Significance
Carcinoembryonic antigen-related cell adhesion molecules (CEACAMs) are cell surface proteins that regulate cell adhesion and signaling in cancer, infection, and immunity through their diverse oligomeric states. Although X-ray crystal structures of CEACAM homodimers have been described, how they form heterodimers or remain monomers is poorly understood. Here we present the crystal structures of homodimeric CEACAM6, monomeric CEACAM8, and the heterodimeric CEACAM6–CEACAM8 complex. Our crystallographic and biophysical data suggest ways in which CEACAM6 and CEACAM8 regulate the biological functions of one another.
Keywords: CEACAM, X-ray crystallography, isothermal titration calorimetry, analytical ultracentrifugation
Abstract
Carcinoembryonic antigen-related cell adhesion molecules (CEACAMs) comprise a large family of cell surface adhesion molecules that bind to themselves and other family members to carry out numerous cellular functions, including proliferation, signaling, differentiation, tumor suppression, and survival. They also play diverse and significant roles in immunity and infection. The formation of CEACAM oligomers is caused predominantly by interactions between their N-terminal IgV domains. Although X-ray crystal structures of CEACAM IgV domain homodimers have been described, how CEACAMs form heterodimers or remain monomers is poorly understood. To address this key aspect of CEACAM function, we determined the crystal structures of IgV domains that form a homodimeric CEACAM6 complex, monomeric CEACAM8, and a heterodimeric CEACAM6–CEACAM8 complex. To confirm and quantify these interactions in solution, we used analytical ultracentrifugation to measure the dimerization constants of CEACAM homodimers and isothermal titration calorimetry to determine the thermodynamic parameters and binding affinities of CEACAM heterodimers. We found the CEACAM6–CEACAM8 heterodimeric state to be substantially favored energetically relative to the CEACAM6 homodimer. Our data provide a molecular basis for the adoption of the diverse oligomeric states known to exist for CEACAMs and suggest ways in which CEACAM6 and CEACAM8 regulate the biological functions of one another, as well as of additional CEACAMs with which they interact, both in cis and in trans.
Carcinoembryonic antigen-related cell adhesion molecules (CEACAMs) are Ig-related proteins encoded by 12 genes on human chromosome 19q13 (1). Expression patterns of each CEACAM have been observed to be distinct (2). Several CEACAMs are expressed and anchored predominantly on the surfaces of epithelial, endothelial, lymphocyte, myeloid, and granulocyte cells. Certain CEACAMs, however, are expressed only on one cell type or tissue, including CEACAM3, CEACAM8, and CEACAM16, which are expressed on phagocytes, granulocytes, and in the inner ear, respectively (3–5). With distinct expression patterns and localizations, CEACAMs are typically observed to be involved in numerous and diverse cellular functions, including cell adhesion, proliferation, signaling, differentiation, tumor suppression, and survival (6–10). Certain CEACAMs, however, such as CEACAM3 and CEACAM16, have specific roles in phagocytosis and hearing, respectively (3, 4). Several pathogenic bacteria, such as Neisseria meningtidis, Escherichia coli, and Haemophilus influenza use adhesins to interact and anchor themselves to host cell surfaces through CEACAM recruitment (11–13). Because CEACAMs are involved in proliferation, tumor suppression, and survival, CEACAM dysregulation is frequently observed in tumor growth and metastasis (8, 14–17). All CEACAMs, except CEACAM16, are tethered to the surfaces of cells through either a single transmembrane domain or a glycophosphatidylinositol (GPI) anchor at their C terminus (7). Each CEACAM contains an N-terminal Ig variable-region–like (IgV) domain (6, 7), which is separated from its C-terminal domain by a variable number of Ig constant region-type 2-like (IgC2-like) domains, ranging in number from zero to six. One function of CEACAMs is cell adhesion (10). CEACAMs achieve this primarily through the N-terminal IgV domain, which can dimerize either through homo- or heterophillic interactions (18–20). Dimerization of CEACAMs can be either in cis or in trans, with the latter allowing for cell–cell adhesion (21, 22).
CEACAM6 and CEACAM8 are both anchored to cells by a GPI motif, with two interspersing IgC2 domains (6). They are thus unable to directly signal like CEACAM1, which is anchored by a transmembrane domain and contains a C-terminal cytosolic immunoreceptor tyrosine-based inhibitory motif (6, 23). CEACAM8 is exclusively expressed on granulocytes, whereas CEACAM6 is expressed on the epithelial cells of the gastrointestinal tract and granulocytes (24). High levels of CEACAM6 expression are typically observed in several different cancers (17, 25, 26). Studies have shown that CEACAM6 inhibits anoikis, resistance to apoptosis in the absence of adhesion to the extracellular matrix, thereby promoting metastasis (27). Although less is known about CEACAM8, mRNA of both CEACAM6 and CEACAM8 are up-regulated in acute lymphoblastic leukemia (28).
The homodimerization of CEACAM N-terminal IgV domains, in particular those of CEACAM1 and CEACAM5, has been described previously (18, 19). However, the molecular mechanisms by which CEACAMs can heterodimerize have yet to be elucidated. It has been reported that CEACAM6 can form heterodimers with CEACAM1, CEACAM5, and CEACAM8, whereas CEACAM8 can heterodimerize with CEACAM1 and CEACAM6 (20, 29). Like homodimerization, CEACAM heterodimerization appears to require N-terminal IgV domains (24). In this study, we present X-ray crystallographic and biophysical data showing that, in both crystals and in solution, CEACAM6 homodimerizes, CEACAM8 is monomeric, and the CEACAM6–CEACAM8 heterodimer represents the energetically preferred state.
Results
CEACAM6 Forms a Homodimer Distinct from Those Observed Previously.
To gain further insight to CEACAM homo- and heterodimerization, we solved the X-ray crystal structure of the N-terminal IgV domain of human CEACAM6 to a resolution of 1.83 Å (Tables S1 and S2). As expected, CEACAM6 exhibited the typical IgV-like fold of two β-sheets, consisting of strands ABDE and A′GFCC′C′′ (Fig. 1A) and forms a homodimer (Fig. 1B), similar but with distinctions from those observed previously for CEACAM1 and CEACAM5 (18, 19). Superposition of CEACAM6 onto CEACAM5 using one pair of chains (i.e., one from each dimer) results in an RMSD of 0.7 Å, with a substantially larger RMSD of 2.5 Å observed for the other pair of chains (Fig. 1C), resulting in a twist in the dimerization interface and, thus, the entire protein. Similar regions in each monomer are buried upon dimerization and the interfaces are both symmetrical. Specifically, the GFCC′C′′ strands of each CEACAM form the dimerization interface. The sequence identity between CEACAM5 and CEACAM6 is high (88.9%), with only 11 residues differing between the two proteins, of which only 3 (I30, L45, and Q90) are found in the dimerization interface (Fig. S1). Relative to CEACAM5, these differences result in the loss of π-π stacking and a hydrogen bond at positions 30 and 45, respectively, but the gain of a salt bridge at position 90 (Fig. 1D). Mutation of these residues individually in CEACAM5 (F29I, Q44L, and H89Q, with sequence numbering as for CEACAM5; there is a numbering discrepancy of one position in CEACAM6, such that position 44 in CEACAM5 is position 45 in CEACAM6) has shown only position 44 to be critical for dimerization, although the other two positions may regulate dimerization (19). In CEACAM5, Q44 forms a hydrogen bond to the carbonyl of the main chain at L95. As this contact is asymmetric, the contact is made twice by each molecule in the dimer, whereas residues at positions 30 and 90 form symmetrical interactions and, therefore, only one contact each in the dimer interface. The unique surface chemistry of CEACAM6 compared with CEACAM1 and CEACAM5 results in a difference in buried surface area. CEACAM6 buries 1,695 Å2 of surface area in the dimerization interface, whereas CEACAM1 and CEACAM5 bury 1,600 Å2 and 1,460 Å2 of surface area, respectively. Shape complementarity in the CEACAM6 homodimer interface (SC = 0.70) is lower than that of CEACAM5 (SC = 0.72) and CEACAM1 (SC = 0.81).
