Abstract
The technique of site-directed spin labeling (SDSL) provides unique information on biomolecules by monitoring the behavior of a stable radical tag (i.e., spin label) using electron paramagnetic resonance (EPR) spectroscopy. In this chapter, we describe an approach in which SDSL is integrated with computational modeling to map conformations of nucleic acids. This approach builds upon a SDSL tool kit previously developed and validated, which includes three components: (i) a nucleotide-independent nitroxide probe, designated as R5, which can be efficiently attached at defined sites within arbitrary nucleic acid sequences; (ii) inter-R5 distances in the nanometer range, measured via pulsed EPR; and (iii) an efficient program, called NASNOX, that computes inter-R5 distances on given nucleic acid structures. Following a general framework of data mining, our approach uses multiple sets of measured inter-R5 distances to retrieve “correct” all-atom models from a large ensemble of models. The pool of models can be generated independently without relying on the inter-R5 distances, thus allowing a large degree of flexibility in integrating the SDSL-measured distances with a modeling approach best suited for the specific system under investigation. As such, the integrative experimental/computational approach described here represents a hybrid method for determining all-atom models based on experimentally-derived distance measurements.
1. INTRODUCTION
Rapid advances in a number of areas of biology, such as the discovery of ribozymes, riboswitches, noncoding RNAs, and noncanonical DNA structures, have clearly established that nucleic acids, including both DNA and RNA, are not just passive information carriers; instead, they play active and crucial roles as regulators and executors of biological functions. Similar to that of proteins, the ability of nucleic acids to fold into complex and compact three-dimensional structures is crucial for their functions. As such, information on tertiary structures of nucleic acids, as well as their complexes with proteins and small-molecule ligands, is essential for understanding many biological processes, and methodologies for obtaining such information are of great importance.
In this chapter, we describe an integrated approach in which long-range distances measured via site-directed spin labeling (SDSL) are combined with computational modeling to obtain structural models of nucleic acids. In SDSL, chemically stable radicals (i.e., spin labels) are covalently attached at specific sites of a macromolecule. The behavior of the spin labels is monitored using electron paramagnetic resonance (EPR) spectroscopy, from which local information on the macromolecule is obtained (Fig. 1; Hubbell & Altenbach, 1994). SDSL has been demonstrated as a powerful tool for investigating structure and dynamics of biomolecules, as evidenced by accompanying articles in this volume, as well as a number of excellent reviews (Ding et al., 2015; Fedorova & Tsvetkov, 2013; Hubbell & Altenbach, 1994; Hubbell, Cafiso, & Altenbach, 2000; Hubbell, López, Altenbach, & Yang, 2013; Krstic, Endeward, Margraf, Marko, & Prisner, 2012; Shelke & Sigurdsson, 2012; Sowa & Qin, 2008).
Figure 1.
The general strategy of site-directed spin labeling (SDSL). Step 1: Attach a spin label (i.e., a nitroxide) to specific site(s) of a macromolecule. Step 2: Monitor behavior of the spin label using EPR spectroscopy. Step 3: Derive information about the target molecule. The table shows the two primary types of information that can be derived from EPR measurements on nucleic acids. Adapted from Sowa and Qin (2008) with permission.
One of the main EPR observables used in SDSL studies is the distance between a pair of spin labels, which can be obtained by measuring magnetic dipolar coupling using either continuous-wave (cw) or, more recently, pulsed EPR techniques (Cafiso, 2012; Jeschke, 2012; Krstic et al., 2012; Polyhach, Bordignon, & Jeschke, 2011; Schiemann & Prisner, 2007). The measurable range of distances spans from 5 to up to 100 A °, providing direct structural constraints for monitoring conformational change and for mapping biomolecular structure. Compared to X-ray crystallography, SDSL distance measurements do not require crystalline samples, and therefore are suitable for studying systems that do not yield crystals or are susceptible to artifacts due to crystal packing. Compared to solution NMR measurements, SDSL is not limited by the molecular weight of the system, requires a smaller amount of sample, and provides longer-range distances. Last, but not least, in contrast to techniques, such as fluorescence resonance energy transfer, which generally use two chemically distinct fluorophores, with SDSL, distances can be measured using a pair of chemically identical spin labels (e.g., nitroxides), which simplifies the labeling procedure. In addition, in the majority of spin labels, the unpaired electron(s) is localized at a known position(s), thereby facilitating modeling that explicitly accounts for the label.
One should also take into account “limitations” when designing studies for SDSL distance measurements. Most spin labels are extrinsic probes tagged onto the target molecules, so it is important to assess perturbations to the native system due to the labels. The measured interspin distances (rspin) are generally different from the actual distances at the parent molecule (rtarget) and a number of methods are reported to correlate the two (e.g., Hagelueken, Ward, Naismith, & Schiemann, 2012; Hatmal et al., 2012; Polyhach et al., 2011; Qin et al., 2007; see also Section 4). Currently, most long-range SDSL distance measurements require the use of cryogenic temperatures, due to limitations imposed by spin relaxation, and one should assess potential impact on the particular system being studied (e.g., see Georgieva et al., 2012). It is also important to note that, in SDSL studies, the number of distances measured is relatively small. However, a number of studies have shown that with careful design of experiments these “sparse” long-range constraints can be highly informative (Duss, Michel, et al., 2014; Duss, Yulikov, Allain, & Jeschke, 2015; Duss, Yulikov, Jeschke, & Allain, 2014; Hirst, Alexander, McHaourab, & Meiler, 2011; Zhang et al., 2014, 2012).
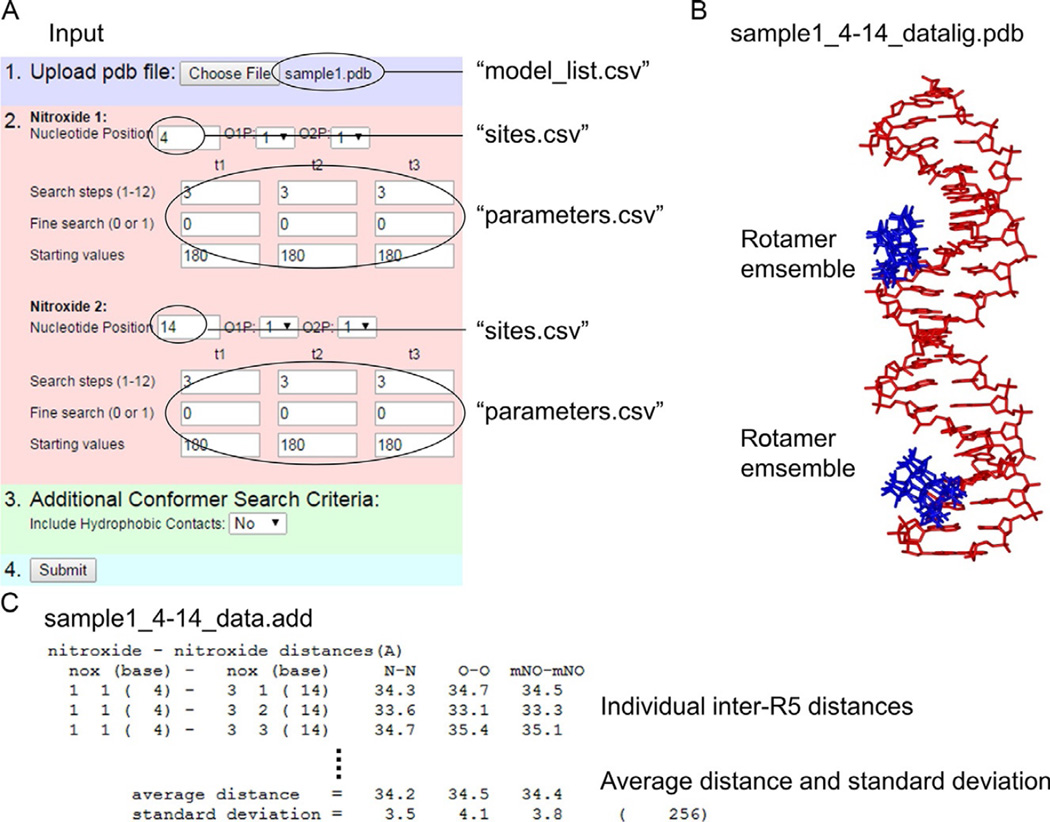
In this chapter, we describe an integrated SDSL/computational-modeling approach that we have developed for mapping an RNA junction (Zhang et al., 2012) and examining sequence-dependent conformations of DNA duplexes (Chen et al., 2013; Zhang et al., 2014). This approach builds upon an SDSL tool kit previously developed and validated (Qin et al., 2007), which includes three components: (i) a nucleotide-independent nitroxide probe, designated as R5, that can be efficiently attached at defined sites within arbitrary nucleic acid sequences (Fig. 2); (ii) inter-R5 distances measured via EPR (Fig. 3); and (iii) a web-accessible program, called NASNOX, that efficiently computes inter-R5 distances on given nucleic acid structures (Fig. 4). The underlying philosophy of our approach is to use multiple measured inter-R5 distances to retrieve “correct” all-atom models from a large ensemble of models. This pool of models can be generated independently without relying on the measured inter-R5 distances, thus allowing a large degree of flexibility in integrating the SDSL-measured distances with a modeling approach that is best suited for the specific system under investigation. The use of experimentally-measured distance constraints in selecting an all-atom model from a large ensemble of pregenerated putative models represents a “hybrid” approach that combines experimental with computational methods in structure determination.
