Abstract
The aim of this study is to clarify the clinical implication and functional role of structure specific recognition protein 1 (SSRP1) in hepatocellular carcinoma (HCC) and explore the underlying mechanism of aberrant high expression of SSRP1 in cancers. In the present investigation, we validated that SSRP1 was upregulated in HCC samples. We also demonstrated that its upregulation was associated with several clinicopathologic features such as higher serum AFP level, larger tumor size, and higher T stage of HCC patients; and its high expression indicated shorter overall survival and faster recurrence. To investigate the role of SSRP1 in HCC progression, both loss- and gain-function models were established. We demonstrated that SSPR1 modulated both proliferation and metastasis of HCC cells in vitro and vivo. Furthermore, we demonstrated that SSRP1-modulated apoptosis process and its knockdown increased the sensitivity of HCC cells to doxorubicin, 5-Fluorouracil, and cisplatin. We also identified microRNA-497 (miR-497) as a posttranscriptional regulator of SSRP1. Ectopic expression of miR-497 inhibited 3'-untranslated-region–coupled luciferase activity and suppressed endogenous SSRP1 expression at both messenger RNA and protein levels. For the first time, we proved that SSRP1 upregulation contributed to HCC development and the tumor-suppressive miR-497 served as its negative regulator.
Introduction
Hepatocellular carcinoma (HCC) is a major health problem worldwide, especially in East Asia including China, Korea, and Japan.1 The mechanism of HCC is highly heterogeneous and complicated that is accompanied by various genetic abnormalities. Even though many potential therapeutic targets and diagnostic biomarkers of this disease have been discovered, its detailed mechanism is still obscure. It is well believed that the elucidation of molecular mechanisms underlying HCC tumorigenesis and progression is important for the development of novel treatments.
Structure-specific recognition protein 1 (SSRP1), a subunit of the facilitates chromatin transcription (FACT) complex, is involved in transcriptional regulation, DNA damage repair, and cell cycle regulation.2,3,4 It is reported that high SSRP1 expression is associated with stem or less-differentiated cells, while low SSRP1 levels are seen in more differentiated cells.5 What's more, SSRP1 expression is upregulated in multiple cancers such as lung cancer, colon cancer, breast cancer, pancreatic cancer, and strongly associated with adverse clinicopathological features and worse overall survival in these diseases.6 In vitro, knockdown of SSRP1 decreases cell growth of non–small-cell lung cancer7; in vivo, FACT expression level is found to be increased during tumorigenesis of mammary carcinoma in transgenic mice expressing the Her2/neu protooncogene, and the inhibition of FACT blocks tumor onset, delays tumor progression, and prolongs survival of mice in a dose-dependent manner.8
The discoveries above indicate that SSRP1 may have roles as diagnostic biomarker and anticancer target in aforementioned cancers. However, to date, no studies have focused on the clinicopathologic significance and functional role of SSRP1 in HCC. Furthermore, although SSRP1 has been shown to be an oncogene in many human cancers, previous studies only focused on its role in stimulating proliferation via promoting cell cycle progress, promoting malignant transformation via inhibiting differentiation, its function in metastasis and chemotherapeutical resistance has rarely been addressed. Another issue remains to be solved is the high expression mechanism of SSRP1 in cancer tissues. MicroRNAs are a class of short noncoding RNA sequences containing 19–22 nucleotides. Various aberrantly expressed miRNAs play crucial roles in malignant transformation by regulating downstream oncogenes and tumor suppressors.9 We made a hypothesis that the aberrant expression of SSRP1 is partially due to upstream dysregulation of microRNAs.
In the current study, we presented the first evidence that the expression pattern of SSRP1 in human HCC and its clinical significance. We demonstrated SSRP1 modulates not only proliferation, but also metastasis and chemosensitivity of HCC. We also demonstrated the negative regulation on SSRP1 level by microRNA-497(miR-497). In conclusion, we identified SSRP1 as an important oncogene in HCC, and aberrant SSRP1 upregulation was partly attributed to the underexpression of miR-497.
Results
SSRP1 is upregulated in HCC on both mRNA and protein levels
Firstly, we observed SSRP1 expression on mRNA level in the large cohorts of HCC patients available from two independent GEO datasets GSE14520 (ref. 10) and GSE22058.11 As shown (Figure 1a), SSRP1 expression was significantly upregulated in these two different HCC cohorts. We also examined the expression levels of SSRP1 in 73 HCC and corresponding noncancerous liver tissues using immunohistochemical staining (Supplementary Data). SSRP1 was expressed in both nucleus and cytoplasm (Figure 1b,c), which was consistent with previous reports.5,6 Our results indicated that SSRP1 was upregulated in the majority of (83.6%, 61/73) of HCC patients. SSRP1 expressions of 10 randomly selected HCC tissues paired with adjacent noncancerous liver tissues were investigated by quantitative real-time PCR (qRT-PCR). From this, all 10 HCC tissues exhibited significantly upregulation of SSRP1 in HCC (Figure 1d). Additionally, expression of SSRP1 was investigated by western blot in different HCC cell lines, namely HepG2, SMMC7721, Huh7, Bel7402, PLC, LM3, 97L, 97H and the normal liver cell line L02. In accord with the results from tissues, significantly higher expression of SSRP1 was detected in most hepatocellular carcinoma cells, compared with L02 cells (Figure 1e). These findings evidently demonstrated that SSRP1 was upregulated in HCC on both mRNA and protein level, implying the importance of it in HCC pathogenesis.
Figure 1.
The expression of SSRP1 is increased in hepatocellular carcinoma (HCC) tissue. (a) SSRP1 expression level in HCC tissue and adjacent/normal tissue in two independent cohorts (GSE14520 and GSE22058). (b) The expression of SSRP1 was upregulated in 83.6% of HCC patients examined by immunohistochemical staining. (c) Representative photographs of staining of SSRP1 protein in a pair of HCC and adjacent tissues. (d) Lysates from paired tissues of HCC and adjacent tissue were analyzed by real-time PCR for the detection of SSRP1. (e) Western blotting analysis of SSRP1 expression in immortalized normal liver L02 cell and 8 HCC cell lines. β-actin was used as loading control.
High expression level of SSRP1 accumulates more copy number variations and mutation counts
Genetic alterations resulting from dysfunctional DNA damage repair, such as somatic copy number alteration and mutations, are recognized as common features of human cancers, and the primary cause of cancer development including HCC was initially attributed to genetic alterations.12 HCC is characterized by a wide spectrum of genomic alterations, some of which might be resulted from defects in homologous recombination (HR).13 It is reported that overexpression of the SSRP1 markedly reduced Rad51 expression, the crucial factor in HR.3 We made a hypothesis that persistent downregulation of HR function by the high expression of SSRP1 could disrupt DNA repair and result in genetic instability in HCC. By analyzing the data of TCGA database,14,15 we demonstrated that greater fraction of copy number altered genome and higher p53 and RB1 mutation frequencies were detected in HCC patients with higher SSRP1 expression than in those with lower SSRP1 expression (Supplementary Figure S1), suggesting that upregulation of SSRP1 may promote HCC progression via accumulating of extra copies of DNA and gene mutations in HCC cells.
