NUT midline carcinoma (NMC), a subtype of squamous cell cancer, is driven by the creation of a translocation oncoprotein, BRD4-NUT. Alekseyenko et al. found that BRD4-NUT forms distinctive nuclear foci in patient tumors, which correlate with ∼100 unprecedented, hyperacetylated expanses of chromatin that reach up to 2 Mb in size. These “megadomains” appear to drive transcription of underlying DNA in NMC patient cells and naïve cells induced to express BRD4-NUT.
Keywords: chromatin hyperacetylation, BRD4, topological domains
Abstract
NUT midline carcinoma (NMC), a subtype of squamous cell cancer, is one of the most aggressive human solid malignancies known. NMC is driven by the creation of a translocation oncoprotein, BRD4-NUT, which blocks differentiation and drives growth of NMC cells. BRD4-NUT forms distinctive nuclear foci in patient tumors, which we found correlate with ∼100 unprecedented, hyperacetylated expanses of chromatin that reach up to 2 Mb in size. These “megadomains” appear to be the result of aberrant, feed-forward loops of acetylation and binding of acetylated histones that drive transcription of underlying DNA in NMC patient cells and naïve cells induced to express BRD4-NUT. Megadomain locations are typically cell lineage-specific; however, the cMYC and TP63 regions are targeted in all NMCs tested and play functional roles in tumor growth. Megadomains appear to originate from select pre-existing enhancers that progressively broaden but are ultimately delimited by topologically associating domain (TAD) boundaries. Therefore, our findings establish a basis for understanding the powerful role played by large-scale chromatin organization in normal and aberrant lineage-specific gene transcription.
Defective chromatin regulators are a common feature of cancer. However, understanding the relevant consequences in most malignancies has been difficult because the disrupted pathways are complex and poorly understood. NUT midline carcinoma (NMC) provides a powerful model in which a translocation-derived fusion protein plays a defining role in the oncogenic blockade of squamous cell differentiation (French et al. 2003; Bauer et al. 2012).
NMC is characterized by the presence of NUT fusion oncogenes (also known as NUTM1), the most common being BRD4-NUT. The NUT fusion oncogenes are as yet the only genetic abnormality found in this cancer, which otherwise typically reveals simple cytogenetics (Kees et al. 1991; Kubonishi et al. 1991; Lee et al. 1993; Toretsky et al. 2003; Thompson-Wicking et al. 2013). Moreover, the years of acquired mutations that form more common squamous cell carcinomas are not required for NMC, which frequently occurs in children and has been reported in neonates (French et al. 2004; Shehata et al. 2010; Bauer et al. 2012). The existence of this single oncogene in an extraordinarily aggressive subtype of squamous cell carcinoma (median survival, 6.7 mo) (Bauer et al. 2012) suggests that BRD4-NUT is a powerful driving oncoprotein with defined targets that should be amenable to chromatin-based analyses. Indeed, knockdown of BRD4-NUT leads to terminal squamous differentiation and arrested proliferation of NMC cells, indicating that it plays a critical role in proliferation through a blockade of differentiation (French et al. 2008).
All known NUT fusion oncogenes (BRD3-NUT [French et al. 2008], BRD4-NUT, or NSD3-NUT [French et al. 2014]) result in the association of NUT with a BET protein, BRD4 or BRD3, defined by the presence of dual bromodomains and an ET domain. The BET protein tethers NUT to acetylated chromatin via its dual bromodomains, forming ∼80–100 large, hyperacetylated nuclear foci (Fig. 1A; French et al. 2008; Reynoird et al. 2010; Yan et al. 2011; data not shown). The function of these foci is not known; however, they have been observed in all of the BRD-NUT tumor tissue and patient cells examined to date (French et al. 2008; Haack et al. 2009; Reynoird et al. 2010; Yan et al. 2011). The critical nature of the bromodomain–acetyl–lysine interaction has been exploited by the development of acetyl lysine mimetic molecules, termed BET inhibitors, that rapidly induce differentiation of NMC cells both in vitro and in vivo (Filippakopoulos et al. 2010) with concomitant disappearance of the hyperacetylated nuclear foci. The effect of BET inhibitors on NMC growth has led to clinical trials treating NMC (ClinicalTrials.gov identifiers NCT01587703, NCT01987362, NCT02259114) and numerous new studies indicating that many other cancer types, including more common hematopoietic and solid malignancies, are dependent on endogenous, nonmutant BRD4 for growth (Delmore et al. 2011; Mertz et al. 2011; Zuber et al. 2011; Henssen et al. 2013; Puissant et al. 2013; Asangani et al. 2014). In these non-NMC cancers, it appears that an important function of BRD4 is its association with genes that define cell identity and encode critical oncogenic driver proteins, such as MYC. MYC has also been implicated as a critical target of BRD4-NUT (Grayson et al. 2014).
Figure 1.
The BRD4-NUT complex forms chromosomal megadomains driving ectopic transcription. (A) Endogenous BRD4-NUT localization in interphase and metaphase nuclei, detected with rabbit monoclonal anti-NUT antibody clone C52. (Left) Immunofluorescence confocal microscopy of endogenous BRD4-NUT expressed in TC-797 cells. (Middle) A human NMC biopsy immunohistochemically stained with anti-NUT. (Right) Immunofluorescence of a metaphase preparation of TC-797 NMC cells stained with anti-NUT. (Inset) Magnified view of chromosomes from the same image. (B) Immunofluorescence of BioTAP-tagged BRD4-NUT. 797TRex and 293TRex Flp-in cells were induced to express C-terminal-tagged or N-terminal-tagged BRD4-NUT. (C) Example of a BRD4-NUT megadomain in 293T cells. An ∼900-kb region of chromosome 20 is shown, illustrating extensive enrichment for BioTAP-tagged BRD4-NUT (16 h), N-Flag-C-HA-BRD4-NUT (7 h), and H3K27ac (7 h) following induction as indicated. The calculated extent of the megadomain is shown as a bar below the H3K27ac track. Transcriptional changes ([red] + strand; [blue] − strand) accompanying induction of BRD4-NUT in 293T cells are illustrated by the 0-h (before induction) and 7-h (following induction) nascent RNA sequencing (RNA-seq) reads. (D) The extent of BRD4-NUT megadomains is an order of magnitude larger than superenhancer regions. The size distribution of H3K27ac regions is shown for 293T cells before (blue) and after (red) induction of BRD4-NUT. (E) Average log2 enrichment profiles of H3K27ac are shown for enhancer regions, superenhancers, and BRD4-NUT megadomains detected in 293T cells. The profiles are scaled to their corresponding median sizes (8 kb, 28 kb, and 177 kb, respectively). The gray area around the average enrichment curve shows the 99.9% confidence interval. (F) Example of a BRD4-NUT megadomain in patient-derived TC-797 cells. A representative ∼900-kb region of chromosome 10q21.1 is shown. The megadomains and transcripts within them are sensitive to JQ1 treatment as measured by nascent RNA-seq. The “IP BRD4-NUT (4 h JQ1 vs. no JQ1)” track shows log2 fold change in BRD4-NUT enrichment with and without JQ1 treatment ([blue] depleted at 4 h; [brown] enriched), illustrating significant reduction of BRD4-NUT enrichment 4 h after JQ1 treatment. Most transcripts within the domain are also attenuated at 4 h (see also Supplemental Fig. S1).
Studies investigating NUT, which is not expressed in normal squamous cells, have provided an important clue to understanding BRD4-NUT function. The NUT portion of BRD-NUT oncoproteins contains a putative transcriptional activation domain that can recruit p300 and activate its histone acetyltransferase (HAT) activity (Reynoird et al. 2010). It is hypothesized that a feed-forward loop of p300 recruitment, acetylation, and further BRD-NUT recruitment to newly acetylated chromatin leads to the sequestration of p300 into BRD-NUT nuclear foci (Reynoird et al. 2010; French 2012). This has led to two nonmutually exclusive models for how BRD-NUT drives oncogenesis. In one, BRD-NUT represses the expression of prodifferentiation genes through its sequestration of p300 away from these genes (Reynoird et al. 2010; Yan et al. 2011; French 2012, 2014). In the other, BRD-NUT directs p300-dependent transcriptional activity to genes required for proliferation and the blockade of differentiation (French 2014; Wang and You 2015).
To define the molecular mechanisms underlying this extraordinarily potent oncoprotein, we sought to identify the genomic loci with which BRD4-NUT associates and define its effects on transcription. Strikingly, we discovered that the BRD4-NUT protein occupies ∼100–200 extremely broad, cell type-specific hyperacetylated domains in the genome. These “megadomains” are much larger than typical activated regions, ranging from 100 kb to 2 Mb. In NMC patient cells, megadomains are associated with the induction of BET inhibitor-sensitive transcription of underlying DNA, leading to dysregulation of MYC, p63, MED24, and additional potential targets. Our ability to induce megadomains in naïve cells revealed initiation at seed sites, typically enhancers but surprisingly not biased toward superenhancers. Most strikingly, megadomains spread to selectively fill entire topologically associating domains (TADs). These results implicate a dramatic misregulation of chromatin and transcription within TAD-delimited regulatory domains in BRD-NUT-driven oncogenesis.
Results
BRD4-NUT nuclear foci correspond to chromatin-bound ‘megadomains’ that drive ectopic transcription in naïve cells
BRD4-NUT associates with chromatin via the dual bromodomains of BRD4 (Dey et al. 2003; French et al. 2008; Grayson et al. 2014), forming ∼80–100 large foci that can be visualized in interphase nuclei and metaphase chromosomes (Fig. 1A). The number of BRD4-NUT foci are similar in cultured NMC cell lines and primary NMC patient tissue. The size of the nuclear foci suggests that BRD4-NUT protein interactions occur in large aggregates (Reynoird et al. 2010; Yan et al. 2011). BRD4-NUT binding might aggregate acetylated chromatin in three dimensions or, alternatively, encompass large contiguous stretches of the genome. To investigate whether expression of BRD4-NUT is sufficient to form foci in non-NMC cells, we expressed transgenic BioTAP-tagged BRD4-NUT in 293TRex, a derivative of the 293T cell line originating from embryonic kidneys (Supplemental Fig. S1A). The BioTAP tag consists of a protein A moiety and a biotin-accepting sequence that is recognized by endogenous mammalian biotin ligases and has been successfully applied to the analysis of diverse chromatin proteins (Alekseyenko et al. 2014a,b). We found that expression of either N-terminal-tagged or C-terminal-tagged BioTAP-BRD4-NUT caused the formation of hyperacetylated nuclear foci in 293T cells, similar in number and appearance to those of endogenous BRD4-NUT in cultured patient-derived cells or primary tumor tissue (Fig. 1B; data not shown).
