proportion of contrast-enhancing tumor and longest axis length of tumor were found to be significantly associated with poor survival. Similarly, Nicolas-Jilwan et al30 reported qualitative imaging features associated with overall survival (OS) and found that the proportion of contrast-enhancing tumor significantly correlated with poor patient outcomes. No quantitative or volume-based parameters were analyzed; although some qualitative imaging parameters have been correlated with OS, progression-free survival (PFS) was not assessed; quantitative imaging metrics can increase the accuracy and reproducibility of tumor assessments, as these tumors are typically highly asymmetrical in morphology.29 Further, although neuroimaging plays a pivotal role in diagnosis and monitoring of therapeutic response, the prognostic and predictive role of imaging characteristics of GBM has remained largely unmined.
Thus, the purpose of this study was to determine the significance of preoperative imaging variables, both quantitative and qualitative, with regard to survival outcomes (OS and PFS) in GBM patients; this study serves as the exhaustive imaging-survival analysis study of The Cancer Genome Atlas (TCGA) GBM patient cohort using The Cancer Imaging Archive (TCIA). TCIA is a multi-institutional imaging repository, funded by the National Cancer Institute (NCI), corresponding to the GBM patient cohort (and other cancer cohorts) of TCGA and provides standardized imaging annotations.26,31,32 In this study, we assess the OS and PFS significance in the 26 standardized preoperative qualitative and semi-quantitative imaging variables provided by the Visually Accessible Rembrandt Images (VASARI) research project, the largest preoperative imaging parameters to date,33 and quantitative volume-based parameters as defined by Zinn et al.34 This study nonselectively reports the entire standardized qualitative imaging parameters of the dataset of TCIA and associated volumetrics with regard to correlates and predictors of both OS and PFS.
Methods
Patient Population and Clinical Variables
In this study, we used the original material and data provided by TCGA, which is a publicly available resource containing multidimensional genomic and clinical information on GBM and other cancers.31 The project in TCGA was conducted in compliance with regulations and policies for the protection of human subjects, and approvals by institutional review boards were appropriately obtained. The preoperative MRIs of the corresponding patients of the project in TCGA were made available for public download from TCIA, which was established by the collaboration between NCI and multiple institutions in the United States.32 We retrospectively identified 94 treatment-naïve GBM patients from TCGA who had both clinical and imaging data available. The clinical variables consisted of age, gender, and Karnofsky performance status (KPS).
Qualitative and Semi-quantitative Imaging Analysis
The qualitative and semi-quantitative imaging dataset annotations were based upon the VASARI feature set for human glioma. This comprehensive feature set contains standardized terminologies of the most common features used to describe primary cerebral neoplasia on standard pre- and postcontrast enhanced MRI.25,31–33 The open-source PACS (picture archiving and communication system) workstation, the ClearCanvas platform (http://www.clearcanvas.ca/), was used for imaging assessments. Board-certified neuroradiologists (C.A.H., 16 y experience; S.N.H., 6 y; M.W., 7 y; P.R., 5 y; R.R.C., 3 y; and M.J., 4 y) were recruited and trained in the use of the feature set. A minimum of 3 different VASARI scores were obtained for each patient. The scores were then collected centrally in the NCI system and subsequently analyzed. The lists of VASARI imaging features, scoring values, and their definitions are summarized in Table 1.33
Table 1.
VASARI imaging features
| Imaging Features | Scoring Value | Definition |
|---|---|---|
| Major axis | NA | The longest diameter of the tumor which is based upon measurement of the FLAIR abnormality on a single axial image that demonstrates the largest cross-sectional area. |
| Minor axis | NA | The diameter of the FLAIR abnormality which is perpendicular to the longest diameter. The measurement is performed on a single axial image that demonstrates the largest cross-sectional area. |
| Tumor location | Frontal lobe Temporal lobe Parietal lobe Occipital lobe Insular Basal ganglia Thalamus Brainstem Cerebellum Corpus callosum |
Location of lesion geographic epicenter, the largest component of the tumor, either contrast-enhancing tumor (CET) or non-contrast-enhancing tumor (nCET). Nonenhancing tumor (nCET) is defined as regions of T2W hyperintensity (less than the intensity of cerebrospinal fluid, with corresponding T1W hypointensity) that are associated with mass effect and architectural distortion, including blurring of the gray-white interface. |
| Side of tumor epicenter | Right Center/Bilateral Left |
Side of lesion epicenter irrespective of whether the lesion crosses into the contralateral hemisphere. |
| Eloquent brain | No eloquent brain Speech motor Speech receptive Motor Vision |
Presence of tumor involvement of the eloquent cortex or immediate subcortical white matter of eloquent cortex. |
| Enhancement quality | No contrast enhancement Mild (when barely discernible but unequivocal degree of enhancement is present relative to precontrast images) Marked (obvious tissue enhancement). |
Qualitative degree of contrast enhancement defined as having all or portions of the tumor that demonstrate significantly higher signal on the postcontrast T1W images compared with precontrast T1W images. |
| Proportion enhancing | <5%, 6%–33%, 34%–67%, 68%–95%, >95% | Visually estimated proportion of enhancing component to the entire tumor |
| Proportion nCET | <5%, 6%–33%, 34%–67%, 68%–95%, >95% | Visually estimated proportion of nonenhancing component to the entire tumor |
| Proportion necrosis | <5%, 6%–33%, 34%–67%, 68%–95%, >95% | Visually estimated proportion of necrosis to the entire tumor. Necrosis is defined as a region within the tumor that does not enhance, is high on T2W and proton density images, is low on T1W images, and has an irregular border. |
| Proportion of edema | <5%, 6%–33%, 34%–67%, 68%–95%, >95% | Visually estimated proportion of edema to the entire tumor. Edema is defined as a region which is greater in signal than nCET and somewhat lower in signal than CSF. Pseudopods are characteristic of edema. |
| Cyst(s) | Absent OR present | Well-defined, rounded, often eccentric regions of very bright T2W signal and low T1W signal essentially matching CSF signal intensity, with very thin, regular, smooth, nonenhancing or regularly enhancing walls, possibly with thin, regular, internal septations. |