The expression levels of SSRP1 in HCC patients correlates with several clinicopathologic characteristics and HCC patients' survival
In order to confirm the correlation between the expression level of SSRP1 and clinicopathologic factors in HCC, we downloaded the clinical information from GSE14520 and analyzed them statistically. Then the samples pooled in the dataset were classified into two groups according to the expression level of SSRP1 in tumor tissue and χ2 test was used. As shown (Table 1), SSRP1 expression was closely associated with higher AFP (P = 0.013), tumor size (P = 0.029), AJCC T stage (P = 0.026), BCLC stage (P = 0.032), and CLIP stage (P = 0.028). The results indicated that high expression of SSRP1 hinted faster carcinoma enlarging and spreading, more severe liver damage, and worse physical conditions. Current analysis also revealed that higher SSRP1 expression significantly correlated with high predicted risk metastasis gene signature (P < 0.0001), which strongly indicated that SSRP1 could influence the expression profile of metastasis-related gene. Additionally, we also use our immunohistochemical results to analyze the correlation between SSRP1 protein expression and tissue differentiation status of HCC. We demonstrated that higher SSRP1 expression is significantly associated with poorer differentiation of cancer tissue (Supplementary Table S1), which is consistent with previous report,5 implying its potential role as an anti-HCC target. Importantly, Kaplan-Meier survival analysis showed HCC patients with tumors displaying high SSRP1 expression levels had significantly shorter overall survival (OS) (P < 0.001, hazard ratio = 2.048, 95% CI = 1.365–3.072, Figure 2a) and recurrence-free survival (RFS) (P = 0.013, hazard ratio = 1.754, 95% CI = 1.247–2.468, Figure 2b) compared to those with high SSRP1 expression tumors. Similar results were obtained in another independent cohort (TCGA cohort), showing patients with higher SSRP1 expression suffered from shorter OS and RFS (Supplementary Figure S2). These results strongly suggested that SSRP1 functioned as an oncogene in HCC and could represent a potential new prognostic factor for HCC after curative hepatectomy. Interestingly, as another subunit of FACT complex, the expression of SPT16 was not significantly associated with the prognosis of HCC patients (Supplementary Figure S3), implying that during the process FACT taking part in HCC progression, the dominator is SSRP1 but not SPT16.
Table 1. Correlation between the SSRP1 expression and the clinicopathologic features of hepatocellular carcinoma (GSE14520).

Figure 2.
High expression of SSRP1 is associated with poor prognosis of hepatocellular carcinoma (HCC). (a) High SSRP1 mRNA levels reduce overall survival of HCC patients in dataset GSE14520. (b) High SSRP1 mRNA levels reduce recurrence-free survival of HCC patients in dataset GSE14520.
SSRP1 modulates HCC cell proliferation in vitro and in vivo
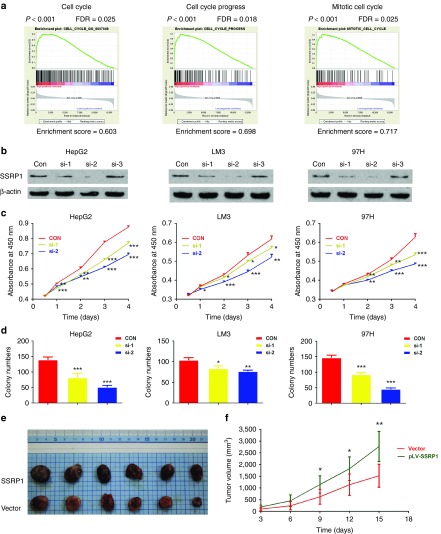
Then SSRP1 was subjected to GSEA16 using the mRNA expression profiling data from GSE14520, in order to identify biological process potentially modulated by SSRP1. GSEA analysis of GO terms demonstrated a lot of gene sets were significantly enriched by SSRP1. As shown, there were enriched expression of gene sets involved in cell cycle and mitosis (Figure 3a). Sustaining mitogenic signaling and abnormal process of cell cycle are crucial for tumor growth and enlarging,12 so our bioinformatic results gave a possible explanation of why high expression of SSRP1 was associated with unfavorable clinicopathological characteristics of HCC such as tumor size and T stage (Table 1).
Figure 3.
SSRP1 modulates proliferation of hepatocellular carcinoma (HCC) cells in vitro and in vivo. (a) GSEA analysis of GO terms showed SSRP1 might regulate gene sets associated with cell cycle checkpoint (left), cell cycle progress (middle), and mitosis process (right). (b) Knockdown of SSRP1 in HepG2 (left), LM3 (middle), and 97H (right) cell lines by siRNAs targeting SSRP1 was confirmed by western blotting; β-actin was used as a loading control. (c) Effect of SSRP1 knockdown on the proliferation on HepG2 (left), LM3 (middle), and 97H (right) cells was determined by CCK-8 assay. (d) Effect of SSRP1 knockdown on colony numbers was determined by colony formation assay in HepG2 (left), LM3 (middle), and 97H (right) cells. (e) Tumors were dissected and each tumor in two groups was exhibited. (f) Representative data showed that overexpression of SSRP1 significantly promoted tumor growth in nude mice xenograft model (n = 6). *, **, *** represents P < 0.05, P < 0.01, P < 0.001 respectively.
Given the findings above, to validate the biological role of SSRP1 in proliferation of HCC, SSRP1 was depleted using two siRNAs in HepG2, LM3 and 97H cells, which exhibit a higher expression of SSRP1. Additionally, SSRP1 was also stably overexpressed by lentivirus-mediated packed pLV-SSRP1 vector in the SMMC7721 cell line, which exhibits a relatively lower level of SSRP1 expression. The knockdown and ectopic expression of SSRP1 in cells were affirmed by western blot (Figure 3b and Supplementary Figure S4a).
