Abstract
Autophagy is a process tightly regulated by various autophagy-related proteins. It is generally classified into non-selective and selective autophagy. Whereas non-selective autophagy is triggered when the cell is under starvation, selective autophagy is involved in eliminating dysfunctional organelles, misfolded and/or ubiquitylated proteins, and intracellular pathogens. These components are recognized by autophagy receptors and delivered to phagophores. Several selective autophagy receptors have been identified and characterized. They usually have some common domains, such as ![]() motif, a specific cargo interacting (ubiquitin-dependent or ubiquitin-independent) domain. Recently, structural data of these autophagy receptors has been described, which provides an insight of their function in the selective autophagic process. In this review, we summarize the most up-to-date findings about the structure-function of autophagy receptors that regulates selective autophagy. [BMB Reports 2016; 49(2): 73-80]
motif, a specific cargo interacting (ubiquitin-dependent or ubiquitin-independent) domain. Recently, structural data of these autophagy receptors has been described, which provides an insight of their function in the selective autophagic process. In this review, we summarize the most up-to-date findings about the structure-function of autophagy receptors that regulates selective autophagy. [BMB Reports 2016; 49(2): 73-80]
Keywords: Autophagy, LIR motif, Receptor, Selective autophagy, Ubiquitin binding domain
INTRODUCTION
The term ‘autophagy’ comes from the Greek words ‘auto’ and ‘phagy’ which means ‘self’ and ‘eating’, respectively. The name was coined by Dr. Christian de Duve for the first time in 1963 (1, 2). For several decades, there were few publications on autophagy. However, the identification of autophagy-related genes (atg) in yeast by Dr. Oshumi’s group in the late 1990s invoked an interest in this field (3). The core machinery involved in the major steps of autophagy, such as autophagy initiation, autophagosome elongation and maturation, and lysosomal fusion, have been extensively studied for the last 10 years (2, 4-16). Initially, autophagy was thought to be the non-selective and bulk degradation process, which is a main pathway induced in response to starvation. However, it is obvious that the elimination of pathogens and the removal of damaged organelles or proteins, are highly selective processes that require specific cargo recognition by the autophagy machinery (17-20). Therefore, autophagy can be divided into two groups: non-selective and selective autophagy. Some specific proteins, known as autophagy receptors, recognize such target cargos and move them to the autophagosomal membrane. The selective autophagy is further divided into several sub-groups depending on its cargos: degradation of aggregated proteins (aggrephagy) (21-24), mitochondria (mitophagy) (25-32), peroxisomes (pexophagy) (33-35), ribosomes (ribophagy) (36), endoplasmic reticulum (ERphagy) (37-39), lipid (lipophagy) (40), nuclear envelope (nucleophagy) (39), and pathogens (xenophagy) (41-44). Different autophagy receptors are involved in the recognition steps of particular cargos and thus, structural studies on autophagy receptors are necessary for understanding their specificities. In this review, we will introduce the domain architecture of several autophagy receptors and structural information of the domains. In addition, the structural basis for the interaction between these domains and its interacting partners will also be reviewed.
DOMAIN ARCHITECTURE OF AUTOPHAGY RECEPTORS
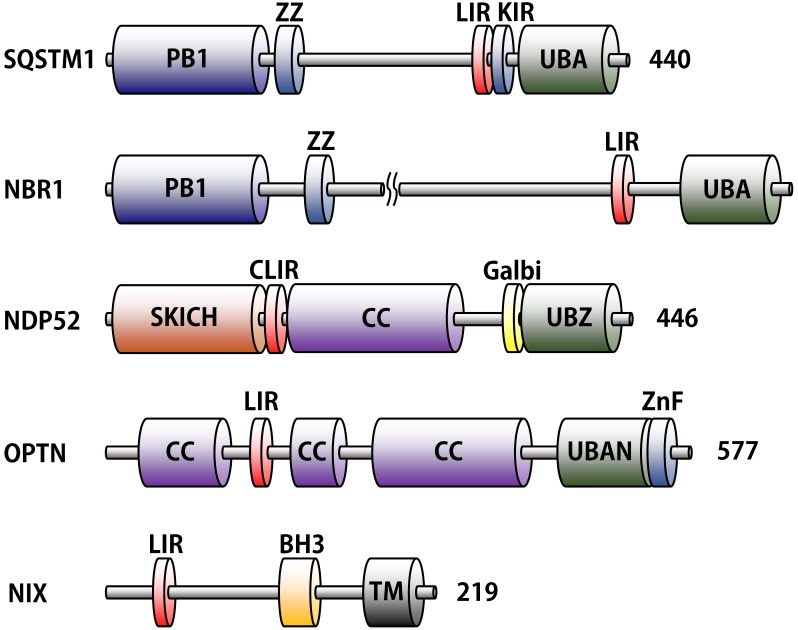
Autophagy receptors are identified by the yeast two-hybrid system and proteomic approaches in mammalian cells (22, 45). The five well-known autophagy receptors, SQSTM1, NBR1, NDP52, OPTN, and NIX, are introduced in this review (Fig. 1 and Table 1). Most of them harbor both LC3-interacting region (LIR) and ubiquitin-binding domain (UBD) (46, 47). The LIR motifs interact with ubiquitin-like proteins like LC3s or GABARAPs in autophagy, and UBD (diverse ubiquitin binding domains in each receptors) with the ubiquitin attached to cargos (19, 48).
Fig. 1. Domain structures of autophagy receptor proteins. LC3-interacting motifs (LIR) and ubiquitin binding domains (UBA, UBZ, and UBAN) are distinguished by red and green color, respectively. The other unique domains represent different colors. The following abbreviations are used for each domain: PB1, Phox and Bem1p; ZZ, ZZ-type zinc finger; LIR, LC3-interacting region; KIR, Keap1-interacting region, UBA, ubiquitin-associated; SKICH, skeletal muscle and kidney-enriched inositol phosphatase carboxyl homology; CLIR, non-canonical LIR; CC, coiled-coil; Galbi, galectin-8 binding region; UBZ, ubiquitin-binding zinc finger; UBAN, ubiquitin-binding in ABIN and NEMO; ZnF, Zn-finger; BH3, Bcl-2 homology region 3; TM, transmembrane.
