Significance
The role of the microRNA (miR) 15/16 family in oncogenesis and tumor progression has been intensively studied. The miR-15a/16-1 cluster is extensively described in B-cell chronic lymphocytic leukemia, characterized by the deletion of the 13q14 locus. The role of the miR-15b/16-2 cluster on chromosome 3q25 is still far from being elucidated. Because miR-15a is highly similar to miR-15b and miR-16-1 is identical to miR-16-2, we generated miR-15b/16-2 knockout mice to better understand the cluster’s role in vivo. These knockout mice developed B-cell lymphomas by age 15–18 mo, modulating the CCND2 (Cyclin D2), CCND1 (Cyclin D1), and IGF1R (insulin-like growth factor 1 receptor) genes involved in proliferation and antiapoptotic pathways in mouse B cells. Our results suggest a tumor suppressor role for the miR-15b/16-2 cluster in animal models of B-cell lymphomas.
Keywords: miRNAs, miR-15b, chronic lymphocytic leukemia, B cells, murine models
Abstract
The central role of the microRNA (miR) 15a/16-1 cluster in B-cell oncogenesis has been extensively demonstrated, with over two-thirds of B-cell chronic lymphocytic leukemia characterized by the deletion of the miR-15a/16-1 locus at 13q14. Despite the well-established understanding of the molecular mechanisms occurring during miR-15a/16-1 dysregulation, the oncogenic role of other miR-15/16 family members, such as the miR-15b/16-2 cluster (3q25), is still far from being elucidated. Whereas miR-15a is highly similar to miR-15b, miR-16-1 is identical to miR-16-2; thus, it could be speculated that both clusters control a similar set of target genes and may have overlapping functions. However, the biological role of miR-15b/16-2 is still controversial. We generated miR-15b/16-2 knockout mice to better understand the cluster’s role in vivo. These mice developed B-cell malignancy by age 15–18 mo with a penetrance of 60%. At this stage, mice showed significantly enlarged spleens with abnormal B cell-derived white pulp enlargement. Flow cytometric analysis demonstrated an expanded CD19+ CD5+ population in the spleen of 40% knockout mice, a characteristic of the chronic lymphocytic leukemia-associated phenotype found in humans. Of note, miR-15b/16-2 modulates the CCND2 (Cyclin D2), CCND1 (Cyclin D1), and IGF1R (insulin-like growth factor 1 receptor) genes involved in proliferation and antiapoptotic pathways in mouse B cells. These results are the first, to our knowledge, to suggest an important role of miR-15b/16-2 loss in the pathogenesis of B-cell chronic lymphocytic leukemia.
MicroRNAs (miRNAs) are a class of small noncoding RNAs that modulate gene expression in many physiological and pathological conditions (1). Altered miRNA expression has been reported in several human cancers, and miRNA expression profiles vary according to the considered tumor (2).
A role for miRNAs in tumorigenesis and progression was originally documented for the miR-15/16 family (2–5). This group of miRNAs encompasses the miR-15a/16-1 cluster (on chromosome 13q14,) the miR-15b/16-2 cluster (on chromosome 3q25), and the miR-195/497 cluster (on chromosome 17p13).
The role of the miR-15a/16-1 cluster in B-cell pathology has been extensively demonstrated (5). The deletion of the miR-15a/16-1 cluster has been reported in over two-thirds of B-cell chronic lymphocytic leukemias (B-CLLs) (5). Our group has demonstrated that the loss of miR-15a/16-1 expression induces higher levels of the antiapoptotic proteins BCL2 and myeloid cell leukemia sequence 1 (BCL2-related) (MCL1) (3, 6). Moreover, this deletion promotes mature B-cell expansion by deregulating the transition from G1 to S phase (7).
On the other hand, the biological role of miR-15b/16-2 is still controversial, as this cluster has been reported to behave as either a tumor suppressor [acute promyelocytic leukemia (8, 9) and osteosarcoma (10)] or an oncogene [melanoma (11), up-regulated in the plasma of colorectal cancer (12) and head and neck carcinoma (13)].
Because the miR-15a/16-1 and miR-15b/16-2 clusters share miRNAs that are highly similar or, in the case of miR-16, identical, it is possible that they control a similar set of target genes and have overlapping functions.
To better characterize the role of miR-15b/16-2 in tumorigenesis and tumor progression, we generated a conventional miR-15b/16-2 knockout mouse model. By the age of 15–18 mo, miR-15b/16-2 knockout mice developed lymphoproliferative disorders closely resembling human B-CLL, with diffuse lymph node enlargement and severe splenomegaly due to the expansion of a CD19+ CD5+ double positive population of neoplastic B cells.
Results
Generation of miR-15b/16-2 Knockout Mice.