As expected (Figure 3c), markedly, HepG2, LM3, and 97H cells displayed a lower cell proliferation rate than control cells after SSRP1 knockdown. Correspondingly, SMMC7721 cells showed a significantly higher cell growth rate after ectopic expression of SSRP1 than that observed in the controls with the empty vector (Supplementary Figure S4b). What's more, cell proliferation was also measured using a plate colony formation assay. Compared with the control cells, SSRP1 knockdown in HepG2, 97H, and LM3 cells led to markedly decreased colony formation ability (Figure 3d). Consistent with these observations, SSRP1-overexpressing SMMC7721 cells displayed significantly increased colony formation (Supplementary Figure S4c). To verify the positive role of SSRP1 in HCC progression in vivo, we performed xenograft tumor assays using SMMC7721 cells stably transfected by SSRP1-overexpression lentiviruses. We found that SSRP1 overexpression significantly promoted xenograft tumor growth in nude mice (Figure 3e,f). Collectively, these findings indicate that SSRP1 is closely associated with the proliferation of HCC cell.
Downregulation of SSRP1 promotes cell cycle arrest and causes apoptosis in HCC cells
Enhancing cell cycle progression and inhibiting apoptosis are two crucial causes of sustaining proliferation. To explore the potential cause of SSRP1 in regulating proliferation of HCC cells, we examined the cell cycle in HCC cells through flow cytometry. After treatment with si-SSRP1 or control siRNA for 48 hours, SSRP1 knockdown led to a significant accumulation of cells at the G0/G1-phase and a significant decrease in cells in the S/G2/M-phase in HepG2, 97H, and LM3 cells (Figure 4a), whereas overexpression of SSRP1 promoted the cell cycle progression of SMMC7721 cells (Supplementary Figure S4d). We then detected the effects of SSRP1 on apoptosis under normal condition or oxidative stress. In both conditions, overexpression of SSRP1 significantly decreased the number of apoptotic cells in SMMC7721 (Supplementary Figure S4e), whereas SSRP1 loss of function significantly increased the numbers of apoptotic cells in HepG2, 97H and LM3 cells (Figure 4b, Supplementary Figure S5a,b). Collectively, these results reveal that SSRP1 modulates both cell cycle and apoptosis in HCC cells.
Figure 4.
SSRP1 regulates hepatocellular carcinoma (HCC) cells proliferation via both cell cycle and apoptosis. (a) Effects of SSRP1 knockdown on the percentage of cells in S/G2/M phase were determined by fluorescence activated cell sorting (FACS) analysis in HepG2 (left), LM3 (middle), and 97H (right) cells. (b) After Annexin V-FITC/PI staining of the indicated cells after treatment without or with H2O2 (1mmol) for 12 hours, effects of SSRP1 knockdown on HepG2 cells apoptosis were determined by FACS analysis (upper), and quantitative analysis of apoptotic cell numbers is shown (bottom). Each bar represents the mean ± SD of three independent tests. **, *** represents P < 0.01 and P < 0.001 respectively.
SSRP1 modulates cell migration and invasion of HCC cells in vitro and in vivo
The role of SSRP1 in promoting HCC cell proliferation is in agreement with the previous reports in breast cancer and non–small-cell lung cancer.6,7 In addition to cell proliferation, our clinicopathological analysis also revealed that higher SSRP1 expression was dramatically associated with a gene expression signature of higher metastasis potential (P < 0.001, Table 1), and the role of SSRP1 in cancer metastasis has not been well characterized. We examined whether SSRP1 was a critical molecular having impact on cell migration and invasion by transwell assays. As shown, knockdown of SSRP1 suppressed the migration and invasion rates of HepG2, 97H, and LM3 cells (Figure 5a,b) whereas forced expression of SSRP1 had the opposite effect on SMMC7721 cells (Supplementary Figure S4f,g). To further substantiate the result, wound healing assay was also employed to evaluate the impact of SSRP1 on cell movement. In line with our previous observations, SSRP1 overexpression increased while SSRP1 knockdown inhibited the mobility of HCC cells (Figure 5c, Supplementary Figure S4h). These results demonstrated that overexpression of SSRP1 enhanced cell migration and invasion while suppression of SSRP1 reduced cell migration and invasion.
Figure 5.
SSRP1 modulates migration, invasion and metastasis ability in hepatocellular carcinoma cells in vitro and in vivo. (a) Cell migration was assessed by transwell assay in HepG2 (left), LM3 (middle), and 97H (right) cells after the cells were transfected with SSRP1 siRNA for 48 hours. The cells which migrated into the bottom surface of the filters were stained. (b) Cell invasion was assessed by transwell assay with matrigel in HepG2 (left), LM3 (middle), and 97H (right) cells after the cells were transfected with SSRP1 siRNA for 48 hours. The cells which invaded into the bottom surface of the filters were stained. The bars represent the mean values of six independent tests (mean ± SD). *, **, *** represents P < 0.05, P < 0.01, P < 0.001 respectively. (c) Movement ability was detected by scratch wound healing assays in HepG2 (left), 97H (middle), and LM3 (right) cells after the cells were transfected with SSRP1 siRNA for 48 hours. (d) Representative photographs showing the colonization of SMMC7721SSRP1 and SMMC7721vector cells in the lung of recipient mice. (e) Incidence and severity of lung metastasis in pulmonary metastasis model with SMMC7721SSRP1 and SMMC7721vector cells.
Colonization at a distant site is the last key step in the metastatic cascade.17 Having observed that SSRP1 modulates HCC cells migration and invasion in vitro, to determine if SSRP1 would stimulate distant colonization of HCC cells, a tail vein injection model was used to imitate the pathophysiological process. Two weeks after injection, mice were killed by cervical decapitation, and lung colonization was quantified by pathological examination. In accordance with this observation in vitro study, in SSRP1 overexpression group, 8 of 10 mice showed severe lung metastasis, whose incidence is significantly higher than that in the empty vector group (1/10) (P = 0.003, Figure 5d,e). These results suggest that upregulation of SSRP1 may have important consequences in metastasis of HCC.
Knockdown of SSRP1 reduces the sensitivity of hepatocellular carcinoma cells to chemotherapeutic drugs
After exploring the role of SSRP1 in HCC cell growth and metastasis, we therefore sought to determine if SSRP1 has the potential to be applied in clinical HCC treatment. Chemotherapy is an important tool in the treatment of HCC. However, for HCC, drug resistance often makes chemotherapy unsatisfactory.18 The negative effect of SSRP1 on HCC cells' apoptotic status prompted us to make a hypothesis that it may also contribute to the drug resistance of HCC cells. We downloaded the pharmacological data set from the NCI website (http://dtp.nci.nih.gov), where the -logGI50 of thousands small molecules and natural products, as well as mRNA expression microarray data were available for 58 human tumor cell lines. Then we conducted a pharmacologic data analysis as previously described.19 Intriguingly, higher SSRP1 mRNA level is associated with a significantly higher mean GI50 (the drug concentration for 50% growth inhibition) for 1,449 compounds, as compared to 134 ± 254 compounds expected by random permutation (P = 0.007; Figure 6a).
Figure 6.