Table 1. Summary of the determined structures of autophagy receptors.
| Name | Domain/motif | Functions | PDB IDs | References |
|---|---|---|---|---|
|
| ||||
| SQSTM1 | PB1 | Oligomerization and binding to PKCξ | 2KKC, 4UF8, 4UF9, 2KTR, 4MJS, | (82-84, 96) |
| ZZ | Interaction with RIPK1 and R-Bip | - | (86) | |
| LIR | LC3 binding | 2K6Q, 2ZJD | (71) | |
| KIR | Keap1 binding | 3ADE, 3WDZ | (53, 90) | |
| UBA | Ubiquitin binding | 1Q02, 2RRU, 3B0F, 2K0B, 2KNV, 2JY7 | (76, 78, 97, 98) | |
| NBR1 | PB1 | Oligomerization | 1WJ6, 2BKF, 2G4S | (85, 99) |
| ZZ | Unknown | - | - | |
| LIR | LC3 binding | 2L8J | (70) | |
| UBA | Ubiquitin binding | 2CP8, 2MJ5, 2MGW | (79) | |
| NDP52 | SKICH | Unknown | 3VVV | (58) |
| CLIR | LC3C binding | 3VVW | (58) | |
| Coiled-Coil | Homo-dimerization | - | (60) | |
| Galbi | Galectin-8 binding | 4GXL,4HAN | (60, 93) | |
| UBZ | Ubiquitin binding | 2MXP, 4XKL | (59) | |
| OPTN | Coiled-Coil | Oligomerization and binding to protein aggregates | - | (23) |
| LIR | LC3 binding | 2LUE, 3VTV, 3VTW | (73) | |
| UBAN | Ubiquitin binding | - | (66) | |
| ZnF | Unknown | 2LO4 | - | |
| NIX | LIR | GABARAP binding | 4WAA | (28) |
| BH3 | Cell death and autophagy pathway | - | (67) | |
| TM | Membrane insertion | - | - | |
The first identified selective autophagy receptor is SQSTM1 (sequestosome 1; also known as p62) (49). SQSTM1 is not only known to act as a scaffold protein in NF-ᴋB pathway, but also interacts to ubiquitin-containing proteins related to protein aggregation diseases (50-52). It has 5 domains: Phox and Bem1p domain (PB1), ZZ-type zinc finger domain (ZZ), LIR, Keap1-interacting region (KIR), and ubiquitin-associated (UBA) domain (53, 54).
NBR1 (Neighbor of BRCA1 gene 1) was cloned as a candidate gene for the ovarian cancer antigen originally (55). Quite similar to SQSTM1, it also contains PB1, ZZ, LIR, and UBA(Fig. 1). Due to a PB1 domain, NBR1 oligomerizes itself or with SQSTM1, and acts independently or cooperatively to degrade the cargos (56).
NDP52 (also known as CALCOCO2), expressed in all human tissues and cells, was originally found in nuclear promyelocytic leukemia bodies (57). It comprises an N-terminal skeletal muscle and kidney-enriched inositol phosphatase carboxyl homology (SKICH) domain, non-canonical LIR (CLIR) motif that can interact with the LC3C specifically, coiled-coil (CC) domain, galectin-8 binding region (Galbi), and ubiquitin-binding zinc finger (UBZ) domain (58-60). It is especially involved in xenophagy for clearing pathogens such as Salmonella, Listeria, Shigella, and Mycobacterium (42, 61-62).
OPTN (optineurin) is the most recently identified autophagy receptor. It has LIR motif, MYO6 binding region, CC, ubiquitin-binding in ABIN and NEMO (UBAN) domain, and Zn-finger (ZnF) domain (23, 63-66). Interestingly, many other receptors have common LIR motif, but OPTN has a unique serine residue at the upstream LIR sequence, which is modified to phosphoserine by Ser/Thr TANK-binding kinase 1 (TBK1) (43).
NIX (also known as BNIP3L) protein, one of BH3-only members of the Bcl-2 family, has LIR motif, BH3 domain, and a transmembrane region (67, 68). As the sole mitophagy receptor, it has only an LIR motif, but no ubiquitin interacting domain (Fig. 1). Table 1 summarizes the function and structural information of each domain in different autophagy receptors.
STRUCTURAL BASIS FOR THE RECOGNITION OF LIR-MOTIF
There are 6 mammalian homologs of Atg8 comprising the microtubule-associated protein (MAP) light chain 3 (LC3) and γ-aminobutyric acid (GABA)-receptor-associated proteins (GABARAP). Therefore, mammalian LIR motif is similar to AIM (Atg8-interacting motif) in yeast (69). These proteins are involved in the elongation and maturation of autophagosome, respectively (70). All autophagy receptors possess the LIR motif for interacting with LC3 (Fig. 1 and Table 1). A general definition of the LIR sequence is Ω-x-x-Ψ, where Ω and Ψ are aromatic and hydrophobic residues respectively, and the two residues in between are any other amino acids. Preferentially, one or more acidic residues are needed before the LIR sequence (69).
Thus far, detailed structural information of several LC3-LIR complexes has been reported. The first complex structure in mammals is LC3B-SQSTM1 LIR peptide [PDB ID: 2ZJD] (Fig. 2A) (71). This complex shows how canonical LIR motif (W-x-x-L) binds to LC3. The N-terminal aromatic tryptophan residue (Ω) of LIR sequence plugs into the hydrophobic pockets and the downstream hydrophobic leucine residue (Ψ) binds to another hydrophobic pocket of LC3B. The acidic residues prior to LIR sequence bind to the basic side chains of LC3B (Fig. 2A) (21, 71). Structure of NIX LIR-fused LC3B [PDB ID: 4WAA] is quite similar to that of LC3B- SQSTM1 LIR (Fig. 2B). The LIR sequence of NBR1 is ‘Y-I-I-I’. The structure of GABARAPL1-NBR1 LIR peptide [PDB ID: 2L8J] shows that all four residues in the peptide interact with GABARAPL1, and the tyrosine and isoleucine residues are in the same Ω-x-x-Ψ manner as the LC3B- SQSTM1 LIR (Fig. 2C) (72).
