Abstract
Arsenic is the most prevalent environmental toxic element and causes health problems throughout the world. The toxicity, mobility, and fate of arsenic in the environment are largely determined by its speciation, and arsenic speciation changes are driven, at least to some extent, by biological processes. In this article, biotransformation of arsenic is reviewed from the perspective of the formation of Earth and the evolution of life, and the connection between arsenic geochemistry and biology is described. The article provides a comprehensive overview of molecular mechanisms of arsenic redox and methylation cycles as well as other arsenic biotransformations. It also discusses the implications of arsenic biotransformation in environmental remediation and food safety, with particular emphasis on groundwater arsenic contamination and arsenic accumulation in rice.
Keywords: arsenic, methylation, degradation, oxidation, reduction, MSMA, roxarsone, rice
INTRODUCTION
One generation passes away, and another generation comes: but earth abides for ever.
— Ecclesiastes 1:4
Our planet is a living organism. Life evolves in response to environmental changes and, in turn, remodels the environment. In this review we speculate on the origin of genes involved in arsenic biology and describe the biological cycles of arsenic biotransformations, in particular redox and methylation cycles. Why does arsenic perturb life? At one level the answer is related to the toxic effects of arsenic on living systems. At a deeper level it is the difference between the chemical properties of arsenic and phosphorus that creates biological problems and prevents arsenic from being utilized in DNA and other high-energy biological molecules in place of phosphorus (see below).
Arsenic is the most ubiquitous environmental toxin and carcinogen, endangering human health (Abernathy et al. 2003). The US Environmental Protection Agency (EPA) and Agency for Toxic Substances and Disease Registry (ATSDR) rank arsenic at the top of the US Priority List of Hazardous Substances (http://www.atsdr.cdc.gov/SPL/index.html). The EPA asserts that it pervades drinking water (National Research Council 2001) and imperils the safety of our food supply (Stone 2008). It enters our water and food supply in many ways, including natural sources, herbicides, wood preservatives, and burning of coal. Arsenic exposure is related to cardiovascular disease (States et al. 2009, Tseng et al. 2003), diabetes (Drobná et al. 2012, Tseng et al. 2002), peripheral neuropathy (Mukherjee et al. 2003) and peripheral vascular (blackfoot) disease (Tseng et al. 1995). Exposure during pregnancy contributes to low birth weight and fetal loss, as well as delayed infant development (Tofail et al. 2009). Long-term exposure to arsenic in drinking water and the food supply, as well as occupational exposure, causes cancer (Abernathy et al. 2003, Chen et al. 1992, Mushak & Crocetti 1995, Naujokas et al. 2013, Tseng et al. 1968). Arsenic contamination of groundwater in Bangladesh and West Bengal, India, has been called the largest mass poisoning of a population in history (Smith et al. 2000). In humans exposure is a major contributor to arsenic-related diseases (Abernathy et al. 2003, Tchounwou et al. 2003), including bladder (Chen et al. 2003), lung (Putila & Guo 2011), and skin cancers (Rossman et al. 2001). The primary sources of dietary arsenic are food (Xue et al. 2010) and drinking water (Kile et al. 2007; NRC 1999, 2001). In 2002 the EPA’s Maximum Containment Level (MCL) was lowered from 50 to 10 μg/L, but in the United States more than 6 million people have drinking water with arsenic levels above 10 μg/L and 2.5 million people with levels above 25 μg/L. Globally, an estimated 150 million people are exposed to unsafe levels of As in the drinking water (Brammer & Ravenscroft 2009).
ARSENIC BIOGEOCHEMISTRY
The average arsenic concentrations (by weight) in the continental crust, soil, and ocean water are estimated to be 2.1, 5, and 0.0023 mg/kg, respectively (http://www.webelements.com/arsenic/geology.html). Baseline arsenic concentrations in freshwater and in soil are in the range of 0.0001–0.002 mg/L and <10 mg/kg, respectively, although much higher concentrations can be found where the environment has been impacted by geothermal sources, mining, smelting, and other anthropogenic activities. There are more than 200 arsenic-containing minerals; many of them exist as sulfide minerals [e.g., arsenopyrites (FeAsS), realgar (As4S4), and orpiment (As2S3)], which are thought to be formed under the high-temperature conditions in Earth’s crust (Smedley & Kinniburgh 2002). However, microbe-mediated formation of orpiment has also been reported (Newman et al. 1998).
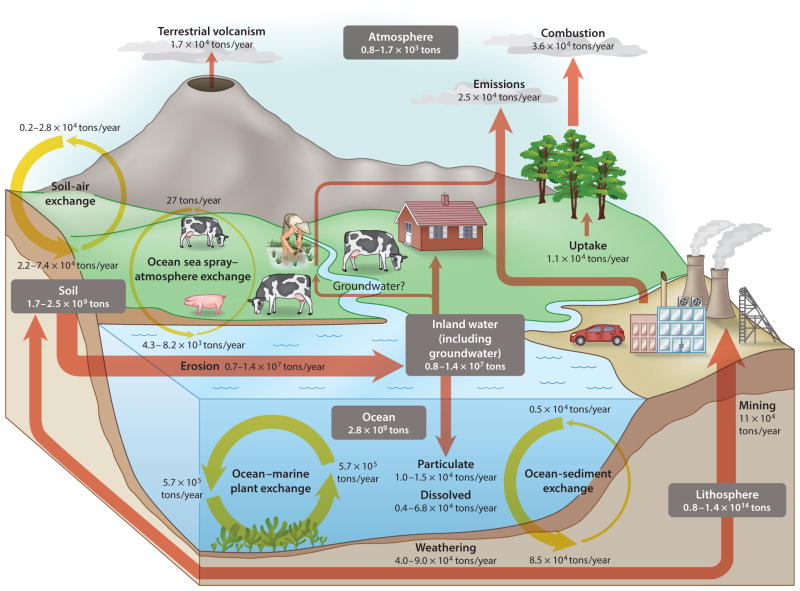
Not surprisingly, the majority of arsenic in Earth’s crust is present in the lithosphere; its pool size is approximately five orders of magnitude larger than that in the ocean or the terrestrial soil (Wenzel 2013) (Figure 1). Weathering of rocks, geothermal and volcanic activities, mining, and smelting release arsenic from the lithosphere to the terrestrial and oceanic environments. The amounts of arsenic in the ocean and terrestrial soil are comparable. Arsenic also enters the biosphere and can be transferred through the food chain. Although arsenic is not essential to living organisms, it is taken up by the uptake pathways for analogous essential or beneficial nutrients, e.g., arsenate via the phosphate transporters and arsenite via the aquaglyceroporin channels (Bhattacharjee et al. 2008, Yang et al. 2012, Zhao et al. 2010). The total amount of arsenic accumulated in terrestrial plants is estimated to be 1.8 × 105 tons, which is approximately four orders of magnitude less than that in soil (Wenzel 2013). This difference in the pool size reflects a generally limited accumulation of arsenic in terrestrial plants, likely due to a low bioavailability of arsenic in soil or phytotoxicity when bioavailable arsenic in soil is elevated. There are exceptions—for example, a number of ferns can accumulate well over 1,000 mg/kg of arsenic in their above-ground tissues without suffering from phytotoxicity (Ma et al. 2001, Zhao et al. 2002); such plants are termed arsenic hyperaccumulators. The ratios of plant arsenic to soil concentrations are often >10 in the hyperaccumulators, compared with the <0.1 ratios typically found in nonhyperaccumulators. It is interesting that arsenic hyperaccumulators appear to be confined to a few pteridophyte species, which are early land plants. This may have something to do with the loss of ACR3 orthologous genes during the evolution of flowering plants (Indriolo et al. 2010).
