A phylogenetically conserved group of transicription factors form trimers to regulate root nodule development and early gene expression during the legume-Rhizobium symbiosis.
Abstract
The endosymbiotic association between legumes and soil bacteria called rhizobia leads to the formation of a new root-derived organ called the nodule in which differentiated bacteria convert atmospheric nitrogen into a form that can be assimilated by the host plant. Successful root infection by rhizobia and nodule organogenesis require the activation of symbiotic genes that are controlled by a set of transcription factors (TFs). We recently identified Medicago truncatula nuclear factor-YA1 (MtNF-YA1) and MtNF-YA2 as two M. truncatula TFs playing a central role during key steps of the Sinorhizobium meliloti-M. truncatula symbiotic interaction. NF-YA TFs interact with NF-YB and NF-YC subunits to regulate target genes containing the CCAAT box consensus sequence. In this study, using a yeast two-hybrid screen approach, we identified the NF-YB and NF-YC subunits able to interact with MtNF-YA1 and MtNF-YA2. In yeast (Saccharomyces cerevisiae) and in planta, we further demonstrated by both coimmunoprecipitation and bimolecular fluorescence complementation that these NF-YA, -B, and -C subunits interact and form a stable NF-Y heterotrimeric complex. Reverse genetic and chromatin immunoprecipitation-PCR approaches revealed the importance of these newly identified NF-YB and NF-YC subunits for rhizobial symbiosis and binding to the promoter of MtERN1 (for Ethylene Responsive factor required for Nodulation), a direct target gene of MtNF-YA1 and MtNF-YA2. Finally, we verified that a similar trimer is formed in planta by the common bean (Phaseolus vulgaris) NF-Y subunits, revealing the existence of evolutionary conserved NF-Y protein complexes to control nodulation in leguminous plants. This sheds light on the process whereby an ancient heterotrimeric TF mainly controlling cell division in animals has acquired specialized functions in plants.
Under nitrogen deficiency, legume plants are able to mutually interact with soil-borne bacteria collectively called rhizobia (Oldroyd et al., 2011). This endosymbiotic association results in the formation of a new root organ, named nodule, in which differentiated rhizobia fix atmospheric nitrogen for the benefit of the host plant. Nodule organogenesis is activated upon perception of rhizobia and involves a molecular dialogue between the two organisms (Oldroyd, 2013). Lipochitooligosaccharidic signals called Nod factors, produced by the bacteria, are key components of this dialogue, and these molecules can induce a number of early molecular, physiological, and morphological responses mimicking the first stages of the host plant-bacteria interaction (Lerouge et al., 1990). Morphological responses to rhizobia include local root hair deformations in which bacterial microcolonies become entrapped in curled root hairs before rhizobial infection is initiated via infection threads. Beneath the infection site, cells of the central and inner root cortex, endodermis, and pericycle tissues start to divide to form a nodule primordium from which a nodule meristem will originate (Xiao et al., 2014). Concomitant to nodule organogenesis, rhizobia-containing infection threads progress from the root hair to the newly formed nodule primordia where bacteria will be released to form a mature nodule structure (Fournier et al., 2008; Oldroyd and Downie, 2008). Depending on the legume host plant, a determinate nodule (ball-shaped organ with a time-limited growth as in common bean [Phaseolus vulgaris] or Lotus japonicus) or an indeterminate nodule (cylinder-shaped organ with a persistent meristem allowing indeterminate growth as in Medicago truncatula) is formed. Mature indeterminate nodules are composed of distinct zones characterized by specialized tissues and functions (Vasse et al., 1990). In the most apical part of the nodule, zone 1 consists of a meristematic zone that drives nodule growth. Below the meristem, in the infection zone (zone 2), plant cells start to differentiate, and rhizobia are released from infection threads into cytoplasmic vesicles called symbiosomes. Zone 3 (the fixation zone) is the central and most extensive region of the nodule where nitrogen fixation occurs by terminally differentiated nitrogen-fixing bacteroids. In older nodules, a senescent zone (zone 4) can eventually be observed at the base of the elongated nodules.
Genetic, molecular, and biochemical approaches have led to the identification of several genes involved in the regulation of nodulation (Desbrosses and Stougaard, 2011). Most of these genes are conserved in the Leguminosae family, in particular, members of the common symbiotic signaling pathway involved in both the rhizobial and arbuscular mycorrhizal root symbioses (Oldroyd, 2013). A central response in this common symbiotic signaling pathway is the activation of oscillatory nuclear calcium signatures that are decoded by the central calcium calmodulin kinase CCaMK/DMI3 (for Calcium Calmodulin-dependent kinase/Doesn’t Make Infections) regulator that, together with the IPD3 (for Interacting Protein 3 of DMI3)/CYCLOPS transcription factor (TF; Singh et al., 2014), regulate symbiotic gene expression. A number of other symbiotic TFs, such as NIN (for Nodule Inception; Schauser et al., 1999), NSP1 (for Nod factor Signalling Protein1; Smit et al., 2005), NSP2 (Kaló et al., 2005), and ERN1 (for ethylene Response factor involved in Nodulation1)/ERN2 (Andriankaja et al., 2007; Middleton et al., 2007; Cerri et al., 2012), have been involved in the regulation of early symbiotic gene expression. Among genes that are up-regulated in M. truncatula during symbiosis are the nuclear factor NF-YA1 and NF-YA2 TFs (Breakspear et al., 2014; Laloum et al., 2014; Laporte et al., 2014; Roux et al., 2014; Larrainzar et al., 2015). MtNF-YA1 and MtNF-YA2 play partially overlapping roles during NF signaling by directly regulating the transcription of the ERN1-encoding gene (Laloum et al., 2014). In addition, MtNF-YA1 is required for subsequent steps of rhizobial infection and nodule meristem formation and maintenance in M. truncatula (Combier et al., 2006, 2008; Laporte et al., 2014; Xiao et al., 2014). MtNF-YA1 and its close homolog MtNF-YA2 belong to the NF-Y TF family, also called heme activator protein (HAP) or CCAAT box binding factor (CBF), found in all eukaryotic organisms (Mantovani, 1999; Dolfini et al., 2012). This TF family is composed of three subunits, NF-YA, NF-YB, and NF-YC, which form a heterotrimeric complex that binds to the core CCAAT box cis-element found on target promoter sequences. In the NF-Y trimeric complex, the NF-YA subunit is involved in the specific recognition and binding of the CCAAT box, but can only achieve this binding in the presence of the unrelated NF-YB and NF-YC proteins, which contain histone fold motifs. These deviant histone proteins are involved in promoting chromatin accessibility to NF-YA and other specific TFs, as well as the recruitment of additional interacting proteins (Nardini et al., 2013; Oldfield et al., 2014). In animal cells, each subunit is encoded by a single gene, and their molecular assembly has been well characterized: whereas the NF-YA subunit with its nuclear localization signals is targeted to the nucleus, the NF-YB and NF-YC subunits first form a dimer in the cytoplasm, before being transported to the nucleus to form a heterotrimer with NF-YA that binds to the CCAAT box (Mantovani, 1999). In contrast to the single-gene situation in mammals, plant NF-Y genes are encoded by small gene families of about 10 members for each subunit, resulting in a high combinatorial potential for NF-Y complexes (Petroni et al., 2012; Laloum et al., 2013). This raises the question of whether the diversification of the NF-Y family in plants is related to increased functional redundancy and/or functional specialization associated with different plant-related processes.
In legumes, a number of NF-Y subunits were independently reported to participate in nitrogen-fixing rhizobia symbiosis. This includes the M. truncatula MtNF-YA1 and A2 subunits (Combier et al., 2006; Laloum et al., 2014; Laporte et al., 2014), but also the two L. japonicus NF-Y subunit-encoding genes LjNF-YA1 and LjNF-YB1 (Soyano et al., 2013). Another symbiotic NF-Y subunit, PvNF-YC1, was described in the common bean as required for both rhizobial infection and nodule development (Zanetti et al., 2010). However, the physical interaction between the different symbiotic NF-Y subunits that leads to the formation of a functional heterotrimeric complex has not been described so far. In this study, we used a yeast two-hybrid (Y2H) approach to identify NF-YB and NF-YC subunits interacting with MtNF-YA1 and MtNF-YA2 during the rhizobial symbiotic process. We validated these protein interactions and demonstrated NF-Y trimer formation both in yeast (Saccharomyces cerevisiae) and in planta. In addition, we demonstrated the functional implication of these subunits during nodulation, and showed that there is a conservation in NF-Y subunits involved in rhizobial symbiosis across legume plants.
RESULTS
The Symbiotic Expression Pattern of NF-Y Subunits in M. truncatula
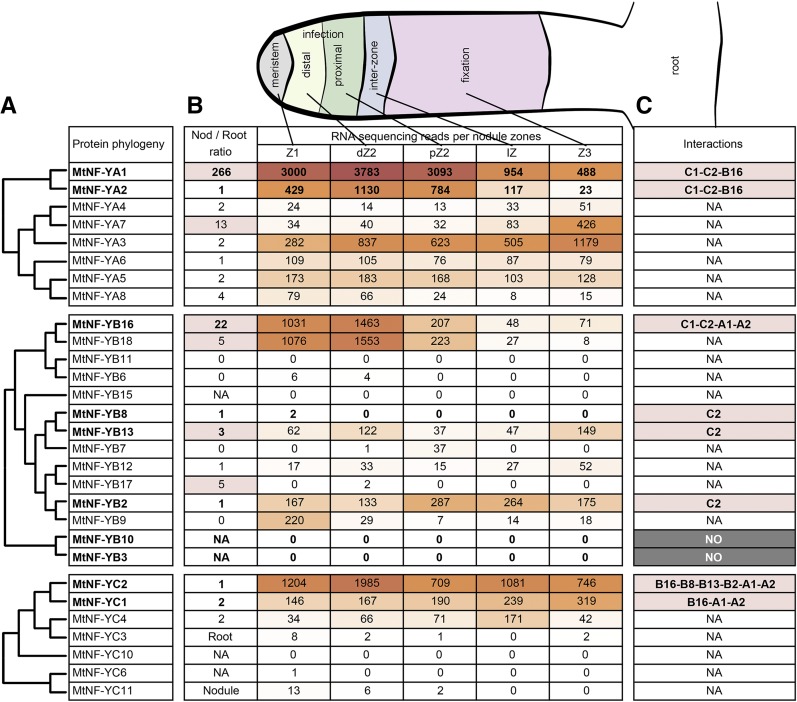
To get an overall picture of the interaction potential of the 8 NF-YA, the 14 NF-YB, and the 7 NF-YC proteins of M. truncatula during symbiotic nodule development, we integrated transcriptomic data produced by laser-captured microdissected nodule zones (Roux et al., 2014) with a phylogenetic analysis of all M. truncatula NF-Y proteins (Fig. 1). Strikingly, for all three NF-Y subunits, a group of two closely related genes represent most of the NF-Y transcripts found in nodules, namely, MtNF-YA1 and MtNF-YA2, MtNF-YB16 and MtNF-YB18, and MtNF-YC1 and MtNF-YC2. MtNF-YA1 and MtNF-YA2 are mainly expressed in the apex of the nodule (meristematic region [Z1] and infection zone [dZ2 and pZ2]). In addition, MtNF-YA1 is much more strongly expressed in nodules compared with roots, where its expression is almost undetectable. The other MtNF-YA genes that are grouped in distinct phylogenic clades are significantly less expressed in symbiotic tissues than MtNF-YA1 and MtNF-YA2. One exception is MtNF-YA3, which is mainly expressed in proximal nodule tissues (i.e. the nodule interzone and fixation zone [Z3]) in which MtNF-YA1 and MtNF-YA2 are only barely expressed (Combier et al., 2006; Laloum et al., 2014). Concerning the NF-YB subunits, MtNF-YB16 and MtNF-YB18 are the only two genes that are expressed in nodules at very high levels. Other NF-YB members, namely, MtNF-YB13, B2, and B9, can also be detected in nodules but at much lower levels (Fig. 1). Interestingly, MtNF-YB16 and the closely related MtNF-YB18 are also predominantly expressed in the apical region of the nodule and at higher levels in symbiotic tissues compared with roots, similar to what is observed for MtNF-YA1 and A2. In the NF-YC family, only MtNF-YC2 and MtNF-YC1 show a significant expression level in nodules. However, whereas MtNF-YC2 is expressed predominantly in the nodule apex, MtNF-YC1 is expressed in all nodule zones. Based on their expression in the same symbiotic nodule tissues, four NF-Y subunits, MtNF-YB16, MtNF-YB18, MtNF-YC1, and MtNF-YC2, thus are the best likely candidates to form heterotrimeric NF-Y complexes with MtNF-YA1 and/or MtNF-YA2 in nodules.