Based on these results, we proceeded to perform ChIP-seq (chromatin immunoprecipitation [ChIP] combined with deep sequencing) from the 293T cells expressing tagged BRD4-NUT via tandem affinity pull-down of the BioTAP tag. We found that both N-terminal-tagged and C-terminal-tagged BioTAP-BRD4-NUT were bound at highly enriched and very broad domains in the 293TRex genome (262 regions at a size cutoff of 96 kb) (example shown in Fig. 1C). Based on their unprecedented size, we named these BRD4-NUT regions “megadomains.”
To confirm that these results were independent of the epitope tag used, we expressed Flag-BRD4-NUT-HA and found that it bound to the same set of broad domains in 293T cells. Consistent with previous imaging data of nuclear foci, these BRD4-NUT megadomains also showed marked enrichment for H3K27 acetylation by ChIP-seq (Fig. 1C; Supplemental Fig. S1B). While H3K27ac normally marks active enhancers and other regulatory regions, the spatial extent of the H3K27ac enrichment at the BRD4-NUT domains is an order of magnitude larger than recently defined “superenhancers” (Fig. 1D,E) and displays a more continuously enriched profile rather than appearing as a cluster of individual peaks (Fig. 1C,E). As the H3K27ac antibody provides a particularly robust signal, we used it to optimize the parameters for uniform calling of megadomain boundaries in different cell types going forward (see the Materials and Methods). The megadomains in 293T cells ranged in size from 96 kb to 1.4 Mb (mean size of 247 kb).
We next sought to evaluate the effect of BRD4-NUT binding on transcription of underlying DNA. Nascent RNA sequencing (RNA-seq) performed to capture immediate changes in transcription revealed that BioTAP-BRD4-NUT megadomains were associated with an overall increase in transcription of genes and noncoding RNAs (ncRNAs) within the domain and adjacent regions (Fig. 1C; Supplemental Fig. S2A). The fact that this occurs in naïve cells that do not harbor an endogenous BRD4-NUT oncoprotein supports a model in which expression of BRD4-NUT leads to de novo megadomain formation and increased transcription.
Cell type-specific megadomains drive transcription in NMC patient cells
To determine whether the results in 293T cells are representative of BRD4-NUT localization and activity in patient NMC cells, we examined 797TRex Flp-in cells (Grayson et al. 2014) induced to express BioTAP-tagged BRD4-NUT. 797TRex Flp-in cells are derived from TC-797 cells from a patient with NMC (Toretsky et al. 2003) and express endogenous oncogenic BRD4-NUT. Analysis of DNA from the BioTAP-BRD4-NUT pull-downs revealed a number and size of megadomains similar to those in 293TRex cells (Fig. 1F), but, interestingly, there was very little overlap between 797TRex and 293TRex megadomains (Supplemental Fig. S1C). To test whether the megadomains could be an artifact of overexpression of tagged BRD4-NUT, we performed ChIP of endogenous BRD4-NUT in TC-797 cells using a highly specific monoclonal antibody to NUT (Haack et al. 2009). Anti-NUT ChIP-seq revealed that endogenous BRD4-NUT occupied similar-sized megadomains that mostly overlapped those in 797TRex-BioTAP-BRD4-NUT cells (73.2% based on overall sequence overlap; 56% based on the number of domains), confirming our original result with recombinant BioTAP-BRD4-NUT (Fig. 1F).
Despite the lack of overlap between most regions bound in TC-797 and 293T cells, BRD4-NUT megadomains in TC-797 cells are also strongly enriched for H3K27ac (Fig. 1F) and show other chromatin features associated with active transcription, as previously noted for BRD4-NUT nuclear foci (Reynoird et al. 2010; Yan et al. 2011). Specifically, ChIP-seq in TC-797 cells revealed concurrent enrichment of megadomains for H3K9Ac, H3K14Ac, and H3K36Me3 (Fig. 2A).
Figure 2.
Transcriptionally active, BET inhibitor-sensitive BRD4-NUT megadomains arise from seed regions. (A) BRD4-NUT megadomains are enriched for histone marks associated with active transcription. Average enrichment profiles within the megadomains (scaled to the same size) and 100-kb flanking regions are shown for TC-797 cells. (B) Average BRD4-NUT enrichment within the megadomains is reduced following JQ1 treatment of TC-797 cells. Scaled average profiles of BRD4-NUT enrichment are shown for TC-797 cells without JQ1 and 4 h following JQ1 treatment. (C) The average level of transcriptional activity is reduced within the megadomains and their flanking regions following JQ1 treatment of TC-797 cells. Similar to Figure 1F, the scaled average profile of nascent RNA-seq is shown for cells without JQ1 treatment and for cells 1 h, 2.5 h, and 4 h following JQ1 treatment. (D) Expression levels (log2[FPKM]) of all genes within the megadomain regions in TC-797 cells show broad reduction of transcriptional activity following JQ1 treatment. Each column corresponds to a gene within the TC-797 megadomain. Each row indicates JQ1 treatment condition: no JQ1 or 1 h, 2.5 h, or 4 h following treatment. (FPKM) Fragments per kilobase of exon per million reads mapped. (E) BRD4-NUT and H3K27ac enrichment in a 293T BRD4-NUT induction time course. The resulting domain calls (based on H3K27ac profiles) are shown as bars below the enrichment profiles. The domain border expansion is traced using green (expanding to the left) and red (expanding to the right) arrows. Seed regions are indicated by brown dotted circles, and an interseed region that will later “fill in” with BRD4-NUT and H3K27ac is indicated with a dotted green circle. For illustrative purposes, dotted arrows from seed and interseed regions point to corresponding regions in the violin plot (F) of all seed and interseed regions. (F) BRD4-NUT megadomain seed regions are enriched with H3K27ac. The violin plots show H3K27ac enrichment in 293T cells prior to BRD4-NUT induction for the seed regions that will show BRD4-NUT enrichment within 2 h following induction (early domains; brown) and the interseed regions that will be bound by BRD4-NUT only after 2 h (late domains; green). Colored dotted arrows pointing from the corresponding examples of these regions in Figure 1E are shown for illustrative purposes. The H3K27ac enrichment level found at regular enhancers within 293T cells is shown as a reference (gray). Genome-wide average enrichment of H3K27ac is shown with the dashed brown line. (G) Examples of the three classes of BRD4-NUT domain formation. Domains of the first class are formed by expanding from a single pre-existing H3K27ac region. Class 2 domains form by expanding from multiple pre-existing H3K27ac regions. Class 3 domains start with one or more pre-existing H3K27ac regions and activate additional enhancer-like regions during formation (see also Supplemental Fig. S2).
If BRD4-NUT megadomains require bromodomain interaction with acetylated chromatin and are important for the block in differentiation of NMC cells in culture, then treatment with the BET inhibitor JQ1, which leads to differentiation of NMC cells (Filippakopoulos et al. 2010), should reverse their formation. Indeed, we found that megadomain occupancy by endogenous BRD4-NUT diminished rapidly (Figs. 1F, 2B) within 4 h of JQ1 treatment.
To determine the impact of the BRD4-NUT megadomains on transcription in TC-797 cells, we performed nascent RNA-seq over a time course following JQ1 treatment of TC-797 cells (Figs. 1F, 2C,D). Transcription decreased with similar kinetics in both genic and intergenic regions, with some small degree of recovery observed by 4 h (Supplemental Fig. S2B). At 4 h following JQ1 treatment, the genes within megadomains were significantly more affected than those outside (>1 Mb away; P = 2.0 × 10−28, Wilcoxon rank-sum test), with the majority exhibiting down-regulation of expression (85.8%) (Fig. 2D). These results confirmed the strong link between BRD4-NUT targeting, histone acetylation, and increased transcription.
Megadomains initiate from a cell-specific subset of enhancers
It has been hypothesized that a feed-forward loop of p300 recruitment, acetylation, and further BRD4-NUT recruitment to newly acetylated chromatin is a primary feature of BRD4-NUT potency (Reynoird et al. 2010; French 2012; Wang and You 2015). To determine whether this model might be relevant to the formation of BRD4-NUT megadomains, we asked whether p300 was broadly colocalized with BRD4-NUT in NMC patient cells. We found a strong correlation between BRD4-NUT, p300, and active histone marks, particularly H3K27ac, in megadomains (Figs. 1F, 2A). These results support the feed-forward model in which the NUT portion of the fusion oncoprotein attracts HATs to reinforce and spread BRD4 binding through its bromodomains.
To further test the model, we asked whether induction of BRD4-NUT in naïve cells might show a time course of progression from initial binding sites to full megadomain targeting. We performed ChIP-seq for H3K27ac over a time course of BRD4-NUT induction in 293TRex Flp-in cells, aiming to identify the elements within the megadomains from which the BRD4-NUT domain formation initiates. We found that BRD4-NUT induction results in the gradual spread of BRD4-NUT from relatively narrow peaks into megadomains, accompanied by simultaneous coexpansion of H3K27Ac (Fig. 2E). Focusing on H3K27ac, we examined the earliest (2-h) time point to determine a set of “early domains” within the regions that will give rise to megadomains. We found that before BRD4-NUT induction, the regions that will become these “early domains” (or seed regions) are strongly enriched with H3K27ac. The H3K27ac enrichment of these seed regions is comparable in magnitude with enrichment observed at enhancers and is notably higher than the interseed regions that will eventually become filled in by the megadomains (Fig. 2F). This indicates that, in most cases, the megadomain initiates from a “seed” enhancer or regulatory region already existing in the cell of origin. Despite the association of wild-type BRD4 and H3K27ac occupancy with “superenhancers” implicated in various cancers (Loven et al. 2013), it should be noted that the seed enhancers in our analyses are not the most active set of enhancers, and, perhaps surprisingly, superenhancers are not overrepresented among “seed” regions (P = 0.988) (Supplemental Fig. S2C).