Table S1.
Crystallographic data collection and refinement statistics
| Data collection and refinement | CEACAM6 | CEACAM8 | CEACAM6/8 |
| Data collection | |||
| Space group | P63 | P4132 | P212121 |
| Unit-cell parameters (Å) | a = b = 96.66 c = 54.36 90.0°, 90.0°, 120.0° | a = b = c = 94.34 90.0°, 90.0°, 90.0° | a = 62.47 b = 97.19 c = 97.69 90.0°, 90.0°, 90.0° |
| Resolution (Å) | 83.71–1.83 (1.87–1.83) | 66.71–1.45 (1.47–1.45) | 68.90–1.85 (1.89–1.85) |
| No. of reflections | 73,551 | 1,274,216 | 342,059 |
| No. of unique reflections | 24,029 | 26,074 | 51,361 |
| Multiplicity | 3.1 (3.1) | 48.9 (49.0) | 6.7 (5.9) |
| Completeness (%) | 94.5 (95.1) | 100.0 (100.0) | 99.7 (99.1) |
| Rmerge | 0.125 (1.134) | 0.135 (4.627) | 0.091 (1.341) |
| Rmeas | 0.160 (1.431) | 0.138 (4.719) | 0.107 (1.618) |
| Rp.i.m | 0.099 (0.852) | 0.027 (0.927) | 0.056 (0.650) |
| <I>/<σ(I)> | 7.3 (1.50) | 27.0 (1.40) | 11.3 (1.2) |
| CC1/2 | 0.990 (0.516) | 1.000 (0.535) | 0.999 (0.482) |
| Wilson B factor (Å) | 16.4 | 15.1 | 25.9 |
| Refinement | |||
| Resolution (Å) | 83.71–1.83 | 66.71–1.45 | 68.90–1.85 |
| Rcryst (%) | 16.7 | 14.5 | 18.6 |
| Rfree (%) | 19.5 | 18.0 | 21.3 |
| No. of reflections | |||
| Work set | 22,783 | 24,772 | 48,753 |
| Test set | 1,226 | 1,250 | 2,549 |
| No. of molecules/atoms | |||
| Protein | 2/1691 | 1/881 | 4/3404 |
| Water | 117 | 111 | 205 |
| RMSD | |||
| Bond lengths (Å) | 0.012 | 0.011 | 0.008 |
| Bond angles (°) | 1.530 | 1.510 | 1.230 |
| Mean B factor (Å2) | 22.8 | 21.6 | 18.0 |
Table S2.
Hydrogen bonds in CEACAM homo- and heterodimer complexes
| CEACAM | Atom (chain 1) | Distance (Å) | Atom (chain 2) |
| CEACAM1 | |||
| 1 | B:SER 32[ OG ] | 2.84 | A:LEU 95[ O ] |
| 2 | B:GLY 41[ N ] | 2.63 | A:GLU 99[ OE2 ] |
| 3 | B:GLN 44[ NE2 ] | 2.44 | A:LEU 95[ O ] |
| 4 | B:GLN 89[ NE2 ] | 2.68 | A:GLN 89[ OE1 ] |
| 5 | B:GLN 89[ NE2 ] | 3.48 | A:TYR 34[ OH ] |
| 6 | B:ASN 97[ ND2 ] | 3.14 | A:SER 32[ OG ] |
| 7 | B:ASN 97[ ND2 ] | 3.7 | A:TYR 34[ OH ] |
| 8 | B:LEU 95[ O ] | 2.73 | A:SER 32[ OG ] |
| 9 | B:GLU 99[ OE2 ] | 2.66 | A:GLY 41[ N ] |
| 10 | B:ASN 97[ O ] | 3.45 | A:GLN 44[ NE2 ] |
| 11 | B:ASP 94[ O ] | 3.71 | A:THR 56[ OG1 ] |
| 12 | B:ASP 94[ OD1 ] | 2.82 | A:THR 56[ OG1 ] |
| 13 | B:TYR 34[ OH ] | 3.72 | A:GLN 89[ NE2 ] |
| 14 | B:GLN 89[ OE1 ] | 2.82 | A:GLN 89[ NE2 ] |
| 15 | B:GLN 44[ OE1 ] | 3.07 | A:ASN 97[ N ] |
| 16 | B:SER 32[ OG ] | 3.7 | A:ASN 97[ ND2 ] |
| CEACAM5 | |||
| 1 | B:GLN 44[ NE2 ] | 2.97 | A:LEU 95[ O ] |
| 2 | B:ASN 97[ N ] | 2.92 | A:GLN 44[ OE1 ] |
| 3 | B:ASN 97[ ND2 ] | 3.1 | A:SER 32[ OG ] |
| 4 | B:LEU 95[ O ] | 2.92 | A:SER 32[ OG ] |
| 5 | B:LEU 95[ O ] | 2.72 | A:GLN 44[ NE2 ] |
| 6 | B:GLU 99[ OE2 ] | 2.75 | A:GLY 41[ N ] |
| CEACAM6 | |||
| 1 | B:LYS 2[ NZ ] | 3.04 | A:ASN 43[ OD1 ] |
| 2 | B:SER 33[ OG ] | 3 | A:LEU 96[ O ] |
| 3 | B:ARG 39[ NH1 ] | 3.44 | A:GLU 38[ O ] |
| 4 | B:GLY 42[ N ] | 2.84 | A:GLU 100[ OE2 ] |
| 5 | B:GLN 90[ NE2 ] | 2.86 | A:GLN 90[ OE1 ] |
| 6 | B:GLN 90[ NE2 ] | 3.46 | A:TYR 35[ OH ] |
| 7 | B:ASN 98[ ND2 ] | 3.25 | A:SER 33[ OG ] |
| 8 | B:ASN 98[ ND2 ] | 3.5 | A:GLN 90[ OE1 ] |
| 9 | B:LEU 96[ O ] | 2.67 | A:SER 33[ OG ] |
| 10 | B:GLU 100[ OE1 ] | 2.62 | A:GLY 42[ N ] |
| 11 | B:GLN 90[ OE1 ] | 2.99 | A:GLN 90[ NE2 ] |
| 12 | B:SER 33[ OG ] | 3.52 | A:ASN 98[ ND2 ] |
| CEACAM6/8 | |||
| 1 | D:ARG 39[ NH2 ] | 3.27 | C:GLU 38[ O ] |
| 2 | D:GLY 42[ N ] | 3.05 | C:GLU 100[ OE1 ] |
| 3 | D:GLN 90[ NE2 ] | 2.93 | C:GLN 90[ OE1 ] |
| 4 | D:GLN 90[ OE1 ] | 2.93 | C:GLN 90[ NE2 ] |
| 5 | D:LEU 96[ O ] | 2.66 | C:ASN 33[ ND2 ] |
| 6 | D:GLU 100[ OE1 ] | 2.83 | C:ALA 42[ N ] |
Fig. 1.