Figure 1.
Symbiotic expression pattern and phylogenetic tree of M. truncatula NF-Y subunits. A, The 8 NF-YA, 14 NF-YB, and 7 NF-YC subunits of M. truncatula are classified following a protein-based phylogenetic analysis using the maximum-likelihood method. B, The first column illustrates the expression ratio of 10-d-old entire nodules compared with roots based on RNA sequencing. The middle columns represents normalized expression data based on laser caption followed by RNA sequencing analysis in different nodule tissues at 15 d postinoculation as illustrated in the drawing above (B). The columns correspond to the meristematic zone (Z1), the apical and distal infection zone (dZ2 and pZ2), the nodule interzone (IZ), and the nitrogen-fixation zone (Z3; Roux et al., 2014). The dark-brown to white color scale indicates the highest to lowest expression, respectively. C, Summary of the interaction data obtained in Y2H using NF-Y subunits. NA, Not available.
MtNF-YA1 and MtNF-YA2 Interact with Two Members of the MtNF-YC Family
To identify components of the multimeric NF-Y protein complex that actually interact with MtNF-YA1 during the symbiotic interaction between M. truncatula and Sinorhizobium meliloti, we performed a Y2H screen using MtNF-YA1 as a bait. Nearly 4 million clones were screened, leading to the identification of 42 putative interacting proteins. Among the positive clones, we identified MtNF-YC1 and MtNF-YC2. A phylogenetic analysis using the seven NF-YC proteins of M. truncatula and common bean indicated that MtNF-YC2 is the closest homolog of PvNF-YC1, an NF-YC subunit identified in common bean as a protein playing roles comparable with MtNF-YA1 during rhizobial infection and nodule development (Zanetti et al., 2010), and that MtNF-YC2 is the closest homolog of PvNF-YC6 (Supplemental Fig. S1). We then tested the microsynteny between the M. truncatula and common bean genes coding for these NF-YC proteins using the Locus search tool of the Plant Genome Duplication Database (Lee et al., 2013). The synteny analysis revealed that both MtNF-YC1 and MtNF-YC2 exhibited large syntenic blocks with PvNF-YC1 and PvNF-YC6, indicating that they are most likely orthologs (Supplemental Fig. S2). A similar analysis using MtNF-YA subunits strongly suggests that MtNF-YA1 and MtNF-YA2 are orthologs of PvNF-YA1 and PvNF-YA9 (Supplemental Figs. S1 and S2).
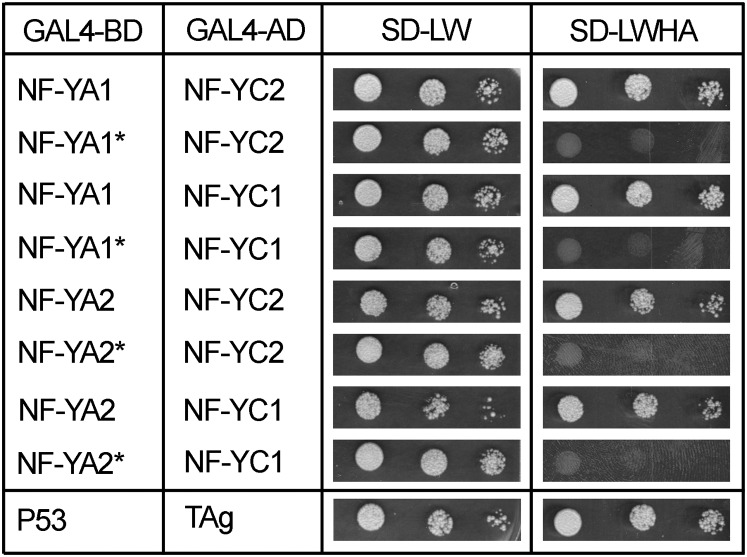
MtNF-YC1 and C2 were then recloned in the appropriate vectors, and pairwise Y2H experiments confirmed the interaction of MtNF-YA1 with MtNF-YC1 and MtNF-YC2 (Fig. 2). Similar results were also obtained with the closely related MtNF-YA2 protein (Fig. 2).
Figure 2.
MtNF-YA1 and MtNF-YA2 interact with MtNF-YC1 and MtNF-YC2 in yeast. Interaction of BD (for Gal4 Binding domain)-NF-YA1, BD-NF-YA2, BD-NF-YA1*, or BD-NF-YA2* with AD (for Gal4 Activation Domain)-NF-YC1 or AD-NF-YC2 using Y2H. BD-P53 and AD-TAg were used as positive interacting controls. Yeast suspensions of optical density (OD) of 1, 0.1, and 0.01 were spotted on selective medium for plasmid only (SD-LW) or plasmid plus interaction (SD-LWHA).
Taking advantage of the high degree of conservation between the protein interaction domain of NF-YA proteins across kingdoms (Laloum et al., 2014), we then created Lys (K) to glutamic (E) mutations in conserved K171 (of NF-YA1) and K173 (of NF-YA2) residues, previously shown in animals to be essential for NF-Y subunit interaction (Mantovani et al., 1994; Supplemental Fig. S3). Using yellow fluorescent protein (YFP)-tagged proteins, we showed that both the native and the mutated MtNF-YA subunits localized to the nucleus of Nicotiana benthamiana cells, indicating that this mutation did not affect the subcellular localization of NF-YA subunits (Supplemental Fig. S4). Interestingly, the mutated proteins, called MtNF-YA1* and MtNF-YA2*, lost the ability to interact with MtNF-YC1 and C2 in yeast (Fig. 2). These results indicate that the highly conserved Lys residue is also important for NF-Y subunit interaction in plant cells. These mutant proteins therefore represented ideal negative controls for further interaction experiments. In an attempt to confirm these interactions in planta, we used the bimolecular-fluorescence complementation (BiFC) approach (Hu et al., 2002). Whereas the interaction between positive control proteins cYFP-NSP1 and nYFP-NSP2 (Hirsch et al., 2009) was clearly detected by the appearance of the YFP fluorescent protein signal in the nuclei of N. benthamiana-expressing cells, we were unable to detect any clear interaction between MtNF-YA1 or A2 with respective MtNF-YC1 or C2, as previously observed in yeast (Supplemental Fig. S5).
MtNF-YC1 and MtNF-YC2 Interact with the MtNF-YB16 Subunit
After identifying two NF-YC subunits capable of interacting with MtNF-YA1 and A2, we then performed a new Y2H screen using MtNF-YC2 as a bait to identify interacting NF-YB subunits. MtNF-YC2 was preferred to MtNF-YC1 because of its higher expression levels in nodules and its expression pattern, which is closer to that observed for MtNF-YA1 (Fig. 1). About 1.6 million clones were screened from a Y2H complementary DNA (cDNA) library (see “Materials and Methods”) leading to the identification of 26 putative interacting proteins. Those included two different NF-YB proteins: MtNF-YB13 (two clones) and MtNF-YB16 (eight clones). A phylogenetic analysis of the M. truncatula NF-YB proteins showed that MtNF-YB13 and MtNF-YB16 belong to two subgroups of NF-YB proteins that are related but distinct from the divergent and seed-specific LEC (for Leafy Cotyledon) group (Supplemental Fig S1). We then performed a second phylogenetic analysis using the 14 NF-YB proteins of M. truncatula and common bean together with LjNF-YB1, an L. japonicus subunit that, together with LjNF-YA1, was shown to play an important role during nodule organogenesis (Soyano et al., 2013; Supplemental Fig. S1). Interestingly, this analysis indicates that MtNF-YB16 is the closest homolog of LjNF-YB1. We then tested the microsynteny between the M. truncatula, L. japonicus, and common bean genes coding for these NF-YB proteins using the Plant Genome Duplication Database (Lee et al., 2013), but no syntenic blocks were detected.