| Multifocal or multicentric | Focal Multifocal or multicentric Gliomatosis |
Multifocal is defined as having at least one region of tumor, either enhancing or nonenhancing, which is not contiguous with the dominant lesion and is outside the region of signal abnormality (edema) surrounding the dominant mass. This can be defined as those resulting from dissemination or growth by an established route, spread via commissural or other pathways, or via CSF channels or local metastases. Multicentric are widely separated lesions in different lobes or different hemispheres that cannot be attributed to one of the previously mentioned pathways. Gliomatosis refers to generalized neoplastic transformation of the white matter of most of a hemisphere. |
| T1/FLAIR ratio | Expansive (size of precontrast T1 abnormality approximates size of FLAIR abnormality) Mixed (size of T1 abnormality moderately less than FLAIR envelope) Infiltrative (size of precontrast T1 abnormality much smaller than size of FLAIR abnormality) |
Gross comparison in the overall lesion size between precontrast T1 and FLAIR (in the same plane). Use T2 if FLAIR is not provided. |
| Thickness of enhancing margin | Thin (<3 mm) Thick/nodular (>3 mm) Solid (only solid enhancement, no rim) |
The thickness of enhancing margin of the tumor. The scoring is not applicable if there is no contrast enhancement. |
| Definition of the enhancing margin | Well defined Poorly defined |
The definition of the outside margin of the enhancement. The scoring is not applicable if there is no contrast enhancement. |
| Definition of the nonenhancing margin | Well defined Poorly defined |
The definition of the outside margin of the nonenhancing margin of the tumor. |
| Hemorrhage | Yes OR No | Intrinsic hemorrhage anywhere in the tumor matrix. Any intrinsic foci of low signal on T2WI or high signal on T1WI. |
| Diffusion characteristics | Facilitated Restricted Mixed Indeterminate |
Predominantly facilitated or restricted diffusion in the enhancing or nCET portion of the tumor. (Based on apparent diffusion coefficient [ADC] map) (Rate CET alone when present, otherwise use nCET) Indeterminate = unsure. Mixed = relatively equal proportion of facilitated and restricted. No ADC maps = use no images. Proportion of tissue not relevant. |
| Pial invasion | Yes OR No | Enhancement of the overlying pia in continuity with enhancing or nonenhancing tumor. |
| Ependymal extension | Yes OR No | Invasion of any adjacent ependymal surface in continuity with enhancing or nonenhancing tumor matrix. |
| Cortical involvement | Yes OR No | Nonenhancing or enhancing tumor extending to the cortical mantle or cortex is no longer distinguishable relative to subjacent tumor. |
| Deep white matter invasion | Yes OR No | Enhancing or nCET tumor extending into the internal capsule, corpus callosum, or brainstem. |
| nCET crosses midline | Yes OR No | nCET crosses into contralateral hemisphere through white matter commissures (exclusive of herniated ipsilateral tissue). |
| CET crosses midline | Yes OR No | Enhancing tissue crosses into contralateral hemisphere through white matter commissures (exclusive of herniated ipsilateral tissue). |
| Satellites | Yes OR No | An area of enhancement within the region of signal abnormality surrounding the dominant lesion but not contiguous in any part with the major tumor mass. |
| Calvarial remodeling | Yes OR No | Erosion of inner table of skull (possibly a secondary sign of slow growth). |
Quantitative Volumetric Imaging Analysis
Image acquisition, volume selection, and sequence definition
Preoperative fluid-attenuated inversion recovery (FLAIR) and postcontrast T1-weighted imaging (T1WI) data were downloaded from TCIA and used for segmentation of the 3 different GBM compartments, namely edema/tumor invasion, tumor, and necrosis. The area of peritumoral T2/FLAIR hyperintensity in GBM reflects an admixture of infiltrative tumor and vasogenic edema. For nomenclature purposes, this area is referred to as “edema/tumor invasion” in this study. Enhancement identified on postcontrast T1WI corresponds to an area of active viable tumor with disrupted blood–brain barrier; therefore, this area is defined as “tumor.” The region within the tumor that does not show enhancement was defined as “necrosis”; and, thus, its volume was also quantified on postcontrast T1WI.
Software and Image Analysis
The open-source software platform, 3D Slicer software 3.6.3 (http://www.slicer.org), developed between the Surgical Planning Laboratory at the Brigham and Women's Hospital and the MIT Artificial Intelligence Laboratory in 1998, was used as described by Zinn et al34 for image visualization, analysis, and interactive segmentation.35–37 Tumor segmentation was performed in consensus by a neuroradiologist (R.R.C.) and a neurosurgery resident (P.O.Z.) with more than 6 years of experience in image segmentation.
Image Registration and Segmentation
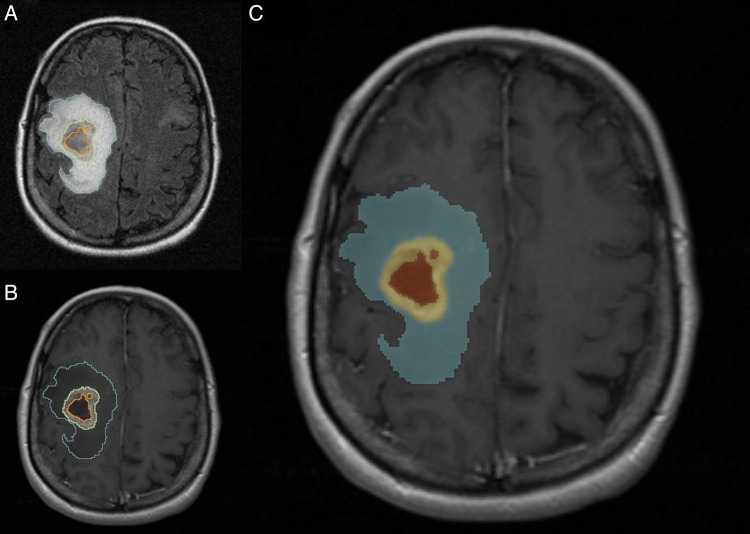
First, FLAIR and postcontrast T1WI sequences were aligned and registered to each other to overcome the differences in imaging parameters such as scanning angles and voxel thickness. Resampling of the FLAIR volume to the matrix of the T1WI series was performed in those cases where the voxel size of the FLAIR and T1WI were different. The images were adequately registered in most scans; and an error of registration <2 mm was deemed acceptable. We then performed tumor segmentation in a simple hierarchical model of anatomy from peripheral to central (Fig. 1). The volumes of each tumor compartment were subsequently computed automatically.
Fig. 1.
A 51-year-old female patient with right frontoparietal GBM. Representative case of tumor segmentation. (A) Axial FLAIR image shows segmentation of the FLAIR hyperintensity region defined as edema/tumor invasion (blue) (B) Axial postcontrast T1WI demonstrates segmentation of the enhancing tumor (yellow) and area of necrosis (orange). (C) Label map image demonstrating the segmented tumor.
Statistical Modeling for Survival Analysis and Survival Prediction
OS was defined as the time between the date of pathological diagnosis and the date of death or the date of last clinical follow-up. PFS was measured from the date of the first surgery to the first recurrence of the disease. The univariable Cox proportional hazards model was used to determine hazard ratios (HRs) of each variable as a predictor of OS and PFS. Each variable that had a P-value <.2 in the univariable analysis was selected to be included in a stepwise forward multivariable Cox regression analysis. All of the above steps were done for the VASARI features and volumetric features separately, as well as for the VASARI and volumetric features together (combined model). To evaluate the effect of age and KPS on each imaging feature in the combined multivariable model, we calculated the age- and KPS-adjusted HR.