The BRD4-NUT induction time course suggests three types of domains, the simplest one containing a single seed enhancer (class 1), more complex cases containing multiple seed enhancers (class 2), and some in which additional enhancer-like regions arise, including some without pre-existing enhancers (class 3) (Fig. 2G). These three classes account for 46.5% of megadomains, while the remaining megadomains form without clearly defined seed regions, although the exact fraction depends on the stringency of the enhancer calls (Supplemental Fig. S2D).
The lack of overlap between megadomains observed in 293T and TC-797 cells (Supplemental Fig. S1D) and the observation that many megadomains arise from pre-existing enhancers are consistent with a model in which localization of the BRD4-NUT megadomains is determined by the initial transcriptional and regulatory configuration of the cell. By that logic, megadomains in cells of similar derivation should demonstrate more overlap. Furthermore, the location of megadomains in NMC cells may reveal common gene targets that are critical to the blockade of differentiation and growth of NMC cells. Thus, we compared, by ChIP-seq, the localization of endogenous BRD-NUT megadomains in several different NMCs, including TC-797 (BRD4-NUT+, thymus), PER-403 (BRD4-NUT+, lung), 1015 (BRD4-NUT+, lung), and tumor tissue from which 1015 was derived (Fig. 3A–C). Consistent with our expectations, the non-NMC 293T cells showed the most distinct megadomain placement compared with the NMC-originating cells. Similarly, we found the highest domain number overlap between the 1015 cell line and the patient tumor tissue of origin (72%), demonstrating that megadomains are not artifacts of cells in culture but are also observed in NMC patient tissue. This is also consistent with the characteristic detection of BRD4-NUT nuclear foci in both NMC tumors and patient-derived cell lines. A notable percentage (32.2%) of megadomains did overlap in at least two NMC cell lines (Fig. 3A,B; Supplemental Fig. S3), and 10.4% overlapped in three of the five cell lines and derivatives. Only 1.74% overlapped in all NMCs (Fig. 3A; Supplemental Table 1S); however, the mean size of megadomains was within the same magnitude (247–565 kb) in all of the examined cell lines.
Figure 3.
BRD4-NUT domain placement in different cells reveals a limited number of common regions. (A) Overview of BRD4-NUT megadomain positions within the genome. Positions of the BRD4-NUT megadomains detected in the five examined NMC cell types and derivatives (TC-797, 797TRex, Per403, 1015 tissue, and 1015 cell line) are shown as black rectangles. Loci at which domains were observed in three or more cell types are highlighted with red. (B) Related cell types show higher similarity of megadomain positions. A clustered matrix shows pair-wise similarity of BRD4-NUT megadomain positions detected in different cell types. The number of megadomains in each cell type (row) overlapped by another cell type (column) is shown by the numbers in each cell, and the percentage overlap is shown with color. (C) An example of BRD4-NUT megadomains within the MYC locus in different cell types. Tracks shown in brown represent enrichment and the calculated megadomain of BRD3-NUT in this region. The plot also shows the effect of JQ1 treatment on the transcriptional activity (as measured by nascent RNA-seq) over time in TC-797 cells as described above (see also Supplemental Fig. S3).
Megadomains often fill TADs
As many megadomains exhibit well-defined boundaries, we sought to identify genomic elements that may determine the extent of the megadomains. Recently published high-resolution Hi-C data have demonstrated that TADs are chromatin compartments that are largely conserved in their positions between cell types (Dixon et al. 2012). Therefore, we used high-resolution mapping data from the GM12878 cell line (Rao et al. 2014) to determine whether megadomain boundaries in NMC cells commonly coincide with the boundaries of TADs (Fig. 4A). Indeed, the average Hi-C signal (observed over expected) across all megadomains (Fig. 4B) as well as TAD boundary scores (Fig. 4C; see the Materials and Methods) demonstrate that most of the megadomains are bounded by the extent of a TAD. As was previously observed for TAD boundaries (Dixon et al. 2012; Rao et al. 2014), the megadomain boundaries also show significant enrichment in occurrence of active enhancers (Fig. 4D), CTCF binding (Fig. 4E), and transcription start sites (P < 0.001) (Supplemental Fig. S2E).
Figure 4.
BRD4-NUT megadomains expand from a select set of pre-existing enhancers to fill topological domains. (A) The extent of the BRD4-NUT domains tends to correspond to topological domains. Hi-C observed/expected signal (from GM12878 cells) is shown for the c-MYC region. While the extent of BRD4-NUT megadomains (gray bars on the margins) varies between the cells, their boundaries tend to correspond to the boundaries of different TADs. (B) BRD4-NUT megadomains tend to coincide with the topological domains. An average observed/expected Hi-C interaction signal is shown for the domains identified in all of the examined cell lines. For the regions where domains were observed in multiple cell types, a union of all domain regions was used. The profile reveals strong interactions between domain ends and within the domain and a lack of interaction of the domain with the neighboring regions. (C) Most of the BRD4-NUT megadomain boundaries coincide with topological domain boundaries. A Hi-C topological domain boundary score (see the Materials and Methods) is shown for all of the detected megadomains (heat map at the bottom), with the average profile shown on the top. (D) BRD4-NUT megadomain boundaries tend to coincide with enhancer positions. The average frequency of occurrence of annotated enhancer positions (across 86 cell and tissue types) (Hnisz et al. 2013) is shown. The orange curve and light-orange area show the mean and standard error of the occurrence frequency of megadomain-sized blocks randomly positioned within the genome. (E) BRD4-NUT megadomain boundaries tend to coincide with CTCF-binding sites. The orange curve and light-orange area show the mean and standard error of the occurrence frequency of randomly placed megadomain-sized blocks (see also Supplemental Fig. S4).
Examining the genomic regions where megadomains occur in more than one cell type, we found that the exact boundaries of megadomains can vary between cell types, with their positions coinciding with secondary minor TAD boundaries within larger major TADs (Fig. 4A). We also found that, in some cases, the megadomains do not exhibit well-defined boundaries, with the enrichment of BRD4-NUT and H3K27ac tapering off without reaching the TAD boundary (Supplemental Fig. S4). This suggests that the extent of megadomains is determined by a dynamic balance of acetylation and deacetylation, with more prominent domains being able to extend the entire length of a TAD and, in some cases, overcome intermediate TAD boundaries. These observations are consistent with important roles for TADs in delimiting chromatin and regulatory domains within the genome (Dixon et al. 2012; Lupianez et al. 2015). The ability to fill whole megabase-scale regulatory regions is an exceptional characteristic of BRD4-NUT that is likely to help fuel its oncogenic properties.
Megadomains overlap at oncogenic loci in multiple NMC cell lines
Because megadomains are associated with increased transcription of underlying and nearby genes, the common megadomains may deregulate the expression of genes critical to NMC oncogenic growth. We therefore identified genes within megadomains common to multiple (at least three of four) NMCs whose expression decreased most significantly upon treatment with JQ1. We found 1826 annotated genes within or adjacent to (±1 Mb from the edge of the megadomain) overlapping megadomains in at least three of four cell lines. Of these genes, 103 demonstrated significant reduction in expression in response to JQ1 treatment (twofold reduction; P < 0.001), but only 33 exhibited relatively abundant (fragments per kilobase of exon per million reads mapped [FPKM] > 0.5) baseline expression (Table 1). Within this group of 33 genes, there are 13 long ncRNAs (lncRNAs), among which were three—PVT1, CCAT1 (CARLo-5), and CASC19 (CARLo-6)—that map near the MYC locus and are reported to play oncogenic roles in its up-regulation (Fig. 3C; Nagoshi et al. 2012; Kim et al. 2014; Riquelme et al. 2014; Tseng et al. 2014; Xiang et al. 2014; Zhang et al. 2014). The identification of these lncRNA loci associated with BRD-NUT megadomains is of particular interest given the known critical role of MYC in the blockade of differentiation and maintenance of NMC growth (Grayson et al. 2014). Other cancer-related genes in this group include the squamous carcinoma-associated oncogene TP63 (Stransky et al. 2011; Ramsey et al. 2013; Venkatanarayan et al. 2015), SOX5 (Zafarana et al. 2002; Ma et al. 2009), MYB (Yokota et al. 1986; Miao et al. 2011; Gao et al. 2014), and MED24 (Clark et al. 2002; Luoh 2002; Hasegawa et al. 2012). The identification of several JQ1-sensitive cancer-associated genes underlying megadomains common to multiple NMC cell lines and tissue provides support for our hypothesis that BRD-NUT megadomains may deregulate expression of critical progrowth genes.
Table 1.