Crystal structure of CEACAM6. (A) The overall structural fold of the IgV domain of CEACAM6 with secondary elements labeled. (B) Structure of the CEACAM6 dimer. (C) Superposition of the CEACAM6 dimer (red chains) onto the CEACAM5 dimer (blue chains). (D) Differences in the dimerization interface result in the loss of π-π stacking (Left), gain of a salt bridge (Center), and loss of a hydrogen bond (Right) for CEACAM6 relative to CEACAM5.
Fig. S1.
Sequence alignment and interface residues of CEACAMs. Sequence alignment of CEACAMs using ClustalW. Secondary structure of CEACAMs are depicted above the alignments. Residues in red are buried in the dimerization interface (CEACAM6/8 and CEACAM8/6 describing residues of CEACAM6 and CEACAM8 in the CEACAM6–CEACAM8 complex, respectively). The number of amino acid exchanges relative to CEACAM5 are shown at the right. Potential glycosylation sites are shown in green.
CEACAM6 Is a Weak Homodimer in Solution.
To determine whether CEACAM6 homodimerizes in solution as well as in crystals, we measured the CEACAM6 dimerization constant (Kdimerization) by sedimentation equilibrium analysis using analytical ultracentrifugation (AUC) (Fig. 2A). By globally fitting these data to a monomer-dimer self-association model, we estimated an average molecular weight of 15.7 ± 0.7 kDa [theoretical molecular weight (MW) = 12,035 Da] and a Kdimerization of 60 ± 15 μM, demonstrating that CEACAM6 dimerizes in solution, albeit weakly. Using the same AUC methods, we found that CEACAM1 and CEACAM5 form 130- and 50-fold tighter homodimers, respectively, than does CEACAM6; CEACAM1 exhibited an average molecular weight of 23.0 ± 0.9 kDa (theoretical MW = 12,093 Da) and a Kdimerization of 450 nM (+370/−230 nM), whereas CEACAM5 exhibited an average molecular weight of 23.0 ± 0.8 kDa (theoretical MW = 12,581 Da) and a Kdimerization of 1.3 ± 0.6 μM (Fig. 2 B and C). These data indicate that small differences in the dimerization interfaces among CEACAMs can significantly alter their propensities to homodimerize. A difference of just three amino acids between CEACAM6 and CEACAM5 is sufficient to weaken the Kdimerization by 50-fold, and results in a CEACAM6 homodimer that is structurally distinct from those formed by CEACAM1 and CEACAM5. We made alanine mutations of the three differing amino acids in the homodimerization interface (I30A, L45A, and Q90A) as well as the L96A mutation, which has been shown to be important for dimerization (19) and measured their dimerization constants by AUC (Fig. S2 and Table S3). We found that the L45A and Q90A mutation had little effect on dimerization with a Kdimerization of 61 μM (+24/−17 μM) and 48 μM (+12/−12 μM), respectively. However, the L96A mutation results in an ∼fivefold weakening of dimerization with a Kdimerization of 330 μM (+200/−120). The I30A mutation abolishes dimerization. Our mutational studies show that I30 is critical for CEACAM6 dimerization, whereas L96 is also important for self-association.
Fig. 2.
Oligomeric states of homotypic CEACAM preparations. Sedimentation equilibrium analyses (Upper) and residuals of the fits for each curve (Lower) for (A) CEACAM6, (B) CEACAM1, (C) CEACAM5, and (D) CEACAM8. (E) Kdimerization values and SDs for all homotypic CEACAM interactions.
Fig. S2.
Sedimentation equilibrium analyses (Upper) and residuals of fitted data for each curve (Lower) for CEACAM6 mutants (A) I30A, (B) L45A, (C) Q90A and (D) L96A, and CEACAM8 mutants (E) R45A, (F) Q90A, and (G) L96A.
Table S3.
Dimerization constants of CEACAM homo- and heterodimer complexes as measured by AUC or ITC, respectively
| Proteins | Method | Kdimerization (μM) | ΔH (kcal/mol−1) | TΔS (kcal/mol−1) | N |
| WT CEACAM6-WT CEACAM6 | AUC | 60 +15/−15 | |||
| WT CEACAM8-WT CEACAM8 | AUC | 650+350/−300 | |||
| CEACAM6 I30A-CEACAM6 I30A | AUC | Monomer | |||
| CEACAM6 L45A-CEACAM6 L45A | AUC | 61 +24/−17 | |||
| CEACAM6 Q90A-CEACAM6 Q90A | AUC | 48 +12/−12 | |||
| CEACAM6 L96A-CEACAM6 L96A | AUC | 330 +200/−120 | |||
| CEACAM8 R45A-CEACAM8 R45A | AUC | Monomer | |||
| CEACAM8 Q90A-CEACAM8 Q90A | AUC | Monomer | |||
| CEACAM8 L96A-CEACAM8 L96A | AUC | Monomer | |||
| WT CEACAM6-WT CEACAM8 | ITC | 2.0 ± 0.8 | −4.6 ± 0.3 | +3.2 | 1:1.03 ± 0.04 |
| WT CEACAM6-WT CEACAM8 glyco | ITC | 4.5 ± 0.5 | −6.4 ± 0.1 | +0.9 | 1:0.72 ± 0.02 |
| CEACAM6 I30A-WT CEACAM8 | ITC | No binding | |||
| CEACAM6 L45A-WT CEACAM8 | ITC | 12 ± 2 | −5.1 ± 0.4 | +1.6 | 1:1.12 ± 6 |
| CEACAM6 Q90A-WT CEACAM8 | ITC | 22 ± 6 | −3.3 ± 0.6 | +3.0 | 1:1.23 ± 0.12 |
| CEACAM6 L96A-WT CEACAM8 | ITC | No binding | |||
| WT CEACAM6-CEACAM8 R45A | ITC | No binding | |||
| WT CEACAM6-CEACAM8 Q90A | ITC | 0.8 ± 0.08 | −9.0 ± 0.1 | −0.7 | 1:0.9 ± 0.00 |
| WT CEACAM6-CEACAM8 L96A | ITC | No binding | |||
The CEACAM8 Crystal Structure Indicates a Physiological Monomer.
Relative to all other CEACAM crystal structures, we observed distinct crystallographic packing properties for CEACAM8 (Table S1), in which the interface was comprised of the GFCC′C′′ strands from one molecule of CEACAM8 and the AE strands of the second molecule (Fig. 3A), forming an asymmetrical dimer, unique for CEACAMs. This interface buries only 1,140 Å2 of surface area between two CEACAM8 molecules, significantly smaller than the other dimerization interfaces, suggesting an interaction dependent on crystal packing rather than a physiological complex. CEACAM8 has only 71% sequence identity to CEACAM5 (Fig. S1). Of the 30 differing residues, 9 are in the dimerization interface. We created a CEACAM8 homodimer model by superimposing two copies of CEACAM8 onto the CEACAM5 dimer and identified two residues, R45 and M97, which exhibit a steric clash (Fig. 3B), suggesting at least one structural incompatibility with the typical CEACAM dimerization interface. A previous study showed that the L44R mutation in CEACAM5 (position 45 in CEACAM8) results in a CEACAM5 monomer (19). Although CEACAM8 can homodimerize, albeit weakly, it is unknown whether the crystal structure of CEACAM8 represents the measured dimer as seen by AUC or is in fact a monomer. To test this we made two mutations, R45A and L96A, and measured their Kdimerization (Fig. S2). If CEACAM8 forms a canonical dimer like those observed in the CEACAM1, -5, and -6 structures, in which both R45A and L96A mutations make important dimerization contacts, it therefore would be monomeric as measured by AUC. However, if dimerization of CEACAM8 forms an asymmetrical dimer as described above, R45A would have no effect on dimerization as it is not buried in this dimerization interface. We found that both the R45A and L96A mutations resulted in monomeric CEACAM8, demonstrating that the observed asymmetrical dimer described above is actually monomeric and that dimeric CEACAM8 most likely forms a canonical CEACAM dimer similar to those observed for CEACAM1, -5, and -6. We made a further mutation in the CEACAM8 dimerization interface, Q90A and also found it to form monomers (Fig. S2).