Figure 1.
Stocks and fluxes of arsenic in various Earth components. Gray boxes indicate stocks of arsenic in a given component, and arrows indicate the fluxes of arsenic toward a given component. Data presented in this diagram are summarized from Wenzel (2013), in which detailed references are cited regarding the estimation of these stocks and fluxes.
Arsenic occurs in different chemical forms in the environment. It has four oxidation states: +5, +3, 0, and −3, represented by arsenate, arsenite, elemental arsenic, and arsine, respectively (http://www.webelements.com/arsenic/compounds.html). In living organisms arsenic is found only in the pentavalent and trivalent oxidation states. The valence state of arsenic in arsine (AsH3) is not clear, as the electronegativities of arsenic and hydrogen are almost identical; the +3 oxidation state has also been assumed for arsenic in arsine (Cullen & Reimer 1989). Arsenate is the predominant form in the oxic environment, such as in the surface water and aerobic soils. Reduction of arsenate to arsenite occurs under suboxic conditions (Eh < 100 mV at neutral pH); thus, arsenite becomes the predominant form under reducing conditions, e.g., flooded paddy soils or anaerobic sediments. Reduction of arsenate to arsenite in soil or sediment generally leads to markedly increased mobility because the latter is less strongly adsorbed by iron oxides/oxyhydroxides in the solid phase (Fendorf et al. 2008). This process has important consequences manifested by arsenic mobilization in groundwater and increased arsenic transfer from paddy soil to rice (see sections below). Some arsenite may be present even in oxic environments as a result of biological transformation and redox disequilibrium. For example, the As(V) to As(III) ratio in open seawater is typically 10–100, not the 1015–1026 that would be expected on the basis of thermodynamic equilibrium (Andreae 1979, Smedley & Kinniburgh 2002). Inside living cells, arsenic is predominantly in the As(III) oxidation state due to the prevailing redox conditions (Eh considerably lower than 0 mV). While arsenate behaves as a phosphate analog competing for the phosphate transporters, the two oxyanions differ in their propensity for redox reactions. Phosphate is much more difficult than arsenate to reduce, and, therefore, subsequent transformations of arsenate and phosphate can be different.
THE PRIMORDIAL RISE OF ARSENIC BIOLOGY
Arsenic has posed a dilemma for the physiology of microorganisms since the origin of life. Primordial Earth is defined as the first billion years, or gigayear. On the geologic timescale, this comprises all of the Hadean eon, as well as the Eoarchean and part of the Paleoarchean eras of the Archean eon. It is also defined by the emergence of life and, later, photosynthesis (Hohmann-Marriott & Blankenship 2011). It is well accepted that primordial oceans formed in the Hadean eon approximately 700 million years after the crust formed. Although we can only speculate on how and where life first arose, the first organisms probably evolved approximately 3.5–3.8 billion years ago, when the temperature of the oceans was higher than today (estimated variously at between 40°C and 70°C, compared with 20°C today), the pH lower (4.8 to 6.5, compared with 8.2 today) and the atmosphere devoid of oxygen. Thus the earliest ecosystems existed in an anoxic world, and their activities were driven by anaerobic metabolism (Canfield et al. 2006). Whereas iron in present-day oceans precipitates as Fe(OH)3, in the original anoxic oceans it would have been dissolved as Fe2+. Arsenic, which presently forms insoluble ferric arsenate salts, would have existed as the highly mobile arsenite, so soluble arsenic concentrations were likely relatively high in the early Earth environments. There was much more volcanic activity than at present. In fact, in many areas of the world today the amount of arsenic produced in volcanic areas and hot springs, often as insoluble sulfides, is very high. In oceanic hydrothermal vents, which may have conditions similar to those of primordial Earth, the concentration of dissolved arsenic is frequently several orders of magnitude higher than the 2–4 μg/L found in seawater and sometimes even higher. For example, in the marine shallow-water hydrothermal vents in Tutum Bay, eastern Papua New Guinea, arsenic concentrations in the hydrothermal fluid are approximately 900 μg/L, with arsenite being the dominant species (Price et al. 2007). Diffusion of vent fluid to the surrounding environment leads to accumulation of arsenic in marine sediments. In Tutum Bay, the average arsenic concentration in surface sediments ranges between 52 and 1,483 mg/L, with an average of 527 mg/L, excluding the vent precipitates (Price & Pichler 2005).
Hot springs also often contain high levels of arsenic. For example, in hot spring water from Yellowstone National Park, the arsenic concentration is usually approximately 2 mg/L (Langner et al. 2001). In Meager Creek hot springs, Canada, the arsenic concentration is over 0.2 mg/L (Koch et al. 1999). Arsenic concentrations commonly range from a few milligrams per liter to tens of milligrams per liter in geothermal fluids from Latin America (Lopez et al. 2012). A survey of 11 hot springs in Yunnan, Southwest China, showed that arsenic concentrations are around 0.5 mg/L (Zhang et al. 2008). Arsenic exists mainly as trivalent inorganic arsenite in hot springs and is rapidly oxidized upon reaching the surface and in streams/creeks, either by air or mediated by microbes (Wilkie & Hering 1998). With the advent of novel analytical capabilities, investigators have found that a large proportion of inorganic soluble arsenic in geothermal waters (particularly sulfidic waters) is actually thiolated and that, under alkaline conditions, thioarsenates account for over 80% of total arsenic (Planer-Friedrich et al. 2007). However, thiolated arsenicals exist in situ as arsenites, which are rapidly oxidized to thioarsenates when exposed to oxygen during analysis (Planer-Friedrich et al. 2010).