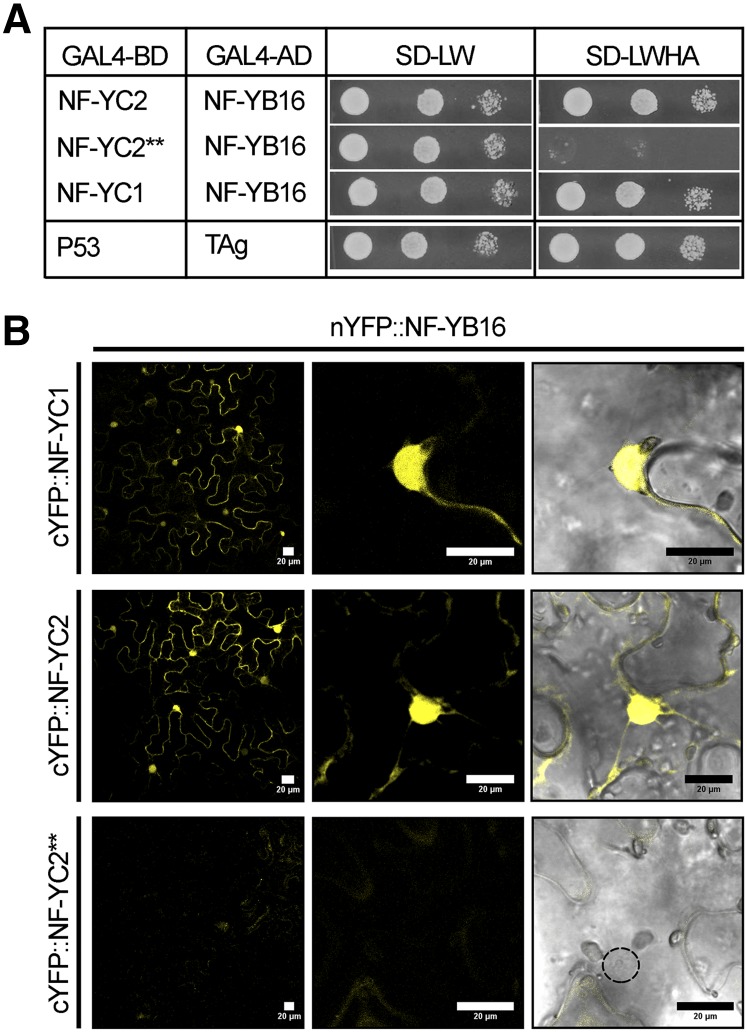
The interaction between MtNF-YC2 and MtNF-YB16 was confirmed using pairwise Y2H experiments, and similar results were obtained with the closely related MtNF-YC1 (Fig. 3A). As for MtNF-YA1, we created an MtNF-YC2 protein mutated in two conserved amino acids shown to be essential in animal NF-YC proteins for the interaction with NF-YB subunits (Kim et al., 1996; Fig. 3A), and we called it MtNF-YC2**. Here again, the interaction between the mutated protein and MtNF-YB16 was lost, reinforcing our interaction results and showing that, as in animal NF-YC proteins, the interaction between B and C subunits depends upon the conserved Ile-136 and Asp-139 residues within the protein interaction domain of NF-YC (Supplemental Fig. S3).
Figure 3.
MtNF-YC1 and MtNF-YC2 interact with MtNF-YB16 in yeast and in planta. A, Interaction of BD-MtNF-YC2, BD-MtNF-YC2**, or BD-MtNF-YC1 with AD-MtNF-YB16 using Y2H. BD-P53 and AD-TAg were used as positive interacting controls. Yeast suspensions of OD of 1, 0.1, and 0.01 were spotted on selective medium for plasmid only (SD-LW) or plasmid plus interaction (SD-LWHA): MtNF-YC2** is an NF-YC2 protein with two I136 to D and L139 to E mutations in conserved amino acids, serving as a negative control for interaction. B, In planta interaction of MtNF-YB16 with MtNF-YC1, MtNF-YC2, or MtNF-YC2** using BiFC. N. benthamiana leaves were agroinfiltrated with nYFP-MtNF-YB16 together with cYFP-MtNF-YC1 (top), cYFP-MtNF-YC2 (middle), or cYFP-MtNF-YC2** (negative control, bottom) translational fusions. Confocal images of the YFP fluorescence (left), a magnification (middle), and a merge between YFP and bright-field images (right) are shown. Bar = 20 μm.
To evaluate these interactions in planta, we performed BiFC experiments. Expression of cYFP-MtNF-YC1 or cYFP-MtNF-YC2 with nYFP-MtNF-YB16 led to a strong fluorescent signal at the expected YFP wavelength (Supplemental Fig. S6) both in the nucleus and in the cytoplasm of N. benthamiana cells. By contrast, no fluorescent signal was observed when the c-YFP-MtNF-YC2** (I136D/L142E) was cotransformed with nYFP-MtNF-YB16 (Fig. 3B). The observed interactions between MtNF-YC1 or C2 and MtNF-YB16 were confirmed by using the reverse combination of C- and N-terminal YFP domains fused to MtNF-YC and MtNF-YB, respectively (Supplemental Fig. S6). Therefore, we concluded that MtNF-YB16 and the two MtNF-YC1 and C2 proteins interact in planta, and that this interaction depends on the conserved (I136 and D139) residues.
Specificity of NF-Y Interactions
To test the specificity of the interaction between B and C subunits, we tested respective interactions of MtNF-YC2 with six different NF-YB subunits belonging to different phylogenetic subgroups that are either strongly expressed (B16), weakly expressed (B2, B13), or not expressed in nodules (B3, B8, B10; Fig. 1). With the notable exception of the divergent and seed-specific LEC group (i.e. MtNF-YB3 and B10), all NF-YB subunits tested interact with MtNF-YC1 and MtNF-YC2, regardless of their phylogenetic distance to MtNF-YB16 (Supplemental Fig. S7). As expected, MtNF-YC2** used as a negative control did not interact with any of the NF-YB proteins tested (Supplemental Fig. S7). These results suggest that, as was already demonstrated in Arabidopsis (Arabidopsis thaliana; Calvenzani et al., 2012), the interaction specificity between NF-YB and NF-YC subunits is weak in M. truncatula.
To evaluate the specificity of interaction between A and B subunits, using Y2H, we tested the respective interaction of MtNF-YA1 with different NF-YB proteins, namely, NF-YB2, B3, B8, B10, B13, and B16 (Fig. 1). Strikingly, no interaction could be detected, even for MtNF-YB2, B8, B13, and B16, which are able to interact with MtNF-YC2 (Supplemental Fig. S7). This suggests that the interactions between MtNF-YA and MtNF-YB proteins cannot be detected in yeast.
MtNF-YB16 and MtNF-YC1/MtNF-YC2 Interact with MtNF-YA1 and MtNF-YA2 to Form NF-Y Trimers in Yeast and in Planta
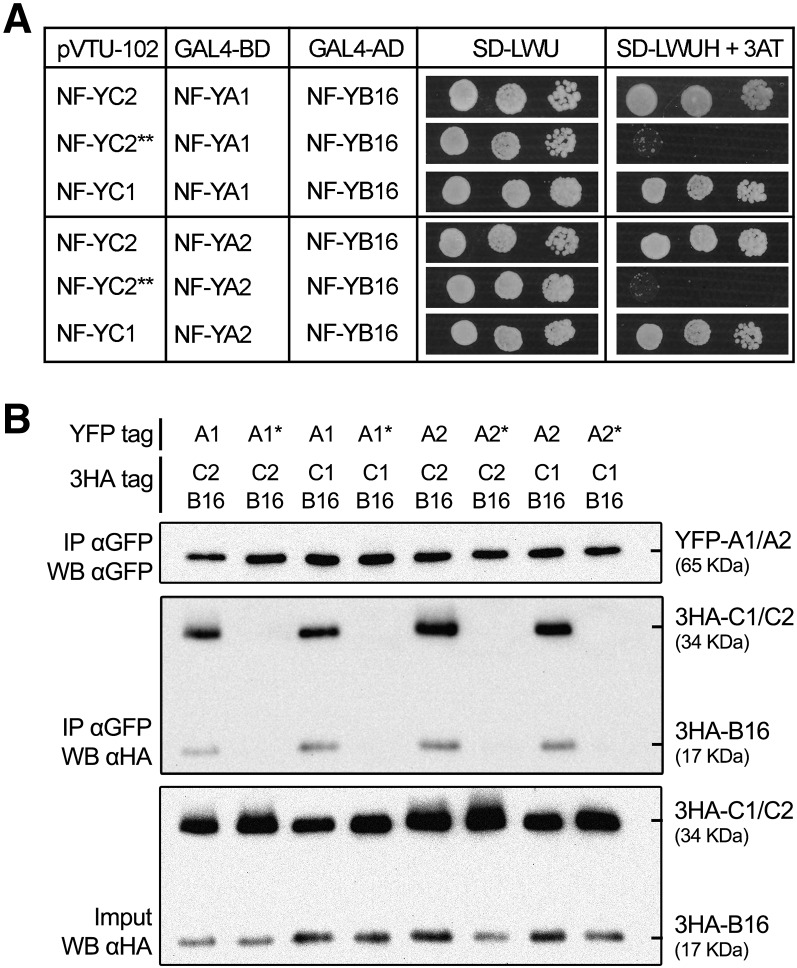
After demonstrating interactions between, respectively, A and C (MtNF-YA1 and A2 with MtNF-YC1 and C2) and C and B (MtNF-YC1 and C2 with MtNF-YB16/B13) subunits, we aimed to evaluate whether a YA-YB-YC NF-Y trimer could be formed with the above-mentioned subunits. To evaluate this question, we set up yeast three-hybrid experiments based on the coexpression of the three subunits simultaneously. Coexpressing A subunits (BD-MtNF-YA1 or BD-NF-YA2) with B (AD-MtNF-YB16) together with C subunits (MtNF-YC2 or MtNF-YC1) enabled strong yeast growth in selective medium, indicating the formation of a stable NF-Y heterotrimer. Yeast growth could not be observed when the C subunit was replaced by the mutated MtNF-YC2** version, in agreement with our previous pairwise experiments (Fig. 4A; Supplemental Fig. S7).
Figure 4.
MtNF-YA1 and A2 form NF-Y trimers with MtNF-YB16 and MtNF-YC1 and C2. A, Tripartite interaction test of MtNF-YB16. MtNF-YA1 or MtNF-YA2, and MtNF-YC1, MtNF-YC2, or MtNF-YC2** by yeast triple hybrid. The interaction between BD-MtNF-YA1 or BD-MtNF-YA2 and AD-MtNF-YB16 was tested after expression of the third MtNF-YC1, MtNF-YC2, or MtNF-YC2** (negative control) subunit. Yeast suspensions at OD of 1, 0.1, and 0.01 were spotted on selective medium for plasmid only (SD-LWU) or plasmid plus interaction (SD-LWUH + 5 mm 3-amino-1,2,4-triazole). MtNF-YC2** is an MtNF-YC2 protein with two I136 to D and L139 to E mutations. B, Coimmunoprecipitation (CoIP) assay, MtNF-YA1, MtNF-YA2, and their corresponding control (MtNF-YA1* and MtNF-YA2* with a K171 or 173 to E mutation) tagged with YFP were coexpressed in N. benthamiana with MtNF-YB16 and MtNF-YC1 or MtNF-YC2 fused to the 3HA tag and affinity bound with GFP magnetotrap. The crude extracts (input) and immunoprecipitated (IP) fractions were subjected to protein western blots (WB). Tagged proteins were detected with anti-GFP antibodies: α-GFP (YFP-tagged proteins) or anti-HA antibodies: α-HA (3HA-tagged proteins).
To confirm the formation of these heterotrimers in planta, we performed CoIP experiments in N. benthamiana. For this purpose, we coexpressed YFP-tagged MtNF-YA subunits (MtNF-YA1 or MtNF-YA2) or the corresponding mutated versions (MtNF-YA1* and MtNF-YA2*) together with 3HA (for Haemagglutinin)-tagged C-subunits (MtNF-YC1 or MtNF-YC2) and the B-subunit MtNF-YB16 in N. benthamiana leaf epidermal cells. Immunoprecipitations were performed using anti-GFP antibodies, and the presence of NF-YB and NF-YC subunits was detected by immunoblot using anti-HA antibodies. Trimer formation was observed only when native MtNF-YA1 or MtNF-YA2 proteins, but not the mutated forms of MtNF-YA, were coexpressed with MtNF-YC1 or C2 and MtNF-YB16 (Fig. 4B). In summary, we showed that specific NF-Y trimeric complexes are formed both in yeast and in planta when coexpressing compatible NF-Y subunits.