Figure 2.
Nucleic acid labeling using the nucleotide-independent R5 nitroxide probe. (A) Synthesis of compound 2, the reactive R5 precursor. (B) An example of reverse-phase HPLC characterization of compounds 1 (shown in black) and 2 (shown in red, light gray in the print version). Data shown were collected using a C18 column (Prosphere™ C18, Grace Davidson, Inc.) and a linear gradient generated with buffer A: 0.1 M triethylammonium acetate (TEAA, pH 7.0) and 5% (v/v) acetonitrile; and buffer B: 100% acetonitrile. (C) Site-specific attachment of compound 2 to a phosphorothioate-modified nucleic acid strand. As shown, R5 is attached to the Sp diastereomer of the nth nucleotide, and the three torsion angles about the single bonds connecting the pyrroline ring to the nucleic acid are indicated. Adapted from Qin et al. (2007) with permission.
Figure 3.
Measuring nanometer distances using DEER spectroscopy. (A) Pulse sequences for the four-pulse DEER. Three pulses are applied to the observe frequency and generate a refocused echo (shown as “Observe”). Another pulse is applied to a different frequency (pump frequency) to flip the pump spin. Shown on the right is a field-sweep spectrum acquired at the observe frequency with pump spin and observe spin indicated. (B) An example of DEER data acquired on a DNA duplex. Left: Measured normalized echo decay (black) overlaid with the simulated background decay (red, light gray in the print version); center: Measured background-corrected echo decay (black) overlaid with the simulated trace computed from the optimized distance distribution (red, light gray in the print version); right: The corresponding optimized distance distribution computed using Deer Analysis 2013. Adapted from Qin et al. (2007), Sowa and Qin (2008), and Zhang et al. (2014) with permission.
Figure 4.
The NASNOX program for computing expected inter-R5 distances on given nucleic acid structures. (A) Examples of input parameters. The previously reported web interface of NASNOX_W (Qin et al., 2007) is shown to illustrate the organization of input information in the parameter files used to execute NASNOX in the batch mode. (B) An example of structure output in which a pair of R5’s were modeled onto nucleotides 4 and 14 of the sample1.pdb input structure. The DNA is shown in red (light gray in the print version), and the allowed R5 rotamers at each site are shown in blue (black in the print version). (C) An example of text output showing the individual inter-R5 distances and the average distance and standard deviation for the entire ensemble. Adapted from Qin et al. (2007) with permission.
2. SDSL OF NUCLEIC ACIDS USING A NUCLEOTIDEINDEPENDENT NITROXIDE PROBE
A variety of methods for attaching a spin label at a specific site of a target DNA or RNA have been reported and are summarized in a number of recent reviews (Fedorova & Tsvetkov, 2013; Shelke & Sigurdsson, 2012; Sowa & Qin, 2008), as well as in a number of chapters within this volume. Here, we focus on a family of nitroxides, called the R5-series, which can be attached to a phosphorothioate group introduced at a defined location of the nucleic acid backbone during solid-phase chemical synthesis (Qin, Butcher, Feigon, & Hubbell, 2001; Qin et al., 2007). Among this family of probes, R5 (Fig. 2) has been primarily used for mapping nucleic acid structures and will be the focus of the method described herein. The other probes, R5a (Popova, Kálai, Hideg, & Qin, 2009) and R5c (Nguyen, Popova, Hideg, & Qin, 2015), are attached based on the same chemistry, but include substituent(s) that modulate motions of the nitroxide moiety with respect to the parent molecule. When used as single labels, R5a and R5c have been shown to be advantageous in probing local environments (Ding, Zhang, Tham, & Qin, 2014; Grant, Boyd, Herschlag, & Qin, 2009; Nguyen et al., 2015; Popova et al., 2012, 2009; Popova & Qin, 2010). It will be of interest to further explore whether they also offer enhancement in pairwise distance measurements, e.g., narrowing the distribution of the measured distances.
The R5 label has a number of advantages for mapping nucleic acid structures. Most importantly, the labeling site is not restricted by base identity, and the labeling method is highly effective and cost efficient (Qin et al., 2001, 2007). This enables scanning of the target molecule to obtain multiple distance measurements (Chen et al., 2013; Zhang et al., 2014, 2012), which is highly desirable in structural mapping. In addition, R5 has been shown to minimally perturb native nucleic acid duplexes (Cai et al., 2006, 2007), and an efficient program, called NASNOX (Price et al., 2007; Qin et al., 2007), has been developed to accurately correlate the measured inter-R5 distances to a given target structure.
In this section, we describe methods for attaching R5 to a specific site of a nucleic acid strand. The overall protocol described here largely follows what has been reported in Qin et al. (2007), with improvements noted where appropriate.
2.1 Oligonucleotides with Site-Specific Phosphorothioate Modifications
To achieve site-specific covalent attachment of a spin label(s), the phosphorodiester group(s) at the intended labeling site(s) is modified to a chemically more reactive phosphorothioate (Fig. 2). In this chapter (and throughout our work), we follow the convention that each phosphate belongs to the ribose on the 3′ -side of the P atom, such that, for an R5 labeled at the nth nucleotide, the phosphorothioate substitution is located between the (n−1)th and nth nucleotide (Fig. 2C). Furthermore, for RNA labeling, the 2′ -OH group at the (n−1)th position (i.e., 5′ to the phosphorothioate modification, Fig. 2C) must be substituted with a nonnucleophilic functional group in order to prevent strand scission upon labeling (Qin et al., 2001). The simplest substitution is a 2′-H (Fig. 2C), although others such as 2′-O-methyl and 2′-F, also suffice. It is worth noting that one, two, or even more phosphorothioates can be installed simultaneously within an oligonucleotide strand (Zhang et al., 2012, 2014), depending on the desired number of labels.
Oligonucleotides with site-specific phosphorothioate modifications are most easily obtained via chemical synthesis using the well-established phosphoramide scheme and are available from many commercial sources. In our work, we have been using custom-synthesized oligonucleotides from vendors such as Integrated DNA Technology (http://www.idtdna.com/). In general, a synthesis scale of 100 nmol usually yields greater than 50 nmol of products for oligonucleotides less than 30 nucleotides (nt) in length, which is sufficient for EPR studies. It is important to recognize that chemical synthesis results in two possible diastereomers (Rp and Sp) at each phosphorothioate-modified nucleotide. Once properly accounted for (see NASNOX modeling, Section 4.2.1), the presence of the diastereomers does not impact the use of the R5-series of probes (Qin et al., 2007).