We can therefore extrapolate that life evolved in an environment rich in trivalent inorganic As(III) given that the atmosphere did not contain oxygen for at least another billion years. As a consequence, these Archean ancestors must have developed mechanisms to avert arsenic toxicity quite early. As(III) is toxic primarily because of its strong interactions with vicinal thiols such as cysteine pairs in proteins (Hughes 2002). It binds to monothiols with lower affinity, but sufficiently high concentrations of As(III) would tie up critical monothiols such as reduced glutathione (GSH) (Delnomdedieu et al. 1994) and prevent cells from being able to regulate their intracellular redox state. It is not surprising, then, that nearly every organism has one or more As(III) detoxification pathways (Bhattacharjee & Rosen 2007). In bacteria and archaea, which are closer to the direct line with the last common ancestor, arsenic resistance (ars) genes are organized in operons that confer arsenic tolerance. Removal of As(III) from the cell is the most straightforward pathway to resistance, and nearly every prokaryote has either ArsB or Acr3, two unrelated membrane transport proteins that catalyze As(III) efflux from the cytosol to the medium (Yang et al. 2012). ArsB is mainly restricted to prokaryotes, while members of the Acr3 family are found in bacteria, archaea, fungi, and nonflowering plants (Castillo & Saier 2010, Indriolo et al. 2010, Yang et al. 2012). Eukaryotes also have efflux pumps for the triglutathione conjugates of As(III). The Mrp members of the ABC (ATP-binding cassette) superfamily of ATP-coupled pumps transport As(GS)3 out of the cytosol, sometimes to sequester it in intracellular compartments, as in the case of the vacuolar YCF1 pump of Saccharomyces cerevisiae (Ghosh et al. 1999) or the mammalian MRP2, which extrudes As(GS)3 from liver to bile (Leslie 2011). Other members of this family transport different thiol conjugates, including phytochelatins in plants (Song et al. 2010) and trypanothione in trypanosomatids (Mukhopadhyay et al. 1996).
The rise of photosynthetic microorganisms approximately 2.4 billion years ago is documented by the presence of metal sulfides preceding the Great Oxidation Event and the banded iron formation in the Mount McRae Shale (Kaufman et al. 2007). Once the atmosphere became oxidizing, the majority of aqueous arsenite would have been oxidized to arsenate [As(V)]. This was a crisis for organisms that could detoxify As(III) but not As(V). By that time life had diversified from the last common ancestor, so numerous types of microbes had to independently evolve strategies to handle inorganic arsenate. Although it might appear overly complicated and paradoxical to convert the less harmful arsenate into the more toxic arsenite, the simplest solution was to reduce the intracellular As(V) to As(III), for which the cells already had mechanisms for cytosolic removal. This selective pressure led to the evolution of at least three families of small arsenate reductase enzymes whose sole function is to detoxify arsenate (Mukhopadhyay & Rosen 2002) (see below).
ARSENIC AND SHADOW LIFE
The idea of a shadow life with organisms that use a biochemistry unrelated to ours is attractive and intriguing (Davies et al. 2009). Earthly life primarily uses the six elements hydrogen, carbon, nitrogen, oxygen, phosphorus, and sulfur. They are abundant light elements that form strong chemical bonds with one another. Carbon forms long-chain polymers that are the building blocks of life. Why doesn’t biology use the less abundant elements as substitutes in biological molecules, especially the heavier elements in the same group of the periodic table that share similar chemical properties with the lighter elements?
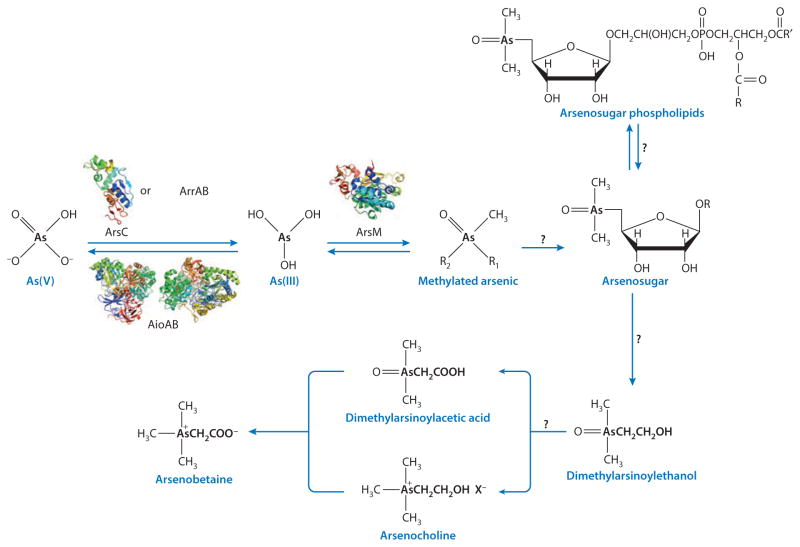
Recently microbes that use arsenate in place of phosphate have been suggested to constitute a shadow life (Wolfe-Simon et al. 2010). This controversial idea has been contested (Rosen et al. 2011), and there is little support for the existence of arsenic-dependent organisms. Biologically, phosphorus is found in phosphate and phosphate esters in the P(V) oxidation state. In contrast, arsenic has two oxidation states, As(III) and As(V), that biologically affect organisms (see below). Arsenite is the more toxic because it is reactive and forms strong metallic-like bonds with biological molecules. Arsenate is much less toxic, is chemically similar to phosphate, and can form esters similar to phosphate esters. So could earthly life have used arsenate interchangeably with phosphate? Why aren’t organisms that can use arsenate in place of phosphate common today? Arsenate is in fact reduced and methylated and then incorporated into a number of organic molecules in marine organisms for detoxification, including arsenosugars, arsenolipids, and arsenobetaine (Francesconi & Kuehnelt 2002) (Figure 2). However, arsenate esters are less stable than phosphate esters in part because of the shorter P-O bond length of approximately 1.5 Å compared with the As-O bond length of approximately 2 Å. Longer bonds are more easily broken, and, consequently, arsenate esters hydrolyze much faster than phosphate esters. In DNA an arsenodiester bond would be at least 1,000-fold less stable than the phosphodiester bond, and DNA with arsenate incorporated in place of phosphate would hydrolyze so fast that life would be impossible on Earth (Rosen et al. 2011). That doesn’t mean that there couldn’t be organisms in which arsenic replaces phosphorus, but those organisms would have fragile As-O polymers. Such life might be able to evolve in the frigid nonaqueous oceans of the moons of planetary gas giants, but the hypothesis that arsenate could replace phosphate in DNA is unlikely to be true on Earth.