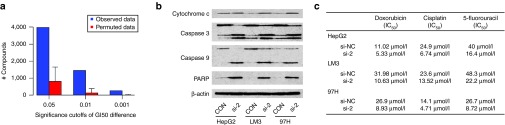
SSRP1 is associated with chemoresistance of hepatocellular carcinoma. (a) SSRP1 is associated with cellular sensitivity to small molecules and natural products. (b) Knockdown of SSRP1 activates apoptotic pathways. (c) siRNA-mediated SSRP1 knockdown increases the sensitivity of hepatocellular carcinoma cells to chemotherapeutics.
We then examined the expressions of apoptosis-related proteins 48 hours post-knockdown of SSRP1 in HepG2, 97H, and LM3 cells by western blot. As shown (Figure 6b), si-SSRP1 transfection induced the cleavage of caspase-9, 3 and PARP and the release of mitochondrial cytochrome c into the cytosol in all the three cell lines. These results reveal that SSRP1 exerts an inhibitory effect on mitochondrial apoptotic pathway.
Prompted by these results, we assayed the chemosensitivity to the three most common chemotherapy drugs, doxorubicin, cisplatin and 5-fluorouracil, to compare SSRP1 knockdown groups and the according control groups. The results showed that the IC50s of all the chemotherapy drugs were remarkably decreased by the knockdown of SSRP1 in all the three HCC cell lines (Figure 6b). These data collectively indicated that SSRP1 contributed to reducing the sensitivity of HCC cells to chemotherapeutic drugs.
Identification of SSRP1 as a novel target for miR-497
Another issue remained to be solved, in not only HCC but also other cancers, is the high expression mechanism of SSRP1 in cancer tissues. Amplification of genes is an important mechanism of overexpression.12 To gain further insight, we have investigated whether SSRP1 locus is amplified in human cancers using TCGA data available on cBioPortal. However, we found that the incidence of homozygous gain of SSRP1 locus was quite low (Supplementary Figure S6). In addition to genomic gain, miRNAs contribute importantly to gene expression regulation. It would be interesting to determine whether or not SSRP1 expression could be regulated by a specific miRNA in HCC.
Three databases, including TargetScan, miRanda, and miRwalk were searched computationally for potential microRNAs that are complementary to the 3'-UTR of SSRP1. To reduce false positives, candidates were only considered if they were predicted by all the three methods. One candidate identified by this approach was miR-497, which has two complementary sites to 3'-UTR of SSRP1 (located at 158~178 and 268~287 respectively, Figure 7a). It has been proved that miR-497 is downregulated in HCC,20,21 and we also validated this by analyzing the GEO datasets GSE21362 (ref. 22) (Figure 7b). So, we supposed that the overexpression of SSRP1 was partially attributed to the downregulation of miR-497. After transfection with miR-497, we observed that the SSRP1 was reduced on both mRNA and protein levels in HepG2 and LM3 cells (Figure 7c,d). Then wild-type or mutant (MUT1, MUT2, and MUT1 and 2) 3'-UTR constructs of SSRP1 were cloned into a psi-CHECK2 vector, respectively, and cotransfected with miR-497 mimics in HEK293 cells. It was also found that miR-497 was able to suppress reporter gene activity of SSRP1 in HEK293T cells, whereas MUT2 plasmid, but not MUT1 showed no change in the reporter gene activity (Figure 7e). These data suggested that SSRP1 was a direct target of miR-497 and the effective binding site is the second one (268~287) but not the first one (158~178). A negative correlation between miR-497 and the SSRP1 expression levels phenomenon was also seen in HCC samples (Figure 7f; P = 0.0025, R = −0.6383). These data support the idea that SSRP1 expression is negatively regulated by miR-497 in HCC.
Figure 7.
miR-497 posttranscriptionally regulated SSRP1 expression by directly targeting its 3' UTR. (a) The target sites of miR-497 in 3'-UTR of SSRP1 are shown as a schematic representation. (b) miR-497 expression level in hepatocellular carcinoma (HCC) tissue and adjacent/normal tissue from the mRNA microarray data obtained from GSE21362. (c) Western blotting assays showed the expression of SSRP1 protein in HepG2 and LM3 cells transfected with miR-497. (d) Real-time quantitive PCR showed the expression of SSRP1 mRNA in HepG2 and LM3 cells transfected with miR-497. (e) Wild-type or mutant 3'-UTR constructs of SSRP1 were cloned into a psi-CHECK2 vector, respectively, and cotransfected with miR-497 mimics in HEK293 cells. Renilla luciferase activities were normalized to firefly luciferase activities. All assays were performed in triplicates and repeated at least three times. (f) An inverse correlation was found between miR-497 expression and SSRP1 in HCC samples (Spearman's correlation, P = 0.0025, R = −0.6383). Data are given as the mean ± SD of three independent experiments; **P < 0.01, ***P < 0.001.
Discussion
Our results strongly proved that SSRP1 promote HCC progression, which is similar with the previous studies in other cancer types such as NSCLC, colon cancer and breast cancer.6,7 Our data showed knockdown of SSRP1 inhibited cell cycle process and promoted apoptosis in HCC cells in vitro, which may result in slower cancer cell proliferation. However, the underlying mechanism still remains unclear, and here several possible explanations are proposed.
Firstly, a hallmark of cancer cell is sustaining proliferative signaling, accompanying enhancing DNA replication.12 SSRP1 is a crucial regulator for maintaining the normal process of DNA replication for that FACT associates with the MCM helicase, promoting the DNA unwinding activity of the MCM helicase on nucleosomal template,23,24 and disruption of the FACT-MCM complex triggers delayed DNA replication initiation. For example, in chicken (Gallus gallus) DT40 cells, SSRP1 knockdown inhibited cell growth and caused a delay in S-phase cell cycle progression due to the inhibition of replication fork progression.23 Secondly, SSRP1 colocalizes with the spindle and midbody microtubules, facilitating tubulin polymerization and microtubules bundling; so its knockdown will result in disorganized spindle structures and block mitosis.25 Furthermore, FACT, a large heterodimeric complex consisting of SSRP1 and SPT16, is categorized as a histone chaperone crucial for nucleosome reorganization during transcription.26 FACT interacts with DNA-binding surfaces of H2A/H2B dimers, decreasing the lifetime of nonproductive RNA polymerase II -nucleosome complexes and facilitating the formation of productive complexes containing nucleosomal DNA partially uncoiled from the histone octamer during transcription.26 High expression level of SSRP1 may activate the transcription of some crucial downstream oncogenes, in turn promotes the progression of cancers. Additionally, it is reported that FACT modulates NF-κB and p53 pathways,7,8,27 both of which are dysregulated in nearly all tumors and considered as driver genes of cancers. We make a hypothesis that high expression SSRP1 will activate NF-κB pathway but suppress p53 pathway in HCC, which remains to be verified. Lastly, in this work, we also observed that there is a correlation between SSRP1 levels and DNA instability, including extra copies of DNA and mutations in specific genes such as TP53 and RB1 (Supplementary Figure S1). As previous studies reported, SSRP1 is involved in the regulation of DNA damage response and repair process; deficiency of SSRP1 might result in aberrant homologous recombination.3 So, it is reasonable to speculate that SSRP1 dysregulation may take part in the formation of instability of cancer genome, promoting HCC progression via accumulating of extra copies of DNA and gene mutations in cells. Whether this assumption is true and which genes are affected remain to be validated by further experiments.