Prioritization of genes and ncRNAs potentially important to NMC growth

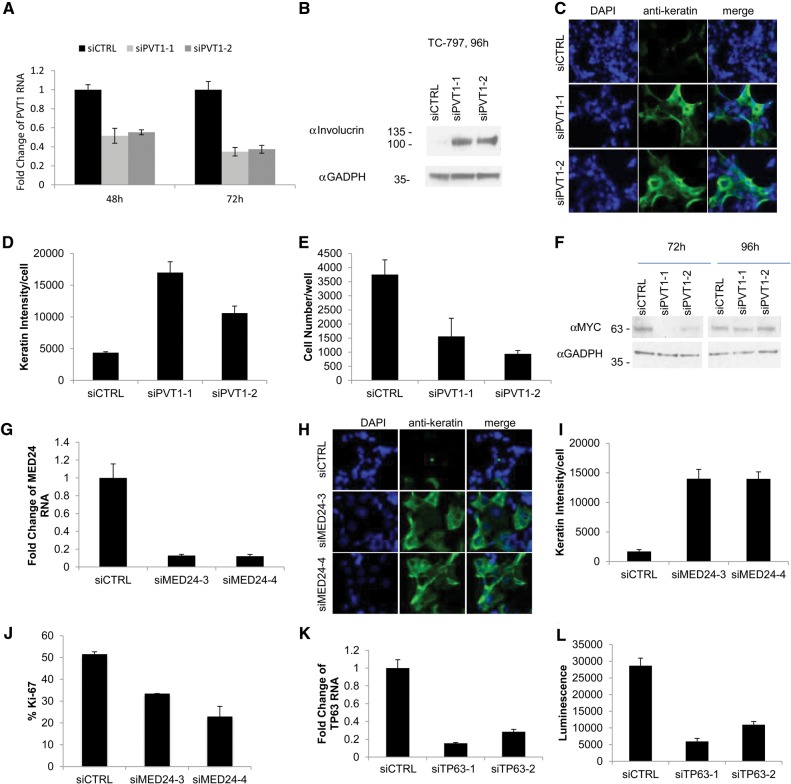
Our next set of experiments was designed to determine whether the expression of at least a subset of these megadomain/cancer-associated genes is required for NMC growth. We first sought to determine the importance of PVT1 and CCAT1 lncRNAS in the blockade of differentiation and proliferation of NMC cells given (1) their strong JQ1 sensitivity in our nascent-seq and quantitative RT–PCR (qRT–PCR) profiles (Fig. 3C; Supplemental Fig. S5A), (2) their linkage and prior relationships to MYC regulation, and (3) their association with megadomains that overlap in all NMC cell lines and tissue analyzed, including a BRD3-NUT NMC cell line, 10326 (Fig. 3A,C). CCAT1 has been implicated in up-regulation of MYC, but we were unable to achieve sufficient knockdown of this lncRNA to adequately evaluate its effect on MYC levels or the growth of NMC cells (data not shown). However, treatment with siRNAs targeting PVT1 (Fig. 5A) resulted in differentiation of TC-797 NMC cells, as scored by expression of the epithelial marker keratin and the terminal squamous differentiation marker involucrin (Fig. 5B–D), morphologic flattening (Fig. 5C), and decreased proliferation (Fig. 5E). In these cells, knockdown of PVT1 resulted in decreased MYC protein but not RNA (Fig. 5F; Supplemental Fig. S5B), supporting a post-transcriptional role for PVT1 RNA in MYC regulation. These results are consistent with studies in mice in which PVT1 RNA has been proposed to stabilize MYC protein through direct interaction (Tseng et al. 2014). Our results support a mechanism by which BRD-NUT megadomains maintain MYC protein levels and thereby blockade of differentiation at least in part through the up-regulation of PVT1 lncRNA.
Figure 5.
Evidence that the BRD4-NUT megadomain-associated genes PVT1, MED24, and TP63 are oncogenic drivers in NMC. (A) qRT–PCR of PVT1 levels following transfection of control or PVT1 siRNAs into TC-797 cells. qRT–PCR was performed in triplicate. Error bars indicate the mean ± SD of the triplicate wells. (B) Immunoblot of involucrin levels (Sigma, catalog no. I9018) in TC-797 cells subjected to siRNA knockdown of PVT1. (C) High-throughput 384-well plate immunofluorescent assay of keratin expression in TC-797 cells 96 h following siRNA knockdown of PVT1. TC-797 cells were stained with anti-keratin (Dako), and nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI). Representative photographs use identical magnification (×400). (D,E) Quantitative analysis of keratin intensity and cell numbers from the high-throughput assay of PVT-1 knockdown in C. Average results from three biological replicates, each performed in triplicate, are shown. Error bars indicate the mean ± SD of the triplicate experiments. (F) Immunoblot of MYC levels (Cell Signaling, catalog no. 9402) in TC-797 cells subjected to siRNA knockdown of PVT1. Protein levels are decreased after 72 h but appear to recover at 96 h. However, at both time points, differentiation has already commenced, beginning at 48 h. (G) qRT–PCR of MED24 mRNA levels following transfection of control or MED24 siRNAs into TC-797 cells. qRT–PCR was performed in triplicate. Error bars indicate the mean ± SD of the triplicate wells. (H) Immunofluorescent assay of keratin expression in TC-797 cells 96 h following MED24 knockdown as in C. Representative photographs use identical magnification (×400). (I) Quantitative analysis of keratin intensity after MED24 knockdown in TC-797 cells in H. Results are from triplicate experiments, as in D. (J) Ki-67 fraction (marker of cell division) quantified from MED24 knockdown in TC-797 cells in H. Results are from triplicate experiments, as in D. (K) qRT–PCR of TP63 mRNA levels following transfection of control or TP63 siRNAs into TC-797 cells. qRT–PCR was performed in triplicate. Error bars indicate the mean ± SD of the triplicate wells. (L) Cell viability assay (CellTiter-Glo) of TC-797 cells 96 h following siRNA knockdown of TP63 versus control. Results are from three biological replicates, each performed in triplicate. Error bars indicate the mean ± SD of the triplicate wells (see also Supplemental Fig. S5).
We next turned our attention to genes not previously implicated in NMC pathogenesis whose expression is coupled to megadomains. Identification of the Mediator subunit MED24 as a BRD4-NUT target gene (Table 1) suggested an additional positive reinforcement loop for BRD4-NUT oncogenesis, as Mediator is known to interact physically with BRD4 (Jiang et al. 1998; Houzelstein et al. 2002; Dawson et al. 2011; Rahman et al. 2011). While MED24 serves a broad role in transcriptional regulation during embryonic development (Ito et al. 2002), its post-embryonic role appears to be tissue-specific coactivation of gene expression (Gu et al. 1999; Zhang and Fondell 1999). It is essential for pubertal breast development and in vitro breast cancer growth and is amplified in breast cancer (Clark et al. 2002; Luoh 2002; Hasegawa et al. 2012). The known physical association of MED24 and BRD4 (Dawson et al. 2011; Rahman et al. 2011) as well as BRD4's well-documented role in breast and other epithelial cancers (Crawford et al. 2008; Kadota et al. 2009; Alsarraj and Hunter 2012; Asangani et al. 2014; Shi et al. 2014, 2015) made MED24 a particularly attractive novel BRD4-NUT target to evaluate for its oncogenic function. Therefore, we tested the effect of MED24 knockdown on TC-797 cells in culture. We found that siRNAs targeting MED24 (Fig. 5G) induced differentiation of TC-797 NMC cells, as scored by expression of keratin (Fig. 5H,I), morphologic flattening (Fig. 5H), and decreased proliferation (Fig. 5J). These results support a model in which MED24 participation as both a target and a cofactor of BRD4-NUT protein complexes is likely to provide another positive reinforcement loop for the establishment of megadomains and their transcriptional activity.
Finally, we noted that TP63 maps to a megadomain region in all NMC cells tested and is also sensitive to JQ1 treatment (Table 1). TP63 is an epithelial-specific developmental gene (Yang et al. 1998; Koster and Roop 2004) that encodes a transcription factor with an established role in squamous cell carcinoma (Stransky et al. 2011; Ramsey et al. 2013; Venkatanarayan et al. 2015). Furthermore, p63 is a p53 family member whose oncogenic ΔN isoform is expressed in the majority of NMCs (Bishop and Westra 2012; Tilson and Bishop 2013). Therefore, to test for a potential role in NMC, we treated TC-797 cells with siRNAs against TP63. We found that knockdown of p63 led to markedly decreased viability of NMC cells (Fig. 5K,L) but, interestingly, did not lead to differentiation (Supplemental Fig. S5D). These results suggest that TP63 is yet another megadomain-associated gene required for NMC cell growth and viability but lacks a detectable role in the blockade of differentiation. Taken together, the findings further support a mechanism by which BRD4-NUT maintains NMC growth and arrested differentiation through not only the regulation of MYC but also the positive regulation of MED24 and p63. Thus, even in this “simple” genetic cancer model, our results suggest that multiple BRD4-NUT megadomain-regulated factors conspire to block differentiation and/or maintain cancer growth.
Discussion
A shortcut to epithelial cancer
Normal cells possess multiple mechanisms to protect against events that may lead to abnormal proliferation. Cancer cells navigate around these safeguards through the acquisition of numerous aberrations, including the development of immortalization, resistance to apoptosis, alteration of cellular metabolism, maintenance of growth, and avoidance of growth suppression. To attain these “hallmarks of cancer” (Hanahan and Weinberg 2000), epithelial cancer cells characteristically evolve complex mutational backgrounds. NMC is one rare epithelial cancer that has possibly developed a genetic shortcut to acquire this cancer phenotype, as evidenced by its simple cytogenetics and occurrence in infants and young children. Here we demonstrate a likely mechanism for this short evolution: massive disruption of normal gene regulation at key cell type-specific genes in NMC cells by the oncogenic fusion protein BRD4-NUT. Our model of this feed-forward process is illustrated in Figure 6. BRD4-NUT forms enormous (0.1- to 1.5-Mb) contiguous stretches of active chromatin, which we termed “megadomains.” Megadomains are enriched with BRD4-NUT and p300, resulting in increased transcription of underlying ncRNAs and adjacent genes. The expression of at least a subset of megadomain-containing genes is critical to the growth of the host NMC cell.
Figure 6.
Feed-forward model of megadomain formation. BRD4-NUT is tethered to acetylated chromatin by BRD4 bromodomains. The NUT portion of the fusion protein recruits p300, leading to increased local histone acetylation and thus a self-perpetuating recruitment of BRD4-NUT. The ultimate result is uncontrolled spreading of BRD4-NUT across chromatin, forming megadomains that typically are limited only by TAD boundaries.
While the chromatin-association of megadomains is bromodomain-dependent, their genomic localization varies with respect to cell type, showing more similarity between cells of similar origin. BRD4 has been implicated in maintaining cell identity through its interaction with gene regulatory regions (Dey et al. 2003, 2009; Di Micco et al. 2014). In part, the maintenance of cell identity is postulated to occur through stable interaction of BRD4 with chromosomes during mitosis (Dey et al. 2000). Similarly, we found that BRD4-NUT remains constitutively associated with chromosomes during mitosis (Fig. 1A). Thus, it is interesting to speculate that the limited number of BRD4-NUT megadomains may identify critical loci that help define heritable cell states.