Figure 2.
Arsenic species and their biotransformations. Known enzymes for biotransformation are indicated in each step, and representative protein structures are shown for each enzyme: ArsC (Martin et al. 2001), AioAB (Ellis et al. 2001, Warelow et al. 2013), and ArsM (Ajees et al. 2012). Question marks indicate steps for which the molecular mechanism has yet to be unraveled. The diagram is partly adapted from Edmonds & Francesconi (1987).
ARSENIC OXIDATION AND REDUCTION: PATHWAYS FOR ENERGY METABOLISM AND RESISTANCE
Again, early life likely evolved in an environment rich in trivalent inorganic As(III) given that the atmosphere did not contain oxygen for at least another billion years. As a consequence, these Archean ancestors may have developed enzymes that oxidized arsenite to capture energy quite early (Lebrun et al. 2003). Related enzymes that capture the reducing potential of arsenate by dissimilation may have evolved subsequently, recycling the arsenate to arsenite (van Lis et al. 2013). Extant microorganisms catalyze both types of reactions, creating an arsenic redox cycle detailed in the following sections.
Arsenite Oxidation
In addition to arsenite efflux, rapid arsenite oxidation by microbes has been observed to occur widely, for example, in geothermal/hot spring waters, and these studies have indicated that the oxidation process is largely mediated by microbial activities (Mukhopadhyay et al. 2002, Oremland & Stolz 2003). The first arsenite oxidase was purified from the periplasm of the β-proteobacterium Alcaligenes faecalis (Anderson et al. 1992). All aerobic arsenite oxidation discovered to date involves arsenite oxidases that contain two heterologous subunits: AioA (AoxB) and AioB (AoxA) (Stolz et al. 2006, Zargar et al. 2010) (Figure 2). These subunits have been assigned different names, but Aio is the presently accepted nomenclature. Phylogenetic analysis of the two subunits of the heterodimeric enzyme suggests an early origin of arsenite oxidases before the divergence of archaea and bacteria (Lebrun et al. 2003). It makes sense that such an ancient oxidase function evolved before the atmosphere became oxidizing, allowing arsenite oxidation to couple to energy production. In present-day Mono Lake, California, several strains of bacteria have been shown to be arsenite oxidizers that use nitrate or selenate as electron acceptors, coupling arsenite to ATP production (Fisher & Hollibaugh 2008, Oremland et al. 2002), and have been adapted for use in bioreactors (Sun et al. 2010a). Anoxygenic photosynthesis has also been found to be coupled to arsenite oxidation in microbial mats of cyanobacteria and photosynthetic bacteria from Mono Lake (Kulp et al. 2008). Recently, the arsenite oxidase subunit AioA was shown to be involved in anaerobic oxidation of arsenite coupled to chlorate reduction (Sun et al. 2010b). However, direct evidence showing the involvement of AioA in anoxic arsenite oxidation is still lacking.
Recently, an enzyme (Arx) that is closely related to respiratory arsenate reductase (see below) in amino acid sequence but physiologically performs As(III) oxidation instead of As(V) reduction was isolated as an ArxAB heterodimer from the genome of the arsenite-oxidizing nitrate reducer Alkalilimnicola ehrlichii strain MLHE-1 (a chemolithoautotrophic anaerobe) (Zargar et al. 2010). In fact, structural features of Arx subunits (ArxA and ArxB) are more similar to those of respiratory arsenate reductases than to those of arsenite oxidases (van Lis et al. 2013, Zargar et al. 2012). Intriguingly, the ArxAB enzyme from A. ehrlichii strain MLHE-1 has not only arsenite oxidase but also arsenate reductase activity in vitro, although the strain is incapable of respiratory growth on arsenate (Richey et al. 2009). The Arx enzyme has been demonstrated to act as an alternative arsenite oxidase during anaerobic respiration of nitrate, as in A. ehrlichii strain MLHE-1, and also of selenate (Fisher & Hollibaugh 2008). Arx may also participate in anoxygenic photosynthesis. A photosynthetic purple sulfur bacterium (Ectothiorhodospira sp. strain PHS-1) capable of anoxic arsenite oxidation was isolated from a hot spring biofilm (Kulp et al. 2008). Another arxA gene was identified in this organism. Further analysis revealed a wide distribution of arxA-like genes in arsenic-rich Mono Lake and Hot Creek sediments in California and alkaline microbial mats from Yellowstone National Park (Zargar et al. 2012). This new clade of arsenite oxidase genes has been proposed to have evolved in archaea and may be ancestral to arsenite oxidases and respiratory arsenate reductases (Oremland et al. 2009, Zargar et al. 2012). In contrast, a recent phylogenic study indicates that both Arx and respiratory arsenate reductases originated after the divergence of bacteria and archaea (van Lis et al. 2013).