When ordering a custom-synthesized oligonucleotide, one must adhere to the vendor’s instructions on how to specify the desired location(s) of the internal phosphorothioate modification and check with the vendor regarding any additional in-house workup steps required. Typically, oligonucleotides that have undergone the basic postsynthesis deprotection and desalting procedures (e.g., “standard desalting” at Integrated DNA Technology) can be used for R5 labeling without further purification. However, we recommend first checking the quality of the synthesis products using high-performance liquid chromatography (HPLC) or gel electrophoresis. The phosphorothioate-modified oligonucleotides can be stored in powder form at −20 °C for 3 months or longer.
2.2 Preparation of the R5 Precursor
The R5 precursor, 3-iodomethyl-1-oxyl-2,2,5,5-tetramethylpyrroline (Hankovszky, Hideg, & Lex, 1980; compound 2, Fig. 2A), is available commercially (e.g., Toronto Research Chemicals, cat. no. I709500). Alternatively, because compound 2 is light sensitive and may degrade upon long-term storage, one may desire to prepare compound 2 freshly before each labeling reaction. This can be done using a more stable compound, 1-oxyl-2,2,5,5-tetramethyl-3(methanesulfonyloxymethyl)pyrroline (compound 1, Fig. 2A; Toronto Research Chemicals, cat. no. O872400), as a starting material, following the protocol described below.
Step 2.2.1
Prepare compound 2 from compound 1
Thoroughly mix an equal amount of 1 and NaI in acetone. Incubate this reaction mixture at 37 °C for 60 min.
Notes
The amount of 1 used can vary in a wide range, and is usually set based on the scale of the subsequent nucleic acid labeling reaction. For example, in our work a typical reaction starts with 2.48 mg of 1 (or 20 mM in a reaction volume of 500 µL), which generally yields sufficient product (i.e., compound 2) for a 100 µL labeling reaction as described in Section 2.3.
This reaction should contain equimolar amounts of NaI and 1. An excess of NaI (e.g., >10%) will compete with compound 2 in the subsequent reaction, while an excess of 1 is simply a waste of the reagent.
As the reaction progresses, a significant amount of white precipitate (by-product NaOSO2CH3) should become visible.
Step 2.2.2
Recover compound 2
To separate 2 and by-products, spin down the reaction mixture in a bench-top centrifuge (typically at 14,000 rpm for 10 min at room temperature (20–25 °C)). Recover the supernatant, which contains the solubilized 2. Wash the precipitate with acetone, and repeat the spin-down procedure again, as described above. Combine all the supernatant fractions, then remove the solvent (e.g., using a benchtop speed vacuum) to obtain 2 as a red-brown oil.
Notes
2 is light sensitive, and from this point forward the tubes should be wrapped with aluminum foil to prevent excessive exposure to light.
One may characterize conversion from 1 to 2 by methods such as reverse-phase HPLC (see example in Fig. 2B) or mass spectrometry. The conversion is usually quantitative.
2 can be stored at −20 or −80 °C for up to 6 months. However, this may result in reduced labeling efficiency.
2.3 Oligonucleotide Labeling
Step 2.3.1
Prepare a stock solution of compound 2 Immediately before the labeling reaction, dissolve 2 (from Step 2.2.2 above or directly obtained from a vendor) in acetonitrile. Typically, a 1 M stock is prepared, with the total amount of compound 2 determined by the labeling scale according to the following Step 2.3.2.
Notes
A solvent such as acetonitrile is used here and in the subsequent step to ensure solubility of 2 at the concentration required for the labeling reaction. We have found that formamide may also be used and may provide an additional benefit to denature the nucleic acids and enhance labeling.
Step 2.3.2
Assemble the labeling reaction
A typical reaction mixture contains 100 mM compound 2, 100–150 µM oligonucleotide, 0.1 M 2-(N-morpholino)ethanesulfonic acid pH 5.8, and 20% (v/v) acetonitrile. Incubate the reaction mixture in the dark (wrap with aluminum foil) at room temperature for 12–24 h. Apply constant and gentle mixing during incubation.
Notes
Oligonucleotides are generally dissolved in sterile ultrapure water or aqueous buffer (e.g., ME buffer: 10 mM MOPS, 1 mM EDTA, pH 6.5). As discussed above, crude oligonucleotides (deprotected and desalted) can be labeled directly.
Studies have shown that 2 remains intact throughout the reaction period under conditions described here; however, the reaction rate is slow, on the order of 10−2 M−1 min−1 (data not shown). Conversely, prolonged incubation may lead to oligonucleotide degradation and/or off-target labeling. As such, we have found in most cases that using a concentration of 2 above 60 mM with an incubation time between 12 and 24 h gives the best results.
Oligonucleotide concentration can be increased up to 1 mM, as needed. If a very large amount of labeled oligonucleotide is required, multiple reactions can be performed in tandem.
One may reduce the reaction volume (e.g., 20 µL) in order to accommodate a smaller amount of 2. There are no particular restrictions on reaction volume, as long as mixing of reagents is not impeded.
No special accommodations are necessary to attach two labels, rather than one, to a single oligonucleotide strand containing two phosphorothioate modifications.
The reaction mixture can be stored at −20 or −80 °C for up to 1 week. However, it is recommended that purification is performed as soon as possible after labeling.
2.4 Purification of Labeled Oligonucleotides
In choosing the appropriate purification scheme, multiple objectives should be considered, including: (i) removing excess unattached spin labels; (ii) separating labeled and unlabeled oligonucleotides; and (iii) in case crude oligonucleotides are used, separating full-length species from synthesis failures. In a majority of cases, we have found that HPLC or denaturing gel electrophoresis can achieve the desired degree of purification.
2.4.1 High-Performance Liquid Chromatography
We have successfully used anion-exchange HPLC to purify many oligonucleotides that are 10–55 nt long (Grant, Popova, & Qin, 2008; Qin et al., 2007). In particular, we have used a DNAPac PA-100 column from Dionex Inc. Upon successful anion-exchange HPLC purification, the samples usually need to be desalted, for example, using reverse-phase or size-exclusion chromatography. Following desalting, samples are generally lyophilized and stored at −20 or −80 °C.
Note that attachment of R5 results in the loss of one or more negative charges on the oligonucleotide strand and, in most cases, the labeled sample will elute earlier than the unlabeled one on anion-exchange HPLC (Qin et al., 2007). This is a very useful diagnostic feature for assessing the success of the labeling reaction. The PA-100 column also provides some degree of sample separation based on polarity. This enables separation of the Rp and Sp diastereomers in some oligonucleotides, which are manifested as splitting of the sample peak in the HPLC trace (Grant et al., 2008; Popova & Qin, 2010).
A potential problem is that the HPLC trace shows broadly overlapping peaks, which indicates lack of separation between different species. This is particularly prone to occur with oligonucleotides that form stable secondary structures. In such cases, one should consider an alternative to HPLC, such as denaturing electrophoresis, discussed below.
2.4.2 Denaturing Polyacrylamide Gel Electrophoresis
Products of the labeling reaction can be purified with denaturing polyacrylamide gel electrophoresis (PAGE) using standard protocols for gel preparation, electrophoresis, and sample recovery. We have found that R5 is prone to degradation at elevated temperatures in the polyacrylamide gel matrix, possibly due to the involvement of nitroxides in radical-catalyzed polymerization reactions at an elevated temperature (Hawker, Bosman, & Harth, 2001). Thus, it is important to maintain a low temperature (e.g., 4 °C) during electrophoresis (Zhang et al., 2012).