For all categorical and continuous variables that had more than 2 groups, a decision tree approach based on recursive partitioning analysis was applied to establish subgroups and cutoff values. This is a decision tree method for defining predictors. Each variable was examined for the best split, which was chosen to maximize the difference in the responses between the 2 branches of the split.
Receiver operating characteristic (ROC) analysis and area under the curve (AUC) were used to estimate the accuracy of the Cox models. All statistical analyses were performed using Stata v10. P ≤ .05 was considered statistically significant.
Results
Patient Population and Characteristics
Enrolled in the study were a total of 94 TCGA patients with GBM (31 female, 63 male; mean age 57.5; median age 59.5; range 14–84 y). The demographic and other characteristics of the included patients are summarized in Table 2. KPS parameters were available in 73 patients.
Table 2.
Demographic and clinical characteristics of the patients with glioblastoma (n = 94)
| Age, y (range) | 59.5 (14–84) |
| Sex | |
| Female | 31 (29%) |
| Male | 63 (71%) |
| KPS, n = 73 | |
| ≥80 | 59 (80%) |
| <80 | 14 (20%) |
| Overall survival time, mo (range) | 13.0 (0.5–58.5) |
| Progression-free survival, mo (range) | 6.4 (0.5–47) |
Univariable and Kaplan–Meier Survival Analysis
With respect to clinical variables, age was a statistically significant prognostic factor for both OS (HR = 1.025, 95% CI: 1.009–1.043, P = .002) and PFS (HR = 1.021, 95% CI: 1.003–1.039, P = .036), whereas KPS was significant for only OS (HR = 0.202, 95% CI: 0.604–0.654, P = .008) (Table 3).
Table 3.
Univariable Cox regression analysis of survival in patients with glioblastoma
| Overall Survival |
Progression-free Survival |
|||
|---|---|---|---|---|
| HR (95% CI) | P-value | HR (95% CI) | P-value | |
| Age, y | 1.25 (1.009–1.043) | .002 | 1.021 (1.003–1.039) | .036 |
| KPS | 0.202 (0.604–0.654) | .008 | 0.992 (0.972–1.013) | .457 |
| Sex (male vs female) | 0.78 (0.50–1.26) | .3 | 1.26 (0.75–2.12) | .4 |
| VASARI features | ||||
| Major axis (mm) | 1.015 (1.004–1.027) | .009 | 1.015 (1.002–1.027) | .027 |
| Major axis (median cutoff) (≥75 vs <75) | 1.708 (1.106–2.644) | .016 | 1.933 (1.181–3.179) | .009 |
| Major axis (mean cutoff) (≥77 vs <77) | 1.902 (1.225–2.957) | .004 | 1.791 (1.093–2.927) | .021 |
| Major axis (DT cutoff) (≥80 vs <80) | 1.986 (1.268–3.087) | .003 | 2.099 (1.254–3.473) | .005 |
| Minor axis (mm) | 1.010 (0.009–1.029) | .283 | 1.017 (0.996–1.040) | .118 |
| Minor axis (median cutoff) (≥48 vs <48) | 1.216 (0.794–1.870) | .369 | 1.278 (0.796–2.046) | .307 |
| Minor axis (mean cutoff) (≥50 vs <50) | 1.203 (0.784–1.845) | .395 | 1.269 (0.786–2.030) | .325 |
| Minor axis (DT cutoff) (≥60 vs <60) | 1.023 (0.596–1.674) | .932 | 1.706 (0.919–2.991) | .088 |
| Tumor location (frontal/parietal/temporal vs rest) | 2.278 (1.174–4.418) | .015 | 1.825 (0.864–3.853) | .115 |
| Side of tumor epicenter (left vs right) | 0.746 (0.486–1.150) | .183 | 0.844 (0.526–1.356) | .480 |
| Eloquent brain (eloquent vs noneloquent) | 1.453 (0.946–2.209) | .087 | 1.130 (0.708–1.775 | .603 |
| Enhancement quality (marked vs mild/no enhancement) | 1.397 (0.627–3.974) | .446 | 0.889 (0.417–2.306) | .788 |
| Cyst (yes vs no) | 0.661 (0.162–1.768) | .453 | 0.951 (0.288–2.315) | .923 |
| Distribution (multifocal/multicentric/gliomatosis vs focal) | 2.535 (1.109–5.064) | .029 | 2.255 (0.919–4.759) | .073 |
| T1/FLAIR ratio (infiltrative/mixed vs expansive) | 1.600 (1.013–2.486) | .044 | 1.791 (1.081–2.915) | .024 |
| Thickness of enhancing margin (thick/nodular/solid vs thin) | 1.412 (0.585–4.637) | .480 | 1.187 (0.486–3.923) | .734 |
| Definition of the enhancing margin (poorly defined vs well defined) | 1.146 (0.601–2.477) | .697 | 1.452 (0.733–3.307) | .303 |
| Definition of the nonenhancing margin (poorly defined vs well defined) | 1.521 (0.947–2.390) | .081 | 1.468 (0.873–2.398) | .144 |
| Hemorrhage (yes vs no) | 1.094 (0.692–1.695) | .695 | 0.723 (0.423–1.191) | .207 |
| Diffusion characteristics (restricted/mixed vs facilitated) | 1.070 (0.698–1.657) | .757 | 0.686 (0.428–1.112) | .125 |
| Pial invasion (yes vs no) | 0.895 (0.578–1.374) | .615 | 0.760 (0.465–1.221) | .259 |
| Ependymal extension (yes vs no) | 1.883 (1.217–2.949) | .004 | 0.982 (1.225–3.229) | .005 |
| Cortical involvement (yes vs no) | 0.664 (0.338–1.504) | .302 | 0.803 (0.371–2.106) | .624 |
| Deep white matter invasion (yes vs no) | 1.823 (1.175–2.871) | .007 | 2.510 (1.519–4.227) | <.001 |