Fig. 2. Structures of LC3-LIR complex. (A) LC3B/SQSTM1-LIR peptide complex [2ZJD], (B) NIX-LIR fused LC3B [4WAA], (C) GABARAPL1/NBR1-LIR peptide complex [2L8J], (D) LC3C/NDP52 SKICHLIR region complex [3VVW], and (E) LC3B/OPTN-phosphoLIR peptide complex [2LUE]. Surface of LC3 or GABARAP protein is shown in dark gray color, and the ribbon of LIR motifs in green. The side chain of LC3 interacting residues is also shown in stick model, colored green, red, blue, and orange for carbon, oxygen, nitrogen, and phosphorous, respectively.
Deviated LIR sequences have also been identified (43, 58). The NDP52 and OPTN of xenophagy receptors, especially those related to Salmonella, have unique and non-canonical LIR motif. The LIR sequence of NDP52 comprises the ‘I-L-V-V’ and lacks the aromatic residue. The crystal structure of LC3C-NDP52 complex [PDB ID: 3VVW] revealed that the non-canonical LIR (termed CLIR) binds to the β2 strand of LC3C, but does not reach the hydrophobic pocket that normally binds the tryptophan in a canonical LIR (Fig 2D). The Asp132 residue of NPD52 outside CLIR interacts with Lys57 of LC3C (58).
Another xenophagy receptor protein, OPTN, is activated by phosphorylation. The LIR sequence of OPTN is ‘F-V-E-I’ which is a canonical LIR motif, but the binding affinity of LC3BOPTN LIR is only 64.5-40.0 μM, which is lower than that of LC3B-OPTN phospho-LIR (4 μM). The NMR structure of LC3B-OPTN phospho-LIR [PDB ID: 2LUE] revealed that phospho-Ser177 upstream of the LIR motif in OPTN tightly binds to Arg11 and Lys51 of LC3B (Fig. 2E) (43, 73). These results clearly show that the acidic residue or phosphorylation site upstream of the LIR region also needs to be taken into account.
UBIQUITIN BINDING DOMAIN OF AUTOPHAGY RECEPTORS
According to recent papers, modification of targets with ubiquitin relates not only to proteasome degradation systems, but also degradation signals in selective autophagy (19, 74). When invaded pathogens in cytosol are decorated by a poly-ubiquitin, like K48- or K63-linked chain, they are recognized by the UBD of autophagy receptors and removed to phagophores (61, 75). SQSTM1/p62, NBR1, NDP52, and OPTN each have unique UBD (Table 1). Three UBD structures of SQSTM1, NBR1 and NDP52 are currently elucidated, and the details are as follows.
The solution and crystal structure of UBA domain of SQSTM1 were determined in 2003 and 2011, respectively (Fig. 3A) (76, 77). Although the complex structure between SQSTM1 UBA and ubiquitin is still unknown, conformational changes such as α3 helix extension and side chain flipping at Gln434 and Tyr435 upon ubiquitin binding are revealed through NMR titration (77). Interestingly, SQSTM1 UBA domain prefers to bind K48- or K63-liked di-ubiquitin rather than mono-ubiquitin (78). The structure of NBR1 UBA domain is very similar to that of UBA domain of SQSTM1 (Fig. 3B). Although NBR1 UBA domain also consists of three helixes, the main difference is in the position of helix-3, which encloses a larger angle with helix-2 (16° difference) and is extended by approximately one turn (4 residues) in NBR1 as compared with SQSTM1 (79). The alanine mutation of Phe929 and Leu954 could completely abolish ubiquitin interaction. Substitution of Glu926 for alanine decreases the affinity to ubiquitin about 3-fold, thereby highlighting the contribution of the electrostatic interaction with ubiquitin. The overall structures of UBA domain of SQSTM1-Ub and NBR1-Ub complex resemble the well-known canonical UBA domain of DSK2 and ubiquitin complex, in which the UBA helices adopt a similar geometry, and interfacial residues are involved in forming similar hydrophobic contacts (80).
Fig. 3. Overall structures of ubiquitin binding domain (UBD) and ubiquitin complex. (A) Ubiquitin (model) with SQSTM1 UBA domain [1Q02] and (B) Ubiquitin with NBR1 UBA domain [2MJ5] are shown in ribbon. The ubiquitin interacting Gln434 and Tyr435 of SQSTM1 UBA, and Glu926, Phe929, and Leu954 of NBR1 UBA are shown in stick model. (C) Ubiquitin with NDP52 ZF2 domain [4XKL]. A bound zinc atom is shown as a gray ball. UBD and ubiquitin are colored green and cyan, respectively.
The structures of NDP52 UBZ domain with or without ubiquitin [PDB IDs: 2MXP and 4XKL] have recently been determined (Fig. 3C) (59). The UBZ domain has two zinc-finger domains, a dynamic unconventional zinc finger (ZF1) and a canonical C2H2-type zinc finger (ZF2). Surprisingly, only one of two zinc-fingers, ZF2, can interact with 3 types of poly-ubiquitin chains (K48- and K63-linked as well as M1 (methionine 1)-linked) (59).
UBIQUITIN-INDEPENDENT INTERACTION DOMAINS OF AUTOPHAGY RECEPTORS
PB1 domain is a scaffold module that interacts with each other in a front-to-back mode to arrange heterodimers or homooligomers. PB1-containg proteins display the topology of an ubiquitin-like β-grasp fold, including six-stranded β-sheets and two α-helices (Fig. 4A) (81). The folding pattern of PB1 domain of SQSTM1 is quite similar with that of other PB1-containing proteins. The p62 dimer or oligomers are formed by the electrostatic interaction of two opposite charged surface patches with conserved acidic and basic residues (Fig. 4A) (82). The PB1 (residues 1-122) domain of SQSTM1 assembles to form flexible helical polymers (83). On interacting with K63-linked octa-ubiquitin chain (but not with mono- and di-ubiquitin, or LC3), the SQSTM1 helical filament could dissociate by induced conformational changes (83, 84). The structure of the PB1 domain of NBR1 is also similar and reminiscent of those from previously characterized PB1 domains, including SQSTM1 (85).