2.5 Characterization of Labeled Oligonucleotides
The purified labeled oligonucleotides are generally resuspended in ME buffer or sterile ultrapure water, and their concentrations are determined by absorbance at 260 nm. Note that at 260 nm, absorption of the R5 label (extinction coefficient −1000 M1 cm−1; Qin et al., 2007) is negligible compared to that of nucleic acids (average extinction coefficient −10,000 M−1 cm−1 per nucleotide). Therefore, UV–vis spectroscopy is not capable of characterizing the degree of labeling. Instead, one may use mass spectrometry, HPLC, or gel electrophoresis to characterize the labeling efficiency.
In addition, the labeled sample should be characterized using cw-EPR spectroscopy. The cw-EPR spectral lineshape, which reports rotational motions of the radical, is very informative in diagnosing whether the radical signal originates from a spin label attached to a macromolecule (Zhang, Cekan, Sigurdsson, & Qin, 2009). In addition, by comparing the observed signal to a standard (e.g., TEMPO) of known concentration, one can carry out a spin-counting procedure (Zhang et al., 2009) to quantify the amount of radical signal present within the sample, thereby determining the degree of labeling.
3. MEASURING INTER-R5 DISTANCES USING DOUBLE ELECTRON–ELECTRON RESONANCE SPECTROSCOPY
Interspin distances are measured by determining the strength of magnetic dipolar interaction by using cw-EPR (for interspin distances <20 A °) or pulsed-EPR (for distances from 20 to −100 A °). In particular, developments of pulsed-EPR methodologies have been one of the major advances in SDSL and have been extensively reviewed (Cafiso, 2012; Jeschke, 2012; Krstic et al., 2012; Polyhach et al., 2011; Schiemann & Prisner, 2007), including in several chapters in this volume.
In our work, we have primarily used a four-pulse double electron– electron resonance (DEER) scheme (Pannier, Veit, Godt, Jeschke, & Spiess, 2000) to measure inter-R5 distances that are >20 A °. In this scheme (Fig. 3A), a three-pulse sequence is applied to a population of “observe spins,” which generates a fixed refocused echo for detection. A fourth pulse, which is set to invert a different population of “pump spins,” is applied at a time that is varying. Dipolar coupling between the observe and pump spins results in modulations of refocused echo amplitude, with the modulation frequency being a function of the interspin distance. Acquisition and analysis of DEER data on nucleic acids and protein–nucleic acid complexes follow the same methodologies used for protein and membrane studies, which are extensively described in other chapters in this volume. In the following section, we outline the key steps, and note issues that are specific to nucleic acid studies.
3.1 DEER Sample Preparation
Our DEER measurements were generally carried out on a Bruker ELEXSYS E580 X-band spectrometer equipped with a MS3 or a MD4 resonator, and a typical DEER sample is approximately 25 µL, with the concentration of the doubly labeled sample approximately 100 µM. Note that the signal-to-noise (S/N) ratio of the data depends on the number of spins within the active measuring volume, and it is possible to use smaller amounts of sample (e.g., lowering concentration to −60 µM or reducing volume to −20 µL), although a longer acquisition time may be required. On the other hand, sample concentration exceeding 250 µM should be avoided, as it could potentially lead to intermolecular spin–spin interactions that complicate data acquisition and analysis.
In the absence of sample degradation, the R5 label is usually stably attached to the target molecule during typical folding and preparation procedures, such as dialysis and overnight incubation. In particular, we have found that a majority of the R5 label (i.e., >95%) remains attached during typical heat denaturing (e.g., incubation at 95 °C for up to 5 min), used to ensure proper annealing or folding of nucleic acids.
Another important issue is the use of cryoprotective agents, such as glycerol, sucrose, or ficoll. In order to achieve the required spin phase memory time to measure distances >20 A ° using a nitroxide label such as R5, DEER measurements are carried out at cryogenic temperatures (80 K or below), with the sample in a glassy state, in which the molecules are homogeneously distributed without aggregation (Zecevic, Eaton, Eaton, & Lindgren, 1998). As such, a sample is usually first assembled at physiological temperatures, then flash-frozen in the presence of cryoprotective agents. In our studies with nucleic acids, we typically use glycerol (in the range of 20–50%v/v) as a cryoprotectant, and flash-freeze the sample by directly immersing the capillary in which it is contained into liquid nitrogen. There are extensive investigations of the effects of various cryoprotectants and freezing procedures (e.g., Freed, Khan, Horanyi, & Cafiso, 2011; Galiano, Blackburn, Veloro, Bonora, & Fanucci, 2009; Georgieva et al., 2012; Kirilina, Grigoriev, & Dzuba, 2004; Sato et al., 2008), which readers are strongly advised to consult.
In assessing a specific procedure for preparing DEER samples, one particularly useful control is to measure single-labeled samples prepared by following identical procedures used for double-labeled samples. In a single-labeled sample, one expects to detect only intermolecular dipolar interaction, but not intramolecular interactions. Therefore, in a properly prepared sample, the resulting background-corrected DEER trace should show no (or very little) decay of the refocused echo. However, if one does observe a decay signal on the single-labeled sample, it could be due to excess crowding (i.e., sample concentration >250 µM) or local aggregation.
3.2 DEER Data Acquisition
DEER acquisition generally proceeds through three stages: (i) sample insertion and resonator and bridge tuning; (ii) field-sweep spectrum acquisition; and (iii) DEER setup and acquisition. As DEER acquisition procedures are described extensively within other chapters in this volume as well as in the Bruker manuals (Weber, 2005, 2006), only a summary of each of these steps is presented here. Operation procedures may vary between different spectrometers, and readers are also strongly encouraged to consult instructions provided by the spectrometer vendor, which contain detailed descriptions of necessary protocols as well as tips and safety checks that are important for the handling of the spectrometer.
Step 3.2.1
Sample insertion and resonator and bridge tuning
Following vendor instructions, turn on the spectrometer, cool down the cryostat to the desired temperature (e.g., 80 or 50 K), and insert the flash-frozen sample. Care should be taken to minimize temperature fluctuation and air exposure to avoid water condensation within the resonator. Furthermore, to achieve optimal positioning of the sample within the active measuring volume, a common practice is to set the spectrometer to the cw-EPR “tune” mode, and follow the shift of the resonating frequency (i.e., the “dip”). Typically, when sample is optimally positioned, the dip is located farthest away from position observed for the empty resonator. Over-couple the resonator before switching the spectrometer to “operate” mode.
Step 3.2.2
Field-sweep spectrum acquisition
Following vendor instructions, over-couple the resonator, turn on the pulse traveling wave tube (TWT) amplifier, and switch the spectrometer to the “pulse” mode. Be sure to carry out the required safety tests. In particular, check the presence of the defense pulses before switching the TWT from “standby” to “operate,” then set the TWT to “operate,” slowly increasing the microwave (MW) power (decreasing MW attenuation) and checking cavity ringdown. Note: set the magnetic field away from the resonating field, so that the actual signal of the sample will not be mistaken for the ringdown.
Next, identify and optimize the refocused echo (Fig. 3A), which will be used as the “reporter” in DEER acquisition. Note that in a Bruker E580 system, the frequency of the detection channel (νobserve, as this channel is used for monitoring the observe spin) is not a user-controlled variable, and one should identify the proper sample signal by tuning magnetic field H (based on the relationship h·νobserve = g · β · H; where h is Planck’s constant, β is the Bohr magneton, and the electron g value is approximately 2 for a nitroxide sample). As such, the general procedure is to set up the proper three-pulse sequence for the observe spin (Fig. 3A), set the magnetic field position to the expected center field of the signal, and find the refocused echo. Note that phase cycling can be used to identify the correct refocused echo.
Once the refocused echo is identified, maximize its signal by adjusting acquisition parameters (e.g., field position, microwave power, signal phase). Then, obtain the field-sweep spectrum by scanning the magnetic field spanning the desired range. Note that phase cycling can be used to suppress artifacts and to eliminate unwanted echoes.