Arsenate Reduction
As the atmosphere became oxidizing, and arsenite was oxidized to arsenate, respiratory reductases evolved to use arsenate as a terminal electron acceptor in energy-generating respiratory chains. Thermodynamic calculations indicate that dissimilatory arsenate reduction can provide enough reducing potential to sustain microbial life. A microorganism isolated from arsenic-contaminated sediments of the Aberjona watershed in eastern Massachusetts was demonstrated to grow by reducing (respiring) arsenate (Ahmann et al. 1994). Such anaerobic dissimilatory As(V) reduction is catalyzed by the arsenate respiratory reductase (Arr) complex, which consists of a large catalytic subunit (ArrA) and a small subunit (ArrB) (Afkar et al. 2003, Krafft & Macy 1998, Saltikov & Newman 2003, van Lis et al. 2013) (Figure 2). Like Arx, the Arr enzyme also has been demonstrated to be bidirectional; it shows both arsenate reductase and arsenite oxidase activity in vitro (Richey et al. 2009). It has been difficult to monitor the existence and activity of As(V)-respiring bacteria in diverse environments. The arrA gene encodes the large subunit of the reductase, and this gene can be used as a reliable marker for arsenate respiration (Malasarn et al. 2004). For example, in arsenic-rich soda lakes in California, arrA was detected in sediment samples, and its abundance corresponded with in situ rates of arsenate reduction (Kulp et al. 2006). Multiple alignment of sequences related to ArrA reveals the phylogeny of Arr and closely related enzymes (Duval et al. 2008). This analysis confirms the previously proposed proximity of Arr to the cluster of polysulfide/thiosulfate reductases and unravels a hitherto unrecognized clade even more closely related to Arr. The resulting phylogeny strongly suggests that Arr originated in bacteria subsequent to the generation of an oxidizing atmosphere after the bacteria/archaea divergence and was subsequently laterally distributed within the domain Bacteria. It furthermore suggests that an enzyme related to polysulfide reductase rather than arsenite oxidase may be the precursor of Arr.
Dissimilatory arsenate reduction has been associated with contamination of groundwater. Arsenate is generally less mobile than arsenite, as arsenate can be strongly adsorbed by iron and aluminum oxides in the environment (Fendorf et al. 2008). Reduction of arsenate is therefore considered to be a key mechanism of its mobilization, such as in aquifer sediments causing arsenic contamination in groundwater in south and southeast Asia (Fendorf et al. 2010). Reduction of Fe(III) can also contribute to the release of arsenic to groundwater. The role of Fe(III) reduction in arsenic release to groundwater has been extensively studied (Dhar et al. 2011, Islam et al. 2004, Tufano et al. 2008). For iron-reducing bacteria capable of dissimilatory arsenate reduction, iron and arsenic release can be simultaneous (Oremland & Stolz 2005), but these two processes can also be uncoupled. The coupling/uncoupling of iron and arsenic reduction has been elegantly tested with mutants of Shewanella sp. ANA-3 (Tufano et al. 2008). When uncoupled from iron reduction, arsenate reduction becomes the dominant process controlling arsenic release. Whether these processes are uncoupled seems to be determined by the nature of the microbial community involved and the mineralogy of solid-phase sequestering of arsenic. It is therefore not surprising that in a microcosm study, arsenate reduction lagged behind iron reduction, which means that arsenate was released concomitant with iron reduction, and soluble arsenate was subsequently reduced to arsenite (Islam et al. 2004). Analysis of this microbial community, which is dominated by metal-reducing bacteria such as members of the family Geobacteraceae, demonstrates that this group of microbes do not possess dissimilatory arsenate respiration machinery (Oremland & Stolz 2005), so perhaps arsenic release as arsenite under these conditions is the result of the action of an ArsC-like arsenate reductase (see below).
To cope with the rise of arsenate produced by an oxidizing atmosphere, multiple microbial species independently evolved strategies to detoxify inorganic arsenate by reduction using an entirely different biochemical mechanism than dissimilatory arsenate reduction (Mukhopadhyay & Rosen 2002). Small arsenate reductase enzymes evolved a number of times from the ancestors of proteins such as redoxins and phosphatases. One family of bacterial arsenate reductases, typified by the ArsC enzyme of plasmid R773 (Gladysheva et al. 1994, Oden et al. 1994), is closely related to the glutaredoxin family (Figure 2). This enzyme uses a cysteine residue in the protein and a GSH molecule to reduce As(V) to As(III), producing oxidized glutathione that is rereduced by the enzyme glutathione reductase using NADPH as the source of electrons. A second family of bacterial arsenate reductases, typified by the ArsC of Staphylococcus aureus ( Ji et al. 1994), uses internal cysteine residues during the reduction of arsenate (Messens & Silver 2006). This generates a disulfide bond in the enzyme that is rereduced by thioredoxin, which is then regenerated with thioredoxin reductase and NADPH. These proteins form a branch of low-molecular-weight phosphatases and in fact exhibit phosphatase activity. Although the reactions of the two types of reductases (which, unfortunately and confusingly, were both named ArsC) have similarities, they are the products of independent evolution that occurred after the initial bacterial radiation. The third family of arsenate reductases is found in eukaryotes and appears to be a branch of the CDC25 family of dual-specific phosphatases. The first identified member, termed Acr2, was found in an arsenic resistance cluster of genes in the yeast Saccharomyces cerevisiae (Bobrowicz et al. 1997). Like the R773 ArsC, these proteins use glutaredoxin and GSH as electron donors (Mukhopadhyay & Rosen 1998). An ortholog, LmAcr2, from the tropical parasite Leishmania major, reduces both As(V) and Sb(V) and is required for both resistance to arsenate and activation of the pentavalent antimonial drug sodium stibogluconate (Zhou et al. 2004). LmAcr2 also exhibits phosphatase activity, whereas yeast Acr2 does not. However, yeast Acr2 can be converted to a phosphatase by three mutations (Mukhopadhyay et al. 2003). Both the respiration and detoxification machinery facilitate reduction of arsenate by microbes, but it is not known whether one is more prevalent in the environment than the other, nor is it clear which environmental factors modulate the predominance of the two mechanisms. The facts that arsenate reductases arose at least three times and that some are both reductases and phosphatases or can easily be changed from one activity to the other suggest that evolution of arsenate reductases is relatively rapid and uncomplicated.
ARSENIC METHYLATION CYCLE
There is extensive biotransformation of arsenic by enzymatic processes such as methylation. Methylated arsenic species such as methylarsenate [MAs(V)], dimethylarsenate [DMAs(V)], and trimethylarsine oxide [TMAs(V)O] are found in low concentrations in some soils (Huang et al. 2011). DMAs(V), and occasionally MAs(V) and the tetramethylarsonium (CH3)4As+ cation, is found in rice grain (Hansen et al. 2011, Meharg & Zhao 2012). DMAs(V) and MAs(V) are also found in some lake water, especially in eutrophic lakes, with concentrations usually peaking in the summer coincident with algal and microbial growth (Hasegawa et al. 2010, Smedley & Kinniburgh 2002). Methylation of arsenic can lead to volatilization of methylarsines, mainly trimethylarsine and some mono- and dimethylarsine (Mestrot et al. 2011a). These arsine gases are oxidized in the atmosphere by UV light, forming nonvolatile arsenic species (Mestrot et al. 2011b), which are deposited on land or ocean surfaces.