Fig. 3.
Crystal structure of CEACAM8. (A) Crystal structure of CEACAM8 depicting an asymmetrical dimer due to crystal packing relative to CEACAM5 (superimposed onto the CEACAM8 structure shown in gray). (B) An artificial CEACAM8 homodimer modeled by superposition of CEACAM8 onto the CEACAM5 homodimer shows that residues R45 and M97 of CEACAM8 clash in the dimerization interface.
CEACAM8 Exists Predominately as a Monomer in Solution.
To confirm the monomeric state of CEACAM8 in solution, we performed sedimentation equilibrium analysis, by which we estimated a molecular weight of 13.0 ± 0.8 kDa (theoretical MW = 12,192 Da) and a Kdimerization of 650 μM (+350/−300 μM) for CEACAM8 (Fig. 2D). These data indicate that although CEACAM8 homodimerization can occur at the likely nonphysiologically high concentrations that we tested in our AUC experiments, it is most probable that it is found in an exclusively monomeric state. Furthermore, CEACAM8 exhibits the weakest homodimerization constant compared with the measured values of other CEACAMs, a 1,300-fold difference compared with the strongest, CEACAM1 (Fig. 2E).
CEACAM6 and CEACAM8 Form a Heterodimer.
Cell-based studies have shown that CEACAM6 and CEACAM8 can form heterodimers (20, 24). With such weak homodimerization constants as we measured for CEACAM6 and CEACAM8, a CEACAM6/8 heterodimer may represent an energetically more favorable interaction, which we tested using isothermal titration calorimetry (ITC). Upon titrating CEACAM8 into CEACAM6, we observed that this interaction is both enthalpically (ΔH = −4.6 kcal/mol−1) and entropically (−TΔS = −3.2 kcal/mol−1) driven, with a KD of 2.0 ± 0.8 μM (Fig. 4A), indicating that the CEACAM6/8 heterodimer is 30-fold tighter than the CEACAM6 homodimer. Several studies have noted that other CEACAM heterodimers can form, including: CEACAM6/1, CEACAM3/5, and CEACAM8/1 (20, 29). Accordingly, we also measured the affinities of these interactions by ITC but observed no heterodimer formation (Fig. S3 A–D). As CEACAMs are highly glycosylated proteins and our studies thus far had been conducted with unglycosylated proteins, we expressed, secreted, and purified CEACAM6 and CEACAM8 from HEK293 cells to investigate whether glycosylation has an effect on the CEACAM6–CEACAM8 interaction. Both N-terminal domains are predicted to have three N-linked glycosylation sites (Fig. S1). A silver-stained SDS/PAGE gel showed that both proteins are glycosylated and polydisperse (Fig. S3E). Titration of glycosylated CEACAM8 into glycosylated CEACAM6 only results in a twofold reduction in affinity, with a KD of 4.5 ± 0.5 μM, and remains both enthalpically (ΔH = −6.4 kcal/mol−1) and entropically (TΔS = 0.9 kcal/mol−1) driven (Fig. 4B). However, the stoichiometry of the interaction changed from 1:1 to 1:0.7, suggesting some inactive species are present.
Fig. 4.
ITC binding curve of (A) nonglycosylated CEACAM8 titrated into nonglycosylated CEACAM6 and (B) glycosylated CEACAM8 titrated in to glycosylated CEACAM6.
Fig. S3.
ITC binding curve of (A) CEACAM1 titrated into CEACAM6, (B) CEACAM1 titrated into CEACAM8, (C) CEACAM3 titration into CEACAM6, and (D) CEACAM5 titrated into CEACAM6. (E) Silver-stained SDS/PAGE gel of glycosylated CEACAM6 and CEACAM8 either treated (+) or not treated (−) with PNGaseF.
To gain further insight to the CEACAM6/8 heterodimer, we solved its crystal structure to a resolution of 1.85 Å (Fig. 5A and Table S1). The heterodimer complex superimposes onto the homodimers of CEACAM1, -5, and -6 (CEACAM6 of the heterodimer onto a single chain of the homodimer) with low RMSDs of 0.5, 0.4, and 0.4 Å, respectively. The RMSD of CEACAM8 with the second chain of the homodimers is much higher, with values of 3.0, 4.6, and 2.6 Å, for CEACAM1, -5, and -6, respectively (Fig. 5B). This deviation is greater than that observed for any of the CEACAM homodimers, although the heterodimerization interface remains similar to them. It buries a smaller surface of 1,450 Å2 and exhibits an SC value of 0.58, smaller than the homodimers. The dimerization interface is still comprised of the GFCC′C′′ strands from both CEACAMs, which is surprising as CEACAM8 does not homodimerize using these strands. Unlike the modeled CEACAM8 homodimer (Fig. 3B) in which R45 and M97 exhibit steric clashes, however, we observed in the CEACAM6/8 heterodimer that R45 and M97 of CEACAM8 pack against V97 and L45 of CEACAM6, respectively, forming van der Waals interactions (Fig. 5C).
Fig. 5.
Crystal structure of the CEACAM6/8 complex. (A) Structure of the CEACAM6–CEACAM8 heterodimer. CEACAM6 and CEACAM8 are cyan and green, respectively. (B) Superposition of the CEACAM6–CEACAM8 complex (cyan and green, respectively), CEACAM6 dimer (magenta), and onto the CEACAM5 dimer (gray). (C) 2Fo–Fc composite omit maps of (Left) residues 42–48 of CEACAM6 (cyan), and 93–99 of CEACAM8 (yellow), and (Right) of residues 93–99 of CEACAM6 (cyan) and 42–48 of CEACAM8 (yellow).
All seven mutations that we made to investigate their role in homodimerization (I30A, L45A, Q90A, and L96A for CEACAM6 and R45A, Q90A, and L96A for CEACAM8) are found buried in the heterodimeric interface as well. We tested the effect that these mutations had on the formation of the CEACAM6–CEACAM8 complex by ITC (Fig. S4). We found that the I30A and L96A mutations in CEACAM6, and the R45A and L96A mutations in CEACAM8, abolish complex formation. We also found that the Q90A mutation in CEACAM8 results in 2.5-fold increase in affinity with a Kdimerization of 800 nM, whereas the L45A and Q90A mutations in CEACAM6 result in a weakening of affinity with a Kdimerization of 12 μM and 22 μM, respectively. Mutations in CEACAM6 that disrupt (I30A) or weaken (L96A) homodimerization also prevented heterodimerization. The two CEACAM6 mutations that had little effect on homodimerization (L45A and Q90A) weakened heterodimerization only slightly. All CEACAM8 mutations produced monomeric proteins. However, one mutation (Q90A) increases the affinity of heterodimerization, whereas the other two (R45 and L96A) results in no heterodimer formation. Taken together, these data suggest that the chemistries of both homo- and heterodimerization interfaces are the same.
Fig. S4.