Megadomain formation
When we investigated the formation of megadomains in a heterologous cell line (293T) that does not ordinarily express BRD4-NUT, we found that megadomains initially appear as small peaks at seed regions corresponding to a select set of enhancers and gene promoters. The megadomains spread gradually either unidirectionally or bidirectionally, reminiscent of other “epigenetic” processes in biology such as position effect variegation (PEV), mating type silencing, and dosage compensation. That BRD4-NUT seed regions are located at enhancers or promoters is expected because BRD4 is known to target these highly acetylated regions (Loven et al. 2013; Kanno et al. 2014). What is unclear is why not all promoter/enhancer regions targeted by BRD4 act as seed regions for megadomains. Interestingly, no bias toward the expansion of superenhancers was detected. The distinct patterns of BRD4-NUT domains observed in cells of different patients suggest that multiple alternative factors or stochastic processes may be involved.
In contrast, a common characteristic among different NMC cell types is the large size of megadomains. Although the feed-forward model would predict the possibility of a runaway “chromatin-walking machine” that traverses the entire chromosome, the size of megadomains is limited by TADs or their underlying genomic content. In some cases, megadomains may fill entire regulatory regions for a single functional unit or gene, such as those near MYC, that contain multiple distant enhancers and lncRNAs implicated in the regulation of its expression. Thus, it is possible that megadomain breadth highlights the size and complexity of key, cell type-defining regulatory units structurally organized by TADs. The ability of BRD4-NUT to spread and fill whole regulatory compartments for genes like MYC is likely to explain the extremely aggressive nature of NMC.
While the size of megadomains appears unprecedented, it is possible that similar mechanisms may be used in other biological systems where spreading of acetylation could serve to remodel large chromatin domains. In this regard, it may be significant that wild-type NUT and BRDT, a family member of BRD4, are coexpressed in post-meiotic spermatids (Haack et al. 2009; Goudarzi et al. 2014). Since histone replacement during spermatid chromatin compaction requires hyperacetylation of the entire genome (Boussouar et al. 2008; Rousseaux et al. 2008; Goudarzi et al. 2014), one could speculate that BRDT and NUT could be involved in a similar feed-forward process.
Finally, in addition to their relevance to BRD4-NUT-driven disease, the ability to induce megadomain formation and visualize select TADs as discrete nuclear foci in interphase cells may assist in future investigation of the behavior of these fundamental units of the genome during cellular transitions and differentiation.
Importance of the target cell
NMC is a rare (French et al. 2004; Evans et al. 2012; French 2012; Petrini et al. 2012; Fang et al. 2013), poorly differentiated carcinoma that almost invariably exhibits features of squamous cell origin (French et al. 2004; Bauer et al. 2012; Bishop and Westra 2012). Given that oncogenic chromosomal translocations can be relatively frequent (Shtivelman et al. 1987; Tomlins et al. 2005; Soda et al. 2007), it is unlikely that the rarity of NMC is entirely due to the infrequency of the translocation but rather is due to the inability of most cells in the body to survive the consequences of BRD-NUT expression. Indeed, no cell type tested can survive prolonged overexpression of recombinant BRD4-NUT in culture (CA French, unpubl.). Perhaps BRD4-NUT can only circumvent the pathways that guard against its progrowth, anti-differentiation effects in a specific subtype of primordial squamous cell that is most abundant during young adulthood, the peak age of patients with NMC (Bauer et al. 2012; French 2012). What characteristic does a squamous precursor cell have that may allow circumvention of guardian pathways by BRD4-NUT? We propose that the chromatin landscape of the squamous precursor cell may have the “right” set of active seed sites for full transformation by BRD4-NUT. Other cell types may not survive because they do not have the set of poised regions necessary for survival.
In addition, squamous cell carcinoma nearly always has loss-of-function mutations of TP53, a powerful tumor suppressor that guards against mutagenesis through DNA repair and induction of apoptosis (Stransky et al. 2011; The Cancer Genome Atlas Network 2015). There is evidence that BRD4-NUT facilitates acetylation of p53 through p300, leading to its sequestration and inactivation within BRD4-NUT foci (Reynoird et al. 2010). Intriguingly, we also found a TP53-related squamous cell-expressed gene, TP63, to be regulated by a BRD4-NUT megadomain in all NMCs tested, resulting in JQ1-sensitive TP63 expression (Supplemental Fig. S5C). The ΔN isoform of p63 has been shown to act as a dominant-negative protein against p53 (Yang et al. 1998, Venkatanarayan et al. 2015); thus, its expression in NMC may further inhibit an already compromised p53. These findings support a possible mechanism by which BRD4-NUT can evade gatekeeper functions of p53.
Identification of therapeutic vulnerabilities in chromatin-driven cancers
Even as we propose that NMC driven by BRD4-NUT translocation is a “simple” model for chromatin-driven carcinogenesis, we provide evidence for the perturbation of multiple factors leading to oncogenic disruption in squamous cell differentiation by BRD4-NUT. The feed-forward loops that may perpetuate the aggressive nature of this disease are a major concern. Bromodomain inhibitor therapy shows great promise because it can break the cycle between excess acetylation and BRD4-NUT interaction with those acetylated lysines. Although additional potential targets may not be amenable to chemical inhibition (e.g., MYC), identification of the downstream targets of such factors via iterative, chromatin-based analyses may ultimately reveal combinations of vulnerabilities to be exploited in future therapies.
Materials and methods
Cell culture
NMC cell lines TC-797 (Toretsky et al. 2003), 1015 (Grayson et al. 2014), and 10326 (French et al. 2008) were maintained in Dulbecco modified Eagle medium (DMEM) (Invitrogen) supplemented with 10% (v/v) fetal bovine serum (FBS) (Atlanta Biologicals, S10350), 1% penicillin–streptomycin, and 1× GlutaMAX (Invitrogen). PER-403 (Kees et al. 1991) was maintained in DMEM with 20% FBS and supplemented as above.
797TRex and 293TRex cells, which contain a single genomic FRT recombination site, were maintained as above with blasticidin and zeocin (Invitrogen). The 797TRex and 293-TRex/N-BioTAP-BRD4-NUT, TRex/C-BioTAP-BRD4-NUT, and Flag-BRD4-NUT-HA cell lines were generated by recombination with the plasmid pcDNA5 FRT/TO/N-BioTAP BRD4NUT, FRT/TO/C-BioTAP BRD4NUT, or Flag-BRD4-NUT-HA (details on vectors and constructs will be provided on request) using the Flp-In technology manufacturer's instructions (Invitrogen). The resulting cell lines were maintained in medium (above) supplemented with blasticidin (Invitrogen) and hygromycin (Sigma). pEGFPC1-BRD4-NUT was created as described (French et al. 2008) and transfected into 293T cells using Lipofectamine 2000 according to the manufacturer's instructions (Life Technologies).
ChIP-seq procedure and data analysis
Cells (1.5 × 109) grown as monolayer cultures in 150-mm dishes (100 plates total) were used for BioTAP-XL purification. Tetracycline (1 μg/mL) was added to 60%–70% confluent culture to induce transcription of the N-BioTAP-tagged or C-BioTAP-tagged BRD4-NUT cDNA clones for 24 h. The main steps of BioTAP-XL procedure were performed as described (Alekseyenko et al. 2014a, 2015), as follows: harvesting cells, formaldehyde cross-linking, chromatin preparation, affinity purification, input and immunoprecipitation DNA recovery, and ChIP-seq library preparation.
Formaldehyde cross-linked chromatin for ChIP-seq experiments with antibodies against proteins and histone modifications was prepared from 1.5 × 108 cells of each NMC cell line and 293TRex-Flag-BRD4NUT-HA as described (Alekseyenko et al. 2015). Chromatin from 0.8 g of 1015 patient tissue (Grayson et al. 2014) was prepared as follows: Frozen tissue was ground to a powder with a ceramic mortar and pestle chilled with liquid N2. The powder was immediately transferred into a 50-mL Falcon tube filled with 9 mL of NEB buffer (10% sucrose, 20 mM HEPES at pH 7.6, 10 mM NaCl, 3 mM MgCl2, 0.5% Triton, 0.1 mM PMSF). The mixture was vortexed for 15 sec to break up the clumps, and, immediately after, 45 mL of PBS and 0.5 mL of 37% formaldehyde were added to the tube. The resulting mixture was incubated for 25 min at 25°C on an orbital shaker platform with gentle mixing (150 rpm). Washing and chromatin-shearing steps were performed as described (Alekseyenko et al. 2015).
Affinity purification was performed as described (Kharchenko et al. 2011). Input and immunoprecipitation, DNA recovery, and ChIP-seq library preparation were performed as described (Alekseyenko et al. 2015). The following antibodies against proteins and histone modifications were used for ChIP-seq experiments: 10 µL of anti-H3K27ac (Active Motif, catalog no. 39133) per immunoprecipitation, 15 µL of anti-H3K9ac (Abcam, catalog no. ab12179) per immunoprecipitation, 2 µL of anti-H3K14ac (Abcam, catalog no. ab52946) per immunoprecipitation, 3 µL of anti-H3K36me3 (Abcam, catalog no. ab9050) per immunoprecipitation, 10 µL of anti-P300 (Bethyl Laboratories, catalog no. A300-358A), 5 µL of anti-BRD4-SL (Bethyl Laboratories, catalog no. A301-985A) per immunoprecipitation against short and long isoforms of BRD4, 15 µL of anti-BRD4-L (Active Motif, catalog no. 39909) per immunoprecipitation against long isoforms of BRD4, and 15 µL anti-NUT per immunoprecipitation (Cell Signaling, catalog no. 3625).
ChIP-seq reads were aligned to the human reference genome (GRCh38 assembly) using Bowtie (version 0.12.5), retaining only uniquely mapped reads. Smoothed enrichment profiles were generated using the SPP package (Kharchenko et al. 2008) using a smoothing bandwidth of 5 kb for browser screenshot figures and 1 kb for the rest of the analysis.