As some microorganisms evolved mechanisms to methylate arsenic, other microorganisms evolved pathways to do the reverse reaction, demethylation of methylated arsenical herbicides. This pathway is not restricted to methylated arsenicals but catalyzes the breakdown of numerous aromatic arsenicals, including growth promoters and chemical warfare agents. These processes are depicted in the following sections.
Arsenic Methylation and Volatilization
While arsenite efflux systems are responsible for the majority of arsenic tolerance in most organisms (Yang et al. 2012), a common means of microbial arsenic detoxification is by methylation catalyzed by a family of As(III) S-adenosylmethionine (SAM) methyltransferase enzymes designated ArsM in microbes (AS3MT in higher eukaryotes) (Qin et al. 2006, 2009) (Figure 2). At latest count there are more than 30,000 entries for ArsM/AS3MT sequences in the NCBI database in members of all kingdoms, with the majority in prokaryotic and eukaryotic microbes. These in turn are members of a superfamily of methyltransferases involved in many physiological functions (Liscombe et al. 2012). ArsMs detoxify arsenic by converting inorganic trivalent arsenic [As(III)] into methylated species that eventually end up as pentavalent MAs(V) and DMAs(V) and small amounts of TMAs(V)O and volatile trimethylarsine [TMAs(III)] (Drobná et al. 2006). Because its expression requires the presence of environmental arsenic, it is reasonable to infer that ArsM evolved to confer arsenic resistance rather than an unrelated function. The widespread distribution of these genes implies that genes for arsenic biotransformation evolved early.
The first microbial arsM gene was identified and cloned from the soil bacterium Rhodopseudomonas palustris (Qin et al. 2006). RpArsM is a 283-residue protein (29,656 Da). The cloned arsM gene was expressed in an As(III)-hypersensitive strain of Escherichia coli constructed by deletion of the chromosomal ars operon (Carlin et al. 1995), and it was shown to confer resistance to arsenite, providing the first demonstration that the function of ArsM is arsenic resistance. The transgenic cells methylated medium arsenic, producing DMAs(V), TMAs(V)O, and volatile TMAs(III), clearly establishing that arsM alone is both necessary and sufficient for arsenic detoxification. Purified RpArsM catalyzed methylation of arsenite to di- and trimethylated species, with a final product of TMAs(III) gas. As(III) SAM methyltransferases may have an additional role in forming the precursors of other organoarsenicals such as arsenosugars, arsenocholine, and arsenobetaine (Figure 2).
The presence of arsM genes in soil organisms indicates that these microbes remodel the arsenic landscape via biotransformation and volatilization of environmental arsenic. In support of this hypothesis, an orthologous arsM gene was cloned from the acidothermophilic eukaryotic red alga Cyanidioschyzon strain 5508, an environmental isolate of Cyanidioschyzon merolae isolated from Yellowstone National Park (Qin et al. 2009) (Figure 3). The Yellowstone caldera is an active volcanic region with one of the highest concentrations of arsenic in soil and water in the world. ArsM-expressing Cyanidiales form the major biomass in hot springs around the Norris Geyser Basin of Yellowstone National Park, where the water temperature can reach 57°C, the pH is 0.5 to 3.5, and the arsenic concentration is in the millimolar region (Figure 3a,b). As the arsenic-containing geothermal waters erupt from fumaroles, arsenic sulfides with realgar-like composition precipitate, and the algae grow in contact with the solid arsenic minerals (Figure 3c). It has been proposed that these algae remodel the arsenic environment in geothermal regions (Qin et al. 2009). Volatile arsines evolve from the geothermal waters of Yellowstone National Park (Planer-Friedrich et al. 2006). Interestingly, the amount of volatile arsenic is not related to the concentration of aqueous arsenic, which indicates that the arsenic gas phase is not due to simple partitioning with the aqueous phase. Rather, the arsenic gas may be the result of biological activity of methylating algae and other microorganisms as they strive to thrive in the presence of environmental arsenic.
Figure 3.
Arsenic geothermal biology in Yellowstone National Park. Mats of Cyanidiales algae, which express the ArsM As(III) S-adenosylmethionine (SAM) methyltransferase, dominate the biomass in acidic geothermal outflow channels in (a) East Fork, Tantalus Creek, which drains the Norris Geyser Basin and (b) Nymph Creek, located in the Norris-Mammoth corridor in Yellowstone National Park. (c) Cyanidiales algae grow adjacent to realgar-like mineral phases and geothermal-derived waters containing 0.76 mM total arsenic. The orange-yellow deposits were determined by scanning electron microscope electron dispersive X-ray spectroscopy to be realgar-like, with an As:S ratio range of 1.1–1.3. Reproduced from Qin et al. (2009).
The CmarsM gene from the environmental isolate was heterologously expressed in the arsenic hypersensitive strain of E. coli and shown to confer resistance (Qin et al. 2009). Purified CmArsM is the most active arsenic methylating enzyme identified to date and has proven to be an excellent model system for elucidation of the molecular mechanism of this class of arsenic-biotransforming enzymes (Marapakala et al. 2012). The structure of the enzyme has been determined at 1.75 Å resolution with arsenic or SAM (Ajees et al. 2012) (Figure 2).