To investigate the consequences of miR-15b/16-2 deletion, we generated an miR-15b/16-2 knockout mouse model by replacing the gene with a neomycin cassette (with two loxP sites) (Fig. 1A). The correct homologous recombination was confirmed by Southern blot (Fig. S1A) and PCR analyses (Fig. S1B).
Fig. 1.
Deletion of miR-15b/16-2 in mice. (A) Schematic representation of the miR-15b/16-2 targeting strategy. (B) Northern blot analysis showing robust down-regulation of miR-15b but not of miR-16 in spleens from knockout mice compared with spleens from wild-type mice. Noncoding small nuclear RNA U6 was used as a loading control. (C) qRT-PCR analysis of spleens from knockout mice showing miR-15b down-regulation but not for miR-16 compared with wild-type mice. (D) Survival curve of the miR-15b/16-2 knockout cohort. Mice were followed for 24 mo (730 days) and events were corresponded to mice that died due to illness or those identified as sick (with palpable tumor mass) and then euthanized (P < 0.001). The number of mice in each group is indicated.
Fig. S1.
Deletion of miR-15b/16-2 in mice. (A) Southern blot analysis of wild-type, knockout, and heterozygous (HET) mice. The expected fragments after BamHI digestion are indicated. (B) PCR analysis of wild-type, knockout, and heterozygous mice. The expected bands are indicated. (C–F) Southern blot analysis of colons (C) and lungs (E) from knockout and wild-type mice. Noncoding small nuclear RNA U6 was used as a loading control. qRT-PCR analysis of colons (D) and lungs (F) from knockout and wild-type mice. Small nucleolar RNAs snoR-292 and snoR-135 were used for normalization.
Mice lacking miR-15b/16-2 were viable, efficiently reproduced, and, at birth, had no macroscopic abnormalities. Correct Mendelian ratios were obtained.
miR-15b and miR-16-2 expression was assayed in various organs (lung, colon, and spleen) using both quantitative reverse transcription–PCR (qRT-PCR) and Northern blotting. These tests confirmed a significant down-regulation of knocked-out miRNAs compared with wild-type mice (Fig. 1 B and C and Fig. S1 C–F). Evidence of miR-16 expression even in miR-15b/16-2 knockout mice is largely attributable to the probe in use, which hybridizes to both miR-16-1 and miR-16-2 (Fig. 1 B and C and Fig. S1 C–F). Of note, miR-16 expression was significantly decreased in lung and colon of knockout mice (40% average reduction) (Fig. S1 C–F), whereas its expression in spleen did not seem to be affected (Fig. 1 B and C).
Knockout mice were followed for 24 mo, and survival rates were compared with those of control (wild-type) mice. miR-15b/16-2 knockout mice showed an unequivocal reduced overall survival (OS) compared with the wild type; over a 24-mo time frame, knockout mice have a 70% decrease in OS (P < 0.001) (Fig. 1D).
miR-15b/16-2 Knockout Mice Develop CLL and Non-Hodgkin Lymphomas.
Aiming to find the causes of the miR-15b/16-2 knockout high mortality, we started the biological and histological characterization of this mouse model.
By the age of 15–18 mo, 60% of miR-15b/16-2 knockout mice disclosed features consistent with a B-cell lymphoproliferative disorder. Visually, the knockout mice presented very enlarged spleens, up to fourfold in comparison with wild-type mice (Fig. 2 A and B). Histologically, all such cases were characterized by some degree of splenomegaly with enlargement of the white pulp secondary to expansion/accumulation of B lymphocytes (Fig. 2C).
Fig. 2.
Histological analysis of spleens from wild-type and knockout mice. (A and B) Spleens from knockout mice were enlarged up to fourfold in volume compared with spleens from wild-type mice. (C) H&E (Top) and B220 (Middle) -stained spleen sections from knockout mice show B cell-derived white pulp enlargement with follicular structure disruption (dashed outline) compared with wild-type mice. Wright–Giemsa-stained blood smear (Bottom) shows an increase of small lymphocytes from a knockout mouse compared with a wild-type mouse. (Scale bars: A, 1 cm; C, Upper Left WT and KO, 250 μm; Upper Right WT and KO, 50 μm; Middle WT and KO, 250 μm.) Error bars represent SD.
The histological diagnosis clearly shows the heterogeneity of the B-cell malignancy developed by this knockout mouse model: 10% of miR-15b/16-2 knockout mice had a monoclonal B-cell lymphocytosis, 35% fulfilled the histological criteria for a diagnosis of B-CLL, and 15% were affected by non-Hodgkin B-cell lymphomas (Fig. 3A).