Subcellular Localization and Nuclear Relocalization of MtNF-YB16, MtNF-YC1, and MtNF-YC2
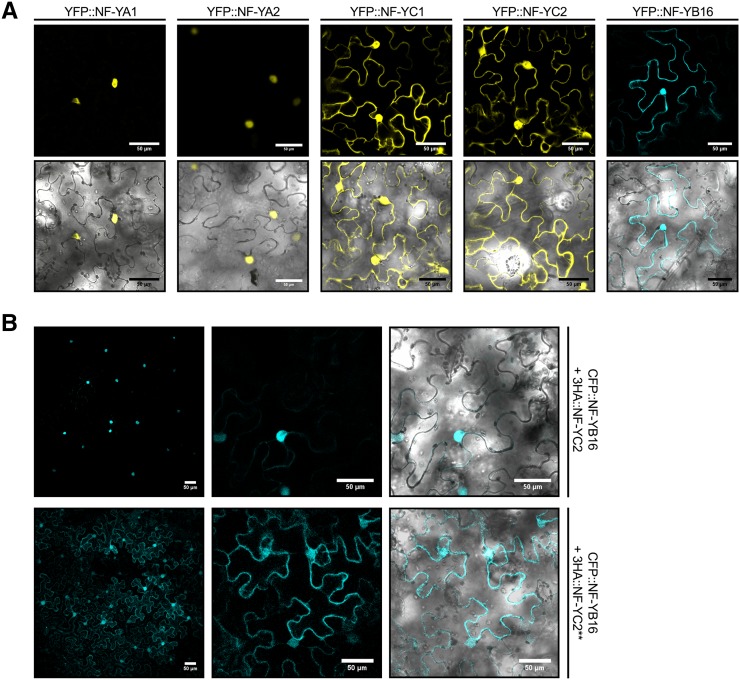
To evaluate the subcellular localization of respective NF-Y subunits, we analyzed YFP or cyan fluorescent protein (CFP) protein fusions expressed in N. benthamiana cells. MtNF-YA1 and MtNF-YA2 localized strictly to the nucleus as previously described (Laloum et al., 2014), whereas MtNF-YC1 and C2 or MtNF-YB16 were found both in the nucleus and in the cytoplasm (Fig. 5A). Western-blot analysis using anti-GFP antibodies confirmed that the respective protein fusions were intact and displayed the expected protein sizes for uncleaved protein fusion versions (58 kD for YFP-NF-YCs and 40 kD for CFP-MtNF-YB16; Supplemental Fig. S8), thus indicating that the dual nuclear/cytoplasmic localization of the NF-Y B and C subunits is not due to cleavage of the fluorescent protein.
Figure 5.
Association dynamics of MtNF-YA1 and A2, MtNF-YB16, And MtNF-YC1 and C2. A, MtNF-YA subunits are nuclear, whereas MtNF-YB and MtNF-YC subunits are nucleocytoplasmic. The YFP-MtNF-YAs or MtNF-YCs and CFP-MtNF-YB16 fusions were introduced in N. benthamiana cells by agroinfiltration. The confocal images of fluorescence channel alone (top) or fluorescence merge to bright-field channel (bottom) were obtained for each construct. Bar = 50 μm. B, MtNF-YB16 nuclear accumulation depends on the presence of MtNF-YC2. The CFP-MtNF-YB16 together with 3HA-MtNF-YC2 or 3HA-MtNF-YC2** were introduced in N. benthamiana cells by agroinfiltration. The confocal images of fluorescence channel alone (top and magnification for the middle) or fluorescence merge to bright-field channel (bottom, magnification) was obtained for each construct. Bar = 50 μm.
To evaluate whether NF-YB subunits can be relocalized to the nucleus via their interaction with NF-YC, as previously suggested for other plant counterparts (Liu and Howell, 2010; Hackenberg et al., 2012), we tested the ability of 3HA-tagged versions of MtNF-YC2 or MtNF-YC2** to modify the subcellular localization of a CFP-MtNF-YB16 fusion. Coexpression of the native MtNF-YC2 led to a strong nuclear concentration of the MtNF-YB16 fluorescent protein, whereas this was not observed when coexpressing the mutated 3HA-MtNF-YC2** version (Fig. 5B). In conclusion, the tested B and C subunit proteins are both distributed in the nucleus and in the cytoplasm, but are concentrated in the nuclear compartment when coexpressed together.
Symbiotic NF-Y Trimers Also Form in Common Bean
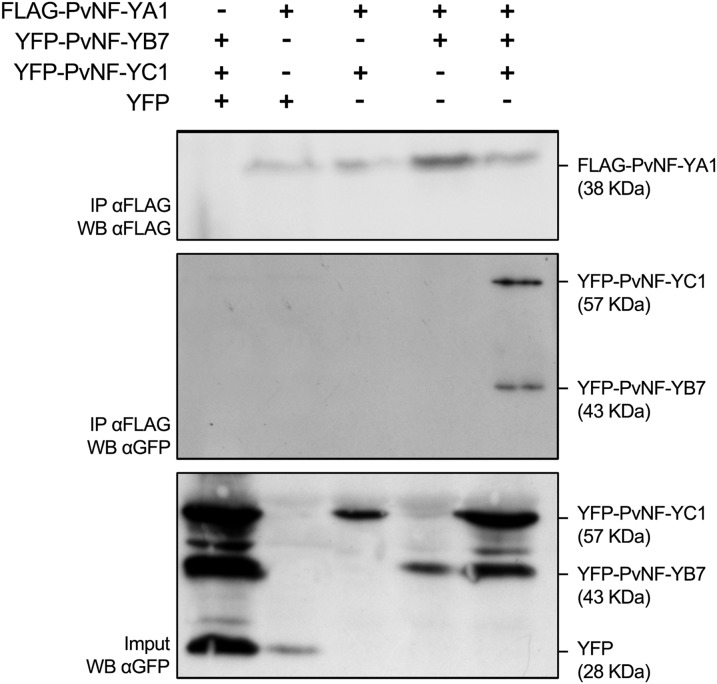
As described above, the MtNF-YC2 subunit characterized as part of the symbiotic NF-Y complex is closely related to the common bean PvNF-YC1, which was previously described as being required for symbiotic infection and nodule development (Zanetti et al., 2010). To evaluate whether PvNF-YC1 could form a heterotrimeric complex with common bean NF-Y subunits phylogenetically related to MtNF-YA1 and A2 and MtNF-YB16 described above, we first selected the common bean counterparts for CoIP experiments. The NF-Y subunits were chosen on the basis of both their phylogenetic position and their similarity to MtNF-YA1 and A2, B16, and C1 and C2 (Supplemental Fig. S1), as well as their expression pattern in common bean (Battaglia et al., 2014; Dalla Via et al., 2015; Ripodas et al., 2015). Tagged versions of PvNF-YA1, PvNF-YB7, and PvNF-YC1 proteins were then coexpressed in N. benthamiana. Using an anti-FLAG (for FLAG octapeptide epitope) antibody, YFP-PvNF-YC1 and YFP-PvNF-YB7 were coimmunopurified together with the FLAG-PvNF-YA1, demonstrating that these three common bean subunits also form a stable NF-Y trimer in planta as shown for M. truncatula NF-Y subunits (Fig. 6).
Figure 6.
PvNF-YA1, PvNF-YB7, and PvNF-YC1 form a heterotrimer in planta. FLAG-PvNF-YA1 fusion was coexpressed with YFP-PvNF-YB7 and/or YFP-PvNF-YC1 in N. benthamiana leaves. FLAG-PvNF-YA1 + free YFP and YFP-PvNF-YB7 + YFP-PvNF-YC1 + free YFP combinations were used as negative controls. CoIPs were performed using αFLAG agarose beads. Crude protein extracts (input) and immunoprecipitated fractions (IP) were analyzed by western blots (WB) using an anti-FLAG antibody (α-FLAG) or an anti-GFP antibody (α-GFP) to detect PvNF-YA1 or PvNF-YB7/PvNF-YC1, respectively.
Nodule Organogenesis Is Affected in MtNF-YC, But Not in MtNF-YB RNA Interference Lines
To investigate the symbiotic roles of MtNF-YB16 and C subunits, RNA interference (RNAi) strategies were use in Agrobacterium rhizogenes-transformed roots. First, an RNAi construct specifically targeting MtNF-YB16 was used to down-regulate (5-fold) the expression of the endogenous MtNF-YB16 gene, but not the closely related MtNF-YB18 (Supplemental Fig. S9). No significant effect on nodulation, either qualitative (morphogenesis) or quantitative (nodule number), was observed in the MtNF-YB16 RNAi roots (Supplemental Fig. S9). As a complementary approach, we designed a less specific RNAi construct, referred to as NF-YB-RNAi, using a conserved domain of MtNF-YB16. All of the closely related MtNF-YB subunit encoding genes (i.e. MtNF-YB16, B18, B6, B11), but not the more distant MtNF-YB2, B8, and B13, were silenced by this construct in transgenic roots (Supplemental Fig. S9). Despite the down-regulation of these MtNF-YB members, no symbiotic phenotype was observed after inoculation of these roots by S. meliloti (Supplemental Fig. S9). This suggests that down-regulation of MtNF-YB16, B18, B6, and B11 subunits is not sufficient to detect major symbiotic phenotypic changes.