Degradation of Organoarsenics
In addition to the microbial biogenesis of methylated arsenicals, massive amounts of organic forms of arsenic are introduced into the environment through anthropogenic activities (Figure 4). Because they have lower toxicity than inorganic arsenicals, pentavalent methylated arsenicals such as MAs(V) and DMAs(V) have been extensively used as herbicides and insect pesticides worldwide and have largely replaced classic inorganic arsenical products (Burló et al. 1999). According to 2006 and 2007 market estimates reported by the US Environmental Protection Agency, 2 to 4 million pounds (900,000 to 1,800,000 kg) of the monosodium salt of MAs(V) (MSMA) was used in the United States during 2007 by industrial, commercial, and governmental market sectors (Grube et al. 2011). During the war in Vietnam, 1.2 million gallons of DMAs(V) were used by the US military as a forest defoliant called Agent Blue—one of the “rainbow herbicides” (Stellman et al. 2003). These relatively benign methylarsenicals, however, are largely degraded into more toxic inorganic forms after they are introduced into the environment. Degradation of methylarsenicals occurs in various environmental settings such as soil (Akkari et al. 1986, Feng et al. 2005, Gao & Buran 1997, Huang et al. 2007, Maki et al. 2006a, Von Endt et al. 1968, Woolson et al. 1982), sludge (Sierra-Alvarez et al. 2006), sediment (Hanaoka et al. 1990), seawater (Sanders 1979), and lakes (Maki et al. 2004, 2006b, 2009). Demethylating microbes have been isolated in soil (Maki et al. 2006a, Von Endt et al. 1968) and lakes (Maki et al. 2004, 2006b). In addition, specific organisms such as Montrachet wine yeast (Crecelius 1977), Candida humicola (Cullen et al. 1979), and Mycobacterium neoaurum (Lehr et al. 2003), have been demonstrated to be capable of demethylating methylarsenicals. All of these reported MAs(V)-demethylating organisms catalyze the reaction alone. In contrast, a novel, recently discovered pathway for demethylation of MAs(V) is composed of two different reaction steps, reduction and demethylation, catalyzed by a soil bacterial community (Yoshinaga et al. 2011). A Burkholderia species capable of reducing MAs(V) to MAs(III) and a Streptomyces species that could demethylate MAs(III) to As(III) were identified in golf course soil treated with the MAs(V) herbicide. Neither organism alone could demethylate the herbicide, but in mixed culture the two bacteria carried out the entire demethylation process from MAs(V) to As(III).
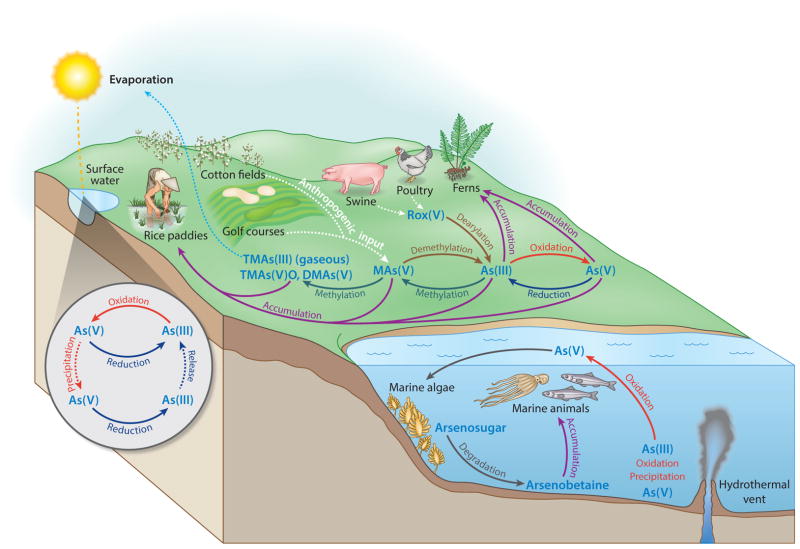
Figure 4.
Arsenic biogeochemical cycles. Various microbial biotransformations of arsenic have roles in cycling of environmental arsenic between various chemical forms. As(V) and As(III) are interconverted by reduction (dark blue) and oxidation (red ), forming an arsenic redox cycle. Reduction of As(V) to As(III) is catalyzed by various microorganisms for arsenic detoxification or anaerobic arsenate respiration. The latter mechanism helps mobilize arsenic in the form of As(III) from As(V)-rich sediments and leads to arsenic contamination of groundwater and/or soil. As(III) is, in turn, oxidized back to As(V) by arsenite-oxidizing microbes. As(III) oxidation results in arsenic precipitation and adsorption to minerals in marine/freshwater sediments. Arsenite is also oxidized through arsenite-dependent anoxygenic photosynthesis. As an alternative detoxification process, arsenic is converted to mono-, di-, and trimethylated arsenicals (aqua). Methylation of arsenic also leads to volatilization of methylarsines, primarily trimethylarsine and some mono- and dimethylarsines (light blue). Inorganic and methylated arsenicals also form a bidirectional methylation cycle, wherein methylarsenicals are demethylated back to inorganic species (brown). Anthropogenic sources such as MAs(V) and roxarsone are also degraded into inorganic arsenicals in the environment and form a part of this cycle (white). In seawater, the major arsenic form As(V) is biotransformed into arsenosugar by algae, a process that is proposed to be completed by the combination of methylation and adenosylation ( gray). Arsenic accumulation is observed widely in many organisms ( purple). Ferns hyperaccumulate inorganic arsenic in above-ground tissues, and rice plants take up not only inorganic arsenic but also methylarsenicals and ultimately accumulate arsenic into the grain. Marine animals, in contrast, accumulate arsenosugar and arsenobetaine. Abbreviations: As(III), arsenite; As(V), arsenate; DMAs(V), dimethylarsenate; MAs(V), methylarsenate; Rox(V), roxarsone; TMAs(III), trimethylarsine; TMAs(V)O, trimethylarsine oxide.
Much information is available about microbial degradation of methylarsenicals in the environment, yet little is known about the molecular mechanisms. From the same golf course soil, a MAs(III)-demethylating Bacillus species was recently isolated, and the gene responsible for MAs(III) demethylation was cloned by selection for herbicide resistance (M. Yoshinaga & B.P. Rosen, submitted for publication). MAs(V) is reduced to MAs(III) by the first step of the novel MAs(V) degradation pathway; its relatively benign toxicity is dramatically increased in the reduced trivalent form, which is even more toxic than inorganic trivalent form As(III) (Petrick et al. 2000, 2001). The molecular mechanisms of MAs(V) reduction, the initial reaction of the two-step breakdown pathway, remain unknown. Reduction of MAs(V) has been also observed in plants such as rice (Li et al. 2009a, Lomax et al. 2011) and castor bean (Ye et al. 2010). A subclass of human glutathione S-transferase (GST) omega, hGSTO1-1, can catalyze in vitro reduction of MAs(V) to MAs(III) (Zakharyan et al. 2001); however, it remains unclear whether hGSTO1-1 contributes significantly to MAs(V) reduction in vivo and whether member(s) of GST are responsible for MAs(V) reduction in bacteria and plants.
