Abstract
Metastatic colorectal cancer (mCRC) carries a poor prognosis with an overall 5-year survival of 13.1%. Therapies guided by tumor profiling have suggested benefit in advanced cancer. We used a multiplatform molecular profiling (MP) approach to identify key molecular changes that may provide therapeutic options not typically considered in mCRC. We evaluated 6892 mCRC referred to Caris Life Sciences by MP including sequencing (Sanger/NGS), immunohistochemistry (IHC) and in-situ hybridization (ISH). mCRC metastases to liver, brain, ovary or lung (n = 1507) showed differential expression of markers including high protein expression of TOPO1 (52%) and/or low RRM1 (57%), TS (71%) and MGMT (39%), suggesting possible benefit from irinotecan, gemcitabine, 5FU/capecitabine and temozolomide, respectively. Lung metastases harbored a higher Her2 protein expression than the primary colon tumors (4% vs. 1.8%, p = 0.028). Brain and lung metastases had higher KRAS mutations than other sites (65% vs 59% vs 47%, respectively, p = 0.07, <0.01), suggesting poor response to anti-EGFR therapies. BRAF-mutated CRC (n = 455) showed coincident high protein expression of RRM1 (56%), TS (53%) and low PDGFR (22%) as compared with BRAF wild-type tumors. KRAS-mutated mCRC had higher protein expression of c-MET (47% vs. 36%) and lower MGMT (56% vs. 63%), suggesting consideration of c-MET inhibitors and temozolomide. KRAS-mutated CRC had high TUBB3 (42% vs. 33%) and low Her2 by IHC (0.5%) and HER2 by FISH (3%, p <0.05). CRC primaries had a lower incidence of PIK3CA and BRAF mutations in rectal cancer versus colon cancer (10% and 3.3%, respectively). MP of 6892 CRCs identified significant differences between primary and metastatic sites and among BRAF/KRAS sub-types. Our findings are hypothesis generating and need to be examined in prospective studies. Specific therapies may be considered for different actionable targets in mCRC as revealed by MP.
Keywords: APC, BRAF, colorectal cancer, c-MET, Cox2, ERCC1, Her2, irinotecan, KRAS, metastasis, molecular profiling, oxaliplatin, peritoneal cancer, precision medicine, TP53, Topo1
Introduction
Colorectal cancer (CRC) is the second leading cause of cancer-related deaths in the United States following lung cancer. While there are more cases of breast cancer or prostate cancer, colorectal cancer contributes to higher mortality. According to the 2015 Surveillance, Epidemiology, and End Results (SEER) program data, there are 132,700 estimated new cases of colorectal cancer in the US and 49,700 estimated deaths. According to SEER, 20% of patients with colorectal cancer will present with distant metastases at diagnosis and 36% have regional spread to lymph nodes. Despite significant progress made over the past 2 decades, metastatic colorectal cancer (mCRC) still carries a poor prognosis. Overall, the 5-year survival is 13.1% in patients with distant metastases from CRC, as compared to 64.9% for all CRC patients, according to the SEER database. A multimodality strategy that combines surgical resection with chemotherapy in neo-adjuvant, adjuvant or conversion settings has resulted in a higher chance of long-term cure. However for mCRC patients, there is need for comprehensive systemic evaluation of individual tumors as a path to development of personalized therapy for mCRC.
Therapies guided by tumor molecular profiling using multiple testing technologies have shown significant clinical benefit in treating patients with advanced cancer.1-4 For example, in CRC, RAS mutation (KRAS, NRAS) status predicts resistance to epidermal growth factor receptor (EGFR) directed therapy and has had a major therapeutic impact.5,6 Similarly, BRAF mutation status has a strong negative prognostic value in mCRC.7
We investigated biomarker profiles of various metastases of colorectal cancer focusing on the potential therapeutic implications if significant differences were observed. Our analysis revealed sub-types of metastases that may potentially benefit from specific targeted or classical therapeutics. This includes the potential to target Her2 in some mCRC with lung metastases, potential preference of FOLFIRI over FOLFOX in some patients with peritoneal metastases, potential to use c-MET targeted therapy in patients with liver metastases, COX2 targeted therapy in patients with bone metastases. The results are hypothesis generating and prompt design of prospective clinical trials as well as preclinical studies. The results are relevant to the field of precision oncology and suggest avenues for future research as well as potential treatment options or approaches for patients with specific molecular profiles.
Methodology
Colorectal tumor samples were submitted to Caris Life Sciences, a CLIA, ISO15189 and CAP certified/accredited laboratory (Phoenix, AZ) for molecular profiling aimed to provide theranostic information based on tumor biomarkers. A multiplatform approach was taken that includes sequencing, IHC, FISH and CISH to investigate targetable biomarker aberrations. Retrospective data analysis was performed to identify biomarker characteristics of the complete cohort as well subgroups of patients. Patient biomarker data, primary tumor site and specimen site were captured and included in the analysis. Association studies were performed by 2-tailed Fisher Exact tests.
Immunohistochemistry
IHC analysis was performed on formalin fixed paraffin embedded tumor samples using automated staining techniques. The primary antibody clones used are as follows: AR (AR441/ AR318), BCRP (6D171), c-KIT (polyclonal), Cox-2 (SP21), EGFR (2-18C9), ER (SP1), p53 (DO-7), PDGFR (polyclonal), Her2 (4B5), ERCC1 (8F1), MGMT (MT23.3), MRP1 (33A6), PGP (C494), PR (1E2/100), PTEN (6H2.1), RRM1 (polyclonal), SPARC monoclonal (122511), SPARC (polyclonal), TOPO1 (1D6), TOPO2A (3F6) and TS (TS106/4H4B1), c-MET (SP44), TUBB3 (Neuronal Class III Beta-Tubulin Polyclonal) and TLE3 (M-201). IHC results were evaluated independently by board-certified pathologists. Results were categorized into positive or negative by defined thresholds specific to each marker based on published evidence. The primary antibody clones used are shown with the cutoffs used to define positive and negative results (supplementary table).
Fluorescent and chromogenic in situ hybridization
FISH assays were used for evaluation of the HER2/neu [HER2/CEP17 probe], EGFR [EGFR/CEP7 probe], TOP2A [TOP2/CEP17 probe], and cMET [cMET/CEP7 probe], cMYC [cMYC/CEP8 probe], PIK3CA [PIK3CA/CEP3 probe] (all Abbott Molecular/Vysis). HER2/CEP17 ratio > 2.2 was considered amplified (CAP/ASCO guideline 2007). EGFR amplification was defined by the presence of EGFR/CEP7 ratio of ≥ 2, or ≥ 15 EGFR copies per cell in ≥ 10% of analyzed cells. TOP2A amplification was defined as TOP2A/CEP17 ratio ≥2.0 while c-MET was considered amplified if ≥ 5 c-MET copies were detected on average. cMYC amplifications was defined as cMYC/CEP8 ratio of >=2.0. PIK3CA amplification was defined as >=3.0 copies of PIK3CA per cell.
CISH assays were used for evaluation of the HER2/neu [INFORM HER2 Dual ISH DNA Probe Cocktail (Ventana)] and cMET status. HER2/CEP17 ratio ≥2 was considered amplified. Laboratory developed assay for cMET gene copy number assessment used commercially available c-MET and chromosome 7 DIG probe (Ventana). cMET was considered amplified if ≥ 5 copies per tumor cells were detected on average.
Next-Gen sequencing
Direct sequence analysis was performed on genomic DNA isolated from formalin-fixed paraffin-embedded tumor samples through the Illumina MiSeq platform using Illumina TruSeq Amplicon Cancer Hotspot panel. All variants that are reported here are detected with >99% confidence based on the frequency of the mutation present and the amplicon coverage. The sequencing included hotspot regions of 45 genes: ABL1, AKT1, ALK, APC, ATM, BRAF, CDH1, CSF1R, CTNNB1, EGFR, ERBB2, ERBB4, FBXW7, FGFR1, FGFR2, FGFR3, FLT3, GNA11, GNAS, GNAQ, HNF1A, HRAS, IDH1, JAK2, JAK3, KDR (VEGFR2), KIT, KRAS, MET, MLH1, MPL, NOTCH1, NPM1, NRAS, PDGFR, PIK3CA, PTEN, PTPN11, RB1, RET, SMAD4, SMARCB1, SMO, STK11, TP53, and VHL.