Recovery and analysis of nascent RNA
Nascent RNA purification was based on the Nascent-seq protocol (Khodor et al. 2011; Ferrari et al. 2013). TC-797 and 1015 cells (2 × 106) were grown in 100-mm dishes. JQ1 was added to the medium to a final concentration of 500 nM for TC-797 cells or 2 µM for 1015 cells. Briefly, cells were harvested and homogenized three times with a 25-guage needle in 1 mL of CSK buffer (15 mM HEPES at pH 7.8, 100 mM NaCl, 3 mM MgCl2, 1 mM EGTA, 500 mM sucrose, 0.5% Triton X-100, Complete protease inhibitor [Roche], 0.5× SUPERaseIN [Life Technologies]) to isolate the nuclei. Nuclei were collected by centrifugation and resuspended in 80 µL of CF buffer (10 mM Tris at pH 8.0, 100 mM NaCl, 25 mM EDTA, 1× SUPERaseIN; Life Technologies). Immediately afterward, 1 mL of NUN buffer (1.5 M UREA, 20 mM HEPES at pH 7.8, 300 mM NaCl, 7.5 mM MgCl2, 500 mM EDTA, 1% NP40) was added, and the sample was vortexed to lyse the nuclei. The mixture was incubated on ice with occasional agitation for 10 min to allow precipitation of genomic DNA and core histones together with engaged RNA polymerase II and nascent transcripts. After the sample was spun 20,000g for 10 min at 4°C, the pellet was washed twice with NUN buffer. Next, the pellet was resuspended in 700 µL of CF buffer supplemented with 1% SDS and 1 mg/mL Proteinase K using a 25-guage needle and incubated for 60 min at 55°C. The lysate was extracted with phenol/chloroform. Nucleic acids were precipitated, dissolved in H2O, and treated with Turbo DNase I (Ambion) for 60 min at 37°C to eliminate DNA contamination. Fifty nanograms of nascent RNA was used to make the RNA library for Illumina following the instructions provided by the manufacturer for the NEBNext ultra directional RNA library preparation kit (New England Biolabs).
Nascent RNA-seq reads were first aligned to the human reference genome using Bowtie (version 0.12.5) with the maximum insert size (-X) set to 100,000, keeping only uniquely mapped reads. HTSeq (Anders et al. 2015) was used to count reads mapped to genes (using Gencode version 21 annotation). DESeq (Anders and Huber 2010) was subsequently employed to estimate nascent transcription levels and analyze differential expression between TC-797 following and prior to JQ1 treatment and between 293TRex following and before BRD4-NUT induction.
Total RNA extraction and quantification by qRT–PCR
Total RNA was extracted from 1 × 106 TC-797 or 1015 cells by a standard TRIzol extraction (Life Technologies) followed by RNA purification and removal of genomic DNA with RNeasy Plus kit (Qiagen). The Vilo cDNA synthesis kit (Invitrogen) was used to reverse-transcribe RNA. To quantify transcript levels, real-time PCR was performed on an ABI 7000 machine as described (Larschan et al. 2007). qPCR primer sequences are provided in Supplemental Table S2.
siRNA transfections
For TC-797, 1015, and PER403 cells, 1.5 × 106 cells were transfected with 50 nM siRNA using the RNAiMAX (Invitrogen, catalog no. 13778) reverse transfection protocol and plated to a 100-mm cell culture dish. siGENOME nontargeting siRNA #4 (Dharmacon, catalog no. D-001210-04) was used as a negative control. All other siRNAs are included in Supplemental Table S2.
Immunofluorescence and immunohistochemistry
Immunofluorescence on TC-797, 797TRex-Flag-BRD4-NUT-HA, and 293TRex-Flag-BRD4-NUT-HA cells was performed as described previously (Reynoird et al. 2010), and nuclei were counterstained with ProLong Gold anti-fade reagent with 4′,6-diamidino-2-phenylindole (DAPI) (Life Technologies, catalog no. P36935). Primary antibodies used were anti-NUT (1:1000; Cell Signaling, catalog no. 3625), anti-H3K27ac (1:2000; Active Motif, catalog no. 39133), and anti-HA (1:1000; Covance Research Products, catalog no. MMS-101R). Secondary antibodies included goat anti-rabbit Alexa fluor 594 and goat anti-mouse Alexa fluor 488 (1:1000; Life Technologies, catalog nos. A-11012 and A-11001). Photographs were taken on a Nikon Eclipse E600 fluorescent microscope using a Spot RTSlider camera (Diagnostic Instruments, Inc.) and Spot Advanced software (Diagnostic Instruments, Inc.). Diagnostic immunohistochemistry of a human NMC was performed as described (Haack et al. 2009).
Immunofluorescence analysis of 384-well plated NMC cells
TC-797 cells were transfected in 384-well format using 50 nM siRNAs, including siPVT1, sip63, and siMED24 (see Supplemental Table S2), as described previously (Grayson et al. 2014). Cells were stained with AE1/AE3 antibody (1:250; Dako, catalog no. M351501-2) to measure keratin intensity and ki-67 antibody (1:600; Cell Signalling, catalog no. 9129S) to quantify cell division, and nuclei were stained with 4 µg/mL Hoechst 33342 (Molecular Probes). Imaging for keratin expression was performed using the ImageXpress high-throughput microscope with MetaXpress software (Molecular Devices) as described previously (Senapedis et al. 2011). Each condition was performed in triplicate in 384-well plate format in three separate experiments. Cell number was calculated using MetaXpress with multiwavelength cell scoring. Keratin expression was scored as average immunofluorescence pixel intensity per cell.
H3K27ac and BRD4-NUT enrichment domain identification
For each cell type, the domains of H3K27ac enrichment were first identified using a three-state hidden Markov model (HMM). Specifically, the Gaussian emission means were set at μ + 2σ, enriched; μ, neutral; μ − 2σ, depleted on log2 scale, where μ and σ denote the genome-wide mean and standard deviation of the genome-wide H3K27ac enrichment profile. The transition probability from the neutral state was set at 10−6, and the return probability was set at 10−3.
To account for variations in the ChIP efficiency, the emission means and transition probabilities for estimating the BRD4-NUT domains were optimized to maximize concordance with the H3K27ac domains detected in the corresponding cell (293TRex, TC-797, and 1015 tissue and 1015 cell lines). For the cell types where H3K27ac ChIP-seq data were not available (PER-403, 10326, and 797TRex), HMM parameters for BRD4-NUT domain estimation were optimized for concordance with H3K27ac in TC-797.
Megadomain identification
An inflection point was identified in the size distribution of enrichment domains in 293T 4 h after BRD4-NUT induction (Fig. 1D). The corresponding domain size at the inflection point was 96 kb and was used as a minimal size cutoff for the subsequent megadomain detection. Enrichment domains exceeding the inflection point were called as “megadomains” in all of the examined cell types.
TAD boundary detection
To detect the boundaries of TADs, we calculated the following score for each 21-bin (bin size = 5 kb) sliding window along the diagonal of the observed/expected Hi-C interaction matrix (from GM12878 cells) (Rao et al. 2014):
where A, B, C, and D are the sum of the upper left, lower left, upper right, and lower right block matrices (10 × 10) of the square Hi-C interaction frequency matrix (21 × 21).
Accession numbers
ChIP-seq and RNA-seq data will be available in Gene Expression Omnibus with accession number GSE70868.
Supplementary Material
Acknowledgments
We are very grateful to Dr. Mel Feany for introducing M.I.K. and C.A.F. and suggesting our collaboration. We thank Dr. Steve Elledge and his laboratory for key DNA reagents, and Dr. Jay Bradner and his laboratory for providing JQ1. We thank Dr. Mel Feany, Dr. Winston Lee, and Kyle McElroy for critically reading the manuscript. Special thanks to Dr. Lucy Jung for her insightful suggestion regarding the megadomain comparison with TADs. This work was supported by grants from the National Institutes of Health (GM101958 to M.I.K. and 2R01CA124633 to C.A.F.), the Ellison Medical Foundation (AG-NS-0965-12 to P.V.K.), and the St. Baldrick's Foundation (to M.I.K. and C.A.F.). Three-hundred-eighty-four well-quantitative immunofluorescence studies were performed at the Institute of Chemistry and Cell Biology, Longwood Campus (ICCB-L, http://iccb.med.harvard.edu).
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.267583.115.