Fig. 3.
miR-15b/16-2 knockout mice develop lymphoproliferative disease. (A) Percentage of B-lymphoid pathologies from knockout mice up to 24 mo old. The number of mice analyzed is indicated. CLL/SLL, chronic lymphocytic leukemia and small lymphocytic lymphoma; MBL, monoclonal B-cell lymphocytosis; NHL, non-Hodgkin B-cell lymphoma. (B) H&E sections of bone marrow and superficial cervical/inguinal lymph node from knockout mice show a nodular infiltration (asterisks) of small lymphocytes. (C and D) Gross pathology of spleen, kidney, and liver (C; scale bar: 1 cm) and H&E sections of salivary gland, kidney, and liver (D) from knockout mice show extensive lymphoma infiltration (asterisks). (E) White blood cells from spleens and bone marrow of knockout and wild-type mice were analyzed by flow cytometry. The plot represents the percentage of CD5+ CD19+ cells among white blood cells still alive. The number of mice analyzed and the P values are indicated. Error bars represent SD. (F) Representative flow cytometry analysis of spleens and bone marrow from knockout and wild-type mice. (Scale bar: C, 1 cm.) SSC-H, side scatter versus FSC-H forward scatter.
Bone marrow and cervical/inguinal lymph nodes were characterized by discrete aggregates of monotonous, small lymphocytes (Fig. 3B). Massive infiltration of nonlymphoid tissues was observed in 12% of knockout cases, with the liver, kidney, and salivary gland being the most severely affected organs (Fig. 3 C and D). In all cases, peripheral blood smear showed increased numbers of small lymphocytes.
Flow cytometry analysis of knockout spleen- and bone marrow-derived lymphocytes provides evidence of a higher proportion of the surface antigens CD19+ and CD5+, markers of B cells and B-cell lymphoproliferative disorders, respectively, compared with wild-type mice (P < 0.02 and P < 0.05, respectively) (Fig. 3 E and F).
Identification of Potential miR-15b/16-2 Target Genes in B Cells.
We began to investigate the effects of miR-15b/16-2 deletion on B-cell biology in this knockout mice model by assessing BCL2 expression, a well-known target of miR-15/16 (3). We found BCL2 faintly up-regulated in knockout mice compared with wild type (Fig. S2A). We continued the research of putative targets able to explain the phenotype previously described. Subsequent consultation of at least one of three computational target prediction algorithms [TargetScan (www.targetscan.org), miRanda (www.microrna.org/microrna/home.do), and PicTar (pictar.mdc-berlin.de)] pinpointed CCND2 (Cyclin D2), CCND1 (Cyclin D1) (14), and IGF1R (insulin-like growth factor 1 receptor) as other putative targets of miR-15b/16-2.
Fig. S2.
Analysis of miR-15b/16-2 targets in miR-15b/16-2–deleted mouse B cells. (A) Western blot analysis of IGF1R and BCL2 expression in B cells from knockout and wild-type mice after LPS stimulation at the indicated time points. Fold change in protein expression is indicated. β-Actin was used as a loading control. (B and C) Western blot analysis of IGF1R expression in B cells from knockout and wild-type mice. β-Actin was used as a loading control. Fold change in protein expression is shown in the histogram. (D) Positions of miR-15b putative binding sites on the CCND2 and IGF1R transcripts and their mutants. (E) psiCHECK-2 vector with the IGF1R WT 3′ UTR insert and Mut miR-15b 3′ UTR containing a deletion of the miR-15b target site in the 3′ UTR was cotransfected with miR-15b or scrambled miR in 293HEK cells. Luciferase activity was recorded after 24 h. Data represent the mean ± SD from at least three independent experiments.
CCND2 and CCND1 are members of the D-type cyclin family and play a pivotal role in the transition from G1 to S phase of the cell cycle (15). IGF1R is a tyrosine kinase regulating cell growth and survival. Considering that CCND2, CCND1, and IGFR1R are frequently up-regulated in B-CLL and diffuse large B-cell lymphoma (DLBCL) (16–21), it makes perfect sense to find them directly targeted by miR-15b/16-2. To validate this putative targeting, we assessed all these three proteins by Western blot (WB). LPS-stimulated miR-15b/16-2–deleted B cells showed higher Cyclin D2 and IGF1R levels compared with wild-type B cells by WB (Fig. 4A and Fig. S2A).
Fig. 4.