Fig. 4. Structures of ubiquitin-independent binding domains in autophagy receptors. (A) PB1 domains of SQSTM1 (magenta) and NBR1 (yellow) are superposed. Basic side chains of Lys7, Arg21, Arg22, and Arg94 residues and acidic side chains of Asp67, Glu68, Asp69, Asp71, and Glu80 residues involved in the oligomerization are shown in stick model. (B) Complex of Keap1 (orange) and SQSTM1-KIR peptide (green). The β-propeller structure of Keap1 recognizes KIR peptide containing phosphorylated Ser351. (C) NDP52 SKICH domain (slate) [3VVV], and (D) Complex between galectin-8 (salmon) and NDP52 Galbi peptide (green) [4HAN]. N-terminal carbohydrate binding domain (N-CRD) and C-terminal carbohydrate binding domain (C-CRD) are labeled. Ribbon diagram of overall structure and stick model of bound peptides are shown.
ZZ-type zinc finger (ZZ domain, residue 122-167) is a mysterious domain in SQSTM1 (Fig. 1 and Table 1). The ZZ domain is related to NF-kB signaling and glucose regulation. This domain could associate with RIPK1. Recently, some studies reveal that the ZZ domain of SQSTM1 could link autophagy to N-end rule pathway (86, 87). However, the mechanism between SQSTM1 and N-end rule is still unclear due to lack of structural information of ZZ domain.
TRAF6 binding (TB) domain of SQSTM1 interacts with TRAF6, an E3 ubiquitin ligase that regulates mTORC1 activity by K63-ubiquitination in a SQSTM1-interaction dependent manner. Through this TB domain, SQSTM1 could function as a crucial mediator for mTORC1 activity regulation (88).
Keap1 interacting region (KIR) is the most well studied domain of SQSTM1. KEAP1-NRF2 system is a response for oxidative stress (89). Under normal conditions, KEAP1 is dimerized and tightly interacts with NRF2. However, stress conditions induce the phosphorylation of S351 in KIR by mTORC1, and this causes the dissociation of KEAP1-NRF2 complex, leading to the formation of the KEAP1-SQSTM1 complex. The phosphorylated SQSTM1 KIR is recognized by β-propeller structure of Keap1 (Fig. 4B). In comparison to the non-phosphorylated state, two additional Keap1 residues (Arg483 and Ser508) were involved in the binding interaction with phosphorylated KIR (90). This phosphorylation of Ser351 in KIR is important for the antioxidant pathway as well as SQSTM1 mediated autophagy.
The SKICH domain of NDP52, which is responsible for the interaction with AZI2/NAP1 and its crystal structure, has been determined at 1.35 Å (Fig. 4C) (58). It shares the immunoglobulin-fold, and its closest structural homolog is the human C3 complement component (42, 91). Recently, the SKICH domain is reported to interact with phosphorylated tau and this suggests that NDP52 plays a role in autophagy-mediated degradation of phosphorylated tau (92).
NDP52 also interacts with galectin-8, a sugar receptor in the region between the CC and UBZ domain (Fig. 1) (44). The galectin-8 has two homologous carbohydrate-recognition domains (N- and C-CRD) linked by a single polypeptide chain. It serves as a danger receptor because it recruits NDP52 to the ruptured Salmonella-containing vesicles by recognizing carbohydrate moiety. The complex structures of galectin-8 and NDP52 peptide (Galbi region, residues 372-380) were determined by two independent groups (60, 93). Only C-CRD of galectin-8, through an opposing site of sugar recognition, directly binds to the Galbi region of NDP52 (Fig. 4D).
CONCLUDING REMARKS
Selective autophagy has attracted a great interest for understanding the autophagic process and the subsequent cellular homeostasis. Different autophagy receptors play a critical role for cargo selection using particular domains or motifs (Table 1). For clear understanding of their selectivity, the structural data of the complex between these recognition domains and target molecules are inevitable. Although many structures of the domain of autophagy receptors are now available, structural information of some domains is still unknown (Table 1). More importantly, it is critical to have the structures of fulllength protein but not a domain structure, because the cooperative action of individual domains occurs for efficient process of selective autophagy. So far, there is no structural information of full-length autophagy receptors at high resolution and it would be necessary to tackle this problem using various other techniques, especially advanced cryo-electron microscopy (94), or using combined methods such as small-angle X-ray scattering with a high resolution structure of each domain (95). These structural works in combination with cell biology, and in vivo data will provide the groundwork for understanding the molecular mechanism of each autophagy receptors.
Acknowledgments
We apologize to the researchers who were not referenced due to space limitations and unintentional missing. This work was supported by grants from the National Research Foundation of Korea (NRF-2011-0028168 and BRL grant: No. 2015041919).