Sanger sequencing
Mutation analysis by Sanger sequencing included selected regions of BRAF, KRAS, cKIT, EGFR, NRAS and PIK3CA genes and was performed by using M13-linked PCR primers designed to flank and amplify targeted sequences. PCR products were bi-directionally cycle sequenced by using the BigDye Terminator v1.1 chemistry (Applied Biosystems) and analyzed using the 3730 DNA Analyzer (Applied Biosystems). Sequence traces were analyzed using Mutation Surveyor software v3.25 (Soft Genetics). A sample was deemed mutated if the same nucleotide change was identified in both the forward and reverse traces as well as being absent in the control sample traces.
Statistics
Odds ratios were calculated as (Biomarker positive N in metastasis/Biomarker negative N in metastasis)/(Biomarker positive N in primary/Biomarker negative N in primary). P values were considered significant at <0.05 and were determined by a 2-tailed Fisher-Exact test.
Results
Distribution frequencies of tumor biomarkers in a large cohort of colorectal cancer patients
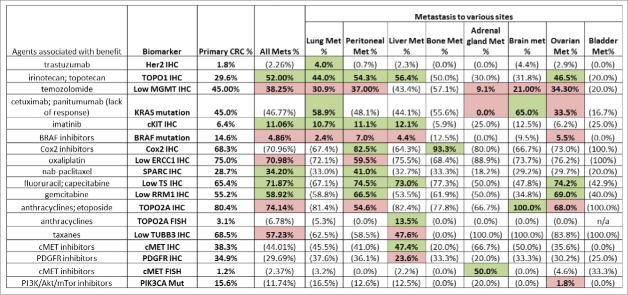
From 2009 to 2013, a total of 6892 colorectal cancer samples were evaluated. The median age of the collective cohort was 60 years, with the majority of patients being males (n = 3690, 54%). Results of the 18 IHC and 4 ISH markers are shown in Table 1. Each biomarker was tested in a different number of cases, which varied from 129 to 5234, with the biomarker frequencies indicating the percent of tumors harboring the specific marker. Potential therapeutic approaches are mentioned in third column. The results suggest that potentially different outcomes may be expected from commonly used chemotherapeutic agents used to treat colorectal cancer. For example, 66.7% of CRC were negative for TS protein expression by IHC suggesting that those tumors may be more sensitive to 5-fluorouracil. The presence of TOPO1 protein in 43.2% of CRC suggests a preference for irinotecan due to predicted sensitivity. In current practice for the population of patients of CRC there is no particular preference for irinotecan over oxaliplatin based combination therapy. Thus for tumors that express TOPO1 protein, particularly those CRCs with peritoneal metastases, investigation of the efficacy of irinotecan vs. oxaliplatin seems warranted. Moreover, in the absence of other information, and given a disease with poor prognosis, the clinician may choose the agent that is more likely to achieve a response based on molecular profiling, i.e. if a tumor with a poor prognosis expresses TOPO1 protein, there may be a preference for irinotecan over oxaliplatin, especially if ERCC1 protein is expressed. The results suggest in the absence of other options, if tumors express low or no MGMT protein, there should be some consideration for use of temozolomide. Similarly, if the cMET gene is amplified or the c-MET protein is overexpressed, some consideration for use of a c-MET inhibitor should be given including through enrollment in an appropriate clinical trial. For mCRC with high Her2 protein expression (1.8%) or HER2 gene amplification (5.7%) there should be some consideration for use of anti-Her2 directed therapies including enrollment in a clinical trial.
Table 1.
IHC/FISH frequencies in 6892 CRC cases analyzed and the associated therapies
| Biomarkers | Total Cases | Biomarker frequencies | Associated therapies |
|---|---|---|---|
| IHC TS (−) | 4907 | 66.7% | capecitabine, fluorouracil |
| IHC RRM1 (−) | 4227 | 58.4% | gemcitabine |
| IHC TOPO1 (+) | 4867 | 43.2% | irinotecan, topotecan |
| IHC MGMT (−) | 4515 | 40.2% | temozolomide |
| IHC cMET (+) | 1375 | 40.8% | cMET inhibitors |
| ISH cMET (+) | 929 | 2.0% | |
| IHC PTEN (−) | 5234 | 62.4% | mTor inhibitors |
| IHC TUBB3 (−) | 876 | 63.0% | paclitaxel, docetaxel |
| IHC TLE3 (+) | 1393 | 11.8% | |
| IHC PDGFR (+) | 1270 | 32.4% | n/a |
| IHC SPARC (+) | 4534 | 32.2% | nab-paclitaxel |
| IHC ERCC1 (−) | 3594 | 71.8% | oxaliplatin |
| IHC cKit (+) | 3103 | 9.0% | cKIT inhibitors |
| IHC PR (+) | 4354 | 1.4% | hormonal therapies |
| IHC AR (+) | 4380 | 0.3% | |
| IHC ER (+) | 4353 | 0.1% | |
| ISH EGFR (+) | 129 | 25.6% | n/a |
| IHC Her2 (+) | 4489 | 1.8% | trastuzumab |
| ISH HER2/neu (+) | 1653 | 5.7% | |
| IHC TOP2A (+) | 3913 | 75.2% | anthracyclines |
| ISH TOP2A (+) | 351 | 3.7% | |
| IHC PGP (−) | 3901 | 53.5% | paclitaxel, docetaxel, anthracyclines |
Table 2 shows the mutation rates of 45 genes. KRAS, BRAF, NRAS, PIK3CA, cKIT and EGFR results are from both Sanger sequencing and NextGen sequencing, while the rest of the mutations were detected by NextGen only. For the genes already known to be commonly mutated in CRC, the results are consistent with present knowledge for APC, TP53, KRAS, PIK3CA, SMAD4, and BRAF. Some findings may be actionable as discussed above, e.g. c-MET, EGFR, ERBB2, cKIT. Of note, HRAS mutations were rarely seen but are not currently screened for as part of “total-Ras” testing in mCRC that includes KRAS and NRAS. PTEN or AKT mutations may suggest clinical trials targeting PI3K or Akt. Others may become actionable in the future such as FGFR1, PDGFRA, ERBB4, IDH1 and the clinician needs to be looking for clinical trials that may offer options for such patients. Although ALK gene mutations were not seen in this cohort, ALK gene fusions have been reported in CRC.8.
Table 2.
Sequencing results of all 6892 CRC cases analyzed
| Mutation rate | Genes |
|---|---|
| 67.9% | APC |
| 58.3% | TP53 |
| 44.0% | KRAS |
| 14.1% | PIK3CA |
| 12.9% | SMAD4 |
| 9.60% | BRAF |
| 7.10% | FBXW7 |
| 2–5% | NRAS, ATM, PTEN, cMET, HNF1A, GNAS |
| 1–2% | ERBB2, CTNNB1, EGFR, AKT1, STK11, KDR, JAK3 |
| 0.5–1% | cKIT, ABL1, PDGFRA, ERBB4, RB1,MLH1 |
| 0.1–0.5% | IDH1, SMO, CSF1R, GNA11, HRAS, VHL, FGFR2, SMARCB1, NPM1, CDH1, FGFR1 |
| 0% | ALK, FLT3, GNAQ, JAK2, MPL, NOTCH1, PTPN11, RET |
Comparison of biomarkers in colon tumors with different sites of metastases reveals significant differences
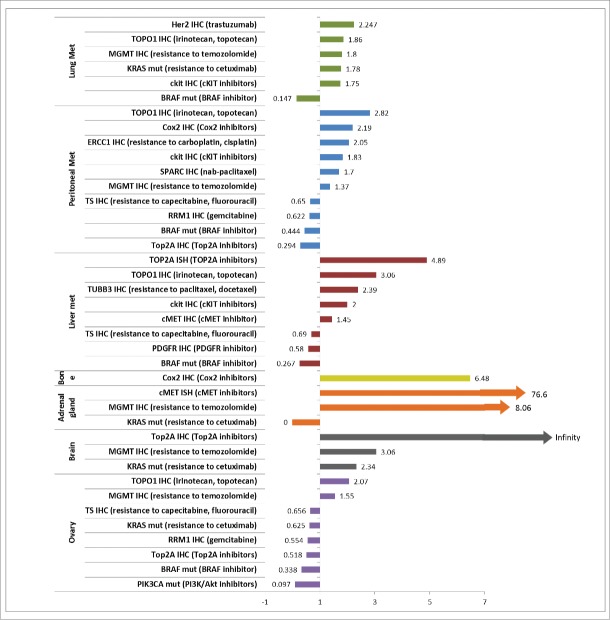
In patients with advanced CRC, the distribution frequencies of IHC, ISH and mutation markers in tumors taken from the colon (primary tumors) (N = 2510) were compared to a mostly unmatched set of tumors taken from distant metastases (43 patients had matched samples). A total of 2010 samples from various distant metastatic sites were included in the analysis, including 918 liver, 465 of peritoneum, 371 of lung, 188 of ovary, 30 of brain, 25 of bone, 13 of adrenal gland as well as 9 bladder samples. The comparative results are detailed in Figures 1 and 2.