Analysis of miR-15b/16-2 targets in miR-15b/16-2–deleted mouse B cells. (A) Western blot analysis of Cyclin D2 expression in B cells from knockout and wild-type mice after LPS stimulation at the indicated time points. β-Actin was used as a loading control. Fold change in protein expression is indicated. hs, hours. (B) Western blot analysis of Cyclin D2 and Cyclin D1 expression in B cells from knockout and wild-type mice. β-Actin was used as a loading control. (C) Relative quantification of Cyclin D1 and Cyclin D2 expression to the β-actin loading control in B cells from knockout and wild-type mice. (D) psiCHECK-2 vector with the CCND2 WT 3′ UTR insert and Mut miR-15b 3′ UTR (mut1, mut2, and mut3) containing a deletion of the miR-15b target site in the 3′ UTR was cotransfected with miR-15b or scrambled miR in 293HEK cells. Luciferase activity was recorded after 24 h. Data represent the mean ± SD from at least three independent experiments.
Moreover, CD19+ B cells from miR-15b/16-2 knockout mice presented higher Cyclin D1, Cyclin D2, and IGF1R expression levels compared with wild-type CD19+ B cells (Fig. 4 B and C and Fig. S2 B and C). To determine whether CCND2 and IGF1R represent a direct target of miR-15b/16-2, we cloned the 3′ UTR miRNA-binding fragments of these genes into the multiple cloning region of the psiCHECK-2 vector and recorded luciferase activity (Fig. S2D). Three putative sites in CCND2 and four putative sites in IGF1R were found using TargetScan (Fig. S2D). 293HEK cells were transfected with the psiCHECK-2 construct containing the 3′ UTR fragment of CCND2 and IGF1R and the miR-15b precursor (pre-miR-15b). Expression of miR-15b significantly decreased luciferase activity (Fig. 4D and Fig. S2E). Conversely, when luciferase assays were performed by using a plasmid harboring the 3′ UTR of CCND2 and IGF1R mRNAs, where the binding sites for miR-15b were deleted by site-directed mutagenesis, we observed a consistent reduction in miR-15b inhibitory effect, mostly in mut3-CCND2 (Fig. 4D) and mut2- and mut4-IGF1R (Fig. S2E) 3′ UTRs. These results supported the bioinformatic predictions confirming the direct targeting of miR-15b/16-2 on the CCND2 and IGF1R 3′ UTR.
miR-15b Expression Is Significantly Down-Regulated in CLL Patients.
We then wanted to translate these exciting findings observed in the miR-15b/16-2 murine model to humans, analyzing the B-CLL patient scenario to evaluate the importance of these discoveries in human lymphomagenesis.
To uncover possible hidden mechanisms of miR-15b “self-regulation,” we aimed to detect expression of both the mature form of miR-15b and its precursor in B-CLL patients. We chose two different cohorts: one with the 13q deletion (22 cases), representing the less aggressive form of CLL, and a second with the 11q deletion (25 cases), associated with a poor clinical outcome and with several adverse prognostic factors. CD19+ B cells from healthy donors were used as controls.
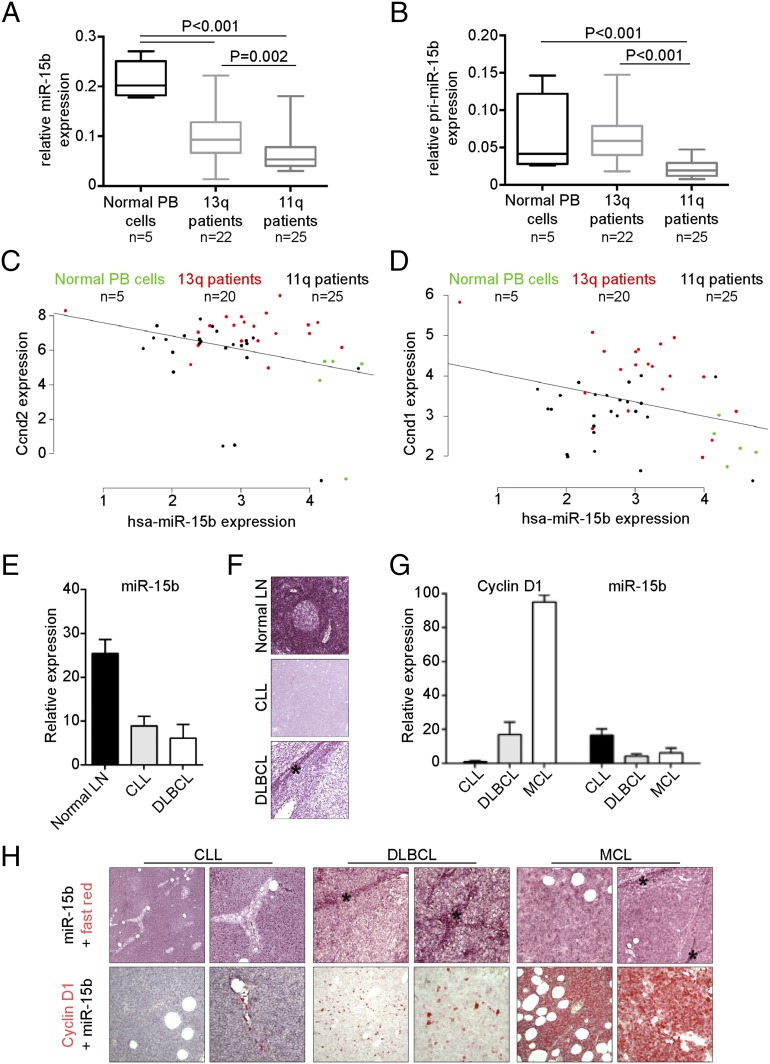
By qRT-PCR, we assayed the expression of the mature form of miR-15b, which was down-regulated in both 13q- and 11q-deleted B-CLL patients compared with normal CD19+ B cells (1.6- and 3.4-fold change, respectively) (Fig. 5A). miR-15b was also significantly down-regulated between 11q-deleted B-CLL patients and 13q-deleted cases (P = 0.002). As for miR-15b, a significant down-regulation of pri-miR-15b was found in 11q-deleted B-CLL compared with both normal B cells and 13q-deleted cases (Fig. 5B). Interestingly, 13q-deleted B-CLL cases seemed to compensate for the absence of miR-15a/16-1 by modulating pri-miR-15b expression, a self-modulating mechanism that will be the target of future investigations.