Step 3.2.3
DEER setup and acquisition
Step 3.2.3.1
Determine the pump frequency
The scheme used in our studies is such that the pump pulse is applied at the center manifold of the nitroxide field-sweep spectrum, while the observe pulse is set at its shoulder at the lower field (Fig. 3A). In a typical DEER measurement carried out on a Bruker E580 spectrometer, the sample is subjected to one magnetic field, and the pump and observe pulses are distinguished by a frequency offset.
Using the field-sweep spectrum obtained above, record the field positions corresponding to the maxima of the center (HC) and low-field manifolds (HL), as well as νobserve, the frequency at which the field-sweep spectrum is obtained. Then compute pump frequency (νpump) as:
| (1) |
Step 3.2.3.2
Optimize the pump pulse
The pump pulse is applied via a separate channel (i.e., the ELDOR pulse channel), and needs to be optimized independently from the observe pulse. With the Bruker E580 system, we generally use the full power available for the ELDOR channel (i.e., ELDOR attenuation at 0 dB), then determine the proper ELDOR pulse length (π(ELDOR)) for a complete inversion of the spins. This can be achieved by performing a “nutation” measurement (Schweiger & Jeschke, 2001; Weber, 2006). An alternative way to optimize the pump pulse (Weber, 2006) is to first follow protocols described in Step 3.2.2 to identify the refocused echo. Then, set an ELDOR pulse at 100 ns after the first observe π pulse (see Fig. 3A) and at the same frequency as νobserve (i.e., the frequency at which the refocused echo is observed). With the ELDOR attenuation set at 0 dB, increase the ELDOR pulse length, which should result in variations in refocused echo intensity. The ELDOR pulse length at which the refocused echo disappears for the first time is the optimized π(ELDOR).
Step 3.2.3.3
Acquire a DEER spectrum
Following steps described above, set the field position to HL identified in Step 3.2.3.1. Set the ELDOR frequency to νpump calculated from Eq. (1) in Step 3.2.3.1, ELDOR attenuation to 0 dB, and π(ELDOR) to the optimized value determined in Step 3.2.3.2. If necessary, readjust acquisition parameters (e.g., location and width of the integrator gate) for optimal detection of the refocused echo. Choose the appropriate phase cycling scheme (e.g., two-step), and set the number of scans as necessary. Acquire the DEER trace until desired S/N is reached, which generally takes 12–24 h depending on sample concentration, spin–spin distance, and other parameters.
3.3 DEER Spectrum Analysis
Two general approaches are used to derive the underlying interspin distance distribution, P(r), from the measured DEER data. In the model-free approach, P(r) is not assumed to adopt a particular functional form, and is retrieved by fitting using, for example, the Tikhonov regularization approach (Bowman, Maryasov, Kim, & DeRose, 2004; Chiang, Borbat, & Freed, 2005; Jeschke, Panek, Godt, Bender, & Paulsen, 2004). The other approach is based on specific model functions of P(r); specifically, one can carry out the fitting with P(r) assumed to be composed of one or more Gaussian functions (Sen, Logan, & Fajer, 2007).
One of the most commonly used programs for such spectral analysis is Deer-Analysis (http://www.epr.ethz.ch/software), developed by Jeschke et al. (2006). A recent version, Deer Analysis 2013, provides options for both model-free and model-based fitting. For details on using Deer-Analysis, readers are encouraged to consult the manuals associated with the program.
4. INTEGRATION OF DEER-MEASURED DISTANCES WITH COMPUTATIONAL MODELING
The DEER-measured distances serve as constraints in computational modeling in order to derive structural features of the target molecule. For this purpose, our approach is to use multiple sets of DEER-measured distances to data-mine a large ensemble of models for candidate structure(s) that are in best agreement with the experimental data (Zhang et al., 2012, 2014). The models are generated independently by any all-atom molecular modeling approach (e.g., Monte Carlo simulations). The viability of each model is then assessed by comparing expected inter-R5 distances in a given model to the corresponding DEER-measured values. Compared to other approaches in which the DEER-measured distances are directly incorporated into model building (e.g., Duss et al., 2015; Duss, Yulikov, et al., 2014), our method provides a large degree of flexibility in choosing the best-suited modeling approach for a specific system.
4.1 Model Generation
A model is defined by the Cartesian coordinates of all heavy atoms of the target molecule in the Protein Data Bank (PDB) file format (Berman et al., 2000). In our approach, any modeling method that outputs PDB files can be used to generate the ensemble of putative structural models. For example, in our work on an RNA three-way junction, the model pool was generated by rigid-body transformation (translation and rotation) of three A-form double helices, with concurrently imposed steric constraints (Zhang et al., 2012). Conversely, in our studies of sequence-dependent DNA duplex shapes, all-atom Monte Carlo simulations were employed to generate a large pool of 10,000 B-form DNA models for subsequent evaluation (Zhang et al., 2014).
4.2 Computing Expected Inter-R5 Distances for the Model Pool
4.2.1 The NASNOX Program
We have established and validated a program, called NASNOX, which computes expected inter-R5 distances in a given nucleic acid structure (Cai et al., 2006; Price et al., 2007; Qin et al., 2007). NASNOX models R5 at a pair of nucleic acid sites using experimentally-determined bond lengths and angles. With the nucleic acid coordinates (i.e., the PDB file) fixed, the program varies, in a stepwise fashion, the torsion angles (t1, t2, and t3, Fig. 2C) around the three single bonds connecting the pyrroline ring to the phosphorothioate, and identifies those allowed R5 conformers that have no steric clash between the nitroxide and the parent molecule. Once the ensemble of allowed conformers is identified, inter-R5 distances are calculated, and the corresponding mean and standard deviation are reported. The NASNOX program has been validated on DNA (Cai et al., 2006), RNA (Cai et al., 2007), and protein–DNA complexes (Chen et al., 2013; Zhang et al., 2014) with known high-resolution structures.
An Internet-accessible version of NASNOX, NASNOX_W (Qin et al., 2007), is available at http://pzqin.usc.edu/NASNOX/. The web server allows a user to upload a target structure (in the proper PDB format) and specify a set of search parameters (see example in Fig. 4), then carry out NASNOX calculations and generate two output files: (i) a PDB file, “data001lig.pdb,” with the allowable R5 conformers modeled onto the original structure, and (ii) a text file, “data.add,” which summarizes the input parameters, lists the allowable conformers, and reports the individual inter- R5 distances, and the average, for the ensemble. A unique feature of NASNOX is its speed—each R5 conformer distribution and corresponding inter-R5 distances can be computed in seconds.
4.2.2 Running NASNOX in a Batch Mode
To examine a large pool of models, NASNOX is run in a batch mode (Zhang et al., 2012, 2014), in which the program automatically computes inter-R5 distances based on user-specified labeling sites and structural models. We describe here procedures for batch-mode operation of NASNOX using a script written in Matlab (The MathWorks, Inc.), which will automatically output one user-specified distance for multiple models.
Step 4.2.2.1
Download and install relevant files
The script and relevant files are compressed as the “NASOX_batch2015. zip” file, which is available for download from http://pzqin.usc.edu/pzqhome/software. Unzip and place the files in a folder. The folder should contain:
Matlab script, “BATCH_NASNOX.m.” The script requires Matlab version 2010b or later.
NASNOX program. An executable file compiled for the Windows operating system that has been successfully tested with Windows XP, 7, and 8.
Nitroxide parameter files for NASNOX.
A set of example models and a readme file.
Step 4.2.2.2
Prepare the input files
Prepare the following input files and place them in the same folder as the Matlab script and NASNOX files:
-
Models of the target nucleic acid molecule:
These are a set of PDB files. Note that the NASNOX program requires a specific format, and the readers are strongly encouraged to consult the “sample.pdb” files provided. Particularly, the program identifies a specific nucleotide based on the order in which it is read; therefore, we suggest that the PDB files list the nucleotides in a consecutive fashion, with the first nucleotide set as “1.” In addition, a “TER” line is required at the end of the PDB file for it to be recognized by the NASNOX program. To check whether the input PDB files are formatted properly, one can use the web-based NASNOX_W (see Section 4.2.1) to execute the program on sample(s) of the input model pool.