In our work, we demonstrated SSRP1 modulates not only proliferation, but also motility, migration, invasion, and chemosensitivity of HCC. More than 90% of HCC-related deaths are resulted from metastasis.28,29 Emergence of metastasis reflects not only the ability of cancer cells to overcome hurdles during the multistep process of metastasis but also the capability to survive chemotherapeutics and other physiological stresses. Additionally, although chemotherapy is important in combined treatment of hepatocellular carcinoma, its effect is unsatisfactory as HCC cells from advanced cancer with vital metastasis often develop resistance to chemotherapeutic drugs.18 Therefore, elucidation of a driver gene of HCC metastasis and chemoresistance is helpful for the improvement of HCC treatment. Our results indicated that SSRP1 is an important target for blocking metastasis and reversing drug resistance in HCC. We propose that quantification of SSRP1 in liver biopsy could be used in combination with pathologic examination to predict biological behaviors of the HCC. The molecular-pathologic diagnosis will be helpful in personalized treatment optimization.
Accumulated evidence has shed light on the importance of miRNAs on regulation of gene expression networks, further involved in multiple signaling pathways. miRNAs can exert positive or negative control over the expression of oncogenes or tumor-suppressor genes to affect tumor growth. Downregulation of miR-497 and its tumor-suppressive role have been reported in multiple cancers, such as lung, breast, cervical, head-and-neck, prostate cancer, and ovarian cancer.30,31,32,33,34,35,36 Increasing evidence indicates that miR-497 negatively regulates numerous well-characterized oncogenic proteins, such as CCND1,30 IGF-1R,32,33 mTOR,34 BCL-2,36 CDC25A, CDK6, and CDK4.20 In HCC, miR-497 is also downregulated and showed significant growth-suppressive activities.19,20 Most cells of adult tissues do not have detectable protein level of SSRP1 while SSRP1 represented a high expression level in undifferentiated or cancerous tissues, however the mechanisms of upregulation of SSRP1 in cancer have yet to be elucidated. Our work showed SSRP1 is a novel direct target of miR-497 and there is a negative correlation between miR-497 and the SSRP1 expression. Intriguingly, a recent research showed that miR-497 decreases cisplatin resistance in ovarian cancer cells.34 Our results demonstrate that SSRP1 is also involved in chemosensitivity of cancer cells, highlighting the potential role of miR-497 downregulation for cancer cells to develop a chemoresistance phenotype. Considering the underexpression of miR-497 is general in many cancers, it may be a good explanation for the upregulation of SSRP1 in cancers.
NF-κB activation will promote the expression of miR-497 and miR-497 exhibits an inhibitory activity through the cognate site in the 3'-UTR of IKKβ gene, a crucial activator in the canonical pathway of NF-κB.37 Intriguingly, overexpression of SSRP1 is involved in the abnormal sustaining activation of NF-κB signaling,26 and herein we demonstrate that it is targeted by miR-497. These suggest that in normal tissues, SSRP1, miR-497, IKKβ, and NF-κB form double negative-feedback loops to maintain homeostasis. It is reported that the downexpression of miR-497 is due to its hypermethylated promoter region in HCC,38 suggesting that aberrant DNA methylation may disrupts the feedback and lead to excessive activation of NF-κB signaling cascade in cancers, lending additional support for a link between miR-497 deregulation and tumourigenesis.
In summary, our data not only have enriched the knowledge of HCC carcinogenesis, but also provide new insight into the function of SSRP1. In addition, we demonstrated SSRP1 is novel target of miR-497, which partially explains the aberrant high expression of SSRP1 in cancers. This observation further suggests the possible interplay between miRNA and abnormal DNA damage repair process during the course of liver carcinogenesis. Further in-depth exploration of the molecular mechanisms of SSRP1 in promoting the proliferation, metastasis, and drug resistance of HCC will be needed to clarify in detail.
Materials and Methods
Cell culture, culture conditions, and antibodies. HepG2, Bel-7402, SMMC7721, Huh7, PLC/PRF-5, LM3, 97L, 97H HCC cells, and L02 human liver cells were grown and routinely maintained in Dulbecco's modified Eagle medium (DMEM; Gibco, Carlsbad, CA) supplemented with 10% FBS, 100 U/ml penicillin, and 100 mg/ml streptomycin. Cells were incubated at 37 °C in 5% CO2 and 95% humidified air. Anti-SSRP1, cytochrome C, Caspase 3, PARP antibodies were purchased from Abcam (Cambridge, UK); anti-Caspase 9 and β-actin antibodies were purchased from Santa Cruz Biotechnology (Dallas, TX). Anti-rabbit and anti-mouse HRP-conjugated secondary antibodies were obtained from Promega Biotechnology (Dallas, TX).
Patients and immunohistochemical staining. The data of mRNA or miRNA expression profile (GSE14520,10 GSE22058,11 and GSE2136222) are available from the NCBI, GEO database. Our study was approved by the ethics review board of The Second People's Hospital of Guangdong Province (Approval number: 2015-KYLL-023). All HCC samples and paired non-neoplastic tissues used in immunohistochemical and real-time PCR were retrieved from the Department of Pathology, Zhongnan Hospital of Wuhan University, China. Before used, all cases were diagnosed by two certificated pathologists without discrepancy. The paraffin-embedded tissues were first stained with hematoxylin and eosin (HE) for histological examination. Subsequently, deparaffinized sections were treated with 3% H2O2 and subjected to antigen retrieval by citric acid (pH 6.0). After overnight incubated with primary antibody (anti-SSRP1 antibody, 1:200) at 4 °C, sections were incubated for 15 minutes at room temperature with horseradish peroxidase-labeled polymer conjugated with secondary antibody (MaxVision Kits) and incubated for 1 minute with diaminobenzidine. The sections were then lightly counterstained with hematoxylin. The sections without primary antibody were served as negative controls. Expression level of SSRP1 was ascertained according to the average score of two pathologists' evaluations.
Transient transfection and establishment of stable expressing cells. siRNAs were designed and purchased commercially (Genepharma, Suzhou, China) as shown in Table 2.
Table 2. The siRNAs used in the study.