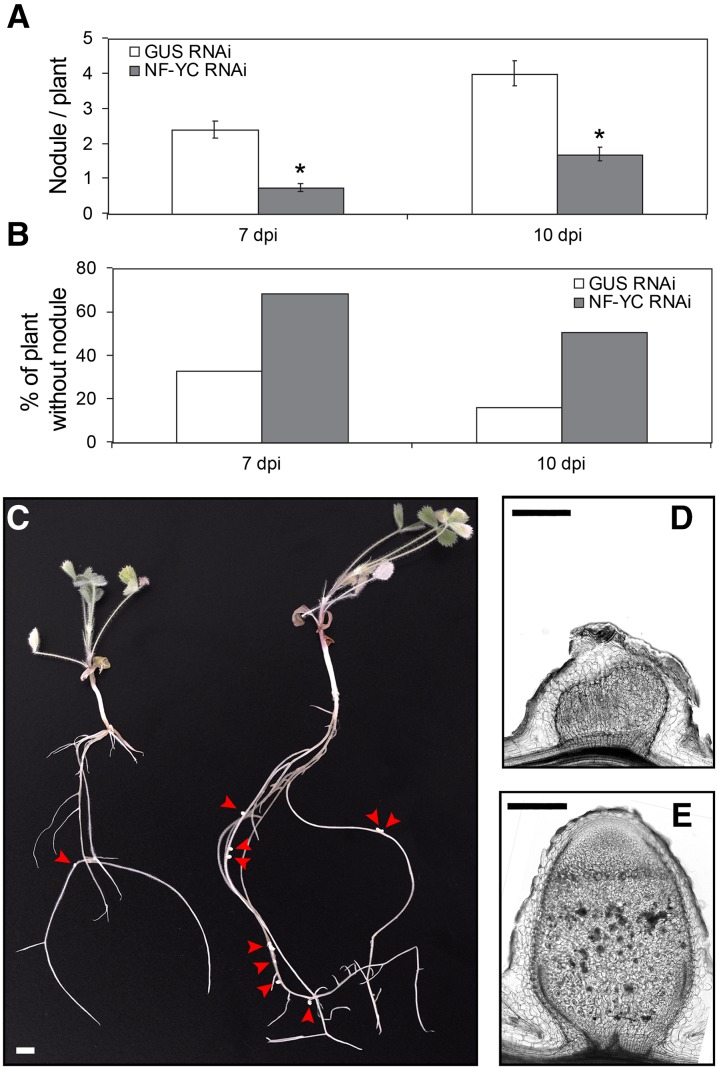
We then investigated the functional importance of MtNF-YC1 and MtNF-YC2 subunits. We designed an RNAi construct that specifically silenced MtNF-YC2 (Supplemental Fig. S10), but as for MtNF-YB16 RNAi constructs, no clear effect on timing of nodulation or on nodule number or morphology was observed. Because we suspected functional redundancy between MtNF-YC1 and MtNF-YC2, we additionally designed an RNAi construct, termed NF-YC-RNAi, that silenced both genes but not the closely related MtNF-YC4 (Supplemental Fig. S10). After transformation of this construct into transgenic roots, we observed a significant (more than 2-fold) reduction in the number of nodules compared with control plants at both 7 and 10 d after inoculation with rhizobia (Fig. 7A). In addition, approximately 50% of the NF-YC RNAi plants showed no nodules at all 10 d after inoculation, whereas only 15% of the control plants showed no nodules at this time point after rhizobia inoculation (Fig. 7B). The NF-YC-RNAi construct also clearly affected nodule development. Most nodules remained small and did not develop the typical nodule organization into differentiated zones found in control plants (Vasse et al., 1990; Fig. 7, C–E). Interestingly, these phenotypes are reminiscent of those observed using MtNF-YA1-RNAi constructs (Combier et al., 2006) or the Mtnf-ya1-1 mutant (Laporte et al., 2014). However, no clear effect on rhizobial infection or NF-induced gene expression (MtNSP1, MtERN1, and MtENOD11 [for Early Nodulation 11]) could be observed (Supplemental Fig. S11).
Figure 7.
The down-regulation of MtNF-YC1 and MtNF-YC2 affects nodule development. A, The number of nodules that developed 7 and 10 d after S. meliloti inoculation were counted in both control GUS RNAi (white boxes) and NF-YC RNAi (gray boxes) composite plants. Results shown are the average number of nodules counted per composite plant from 150 plants in two separate biological replicates. Error bars correspond to the sd. Significant differential expression (*) was determined by Student tests (P ≤ 0.05). B, Percentage of plants showing no nodules in the same data set as in (A). C, Picture of entire NF-YC RNAi (left) and control GUS RNAi plants at 17 d postinoculation. Red arrowheads show the nodules (bar = 5 mm). D and E, Sections of 14-d-old nodules from composite NF-YC RNAi plants (D) and control GUS RNAi plants (E). Longitudinal sections (50 μm thick) were performed on isolated nodules (bar = 0.2 mm).
MtNF-YB16 and MtNF-YC2 Bind the CCAAT Box 3 on the MtERN1 Promoter
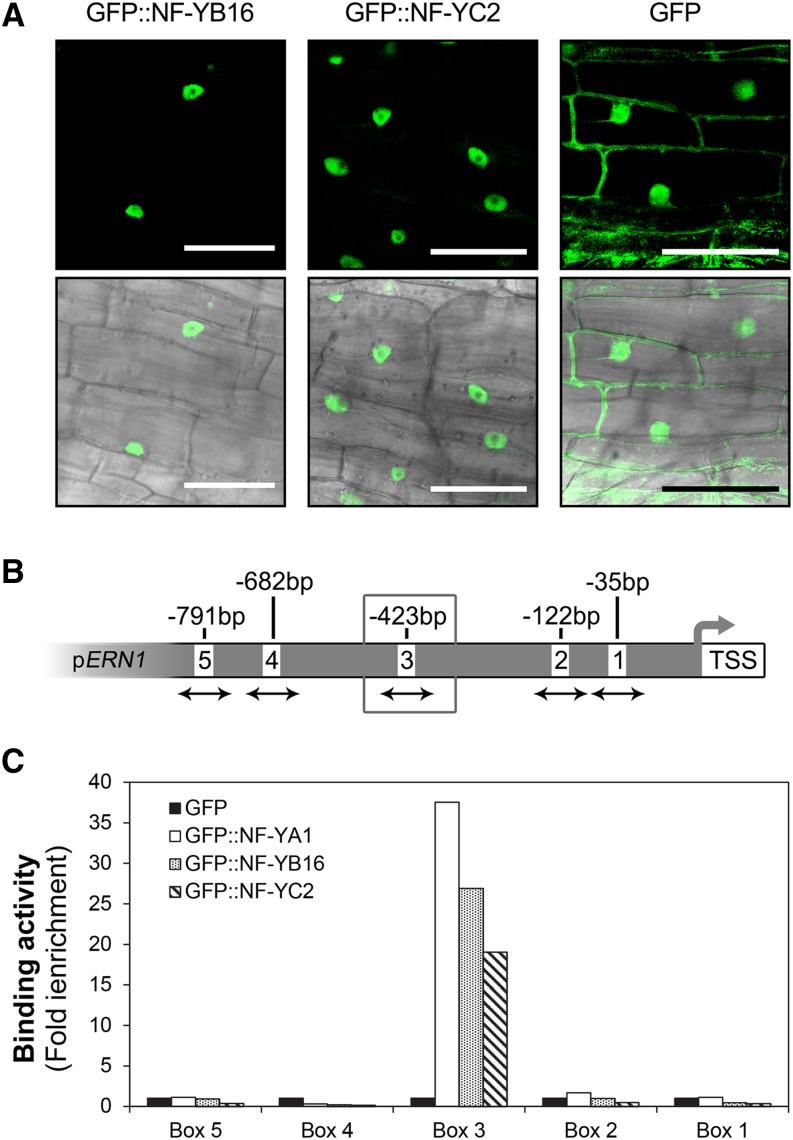
MtNF-YA1 and MtNF-YA2 were recently shown to directly regulate the expression of the symbiosis-associated TF-encoding gene MtERN1 by binding to the third CCAAT box identified within its promoter (Laloum et al., 2014). To evaluate whether the three NF-Y subunits can interact with this target sequence, we performed chromatin immunoprecipitation (ChIP)-PCR experiments using transgenic roots expressing GFP-tagged versions of MtNF-YA1, B16, and C2 proteins (see “Materials and Methods”). As shown in Fig. 8, B and C, similar results were obtained with all three fusion proteins (i.e. strong binding to the third CCAAT box of the ERN1 promoter), showing that MtNF-YB16 and MtNF-YC2 can bind to the same promoter region as MtNF-YA1 and are thus likely to be part of the same transcriptional complex in planta (Fig. 8; Supplemental Figure S12).
Figure 8.
MtNF-YA1, MtNF-YB16 and MtNF-YC2 bind the promoter of MtERN1. A, Overexpression of GFP-MtNF-YB16 (left), GFP-MtNF-YC2 (middle), or GFP alone in M. truncatula transgenic roots. The confocal images of fluorescence channel alone (top) or fluorescence merge to bright-field channel (bottom) were obtained for each construct (bar = 50 μm). B, Schematic representation of CCAAT boxes found in 1.4-kb sequences of MtERN1 upstream of the transcription start site (TSS). The CCAAT boxes are numbered from 1 to 6, and their respective distances from the transcription start site are indicated. Arrows indicate the DNA region amplified by quantitative PCR in the genomic input DNA or in NF-YA1-coprecipitated samples. The preferentially bound box 3 is highlighted by a gray box. C, ChIP-quantitative PCR analysis of MtNF-YA1, MtNF-YB16, and MtNF-YC2 binding to the MtERN1 promoter. Transgenic roots expressing GFP-MtNF-YA1 (white boxes), GFP-NF-YB16 (dot pattern boxes), and GFP-NF-YC2 (stripe pattern boxes) as well as GFP alone (negative control, black boxes) under the control of the 35S promoter were generated, inoculated by S. meliloti, then harvested 4 d postinoculation and used in ChIP experiments using anti-GFP antibodies. Quantitative PCR was subsequently performed using primers surrounding the five CCAAT boxes present in the promoter of MtERN1 (B). Immunoprecipitation values were normalized against those obtained using input genomic DNA, and data represent the fold induction in relation to the control (GFP) samples.
DISCUSSION
One of the main differences between plant and animal NF-Y complexes is the multiplicity of genes encoding each subunit in plants. In M. truncatula, there are 8 NF-YA, 14 NF-YB, and 7 NF-YC subunit-encoding genes, creating the potential for a very modular system with 784 theoretical NF-Y trimers. However, the real number of NF-Y trimers active in a given tissue at a given time depends on both the potential interaction specificity among NF-Y subunits and their respective expression pattern. The diversity of expression patterns of different NF-Y-encoding genes was previously described in the aerial part of the model plant Arabidopsis using promoter-GUS approaches (Siefers et al., 2009). Furthermore, two studies performed in Arabidopsis differ a bit in their conclusions concerning NF-YB-NF-YC interaction specificity. The study by Hackenberg et al. (2012) points toward a significant interaction specificity, with only 31% of proven interactions between NF-YC and NF-YB subunits in yeast, whereas Calvenzani et al. (2012) conclude that the vast majority of NF-YB and NF-YC heterodimerize. Here, even if the present work does not consist of a full interaction study of all M. truncatula NF-YB and NF-YC, our results also point toward the existence of a low level of interaction specificity. Indeed, in the Y2H experiment presented here, only MtNF-YB3 and MtNF-YB10, belonging to the divergent LEC group, were unable to interact with MtNF-YC1 and C2. This interaction was, however, not tested in planta. In addition, the NF-YA, NF-YB, and NF-YC subunits identified here by Y2H screens and shown to form heterotrimers both in yeast and in planta correspond to the subunits whose expression is highest in the symbiotic tissues used to construct the Y2H libraries (nodules or roots treated by NF). This correlation favors the hypothesis that the formation of NF-Y complexes depends more on the relative abundance of each subunit in a given tissue than on the specificity of the interaction. However, in the absence of expression studies at the protein level, we cannot fully assess the impact of NF-Y transcript abundance in trimer specificity, especially as mechanisms of posttranscriptional, translational, and posttranslational control of expression have been reported for NF-Ys (Combier et al., 2006, 2008; Reynoso et al., 2013).