| nCET crosses midline (yes vs no) | 1.572 (0.862–2.684) | 1.134 | 2.182 (1.086–4.080) | .030 |
| CET crosses midline (yes vs no) | 3.593 (1.559–7.261) | .004 | 5.459 (1.985–12.907) | .002 |
| Satellites (yes vs no) | 1.338 (0.801–2.152) | .257 | 2.866 (1.567–5.073) | <.001 |
| Calvarial remodeling (yes vs no) | 0.886 (0.145–2.831) | .84 | 0.922 (0.052–4.224) | .935 |
| Volumetric imaging variables | ||||
| Tumor enhancement volume (mm3) | 1.000018 (1.000005–1.00003) | .006 | 1.00 (0.99–1.00) | .155 |
| Tumor enhancement volume (DT cutoff) (≥35 000 vs <35 000 mm3) | 1.915 (1.55–3.068) | .013 | 1.577 (0.867–2.721) | .131 |
| Tumor enhancement volume (median cutoff) (≥21 046 vs <21 046 mm3) | 1.558 (1.013–2.401) | .043 | 1.536 (0.951–248) | .079 |
| Tumor enhancement volume (mean cutoff) (≥24 850 vs <24 850 mm3) | 1.370 (0.883–2.107) | .158 | 1.628 (0.993–2.640) | .05 |
| Edema/tumor invasion (mm3) | 1.000 (0.999–1.000) | .444 | 1.837 (0.653–4.853) | .242 |
| Edema/tumor invasion (DT cutoff) (≥85 000 vs <85 000 mm3) | 1.783 (1.102–2.816) | .019 | 1.598 (0.992–2.670) | .092 |
| Edema/tumor invasion (median cutoff) (≥59 337 vs <59 337 mm3) | 0.938 (0.604–1.458) | .777 | 0.988 (0.612–1.589) | .960 |
| Edema/tumor invasion (mean cutoff) (≥65 676 vs <65 676 mm3) | 1.036 (0.669–1.592) | .873 | 1.236 (0.761–1.981) | .387 |
| Necrosis (mm3) | 1.000 (0.999–1.000) | .347 | 1.000 (0.999–1.000) | .649 |
| Necrosis (DT cutoff for OS) (≥20 000 vs <20 000 mm3) | 0.975 (0.483–1.784) | .940 | 0.967 (0.423–1.925) | .930 |
| Necrosis (which cutoff PFS) (≥200 vs <200 mm3) | 1.504 (0.738–3.620) | .280 | 1.845 (0.810–5.324) | .156 |
| Necrosis (median cutoff) (≥8261vs <8261 mm3) | 1.037 (0.675–1.589) | .867 | 1.072 (0.666–1.713) | .771 |
| Necrosis (mean cutoff) (≥10 095 vs <10 095 mm3) | 1.060 (0.688–1.624) | .789 | 1.050 (0.651–1.679) | .839 |
| Proportion of edema/tumor invasion | 0.294 (0.080–1.115) | .071 | 0.879 (0.221–3.699) | .858 |
| Proportion of edema/tumor invasion (median cutoff) (≥0.7 vs <0.7) | 1.045 (0.680–1.610) | .841 | 1.162 (0.727–1.863) | .529 |
| Proportion of edema/tumor invasion (mean cutoff) (≥0.65 vs <0.65) | 0.901 (0.583–1.406) | .643 | 0.946 (0.588–1.536) | .819 |
| Proportion of tumor | 8.686 (1.309–53.443) | .026 | 1.599 (0.206–11.344) | .648 |
| Proportion of tumor (median cutoff) (≥0.24 vs <0.24) | 1.592 (1.026–2.482) | .038 | 1.049 (0.651–1.688) | .843 |
| Proportion of tumor (mean cutoff) (≥0.25 vs <0.25) | 1.343 (0.862–2.083) | .190 | 0.950 (0.586–1.527) | .832 |
| Proportion of necrosis | 1.894 (0.140–22.616) | .624 | 0.654 (0.033–11.352) | .773 |
| Proportion of necrosis (median cutoff) (≥0.08 vs <0.08) | 0.816 (0.529–1.255) | .353 | 0.866 (0.541–1.384) | .546 |
| Proportion of necrosis (mean cutoff) (≥0.1 vs <0.1) | 0.827 (0.528–1.277) | .395 | 1.016 (0.626–1.625) | .948 |
| Proportion of tumor and necrosis | 3.399 (0.897–12.477) | .071 | 1.137 (0.270–4.519) | .858 |
| Proportion of tumor and necrosis (median cutoff) (≥0.31 vs <0.31) | 0.957 (0.621–1.470) | .841 | 0.860 (0.537–1.375) | .529 |
| Proportion of tumor and necrosis (median cutoff) (≥0.35 vs <0.35) | 1.109 (0.711–1.715) | .643 | 1.057 (0.651–1.700) | .819 |
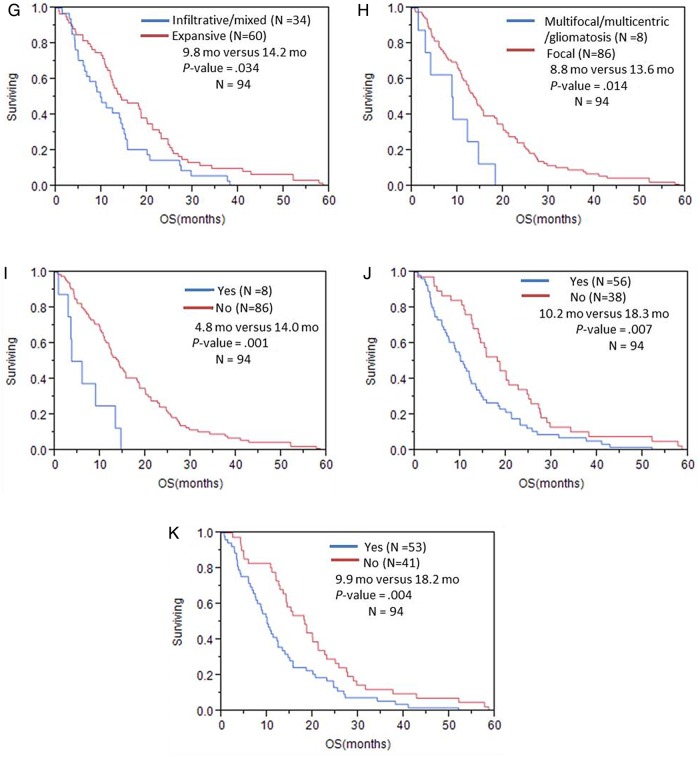
In the univariable analysis of the qualitative imaging variables (Table 3), we found multiple imaging features that were significantly associated with both OS and PFS, namely major axis (longest diameter; OS, P = .009; PFS, P = .027), T1/FLAIR ratio (size of precontrast T1 abnormality compared with size of FLAIR abnormality) (OS, P = .044; PFS, P = .024), presence of ependymal extension (OS, P = .004; PFS, P = .005), deep white matter invasion (OS, P = .007; PFS, P < .001), and contrast-enhancing tumor crossing midline (enhancing tumor invading into contralateral hemisphere through white matter commissures; OS, P = .004; PFS, P = .002). Presence of nonenhancing tumor crossing the midline (nonenhancing tissue crossing into contralateral hemisphere through white matter commissures) and satellite lesions (an area of enhancement within the region of signal abnormality surrounding the dominant lesion but not contiguous in any part with the major enhancing tumor mass) were found to be significant prognostic indicators for decreased PFS (P = .03 and P < .001, respectively), while tumor location (frontal/parietal/temporal vs the rest) and tumor distribution (multifocal/multicentric vs focal) were significant for decreased OS (P = .015 and P = .029, respectively) (Table 3). The survival difference with respect to VASARI features are shown in Figs 2 and 3 (and Supplementary Table S1).