Fig. 5.
miR-15b/16-2 expression in human CLL samples. (A and B) qRT-PCR analysis of miR-15b (A) and pri-miR-15b (B) in CLL samples. The number of patients analyzed and the P values are indicated. (C and D) Correlations between miR-15b and Cyclin D2 (C) or Cyclin D1 (D) expression were determined using the Spearman coefficient in normal and CLL samples (Cyclin D2: n = 50, P = 0.038; Cyclin D1: n = 50, P = 0.021). hsa, human. (E and F) ISH for miR-15b in human normal lymph node (LN) and CLL and DLBCL samples and quantification. (G and H) ISH for miR-15b (H, Upper) and miR-15b and Cyclin D1 (H, Lower) in human CLL, DLBCL, and MCL samples (asterisks indicate normal lymph node remnants showing moderate miR-15b expression). (G) Quantification of miR-15b and Cyclin D1 expression in CLL, DLBCL, and MCL samples. Error bars represent SD.
To further confirm the previously discovered targeting of miR-15b, we assessed CCND2 and CCND1 mRNA levels together with miRNA expression in this same patient cohort (Fig. 5 C and D). miR-15b expression levels were directly and significantly anticorrelated with the CCND2 (Fig. 5C; P = 0.038) and CCND1 (Fig. 5D; P = 0.021) mRNA expression levels. Furthermore, a meta-analysis of gene expression in DLBCL TCGA (The Cancer Genome Atlas) microarray studies pinpointed CCND2 (P = 0.027) and CCND1 (P = 0.004) as significantly anticorrelated with miR-15b expression among the tumor tissues (Fig. S3A).
Fig. S3.
miR-15b/16-2 expression in human CLL samples. (A) Correlations between miR-15b and Cyclin D2 (Left) or Cyclin D1 (Right) expression were determined using Spearman coefficient CLL samples from the TCGA database (Cyclin D2: P = 0.004; Cyclin D1: P = 0.027). (B) ISH for miR-15b in human normal lymph node and CLL and DLBCL samples (asterisks indicate normal lymph node remnants showing moderate miR-15b expression).
In situ hybridization (ISH) confirmed miR-15b down-regulation in CLL and in the most aggressive DLBCL and mantle cell lymphoma (MCL) tumor tissue in comparison with normal lymph node cortex (five cases per tumor class; Fig. 5 E and F and Fig. S3B). Moreover, there was an evident inverse correlation between miR-15b and Cyclin D1 protein expression. In fact, strong and diffuse nuclear Cyclin D1 staining was observed in DLBCL and MCL tissue samples, where miR-15b expression was very low or undetectable (Fig. 5 G and H).
Discussion
This is the first demonstration, to our knowledge, that deletion of miR-15b/16-2 plays an important role in promoting mature B-cell lymphomagenesis. Our in vivo model, describing miR-15b as a tumor suppressor, is substantially consistent with what has previously been observed in other tumors, such as glioma (22), osteosarcoma (10), gastric cancer cells (23), and acute promyelocytic leukemia (8, 9). Moreover, it has been reported that miR-15b overexpression has a protective role of tumor recurrence after resection of hepatocellular carcinoma (24).
We focused our study on the role of miR-15b in vivo and in vitro, because miR-16-1 and miR-16-2 have an identical sequence and exert similar biological effects. The probe for miR-16 hybridizes with both miR-16-1 and miR-16-2, and therefore miR-16 expression is not significantly affected in our miR-15b/16-2–deleted mouse model (Fig. 1 B and C). Of interest, miR-16 expression in the spleen was comparable to that of wild-type mice.