ITC binding curve of WT CEACAM8 being titrated into (A) CEACAM6 I30A, (B) CEACAM6 L45A, (C) CEACAM6 Q90A, and (D) CEACAM6 L96A. (E) CEACAM8 R45A, (F) CEACAM8 Q90A, and (G) CEACAM8 L96A were titrated against WT CEACAM6.
Discussion
CEACAMs are involved in and regulate diverse cellular functions, including cell adhesion and tumor suppression (10, 30). Cell adhesion is achieved through dimerization in a trans arrangement via the N-terminal IgV domains of CEACAMs (21, 22). Thus far, only CEACAM1 and CEACAM5 homodimers have been described structurally, with CEACAM5 forming homodimers with an affinity of 1 μM and CEACAM1 potentially forming higher-order oligomers (19). In this study, we measured the affinity of the homodimerization events for CEACAM1 and CEACAM5 as 450 nM and 1.3 μM, respectively. This is much tighter than what we observed for CEACAM6, which forms a weak homodimer, and CEACAM8, which is effectively monomeric. These homodimerization constants span a range greater than three orders-of-magnitude, despite being modulated by only a few amino acid differences in the interface. All of our studies used only CEACAM N-terminal IgV domains. Notably, several studies have suggested that the presence of multiple IgC2 domains, each separated by flexible linkers, or the transmembrane motif of CEACAM1, GXXXG, increases the affinity of homophilic binding (21, 31, 32). These interactions could potentially strengthen cis engagements. Furthermore, the CEACAM1 ectodomain (IgV+3IgC2) in liposomes form clusters of cis dimers (33). Formation of trans homodimers in these liposomes increases the amount of cis dimer but disrupts the clusters (33), suggesting that the N-terminal domain is flexible enough around the IgC2 domains to allow both cis and trans homodimer interactions.
Using ITC, we observed a heterodimeric interaction between CEACAM6 and CEACAM8 that was energetically favored over any other state of either protein. Glycosylation of CEACAMs occurs in vivo with at least 35 different glycosylation forms identified for CEACAM1 (34). Glycosylation of CEACAM6 and CEACAM6 only slightly weakened the interaction; however, we did observe a change in the stoichiometry. We suspect that certain glycosylation forms of CEACAMs create folded yet inactivate CEACAM molecules that could potentially prevent the formation of the CEACAM6–CEACAM8 interaction and possibly CEACAM homodimers.
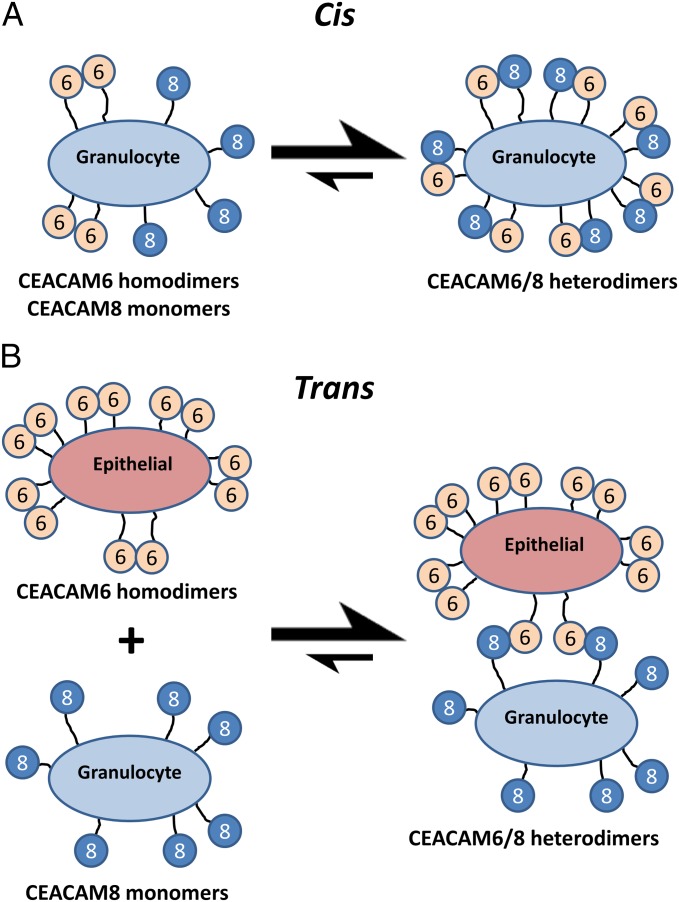
Our data suggest that CEACAM6 and CEACAM8 likely function as regulators of both cis and trans interactions for one another, as well as potentially for other CEACAMs that bind to either or both of them. For example, a granulocyte expressing both CEACAM6 and CEACAM8 would predominately form cis CEACAM6/8 heterodimers (Fig. 6A), inhibiting CEACAM6 from engaging in trans interactions. As CEACAM8 is expressed exclusively on granulocytes and CEACAM6 is expressed additionally on epithelial cells of the gastrointestinal tract, a trans CEACAM6/8 heterodimer (Fig. 6B) could provide a mechanism by which the immune system can engage the epithelia.
Fig. 6.
Effects of CEACAM6 homodimers, CEACAM8 monomers and CEACAM6/8 heterodimers on cis and trans interactions. (A) CEACAM6 and CEACAM8 expressed on the same granulocyte energetically favors formation of cis CEACAM6/8 heterodimers. (B) Expression of CEACAM6 and CEACAM8 on epithelial cells and granulocytes, respectively, energetically favors formation of trans CEACAM6/8 heterodimers.
Disruption of the CEACAM6/8 heterodimer has the potential to increase the pool of free CEACAM6 and CEACAM8 to interfere with other CEACAM functions. Several studies have shown that CEACAM6 also forms heterodimers with CEACAM1, CEACAM5, and CEACAM3 (20, 30), whereas CEACAM8 can form a heterodimer with CEACAM1 (35). Although we observed no formation of these other heterodimers by ITC, we cannot dismiss the possibility that these heterodimers do form, but that they must do so at concentrations higher than the maximum (35 μM) that we used in our ITC experiments. The presence of IgC2 domains, or the formation of clusters and therefore an increase in the local concentration of these proteins, may make the formation of heterodimers more energetically favorable. Several splice variants of CEACAM1 exist, with the long form containing a cytosolic immunoreceptor tyrosine-based inhibitory motif, allowing CEACAM1 to signal and several short forms that cannot signal (7, 8). CEACAM6 and CEACAM8 are anchored in the membrane by a GPI moiety and therefore cannot signal in the same manner as does long-form CEACAM1 (6). Overexpression of the short form of CEACAM1 interferes with CEACAM1 signaling (36). Assuming that CEACAM6 and CEACAM8 can interact with CEACAM1 at high concentrations on the cell surface, they too have the potential to disrupt CEACAM1 signaling through formation of heterodimers. Furthermore, the CEACAM1/8 interaction has been shown to inhibit Toll-like receptor 2-triggered immune responses (37). Thus, CEACAM6 could potentially modulate Toll-like receptor-2 inhibition through its recruitment of CEACAM8.
In summary, we present the X-ray crystal structures of homodimeric CEACAM6, monomeric CEACAM8, and heterodimeric CEACAM6/8. Coupled with our quantitative biophysical analyses of these CEACAMs with themselves and others, we provide a molecular basis for the diverse oligomeric states of CEACAM IgV domains that are important for cell adhesion and signaling in cancer, infection and immunity.
Materials and Methods
For details, see SI Materials and Methods.