Taking into account the MtNF-YB and MtNF-YC expression shown here and in Roux et al. (2014) and the pairwise and trimeric interaction data in yeast and in planta, we can thus conclude that we have identified the MtNF-YB and MtNF-YC proteins that interact with MtNF-YA1 and A2, probably to control symbiotic responses. This conclusion is reinforced by a recent transcriptomic analysis showing that MtNF-YB16 and 18 and MtNF-YC2 are the only NF-Y genes in M. truncatula that, together with MtNF-YA1, show a striking differential expression pattern during the first hours after Rhizobium meliloti inoculation on roots (Larrainzar et al., 2015).
To show that the identified trimeric NF-Y complexes indeed play a biological role during symbiotic nodule development, we performed a combination of functional and phylogenetic approaches. We observed that silencing NF-YB subunits, using either an RNAi construct specifically reducing the expression of MtNF-YB16 alone (NF-YB16 RNAi) or a group of closely related genes (MtNF-YB6, B11, B16, and B18; NF-YB RNAi), had no effect on nodule number or the percentage of nodulated plants. Interestingly, the NF-YB RNAi construct does not affect the expression of two more distantly related NF-YB genes, MtNF-YB2 and MtNF-YB13, which are also expressed at significant levels in nodule tissues. We propose that MtNF-YB2 and MtNF-YB13 might be able to compensate for the effect of silencing the major nodule NF-YB-encoding genes, which would explain the lack of impact of this NF-YB silencing on nodule development. We also used a similar strategy to evaluate the symbiotic role of the NF-YC partners of our NF-Y complex. The silencing of MtNF-YC2 alone had no effect on nodulation; however, the NF-YC RNAi construct silencing both MtNF-YC1 and C2 significantly impacted nodulation. Interestingly, several aspects of this nodulation phenotype were reminiscent of the effects caused by MtNF-YA1 and A2 silencing using RNAi (Laloum et al., 2014). NF-YC RNAi not only significantly reduced the nodule number at several stages of the symbiotic process, but also increased the number of plants not forming any nodules upon inoculation (>50% compared with 15% in control plants), suggesting that nodule initiation is impaired in NF-YC RNAi roots. In addition, silencing of both NF-YC genes significantly affected nodule development when they formed, as these organs stayed small and never showed the organization in five distinct zones typically observed in wild-type adult nodules. The inhibition of rhizobial infection progression described when silencing MtNF-YA1 (Laporte et al., 2014) could not be observed here, possibly as a result of insufficient silencing levels using RNAi strategies. More striking phenotypes might be obtained in the future using insertion mutants rather than RNAi approaches whose effect on gene expression can vary in each experiment and is not a complete knockout. However, multiple mutants may have to be combined to avoid functional complementation by related genes.
To further confirm the presence of both MtNF-YB16 and MtNF-YC2 within protein complexes regulating nodule development, we then performed ChIP-PCR experiments on the promoter of MtERN1, a gene already described as a direct target of MtNF-YA1 (Laloum et al., 2014). Strikingly, the same result was observed whether the experiment was performed with GFP fusions to MtNF-YA1, MtNF-YB16, or MtNF-YC2, but not GFP alone (i.e. strong binding to the third CCAAT box of the MtERN1 promoter). Binding of individual NF-Y subunits to CCAAT boxes has never been observed in any system tested, and it is reasonable to assume that it is also not the case in M. truncatula. Because symbiotic tissues, in which MtNF-YA1, B16, and C2 are strongly expressed, were used for the ChIP assays, it is thus likely that, although only one GFP-tagged NF-Y fusion protein was expressed at the time, in our ChIP experiments, this protein is able to interact with the other endogenous NF-Y subunits to bind the third CCAAT box of the MtERN1 promoter. Together, our results thus strongly suggest that MtNF-YA1 and A2, MtNF-YB16, and MtNF-YC1 and C2 participate in protein complexes that regulate nodulation and early symbiotic gene expression in M. truncatula.
Strikingly, MtNF-YB16 and MtNF-YC2 are the closest homologs of LjNF-YB1 and PvNF-YC1, respectively. These two NF-Y subunits have been characterized in the legume plants L. japonicus and common bean, where they play an important role during the root nodule symbiosis (Zanetti et al., 2010; Soyano et al., 2013). L. japonicus and common bean are two plants that belong to the group of tropical legumes that form determinate nodules, whose development and structure differ greatly from that of indeterminate nodules found in temperate legumes, such as M. truncatula (Sprent, 2007). To further test the conserved role of NF-Y proteins in different legumes, we performed interaction assays using common bean NF-Y proteins, chosen on the basis of their phylogenetic position as well as their expression pattern in common bean nodules (Rípodas et al., 2014). Using similar CoIP experiments, we were able to confirm the formation, in planta, of a stable NF-Y trimer between PvNF-YA1, PvNF-YC1, and PvNF-YB7. We have thus most likely identified an evolutionary-conserved group of interacting NF-Y proteins that have specialized in controlling nodulation in leguminous plants during evolution. The fact that homologous NF-Y trimers from different legume plants control both determinate and indeterminate nodule initiation and development illustrates the conserved and fundamental role played by these heterotrimeric complexes.
The mechanisms of NF-Y heterotrimer formation have been well described in mammals (Mantovani, 1999), and more recently, work has also been initiated in plants that reveals the conservation between the two systems (Calvenzani et al., 2012; Hackenberg et al., 2012). Surprisingly, our first Y2H screen revealed an interaction between MtNF-YA1 and the two NF-YC subunits MtNF-YC1 and C2. In the literature, there are very few reports of plant NF-YA subunits interacting pairwise with NF-YB or NF-YC subunits (Thirumurugan et al., 2008; Hackenberg et al., 2012; Soyano et al., 2013). However, all of these pairwise interactions with NF-YA proteins were only found in systems that possess endogenous NF-Y subunits, and never in vitro. Given the apparent weak interaction specificity between plant NF-YB and NF-YC subunits (Calvenzani et al., 2012), the tested NF-YB or NF-YC subunits are probably capable of forming YB-YC dimers with the endogenous proteins from the expression system used (yeast or N. benthamiana leaves), thereby allowing the studied NF-Y protein to interact. In yeast, this hypothesis is supported by the fact that the yeast ScHAP2 and 3 (NF-YA and B) are capable of interacting with the rat CBF-C (NF-YC) to form a stable NF-Y complex that is able to bind CCAAT boxes (Sinha et al., 1995). We thus suggest that the interaction between MtNF-YA1 and MtNF-YC1 and C2 observed in yeast was possible because of the presence of the yeast NF-YB subunit. However, complex formation between NF-Y subunits from different organisms is not always optimal, and the stoichiometry of the trimer might be affected when heterologous proteins are used in the assays. This might explain why we were unable to confirm the interaction between MtNF-YA1 and MtNF-YC1 and C2 in N. benthamiana using the BiFC system.
In mammals, the NF-Y complex is required to activate developmentally regulated genes, and is described as a key regulator of cell cycle progression (Bhattacharya et al., 2003; Benatti et al., 2011; Bungartz et al., 2012). This study illustrates the fact that, during evolution, NF-YA, NF-YB, and NF-YC families of TFs have diversified and some members have specialized to control plant-specific pathways, such as root nodule development, a process that is only found in very few plant families, including the legumes.
MATERIALS AND METHODS
Plasmid Construction
All DNA fragments used were first introduced in the pK207 vector (modified pDONR207 allowing the insertion of DNA fragments by an Asc1 (for Arthrobacter sp.1)/Pme1 (for Pseudomonas mendocina1) enzymatic digestion; L. Deslandes, Laboratoire des Interactions Plantes-Microorganismes) and then recombined into the appropriate destination vectors, according to the Gateway manufacturer's protocol. For the Y2H experimentations, we used modified pBD and pAD vectors for Gateway recombination (L. Deslandes). The yeast three-hybrid assays were performed with the pVT-U102 (Nemo Peeters, Laboratoire des Interactions Plantes-Microorganismes; Pazhouhandeh et al., 2006). RNAi fragments were cloned into the pFRN destination vector (derived from pFGC5941; National Center for Biotechnology Information accession no. AY310901; de Zélicourt et al., 2012). For subcellular and CoIP approaches, open reading frames (ORFs) were recombined into the PAM-PAT p35S:YFP:Gw or p35S:3HA:Gw destination vectors (L. Deslandes). Finally, for BiFC assay, ORFs were cloned in pdonor207 and then recombined into the pGPTVII-Bar-YN-GW and pGPTVII-Bar-YC-GW for the fusion of the N- and C-terminal fragments of split YFP, respectively, following the gateway protocol as in Hirsch et al. (2009). For CoIP assays in common bean (Phaseolus vulgaris), pENTR/D-TOPO vectors containing the ORFs of PvNF-YA1, PvNF-YB7, and PvNF-YC1 were recombined into the destination vectors GATEWAY p35S:HF-GATA for PvNF-YA1 and PAM-PAT p35S:YFP: Gw for PvNF-YB7 and PvNF-YC1.
Plant Material and Transformation
Medicago truncatula ‘Jemalong A17’ plant seeds were scarified and surface sterilized prior to germination, and were used for Agrobacterium rhizogenes-mediated transformation as described in http://www.noble.org/medicagohandbook. pFRN constructs were electroporated in A. rhizogenes ARquA1 strain (Quandt et al., 1993). Composite plants harboring RNAi constructs were selected in Fahraeus medium supplemented with 25 mg mL−1 kanamycin. After 3 weeks, kanamycin-resistant plants were transferred to pouch paper/agar plates (nitrogen and antibiotic free) as described by Cerri et al. (2012) or to attapulgite medium-containing pots as described by Laporte et al. (2014). After 5 to 7 d, root systems of individual composite plants were inoculated with the Sinorhizobium meliloti strain. For Nicotiana benthamiana agroinfiltration, the Agrobacterium tumefaciens strains GV3101 (for BiFC vectors) or GV3103 (for PAM-PAT vector) were electroporated with the corresponding plasmids. Agroinfiltration and BiFC were then performed as in Battaglia et al. (2014).
Y2H and Yeast Three-Hybrid
MtNF-YA1 or MtNF-YC2 was recombined from the pENTRY vector into the pBD vector (Clontech) using the Gateway technology. The baits were then transformed into the yeast (Saccharomyces cerevisiae) strain AH109 using the LiAc-mediated yeast transformation protocol as described in the Yeast Protocols Handbook (Clontech). The resulting yeast strains were then transformed by the respective cDNA library. The Y2H cDNA library from NF-treated root hairs is described in Andriankaja et al. (2007) and was screened using NF-YA1 as a bait. The nodule Y2H cDNA library was made by the Dualsystems company (http://www.dualsystems.com/) from cDNAs coming from 4- and 19-d-old nodules and screened using MtNF-YC2 as bait. Respectively, 4 × 106 and 1.6 × 106 clones were screened on minimal medium lacking His, Trp, and Leu and supplemented with 5 mm 3-amino-triazol. The HIS3+ yeast colonies were recovered after 3 or 4 d of growth at 28°C and retested for growth on selective medium. The cDNA inserts of positive clones were amplified by PCR from yeast cell extracts and sequenced after a PCR product purification step. Plasmid rescue of nonredundant candidates was then performed. Pairwise targeted Y2H approaches were performed by a double transformation of AH109 yeast with pBD and pAD vectors. The drop tests for the interaction assays were performed using 4 µL of yeast resuspended in water and spotted on selective medium. The Leu−, Trp−, His−, Ade−, Ura− AH111 yeast strain was generated using a 5FOA selection. A liquid rich-medium culture of AH109 was plated on 5FOA-containing yeast peptone dextrose adenine plates, and Ura minus colonies were selected. After verification, these strains were then used as recipient strains for the URA pVT-U102 plasmids (Pazhouhandeh et al., 2006). The AH111 was first transformed with pAD-MtNF-YB16 using the LiAc-mediated yeast transformation. The resulting yeast strains were then double transformed by the pBD and pVTU-102 constructions, and the interactions were tested on minimal medium lacking His, Trp, Leu, and Ura and supplemented with 5 mm 3-amino-triazol.