References
- Alekseyenko AA, Gorchakov AA, Kharchenko PV, Kuroda MI. 2014a. Reciprocal interactions of human C10orf12 and C17orf96 with PRC2 revealed by BioTAP-XL cross-linking and affinity purification. Proc Natl Acad Sci 111: 2488–2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alekseyenko AA, Gorchakov AA, Zee BM, Fuchs SM, Kharchenko PV, Kuroda MI. 2014b. Heterochromatin-associated interactions of Drosophila HP1a with dADD1, HIPP1, and repetitive RNAs. Genes Dev 28: 1445–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alekseyenko AA, McElroy KA, Kang H, Zee BM, Kharchenko PV, Kuroda MI. 2015. BioTAP-XL: cross-linking/tandem affinity purification to study DNA targets, RNA, and protein components of chromatin-associated complexes. Curr Protoc Mol Biol 109: 21.30.21–21.30.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsarraj J, Hunter KW. 2012. Bromodomain-containing protein 4: a dynamic regulator of breast cancer metastasis through modulation of the extracellular matrix. Int J Breast Cancer 2012: 670632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Huber W. 2010. Differential expression analysis for sequence count data. Genome Biol 11: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W. 2015. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asangani IA, Dommeti VL, Wang X, Malik R, Cieslik M, Yang R, Escara-Wilke J, Wilder-Romans K, Dhanireddy S, Engelke C, et al. 2014. Therapeutic targeting of BET bromodomain proteins in castration-resistant prostate cancer. Nature 510: 278–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer D, Mitchell C, Strait K, Lathan C, Stelow E, Lueer S, Muhammed S, Evans A, Sholl L, Rosai J, et al. 2012. Clinicopathologic features and long-term outcomes of NUT midline carcinoma. Clin Cancer Res 18: 5773–5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop JA, Westra WH. 2012. NUT midline carcinomas of the sinonasal tract. Am J Surg Pathol 36: 1216–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boussouar F, Rousseaux S, Khochbin S. 2008. A new insight into male genome reprogramming by histone variants and histone code. Cell Cycle 7: 3499–3502. [DOI] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Network. 2015. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature 517: 576–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark J, Edwards S, John M, Flohr P, Gordon T, Maillard K, Giddings I, Brown C, Bagherzadeh A, Campbell C, et al. 2002. Identification of amplified and expressed genes in breast cancer by comparative hybridization onto microarrays of randomly selected cDNA clones. Genes Chromosomes Cancer 34: 104–114. [DOI] [PubMed] [Google Scholar]
- Crawford NP, Alsarraj J, Lukes L, Walker RC, Officewala JS, Yang HH, Lee MP, Ozato K, Hunter KW. 2008. Bromodomain 4 activation predicts breast cancer survival. Proc Natl Acad Sci 105: 6380–6385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson MA, Prinjha RK, Dittmann A, Giotopoulos G, Bantscheff M, Chan WI, Robson SC, Chung CW, Hopf C, Savitski MM, et al. 2011. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature 478: 529–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, Kastritis E, Gilpatrick T, Paranal RM, Qi J, et al. 2011. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell 146: 904–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey A, Ellenberg J, Farina A, Coleman AE, Maruyama T, Sciortino S, Lippincott-Schwartz J, Ozato K. 2000. A bromodomain protein, MCAP, associates with mitotic chromosomes and affects G2-to-M transition. Mol Cell Biol 20: 6537–6549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey A, Chitsaz F, Abbasi A, Misteli T, Ozato K. 2003. The double bromodomain protein Brd4 binds to acetylated chromatin during interphase and mitosis. Proc Natl Acad Sci 100: 8758–8763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey A, Nishiyama A, Karpova T, McNally J, Ozato K. 2009. Brd4 marks select genes on mitotic chromatin and directs postmitotic transcription. Mol Biol Cell 20: 4899–4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Micco R, Fontanals-Cirera B, Low V, Ntziachristos P, Yuen SK, Lovell CD, Dolgalev I, Yonekubo Y, Zhang G, Rusinova E, et al. 2014. Control of embryonic stem cell identity by BRD4-dependent transcriptional elongation of super-enhancer-associated pluripotency genes. Cell Rep 9: 234–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. 2012. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485: 376–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans AG, French CA, Cameron MJ, Fletcher CD, Jackman DM, Lathan CS, Sholl LM. 2012. Pathologic characteristics of NUT midline carcinoma arising in the mediastinum. Am J Surg Pathol 36: 1222–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang W, French CA, Cameron MJ, Han Y, Liu H. 2013. Clinicopathological significance of NUT rearrangements in poorly differentiated malignant tumors of the upper respiratory tract. Int J Surg Pathol 21: 102–110. [DOI] [PubMed] [Google Scholar]
- Ferrari F, Plachetka A, Alekseyenko AA, Jung YL, Ozsolak F, Kharchenko PV, Park PJ, Kuroda MI. 2013. ‘Jump start and gain’ model for dosage compensation in Drosophila based on direct sequencing of nascent transcripts. Cell Rep 5: 629–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, et al. 2010. Selective inhibition of BET bromodomains. Nature 468: 1067–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French CA. 2012. Pathogenesis of NUT midline carcinoma. Annu Rev Pathol 7: 247–265. [DOI] [PubMed] [Google Scholar]
- French C. 2014. NUT midline carcinoma. Nat Rev Cancer 14: 149–150. [DOI] [PubMed] [Google Scholar]
- French CA, Miyoshi I, Kubonishi I, Grier HE, Perez-Atayde AR, Fletcher JA. 2003. BRD4-NUT fusion oncogene: a novel mechanism in aggressive carcinoma. Cancer Res 63: 304–307. [PubMed] [Google Scholar]
- French CA, Kutok JL, Faquin WC, Toretsky JA, Antonescu CR, Griffin CA, Nose V, Vargas SO, Moschovi M, Tzortzatou-Stathopoulou F, et al. 2004. Midline carcinoma of children and young adults with NUT rearrangement. J Clin Oncol 22: 4135–4139. [DOI] [PubMed] [Google Scholar]
- French CA, Ramirez CL, Kolmakova J, Hickman TT, Cameron MJ, Thyne ME, Kutok JL, Toretsky JA, Tadavarthy AK, Kees UR, et al. 2008. BRD-NUT oncoproteins: a family of closely related nuclear proteins that block epithelial differentiation and maintain the growth of carcinoma cells. Oncogene 27: 2237–2242. [DOI] [PubMed] [Google Scholar]
- French CA, Rahman S, Walsh EM, Kuhnle S, Grayson AR, Lemieux ME, Grunfeld N, Rubin BP, Antonescu CR, Zhang S, et al. 2014. NSD3-NUT fusion oncoprotein in NUT midline carcinoma: implications for a novel oncogenic mechanism. Cancer Discov 4: 928–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao R, Cao C, Zhang M, Lopez MC, Yan Y, Chen Z, Mitani Y, Zhang L, Zajac-Kaye M, Liu B, et al. 2014. A unifying gene signature for adenoid cystic cancer identifies parallel MYB-dependent and MYB-independent therapeutic targets. Oncotarget 5: 12528–12542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudarzi A, Shiota H, Rousseaux S, Khochbin S. 2014. Genome-scale acetylation-dependent histone eviction during spermatogenesis. J Mol Biol 426: 3342–3349. [DOI] [PubMed] [Google Scholar]
- Grayson AR, Walsh EM, Cameron MJ, Godec J, Ashworth T, Ambrose JM, Aserlind AB, Wang H, Evan G, Kluk MJ, et al. 2014. MYC, a downstream target of BRD-NUT, is necessary and sufficient for the blockade of differentiation in NUT midline carcinoma. Oncogene 33: 1736–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Malik S, Ito M, Yuan CX, Fondell JD, Zhang X, Martinez E, Qin J, Roeder RG. 1999. A novel human SRB/MED-containing cofactor complex, SMCC, involved in transcription regulation. Mol Cell 3: 97–108. [DOI] [PubMed] [Google Scholar]
- Haack H, Johnson LA, Fry CJ, Crosby K, Polakiewicz RD, Stelow EB, Hong SM, Schwartz BE, Cameron MJ, Rubin MA, et al. 2009. Diagnosis of NUT midline carcinoma using a NUT-specific monoclonal antibody. Am J Surg Pathol 33: 984–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. 2000. The hallmarks of cancer. Cell 100: 57–70. [DOI] [PubMed] [Google Scholar]
- Hasegawa N, Sumitomo A, Fujita A, Aritome N, Mizuta S, Matsui K, Ishino R, Inoue K, Urahama N, Nose J, et al. 2012. Mediator subunits MED1 and MED24 cooperatively contribute to pubertal mammary gland development and growth of breast carcinoma cells. Mol Cell Biol 32: 1483–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henssen A, Thor T, Odersky A, Heukamp L, El-Hindy N, Beckers A, Speleman F, Althoff K, Schafers S, Schramm A, et al. 2013. BET bromodomain protein inhibition is a therapeutic option for medulloblastoma. Oncotarget 4: 2080–2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-Andre V, Sigova AA, Hoke HA, Young RA. 2013. Super-enhancers in the control of cell identity and disease. Cell 155: 934–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houzelstein D, Bullock SL, Lynch DE, Grigorieva EF, Wilson VA, Beddington RS. 2002. Growth and early postimplantation defects in mice deficient for the bromodomain-containing protein Brd4. Mol Cell Biol 22: 3794–3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito M, Okano HJ, Darnell RB, Roeder RG. 2002. The TRAP100 component of the TRAP/Mediator complex is essential in broad transcriptional events and development. EMBO J 21: 3464–3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang YW, Veschambre P, Erdjument-Bromage H, Tempst P, Conaway JW, Conaway RC, Kornberg RD. 1998. Mammalian mediator of transcriptional regulation and its possible role as an end-point of signal transduction pathways. Proc Natl Acad Sci 95: 8538–8543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadota M, Sato M, Duncan B, Ooshima A, Yang HH, Diaz-Meyer N, Gere S, Kageyama S, Fukuoka J, Nagata T, et al. 2009. Identification of novel gene amplifications in breast cancer and coexistence of gene amplification with an activating mutation of PIK3CA. Cancer Res 69: 7357–7365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno T, Kanno Y, LeRoy G, Campos E, Sun HW, Brooks SR, Vahedi G, Heightman TD, Garcia BA, Reinberg D, et al. 2014. BRD4 assists elongation of both coding and enhancer RNAs by interacting with acetylated histones. Nat Struct Mol Biol 21: 1047–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kees UR, Mulcahy MT, Willoughby ML. 1991. Intrathoracic carcinoma in an 11-year-old girl showing a translocation t(15;19). Am J Pediatr Hematol Oncol 13: 459–464. [DOI] [PubMed] [Google Scholar]