Figure 1.
Percentage breakdowns by primary and metastasis. Green: biomarker expressions associated with benefit of therapies that are statistically significantly higher than the primary CRC; Red: biomarker expressions associated with benefit of therapies that are statistically significantly lower than the primary CRC. The numbers in parentheses are NOT statistically significantly different from the primary CRC.
Figure 2.
Odds ratio of biomarker comparisons in 7 metastases. Shown are biomarkers that are significantly differentially distributed between the metastases and primary (all p < 0.05 by 2-tailed Fisher-Exact test.) Bladder metastasis was also investigated however no significant differences in biomarker distribution were found and is therefore not included here. Odds ratios were calculated as (Biomarker positive N in metastasis / Biomarker negative N in metastasis) / (Biomarker positive N in primary/Biomarker negative N in primary).
Increased Her2, TOPO1, MGMT, and c-Kit protein expression, and KRAS mutations observed in lung metastases from CRC
Lung metastases harbored a higher Her2 protein expression than was observed in primary colon tumors (4% vs. 1.8%, p = 0.028). Among all eight metastases considered, lung was the only metastatic site presenting a significantly different Her2 expression as compared to primary site tumors. Higher TOPO1, MGMT, c-Kit protein expression and KRAS mutation and a lower rate of BRAF mutation were also seen in lung metastasis. The finding of increased Her2 protein expression in mCRC may prompt the use of anti-Her2 directed therapy such as trastuzumab or lapatinib. Some data has been emerging from the HERACLES trial with preliminary evidence for efficacy in patients. Of note, Her2 has been described as a resistance mechanism for anti-EGFR targeted therapy in mCRC and therefore can also be examined and considered for therapeutic targeting under such circumstances. Higher TOPO1 protein expression by IHC in lung metastases from CRC may prompt some preference for FOLFIRI over FOLFOX, and this certainly merits further consideration in terms of analysis of patient outcomes with these profiles, and consideration in future clinical trial design. The use of MGMT and c-Kit proteins as biomarkers may prompt selective use or avoidance of temozolomide, and consideration of a c-Kit inhibitor barring other actionable targets or appropriate clinical trials.
Increased TOPO1, Cox2, ERCC1 and reduced RRM1 protein expression in peritoneal metastases from CRC
Peritoneal metastases presented significant differences in 10 biomarkers including higher protein expression of TOPO1, c-Kit, Cox2 as well as a significantly lower protein expression of RRM1. Peritoneum was the only metastatic site presenting significantly higher SPARC (41% vs. 29%, p < 0.0001) and ERCC1 protein expression (40.5% vs. 25%, p < 0.0001) compared to colon tumors. The altered patterns in peritoneal metastases are hypothesis generating and possibly actionable for this difficult-to-treat subgroup of patients with mCRC. The finding of increased TOPO1 protein expression may steer the choice of therapy toward an irinotecan containing combination therapy regimen and away from an oxaliplatin containing regimen. This hypothesis is further supported by our discovery that some peritoneal metastases from mCRC also have increased ERCC1 protein expression that predicts oxaliplatin resistance. These observations suggest that samples of peritoneal metastasis from CRC should be tested for TOPO1 and ERCC1 protein expression, and that therapy may be appropriately chosen to include irinotecan or oxaliplatin based on the results. It is possible to test the hypothesis that high TOPO1 and ERCC1 protein expression will favor response to irinotecan over oxaliplatin and needs to be further tested in a clinical trial setting. It may be predicted that patients whose peritoneal metastases from CRC with high TOPO1 and ERCC1 protein expression will be more responsive to irinotecan-based combination therapy while those with low TOPO1 and ERCC1 protein expression may be more responsive to an oxaliplatin-based combination therapy. Reduced RRM1 protein expression in peritoneal metastases may direct some patients toward gemcitabine that is not commonly used in CRC.
TOPO2A gene amplification, and increased TOPO1, c-Kit and c-MET protein expression in liver metastases from CRC
Liver metastases showed a high TOPO2A amplification (14% vs. 3%, p = 0.013) and cMET expression by IHC (47.40% vs 38.30%, p = 0.02) compared with colon tumors. Since TOPO2A and HER2 are on the same chromosome, it should be noted that the TOPO2A amplification result was observed using TOPO2A FISH, while no evidence for HER2 amplification or TOPO2A/HER2 amplification was found by HER2 FISH testing. Significantly higher protein expression of TUBB3 and lower expression of PDGFR was also seen in liver metastasis. The highest TOPO1 (56.4%) and c-Kit (12.1%) protein expression was observed in liver metastases. These observations point toward use of irinotecan, c-MET or c-Kit inhibitors in the appropriate clinical setting or through enrollment in a molecular profiling directed clinical trial.
Increased Cox2 protein expression in bone metastases from CRC
The only significant biomarker difference between bone metastases and primary CRC was a significantly higher Cox2 IHC expression, while 14 out of 15 samples presented positive Cox2 protein expression (93%), 68% (820 out of 1200 samples) showed Cox2 protein expression in colon tumors (p = 0.047). This observation suggests that in the appropriate clinical setting, there may be some consideration to the use of a Cox2 inhibitor in patients with mCRC and bone metastases. This observation merits further follow-up in the context of a clinical trial in the appropriate patient population, and investigation of any epidemiological evidence or clinical outcomes data in databases that have used molecular profiling as the data may be relevant, i.e., if patients received Cox2 inhibitors.
Increased c-MET and MGMT protein expression and absence of KRAS mutations in adrenal metastases from CRC
In adrenal gland metastases, KRAS mutation was not found in any of the 9 samples for which the test was performed, by contrast to the 45% mutation rate observed in primary CRC (p = 0.006). c-MET amplification was observed in 1 out of the 2 samples tested (50%) compared to a 1.2% (amplification rate seen in colon tumors (p = 0.029). While these data are certainly preliminary, they do point to potentially significant differences among adrenal metastases and other metastatic sites from CRC. It is not clear why absence of KRAS may select for adrenal metastases from CRC. It should be noted that in cases where multiple technologies were used, the results were combined, i.e. for KRAS gene sequencing, information was combined from both Sanger or NGS, while for cMET amplification, both CISH and FISH data were included.
TOPO2A protein expression and KRAS mutation in brain metastases from CRC
Brain metastases presented a 100% over-expression of TOPO2A protein in our cohort (19 out of 19 samples), as compared to the 80% observed in colon tumors (1175 out of 1161 samples). Brain metastases also showed the highest KRAS mutation rate (65%) among all metastases and when compared to colon tumors (45%) (p = 0.046). The observations point to some potential differences between brain metastases and other metastatic sites, and also suggest that perhaps KRAS mutations’ influence on the biology of CRC may somehow predispose them to the rare site of brain metastasis.
Lower RRM1 protein expression and PIK3CA mutations in ovarian metastases from CRC
Ovarian metastases from CRC showed a significantly lower mutation rate of PIK3CA, in that only 1 mutation is found in 57 samples (1.8%) compared to a 15.6% mutation rate (92 out of 589 samples) in primary CRC (p = 0.002). Lower RRM1 protein expression may prompt use of gemcitabine therapy. It is unclear why a lower PIK3CA mutation rate might be selected for in ovarian metastases from CRC.
Differences in biomarker profiles based on KRAS/BRAF mutation status in CRC
We examined differences in biomarker profiles of KRAS mutant versus wild-type and BRAF mutant vs. wild-type tumors (Fig. 3a/b). HER2 by ISH was more prevalent in the KRAS wild-type subgroup (p<0.001). On the other hand, PIK3CA mutations and c-MET overexpression by IHC were more prevalent in KRAS mutated tumors (p < 0.001). Similarly, comparison between BRAF wild-type (n = 4366) and BRAF mutant tumors (n = 455) revealed many biomarker differences (Fig. 3B) such that higher rates of ABL1 mutation with lower PDGFR expression (22% vs 36%, p < 0.001) was seen preferentially in BRAF mutant tumors. These findings may have implications in terms of clinical trial development and consideration of alternative therapies as shown in Figures 3B. Mutation in other genes (APC, PTEN, HNF1A, ABL1, and RB1) are also reported and may have new drugs in pipeline that can be tested. KRAS-mutated CRC had higher c-MET (47% vs. 36%) and lower MGMT (56% vs. 63%) protein expression as compared to non-KRAS mutated tumors.