Protein Production.
All nonglycosylated CEACAM N-terminal IgV domains were expressed in E. coli and refolded in vitro from inclusion bodies, and were purified to homogeneity by ion exchange and size-exclusion chromatography. Glycosylated CEACAM N-terminal IgV domains were expressed in HEK cells and were purified to homogeneity by nickel affinity and size-exclusion chromatography.
Structural Analysis.
CEACAM6 and CEACAM8 N-terminal domains were concentrated individually to 6 mg/mL or together to 9 mg/mL and crystallized by vapor diffusion in conditions optimized from the JCSG+ screen (Qiagen). Single crystals were flash-cooled in liquid nitrogen in mother liquor containing 20–30% (vol/vol) glycerol. X-ray diffraction data were collected at 100K at the Advanced Photon Source beamline 23-ID-B and the Stanford Synchrotron Radiation Laboratory beamlines 7-1 and 12-2. CEACAM6 and CEACAM8 structures were determined by molecular replacement using a monomer of CEACAM5 (PDB ID code 2QSQ) as a search model and final models built by iterative cycles of manual building in Coot (38) and refinement in Refmac5 (39). The CEACAM6/8 structure was determined by molecular replacement using the refined CEACAM6 and CEACAM8 structures as search models; refinement proceeded as above.
Biophysical Analysis.
Sedimentation equilibrium measurements of CEACAMs were performed using a Beckman-Coulter XL-I analytical ultracentrifuge at 20 °C. Each CEACAM protein sample was prepared at three concentrations and centrifuged at three rotor speeds. Scans were acquired at 280 nm with a step size of 0.001 and five averages per step. The data were globally analyzed using the program WinNonLin (40). ITC experiments were performed using an iTC200 instrument (Malvern). A typical experiment was carried out by titrating CEACAM6 or CEACAM8 loaded in the syringe into a cell loaded with CEACAM1, CEACAM3, CEACAM5, or CEACAM6 at ∼10:1 molar ratios at 25 °C. Heats of dilutions were measured and subtracted from each data set. When no binding was observed at 25 °C measurements were also made at 10 °C. All data were analyzed using Origin 7.0 software.
SI Materials and Methods
Protein Expression and Purification.
Nonglycosylated CEACAM N-terminal domain genes were synthesized as codon-optimized GeneArt Strings (Life Technologies), digested and cloned into an NcoI/XhoI cut pET21d vector with no purification tag. All CEACAM N-terminal domain proteins were expressed in Escherichia coli BL21(DE3) pLysS cells. Cells were grown at 37 °C until an A600nm of 0.6. Protein expression was induced with 1 mM isfopropyl-β-d-thiogalactopyranoside and cells were grown for an additional 4 h at 37 °C. Cells were harvested and resuspended in 50 mM Tris, 500 mM NaCl, 1% (vol/vol) Triton-X 100, pH 7.5 (ice-cold) and lysed by sonication. The soluble fraction was discarded and inclusion bodies were resuspended, sonicated, and pelleted by centrifugation in the same buffer. The pelleted inclusion bodies were then washed with 50 mM Tris, 1,000 mM NaCl, pH 8.0, followed by 50 mM Tris, 500 mM NaCl, pH 7.5. After solubilization in 30 mM Tris, 150 mM NaCl, 8.0 M Urea, pH 8.3 (∼5 mL per liter of cell pellet), the protein was refolded 4 °C by dilution (1:12) into 50 mM CHES, 500 mM l-Arginine, pH 9.2. Refolding was left overnight before dialysis against 10 mM Tris, pH 8.0. Protein was concentrated by anion exchange and purified by size-exclusion chromatography using a MonoQ and Superdex 200 column (GE Healthcare), respectively. For the production of glycosylated CEACAM N-terminal domain proteins, the genes were cloned with an N-terminal hexahistidine tag into an AgeI/NotI cut pSG160 vector. pSG160 is a modified pcDNA4/to vector containing a μ-phosphatase secretion peptide cloned from pHLsec (41). For protein expression of glycosylated CEACAMs, HEK cells grown in FreeStyle 293 with GlutaMAX (Life Technologies) and Gentamicin were transfected with plasmid using polyethyleneimine. HEK cells were grown for 5 d at 37 °C with 8% CO2 before harvesting of the supernatant. Protein was purified by nickel affinity and size-exclusion chromatography using a 5-mL HisTrap Excel and Superdex 200 column (GE Healthcare).
Crystallization.
Nonglycosylated CEACAM N-terminal domains were dialyzed against 20 mM Tris, 100 mM NaCl, pH 7.5 and were screened against the JCSG+ screen (Qiagen) at 6 mg/mL−1 with a protein:reservoir ratio of 150 nL:150 nL at room temperature. Crystals for CEACAM6 grew in 10% (wt/vol) PEG 3000, 0.1 M sodium acetate, pH 4.6, 0.2 M zinc acetate; and CEACAM8 grew in 0.8 M ammonium sulfate, 0.1 M citric acid, pH 3.5. Crystals of CEACAM6 and CEACAM8 were optimized and grew in final conditions of 10% (wt/vol) PEG 8000, 0.1 M sodium acetate, pH 4.6, 0.2 M zinc acetate; and 1.2 M ammonium sulfate, 0.1 M citric acid, pH 3.5, respectively. Single crystals of CEACAM6 were washed in the same condition containing 20% (vol/vol) glycerol before being flash-cooled in liquid nitrogen. Single crystals of CEACAM8 were washed in increasing concentrations of ammonium sulfate and glycerol in increments of 10% to a final concentration of 30% (vol/vol) glycerol before being flash-cooled in liquid nitrogen. The CEACAM6–CEACAM8 complex was prepared by mixing the individual CEACAMs in a 1:1 ratio and dialyzed against 20 mM Tris, 100 mM NaCl, pH 7.5. The complex was screened against the JCSG+ screen (Qiagen) at 9 mg/mL−1 with a protein:reservoir ratio of 150 nL:30 nL. Crystals grew in 20% (wt/vol) PEG 8000, 0.1 M sodium phosphate/citric acid, pH 4.2, 0.2 M NaCl. Crystals were not further optimized. Single crystals were washed in 5% (wt/vol) PEG 8000, 20 mM sodium phosphate/citric acid, pH 4.2, 40 mM NaCl, 5% (vol/vol) glycerol before transferring to the same solution with 20% (vol/vol) glycerol and flash-cooled in liquid nitrogen. Datasets were collected at 100 K on either beamline 23-ID-B at the Advanced Photon Source (wavelength = 1.0332 Å) using a MAR 300 CCD detector, beamline 7-1 at the Stanford Synchrotron Radiation Laboratory (wavelength = 1.0331 Å) using an ADSC Quantum 315r CCD detector or beamline 12-2 at the Stanford Synchrotron Radiation Laboratory (wavelength = 0.97950 Å) using a PILATUS 6M PAD detector.
CEACAM Structure Determination.
Datasets were either processed with HKL2000, iMOSFLM, or XDS and scaled and truncated with either HKL2000 or Aimless (42–44). The program CHAINSAW was used to prune nonconserved residues to the gamma atom of a monomer of CEACAM5 (PDB ID code 2QSQ) to generate a search model for CEACAM6 and CEACAM8 (45). The structures of CEACAM6 and CEACAM8 were solved by molecular replacement using MOLREP and the search model (46). The models were manually rebuilt in Coot (38) and refined with Refmac5 (39), with final Ramachandran statistics of 95.8% favored and 4.2% allowed, and 96.4% favored and 3.6% allowed, for CEACAM6 and CEACAM8, respectively. The structure of the CEACAM6–CEACAM8 complex was solved by molecular replacement using the solved structures of CEACAM6 and CEACAM8. The model was manually rebuilt in Coot and refined with Refmac5 with final Ramachandran statistics of 97.0% favored and 3.0% allowed. Data collection and refinement statistics are shown in Table S1. Structures have been deposited at the protein data bank with PDB ID codes of 4Y8A, 4Y88, and 4YIQ for CEACAM6, CEACAM8, and the CEACAM6–CEACAM8 complex, respectively.