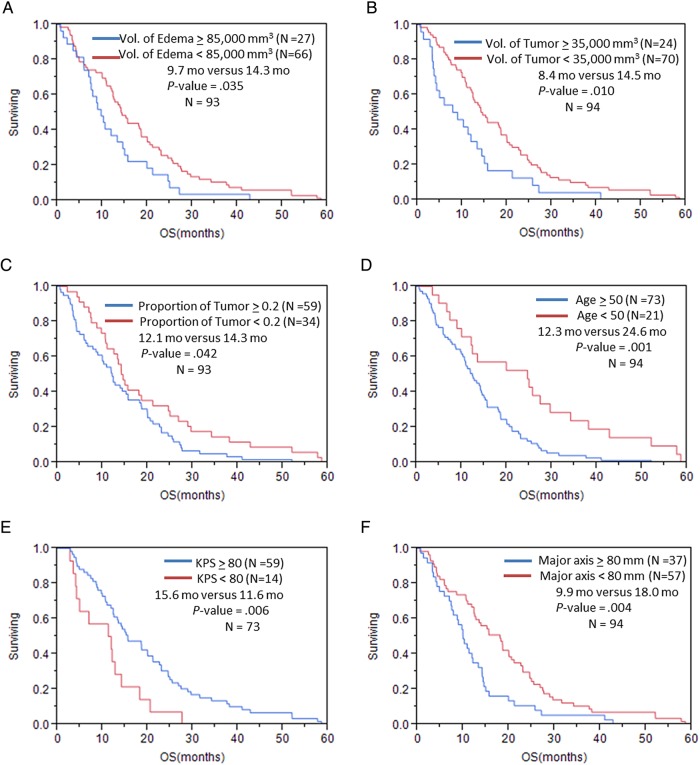
Fig. 2.
Overall survival in patients with GBM stratified by (A) volume of edema/tumor invasion; (B) volume of enhancing tumor; (C) proportion of volume of enhancing tumor; (D) age; (E) KPS; (F) major axis based on VASARI feature set; (G) T1/FLAIR ratio; (H) distribution/focality; (I) enhancement across the midline; (J) deep white matter invasion; (K) ependymal extension.
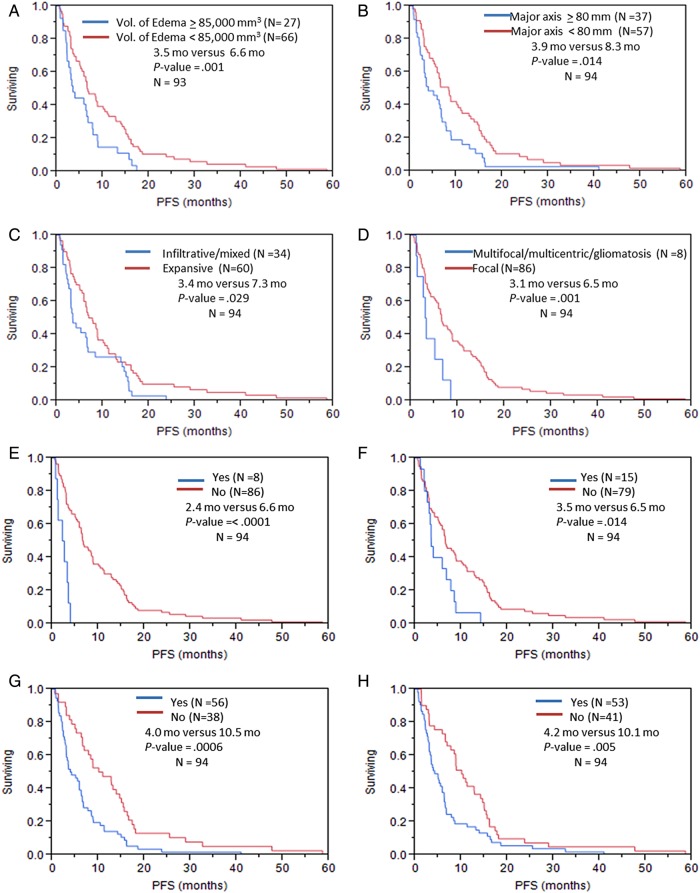
Fig. 3.
Progression-free survival in patients with GBM stratified by (A) volume of edema/tumor invasion; (B) major axis; (C) T1/FLAIR ratio; (D) distribution/focality; (E) enhancement across the midline; (F) nonenhancing tumor across the midline; (G) deep white matter invasion; (H) ependymal extension.
The univariable analysis of the volumetric imaging features (Table 3) showed that volume of the enhancing tumor with cutoff of 35 000 mm3 was a significant predictor of OS (P = .013); the OS for patients with tumor volume of <35 000 mm3 was 14.3 months versus 8.4 months for patients with higher tumor volume (OS benefit = 5.9 mo) (Fig. 2; Supplementary Table S1). The volume of edema/tumor invasion was also significantly associated with OS (P = .019); patients with edema/tumor invasion <85 000 mm3 had significantly longer OS (14.2 mo) in comparison with those who had edema/tumor invasion >85 000 mm3 (9.7 mo). The proportion of enhancing tumor was the other volumetric feature that was significantly (P = .038) associated with OS. Patients whose enhancing portion of the whole tumor was ≥24 had significantly lower OS (11.8 mo) versus the patients with enhancing portion of <24% (14.7 mo). Regarding PFS, volume of enhancing tumor with cutoff of 24 850 mm3 was a significant predictor for PFS (P = .05) (Fig. 3; Supplementary Table S2).
Multivariable Survival Analysis
Two separate sets of multivariable models, one for VASARI features and the other for volumetric features, were analyzed (Table 4). In the VASARI feature model alone, T1/FLAIR ratios, ependymal extension, and deep white matter invasion of tumor were significant independent prognostic indicators of OS (Ps = .03, .02, and .006, respectively). Deep white matter invasion and distribution of tumor were significant predictors of PFS (P = .007 and .003, respectively). Deep white matter invasion was significantly associated with 89% increased risk of death (HR = 1.89, 95% CI: 1.20–2.97) and 95% increased risk of disease progression (HR = 1.95, 95% CI: 1.20–3.16).
Table 4.