The obtained phenotype is comparable to that observed in the miR-15a/16-1 model (7). Both mouse models showed a lymphoproliferative phenotype indicating that miR-15a/16-1– and miR-15b/16-2–deleted mice could represent a model of human CLL. Then, those mice developed lymphoproliferative diseases by age 15–18 mo, following the indolent form of human CLL. Moreover, both miR clusters seemed to control mostly cell proliferation, targeting CCND1, CCND3, CCNE1, CDK6, and others. Here we attempted to identify and verify other mRNA targets and pathways that mediate miR-15b regulation in our in vivo model. Considering that miR-15a and miR-15b share the same seed region but belong to different chromosome loci, miR-15b does not seem to strongly affect the BCL2 gene in this B-cell model (Fig. S2A). Conversely, other proteins are affected by miR-15b/16-2 deletion, including Cyclin D2 and IGF1R (Fig. 4 A and B and Fig. S2 A and B). Indeed, we demonstrated that CCND2 and IGF1R represent a direct target of miR-15b (Fig. 4D and Fig. S2E), suggesting that miR-15b is able to influence and control proliferation and survival of B cells by affecting the expression of these two important players in proliferation and apoptosis.
In humans, we noticed that patients with 13q− CLLs (lacking the miR-15a/16-1 cluster) seem to compensate for the absence of miR-15a/16-1, modulating pri-miR-15b and miR-15b expression (Fig. 4 A and B), an intriguing possible level of miRNA “regulation.” Because miRNAs sharing the same cluster can emphasize their functional cooperation on the same cellular pathways (25, 26), further investigations are needed to understand the role of the 3q25 locus in human lymphoproliferative diseases and its effect on the 13q14 locus.
Taken together, these observations suggest a tumor suppressor role for the miR-15b/16-2 cluster in animal models of B-cell lymphomas. The mouse model described here could be useful for the study of CLL progression. Further studies are needed to investigate the combined effects of miR-15b/16-2 and miR-15a/16-1 on B-cell biology in vivo.
Materials and Methods
Generation of miR-15b/16-2 Knockout Mice.
The miR-15b/16-2 hairpin spans ∼430 bp. To generate the miR-15b/16-2 KO targeting vector, DNA fragments flanking miRNAs, with a NotI-BamHI fragment (3.4 kb) as the 5′ arm and a HindIII-EcoRI fragment (1.3 kb) as the 3′ arm, were isolated from a 129/Sv genomic library and subcloned into a pflox vector. Two selectable markers are incorporated into this vector. The neomycin resistance gene (neo) is flanked by loxP sites and located between the two arms. The herpes simplex virus (HSV) thymidine kinase (tk) gene is placed outside of the 3′ arms. The miR-15b/16-2 KO targeting vector was linearized and transfected into embryonic stem cells (ESCs). After the selection of transfected embryonic stem cells with G418, individual colonies of ESC clones were collected and analyzed for homologous recombination by Southern blot. Three targeted ESC clones were injected into C57BL/6J blastocysts to obtain chimeric mice. Chimeric males that transmitted the targeted allele to the germ line were mated with C57BL/6 females, and germ-line transmission of the targeted allele was confirmed by Southern blotting and PCR analysis. Chimeric mice were produced in the University of Michigan Biomedical Research Core Facilities. All experiments with mice were approved by the Institutional Animal Care and Use Committee (IACUC) and University Laboratory Animal Resources at The Ohio State University.
Flow Cytometry, Histopathology, and Immunohistochemical Analysis of miR-15b/16-2 Knockout Mice.
Immediately after death, the thoracic and abdominal cavities were examined. Spleen, lungs, liver, kidneys, bone marrow, and neoplastic masses (when available) were collected for flow cytometry and histological analysis. Lymphocytes from spleen and bone marrow were isolated for phenotypic analysis by flow cytometry. Cells were stained with the fluorescently labeled antibodies CD19 PerCP Cy5.5 (1D3; BD Pharmingen) and CD5-PE (53-7.3; BD Pharmingen). Tissue samples not used in cytometry analysis were fixed in 10% (vol/vol) neutral-buffered formalin for 24–48 h and processed routinely. Serial histological sections (4- to 6-μm-thick) were obtained from each paraffin block and stained with hematoxylin and eosin (H&E) for histology analysis. For immunophenotyping, representative sections were stained using antibodies against CD45R/B220, CD3, F4/80, Ter119, MPO, and Igκ (BD Pharmingen). Lymphoid lesions were classified according to the Bethesda classification (27).
Analysis of Human CLL Samples.