-
“model_list.csv”:
This is a csv-format file that lists the names of the models described above (Fig. 4). Note that the model names in this file should not contain the “.pdb” suffix.
-
“sites.csv”:
This is a csv-format file listing positions of the two nucleotides at which the inter-R5 distance will be computed (Fig. 4). In the current version, only one pair of positions should be listed in this file. In addition, the program will model R5 onto both the Rp and Sp diastereomers at each selected nucleotide.
-
“parameter.csv”:
This is a csv-format file listing the search parameters for the torsion angles (Fig. 4).
Step 4.2.2.3
Batch execution
Run the “BATCH_NASNOX.m” program in Matlab. This will execute NASNOX based on information specified above.
Step 4.2.2.4
Retrieve output files
For each entry in “model_list.csv,” corresponding “modelname_site1-site2_data.add” and “modelname_site1-site2_datalig.pdb” files are produced. These follow the same output format as those provided from the web-based NASNOX_W program (see Section 4.2.1), but with identifiers added for the respective model names and sites. In particular, for each model, the average inter-R5 distance is listed at the end of the “.add” file (see Fig. 4), which can be extracted and used for model characterization as described below. For more details, one should consult Qin et al., 2007 and the tutorial file provided for NASNOX_W.
4.3 Model Selection and Characterization
Here, the goal is to identify those models in which the expected inter-R5 distances match, collectively, to the set of DEER-measured distances. There are a variety of approaches to achieve this goal, and below we summarize criteria used in our previous work.
4.3.1 The RMSD Metric
One of the simplest approaches is to compute the root-mean-square-deviation (RMSD) between the measured and expected distances (Zhang et al., 2012):
| (2) |
where N is the total number of distances measured, the summation index i designates a particular distance set in a model, rmodel−j is the NASNOX-computed expected distance for the model j, and rdeer is the DEER-measured distance. The lower the RMSDdeer value, the better the model fits to the DEER measurement.
4.3.2 The Modified RMSD Metric
A DEER measurement not only renders the distance information but also provides the distance distribution profile (see Section 3.3). We have defined a modified RMSD metric (RMSDmod) as (Zhang et al., 2012):
| (3) |
where the variables are defined as those in Eq. (2), with an additional parameter σ corresponding to the width of distance distribution obtained from the DEER measurement (see Section 3.3). The RMSDmod metric gives higher weights to datasets with narrow distributions (which are considered to be better defined and of higher information content), and lower weights to those showing broad distributions. Comparing the ranking obtained between RMSDdeer and RMSDmod provides a means to assess impacts due to datasets with broad distance distributions (Zhang et al., 2012).
4.3.3 A Pseudo-Energy Scoring Function
To evaluate closely related models, such as DNA duplexes falling under the B-form category (Zhang et al., 2014), we further developed a scoring function defined as:
| (4) |
Pt represents, under the assumption of an idealized normal distribution, the effective probability of a given set of rmodel values matching the corresponding rdeer values, with a perfect match resulting in a maximum Pt score of 1. We have used the Pt score to assess B-DNAs with sequence-dependent shape (Zhang et al., 2014).
4.4 Additional Considerations
To enhance the chance of finding model(s) that satisfy the measured distances, one should ensure that the model pool has sufficient coverage of the conformational space allowed for the target molecule. In our work on an RNA junction, we generated the model pool using two different methods: stepwise search and Monte Carlo docking (Zhang et al., 2012). While the two pools were not identical, upon applying the DEER constraints, the resulting top models adopted the same configuration. While this is not definitive proof of a correct structure, it provides assurance, to a certain extent, that each of the model pools had sufficient coverage.
Another consideration is the diversity of the model pool. In our studies of DNA duplexes, we computed the heavy atom RMSD between structural models (RMSDstruct) to assess diversity within the pool of B-DNA models (Zhang et al., 2014). In addition, we devised an RMSDstruct versus Pt plot encompassing all 10,000 entries within a given model pool (Fig. 5A). The plot shows a number of features that indicated the search was productive. First, the data points were spread along both axes of the plot, indicating sufficient diversity of the pool. Second, models that were more different (i.e., larger RMSDstruct) from the best-fit model (i.e., the one with the best Pt score) indeed deviated more from the measured distances (i.e., lower Pt score), indicating that the measured distances were capable of differentiating the models
Figure 5.
An example of analyzing a model pool using DEER-measured distances. Data shown are reproduced from reported work on the “p21-RE” duplex (Zhang et al., 2014). (A) The RMSDstruct versus Pt score plot for the 10,000 models generated from MC simulations. Blue circles represent the top 20 ranked models, obtained using 16 sets of measured distances; red triangles represent the top 20 ranked models, obtained using 14 distances. Note that 14 models, including the best-fit model, are retrieved in both searches. (B) Overlay of the top 20 models of the unbound DNA (blue thin lines) and the bound DNA (red). The unbound DNA models are obtained using the integrated SDSL/MC approach, while the bound DNA is from a reported crystal structure, 3TS8. pdb (Emamzadah, Tropia, & Halazonetis, 2011). The analysis shows that the unbound DNA models converge and identifies the mode of DNA deformation upon protein binding (Zhang et al., 2014). Adapted from Zhang et al. (2014) with permission.
Furthermore, in the model search, one might wonder whether increasing the number of experimentally measured distances may improve the model. This may be addressed by analyzing the models retrieved using different combinations of the measured distances. For example, in our DNA work (Zhang et al., 2014), models with high Pt scores (e.g., those with the top 20 scores) are structurally very similar (RMSDstruct <2.0 A °, Fig. 5B). Given the uncertainty present in both the DEER-measured and NASNOX-derived distances, we concluded that increasing the number of measured distances would not further improve the models. Furthermore, we have shown that searches in which a small number of distance constraints were omitted (i.e., using only 14 distances instead of 16) yield the same best-fit model, and the majority of the top 20 ranked models show little structural deviation (i.e., RMSDstruct <2.0 A °, see Fig. 5A). This indicates that the number of DEER-measured distances (in this case, 16 sets) was sufficient.
5. CONCLUSIONS
We present here an integrated method that combines distances measured using a nucleotide-independent nitroxide probe with computational modeling to derive all-atom models of nucleic acids. This approach allows a large degree of flexibility for interfacing with various molecular modeling methods, and its feasibility has been demonstrated in reported work. The approach presented here is a framework for the integrative modeling of nucleic acids with the goal to derive “hybrid models,” by data mining computationally generated structural ensembles of a target molecule using experimental distance measurements from EPR spectroscopy. These hybrid models represent solution-state all-atom structures, which might provide important biological information to the structural biology community.
ACKNOWLEDGMENT
We gratefully acknowledge support from NSF (MCB-0546529 and CHE-1213673) and NIH (GM069557 and RR028992 to P.Z.Q.; GM106056 to R.R.).