References
- 1.Klionsky DJ. Autophagy revisited: a conversation with Christian de Duve. Autophagy. (2008);4:740–743. doi: 10.4161/auto.6398. [DOI] [PubMed] [Google Scholar]
- 2.Ohsumi Y. Historical landmarks of autophagy research. Cell Res. (2014);24:9–23. doi: 10.1038/cr.2013.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mizushima N, Noda T, Yoshimori T, et al. A protein conjugation system essential for autophagy. Nature. (1998);395:395–398. doi: 10.1038/26506. [DOI] [PubMed] [Google Scholar]
- 4.Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat Rev Mol Cell Biol. (2009);10:458–467. doi: 10.1038/nrm2708. [DOI] [PubMed] [Google Scholar]
- 5.Ohsumi Y. Molecular dissection of autophagy: two ubiquitin-like systems. Nat Rev Mol Cell Biol. (2001);2:211–216. doi: 10.1038/35056522. [DOI] [PubMed] [Google Scholar]
- 6.Hong SB, Kim BW, Lee KE, et al. Insights into noncanonical E1 enzyme activation from the structure of autophagic E1 Atg7 with Atg8. Nat Struct Mol Biol. (2011);18:1323–1330. doi: 10.1038/nsmb.2165. [DOI] [PubMed] [Google Scholar]
- 7.Suzuki H, Kaizuka T, Mizushima N, Noda NN. Structure of the Atg101-Atg13 complex reveals essential roles of Atg101 in autophagy initiation. Nat Struct Mol Biol. (2015);22:572–580. doi: 10.1038/nsmb.3036. [DOI] [PubMed] [Google Scholar]
- 8.Fujioka Y, Suzuki SW, Yamamoto H. Structural basis of starvation-induced assembly of the autophagy initiation complex. Nat Struct Mol Biol. (2014);21:513–521. doi: 10.1038/nsmb.2822. [DOI] [PubMed] [Google Scholar]
- 9.Hong SB, Kim BW, Kim JH, Song HK. Structure of the autophagic E2 enzyme Atg10. Acta Crystallogr D Biol Crystallogr. (2012);68:1409–1417. doi: 10.1107/S0907444912034166. [DOI] [PubMed] [Google Scholar]
- 10.Hurley JH, Schulman BA. Atomistic autophagy: the structures of cellular self-digestion. Cell. (2014);157:300–311. doi: 10.1016/j.cell.2014.01.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klionsky DJ, Schulman BA. Dynamic regulation of macroautophagy by distinctive ubiquitin-like proteins. Nat Struct Mol Biol. (2014);21:336–345. doi: 10.1038/nsmb.2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaiser SE, Mao K, Taherbhoy AM, et al. Noncanonical E2 recruitment by the autophagy E1 revealed by Atg7-Atg3 and Atg7-Atg10 structures. Nat Struct Mol Biol. (2012);19:1242–1249. doi: 10.1038/nsmb.2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaiser SE, Qiu Y, Coats JE, Mao K, Klionsky DJ, Schulman BA. Structures of Atg7-Atg3 and Atg7-Atg10 reveal noncanonical mechanisms of E2 recruitment by the autophagy E1. Autophagy. (2013);9:778–780. doi: 10.4161/auto.23644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim JH, Hong SB, Lee JK, et al. Insights into autophagosome maturation revealed by the structures of ATG5 with its interacting partners. Autophagy. (2015);11:75–87. doi: 10.4161/15548627.2014.984276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Popelka H, Klionsky DJ. Post-translationally-modified structures in the autophagy machinery: an integrative perspective. FEBS J. (2015);282:3474–3488. doi: 10.1111/febs.13356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim JH, Song HK. Swapping of interaction partners with ATG5 for autophagosome maturation. BMB Rep. (2015);48:129–130. doi: 10.5483/BMBRep.2015.48.3.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature. (2011);469:323–335. doi: 10.1038/nature09782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang Z, Klionsky DJ. Eaten alive: a history of macroautophagy. Nat Cell Biol. (2010);12:814–822. doi: 10.1038/ncb0910-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kirkin V, McEwan DG, Novak I, Dikic I. A role for ubiquitin in selective autophagy. Mol Cell. (2009);34:259–269. doi: 10.1016/j.molcel.2009.04.026. [DOI] [PubMed] [Google Scholar]
- 20.Kraft C, Peter M, Hofmann K. Selective autophagy: ubiquitin-mediated recognition and beyond. Nat Cell Biol. (2010);12:836–841. doi: 10.1038/ncb0910-836. [DOI] [PubMed] [Google Scholar]
- 21.Pankiv S, Clausen TH, Lamark T, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. (2007);282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 22.Kirkin V, Lamark T, Sou YS, et al. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol Cell. (2009);33:505–516. doi: 10.1016/j.molcel.2009.01.020. [DOI] [PubMed] [Google Scholar]
- 23.Korac J, Schaeffer V, Kovacevic I, et al. Ubiquitin-independent function of optineurin in autophagic clearance of protein aggregates. J Cell Sci. (2013);126:580–592. doi: 10.1242/jcs.114926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu K, Psakhye I, Jentsch S. Autophagic clearance of polyQ proteins mediated by ubiquitin-Atg8 adaptors of the conserved CUET protein family. Cell. (2014);158:549–563. doi: 10.1016/j.cell.2014.05.048. [DOI] [PubMed] [Google Scholar]