Human CLL samples were obtained from the Chronic Lymphocytic Leukemia Research Consortium after informed consent was obtained from patients diagnosed with CLL. Research was performed with the approval of the Institutional Review Board of The Ohio State University. RNA extraction was performed using TRIzol (Life Technologies) according to the manufacturer’s protocol. Real-time PCR experiments were carried out using miR-15b and pri-miR-15b assays for real-time PCR (Applied Biosystems) according to the manufacturer’s protocol. Control human cord blood CD19+ B cells were purchased from AllCells and Lonza.
miR-15b/Cyclin D1 Coexpression Analysis.
Protein/miRNA coexpression analysis was carried out as previously described with minor modifications (28). In situ hybridization for miR-15b and the positive control U6 was done using the 5′ digoxigenin-tagged LNA probe (Exiqon). Negative controls included omission of the probe and the use of a scrambled LNA probe (Exiqon). Only cytoplasmic miR-15b intensity was retained for scoring. After ISH, we used the Benchmark LT automated system from Leica Microsystems BOND-MAX according to the manufacturer’s specifications to perform the immunohistochemistry for Cyclin D1 (clone EP12; Dako). Protein/miRNA expression was quantified analyzing chromogen-specific intensity by ImageJ (NIH).
Cell Cultures, B-Cell Isolation, Transfection, and Western Blot Analysis.
293HEK cells (American Type Culture Collection) were cultured in DMEM (Gibco) and supplemented with 10% (vol/vol) FBS (Sigma-Aldrich) plus antibiotics. Cells were transfected in 12-multiwell plates using Lipofectamine 2000 (Invitrogen) following the manufacturer’s protocol. B cells were isolated and purified using a B-cell isolation kit (Miltenyi Biotec) according to the manufacturer’s instructions and were cultured in RPMI medium plus 10% (vol/vol) FBS with 1 μg/mL LPS for up to 32 h. B cells were harvested at specific time points after stimulation and washed with phosphate buffer saline, and pellets were lysed using RIPA buffer plus protease inhibitors (Cell Signaling Technology). Western blot analysis was carried out using BCL2 (Santa Cruz Biotechnology), IGF1RB (Cell Signaling Technology), Cyclin D2 (BD Pharmingen), and GAPDH (GeneTex) antibodies.
Luciferase Assay.
The predicted miR-binding sites of CCDN2 and IGF1R were cloned downstream of the firefly luciferase gene amplifying by PCR the gene-specific 3′ UTR. PCR products were digested with XhoI-NotI enzymes (New England Biolabs) and inserted into the psiCHECK-2 vector (Promega) multicloning site.
Acknowledgments
We thank Dr. Vincenza Guzzardo for her technical assistance, Kay Hubner for critical reading of the manuscript, Laura Rassenti for help in collecting human samples, Vincenzo Coppola for helpful scientific discussion, Janae Dulaney for technical support, Bryan McElwain and Katrina Moore of the Flow Lab, University Laboratory Animal Resources staff and veterinarians for help in animal husbandry, and Dr. Riccardo Dalla-Favera for kindly providing miR-15a/16-1 knockout mice. This work was supported by NIH/National Cancer Institute Grant R01 CA081534/CA151319 (to C.M.C.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1514954112/-/DCSupplemental.
References
- 1.Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 2.Calin GA, et al. A microRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353(17):1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 3.Cimmino A, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci USA. 2005;102(39):13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aqeilan RI, Calin GA, Croce CM. miR-15a and miR-16-1 in cancer: Discovery, function and future perspectives. Cell Death Differ. 2010;17(2):215–220. doi: 10.1038/cdd.2009.69. [DOI] [PubMed] [Google Scholar]
- 5.Calin GA, et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 2002;99(24):15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Calin GA, et al. MiR-15a and miR-16-1 cluster functions in human leukemia. Proc Natl Acad Sci USA. 2008;105(13):5166–5171. doi: 10.1073/pnas.0800121105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klein U, et al. The DLEU2/miR-15a/16-1 cluster controls B cell proliferation and its deletion leads to chronic lymphocytic leukemia. Cancer Cell. 2010;17(1):28–40. doi: 10.1016/j.ccr.2009.11.019. [DOI] [PubMed] [Google Scholar]
- 8.Careccia S, et al. A restricted signature of miRNAs distinguishes APL blasts from normal promyelocytes. Oncogene. 2009;28(45):4034–4040. doi: 10.1038/onc.2009.255. [DOI] [PubMed] [Google Scholar]
- 9.Garzon R, et al. MicroRNA gene expression during retinoic acid-induced differentiation of human acute promyelocytic leukemia. Oncogene. 2007;26(28):4148–4157. doi: 10.1038/sj.onc.1210186. [DOI] [PubMed] [Google Scholar]
- 10.Jones KB, et al. miRNA signatures associate with pathogenesis and progression of osteosarcoma. Cancer Res. 2012;72(7):1865–1877. doi: 10.1158/0008-5472.CAN-11-2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Satzger I, et al. 2010. MicroRNA-15b represents an independent prognostic parameter and is correlated with tumor cell proliferation and apoptosis in malignant melanoma. Int J Cancer 126(11):2553–2562.