Cells were transiently transfected with siRNA or vectors using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) and were harvested for the analysis of efficiency of knockdown or overexpression 48 hours post-transfection. After validated, the procedure above was repeated for harvesting the cells for assays. For establishment of stable SSRP1-overexpressing cells, overexpressing plasmid was constructed by Inovogen, China; then the plasmid was packed by a lentivirus system (Inovogen, Beijing, China). After removing the medium of the cells, the lentiviral supernatant was added (with 6 μg/ml polybrene). After 24 hours, the supernatant was removed and cells were selected for 4 weeks using DMEM with 10 μg/ml puromycin (Sigma-Aldrich, St Louis, MO).
Western blot. Cells were lysed in lysis buffer containing 50 mmol/l Tris, pH 8.0, 150 mmol/l NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 50 mmol/l NaF, 1 mmol/l Na3 VO4. Then samples were equally loaded on 10% SDS-PAGE, electrophoresed, and transferred onto nitrocellulose membrane (Millipore, Billerica, MA). After blocking with 5% non-fat milk in Tris-buffered saline with Tween 20 (TBST), the membranes were incubated with the primary antibodies overnight at 4 °C. After the overnight incubation with the primary antibodies, membranes were washed and incubated with horseradish peroxidase (HRP)-labeled secondary antibody in TBST for 1 hour. Then the signals were detected by chemiluminescence phototope-HRP kit (Pierce Biotechnology, Rockford, IL) according to manufacturer's instructions. β-actin was used as a loading control.
RNA isolation and real-time PCR analysis. Total RNA was isolated from cell lines or tissues with TRIzol reagents (Invitrogen) according to the manufacturer's instructions. RT-PCR was performed to quantify mature miRNA expression with the NCode miRNA qRT-PCR analysis (Invitrogen), or mRNA expression with SYBR Green PCR Master Mix (TaKaRa, Ohtsu, Japan). U6 snRNA was used for miR-497 normalization and β-actin was used for SSRP1 normalization. Expression of transcripts was assessed using the primers shown in Table 3.
Table 3. The primers used in the study.

Cell proliferation assay. Cells from each group were plated onto 96-well plates containing complete medium on day 0 and allowed to attach overnight. Then, the growth curves of cells, covering a total of four days of culturing, were plotted with the Cell Counting Kit-8 method. In brief, 10 µl Cell Counting Kit-8 (Dojindo, Japan) was added to each well and cultured at 37 °C and 5% CO2 for 1 hour. Then the absorbance was measured with the multifunctional microplate reader at 490 nm. The growth curve was constructed with time as abscissa and absorbance as ordinate. Experiments were performed in triplicate.
Plate colony formation assay. Cells at a log phase of growth were trypsinized, harvested, resuspended and seeded in six-well plates (1,000 cells/well). Then the cells were cultured for 2 weeks (The medium was changed on the day 7). After that, the cells were washed with PBS for twice, fixed with 4% paraformaldehyde for 20 minutes, and then stained with 0.1% crystal violet for 30 minutes. After these, the colonies were carefully washed with PBS until the background was clear. The colony formation rate = (number of colonies/number of incubating cells) × 100%.
Cell-cycle analysis. Cells at a log phase of growth were harvested with 0.25% trypsin and washed twice with PBS. After centrifugation, cells were fixed in 100% ice-cold methanol overnight at −20 °C. Then the fixed cells were stained with 50 mg/ml propidium iodide (PI) in PBS and 1 mg/ml RNAase in PBS for 30 minutes. In each experiment, 10,000 cells were analyzed using BD FACSAria (BD Biosciences, San José, CA). Experiments were performed in triplicate.
Apoptosis analysis. Annexin V-FITC/PI staining was performed to investigate whether SSRP1 regulates the apoptosis of HCC cells. Cells in each group were seeded into six-well plates. When cells of each group were at a log phase of growth, the cells were harvested. In H2O2-induced groups, H2O2 (1 mmol) was added 12 hours before harvest. Then the cells were stained with ApoScreen Annexin V Apoptosis Kit and PI according to the manufacturer's instructions for labeling of apoptotic cells. In each experiment, 10,000 cells were analyzed using BD FACSAria (BD Biosciences). Experiments were performed in triplicate.
In vitro migration and invasion assays. In the scratch wound healing assay, cells were cultured in serum-free medium for 24 hours and wounded with pipette tips. Then the medium was refreshed. The wound closing procedure was observed 48 hours later and photographs were taken. Cell migration assay was conducted using 8-mm pore size Transwell chambers (Corning, Corning, NY). The lower chamber was filled with DMEM containing 10% FBS. Cells were suspended in serum-free DMEM and plated into the upper chamber. Then the chambers were cultivated in 5% CO2 at 37 °C for 48 hours. After that, the cells in the upper chamber were removed with cotton swabs and the bottom surface of the polycarbonate membranes was counted visually using 0.1% crystal violet dye and a light microscope. The invasion assay was same except that matrigel (Clontech, Madison, WI) was used in the transwell chambers (Corning). Cell migration and invasion were determined by counting six random fields under a microscope and the data are presented as mean ± standard deviation (SD).
Nude mice study. Both subcutaneous xenograft and lung metastasis models were constructed. 4-week-old male BALB/C nude mice were used. In subcutaneous xenograft study, 1 × 106 cells (SMMC7721Vector or SMMC7721SSRP1) were resuspended in 100 ml of phosphate-buffered saline (PBS) and injected subcutaneously to the left or right side of nude mice (n = 6). Tumor size was monitored every three days, and tumor volume was calculated with the following formula: 1/2 length × width2. In pulmonary metastasis study, 10 mice were enrolled in each group. 1 × 106 cells (SMMC7721Vector or SMMC7721SSRP1) were injected into caudal vena. Two weeks after caudal vena injection, mice were sacrificed by cervical decapitation, and lung colonization was quantified by pathological examination.
Chemotherapeutic drugs sensitivity analysis by cell counting kit-8. HCC cells (3 × 103 cells/well) were seeded in 96-well plates and treated with different chemotherapeutics doxorubicin, 5-fluorouracil, and cisplatin in different concentrations for 72 hours. Cell Counting Kit-8 (Dojindo, Japan) was added into each well. After 1 hour incubation at 37 °C, the absorbance was measured with the multifunctional microplate reader at 490 nm. The chemotherapeutic drugs sensitivity was evaluated by IC50 parameter (inhibitory concentration of 50% cells). Experiments were performed in triplicate.
Luciferase assay. Cells were seeded into 12-well plates one day before transfection. After 24 hours of plating, the cells were cotransfected with 200 ng of psiCHECK-2 plasmids and 100 nmol/l of miR-497 (or NC microRNA). After 48 hours, luciferase activities were measured with Dual-Luciferase Reporter Assay system (Promega, Madison, WI). Renilla luciferase activity was normalized to firefly luciferase activity.