Figure 3.
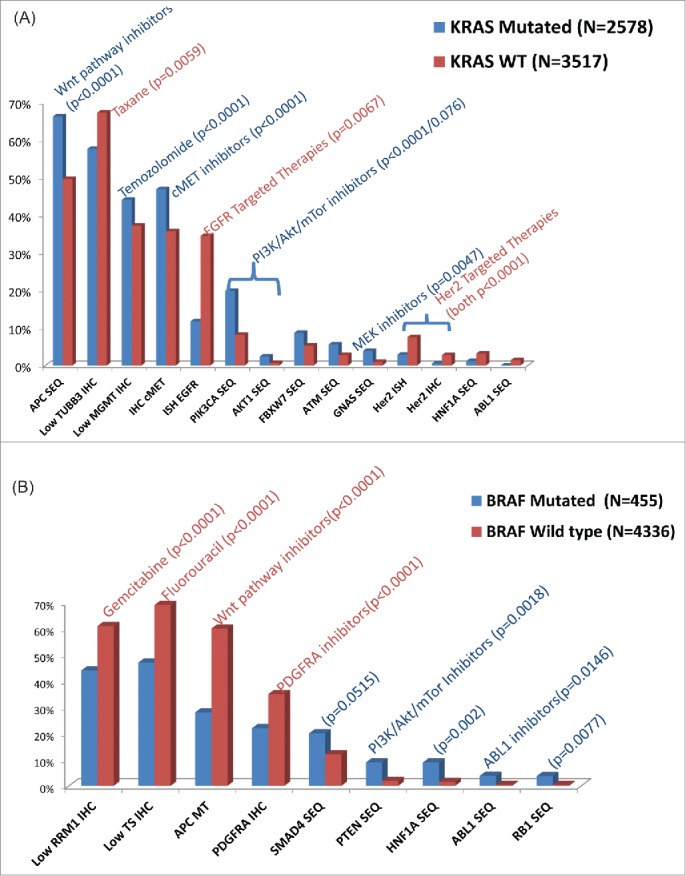
Differential molecular profiling results by: 4A. KRAS mutation status 4B. BRAF mutation status.
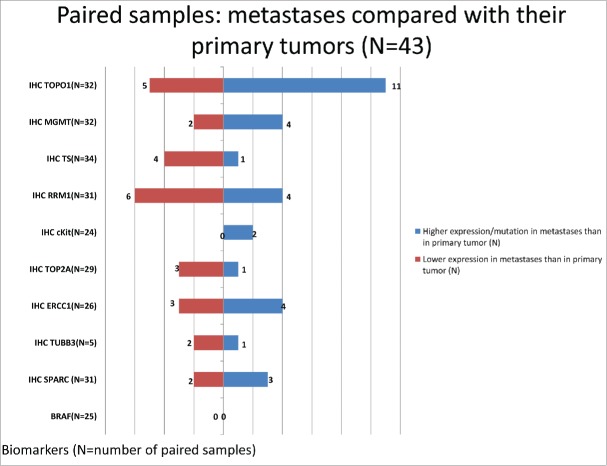
Paired tumor profiles on primary and metastases taken synchronously
A sub-cohort of 43 paired tumor profiles (43 patients received at least 2 tumor profiling tests, one profile performed on the primary CRC and one on the metastasis) on tumor samples taken within 1 month of time frame were analyzed. Figure 4 shows the biomarker differences observed when comparing the metastases to the primary CRC. Gene profiling did not find any differences in the rates of mutations between the primary and synchronous metastasis. In terms of protein expression, TOPO1 and c-Kit proteins were overexpressed in metastatic sites as compared to colonic primaries. A much higher expression of TOPO1 protein was observed in metastases versus primary CRC in the synchronously profiled paired samples: out of the 43 patients, 32 had TOPO1 protein expression tested on both samples, among which 11 showed high TOPO1 protein expression in the metastases and low protein expression in the primary sample, 5 patients showed low protein expression in the metastases while high protein expression in the primary sample, and the other 16 patients showed no change (either both high or both low) in their paired profiles (Fig. 4). This observation overlays with our observation in the large cohort described previously that while 52% of metastases showed positive TOPO1 protein expression, only 30% was observed in tumors taken from the colon (Fig. 4). Similarly, in 24 pairs of c-Kit IHC data available, 2 cases showed higher protein expression in the metastases while no case showed higher expression in the primary tumor, in line with a significantly higher protein expression of c-Kit IHC (11% vs. 6.4%) in the metastases compared with colon tumors. In the large cohort analysis, MGMT protein expression was significantly higher in the metastases than the colon tumors and RRM1, TS and TOP2A proteins had significantly higher expression in the colon tumors (Fig. 1) and the same trend was observed in the paired profiles.
Figure 4.
Comparison of metastases with primary tumors from 43 paired samples.
Discussion
Advancements in therapeutics have significantly improved survival colorectal cancer over the past 2 decades. The addition of oxaliplatin or irinotecan to infusional 5-FU (FOLFOX and FOLFIRI) has significantly improved treatment outcomes for patients with advanced colorectal cancer compared to single agent 5-FU.9-11 Targeting the EGFR pathway, with the antibodies cetuximab and panitumumab and the VEGF pathway with the agents bevacizumab, ziv-aflibercept (anti-VEGF), or ramicirumab (anti-VEGFR2) has further improved the survival of these patients5,6,12-15,16 Regorafenib, a multi-kinase inhibitor has been approved for refractory CRC though the survival benefit is modest and comes with significant toxicities.17 With these therapeutic advancements in mCRC, a higher median survival reaching 2 and half years has been achieved as suggested by the CALBG 80405 study.18 In order to further improve the outcome of this patient population, additional treatment options are needed. Recent efforts have moved away from treating cancers empirically but relying on the biology of each tumor revealed by a multiplatform tumor profiling testing to tailor therapies. This approach has seen exciting results especially in the metastatic and refractory setting.19,1,3
We have employed a comprehensive approach to summarize the biomarker frequencies observed in a large cohort of colorectal cancer patients to gain insight into the potential responders to various therapies in colorectal cancer. Agents that are part of standard of care for colorectal cancer, as mentioned above, including fluorouracil and irinotecan are suggested to benefit a high portion of patients as many patients carry a favorable biomarker profile.20,21 Agents not routinely considered including temozolomide, gemcitabine, taxanes as well as anthracyclines have suggestion of improved activity against tumors carrying low MGMT,22 RRM1,23 TUBB324 and high TOPO2A IHCs respectively.25 Mutation rates vary significantly in colorectal cancer such that APC and TP53 are mutated in more than half of patients, while more than half of the genes tested carry a mutation rate of less than 1%.
Through a comprehensive biomarker profile comparison in a large cohort of CRC samples that is comprised of tumors taken from the colon and 8 metastatic sites, interesting biomarkers features were observed for various metastases and their potential therapeutic implications are discussed below. The rate of HER2/neu amplification was lower in our study compared to published literature but differential amplification was observed in primary colon tumors and lung metastases (1.8% vs 4%). Previous studies have quoted a 5–9% incidence of HER2/neu amplification in CRC26,27 and HER2/neu amplification was shown to be concordant in only 5% of the primary and metastatic lesions.27 In a study by Ramanathan et al., HER2/neu-positive patients with advanced colorectal cancer were treated with trastuzumab (Herceptin® and irinotecan). Of the 138 screened patients, Her2 protein overexpression was only detected in 11 (8%; 2+ in 5 and 3+ in 6 patients), and this resulted in premature termination of the study.28 However, as ∼5% of all colon cancers do overexpress Her2 both in the primary lesion and the corresponding metastasis, Her2-targeted therapy with trastuzumab may still be a treatment option for ∼60,000 patients worldwide each year.29 HER2 amplification status has not shown to affect prognosis in 2 studies.26,30 Her2 overexpression has also been described as a potential mechanism of EGFR inhibitor resistance (cetuximab, panitumumab).31 To date, the preferential Her2 over-expression in CRC with lung metastasis has not been reported. We also reported a predominance of HER2/neu amplification in KRAS wild-type tumors and this may be related to prior EGFR inhibitor exposure in this subgroup. Her2-targeted therapies including trastuzumab have shown marked response in breast and gastric cancer patients presenting with Her2 positivity but due to low prevalence, studies in colorectal cancer have not been decisive.32-34,35 A phase II trial, HERACLES is is currently enrolling chemorefractory patients with Her2 overexpressing mCRC, to dual Her2 targeted therapy (lapatinib+ trastuzumab)36 A larger representation of Her2 positive patients in the lung metastases and KRAS wild-type tumors prompts currently recruiting clinical trials to focus on this subset of patients.