Multivariable Cox models for (a) VASARI and volumetric features, (b) quantitative and qualitative imaging features in patients with glioblastoma, and (c) age- and KPS-adjusted multivariable Cox model for imaging features in patients with glioblastoma
| Overall Survival |
Progression-free Survival |
|||
|---|---|---|---|---|
| HR (95% CI) | P-value | HR (95% CI) | P-value | |
| VASARI features model | ||||
| T1/FLAIR ratio (infiltrative/mixed vs expansive) | 1.64 (1.04–2.6) | .03 | ||
| Ependymal extension (yes vs no) | 1.74 (1.11–2.73) | .02 | ||
| Deep white matter invasion (yes vs no) | 1.89 (1.20–2.97) | .006 | 1.95 (1.20–3.16) | .007 |
| Distribution (multifocal and multicentric vs focal) | 3.6 (1.54–8.43) | .003 | ||
| Volumetric features model | ||||
| Tumor proportion | 17.21 (2.42–122.55) | .004 | ||
| Edema volume (higher vs lower than 85 000 mm3) | 2.09 (1.29–3.39) | .003 | ||
| Tumor volume (higher vs lower than 24 850 mm3) | 1.61 (1.00–2.60) | .050 | ||
| Quantitative and qualitative imaging features | ||||
| Tumor volume (higher vs lower than 35 000 mm3) | 1.74 (1.06–2.86) | 0.03 | ||
| Eloquent brain (eloquent vs noneloquent) | 2.92 (1.72–4.95) | <0.001 | ||
| Deep white matter invasion (yes vs no) | 1.90 (1.15–3.16) | .01 | ||
| Distribution (multifocal and multicentric vs focal) | 2.16 (0.94–4.97) | .07 | ||
| Age- and KPS-adjusted multivariable Cox model | ||||
| Tumor (higher vs lower than 35 000 mm3) | 1.79 (0.94–3.42) | 0.08 | ||
| Eloquent brain (eloquent vs noneloquent) | 2.44 (1.28–4.65) | 0.007 | ||
| Deep white matter invasion (yes vs no) | 1.78 (1.03–3.07) | .04 | ||
| Distribution (multifocal and multicentric vs focal) | 2.07 (0.83–5.15) | .11 | ||
For volumetric features, tumor proportion and edema/tumor invasion volume were strongly associated with prediction of OS. One unit increase in tumor proportion resulted in an increased risk of death by 17 times (HR = 17.21, P = .004). Also, the risk of death was almost doubled in patients with edema/tumor invasion volume >85 000 mm3 compared with those with lower edema/tumor invasion (HR = 2.09, P = .003). Tumor volume with a cutoff value of 24 850 mm3 was significantly associated with PFS (HR = 1.61, P = .050).
The combined (VASARI and volumetric features) multivariable model (Table 4) demonstrated that tumor volume and eloquent brain involvement were independent variables for prediction of OS (Ps = .03 and <.001, respectively). Patients with tumor volume >35 000 mm3 had 74% higher risk of death compared with patients with tumor volume <35 000 mm3 (HR = 1.74, P = .03). Deep white matter invasion remained a significant predictor of progression in the combined model (P = .01). Multifocal/multicentric versus focal tumor was marginally associated with decreased PFS in the combined model (P = .07).
To evaluate the effect of age and KPS on the predictive imaging features, we calculated age- and KPS-adjusted HRs for the final variables in the combined model (Table 4). The HRs and corresponding 95% CIs did not change significantly after adjustment (see Supplementary Fig. S1).
ROC analysis was performed to assess the accuracy of the combined predictive model of OS (see Supplementary Fig. S2A). The AUC was 79%, with CI of 63%–95%. Patients with eloquent brain involvement and tumor volume more than 35 000 mm3 (final combined model variables) had significantly (P = .002) lower OS (4.2 mo) versus the other patients (14.2 mo) (see Supplementary Fig. S5B).
Discussion
This study serves as an exhaustive analysis correlating imaging with patient outcomes to include associations and predictors of OS and PFS in GBM patients, specifically the GBM patient cohort of TCGA. TCIA, resulting from TCGA efforts and funded by the Cancer Imaging Program of the National Institutes of Health, reflects a major step toward the establishment of an imaging database infrastructure, an organized collection of catalogued images, and a standardized imaging feature set that can be used for future similar efforts. The standardized qualitative imaging parameters provided by the VASARI feature set of TCIA, quantitative volumetric parameters provided by Zinn et al,34 and clinical parameters in the GBM cohort of TCGA were exhaustively analyzed. The prognostic role of these variables was systematically investigated.
Importantly, this is the first study investigating the contribution to survival of quantitative volumetric (true 3D volumes) parameters, including specific cutoff values. We demonstrated that the volume of tumor enhancement (ie, active viable tumor with blood-brain barrier breakdown) was an important independent prognostic indicator for prediction of OS, even after adjustment for other imaging features. A larger extent of enhancing disease indicates higher tumor burden compared with lower extent resulting in patient's dismal prognosis. Iliadis et al16 demonstrated that one unit increase in preoperative enhancing tumor was significantly associated with 1% increased risk of mortality (P = .037) and progression (P = .041); but no suitable cutoff value was presented for preoperative tumor volume. Concerning the influence of age and KPS, after adjustment for these factors, tumor volume still showed a marginally significant predictive effect for mortality (P = .08). The patients who had an enhancing tumor volume lower than 35 000 mm3 demonstrated longer OS benefit of 5.1 months compared with the patients who had a higher enhancing tumor volume (P = .013).
With regard to the nonenhancing tumor component, we demonstrated that the volume of edema/tumor invasion with a cutoff value of 85 000 mm3 was a significant predictor factor for mortality (P = .003) among the volumetric features. It is well known that the areas of nonenhancing T2/FLAIR signal abnormality in GBM represent an admixture of bulk tumor invasion and vasogenic edema. These parameters serve as a surrogate marker for tumor invasiveness that partly determines extension of the nonenhancing infiltrative tumor. GBM with infiltrating phenotype also tends to have multifocal disease and poor survival.15 Schoenegger et al22 investigated the prognostic role of peritumoral edema in GBM patients. By using a simple measurement technique (<1 cm or >1 cm from the enhancing tumor margin), the patients were stratified into minor and major edema subgroups, respectively. They found that the major edema subgroup had significantly shorter OS compared with the minor edema subgroup. A number of studies with qualitative assessment of peritumoral edema/nonenhancing disease also reported that the presence and larger extent of peritumoral edema/invasion is a negative prognostic factor.19,21 To the best of our knowledge, our study is the first to identify the cutoff values of the volume-based imaging parameters (ie, enhancing tumor, necrosis, edema/tumor invasion) to predict individual outcome in GBM patients. We found that the volume-based parameter of edema/tumor invasion was an important independent prognostic factor. By using the single optimal cutoff of the edema/tumor invasion volume at 85 000 mm3, we found that the median OS benefit of those patients who had lower edema/invasion volume compared with those who had a higher volume was ∼4.6 months (median OS 14.3 mo vs 9.7 mo; P = .019). These innovative findings introduce a new promising imaging biomarker for patients with GBM derived from volume-based parameters. This new prognostic stratification approach helps provide a more accurate prognosis for individual patients and can be used as a stratification tool in clinical trials.