- 25.Sarraf SA, Raman M, Guarani-Pereira V, et al. Landscape of the PARKIN-dependent ubiquitylome in response to mitochondrial depolarization. Nature. (2013);496:372–376. doi: 10.1038/nature12043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wong YC, Holzbaur EL. Optineurin is an autophagy receptor for damaged mitochondria in parkin-mediated mitophagy that is disrupted by an ALS-linked mutation. Proc Natl Acad Sci U S A. (2014);111:E4439–4448. doi: 10.1073/pnas.1405752111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lazarou M, Sliter DA, Kane LA, et al. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature. (2015);524:309–314. doi: 10.1038/nature14893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Novak I, Kirkin V, McEwan DG, et al. Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep. (2010);11:45–51. doi: 10.1038/embor.2009.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Quinsay MN, Thomas RL, Lee Y, Gustafsson AB. Bnip3-mediated mitochondrial autophagy is independent of the mitochondrial permeability transition pore. Autophagy. (2010);6:855–862. doi: 10.4161/auto.6.7.13005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kanki T, Wang K, Cao Y, Baba M, Klionsky DJ. Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Dev Cell. (2009);17:98–109. doi: 10.1016/j.devcel.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Okamoto K, Kondo-Okamoto N, Ohsumi Y. Mitochondria-anchored receptor Atg32 mediates degradation of mitochondria via selective autophagy. Dev Cell. (2009);17:87–97. doi: 10.1016/j.devcel.2009.06.013. [DOI] [PubMed] [Google Scholar]
- 32.Hanna RA, Quinsay MN, Orogo AM, Giang K, Rikka S, Gustafsson AB. Microtubule-associated protein 1 light chain 3 (LC3) interacts with Bnip3 protein to selectively remove endoplasmic reticulum and mitochondria via autophagy. J Biol Chem. (2012);287:19094–19104. doi: 10.1074/jbc.M111.322933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deosaran E, Larsen KB, Hua R, et al. NBR1 acts as an autophagy receptor for peroxisomes. J Cell Sci. (2013);126:939–952. doi: 10.1242/jcs.114819. [DOI] [PubMed] [Google Scholar]
- 34.Motley AM, Nuttall JM, Hettema EH. Pex3-anchored Atg36 tags peroxisomes for degradation in Saccharomyces cerevisiae. EMBO J. (2012);31:2858–2868. doi: 10.1038/emboj.2012.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang S, Wells CD, Roach PJ. Starch-binding do main-containing protein 1 (Stbd1) and glycogen metabolism: Identification of the Atg8 family interacting motif (AIM) in Stbd1 required for interaction with GABARAPL1. Biochem Biophys Res Commun. (2011);413:420–425. doi: 10.1016/j.bbrc.2011.08.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kraft C, Deplazes A, Sohrmann M, Peter M. Mature ribosomes are selectively degraded upon starvation by an autophagy pathway requiring the Ubp3p/Bre5p ubiquitin protease. Nat Cell Biol. (2008);10:602–610. doi: 10.1038/ncb1723. [DOI] [PubMed] [Google Scholar]
- 37.Khaminets A, Heinrich T, Mari M, et al. Regulation of endoplasmic reticulum turnover by selective autophagy. Nature. (2015);522:354–358. doi: 10.1038/nature14498. [DOI] [PubMed] [Google Scholar]
- 38.Kurth I, Pamminger T, Hennings JC, et al. Mutations in FAM134B, encoding a newly identified Golgi protein, cause severe sensory and autonomic neuropathy. Nat Genet. (2009);41:1179–1181. doi: 10.1038/ng.464. [DOI] [PubMed] [Google Scholar]
- 39.Mochida K, Oikawa Y, Kimura Y, et al. Receptormediated selective autophagy degrades the endoplasmic reticulum and the nucleus. Nature. (2015);522:359–362. doi: 10.1038/nature14506. [DOI] [PubMed] [Google Scholar]
- 40.Singh R, Kaushik S, Wang Y, et al. Autophagy regulates lipid metabolism. Nature. (2009);458:1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zheng YT, Shahnazari S, Brech A, Lamark T, Johansen T, Brumell JH. The adaptor protein p62/SQSTM1 targets invading bacteria to the autophagy pathway. J Immunol. (2009);183:5909–5916. doi: 10.4049/jimmunol.0900441. [DOI] [PubMed] [Google Scholar]
- 42.Thurston TL, Ryzhakov G, Bloor S, von Muhlinen N, Randow F. The TBK1 adaptor and autophagy receptor NDP52 restricts the proliferation of ubiquitin-coated bacteria. Nat Immunol. (2009);10:1215–1221. doi: 10.1038/ni.1800. [DOI] [PubMed] [Google Scholar]
- 43.Wild P, Farhan H, McEwan DG, et al. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science. (2011);333:228–233. doi: 10.1126/science.1205405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thurston TL, Wandel MP, von Muhlinen N, Foeglein A, Randow F. Galectin 8 targets damaged vesicles for autophagy to defend cells against bacterial invasion. Nature. (2012);482:414–418. doi: 10.1038/nature10744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Behrends C, Sowa ME, Gygi SP, Harper JW. Network organization of the human autophagy system. Nature. (2010);466:68–76. doi: 10.1038/nature09204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wild P, McEwan DG, Dikic I. The LC3 interactome at a glance. J Cell Sci. (2014);127:3–9. doi: 10.1242/jcs.140426. [DOI] [PubMed] [Google Scholar]
- 47.Xu Z, Yang L, Xu S, Zhang Z, Cao Y. The receptor proteins: pivotal roles in selective autophagy. Acta Biochim Biophys Sin (Shanghai) (2015);47:571–580. doi: 10.1093/abbs/gmv055. [DOI] [PubMed] [Google Scholar]
- 48.Slobodkin MR, Elazar Z. The Atg8 family: multifunctional ubiquitin-like key regulators of autophagy. Essays Biochem. (2013);55:51–64. doi: 10.1042/bse0550051. [DOI] [PubMed] [Google Scholar]
- 49.Bjorkoy G, Lamark T, Brech A, et al. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. (2005);171:603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moscat J, Diaz-Meco MT, Wooten MW. Signal integration and diversification through the p62 scaffold protein. Trends Biochem Sci. (2007);32:95–100. doi: 10.1016/j.tibs.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 51.Kuusisto E, Salminen A, Alafuzoff I. Ubiquitinbinding protein p62 is present in neuronal and glial inclusions in human tauopathies and synucleinopathies. Neuroreport. (2001);12:2085–2090. doi: 10.1097/00001756-200107200-00009. [DOI] [PubMed] [Google Scholar]
- 52.Zatloukal K, Stumptner C, Fuchsbichler A, et al. p62 Is a common component of cytoplasmic inclusions in protein aggregation diseases. Am J Pathol. (2002);160:255–263. doi: 10.1016/S0002-9440(10)64369-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Komatsu M, Kurokawa H, Waguri S, et al. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat Cell Biol. (2010);12:213–223. doi: 10.1038/ncb2021. [DOI] [PubMed] [Google Scholar]