REFERENCES
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, et al. The protein data bank. Nucleic Acids Research. 2000;28(1):235–242. doi: 10.1093/nar/28.1.235. http://dx.doi.org/10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman MK, Maryasov AG, Kim N, DeRose VJ. Visualization of distance distribution from pulsed double electron–electron resonance data. Applied Magnetic Resonance. 2004;26(1–2):23–39. http://dx.doi.org/10.1007/bf03166560. [Google Scholar]
- Cafiso DS. Taking the pulse of protein interactions by EPR spectroscopy. Biophysical Journal. 2012;103(10):2047–2048. doi: 10.1016/j.bpj.2012.10.005. http://dx.doi.org/10.1016/j.bpj.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Q, Kusnetzow AK, Hideg K, Price EA, Haworth IS, Qin PZ. Nanometer distance measurements in RNA using site-directed spin labeling. Biophysical Journal. 2007;93(6):2110–2117. doi: 10.1529/biophysj.107.109439. http://dx.doi.org/10.1529/biophysj.107.109439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Q, Kusnetzow AK, Hubbell WL, Haworth IS, Gacho GP, Van Eps N. Site-directed spin labeling measurements of nanometer distances in nucleic acids using a sequence-independent nitroxide probe. Nucleic Acids Research. 2006;34(17):4722–4730. doi: 10.1093/nar/gkl546. http://dx.doi.org/10.1093/nar/gkl546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Zhang X, Dantas Machado AC, Ding Y, Chen Z, Qin PZ, et al. Structure of p53 binding to the BAX response element reveals DNA unwinding and compression to accommodate base-pair insertion. Nucleic Acids Research. 2013;41(17):8368–8376. doi: 10.1093/nar/gkt584. http://dx.doi.org/10.1093/nar/gkt584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang YW, Borbat PP, Freed JH. Maximum entropy: A complement to Tikhonov regularization for determination of pair distance distributions by pulsed ESR. Journal of Magnetic Resonance. 2005;177:184–196. doi: 10.1016/j.jmr.2005.07.021. [DOI] [PubMed] [Google Scholar]
- Ding Y, Nguyen P, Tangprasertchai NS, Reyes CV, Zhang X, Qin PZ. Nucleic acid structure and dynamics: Perspectives from site-directed spin labeling. In: Gilbert BC, Ghcechik V, Murphy DM, editors. Electron paramagnetic resonance. Vol. 24. Cambridge, England (Thomas Graham House, Science Park, Cambridge Cb4 4WF): The Royal Society of Chemistry; 2015. pp. 122–147. [Google Scholar]
- Ding Y, Zhang X, Tham KW, Qin PZ. Experimental mapping of DNA duplex shape enabled by global lineshape analyses of a nucleotide-independent nitroxide probe. Nucleic Acids Research. 2014;42(18):e140. doi: 10.1093/nar/gku695. http://dx.doi.org/10.1093/nar/gku695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duss O, Michel E, Yulikov M, Schubert M, Jeschke G, Allain FHT. Structural basis of the non-coding RNA RsmZ acting as a protein sponge. Nature. 2014;509:588–592. doi: 10.1038/nature13271. http://dx.doi.org/10.1038/nature13271. [DOI] [PubMed] [Google Scholar]
- Duss O, Yulikov M, Allain FHT, Jeschke G. Combining NMR and EPR to determine structures of large RNAs and protein–RNA complexes in solution. Methods in Enzymology. 2015;558:279–331. doi: 10.1016/bs.mie.2015.02.005. [DOI] [PubMed] [Google Scholar]
- Duss O, Yulikov M, Jeschke G, Allain FHT. EPR-aided approach for solution structure determination of large RNAs or protein–RNA complexes. Nature Communications. 2014;5(3669) doi: 10.1038/ncomms4669. http://dx.doi.org/10.1038/ncomms4669. [DOI] [PubMed] [Google Scholar]
- Emamzadah S, Tropia L, Halazonetis TD. Crystal structure of a multidomain human p53 tetramer bound to the natural CDKN1A (p21) p53-response element. Molecular Cancer Research. 2011;9(11):1493–1499. doi: 10.1158/1541-7786.MCR-11-0351. http://dx.doi.org/10.1158/1541-7786.mcr-11-0351. [DOI] [PubMed] [Google Scholar]
- Fedorova OS, Tsvetkov YD. Pulsed electron double resonance in structural studies of spin-labeled nucleic acids. Acta Naturae. 2013;5(1):9–32. [PMC free article] [PubMed] [Google Scholar]
- Freed DM, Khan AK, Horanyi PS, Cafiso DS. Molecular origin of electron paramagnetic resonance line shapes on beta-barrel membrane proteins: The local solvation environment modulates spin-label configuration. Biochemistry. 2011;50(41):8792–8803. doi: 10.1021/bi200971x. http://dx.doi.org/10.1021/bi200971x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galiano L, Blackburn ME, Veloro AM, Bonora M, Fanucci GE. Solute effects on spin labels at an aqueous-exposed site in the flap region of HIV-1 protease. The Journal of Physical Chemistry. B. 2009;113(6):1673–1680. doi: 10.1021/jp8057788. http://dx.doi.org/10.1021/jp8057788. [DOI] [PubMed] [Google Scholar]
- Georgieva ER, Roy AS, Grigoryants VM, Borbat PP, Earle KA, Scholes CP, et al. Effect of freezing conditions on distances and their distributions derived from double electron electron resonance (DEER): A study of doubly-spin-labeled T4 lysozyme. Journal of Magnetic Resonance. 2012;216:69–77. doi: 10.1016/j.jmr.2012.01.004. http://dx.doi.org/10.1016/j.jmr.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant GPG, Boyd N, Herschlag D, Qin PZ. Motions of the substrate recognition duplex in a group I intron assessed by site-directed spin labeling. Journal of the American Chemical Society. 2009;131(9):3136–3137. doi: 10.1021/ja808217s. http://dx.doi.org/10.1021/ja808217s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant GPG, Popova A, Qin PZ. Diastereomer characterizations of nitroxide-labeled nucleic acids. Biochemical and Biophysical Research Communications. 2008;371(3):451–455. doi: 10.1016/j.bbrc.2008.04.088. http://dx.doi.org/10.1016/j.bbrc.2008.04.088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagelueken G, Ward R, Naismith JH, Schiemann O. MtsslWizard: In silico spin-labeling and generation of distance distributions in PyMOL. Applied Magnetic Resonance. 2012;42(3):377–391. doi: 10.1007/s00723-012-0314-0. http://dx.doi.org/10.1007/s00723-012-0314-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hankovszky H, Hideg K, Lex L. Nitroxyls; VII1. Synthesis and reactions of highly reactive 1-Oxyl-2,2,5,5-tetramethyl-2,5-dihydropyrrole-3-ylmethyl sulfonates. Synthesis. 1980:914–916. [Google Scholar]
- Hatmal MM, Li Y, Hegde BG, Hegde PB, Jao CC, Langen R, et al. Computer modeling of nitroxide spin labels on proteins. Biopolymers. 2012;97(1):35–44. doi: 10.1002/bip.21699. http://dx.doi.org/10.1002/bip.21699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawker CJ, Bosman AW, Harth E. New polymer synthesis by nitroxide mediated living radical polymerizations. Chemical Reviews. 2001;101(12):3661–3688. doi: 10.1021/cr990119u. http://dx.doi.org/10.1021/cr990119u. [DOI] [PubMed] [Google Scholar]
- Hirst SJ, Alexander N, McHaourab HS, Meiler J. Rosetta EPR: An integrated tool for protein structure determination from sparse EPR data. Journal of Structural Biology. 2011;173(3):506–514. doi: 10.1016/j.jsb.2010.10.013. http://dx.doi.org/10.1016/j.jsb.2010.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbell WL, Altenbach C. Investigation of structure and dynamics in membrane proteins using site-directed spin labeling. Current Opinion in Structural Biology. 1994;4:566–573. [Google Scholar]
- Hubbell WL, Cafiso DS, Altenbach C. Identifying conformational changes with site-directed spin labeling. Nature Structural Biology. 2000;7:735–739. doi: 10.1038/78956. http://dx.doi.org/10.1038/78956. [DOI] [PubMed] [Google Scholar]