Analytical Ultracentrifugation.
Sedimentation equilibrium measurements of CEACAMs were performed using an Optima XL-I analytical ultracentrifuge equipped with a four-hole An-60Ti Rotor (Beckman-Coulter) at 20 °C. Proteins were extensively dialyzed against buffer containing 50 mM Tris, pH 7.5, 50 mM NaCl. SedenTerp (sednterp.unh.edu) was used to calculate protein partial specific volume and solvent density values from the protein amino acid sequences and buffer composition, respectively. Protein samples were prepared at three concentrations (approximately 12, 18, and 30 uM) and loaded into cells equipped with six-hole charcoal-filled epon centerpieces (1.2-cm pathlength) with sapphire windows. Centrifugation was carried out at 29,000, 32,000, and 35,000 rpm or 28,000, 31,000, and 34,000 rpm, and scans were acquired at 280 nm with a step size of 0.001 and five averages per step. The data were globally analyzed to both single species and monomer-dimer self-association models using the program WinNonLin (40).
Isothermal Titration Calorimetry.
Purified CEACAMs were dialyzed against 30 mM Tris, 150 mM NaCl, pH 7.5. ITC experiments were performed using an iTC200 instrument (GE Healthcare). A typical experiment was carried out with the syringe loaded with CEACAM6 or CEACAM8 (360–380 μM) and the cell loaded with CEACAM1, CEACAM3, CEACAM5, or CEACAM6 at 35 μM. Titrations were performed at 25 °C with 16 injections of 2.42-μL aliquots, with 230-s intervals between injections. Heats of dilutions were also measured and subtracted from each dataset. When no binding was observed at 25 °C, measurements were also made at 10 °C. All data were analyzed using Origin 7.0 software.
Acknowledgments
This research was supported in part by the NIH under Grant S10RR15899 (to D.B.). This research used resources of the Advanced Photon Source, a US Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract no. DE-AC02-06CH11357. Use of the Stanford Synchrotron Radiation Lightsource, Stanford Linear Accelerator Center National Accelerator Laboratory, is supported by the DOE, Office of Science, Office of Basic Energy Sciences under Contract no. DE-AC02-76SF00515. The Stanford Synchrotron Radiation Lightsource Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research, and by the NIH, National Institute of General Medical Sciences (including P41GM103393).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Database deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org [PDB ID codes 4Y8A (CEACAM6), 4Y88 (CEACAM8), 4YIQ (CEACAM6–CEACAM8)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1509511112/-/DCSupplemental.
References
- 1.Zebhauser R, et al. Identification of a novel group of evolutionarily conserved members within the rapidly diverging murine Cea family. Genomics. 2005;86(5):566–580. doi: 10.1016/j.ygeno.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 2.Hammarström S. The carcinoembryonic antigen (CEA) family: Structures, suggested functions and expression in normal and malignant tissues. Semin Cancer Biol. 1999;9(2):67–81. doi: 10.1006/scbi.1998.0119. [DOI] [PubMed] [Google Scholar]
- 3.Zheng J, et al. Carcinoembryonic antigen-related cell adhesion molecule 16 interacts with alpha-tectorin and is mutated in autosomal dominant hearing loss (DFNA4) Proc Natl Acad Sci USA. 2011;108(10):4218–4223. doi: 10.1073/pnas.1005842108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schmitter T, Agerer F, Peterson L, Munzner P, Hauck CR. Granulocyte CEACAM3 is a phagocytic receptor of the innate immune system that mediates recognition and elimination of human-specific pathogens. J Exp Med. 2004;199(1):35–46. doi: 10.1084/jem.20030204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Singer BB, et al. Carcinoembryonic antigen-related cell adhesion molecule 1 expression and signaling in human, mouse, and rat leukocytes: Evidence for replacement of the short cytoplasmic domain isoform by glycosylphosphatidylinositol-linked proteins in human leukocytes. J Immunol. 2002;168(10):5139–5146. doi: 10.4049/jimmunol.168.10.5139. [DOI] [PubMed] [Google Scholar]
- 6.Tchoupa AK, Schuhmacher T, Hauck CR. Signaling by epithelial members of the CEACAM family—Mucosal docking sites for pathogenic bacteria. Cell Commun Signal. 2014;12:27. doi: 10.1186/1478-811X-12-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gray-Owen SD, Blumberg RS. CEACAM1: Contact-dependent control of immunity. Nat Rev Immunol. 2006;6(6):433–446. doi: 10.1038/nri1864. [DOI] [PubMed] [Google Scholar]
- 8.Singer BB, et al. Deregulation of the CEACAM expression pattern causes undifferentiated cell growth in human lung adenocarcinoma cells. PLoS One. 2010;5(1):e8747. doi: 10.1371/journal.pone.0008747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Singer BB, et al. CEACAM1 (CD66a) mediates delay of spontaneous and Fas ligand-induced apoptosis in granulocytes. Eur J Immunol. 2005;35(6):1949–1959. doi: 10.1002/eji.200425691. [DOI] [PubMed] [Google Scholar]
- 10.Benchimol S, et al. Carcinoembryonic antigen, a human tumor marker, functions as an intercellular adhesion molecule. Cell. 1989;57(2):327–334. doi: 10.1016/0092-8674(89)90970-7. [DOI] [PubMed] [Google Scholar]
- 11.Virji M, et al. Carcinoembryonic antigens are targeted by diverse strains of typable and non-typable Haemophilus influenzae. Mol Microbiol. 2000;36(4):784–795. doi: 10.1046/j.1365-2958.2000.01885.x. [DOI] [PubMed] [Google Scholar]
- 12.Hill DJ, Virji M. A novel cell-binding mechanism of Moraxella catarrhalis ubiquitous surface protein UspA: Specific targeting of the N-domain of carcinoembryonic antigen-related cell adhesion molecules by UspA1. Mol Microbiol. 2003;48(1):117–129. doi: 10.1046/j.1365-2958.2003.03433.x. [DOI] [PubMed] [Google Scholar]
- 13.Leusch HG, Drzeniek Z, Markos-Pusztai Z, Wagener C. Binding of Escherichia coli and Salmonella strains to members of the carcinoembryonic antigen family: Differential binding inhibition by aromatic alpha-glycosides of mannose. Infect Immun. 1991;59(6):2051–2057. doi: 10.1128/iai.59.6.2051-2057.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Obrink B. On the role of CEACAM1 in cancer. Lung Cancer. 2008;60(3):309–312. doi: 10.1016/j.lungcan.2008.03.020. [DOI] [PubMed] [Google Scholar]
- 15.Litkouhi B, et al. Overexpression of CEACAM6 in borderline and invasive mucinous ovarian neoplasms. Gynecol Oncol. 2008;109(2):234–239. doi: 10.1016/j.ygyno.2008.01.031. [DOI] [PubMed] [Google Scholar]
- 16.Duxbury MS, et al. A novel role for carcinoembryonic antigen-related cell adhesion molecule 6 as a determinant of gemcitabine chemoresistance in pancreatic adenocarcinoma cells. Cancer Res. 2004;64(11):3987–3993. doi: 10.1158/0008-5472.CAN-04-0424. [DOI] [PubMed] [Google Scholar]