- 54.Noda NN, Kumeta H, Nakatogawa H, et al. Structural basis of target recognition by Atg8/LC3 during selective autophagy. Genes Cells. (2008);13:1211–1218. doi: 10.1111/j.1365-2443.2008.01238.x. [DOI] [PubMed] [Google Scholar]
- 55.Campbell IG, Nicolai HM, Foulkes WD, et al. A novel gene encoding a B-box protein within the BRCA1 region at 17q21.1. Hum Mol Genet. (1994);3:589–594. doi: 10.1093/hmg/3.4.589. [DOI] [PubMed] [Google Scholar]
- 56.Lamark T, Kirkin V, Dikic I, Johansen T. NBR1 and p62 as cargo receptors for selective autophagy of ubiquitinated targets. Cell Cycle. (2009);8:1986–1990. doi: 10.4161/cc.8.13.8892. [DOI] [PubMed] [Google Scholar]
- 57.Korioth F, Gieffers C, Maul GG, Frey J. Molecular characterization of NDP52, a novel protein of the nuclear domain 10, which is redistributed upon virus infection and interferon treatment. J Cell Biol. (1995);130:1–13. doi: 10.1083/jcb.130.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.von Muhlinen N, Akutsu M, Ravenhill BJ, et al. LC3C, bound selectively by a noncanonical LIR motif in NDP52, is required for antibacterial autophagy. Mol Cell. (2012);48:329–342. doi: 10.1016/j.molcel.2012.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xie X, Li F, Wang Y, et al. Molecular basis of ubiquitin recognition by the autophagy receptor CALCOCO2. Autophagy. (2015);11:1775–1789. doi: 10.1080/15548627.2015.1082025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim BW, Hong SB, Kim JH, Kwon do H, Song HK. Structural basis for recognition of autophagic receptor NDP52 by the sugar receptor galectin-8. Nat Commun. (2013);4:1613. doi: 10.1038/ncomms2606. [DOI] [PubMed] [Google Scholar]
- 61.Watson RO, Manzanillo PS, Cox JS. Extracellular M. tuberculosis DNA targets bacteria for autophagy by activating the host DNA-sensing pathway. Cell. (2012);150:803–815. doi: 10.1016/j.cell.2012.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mostowy S, Sancho-Shimizu V, Hamon MA, et al. p62 and NDP52 proteins target intracytosolic Shigella and Listeria to different autophagy pathways. J Biol Chem. (2011);286:26987–26995. doi: 10.1074/jbc.M111.223610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Majcher V, Goode A, James V, Layfield R. Autophagy receptor defects and ALS-FTLD. Mol Cell Neurosci. (2015);66:43–52. doi: 10.1016/j.mcn.2015.01.002. [DOI] [PubMed] [Google Scholar]
- 64.Weidberg H, Elazar Z. TBK1 mediates crosstalk between the innate immune response and autophagy. Sci Signal. (2011);4:39. doi: 10.1126/scisignal.2002355. [DOI] [PubMed] [Google Scholar]
- 65.Tumbarello DA, Waxse BJ, Arden SD, Bright NA, Kendrick-Jones J, Buss F. Autophagy receptors link myosin VI to autophagosomes to mediate Tom1-dependent autophagosome maturation and fusion with the lysosome. Nat Cell Biol. (2012);14:1024–1035. doi: 10.1038/ncb2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Heo JM, Ordureau A, Paulo JA, Rinehart J, Harper JW. The PINK1-PARKIN Mitochondrial Ubiquitylation Pathway Drives a Program of OPTN/NDP52 Recruitment and TBK1 Activation to Promote Mitophagy. Mol Cell. (2015);60:7–20. doi: 10.1016/j.molcel.2015.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang J, Ney PA. Role of BNIP3 and NIX in cell death, autophagy, and mitophagy. Cell Death Differ. (2009);16:939–946. doi: 10.1038/cdd.2009.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kanki T. Nix, a receptor protein for mitophagy in mammals. Autophagy. (2010);6:433–435. doi: 10.4161/auto.6.3.11420. [DOI] [PubMed] [Google Scholar]
- 69.Noda NN, Ohsumi Y, Inagaki F. Atg8-family interacting motif crucial for selective autophagy. FEBS Lett. (2010);584:1379–1385. doi: 10.1016/j.febslet.2010.01.018. [DOI] [PubMed] [Google Scholar]
- 70.Weidberg H, Shvets E, Shpilka T, Shimron F, Shinder V, Elazar Z. LC3 and GATE-16/GABARAP subfamilies are both essential yet act differently in autophagosome biogenesis. EMBO J. (2010);29:1792–1802. doi: 10.1038/emboj.2010.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ichimura Y, Kumanomidou T, Sou YS, et al. Structural basis for sorting mechanism of p62 in selective autophagy. J Biol Chem. (2008);283:22847–22857. doi: 10.1074/jbc.M802182200. [DOI] [PubMed] [Google Scholar]
- 72.Rozenknop A, Rogov VV, Rogova NY, et al. Characterization of the interaction of GABARAPL-1 with the LIR motif of NBR1. J Mol Biol. (2011);410:477–487. doi: 10.1016/j.jmb.2011.05.003. [DOI] [PubMed] [Google Scholar]
- 73.Rogov VV, Suzuki H, Fiskin E, et al. Structural basis for phosphorylation-triggered autophagic clearance of Salmonella. Biochem J. (2013);454:459–466. doi: 10.1042/BJ20121907. [DOI] [PubMed] [Google Scholar]
- 74.Shaid S, Brandts CH, Serve H, Dikic I. Ubiquitination and selective autophagy. Cell Death Differ. (2013);20:21–30. doi: 10.1038/cdd.2012.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Manzanillo PS, Ayres JS, Watson RO, et al. The ubiquitin ligase parkin mediates resistance to intracellular pathogens. Nature. (2013);501:512–516. doi: 10.1038/nature12566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ciani B, Layfield R, Cavey JR, Sheppard PW, Searle MS. Structure of the ubiquitin-associated domain of p62 (SQSTM1) and implications for mutations that cause Paget's disease of bone. J Biol Chem. (2003);278:37409–37412. doi: 10.1074/jbc.M307416200. [DOI] [PubMed] [Google Scholar]