Quantitative Reverse Transcription-PCR Analysis
The gene expression analyses were performed as described in Laloum et al. (2014).
CoIP
One hundred milligrams of agroinfiltrated N. benthamiana leaves was used for each experiment. Tissues were fixed in 0.1% (v/v) formaldehyde for 5 min under vacuum for the M. truncatula assay. Protein complexes were then extracted using either the New England Biolabs buffer (25 mm Tris [pH 7.5], 500 mm NaCl, 1 mm dithiothreitol, 10% [v/v] glycerol, 0.2% [v/v] Triton, and 5 mm CaCl2) for the M. truncatula assay or extraction buffer (10 mm Tris [pH 7.5], 150 mm NaCl, 5 mm dithiothreitol, 10% [v/v] glycerol, 1 mm EDTA, 0.1% [v/v] Triton, 5 mm CaCl2) for the common bean assay. Both buffers were supplemented with 2% (w/v) polyvinylpolypyrrolidone and 1.65% (v/v) of protease inhibitors cocktail (Sigma-Aldrich). After incubation at 4°C and centrifugation, the clarified extract was mixed with 40 µL of µMACS anti-GFP microbeads (Miltenyi Biotec) for the M. truncatula assay or 40 µL of α-FLAG affinity gel agarose beads (Sigma-Aldrich) for the common bean assay and incubated in a rocking shaker for 1.5 to 2 h at 4°C. The immunopurification of YFP-tagged NF-Y proteins was performed as described in the user protocol of µMACS epitope tag protein isolation kit (Miltenyi Biotec). The purified protein complexes in the elution buffer were then de-cross linked in the NEB buffer lacking polyvinylpolypyrrolidone (v/v) for the first 2 h at 55°C and then overnight at room temperature. Concerning the FLAG-tagged NF-Y proteins, after centrifugation at 17,000g, the coimmunoprecipitates were washed six times using 1 mL of extraction buffer lacking polyvinylpolypyrrolidone and then incubated with elution buffer (3× FLAG peptide with a final concentration of 200 ng μL−1 in extraction buffer; Sigma-Aldrich) for 10 min at room temperature. The mixtures were centrifuged, and the supernatant was recovered as the eluates of coimmunoprecipitates. Finally, proteins were then separated by SDS-PAGE and detected by immunoblot using an α-GFP monoclonal antibody (Roche) followed by the mouse horseradish peroxidase (HRP) secondary antibody and a high-affinity α-HA HRP-conjugated monoclonal antibody (3F10; Roche) for the M. truncatula assay. In common bean assays, an α-FLAG HRP-conjugated monoclonal antibody (Sigma-Aldrich) or an α-GFP HRP-conjugated monoclonal antibody (Invitrogen). An aliquot of the crude extract was loaded as the input control in the inmunoblot.
Chromatin Immunoprecipitation Analysis
ChIP assays were performed 4 d after rhizobia inoculation M. truncatula-transformed roots bearing the following constructs: 35S:GFP (used as a negative control), 35S:GFP-NF-YA1 or 35S:GFP-NF-YB16 or 35S:GFP-NF-YC2. Nuclei preparation and immunoprecipitation were performed as in Laloum et al. (2014). Two biological replicates were used for each construct, and the results are shown in Figure 8 and Supplemental Figure S11.
Microscopy and Imaging
For BiFC assay and subcellular localization, N. benthamiana leaf disks were infiltrated with water and mounted on microscope slides. Samples were imaged using a Leica TCS SP2 AOBS confocal laser-scanning microscope equipped with a long-distance 40× water-immersion objective (HCX Apo L 0.80). The 458- and 514-nm argon laser lines were used to excite CFP and YFP, respectively. Specific emission windows of 460 to 500 and 530 to 560 nm were used for CFP and YFP, respectively. Images were processed using the Leica confocal and ImageJ (National Institutes of Health) software. Figures 3B, 5, and 8A show maximal projections of selected planes of a Z-stack or three-dimensional reconstructions of confocal image stacks. Nodule tissues were sliced into 50- to 100-mm-thick sections using a vibrating-blade microtome (Leica VT1000 S). Nodule sections (50–100 mm thick) were fixed in 2% glutaraldehyde/0.1 m potassium phosphate buffer and stained for β-galactosidase activities for 1 h. Microscopic observations were performed using a light microscope (Axioplan 2 Imaging; Carl Zeiss) and a CCD camera (AxioCam MRc; Carl Zeiss).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Phylogenetic trees of NF-YA, NF-YB, and NF-YC proteins from M. truncatula, P. vulgaris, and Lotus japonicus.
Supplemental Figure S2. Syntenic analysis between colinear blocks of NF-YA and NF-YC subunits loci of M. truncatula and common bean.
Supplemental Figure S3. Protein binding mutants of MtNF-YA1, MtNF-YA2, and MtNF-YC2.
Supplemental Figure S4. Localization of the YFP-tagged proteins used for coimmunoprecipitation assays.
Supplemental Figure S5. MtNF-YC2 does not interact with MtNF-YA1 in planta bimolecular fluorescence complementation assay in N. benthamiana.
Supplemental Figure S6. MtNF-YB16 interacts with MtNF-YC1 and C2 in planta.
Supplemental Figure S7. Interaction specificity of NF-YB subunits using yeast two-hybrid assays.
Supplemental Figure S8. The YFP-NF-YB16, YFP-NF-YC1, and YFP-NF-YC2 chimeric proteins are not cleaved in N. benthamiana.
Supplemental Figure S9. Nodulation is not affected in NF-YB16 RNAi and NF-YB RNAi roots.
Supplemental Figure S10. Specificity of NF-YC2 RNAi and NF-YC RNAi constructs.
Supplemental Figure S11. Rhizobial infection and NF signaling are not affected by the NF-YC RNAi construct.
Supplemental Figure S12. MtNF-YB16 and MtNF-YC2 bind the promoter of MtERN1 (biological replicate of Figure 8C).
Supplementary Material
Acknowledgments
We thank Nemo Peeters for the AH111 yeast strain and the pVT-U102 vector, Laurent Deslandes for most of the Gateway vectors and technical advice, and Giles Oldroyd for the BiFC vectors.
Glossary
- TF
transcription factor
- Y2H
yeast two-hybrid
- BiFC
bimolecular-fluorescence complementation
- cDNA
complementary DNA
- CoIP
coimmunoprecipitation
- ChIP
chromatin immunoprecipitation
- ORF
open reading frame
- HRP
horseradish peroxidase
Footnotes
This work was supported by the HAPIHUB (grant no. ANR–09–BLAN–0033–01), the French Laboratory of Excellence project “TULIP” (grant nos. ANR–10–LABX–41 and ANR–11–IDEX–0002–02), the Centre National de la Recherche Scientifique-Consejo Nacional de Investigaciones Científicas y Técnicas exchange program Projet International de Cooperation Scientifique (grant no. PICS06688), the French Ministry of Education and Research (grant to T.L.), and the Institut National de la Recherche Agronomique Contrat Jeune Scientifique (to T.L.). C.R, F.A.B., and M.E.Z. are members of the Argentinean National Council of Science and Technology.
Articles can be viewed without a subscription.