Regarding the prognostic role of tumor necrosis, there are various results among the limited numbers of survival studies in GBM.10,19,21,24 Hammoud et al19 found that the amount of tumor necrosis on preoperative scans was the strongest prognostic variable. This is similar to other retrospective studies10,24; however, our study showed no significant association between degree of tumor necrosis and survival. This can be attributed to differences in measurement and grading system, that is, quantitative and qualitative assessment. However, a better explanation can be found based on genomic differences.38 In one of our previous studies, we found different mechanisms and pathways regulating cell death that appears to be sex specific, and in which the mechanism by which cell death occurs in GBM as defined by MRI (necrosis) appears to be driven by a more apoptotic (TP53) pathway in males; while in females, the mechanism by which this occurs appears to be due to a more oncogenic (MYCN) pathway.38 With advancements in the understanding of genetic alterations and biology of GBM, ongoing radiogenomic studies may provide additional information regarding difference in survival and differential tumor responses by providing more comprehensive correlation between specific genetic aberrations and imaging features. Ongoing research in our group is currently investigating these topics.
With regard to the qualitative imaging features, deep white matter invasion was among the most important associated variables with survival, likely reflective of a more biologically aggressive and advanced tumor. In our study, we found that this imaging feature is associated with a shorter PFS, independently and significantly. Even after adjustment for age and KPS, the HR remained significant (P before age and KPS adjustment = 0.01 and after adjustment = 0.04), which showed the strong predictive effect of this invasive imaging feature. A more recent study published by our group39 demonstrated that patients with an invasive imaging signature have a significant decrease in OS and an associated specific genomic expression profile that may explain the mechanism of this spread pattern.
In our study, even though multifocal tumor was not associated with PFS significantly in the final models (P = .07), it was selected in a stepwise model of VASARI features alone as well as in the combined model. The latter demonstrates the predictive power of this feature versus other VASARI features. The clinical significance of this feature has been well established in both the radiology and nonradiology literature.21,39,40 Pope et al21 retrospectively investigated the prognostic significance of certain MRI features in patients with high-grade gliomas and found that the presence of multifocal tumor was one of the significant negative prognostic indicators. Multiple lesions were also found to be associated with lower KPS and inability to perform gross total resection in a retrospective study.39 More recent published literature40 also addressed the importance of this feature and showed that patients with a single focal tumor had a statistically significantly better PFS compared with the multifocal group; however, there was no statistically significant difference in OS, similar to our results.
There are some limitations in our study. Our study is inherently limited by its retrospective design using a preexisting database and images acquired from different scanners from various hospitals involved in data collection by TCGA. However, this dataset as well as the standardized VASARI feature set and validated segmentation technique as described by Zinn et al34 reflects a standardized comprehensive and validated methodology leveraging the concerted efforts of TCIA. Using the database of TCGA, we could not consider some parameters like the extent of tumor resection and the extent of the initial TMZ chemotherapy in our analyses. Despite these limitations, our study demonstrates the value of imaging features in predicting patient outcomes. However, a prospective study design is needed to validate the prognostic utility of these potential imaging biomarkers. Further, additional imaging variables such as radiomic analysis and texture analysis of the tumor can be anticipated to increase the prognostic predictive power of imaging. Additionally, advanced functional MRI such as diffusion imaging and perfusion imaging with quantitative hemodynamic parameters of tumor—for instance, maximum tumor blood volume—are shown to be significantly associated with patient OS27 and could be used as prognostic indicators. Such studies are under way in our group.41–43
Our comprehensive TCGA imaging-outcome analysis establishes the prognostic significance of pretreatment imaging variables and provides specific cutoff values by which clinical decision making can be performed and precision medicine leveraged. This sets the groundwork needed for development of robust imaging biomarkers that can potentially be applied to current classification models and routine clinical practice. The power of TCGA imaging datasets with structured imaging features provided by the VASARI feature set and the GBM volumetrics as defined by Zinn et al34 sets the stage for a new era of personalized qualitative and quantitative feature assessments and reporting; these parameters and cutoff volumetric volumes can serve as a template for future imaging and imaging genomic applications and discoveries as these harbor prognostic significance.
In summary, specific preoperative MRI features and signatures have a significant prognostic role in helping to predict survival in patients with GBM. Given the statistical prognostic significance, specific preoperative MRI brain tumor features can now be considered for prognostic imaging genomic biomarker development and validation. Imaging signatures derived from these features can potentially be used for patient stratification and endpoint prognostic biomarkers in prospective clinical trials.
Supplementary Material
Funding
This research is partially funded by the John S. Dunn Sr. Distinguished Chair in Diagnostic Imaging Fund and MD Anderson Cancer Center startup funding.
Conflict of interest statement. The authors declare that they have no competing interests.
Supplementary Material
References
- 1.Central Brain Tumor Registery of the United States. CBTRUS Statistical Report: Primary Brain and Central Nervous Tumors Diagnosed in the United States in 2004–2008. Hinsdale, IL: CBTRUS; 2012. [Google Scholar]
- 2.Stupp R, Mason WP, van den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987–996. [DOI] [PubMed] [Google Scholar]
- 3.Gehan EA, Walker MD. Prognostic factors for patients with brain tumors. Natl Cancer Inst Monogr. 1977;46:189–195. [PubMed] [Google Scholar]
- 4.Scott GM, Gibberd FB. Epilepsy and other factors in the prognosis of gliomas. Acta Neurol Scand. 1980;61(4):227–239. [DOI] [PubMed] [Google Scholar]
- 5.Gilbert H, Kagan AR, Cassidy F, et al. Glioblastoma multiforme is not a uniform disease! Cancer Clin Trials. 1981;4(1):87–89. [PubMed] [Google Scholar]
- 6.Burger PC, Green SB. Patient age, histologic features, and length of survival in patients with glioblastoma multiforme. Cancer. 1987;59(9):1617–1625. [DOI] [PubMed] [Google Scholar]