- 77.Isogai S, Morimoto D, Arita K, et al. Crystal structure of the ubiquitin-associated (UBA) domain of p62 and its interaction with ubiquitin. J Biol Chem. (2011);286:31864–31874. doi: 10.1074/jbc.M111.259630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Long J, Gallagher TR, Cavey JR, et al. Ubiquitin recognition by the ubiquitin-associated domain of p62 involves a novel conformational switch. J Biol Chem. (2008);283:5427–5440. doi: 10.1074/jbc.M704973200. [DOI] [PubMed] [Google Scholar]
- 79.Walinda E, Morimoto D, Sugase K, Konuma T, Tochio H, Shirakawa M. Solution structure of the ubiquitin-associated (UBA) domain of human autophagy receptor NBR1 and its interaction with ubiquitin and polyubiquitin. J Biol Chem. (2014);289:13890–13902. doi: 10.1074/jbc.M114.555441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Michielssens S, Peters JH, Ban D, et al. A designed conformational shift to control protein binding specificity. Angew Chem Int Ed Engl. (2014);53:10367–10374. doi: 10.1002/anie.201403102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Moscat J, Diaz-Meco MT, Albert A, Campuzano S. Cell signaling and function organized by PB1 domain interactions. Mol Cell. (2006);23:631–640. doi: 10.1016/j.molcel.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 82.Saio T, Yokochi M, Kumeta H, Inagaki F. PCS-based structure determination of protein-protein complexes. J Biomol NMR. (2010);46:271–280. doi: 10.1007/s10858-010-9401-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ciuffa R, Lamark T, Tarafder AK, et al. The selective autophagy receptor p62 forms a flexible filamentous helical scaffold. Cell Rep. (2015);11:748–758. doi: 10.1016/j.celrep.2015.03.062. [DOI] [PubMed] [Google Scholar]
- 84.Saio T, Yokochi M, Inagaki F. The NMR structure of the p62 PB1 domain, a key protein in autophagy and NF-kappaB signaling pathway. J Biomol NMR. (2009);45:335–341. doi: 10.1007/s10858-009-9370-7. [DOI] [PubMed] [Google Scholar]
- 85.Muller S, Kursula I, Zou P, Wilmanns M. Crystal structure of the PB1 domain of NBR1. FEBS Lett. (2006);580:341–344. doi: 10.1016/j.febslet.2005.12.021. [DOI] [PubMed] [Google Scholar]
- 86.Cha-Molstad H, Sung KS, Hwang J, et al. Amino-terminal arginylation targets endoplasmic reticulum chaperone BiP for autophagy through p62 binding. Nat Cell Biol. (2015);17:917–929. doi: 10.1038/ncb3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cha-Molstad H, Kwon YT, Kim BY. Amino-terminal arginylation as a degradation signal for selective autophagy. BMB Rep. (2015);48:487–488. doi: 10.5483/BMBRep.2015.48.9.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Katsuragi Y, Ichimura Y, Komatsu M. p62/SQSTM1 functions as a signaling hub and an autophagy adaptor. FEBS J. (2015);282:4672–4678. doi: 10.1111/febs.13540. [DOI] [PubMed] [Google Scholar]
- 89.Jain A, Lamark T, Sjottem E, et al. p62/SQSTM1 is a target gene for transcription factor NRF2 and creates a positive feedback loop by inducing antioxidant response element-driven gene transcription. J Biol Chem. (2010);285:22576–22591. doi: 10.1074/jbc.M110.118976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ichimura Y, Waguri S, Sou YS, et al. Phosphorylation of p62 activates the Keap1-Nrf2 pathway during selective autophagy. Mol Cell. (2013);51:618–631. doi: 10.1016/j.molcel.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 91.Janssen BJ, Huizinga EG, Raaijmakers HC, et al. Structures of complement component C3 provide insights into the function and evolution of immunity. Nature. (2005);437:505–511. doi: 10.1038/nature04005. [DOI] [PubMed] [Google Scholar]
- 92.Jo C, Gundemir S, Pritchard S, Jin YN, Rahman I, Johnson GV. Nrf2 reduces levels of phosphorylated tau protein by inducing autophagy adaptor protein NDP52. Nat Commun. (2014);5:3496. doi: 10.1038/ncomms4496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Li S, Wandel MP, Li F, et al. Sterical hindrance promotes selectivity of the autophagy cargo receptor NDP52 for the danger receptor galectin-8 in antibacterial autophagy. Sci Signal. (2013);6 doi: 10.1126/scisignal.2003730. ra9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Nogales E, Scheres SH. Cryo-EM: A Unique Tool for the Visualization of Macromolecular Complexity. Mol Cell. (2015);58:677–689. doi: 10.1016/j.molcel.2015.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rambo RP, Tainer JA. Super-resolution in solution X-ray scattering and its applications to structural systems biology. Annu Rev Biophys. (2013);42:415–441. doi: 10.1146/annurev-biophys-083012-130301. [DOI] [PubMed] [Google Scholar]
- 96.Ren J, Wang J, Wang Z, Wu J. Structural and biochemical insights into the homotypic PB1-PB1 complex between PKCzeta and p62. Sci China Life Sci. (2014);57:69–80. doi: 10.1007/s11427-013-4592-z. [DOI] [PubMed] [Google Scholar]
- 97.Evans CL, Long JE, Gallagher TR, Hirst JD, Searle MS. Conformation and dynamics of the three-helix bundle UBA domain of p62 from experiment and simulation. Proteins. (2008);71:227–240. doi: 10.1002/prot.21692. [DOI] [PubMed] [Google Scholar]
- 98.Long J, Garner TP, Pandya MJ, et al. Dimerisation of the UBA domain of p62 inhibits ubiquitin binding and regulates NF-kappaB signalling. J Mol Biol. (2010);396:178–194. doi: 10.1016/j.jmb.2009.11.032. [DOI] [PubMed] [Google Scholar]
- 99.Mueller-Dieckmann C, Panjikar S, Schmidt A, et al. On the routine use of soft X-rays in macromolecular crystallography. Part IV. Efficient determination of anomalous substructures in biomacromolecules using longer X-ray wavelengths. Acta Crystallogr D Biol Crystallogr. (2007);63:366–380. doi: 10.1107/S0907444906055624. [DOI] [PubMed] [Google Scholar]