References
- Andriankaja A, Boisson-Dernier A, Frances L, Sauviac L, Jauneau A, Barker DG, de Carvalho-Niebel F (2007) AP2-ERF transcription factors mediate Nod factor-dependent Mt ENOD11 activation in root hairs via a novel cis-regulatory motif. Plant Cell 19: 2866–2885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battaglia M, Rípodas C, Clúa J, Baudin M, Aguilar OM, Niebel A, Zanetti ME, Blanco FA (2014) A nuclear factor Y interacting protein of the GRAS family is required for nodule organogenesis, infection thread progression, and lateral root growth. Plant Physiol 164: 1430–1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benatti P, Dolfini D, Viganò A, Ravo M, Weisz A, Imbriano C (2011) Specific inhibition of NF-Y subunits triggers different cell proliferation defects. Nucleic Acids Res 39: 5356–5368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya A, Deng JM, Zhang Z, Behringer R, de Crombrugghe B, Maity SN (2003) The B subunit of the CCAAT box binding transcription factor complex (CBF/NF-Y) is essential for early mouse development and cell proliferation. Cancer Res 63: 8167–8172 [PubMed] [Google Scholar]
- Breakspear A, Liu C, Roy S, Stacey N, Rogers C, Trick M, Morieri G, Mysore KS, Wen J, Oldroyd GED, et al. (2014) The root hair “infectome” of Medicago truncatula uncovers changes in cell cycle genes and reveals a requirement for auxin signaling in rhizobial infection. Plant Cell 26: 4680–4701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bungartz G, Land H, Scadden DT, Emerson SG (2012) NF-Y is necessary for hematopoietic stem cell proliferation and survival. Blood 119: 1380–1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvenzani V, Testoni B, Gusmaroli G, Lorenzo M, Gnesutta N, Petroni K, Mantovani R, Tonelli C (2012) Interactions and CCAAT-binding of Arabidopsis thaliana NF-Y subunits. PLoS One 7: e42902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerri MR, Frances L, Laloum T, Auriac MC, Niebel A, Oldroyd GED, Barker DG, Fournier J, de Carvalho-Niebel F (2012) Medicago truncatula ERN transcription factors: regulatory interplay with NSP1/NSP2 GRAS factors and expression dynamics throughout rhizobial infection. Plant Physiol 160: 2155–2172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combier JP, de Billy F, Gamas P, Niebel A, Rivas S (2008) Trans-regulation of the expression of the transcription factor MtHAP2-1 by a uORF controls root nodule development. Genes Dev 22: 1549–1559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combier JP, Frugier F, de Billy F, Boualem A, El-Yahyaoui F, Moreau S, Vernié T, Ott T, Gamas P, Crespi M, et al. (2006) MtHAP2-1 is a key transcriptional regulator of symbiotic nodule development regulated by microRNA169 in Medicago truncatula. Genes Dev 20: 3084–3088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalla Via V, Narduzzi C, Aguilar OM, Zanetti ME, Blanco FA (2015) Changes in the common bean transcriptome in response to secreted and surface signal molecules of Rhizobium etli. Plant Physiol 169: 1356–1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desbrosses GJ, Stougaard J (2011) Root nodulation: a paradigm for how plant-microbe symbiosis influences host developmental pathways. Cell Host Microbe 10: 348–358 [DOI] [PubMed] [Google Scholar]
- de Zélicourt A, Diet A, Marion J, Laffont C, Ariel F, Moison M, Zahaf O, Crespi M, Gruber V, Frugier F (2012) Dual involvement of a Medicago truncatula NAC transcription factor in root abiotic stress response and symbiotic nodule senescence. Plant J 70: 220–230 [DOI] [PubMed] [Google Scholar]
- Dolfini D, Gatta R, Mantovani R (2012) NF-Y and the transcriptional activation of CCAAT promoters. Crit Rev Biochem Mol Biol 47: 29–49 [DOI] [PubMed] [Google Scholar]
- Fournier J, Timmers ACJ, Sieberer BJ, Jauneau A, Chabaud M, Barker DG (2008) Mechanism of infection thread elongation in root hairs of Medicago truncatula and dynamic interplay with associated rhizobial colonization. Plant Physiol 148: 1985–1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackenberg D, Wu Y, Voigt A, Adams R, Schramm P, Grimm B (2012) Studies on differential nuclear translocation mechanism and assembly of the three subunits of the Arabidopsis thaliana transcription factor NF-Y. Mol Plant 5: 876–888 [DOI] [PubMed] [Google Scholar]
- Hirsch S, Kim J, Muñoz A, Heckmann AB, Downie JA, Oldroyd GED (2009) GRAS proteins form a DNA binding complex to induce gene expression during nodulation signaling in Medicago truncatula. Plant Cell 21: 545–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu CD, Chinenov Y, Kerppola TK (2002) Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol Cell 9: 789–798 [DOI] [PubMed] [Google Scholar]
- Kaló P, Gleason C, Edwards A, Marsh J, Mitra RM, Hirsch S, Jakab J, Sims S, Long SR, Rogers J, et al. (2005) Nodulation signaling in legumes requires NSP2, a member of the GRAS family of transcriptional regulators. Science 308: 1786–1789 [DOI] [PubMed] [Google Scholar]
- Kim IS, Sinha S, de Crombrugghe B, Maity SN (1996) Determination of functional domains in the C subunit of the CCAAT-binding factor (CBF) necessary for formation of a CBF-DNA complex: CBF-B interacts simultaneously with both the CBF-A and CBF-C subunits to form a heterotrimeric CBF molecule. Mol Cell Biol 16: 4003–4013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laloum T, Baudin M, Frances L, Lepage A, Billault-Penneteau B, Cerri MR, Ariel F, Jardinaud MF, Gamas P, de Carvalho-Niebel F, et al. (2014) Two CCAAT-box-binding transcription factors redundantly regulate early steps of the legume-rhizobia endosymbiosis. Plant J 79: 757–768 [DOI] [PubMed] [Google Scholar]
- Laloum T, De Mita S, Gamas P, Baudin M, Niebel A (2013) CCAAT-box binding transcription factors in plants: Y so many? Trends Plant Sci 18: 157–166 [DOI] [PubMed] [Google Scholar]
- Laporte P, Lepage A, Fournier J, Catrice O, Moreau S, Jardinaud MF, Mun JH, Larrainzar E, Cook DR, Gamas P, et al. (2014) The CCAAT box-binding transcription factor NF-YA1 controls rhizobial infection. J Exp Bot 65: 481–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larrainzar E, Riely BK, Kim SC, Carrasquilla-Garcia N, Yu HJ, Hwang HJ, Oh M, Kim GB, Surendrarao AK, Chasman D, et al. (2015) Deep sequencing of the Medicago truncatula root transcriptome reveals a massive and early interaction between nodulation factor and ethylene signals. Plant Physiol 169: 233–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TH, Tang H, Wang X, Paterson AH (2013) PGDD: a database of gene and genome duplication in plants. Nucleic Acids Res 41: D1152–D1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerouge P, Roche P, Faucher C, Maillet F, Truchet G, Promé JC, Dénarié J (1990) Symbiotic host-specificity of Rhizobium meliloti is determined by a sulphated and acylated glucosamine oligosaccharide signal. Nature 344: 781–784 [DOI] [PubMed] [Google Scholar]
- Liu J-X, Howell SH (2010) bZIP28 and NF-Y transcription factors are activated by ER stress and assemble into a transcriptional complex to regulate stress response genes in Arabidopsis. Plant Cell 22: 782–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantovani R. (1999) The molecular biology of the CCAAT-binding factor NF-Y. Gene 239: 15–27 [DOI] [PubMed] [Google Scholar]
- Mantovani R, Li XY, Pessara U, Hooft van Huisjduijnen R, Benoist C, Mathis D (1994) Dominant negative analogs of NF-YA. J Biol Chem 269: 20340–20346 [PubMed] [Google Scholar]
- Middleton PH, Jakab J, Penmetsa RV, Starker CG, Doll J, Kaló P, Prabhu R, Marsh JF, Mitra RM, Kereszt A, et al. (2007) An ERF transcription factor in Medicago truncatula that is essential for Nod factor signal transduction. Plant Cell 19: 1221–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nardini M, Gnesutta N, Donati G, Gatta R, Forni C, Fossati A, Vonrhein C, Moras D, Romier C, Bolognesi M, et al. (2013) Sequence-specific transcription factor NF-Y displays histone-like DNA binding and H2B-like ubiquitination. Cell 152: 132–143 [DOI] [PubMed] [Google Scholar]
- Oldfield AJ, Yang P, Conway AE, Cinghu S, Freudenberg JM, Yellaboina S, Jothi R (2014) Histone-fold domain protein NF-Y promotes chromatin accessibility for cell type-specific master transcription factors. Mol Cell 55: 708–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldroyd GE, Murray JD, Poole PS, Downie JA (2011) The rules of engagement in the legume-rhizobial symbiosis. Annu Rev Genet 45: 119–144 [DOI] [PubMed] [Google Scholar]
- Oldroyd GED. (2013) Speak, friend, and enter: signalling systems that promote beneficial symbiotic associations in plants. Nat Rev Microbiol 11: 252–263 [DOI] [PubMed] [Google Scholar]
- Oldroyd GED, Downie JA (2008) Coordinating nodule morphogenesis with rhizobial infection in legumes. Annu Rev Plant Biol 59: 519–546 [DOI] [PubMed] [Google Scholar]
- Pazhouhandeh M, Dieterle M, Marrocco K, Lechner E, Berry B, Brault V, Hemmer O, Kretsch T, Richards KE, Genschik P, et al. (2006) F-box-like domain in the polerovirus protein P0 is required for silencing suppressor function. Proc Natl Acad Sci USA 103: 1994–1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petroni K, Kumimoto RW, Gnesutta N, Calvenzani V, Fornari M, Tonelli C, Holt BF III, Mantovani R (2012) The promiscuous life of plant NUCLEAR FACTOR Y transcription factors. Plant Cell 24: 4777–4792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quandt HJ, Puhler A, Broer I (1993) Transgenic root nodules of Vicia hirsuta: a fast and efficient system for the study of gene expression in indeterminate-type nodules. Mol Plant Microbe Interact 6: 699–706 [Google Scholar]
- Reynoso MA, Blanco FA, Bailey-Serres J, Crespi M, Zanetti ME (2013) Selective recruitment of mRNAs and miRNAs to polyribosomes in response to rhizobia infection in Medicago truncatula. Plant J 73: 289–301 [DOI] [PubMed] [Google Scholar]
- Ripodas C, Castaingts M, Clua J, Blanco F, Zanetti ME (2015) Annotation, phylogeny and expression analysis of the nuclear factor Y gene families in common bean (Phaseolus vulgaris). Front Plant Sci 5: 761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rípodas C, Clúa J, Battaglia M, Baudin M, Niebel A, Zanetti ME, Blanco F (2014) Transcriptional regulators of legume-rhizobia symbiosis: nuclear factors Ys and GRAS are two for tango. Plant Signal Behav 9: e28847–e28847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux B, Rodde N, Jardinaud MF, Timmers T, Sauviac L, Cottret L, Carrère S, Sallet E, Courcelle E, Moreau S, et al. (2014) An integrated analysis of plant and bacterial gene expression in symbiotic root nodules using laser-capture microdissection coupled to RNA sequencing. Plant J 77: 817–837 [DOI] [PubMed] [Google Scholar]
- Schauser L, Roussis A, Stiller J, Stougaard J (1999) A plant regulator controlling development of symbiotic root nodules. Nature 402: 191–195 [DOI] [PubMed] [Google Scholar]
- Siefers N, Dang KK, Kumimoto RW, Bynum WE IV, Tayrose G, Holt BF III (2009) Tissue-specific expression patterns of Arabidopsis NF-Y transcription factors suggest potential for extensive combinatorial complexity. Plant Physiol 149: 625–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S, Katzer K, Lambert J, Cerri M, Parniske M (2014) CYCLOPS, a DNA-binding transcriptional activator, orchestrates symbiotic root nodule development. Cell Host Microbe 15: 139–152 [DOI] [PubMed] [Google Scholar]
- Sinha S, Maity SN, Lu J, de Crombrugghe B (1995) Recombinant rat CBF-C, the third subunit of CBF/NF-Y, allows formation of a protein-DNA complex with CBF-A and CBF-B and with yeast HAP2 and HAP3. Proc Natl Acad Sci USA 92: 1624–1628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit P, Raedts J, Portyanko V, Debellé F, Gough C, Bisseling T, Geurts R (2005) NSP1 of the GRAS protein family is essential for rhizobial Nod factor-induced transcription. Science 308: 1789–1791 [DOI] [PubMed] [Google Scholar]
- Soyano T, Kouchi H, Hirota A, Hayashi M (2013) Nodule inception directly targets NF-Y subunit genes to regulate essential processes of root nodule development in Lotus japonicus. PLoS Genet 9: e1003352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprent JI. (2007) Evolving ideas of legume evolution and diversity: a taxonomic perspective on the occurrence of nodulation. New Phytol 174: 11–25 [DOI] [PubMed] [Google Scholar]
- Thirumurugan T, Ito Y, Kubo T, Serizawa A, Kurata N (2008) Identification, characterization and interaction of HAP family genes in rice. Mol Genet Genomics 279: 279–289 [DOI] [PubMed] [Google Scholar]
- Vasse J, de Billy F, Camut S, Truchet G (1990) Correlation between ultrastructural differentiation of bacteroids and nitrogen fixation in alfalfa nodules. J Bacteriol 172: 4295–4306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao TT, Schilderink S, Moling S, Deinum EE, Kondorosi E, Franssen H, Kulikova O, Niebel A, Bisseling T (2014) Fate map of Medicago truncatula root nodules. Development 141: 3517–3528 [DOI] [PubMed] [Google Scholar]
- Zanetti ME, Blanco FA, Beker MP, Battaglia M, Aguilar OM (2010) A C subunit of the plant nuclear factor NF-Y required for rhizobial infection and nodule development affects partner selection in the common bean-Rhizobium etli symbiosis. Plant Cell 22: 4142–4157 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.