- 7.Kowalczuk A, Macdonald RL, Amid ei C, et al. Quantitative imaging study of extent of surgical resection and prognosis of malignant astrocytomas. Neurosurgery. 1997;41(5):1028–1036; discussion 1036–1038. [DOI] [PubMed] [Google Scholar]
- 8.Hess KR. Extent of resection as a prognostic variable in the treatment of gliomas. J Neurooncol. 1999;42(3):227–231. [DOI] [PubMed] [Google Scholar]
- 9.Kreth FW, Berlis A, Spiropoulou V, et al. The role of tumor resection in the treatment of glioblastoma multiforme in adults. Cancer. 1999;86(10):2117–2123. [PubMed] [Google Scholar]
- 10.Lacroix M, Abi-Said D, Fourney DR, et al. A multivariate analysis of 416 patients with glioblastoma multiforme: prognosis, extent of resection, and survival. J Neurosurg. 2001;95(2):190–198. [DOI] [PubMed] [Google Scholar]
- 11.Stummer W, Reulen HJ, Meinel T, et al. Extent of resection and survival in glioblastoma multiforme: identification of and adjustment for bias. Neurosurgery. 2008;62(3):564–576;discussion 564–576. [DOI] [PubMed] [Google Scholar]
- 12.Chaichana KL, Chaichana KK, Olivi A, et al. Surgical outcomes for older patients with glioblastoma multiforme: preoperative factors associated with decreased survival. Clinical article. J Neurosurg. 2011;114(3):587–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hegi ME, Diserens AC, Gorlia T, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352(10):997–1003. [DOI] [PubMed] [Google Scholar]
- 14.Yan H, Parsons DW, Jin G, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360(8):765–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Belden CJ, Valdes PA, Ran C, et al. Genetics of glioblastoma: a window into its imaging and histopathologic variability. Radiographics. 2011;31(6):1717–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iliadis G, Kotoula V, Chatzisotiriou A, et al. Volumetric and MGMT parameters in glioblastoma patients: survival analysis. BMC Cancer. 2012;12:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aghi M, Gaviani P, Henson JW, et al. Magnetic resonance imaging characteristics predict epidermal growth factor receptor amplification status in glioblastoma. Clin Cancer Res. 2005;11(24 Pt 1):8600–8605. [DOI] [PubMed] [Google Scholar]
- 18.Andreou J, George AE, Wise A, et al. CT prognostic criteria of survival after malignant glioma surgery. AJNR Am J Neuroradiol. 1983;4(3):488–490. [PMC free article] [PubMed] [Google Scholar]
- 19.Hammoud MA, Sawaya R, Shi W, et al. Prognostic significance of preoperative MRI scans in glioblastoma multiforme. J Neurooncol. 1996;27(1):65–73. [DOI] [PubMed] [Google Scholar]
- 20.Steltzer KJ, Sauve KI, Spence AM, et al. Corpus callosum involvement as a prognostic factor for patients with high-grade astrocytoma. Int J Radiat Oncol Biol Phys. 1997;38(1):27–30. [DOI] [PubMed] [Google Scholar]
- 21.Pope WB, Sayre J, Perlina A, et al. MR imaging correlates of survival in patients with high-grade gliomas. AJNR Am J Neuroradiol. 2005;26(10):2466–2474. [PMC free article] [PubMed] [Google Scholar]
- 22.Schoenegger K, Oberndorfer S, Wuschitz B, et al. Peritumoral edema on MRI at initial diagnosis: an independent prognostic factor for glioblastoma? Eur J Neurol. 2009;16(7):874–878. [DOI] [PubMed] [Google Scholar]
- 23.Ramakrishna R, Barber J, Kennedy G, et al. Imaging features of invasion and preoperative and postoperative tumor burden in previously untreated glioblastoma: correlation with survival. Surg Neurol Int. 2010;1:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ekici MA, Bulut T, Tucer B, et al. Analysis of the mortality probability of preoperative MRI features in malignant astrocytomas. Turk Neurosurg. 2011;21(3):271–279. [DOI] [PubMed] [Google Scholar]
- 25.Kaur G, Bloch O, Jian BJ, et al. A critical evaluation of cystic features in primary glioblastoma as a prognostic factor for survival. J Neurosurg. 2011;115(4):754–759. [DOI] [PubMed] [Google Scholar]
- 26.Gutman DA, Cooper LA, Hwang SN, et al. MR imaging predictors of molecular profile and survival: multi-institutional study of the TCGA glioblastoma data set. Radiology. 2013;267(2):560–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jain R, Poisson L, Narang J, et al. Genomic mapping and survival prediction in glioblastoma: molecular subclassification strengthened by hemodynamic imaging biomarkers. Radiology. 2013;267(1):212–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mazurowski MA, Desjardins A, Malof JM. Imaging descriptors improve the predictive power of survival models for glioblastoma patients. Neuro Oncol. 2013;15(10):1389–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cordova JS, Schreibmann E, Hadjipanayis CG, et al. Quantitative tumor segmentation for evaluation of extent of glioblastoma resection to facilitate multisite clinical trials. Transl Oncol. 2014;7(1):40–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nicolasjilwan M, Hu Y, Yan C, et al. Addition of MR imaging features and genetic biomarkers strengthens glioblastoma survival prediction in TCGA patients. J Neuroradiol. 2015;42(4):212–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cancer Genome Atlas Research N. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clark K, Vendt B, Smith K, et al. The Cancer Imaging Archive (TCIA): maintaining and operating a public information repository. J Digit Imaging. 2013;26(6):1045–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Visually Accessible Rembrandt Images (VASARI) feature set for human glioma. September 11, 2014.
- 34.Zinn PO, Mahajan B, Sathyan P, et al. Radiogenomic mapping of edema/cellular invasion MRI-phenotypes in glioblastoma multiforme. PloS One. 2011;6(10):e25451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gering DT, Nabavi A, Kikinis R, et al. An integrated visualization system for surgical planning and guidance using image fusion and an open MR. J Magn Reson Imaging. 2001;13(6):967–975. [DOI] [PubMed] [Google Scholar]
- 36.Pichon E, Tannenbaum A, Kikinis R. A statistically based flow for image segmentation. Med Image Anal. 2004;8(3):267–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Archip N, Jolesz FA, Warfield SK. A validation framework for brain tumor segmentation. Acad Radiol. 2007;14(10):1242–1251. [DOI] [PubMed] [Google Scholar]
- 38.Colen RR, Wang J, Singh SK, et al. Glioblastoma: imaging genomic mapping reveals sex-specific oncogenic associations of cell death. Radiology. 2015;275(1):215–227. [DOI] [PubMed] [Google Scholar]
- 39.Thomas RP, Xu LW, Lober RM, et al. The incidence and significance of multiple lesions in glioblastoma. J Neurooncol. 2013;112(1):91–97. [DOI] [PubMed] [Google Scholar]
- 40.Paulsson AK, Holmes JA, Peiffer AM, et al. Comparison of clinical outcomes and genomic characteristics of single focus and multifocal glioblastoma. J Neurooncol. 2014;119(2):429–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chaddad A, Colen RR. Statistical feature selection for enhanced detection of brain tumor. In: SPIE. Vol 9217: Application of Digital Image Processing XXXVII; 2014. [Google Scholar]
- 42.Chaddad A, Zinn PO, Colen RR. Brain tumor identification using Gaussian Mixture Model features and Decision Trees Classification. In: 48th Annual Conference on Information Sciences and Systems (CISS), 2014:1–4. [Google Scholar]
- 43.Chaddad A, Zinn PO, Colen RR. Quantitative texture analysis for glioblastoma phenotypes discrimination. IEEE CODIT, Biomedical engineering and clinical application; 2014. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.