Peritoneal carcinomatosis from colorectal origin traditionally has been a difficult-to-treat subtype.37 Current standards involve systemic chemotherapy and an increasing role of cytoreductive surgery and HIPEC (Hyperthermic Intraperitoneal Chemotherapy).38 Prior work has highlighted the poor prognosis for CRC patients with peritoneal metastases and potentially improved outcomes with Oxaliplatin based therapy in an unselected population.39 Our findings of higher TOPO1 and ERCC1 protein expression in peritoneal metastases have not been described previously. Hence, clinical trial accrual to irinotecan based therapy in this biomarker driven subset of patients deserves further exploration.20,40
The altered patterns in peritoneal metastases are hypothesis generating and possibly actionable for this difficult to treat subgroup of patients with mCRC. The finding of increased TOPO1 protein expression may steer the choice of therapy toward an irinotecan containing combination therapy regimen and away from an oxaliplatin containing regimen. This is further supported by the finding that some peritoneal metastases from mCRC have increased ERCC1 protein expression that predicts oxaliplatin resistance. These observations suggest that samples of peritoneal metastasis from CRC should be tested for TOPO1 and ERCC1 protein expression, and that therapy may be appropriately chosen to include irinotecan or oxaliplatin based on the results. It is possible to test the hypothesis that high TOPO1 and ERCC1 protein expression will favor response to irinotecan over oxaliplatin in a clinical trial. It may be predicted that patients whose peritoneal metastases from CRC have high TOPO1 and ERCC1 protein expression will be more responsive to irinotecan-based combination therapy while those with low TOPO1 and ERCC1 protein expression may be more responsive to an oxaliplatin-based combination therapy. Reduced RRM1 protein expression in peritoneal metastases may direct some patients toward gemcitabine that is not commonly used in CRC.
SPARC (secreted protein acidic and rich in cysteine) is a member of a family of matricellular glycoproteins, whose function is to modulate cell–matrix interactions and cell function without participating in the structural scaffold of the extracellular matrix. Its role in colorectal cancer has been studied and a survival analysis of 292 colorectal carcinoma patients revealed a poorer prognosis for patients lacking SPARC expression than for patients with normal SPARC protein expression (56.79% vs. 75.83% 5-year survival rate, p = 0.0014).41 We found a higher expression of SPARC protein in peritoneal metastases from colon cancer. The potential role of this protein expression to serve as a biomarker for response to chemotherapy (Nab-Paclitaxel) has been studied in pancreatic cancer and generated conflicting results however the effect in colorectal cancer is unknown.42-44
We found higher c-MET protein expression (by IHC) in liver metastases. Zeng et al. have previously reported cMET amplification by copy number in liver metastases (9%). They also reported poor overall survival in this subset of patients. In conjunction with our findings, this may serve as a rationale for designing clinical trials using c-MET inhibitors in colorectal cancers. There are a few c-MET inhibitor clinical trials accruing patients with advanced CRC, but none are restricted to patients with liver metastases (NCT00788957, NCT01892527).45,46
CRC metastases to the ovary, brain, adrenal gland and bone are rarer events and the incidence of metastases to uncommon sites has increased with improved treatments and survival.47 Given their rarity, these metastases have not been systematically studied and therefore published clinical data is lagging. Ovaries are an uncommon site of metastasis for colorectal cancer but also the most common non-genital tract related metastasis to the ovaries (32%).48,49 Our analysis of 188 ovarian metastases samples did not identify major molecular variations except a lower incidence of PIK3CA mutation, when compared to colonic primaries. Hence, genomic alterations may not explain the poor response of ovarian metastases to systemic therapies as seen in clinical practice making surgical resection the primary treatment strategy, where feasible.50,51 Cox2 expression has been suggested to be involved in bone metastasis and Cox2 inhibitors can effectively halt the process in breast cancer.52,53 In CRC, Cox-2 isoform overexpression has been associated with tumor angiogenesis and growth.54 In advanced lung cancer, Cox-2 protein overexpression has been suggested as a marker of response to Cox-2 inhibitors when combined with chemotherapy.55 The higher incidence of Cox-2 protein expression in bony metastases seen in our cohort, may highlight a potential role for these agents in conjunction with chemotherapy. CRC with KRAS mutation have a higher incidence of brain metastases as reflected by the selective increase in incidence of KRAS mutations in brain metastasis as described in prior studies.56 In contrast, despite a small sample, absence of KRAS mutation in all adrenal metastases can help us make definite conclusions about usefulness of EGFR directed therapies in such tumors.
The large number of analyzed samples is from a heterogeneous population of patients at various stages of diseases and treatment status. Therefore, we also performed a sub-group analysis on a homologous cohort of 43 patients who had both the primary CRC sample and a metastatic sample profiled synchronously (time interval of profiling 1 month). As expected, the biomarker differences observed in this sub-cohort overlay significantly with the observations made in the analysis from the large cohort (liver metastasis of 918 and primary of 2510 samples): significantly higher expression of TOPO1, TUBB3, c-Kit and lower expression of TS proteins was observed in the large cohort of liver metastasis compared to primary CRC, coinciding with the trends observed in the paired samples from the 15 patients with liver metastases. Unfortunately, interesting markers including TOPO2A FISH and c-MET IHC paired data in liver metastases were not available from these 15 patients. Nevertheless, the sub-cohort analysis indicates that the observations made in the large cohort of samples from a commercial database is representative of the biomarker features of different metastatic sites in spite of potentially confounding factors including disease progression and treatments.
We believe our findings provide future direction and insight into the molecular variations of CRC metastases. These findings provide a rationale for biomarker driven proof-of-concept research, including directing research on the impact of potentially actionable molecular targets on individual patient clinical outcomes. These are hypothesis-generating results and should be tested both retrospectively and prospectively in the clinical trial setting. More importantly, the molecular heterogeneity presented highlights the need to obtain the specific information on patient’s particular tumor as an important step in designing patient treatment strategy. Unfortunately, a recent study of molecular profiling matched accrual to phase I clinical trials, failed to a clinical benefit in chemorefractory mCRC patients.57 This points toward another important area of future research as to the timing of introducing targeted therapies, with respect to traditional chemotherapy.
Our study has some limitations. We do not have any clinical outcomes information on the study population and hence, cannot correlate our findings with demographic or treatment related factors as well as survival data. Molecular profiling was limited to the 45 genes representing the most common driver mutations seen in different cancer types rather than a whole exome sequencing and this may limit the accuracy of our conclusions. Clinical utility of the actionable mutations identified through this platform is also limited to some extent by reimbursement issues associated with use of novel or unconventional therapies, as insurance companies do not entertain molecular profiling results (in absence of level I evidence) as rationale for trying new drugs or combinations. Another limitation is that for the analysis of the metastatic sites, most of the samples were not paired samples and so the comparisons were not in most cases between metastases and primary tumors from the same patient, but rather a population of metastases vs. a population of primary tumors.
Overall, our large database of predictive markers in a large cohort of CRC patients with various metastases, both common and uncommon, serves as a reference for clinical trial design and for clinicians when dealing with these difficult-to-treat cancers. Once validated in a prospective randomized fashion, treatment strategies tailored to metastatic sites of colorectal cancer can be designed to account for the distinct biomarker features reported here. More importantly, the molecular heterogeneity presented highlights the need to obtain the specific information on patient’s particular tumor as an important step in designing future clinical trials.