- Hubbell WL, López CJ, Altenbach C, Yang Z. Technological advances in site-directed spin labeling of proteins. Current Opinion in Structural Biology. 2013;23(5):725–733. doi: 10.1016/j.sbi.2013.06.008. http://dx.doi.org/10.1016/j.sbi.2013.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeschke G. DEER distance measurements on proteins. Annual Review of Physical Chemistry. 2012;63(1):419–446. doi: 10.1146/annurev-physchem-032511-143716. http://dx.doi.org/10.1146/annurev-physchem-032511-143716. [DOI] [PubMed] [Google Scholar]
- Jeschke G, Chechik V, Ionita P, Godt A, Zimmermann H, Banham J, et al. Deer Analysis 2006—A comprehensive software package for analyzing pulsed ELDOR data. Applied Magnetic Resonance. 2006;30(3–4):473–498. http://dx.doi.org/10.1007/BF03166213. [Google Scholar]
- Jeschke G, Panek G, Godt A, Bender A, Paulsen H. Data analysis procedures for pulse ELDOR measurements of broad distance distributions. Applied Magnetic Resonance. 2004;26(1–2):223–244. http://dx.doi.org/10.1007/BF03166574. [Google Scholar]
- Kirilina EP, Grigoriev IA, Dzuba SA. Orientational motion of nitroxides in molecular glasses: Dependence on the chemical structure, on the molecular size of the probe, and on the type of the matrix. The Journal of Chemical Physics. 2004;121(24):12465–12471. doi: 10.1063/1.1822913. http://dx.doi.org/10.1063/1.1822913. [DOI] [PubMed] [Google Scholar]
- Krstic I, Endeward B, Margraf D, Marko A, Prisner TF. Structure and dynamics of nucleic acids. Topics in Current Chemistry. 2012;321:159–198. doi: 10.1007/128_2011_300. http://dx.doi.org/10.1007/128_2011_300. [DOI] [PubMed] [Google Scholar]
- Nguyen PH, Popova AM, Hideg K, Qin PZ. A nucleotide-independent cyclic nitroxide label for monitoring segmental motions in nucleic acids. BMC Biophysics. 2015;8(6) doi: 10.1186/s13628-015-0019-5. http://dx.doi.org/10.1186/s13628-015-0019-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pannier M, Veit S, Godt A, Jeschke G, Spiess HW. Dead-time free measurement of dipole-dipole interactions between electron spins. Journal of Magnetic Resonance. 2000;142:331–340. doi: 10.1006/jmre.1999.1944. http://dx.doi.org/10.1006/jmre.1999.1944. [DOI] [PubMed] [Google Scholar]
- Polyhach Y, Bordignon E, Jeschke G. Rotamer libraries of spin labelled cysteines for protein studies. Physical Chemistry Chemical Physics. 2011;13(6):2356–2366. doi: 10.1039/c0cp01865a. http://dx.doi.org/10.1039/c0cp01865a. [DOI] [PubMed] [Google Scholar]
- Popova AM, Hatmal MM, Frushicheva MP, Price EA, Qin PZ, Haworth IS. Nitroxide sensing of a DNA microenvironment: Mechanistic insights from EPR spectroscopy and molecular dynamics simulations. The Journal of Physical Chemistry. B. 2012;116(22):6387–6396. doi: 10.1021/jp303303v. http://dx.doi.org/10.1021/jp303303v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popova AM, Kálai T, Hideg K, Qin PZ. Site-specific DNA structural and dynamic features revealed by nucleotide-independent nitroxide probes. Biochemistry. 2009;48(36):8540–8550. doi: 10.1021/bi900860w. http://dx.doi.org/10.1021/bi900860w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popova AM, Qin PZ. A nucleotide-independent nitroxide probe reports on site-specific stereomeric environment in DNA. Biophysical Journal. 2010;99(7):2180–2189. doi: 10.1016/j.bpj.2010.08.005. http://dx.doi.org/10.1016/j.bpj.2010.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price EA, Sutch BT, Cai Q, Qin PZ, Haworth IS. Computation of nitroxide-nitroxide distances for spin-labeled DNA duplexes. Biopolymers. 2007;87:40–50. doi: 10.1002/bip.20769. http://dx.doi.org/10.1002/bip.20769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin PZ, Butcher SE, Feigon J, Hubbell WL. Quantitative analysis of the GAAA tetraloop/receptor interaction in solution: A site-directed spin labeling study. Biochemistry. 2001;40:6929–6936. doi: 10.1021/bi010294g. http://dx.doi.org/10.1021/bi010294g. [DOI] [PubMed] [Google Scholar]
- Qin PZ, Haworth IS, Cai Q, Kusnetzow AK, Grant GPG, Price EA, et al. Measuring nanometer distances in nucleic acids using a sequence-independent nitroxide probe. Nature Protocols. 2007;2(10):2354–2365. doi: 10.1038/nprot.2007.308. http://dx.doi.org/10.1038/nprot.2007.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato H, Kathirvelu V, Spagnol G, Rajca S, Rajca A, Eaton SS, et al. Impact of electron–electron spin interaction on electron spin relaxation of nitroxide diradicals and tetraradical in glassy solvents between 10 and 300 K. The Journal of Physical Chemistry. B. 2008;112(10):2818–2828. doi: 10.1021/jp073600u. http://dx.doi.org/10.1021/jp073600u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiemann O, Prisner TF. Long-range distance determinations in biomacromolecules by EPR spectroscopy. Quarterly Reviews of Biophysics. 2007;40(1):1–53. doi: 10.1017/S003358350700460X. http://dx.doi.org/10.1017/S003358350700460X. [DOI] [PubMed] [Google Scholar]
- Schweiger A, Jeschke G. Principles of pulse electron paramagnetic resonance. Oxford: Oxford University Press; 2001. Nuclear modulation effect I: Basic experiments; pp. 247–295. [Google Scholar]
- Sen KI, Logan TM, Fajer PG. Protein dynamics and monomer-monomer interactions in AntR activation by electron paramagnetic resonance and double electron–electron resonance. Biochemistry. 2007;46(41):11639–11649. doi: 10.1021/bi700859p. http://dx.doi.org/10.1021/bi700859p. [DOI] [PubMed] [Google Scholar]
- Shelke SA, Sigurdsson ST. Site-directed spin labelling of nucleic acids. European Journal of Organic Chemistry. 2012;12:2291–2301. http://dx.doi.org/10.1002/ejoc.201101434. [Google Scholar]
- Sowa GZ, Qin PZ. Site-directed spin labeling studies on nucleic acid structure and dynamics. Progress in Nucleic Acid Research and Molecular Biology. 2008;82:147–197. doi: 10.1016/S0079-6603(08)00005-6. http://dx.doi.org/10.1016/S0079-6603(08)00005-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber RT. Bruker ELEXSYS E850 User’s Manual (v 2.0) Billerica, MA, USA: Bruker BioSpin Corporation; 2005. [Google Scholar]
- Weber RT. Bruker pulsed ELDOR Option User’s Manual (v 1.0) Billerica, MA, USA: Bruker BioSpin Corporation; 2006. [Google Scholar]
- Zecevic ANA, Eaton GR, Eaton SS, Lindgren M. Dephasing of electron spin echoes for nitroxyl radicals in glassy solvents by non-methyl and methyl protons. Molecular Physics. 1998;95(6):1255–1263. http://dx.doi.org/10.1080/00268979809483256. [Google Scholar]
- Zhang X, Cekan P, Sigurdsson ST, Qin PZ. Studying RNA using site-directed spin-labeling and continuous-wave electron paramagnetic resonance spectroscopy. Methods in Enzymology. 2009;469:303–328. doi: 10.1016/S0076-6879(09)69015-7. http://dx.doi.org/10.1016/S0076-6879(09)69015-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Dantas Machado AC, Ding Y, Chen Y, Lu Y, Duan Y, et al. Conformations of p53 response elements in solution deduced using site-directed spin labeling and Monte Carlo sampling. Nucleic Acids Research. 2014;42(4):2789–2797. doi: 10.1093/nar/gkt1219. http://dx.doi.org/10.1093/nar/gkt1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Tung CS, Sowa GZ, Hatmal MM, Haworth IS, Qin PZ. Global structure of a three-way junction in a phi29 packaging RNA dimer determined using site-directed spin labeling. Journal of the American Chemical Society. 2012;134(5):2644–2652. doi: 10.1021/ja2093647. http://dx.doi.org/10.1021/ja2093647. [DOI] [PMC free article] [PubMed] [Google Scholar]