- 17.Duxbury MS, Ito H, Benoit E, Ashley SW, Whang EE. CEACAM6 is a determinant of pancreatic adenocarcinoma cellular invasiveness. Br J Cancer. 2004;91(7):1384–1390. doi: 10.1038/sj.bjc.6602113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fedarovich A, Tomberg J, Nicholas RA, Davies C. Structure of the N-terminal domain of human CEACAM1: Binding target of the opacity proteins during invasion of Neisseria meningitidis and N. gonorrhoeae. Acta Crystallogr D Biol Crystallogr. 2006;62(Pt 9):971–979. doi: 10.1107/S0907444906020737. [DOI] [PubMed] [Google Scholar]
- 19.Korotkova N, et al. Binding of Dr adhesins of Escherichia coli to carcinoembryonic antigen triggers receptor dissociation. Mol Microbiol. 2008;67(2):420–434. doi: 10.1111/j.1365-2958.2007.06054.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oikawa S, et al. A specific heterotypic cell adhesion activity between members of carcinoembryonic antigen family, W272 and NCA, is mediated by N-domains. J Biol Chem. 1991;266(13):7995–8001. [PubMed] [Google Scholar]
- 21.Watt SM, et al. Homophilic adhesion of human CEACAM1 involves N-terminal domain interactions: Structural analysis of the binding site. Blood. 2001;98(5):1469–1479. doi: 10.1182/blood.v98.5.1469. [DOI] [PubMed] [Google Scholar]
- 22.Zhou H, Fuks A, Alcaraz G, Bolling TJ, Stanners CP. Homophilic adhesion between Ig superfamily carcinoembryonic antigen molecules involves double reciprocal bonds. J Cell Biol. 1993;122(4):951–960. doi: 10.1083/jcb.122.4.951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huber M, et al. The carboxyl-terminal region of biliary glycoprotein controls its tyrosine phosphorylation and association with protein-tyrosine phosphatases SHP-1 and SHP-2 in epithelial cells. J Biol Chem. 1999;274(1):335–344. doi: 10.1074/jbc.274.1.335. [DOI] [PubMed] [Google Scholar]
- 24.Kuroki M, et al. Identification and comparison of residues critical for cell-adhesion activities of two neutrophil CD66 antigens, CEACAM6 and CEACAM8. J Leukoc Biol. 2001;70(4):543–550. [PubMed] [Google Scholar]
- 25.Duxbury MS, et al. Overexpression of CEACAM6 promotes insulin-like growth factor I-induced pancreatic adenocarcinoma cellular invasiveness. Oncogene. 2004;23(34):5834–5842. doi: 10.1038/sj.onc.1207775. [DOI] [PubMed] [Google Scholar]
- 26.Schölzel S, et al. Carcinoembryonic antigen family members CEACAM6 and CEACAM7 are differentially expressed in normal tissues and oppositely deregulated in hyperplastic colorectal polyps and early adenomas. Am J Pathol. 2000;156(2):595–605. doi: 10.1016/S0002-9440(10)64764-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Duxbury MS, Ito H, Zinner MJ, Ashley SW, Whang EE. CEACAM6 gene silencing impairs anoikis resistance and in vivo metastatic ability of pancreatic adenocarcinoma cells. Oncogene. 2004;23(2):465–473. doi: 10.1038/sj.onc.1207036. [DOI] [PubMed] [Google Scholar]
- 28.Lasa A, et al. High expression of CEACAM6 and CEACAM8 mRNA in acute lymphoblastic leukemias. Ann Hematol. 2008;87(3):205–211. doi: 10.1007/s00277-007-0388-1. [DOI] [PubMed] [Google Scholar]
- 29.Skubitz KM, Skubitz AP. Interdependency of CEACAM-1, -3, -6, and -8 induced human neutrophil adhesion to endothelial cells. J Transl Med. 2008;6:78. doi: 10.1186/1479-5876-6-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oikawa S, et al. Cell adhesion activity of non-specific cross-reacting antigen (NCA) and carcinoembryonic antigen (CEA) expressed on CHO cell surface: Homophilic and heterophilic adhesion. Biochem Biophys Res Commun. 1989;164(1):39–45. doi: 10.1016/0006-291x(89)91679-3. [DOI] [PubMed] [Google Scholar]
- 31.Stern N, et al. Carcinoembryonic antigen (CEA) inhibits NK killing via interaction with CEA-related cell adhesion molecule 1. J Immunol. 2005;174(11):6692–6701. doi: 10.4049/jimmunol.174.11.6692. [DOI] [PubMed] [Google Scholar]
- 32.Patel PC, et al. Inside-out signaling promotes dynamic changes in the carcinoembryonic antigen-related cellular adhesion molecule 1 (CEACAM1) oligomeric state to control its cell adhesion properties. J Biol Chem. 2013;288(41):29654–29669. doi: 10.1074/jbc.M113.504639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klaile E, et al. The CEACAM1 N-terminal Ig domain mediates cis- and trans-binding and is essential for allosteric rearrangements of CEACAM1 microclusters. J Cell Biol. 2009;187(4):553–567. doi: 10.1083/jcb.200904149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lucka L, et al. Identification of Lewis X structures of the cell adhesion molecule CEACAM1 from human granulocytes. Glycobiology. 2005;15(1):87–100. doi: 10.1093/glycob/cwh139. [DOI] [PubMed] [Google Scholar]
- 35.Jiang L, Barclay AN. Identification of leucocyte surface protein interactions by high-throughput screening with multivalent reagents. Immunology. 2010;129(1):55–61. doi: 10.1111/j.1365-2567.2009.03153.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Müller MM, Klaile E, Vorontsova O, Singer BB, Obrink B. Homophilic adhesion and CEACAM1-S regulate dimerization of CEACAM1-L and recruitment of SHP-2 and c-Src. J Cell Biol. 2009;187(4):569–581. doi: 10.1083/jcb.200904150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Singer BB, et al. Soluble CEACAM8 interacts with CEACAM1 inhibiting TLR2-triggered immune responses. PLoS One. 2014;9(4):e94106. doi: 10.1371/journal.pone.0094106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Murshudov GN, et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr. 2011;67(Pt 4):355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Johnson ML, Correia JJ, Yphantis DA, Halvorson HR. Analysis of data from the analytical ultracentrifuge by nonlinear least-squares techniques. Biophys J. 1981;36(3):575–588. doi: 10.1016/S0006-3495(81)84753-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aricescu AR, Lu W, Jones EY. A time- and cost-efficient system for high-level protein production in mammalian cells. Acta Crystallogr D Biol Crystallogr. 2006;62(Pt 10):1243–1250. doi: 10.1107/S0907444906029799. [DOI] [PubMed] [Google Scholar]
- 42.Otwinowski Z, Minor W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods in Enzymology, ed Carter CW Jr. (Academic, Amsterdam), Vol 276, pp 307–326.
- 43.Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67(Pt 4):235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stein N. CHAINSAW: A program for mutating pdb files used as templates in molecular replacement. J Appl Cryst. 2008;41(3):641–643. [Google Scholar]
- 46.Vagin A, Teplyakov A. MOLREP: An automated program for molecular replacement. J Appl Cryst. 1997;30(6):1022–1025. [Google Scholar]