Disclosure of Potential Conflict of Interest
J Xiu, Z Gatalica and S Reddy are employees of Caris Life Sciences
Acknowledgments
The results were presented at the 2014 annual American Society of Clinical Oncology meeting (Abstract No. 134628: El-Deiry, W.S., et al. “Molecular profiling of 6,892 colorectal cancer patients to identify potential treatment options.”). W.S.E-D. is an American Cancer Society Research Professor. This work was collaborative with Caris Life Sciences. There was no financial support to academic authors or institutions from Caris Life Sciences. As of 2015, W.S.E-D. serves as co-Chair of the national Steering Committee for the Caris Centers of Excellence Network in Precision Medicine and Chair of the Phase I Committee. In 2015, Fox Chase Cancer Center was designated as a Caris Center of Excellence in Precision Medicine. Neither W.S.E-D. nor Fox Chase Cancer Center receive any financial support for the roles stated above.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website
References
- 1.Jameson GS, Petricoin EF, Sachdev J, Liotta LA, Loesch DM, Anthony SP, Chadha MK, Wulfkuhle JD, Gallagher RI, Reeder KA, et al. A pilot study utilizing multi-omic molecular profiling to find potential targets and select individualized treatments for patients with previously treated metastatic breast cancer. Breast Cancer Res Treat 2014; 147(3):579–88; PMID:25209003; http://dx.doi.org/ 10.1007/s10549-014-3117-1 [DOI] [PubMed] [Google Scholar]
- 2.Tsimberidou AM, Iskander NG, Hong DS, Wheler JJ, Falchook GS, Fu S, Piha-Paul S, Naing A, Janku F, Luthra R, et al. Personalized medicine in a phase I clinical trials program: the MD Anderson Cancer Center initiative. Clin Cancer Res 2012; 18(22):6373–83; PMID:22966018; http://dx.doi.org/ 10.1158/1078-0432.CCR-12-1627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tsimberidou AM, Wen S, Hong DS, Wheler JJ, Falchook GS, Fu S, Piha-Paul S, Naing A, Janku F, Aldape K, et al. Personalized medicine for patients with advanced cancer in the phase I program at MD anderson: validation and landmark analyses. Clin Cancer Res 2014; 20(18):4827–36; PMID:24987059; http://dx.doi.org/ 10.1158/1078-0432.CCR-14-0603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Von Hoff DD, Ramanathan RK, Borad MJ, Laheru DA, Smith LS, Wood TE, Korn RL, Desai N, Trieu V, Iglesias JL, et al. Gemcitabine plus nab-paclitaxel is an active regimen in patients with advanced pancreatic cancer: a phase I/II trial. J Clin Oncol 2011; 29(34):4548–54; PMID:21969517; http://dx.doi.org/ 10.1200/JCO.2011.36.5742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Van Cutsem E, Nowacki M, Lang I, Cascinu S, Shchepotin I, Maurel J, Rougier P, Cunningham D, Nippgen J, Köhneet C. Randomized phase III study of irinotecan and 5-FU/FA with or without cetuximab in the first-line treatment of patients with metastatic colorectal cancer (mCRC): The CRYSTAL trial. J Clin Oncol 2007; 25(18S):4000 [Google Scholar]
- 6.Douillard J-Y, Siena S, Cassidy J, Tabernero J, Burkes R, Barugel M, Humblet Y, Bodoky G, Cunningham D, Jassem J, et al. Randomized, phase III trial of panitumumab with infusional fluorouracil, leucovorin, and oxaliplatin (FOLFOX4) versus FOLFOX4 alone as first-line treatment in patients with previously untreated metastatic colorectal cancer: the PRIME study. J Clin Oncol 2010:JCO. 2009.27 4860; PMID: 20921465; http://dx.doi.org/16024606 10.1200/JCO.2009.27.4860 [DOI] [PubMed] [Google Scholar]
- 7.Samowitz WS, Sweeney C, Herrick J, Albertsen H, Levin TR, Murtaugh MA, Wolff RK, Slattery ML. Poor survival associated with the BRAF V600E mutation in microsatellite-stable colon cancers. Cancer Res 2005; 65(14):6063–9; PMID:16024606; http://dx.doi.org/ 10.1158/0008-5472.CAN-05-0404 [DOI] [PubMed] [Google Scholar]
- 8.Aisner DL, Nguyen TT, Paskulin DD, Le AT, Haney J, Schulte N, Chionh F, Hardingham J, Mariadason J, Tebbutt N, et al. ROS1 and ALK fusions in colorectal cancer, with evidence of intratumoral heterogeneity for molecular drivers. Mol Cancer Res 2014; 12(1):111–8; PMID:24296758; http://dx.doi.org/ 10.1158/1541-7786.MCR-13-0479-T [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Douillard J, Cunningham D, Roth AD, Navarro M, James RD, Karasek P, Jandik P, Iveson T, Carmichael J, Alakl M, et al. Irinotecan combined with fluorouracil compared with fluorouracil alone as first-line treatment for metastatic colorectal cancer: a multicentre randomised trial. The Lancet 2000; 355(9209):1041–7; PMID:10744089; http://dx.doi.org/ 10.1016/S0140-6736(00)02034-1 [DOI] [PubMed] [Google Scholar]
- 10.de Gramont A, Figer A, Seymour M, Homerin M, Hmissi A, Cassidy J, Boni C, Cortes-Funes H, Cervantes A, Freyer G, et al. Leucovorin and fluorouracil with or without oxaliplatin as first-line treatment in advanced colorectal cancer. J Clin Oncol 2000; 18(16):2938–47; PMID:10944126 [DOI] [PubMed] [Google Scholar]
- 11.Tournigand C, André T, Achille E, Lledo G, Flesh M, Mery-Mignard D, Quinaux E, Couteau C, Buyse M, Ganem G, et al. FOLFIRI followed by FOLFOX6 or the reverse sequence in advanced colorectal cancer: a randomized GERCOR study. J Clin Oncol 2004; 22(2):229–37; PMID:14657227; http://dx.doi.org/ 10.1200/JCO.2004.05.113 [DOI] [PubMed] [Google Scholar]
- 12.Hurwitz H, Fehrenbacher L, Novotny W, Cartwright T, Hainsworth J, Heim W, Berlin J, Baron A, Griffing S, Holmgren E, et al. Bevacizumab plus irinotecan, fluorouracil, and leucovorin for metastatic colorectal cancer. New Engl J Med 2004; 350(23):2335–42; PMID:15175435; http://dx.doi.org/ 10.1056/NEJMoa032691 [DOI] [PubMed] [Google Scholar]
- 13.Saltz LB, Clarke S, Díaz-Rubio E, Scheithauer W, Figer A, Wong R, Koski S, Lichinitser M, Yang TS, Rivera F, et al. Bevacizumab in combination with oxaliplatin-based chemotherapy as first-line therapy in metastatic colorectal cancer: a randomized phase III study. J Clin Oncol 2008; 26(12):2013–9; PMID:18421054; http://dx.doi.org/ 10.1200/JCO.2007.14.9930 [DOI] [PubMed] [Google Scholar]
- 14.Van Cutsem E, Köhne CH, Hitre E, Zaluski J, Chang Chien CR, Makhson A, D'Haens G, Pintér T, Lim R, Bodoky G, et al. Cetuximab and chemotherapy as initial treatment for metastatic colorectal cancer. New Engl J Med 2009; 360(14):1408–17; PMID:19339720; http://dx.doi.org/ 10.1056/NEJMoa0805019 [DOI] [PubMed] [Google Scholar]
- 15.Cunningham D, Humblet Y, Siena S, Khayat D, Bleiberg H, Santoro A, Bets D, Mueser M, Harstrick A, Verslype C, et al. Cetuximab monotherapy and cetuximab plus irinotecan in irinotecan-refractory metastatic colorectal cancer. New Engl J Med 2004; 351(4):337–45; PMID:15269313; http://dx.doi.org/ 10.1056/NEJMoa033025 [DOI] [PubMed] [Google Scholar]