Bioinformatics analysis. Gene expression profiles including GSE14520 and GSE22058 were downloaded from the Gene Expression Omnibus (GEO) site and analyzed. JAVA program for GSEA (http://www.broadinstitute.org/gsea)13 was used to analyze the potential genes influenced by SSRP1 low expression. The data from GSE14520 was downloaded and divided into two groups (high versus low) according to the expression of SSRP1 and MsigDB c5 (GO gene sets, 1454 gene sets) was used. Gene sets with a false discovery rate value < 0.25 and normal P < 0.05 after 1,000 permutations were regarded as significantly enrichment. The mutation counts and fraction of copy number altered genome data for each TCGA hepatocellular carcinoma individuals were directly downloaded from the cBioPortal for Cancer Genomics (http://cbioportal.org).14,15 The logGI50 of small molecules and natural products, and the mRNA expression microarray data of human tumor cell lines were downloaded from the NCI website (http://dtp.nci.nih.gov).
Statistical analysis. The correlation between gene expression and the clinicopathologic features was analyzed by Chi-square test. Averaged replicates of three independent experiments were used in cellular studies, and results were statistically analyzed using the two-tailed, unpaired Student's t-test. Results were expressed as mean values with 95% confidence intervals. Error bars represented standard error. P < 0.05 was considered statistically significant.
SUPPLEMENTARY MATERIAL Figure S1. Higher SSRP1 expression is associated with larger somatic copy number variation and more mutations in HCC patients. Figure S2. High expression of SSRP1 is associated with poor prognosis of HCC. Figure S3. SPT16 is not associated with the prognosis of HCC patients. Figure S4. Overexpression of SSRP1 promotes the malignant phenotype of SMMC7721 cells in vitro. Figure S5. Knockdown of SSRP1 increases the apoptotic cells in LM3 (A) and 97H (B) cells with or without H2O2 stimulation. Figure S6. Cross-cancer summary of homozygous mutations and copy number variations in all cancers available on cBioPortal. Table S1. Correlation between the SSRP1 protein expression and differentiation status in 73 HCC samples. Supplementary Data
Acknowledgments
The study was supported by granted from the National Natural Science Foundation of China (No. 81071990) and the Guangdong Province Natural Science Foundation (No.2015A030313725). Conceived and designed the experiments: Q.S.D., K.H. Performed the experiments: Q.S.D., K.H., T.L., H.N.W., J.Q.Z., K.Y.C., J.F.X., X.P.D., R.H., Z.L.X., W.J.Z., and J.L.H. Contributed funds/reagents/materials/samples: G.A.X., X.Y.J., H.L., and H.G.Y. Statastic analysis: Q.S.D. Wrote the paper: Q.S.D., K.H., T.L., and Y.C.D. All authors read and approved the final manuscript. The authors declare that they have no competing interest.
Supplementary Material
Higher SSRP1 expression is associated with larger somatic copy number variation and more mutations in HCC patients.
High expression of SSRP1 is associated with poor prognosis of HCC.
SPT16 is not associated with the prognosis of HCC patients.
Overexpression of SSRP1 promotes the malignant phenotype of SMMC7721 cells in vitro.
Knockdown of SSRP1 increases the apoptotic cells in LM3 (A) and 97H (B) cells with or without H2O2 stimulation.
Cross-cancer summary of homozygous mutations and copy number variations in all cancers available on cBioPortal.
orrelation between the SSRP1 protein expression and differentiation status in 73 HCC samples.
References
- Jemal, A, Bray, F, Center, MM, Ferlay, J, Ward, E and Forman, D (2011). Global cancer statistics. CA Cancer J Clin 61: 69–90. [DOI] [PubMed] [Google Scholar]
- Birch, JL, Tan, BC, Panov, KI, Panova, TB, Andersen, JS, Owen-Hughes, TA et al. (2009). FACT facilitates chromatin transcription by RNA polymerases I and III. EMBO J 28: 854–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari, A, Mazina, OM, Shinde, U, Mazin, AV and Lu, H (2009). A role for SSRP1 in recombination-mediated DNA damage response. J Cell Biochem 108: 508–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundu, LR, Seki, M, Watanabe, N, Murofushi, H, Furukohri, A, Waga, S et al. (2011). Biphasic chromatin binding of histone chaperone FACT during eukaryotic chromatin DNA replication. Biochim Biophys Acta 1813: 1129–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia, H, Fleyshman, D, Kolesnikova, K, Safina, A, Commane, M, Paszkiewicz, G et al. (2011). Expression of FACT in mammalian tissues suggests its role in maintaining of undifferentiated state of cells. Oncotarget 2: 783–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia, H, Miecznikowski, JC, Safina, A, Commane, M, Ruusulehto, A, Kilpinen, S et al. (2013). Facilitates chromatin transcription complex is an “accelerator” of tumor transformation and potential marker and target of aggressive cancers. Cell Rep 4: 159–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dermawan, JK, Gurova, K, Pink, J, Dowlati, A, De, S, Narla, G et al. (2014). Quinacrine overcomes resistance to erlotinib by inhibiting FACT, NF-κB, and cell-cycle progression in non-small cell lung cancer. Mol Cancer Ther 13: 2203–2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koman, IE, Commane, M, Paszkiewicz, G, Hoonjan, B, Pal, S, Safina, A et al. (2012). Targeting FACT complex suppresses mammary tumorigenesis in Her2/neu transgenic mice. Cancer Prev Res (Phila) 5: 1025–1035. [DOI] [PubMed] [Google Scholar]
- Feng, B, Wang, R and Chen, LB (2012). Review of miR-200b and cancer chemosensitivity. Biomed Pharmacother 66: 397–402. [DOI] [PubMed] [Google Scholar]
- Roessler, S, Jia, HL, Budhu, A, Forgues, M, Ye, QH, Lee, JS et al. (2010). A unique metastasis gene signature enables prediction of tumor relapse in early-stage hepatocellular carcinoma patients. Cancer Res 70: 10202–10212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burchard, J, Zhang, C, Liu, AM, Poon, RT, Lee, NP, Wong, KF et al. (2010). microRNA-122 as a regulator of mitochondrial metabolic gene network in hepatocellular carcinoma. Mol Syst Biol 6: 402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan, D and Weinberg, RA (2011). Hallmarks of cancer: the next generation. Cell 144: 646–674. [DOI] [PubMed] [Google Scholar]
- Hsieh, YH, Chang, YY, Su, IJ, Yen, CJ, Liu, YR, Liu, RJ et al. (2015). Hepatitis B virus pre-S2 mutant large surface protein inhibits DNA double-strand break repair and leads to genome instability in hepatocarcinogenesis. J Pathol 236: 337–347. [DOI] [PubMed] [Google Scholar]
- Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013; 269: pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerami, E, Gao, J, Dogrusoz, U, Gross, BE, Sumer, SO, Aksoy, BA et al. (2012). The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2: 401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian, A, Tamayo, P, Mootha, VK, Mukherjee, S, Ebert, BL, Gillette, MA et al. (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 102: 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao, ZH, Wan, JL, Zeng, LY, Xie, L, Sun, HC, Qin, LX et al. (2013). miR-612 suppresses the invasive-metastatic cascade in hepatocellular carcinoma. J Exp Med 210: 789–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, P, Lin, Y, Zhang, Y, Zhu, Z and Huo, K (2013). SSX2IP promotes metastasis and chemotherapeutic resistance of hepatocellular carcinoma. J Transl Med 11: 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, G, Chong, RA, Yang, Q, Wei, Y, Blanco, MA, Li, F et al. (2009). MTDH activation by 8q22 genomic gain promotes chemoresistance and metastasis of poor-prognosis breast cancer. Cancer Cell 15: 9–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuta, M, Kozaki, K, Tanimoto, K, Tanaka, S, Arii, S, Shimamura, T et al. (2013). The tumor-suppressive miR-497-195 cluster targets multiple cell-cycle regulators in hepatocellular carcinoma. PLoS One 8: e60155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, JJ, Zhang, YN, Liao, JZ, Ke, KP, Chang, Y, Li, PY et al. (2015). MiR-497 suppresses angiogenesis and metastasis of hepatocellular carcinoma by inhibiting VEGFA and AEG-1. Oncotarget 6: 29527–29542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato, F, Hatano, E, Kitamura, K, Myomoto, A, Fujiwara, T, Takizawa, S et al. (2011). MicroRNA profile predicts recurrence after resection in patients with hepatocellular carcinoma within the Milan Criteria. PLoS One 6: e16435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan, BC, Chien, CT, Hirose, S and Lee, SC (2006). Functional cooperation between FACT and MCM helicase facilitates initiation of chromatin DNA replication. EMBO J 25: 3975–3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe, T, Sugimura, K, Hosono, Y, Takami, Y, Akita, M, Yoshimura, A et al. (2011). The histone chaperone facilitates chromatin transcription (FACT) protein maintains normal replication fork rates. J Biol Chem 286: 30504–30512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, SX, Li, Y, Jin, Y, Zhang, Q, Keller, DM, McQuaw, CM et al. (2010). Structure-specific recognition protein 1 facilitates microtubule growth and bundling required for mitosis. Mol Cell Biol 30: 935–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh, FK, Kulaeva, OI, Patel, SS, Dyer, PN, Luger, K, Reinberg, D et al. (2013). Histone chaperone FACT action during transcription through chromatin by RNA polymerase II. Proc Natl Acad Sci USA 110: 7654–7659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasparian, AV, Burkhart, CA, Purmal, AA, Brodsky, L, Pal, M, Saranadasa, M et al. (2011). Curaxins: anticancer compounds that simultaneously suppress NF-κB and activate p53 by targeting FACT. Sci Transl Med 3: 95ra74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, JG and Zhang, SW (2011). Liver cancer epidemic in China: past, present and future. Semin Cancer Biol 21: 59–69. [DOI] [PubMed] [Google Scholar]
- El-Serag, HB (2011). Hepatocellular carcinoma. N Engl J Med 365: 1118–1127. [DOI] [PubMed] [Google Scholar]
- Han, Z, Zhang, Y, Yang, Q, Liu, B, Wu, J, Zhang, Y et al. (2015). miR-497 and miR-34a retard lung cancer growth by co-inhibiting cyclin E1 (CCNE1). Oncotarget 6: 13149–13163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, D, Zhao, Y, Liu, C, Chen, X, Qi, Y, Jiang, Y et al. (2011). Analysis of MiR-195 and MiR-497 expression, regulation and role in breast cancer. Clin Cancer Res 17: 1722–1730. [DOI] [PubMed] [Google Scholar]
- Luo, M, Shen, D, Zhou, X, Chen, X and Wang, W (2013). MicroRNA-497 is a potential prognostic marker in human cervical cancer and functions as a tumor suppressor by targeting the insulin-like growth factor 1 receptor. Surgery 153: 836–847. [DOI] [PubMed] [Google Scholar]
- Xu, JW, Wang, TX, You, L, Zheng, LF, Shu, H, Zhang, TP et al. (2014). Insulin-like growth factor 1 receptor (IGF-1R) as a target of MiR-497 and plasma IGF-1R levels associated with TNM stage of pancreatic cancer. PLoS One 9: e92847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, S, Fu, GB, Tao, Z, OuYang, J, Kong, F, Jiang, BH et al. (2015). MiR-497 decreases cisplatin resistance in ovarian cancer cells by targeting mTOR/P70S6K1. Oncotarget 6: 26457–26471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong, XJ, Duan, LJ, Qian, XQ, Xu, D, Liu, HL, Zhu, YJ et al. (2015). Tumor-suppressive microRNA-497 targets IKKβ to regulate NF-κB signaling pathway in human prostate cancer cells. Am J Cancer Res 5: 1795–1804. [PMC free article] [PubMed] [Google Scholar]
- Yadav, S, Pandey, A, Shukla, A, Talwelkar, SS, Kumar, A, Pant, AB et al. (2011). miR-497 and miR-302b regulate ethanol-induced neuronal cell death through BCL2 protein and cyclin D2. J Biol Chem 286: 37347–37357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mechtler, P, Singhal, R, Kichina, JV, Bard, JE, Buck, MJ and Kandel, ES (2015). MicroRNA analysis suggests an additional level of feedback regulation in the NF-κB signaling cascade. Oncotarget 6: 17097–17106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, XX, Kuang, SZ, Liao, JZ, Xu, CR, Chang, Y, Wu, YL et al. (2015). The regulation of microRNA expression by DNA methylation in hepatocellular carcinoma. Mol Biosyst 11: 532–539. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Higher SSRP1 expression is associated with larger somatic copy number variation and more mutations in HCC patients.
High expression of SSRP1 is associated with poor prognosis of HCC.
SPT16 is not associated with the prognosis of HCC patients.
Overexpression of SSRP1 promotes the malignant phenotype of SMMC7721 cells in vitro.
Knockdown of SSRP1 increases the apoptotic cells in LM3 (A) and 97H (B) cells with or without H2O2 stimulation.
Cross-cancer summary of homozygous mutations and copy number variations in all cancers available on cBioPortal.
orrelation between the SSRP1 protein expression and differentiation status in 73 HCC samples.