- 12.Kanaan Z, et al. A plasma microRNA panel for detection of colorectal adenomas: A step toward more precise screening for colorectal cancer. Ann Surg. 2013;258(3):400–408. doi: 10.1097/SLA.0b013e3182a15bcc. [DOI] [PubMed] [Google Scholar]
- 13.Sun L, et al. MiR-200b and miR-15b regulate chemotherapy-induced epithelial-mesenchymal transition in human tongue cancer cells by targeting BMI1. Oncogene. 2012;31(4):432–445. doi: 10.1038/onc.2011.263. [DOI] [PubMed] [Google Scholar]
- 14.Bonci D, et al. The miR-15a-miR-16-1 cluster controls prostate cancer by targeting multiple oncogenic activities. Nat Med. 2008;14(11):1271–1277. doi: 10.1038/nm.1880. [DOI] [PubMed] [Google Scholar]
- 15.Sherr CJ. The Pezcoller Lecture: Cancer cell cycles revisited. Cancer Res. 2000;60(14):3689–3695. [PubMed] [Google Scholar]
- 16.Jaroslav P, et al. Expression of cyclins D1, D2, and D3 and Ki-67 in leukemia. Leuk Lymphoma. 2005;46(11):1605–1612. doi: 10.1080/10428190500215100. [DOI] [PubMed] [Google Scholar]
- 17.Lossos IS, et al. Prediction of survival in diffuse large-B-cell lymphoma based on the expression of six genes. N Engl J Med. 2004;350(18):1828–1837. doi: 10.1056/NEJMoa032520. [DOI] [PubMed] [Google Scholar]
- 18.Paul JT, et al. Cyclin D expression in chronic lymphocytic leukemia. Leuk Lymphoma. 2005;46(9):1275–1285. doi: 10.1080/10428190500158797. [DOI] [PubMed] [Google Scholar]
- 19.Delmer A, et al. Overexpression of cyclin D2 in chronic B-cell malignancies. Blood. 1995;85(10):2870–2876. [PubMed] [Google Scholar]
- 20.Fürstenberger G, Senn HJ. Insulin-like growth factors and cancer. Lancet Oncol. 2002;3(5):298–302. doi: 10.1016/s1470-2045(02)00731-3. [DOI] [PubMed] [Google Scholar]
- 21.Yaktapour N, et al. Insulin-like growth factor-1 receptor (IGF1R) as a novel target in chronic lymphocytic leukemia. Blood. 2013;122(9):1621–1633. doi: 10.1182/blood-2013-02-484386. [DOI] [PubMed] [Google Scholar]
- 22.Zheng X, Chopp M, Lu Y, Buller B, Jiang F. MiR-15b and miR-152 reduce glioma cell invasion and angiogenesis via NRP-2 and MMP-3. Cancer Lett. 2013;329(2):146–154. doi: 10.1016/j.canlet.2012.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xia L, et al. 2008. miR-15b and miR-16 modulate multidrug resistance by targeting BCL2 in human gastric cancer cells. Int J Cancer 123(2):372–379.
- 24.Chung GE, et al. High expression of microRNA-15b predicts a low risk of tumor recurrence following curative resection of hepatocellular carcinoma. Oncol Rep. 2010;23(1):113–119. [PubMed] [Google Scholar]
- 25.Pichiorri F, et al. Downregulation of p53-inducible microRNAs 192, 194, and 215 impairs the p53/MDM2 autoregulatory loop in multiple myeloma development. Cancer Cell. 2010;18(4):367–381. doi: 10.1016/j.ccr.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26.Ventura A, et al. Targeted deletion reveals essential and overlapping functions of the miR-17∼92 family of miRNA clusters. Cell. 2008;132(5):875–886. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morse HC, III, et al. Hematopathology subcommittee of the Mouse Models of Human Cancers Consortium Bethesda proposals for classification of lymphoid neoplasms in mice. Blood. 2002;100(1):246–258. doi: 10.1182/blood.v100.1.246. [DOI] [PubMed] [Google Scholar]
- 28.Nuovo GJ, et al. A methodology for the combined in situ analyses of the precursor and mature forms of microRNAs and correlation with their putative targets. Nat Protoc. 2009;4(1):107–115. doi: 10.1038/nprot.2008.215. [DOI] [PMC free article] [PubMed] [Google Scholar]