- 16.Tabernero J, Takayuki Y, Cohn AL. Ramucirumab versus placebo in combination with second-line FOLFIRI in patients with metastatic colorectal carcinoma that progressed during or after first-line therapy with bevacizumab, oxaliplatin, and a fluoropyrimidine (RAISE): a randomised, double-blind, multicentre, phase 3 study. Lancet Oncol 2015; 16(5):499–508; PMID:25877855; http://dx.doi.org/ 10.1016/S1470-2045(15)70127-0 [DOI] [PubMed] [Google Scholar]
- 17.Grothey A, Van Cutsem E, Sobrero A, Siena S, Falcone A, Ychou M, Humblet Y, Bouché O, Mineur L, Barone C, et al. Regorafenib monotherapy for previously treated metastatic colorectal cancer (CORRECT): an international, multicentre, randomised, placebo-controlled, phase 3 trial. The Lancet 2013; 381(9863):303–12; PMID:23177514; http://dx.doi.org/ 10.1016/S0140-6736(12)61900-X [DOI] [PubMed] [Google Scholar]
- 18.Venook AP, Niedzwiecki D, Lenz H-J, Innocenti F, Mahoney MR, O'Neil BH, Shaw JE, Polite BN, Hochster HS, Atkins JN, et al. CALGB/SWOG 80405: Phase III trial of irinotecan/5-FU/leucovorin (FOLFIRI) or oxaliplatin/5-FU/leucovorin (mFOLFOX6) with bevacizumab (BV) or cetuximab (CET) for patients (pts) with KRAS wild-type (wt) untreated metastatic adenocarcinoma of the colon or rectum (MCRC). J Clin Oncol 32:5s, 2014 (suppl; abstr LBA3) [Google Scholar]
- 19.Von Hoff DD, Stephenson JJ Jr, Rosen P, Loesch DM, Borad MJ, Anthony S, Jameson G, Brown S, Cantafio N, Richards DA, et al. Pilot study using molecular profiling of patients' tumors to find potential targets and select treatments for their refractory cancers. J Clin Oncol 2010; 28:4877–83; PMID:20921468 [DOI] [PubMed] [Google Scholar]
- 20.Braun MS, Richman SD, Quirke P, Daly C, Adlard JW, Elliott F, Barrett JH, Selby P, Meade AM, Stephens RJ, et al. Predictive biomarkers of chemotherapy efficacy in colorectal cancer: results from the UK MRC FOCUS trial. J Clin Oncol 2008; 26(16):2690–8; PMID:18509181; http://dx.doi.org/ 10.1200/JCO.2007.15.5580 [DOI] [PubMed] [Google Scholar]
- 21.Qiu LX, Tang QY, Bai JL, Qian XP, Li RT, Liu BR, Zheng MH. Predictive value of thymidylate synthase expression in advanced colorectal cancer patients receiving fluoropyrimidine-based chemotherapy: evidence from 24 studies. Int J Cancer 2008; 123(10):2384–9; PMID:18729195; http://dx.doi.org/ 10.1002/ijc.23822 [DOI] [PubMed] [Google Scholar]
- 22.Shacham-Shmueli E, Beny A, Geva R, Blachar A, Figer A, Aderka D. Response to temozolomide in patients with metastatic colorectal cancer with loss of MGMT expression: a new approach in the era of personalized medicine? J Clin Oncol 2011; 29(10):e262-5; PMID:21220612; http://dx.doi.org/ 10.1200/JCO.2010.32.0242 [DOI] [PubMed] [Google Scholar]
- 23.Gong W, Zhang X, Wu J, Chen L, Li L, Sun J, Lv Y, Wei X, Du Y, Jin H, et al. RRM1 expression and clinical outcome of gemcitabine-containing chemotherapy for advanced non-small-cell lung cancer: a meta-analysis. Lung Cancer 2012; 75(3):374–80; PMID:21889227; http://dx.doi.org/ 10.1016/j.lungcan.2011.08.003 [DOI] [PubMed] [Google Scholar]
- 24.Zhang HL, Ruan L, Zheng LM, Whyte D, Tzeng CM, Zhou XW. Association between class III beta-tubulin expression and response to paclitaxel/vinorebine-based chemotherapy for non-small cell lung cancer: a meta-analysis. Lung Cancer 2012; 77(1):9–15; PMID:22306125; http://dx.doi.org/ 10.1016/j.lungcan.2012.01.005 [DOI] [PubMed] [Google Scholar]
- 25.O'Malley FP, Chia S, Tu D, Shepherd LE, Levine MN, Huntsman D, Bramwell VH, Andrulis IL, Pritchard KI. Topoisomerase II alpha protein and responsiveness of breast cancer to adjuvant chemotherapy with CEF compared to CMF in the NCIC CTG randomized MA.5 adjuvant trial. Breast Cancer Res Treat 2011; 128(2):401–9; PMID:21519837; http://dx.doi.org/ 10.1007/s10549-011-1511-5 [DOI] [PubMed] [Google Scholar]
- 26.Seo AN, Kwak Y, Kim DW, Kang SB, Choe G, Kim WH, Lee HS. HER2 Status in colorectal cancer: its clinical significance and the relationship between HER2 gene amplification and expression. PloS One 2014; 9(5):e98528; PMID:24879338; http://dx.doi.org/ 10.1371/journal.pone.0098528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee WS, Park YH, Lee JN, Baek JH, Lee TH, Ha SY. Comparison of HER2 expression between primary colorectal cancer and their corresponding metastases. Cancer Med 2014; 3(3):674–80; PMID:24668895; http://dx.doi.org/ 10.1002/cam4.228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramanathan RK, Hwang JJ, Zamboni WC, Sinicrope FA, Safran H, Wong MK, Earle M, Brufsky A, Evans T, Troetschel M, et al. Low overexpression of HER-2/neu in advanced colorectal cancer limits the usefulness of trastuzumab (Herceptin) and irinotecan as therapy. A phase II trial. Cancer Invest 2004; 22(6):858–65; PMID:15641483; http://dx.doi.org/ 10.1081/CNV-200039645 [DOI] [PubMed] [Google Scholar]
- 29.Blok EJ, Kuppen PJ, van Leeuwen JE, Sier CF. Cytoplasmic Overexpression of HER2: a Key Factor in Colorectal Cancer. Clin Med Insights Oncol 2013; 7:41–51; PMID:23471238; http://dx.doi.org/ 10.4137/CMO.S10811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kruszewski WJ, Rzepko R, Ciesielski M, Szefel J, Zieliński J, Szajewski M, Jasiński W, Kawecki K, Wojtacki J. Expression of HER2 in colorectal cancer does not correlate with prognosis. Dis Markers 2010; 29(5):207–12; PMID:21206005; http://dx.doi.org/ 10.1155/2010/109063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bertotti A, Migliardi G, Galimi F, Sassi F, Torti D, Isella C, Corà D, Di Nicolantonio F, Buscarino M, Petti C, et al. A molecularly annotated platform of patient-derived xenografts (“xenopatients”) identifies HER2 as an effective therapeutic target in cetuximab-resistant colorectal cancer. Cancer Disc 2011; 1(6):508–23; PMID:22586653; http://dx.doi.org/ 10.1158/2159-8290.CD-11-0109 [DOI] [PubMed] [Google Scholar]
- 32.Vogel CL, Cobleigh MA, Tripathy D, Gutheil JC, Harris LN, Fehrenbacher L, Slamon DJ, Murphy M, Novotny WF, Burchmore M, et al. Efficacy and safety of trastuzumab as a single agent in first-line treatment of HER2-overexpressing metastatic breast cancer. J Clin Oncol 2002; 20(3):719–26; PMID:11821453; http://dx.doi.org/ 10.1200/JCO.20.3.719 [DOI] [PubMed] [Google Scholar]
- 33.Bang Y-J, Van Cutsem E, Feyereislova A, Chung HC, Shen L, Sawaki A, Lordick F, Ohtsu A, Omuro Y, Satoh T, et al. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): a phase 3, open-label, randomised controlled trial. The Lancet 2010; 376(9742):687–97; PMID:20728210; http://dx.doi.org/ 10.1016/S0140-6736(10)61121-X [DOI] [PubMed] [Google Scholar]
- 34.Sorscher SM, Marked response to single agent trastuzumab in a patient with metastatic HER-2 gene amplified rectal cancer. Cancer Invest 2011; 29(7):456–9; PMID:21696295; http://dx.doi.org/ 10.3109/0735907.2011.590569 [DOI] [PubMed] [Google Scholar]
- 35.Ramanathan RK, Hwang JJ, Zamboni WC, Sinicrope FA, Safran H, Wong MK, Earle M, Brufsky A, Evans T, Troetschel M, et al.. Low overexpression of HER-2/Neu in advanced colorectal cancer limits the usefulness of trastuzumab (Herceptin→) and irinotecan as therapy. A phase II trial #, †. Cancer Invest 2004; 22(6):858-65; PMID:15641483; http://dx.doi.org/ 10.1081/CNV-200039645 [DOI] [PubMed] [Google Scholar]