- Kharchenko PV, Tolstorukov MY, Park PJ. 2008. Design and analysis of ChIP-seq experiments for DNA-binding proteins. Nat Biotechnol 26: 1351–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharchenko PV, Alekseyenko AA, Schwartz YB, Minoda A, Riddle NC, Ernst J, Sabo PJ, Larschan E, Gorchakov AA, Gu T, et al. 2011. Comprehensive analysis of the chromatin landscape in Drosophila melanogaster. Nature 471: 480–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodor YL, Rodriguez J, Abruzzi KC, Tang CH, Marr MT II, Rosbash M. 2011. Nascent-seq indicates widespread cotranscriptional pre-mRNA splicing in Drosophila. Genes Dev 25: 2502–2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T, Cui R, Jeon YJ, Lee JH, Sim H, Park JK, Fadda P, Tili E, Nakanishi H, Huh MI, et al. 2014. Long-range interaction and correlation between MYC enhancer and oncogenic long noncoding RNA CARLo-5. Proc Natl Acad Sci 111: 4173–4178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koster MI, Roop DR. 2004. The role of p63 in development and differentiation of the epidermis. J Dermatol Sci 34: 3–9. [DOI] [PubMed] [Google Scholar]
- Kubonishi I, Takehara N, Iwata J, Sonobe H, Ohtsuki Y, Abe T, Miyoshi I. 1991. Novel t(15;19)(q15;p13) chromosome abnormality in a thymic carcinoma. Cancer Res 51: 3327–3328. [PubMed] [Google Scholar]
- Larschan E, Alekseyenko AA, Gortchakov AA, Peng S, Li B, Yang P, Workman JL, Park PJ, Kuroda MI. 2007. MSL complex is attracted to genes marked by H3K36 trimethylation using a sequence-independent mechanism. Mol Cell 28: 121–133. [DOI] [PubMed] [Google Scholar]
- Lee AC, Kwong YI, Fu KH, Chan GC, Ma L, Lau YL. 1993. Disseminated mediastinal carcinoma with chromosomal translocation (15;19). A distinctive clinicopathologic syndrome. Cancer 72: 2273–2276. [DOI] [PubMed] [Google Scholar]
- Loven J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, Bradner JE, Lee TI, Young RA. 2013. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 153: 320–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luoh SW. 2002. Amplification and expression of genes from the 17q11 approximately q12 amplicon in breast cancer cells. Cancer Genet Cytogenet 136: 43–47. [DOI] [PubMed] [Google Scholar]
- Lupianez DG, Kraft K, Heinrich V, Krawitz P, Brancati F, Klopocki E, Horn D, Kayserili H, Opitz JM, Laxova R, et al. 2015. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell 161: 1012–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S, Chan YP, Woolcock B, Hu L, Wong KY, Ling MT, Bainbridge T, Webber D, Chan TH, Guan XY, et al. 2009. DNA fingerprinting tags novel altered chromosomal regions and identifies the involvement of SOX5 in the progression of prostate cancer. Int J Cancer 124: 2323–2332. [DOI] [PubMed] [Google Scholar]
- Mertz JA, Conery AR, Bryant BM, Sandy P, Balasubramanian S, Mele DA, Bergeron L, Sims RJ III. 2011. Targeting MYC dependence in cancer by inhibiting BET bromodomains. Proc Natl Acad Sci 108: 16669–16674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao RY, Drabsch Y, Cross RS, Cheasley D, Carpinteri S, Pereira L, Malaterre J, Gonda TJ, Anderson RL, Ramsay RG. 2011. MYB is essential for mammary tumorigenesis. Cancer Res 71: 7029–7037. [DOI] [PubMed] [Google Scholar]
- Nagoshi H, Taki T, Hanamura I, Nitta M, Otsuki T, Nishida K, Okuda K, Sakamoto N, Kobayashi S, Yamamoto-Sugitani M, et al. 2012. Frequent PVT1 rearrangement and novel chimeric genes PVT1-NBEA and PVT1-WWOX occur in multiple myeloma with 8q24 abnormality. Cancer Res 72: 4954–4962. [DOI] [PubMed] [Google Scholar]
- Petrini P, French CA, Rajan A, Cameron MJ, Jaffe ES, Zucali PA, Xie J, Wang Y, Giaccone G. 2012. NUT rearrangement is uncommon in human thymic epithelial tumors. J Thorac Oncol 7: 744–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puissant A, Frumm SM, Alexe G, Bassil CF, Qi J, Chanthery YH, Nekritz EA, Zeid R, Gustafson WC, Greninger P, et al. 2013. Targeting MYCN in neuroblastoma by BET bromodomain inhibition. Cancer Discov 3: 308–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman S, Sowa ME, Ottinger M, Smith JA, Shi Y, Harper JW, Howley PM. 2011. The Brd4 extraterminal domain confers transcription activation independent of pTEFb by recruiting multiple proteins, including NSD3. Mol Cell Biol 31: 2641–2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey MR, Wilson C, Ory B, Rothenberg SM, Faquin W, Mills AA, Ellisen LW. 2013. FGFR2 signaling underlies p63 oncogenic function in squamous cell carcinoma. J Clin Invest 123: 3525–3538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SS, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES, et al. 2014. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159: 1665–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynoird N, Schwartz BE, Delvecchio M, Sadoul K, Meyers D, Mukherjee C, Caron C, Kimura H, Rousseaux S, Cole PA, et al. 2010. Oncogenesis by sequestration of CBP/p300 in transcriptionally inactive hyperacetylated chromatin domains. EMBO J 29: 2943–2952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riquelme E, Suraokar MB, Rodriguez J, Mino B, Lin HY, Rice DC, Tsao A, Wistuba II. 2014. Frequent coamplification and cooperation between C-MYC and PVT1 oncogenes promote malignant pleural mesothelioma. J Thorac Oncol 9: 998–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousseaux S, Reynoird N, Escoffier E, Thevenon J, Caron C, Khochbin S. 2008. Epigenetic reprogramming of the male genome during gametogenesis and in the zygote. Reprod Biomed Online 16: 492–503. [DOI] [PubMed] [Google Scholar]
- Senapedis WT, Kennedy CJ, Boyle PM, Silver PA. 2011. Whole genome siRNA cell-based screen links mitochondria to Akt signaling network through uncoupling of electron transport chain. Mol Biol Cell 22: 1791–1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shehata B, Steelman CK, Abramowsky CR, Olson T, French C, Saxe D, Ricketts R, Katzenstein H. 2010. NUT midline carcinoma in a newborn with multiorgan disseminated tumor and a two-year-old with a pancreatic/hepatic primary. Pediatr Dev Pathol 13: 481–485. [DOI] [PubMed] [Google Scholar]
- Shi J, Wang Y, Zeng L, Wu Y, Deng J, Zhang Q, Lin Y, Li J, Kang T, Tao M, et al. 2014. Disrupting the interaction of BRD4 with diacetylated Twist suppresses tumorigenesis in basal-like breast cancer. Cancer Cell 25: 210–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Cao J, Zhou BP. 2015. Twist-BRD4 complex: potential drug target for basal-like breast cancer. Curr Pharm Des 21: 1256–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shtivelman E, Gale RP, Dreazen O, Berrebi A, Zaizov R, Kubonishi I, Miyoshi I, Canaani E. 1987. bcr–abl RNA in patients with chronic myelogenous leukemia. Blood 69: 971–973. [PubMed] [Google Scholar]
- Soda M, Choi YL, Enomoto M, Takada S, Yamashita Y, Ishikawa S, Fujiwara S, Watanabe H, Kurashina K, Hatanaka H, et al. 2007. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature 448: 561–566. [DOI] [PubMed] [Google Scholar]
- Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, Kryukov GV, Lawrence MS, Sougnez C, McKenna A, et al. 2011. The mutational landscape of head and neck squamous cell carcinoma. Science 333: 1157–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson-Wicking K, Francis RW, Stirnweiss A, Ferrari E, Welch MD, Baker E, Murch AR, Gout AM, Carter KW, Charles AK, et al. 2013. Novel BRD4-NUT fusion isoforms increase the pathogenic complexity in NUT midline carcinoma. Oncogene 32: 4664–4674. [DOI] [PubMed] [Google Scholar]
- Tilson MP, Bishop JA. 2013. Utility of p40 in the differential diagnosis of small round blue cell tumors of the sinonasal tract. Head Neck Pathol 8: 141–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, Varambally S, Cao X, Tchinda J, Kuefer R, et al. 2005. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science 310: 644–648. [DOI] [PubMed] [Google Scholar]
- Toretsky JA, Jenson J, Sun CC, Eskenazi AE, Campbell A, Hunger SP, Caires A, Frantz C, Hill JL, Stamberg J. 2003. Translocation (11;15;19): a highly specific chromosome rearrangement associated with poorly differentiated thymic carcinoma in young patients. Am J Clin Oncol 26: 300–306. [DOI] [PubMed] [Google Scholar]
- Tseng YY, Moriarity BS, Gong W, Akiyama R, Tiwari A, Kawakami H, Ronning P, Reuland B, Guenther K, Beadnell TC, et al. 2014. PVT1 dependence in cancer with MYC copy-number increase. Nature 512: 82–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatanarayan A, Raulji P, Norton W, Chakravarti D, Coarfa C, Su X, Sandur SK, Ramirez MS, Lee J, Kingsley CV, et al. 2015. IAPP-driven metabolic reprogramming induces regression of p53-deficient tumours in vivo. Nature 517: 626–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, You J. 2015. Mechanistic analysis of the role of bromodomain-containing protein 4 (BRD4) in BRD4-NUT oncoprotein-induced transcriptional activation. J Biol Chem 290: 2744–2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang JF, Yin QF, Chen T, Zhang Y, Zhang XO, Wu Z, Zhang S, Wang HB, Ge J, Lu X, et al. 2014. Human colorectal cancer-specific CCAT1-L lncRNA regulates long-range chromatin interactions at the MYC locus. Cell Res 24: 513–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Diaz J, Jiao J, Wang R, You J. 2011. Perturbation of BRD4 protein function by BRD4-NUT protein abrogates cellular differentiation in NUT midline carcinoma. J Biol Chem 286: 27663–27675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang A, Kaghad M, Wang Y, Gillett E, Fleming MD, Dotsch V, Andrews NC, Caput D, McKeon F. 1998. p63, a p53 homolog at 3q27-29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol Cell 2: 305–316. [DOI] [PubMed] [Google Scholar]
- Yokota J, Tsunetsugu-Yokota Y, Battifora H, Le Fevre C, Cline MJ. 1986. Alterations of myc, myb, and rasHa proto-oncogenes in cancers are frequent and show clinical correlation. Science 231: 261–265. [DOI] [PubMed] [Google Scholar]
- Zafarana G, Gillis AJ, van Gurp RJ, Olsson PG, Elstrodt F, Stoop H, Millan JL, Oosterhuis JW, Looijenga LH. 2002. Coamplification of DAD-R, SOX5, and EKI1 in human testicular seminomas, with specific overexpression of DAD-R, correlates with reduced levels of apoptosis and earlier clinical manifestation. Cancer Res 62: 1822–1831. [PubMed] [Google Scholar]
- Zhang J, Fondell JD. 1999. Identification of mouse TRAP100: a transcriptional coregulatory factor for thyroid hormone and vitamin D receptors. Mol Endocrinol 13: 1130–1140. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Zhu Z, Zhang B, Li W, Li X, Wu X, Wang L, Fu L, Dong JT. 2014. Frequent mutation of rs13281615 and its association with PVT1 expression and cell proliferation in breast cancer. J Genet Genomics 41: 187–195. [DOI] [PubMed] [Google Scholar]
- Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, Magoon D, Qi J, Blatt K, Wunderlich M, et al. 2011. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature 478: 524–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.