- 36.Siena S. et al. Therapeutic dual inhibition of HER2 pathway for metastatic colorectal cancer (mCRC): The HERACLES trial. J Clin Oncol 33, 2015 (suppl 3; abstr 565) [Google Scholar]
- 37.Klaver YL, Lemmens VE, Creemers GJ, Rutten HJ, Nienhuijs SW, de Hingh IH. Population-based survival of patients with peritoneal carcinomatosis from colorectal origin in the era of increasing use of palliative chemotherapy. Ann Oncol 2011; 22(10):2250–6; PMID:21345939; http://dx.doi.org/ 10.1093/annonc/mdq762 [DOI] [PubMed] [Google Scholar]
- 38.Shen P, Hawksworth J, Lovato J, Loggie BW, Geisinger KR, Fleming RA, Levine EA. Cytoreductive surgery and intraperitoneal hyperthermic chemotherapy with mitomycin C for peritoneal carcinomatosis from nonappendiceal colorectal carcinoma. Ann Surg Oncol 2004; 11(2):178–86; PMID:14761921; http://dx.doi.org/ 10.1245/ASO.2004.05.009 [DOI] [PubMed] [Google Scholar]
- 39.Franko J, Shi Q, Goldman CD, Pockaj BA, Nelson GD, Goldberg RM, Pitot HC, Grothey A, Alberts SR, Sargent DJ. Treatment of colorectal peritoneal carcinomatosis with systemic chemotherapy: A pooled analysis of north central cancer treatment group phase III trials N9741 and N9841. J Clin Oncol 2012; 30(3):263–7; PMID:22162570; http://dx.doi.org/ 10.1200/JCO.2011.37.1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li P, Fang YJ, Li F, Ou QJ, Chen G, Ma G. ERCC1, defective mismatch repair status as predictive biomarkers of survival for stage III colon cancer patients receiving oxaliplatin-based adjuvant chemotherapy. Br J Cancer 2013; 108(6):1238–44; PMID:23481186; http://dx.doi.org/ 10.1038/bjc.2013.83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang E, Kang HJ, Koh KH, Rhee H, Kim NK, Kim H. Frequent inactivation of SPARC by promoter hypermethylation in colon cancers. Int J Cancer 2007; 121(3):567–75; PMID:17397030; http://dx.doi.org/ 10.1002/ijc.22706 [DOI] [PubMed] [Google Scholar]
- 42.Von Hoff, D.D., M. Hidalgo, et. al. (2011) “Gemcitabine plus nabpaclitaxel is an active regimen in patients with advanced pancreatic cancer: a phase I/II trial.” J. Clin. Oncol.DOI: 10.1200/JCO.2011.36.5742.; PMID: 21969517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Von Hoff DD, Ervin T, Arena FP, Chiorean EG, Infante J, Moore M, Seay T, Tjulandin SA, Ma WW, Saleh MN, et al. Increased survival in pancreatic cancer with nab-paclitaxel plus gemcitabine. New Engl J Med 2013; 369(18):1691–703; PMID:24131140; http://dx.doi.org/ 10.1056/NEJMoa1304369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hidalgo M, Plaza C, Musteanu M, Illei P, Brachmann CB, Heise C, Pierce D, Lopez-Casas PP, Menendez C, Tabernero J, et al. SPARC expression did not predict efficacy of nab-paclitaxel plus gemcitabine or gemcitabine alone for metastatic pancreatic cancer in an exploratory analysis of the phase III MPACT trial. Clin Cancer Res 2015; http://dx.doi.org/ 10.1158/1078-0432.CCR-14-3222 [DOI] [PubMed] [Google Scholar]
- 45.Clinical Trials.gov 2015. [cited 2015 Mrch 31]; Available from: https://clinicaltrials.gov/ [Google Scholar]
- 46.Zeng ZS, Weiser MR, Kuntz E, Chen CT, Khan SA, Forslund A, Nash GM, Gimbel M, Yamaguchi Y, Culliford AT 4th, et al. c-Met gene amplification is associated with advanced stage colorectal cancer and liver metastases. Cancer Lett 2008; 265(2):258–69; PMID:18395971; http://dx.doi.org/ 10.1016/j.canlet.2008.02.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sundermeyer ML, Meropol NJ, Rogatko A, Wang H, Cohen SJ. Changing patterns of bone and brain metastases in patients with colorectal cancer. Clin Colorectal Cancer 2005; 5(2):108–13; PMID:16098251; http://dx.doi.org/ 10.3816/CCC.2005.n.022 [DOI] [PubMed] [Google Scholar]
- 48.Birnkrant A, Sampson J, Sugarbaker P. Ovarian metastasis from colorectal cancer. Dis Colon & Rectum 1986; 29(11):767–71; PMID:3533472; http://dx.doi.org/ 10.1007/BF02555331 [DOI] [PubMed] [Google Scholar]
- 49.Moore RG, Chung M, Granai CO, Gajewski W, Steinhoff MM. Incidence of metastasis to the ovaries from nongenital tract primary tumors. Gynecol Oncol 2004; 93(1):87–91; PMID:15047218; http://dx.doi.org/ 10.1016/j.ygyno.2003.12.039 [DOI] [PubMed] [Google Scholar]
- 50.Goéré D, Daveau C, Elias D, Boige V, Tomasic G, Bonnet S, Pocard M, Dromain C, Ducreux M, Lasser P, et al. The differential response to chemotherapy of ovarian metastases from colorectal carcinoma. Euro J Surg Oncol (EJSO) 2008; 34(12):1335–9; PMID:18455357; http://dx.doi.org/ 10.1016/j.ejso.2008.03.010 [DOI] [PubMed] [Google Scholar]
- 51.Huang P, Weber TK, Mendoza C, Rodriguez-Bigas MA, Petrelli NJ. Long-term survival in patients with ovarian metastases from colorectal carcinoma. Ann Surg Oncol 1998; 5(8):695–8; PMID:9869515; http://dx.doi.org/ 10.1007/BF02303479 [DOI] [PubMed] [Google Scholar]
- 52.Singh B, Berry JA, Shoher A, Ayers GD, Wei C, Lucci A. COX-2 involvement in breast cancer metastasis to bone. Oncogene 2007; 26(26):3789–96; PMID:17213821; http://dx.doi.org/ 10.1038/sj.onc.1210154 [DOI] [PubMed] [Google Scholar]
- 53.Klenke FM, Gebhard MM, Ewerbeck V, Abdollahi A, Huber PE, Sckell A. The selective Cox-2 inhibitor Celecoxib suppresses angiogenesis and growth of secondary bone tumors: an intravital microscopy study in mice. BMC Cancer 2006; 6:9; PMID:16409625; http://dx.doi.org/ 10.1186/1471-2407-6-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stoehlmacher J, Lenz HJ. Cyclooxygenase-2 inhibitors in colorectal cancer. Semin Oncol 2003; 30(3 Suppl 6):10–6; PMID:12802790; http://dx.doi.org/ 10.1016/S0093-7754(03)00120-9 [DOI] [PubMed] [Google Scholar]
- 55.Edelman MJ, Watson D, Wang X, Morrison C, Kratzke RA, Jewell S, Hodgson L, Mauer AM, Gajra A, Masters GA, et al. Eicosanoid modulation in advanced lung cancer: cyclooxygenase-2 expression is a positive predictive factor for celecoxib + chemotherapy–Cancer and Leukemia Group B Trial 30203. J Clin Oncol 2008; 26(6):848–55; PMID:18281656; http://dx.doi.org/ 10.1200/JCO.2007.13.8081 [DOI] [PubMed] [Google Scholar]
- 56.Tie J, Lipton L, Desai J, Gibbs P, Jorissen RN, Christie M, Drummond KJ, Thomson BN, Usatoff V, Evans PM, et al. KRAS mutation is associated with lung metastasis in patients with curatively resected colorectal cancer. Clin Cancer Res 2011; 17(5):1122–30; PMID:21239505; http://dx.doi.org/ 10.1158/1078-0432.CCR-10-1720 [DOI] [PubMed] [Google Scholar]
- 57.Dienstmann R, Serpico D, Rodon J, Saura C, Macarulla T, Elez E, Alsina M, Capdevila J, Perez-Garcia J, Sánchez-Ollé G, et al. Molecular profiling of patients with colorectal cancer and matched targeted therapy in phase I clinical trials. Mol Cancer Ther 2012; 11(9):2062–71; PMID:22723336; http://dx.doi.org/ 10.1158/1535-7163.MCT-12-0290 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.