Organoarsenicals introduced anthropogenically into the environment include not only methylated arsenicals but also more complex aromatic arsenicals (Figure 4). Phenylarsenic compounds such as roxarsone (4-hydroxy-3-nitrophenylarsonic acid), nitarsone (4-nitrophenylarsonic acid), and p-arsanilic acid (4-aminophenylarsonic acid) have been used extensively and in large amounts in animal husbandry for disease prevention, growth promotion, enhanced feed utilization, and improved meat pigmentation. The majority of these compounds is not retained by the animals but is excreted into the environment unchanged (Morrison 1969). On the basis of broiler production and roxarsone feed dosage, it is estimated that approximately 2 million pounds (900,000 kg) of roxarsone is released into environment in the United States annually by the poultry industry alone (Covey et al. 2010, Rutherford et al. 2003). Moreover, aromatic arsenicals have been also used for chemical warfare for more than a century. Diphenylated arsenic compounds such as Clark I (diphenylchloroarsine) and Clark II (diphenylcyanoarsine) were produced as chemical warfare agents during World Wars I and II. These were disposed of in land and seas after those wars, but diphenylarsinic acid, which results from chemical transformation in the environment, accumulates in contaminated sites (Harada et al. 2009). As methylated arsenicals, these aromatic arsenic compounds also undergo degradation and eventually become the more toxic inorganic arsenicals in the environment (Cortinas et al. 2006, Garbarino et al. 2003, Köhler et al. 2001, Maejima et al. 2011, Makris et al. 2008, Stolz et al. 2007). A few degrading microbes have been isolated (Harada et al. 2009, Nakamiya et al. 2007, Stolz et al. 2007); however, no molecular details of aromatic arsenical degradation have been reported. In a preliminary investigation of the substrate specificity of the cloned MAs(III) demethylating enzyme, it was found to degrade trivalent roxarsone to As(III) both in vivo and in vitro, which suggests that it might also be involved in breakdown of environmental aromatic arsenical growth promoters (M. Yoshinaga & B.P. Rosen, submitted for publication), Pentavalent aromatic arsenicals such as roxarsone are proposed to undergo a two-step pathway of sequential reduction followed by cleavage of the C-As bond, in analogy with the demethylation of the MSMA herbicide by a microbial community. Since the MAs(V)-reducing bacterial isolates from Florida golf course soils cannot reduce roxarsone (M. Yoshinaga & B.P. Rosen, submitted for publication), there may be as-yet-unidentified bacteria capable of reducing pentavalent phenylarsenicals to the trivalent form. Homologs of the recently cloned MAs(III) demethylase are distributed in many bacterial species, so such organoarsenic degradations are proposed to widely occur in nature, which would impact the arsenic biogeocycle and complete the cycles of arsenic methylation and demethylation.
Other Arsenic Biotransformations
The concentration of arsenic in seawater is approximately 1 to 2 μg/L, almost entirely in inorganic forms. It can be enriched 1,000- to 100,000-fold in marine organisms, with most arsenic in organic forms such as arsenosugars (Edmonds & Francesconi 1981) (Figure 2). Arsenate, the major arsenic form in seawater, is biotransformed into arsenosugars by algae (Miyashita et al. 2011) (Figure 4). The presence of arsenosugars was first confirmed by isolation and identification of two such species from the brown kelp (algae) Ecklonia radiate (Edmonds & Francesconi 1981). At least 15 arsenosugars have been identified from brown algae and other algal families (Edmonds & Francesconi 1981). Among these, the oxo-arsenosugars, which contain a chemically active dimethylarsinoyl group at the C5 position of D-ribose derivatives, are the most common type (Miyashita et al. 2011). Oxo-arsenosugar-glycerol and oxo-arsenosugar-phosphate are the major forms present in almost all marine macroalgae, especially in Chlorophyta (green algae) and Rhodophyta (red algae) (Shibata et al. 1992). Small amounts of these two oxo-arsenosugars have also been found in freshwater green algae and green macroalgae (Koch et al. 1999, Miyashita et al. 2009). Arsenate can be biotransformed into oxo-arsenosugars by the freshwater unicellular green alga Chlamydomonas reinhardtii (Miyashita et al. 2011). To date, no arsenosugar has been identified outside the marine and freshwater environment (Rahman et al. 2012), except in the urine of humans who consumed seafood (Le et al. 1994).
Although arsenosugar is present in many marine and freshwater organisms, the synthesis pathway remains elusive. Arsenosugars have been proposed to be produced as a by-product of the detoxification process (Edmonds et al. 1993). Arsenosugars are probably biosynthesized by methylation and adenosylation (Figure 4), either simultaneously or sequentially. As noted above, the methylation of arsenic is carried out by the SAM-dependent arsenic methyltransferase (Ye et al. 2012). The methyltransferase is not likely to carry out the adenosylation at the same time, however. The enzyme that catalyzes adenosylation has not been identified, and it is not clear whether there are other enzymes involved. As SAM is the methyl donor, it has also been suggested that the adenosyl group derives from SAM as well. However, evidence to support this hypothesis is lacking. Obviously, more research is required.
Seaweed is rich in arsenosugars, with tissue concentrations of arsenic up to 100 μg/g, primarily in the form of arsenosugars (Francesconi & Kuehnelt 2002). Applying seaweed to the land as a soil improver and fertilizer has been common practice in many coastal regions of the world (Castlehouse et al. 2003). This may lead to the buildup of arsenic in soils. Long-term application of seaweed increases arsenic concentrations up to 10-fold in coastal soils currently manured with seaweed. With green agriculture and growing demand for organically farmed product, the use of seaweed as a fertilizer has been revived. The long-term environmental consequences of seaweed application to soils need further investigation. Arsenosugar applied to the soil decomposes primarily to DMAs(V), as well as to arsenate and arsenite. Although a decomposition pathway was proposed, none of the specific enzymes has been identified.
Even in hydrothermal vents, where there are no algae or other photosynthetic organisms, and where arsenic exists primarily in the form of arsenite in the hydrothermal vent fluids, organic arsenicals are predominant in vent biota. For example, the major arsenic species in shrimp from a mid-Atlantic vent is arsenobetaine, and in mussels, arsenic is present mainly as arsenosugars (Taylor et al. 2012). Although little is known about the biosynthesis of these organic arsenicals, they are likely of vent origin, possibly synthesized by chemolithoautotrophic bacteria.
IMPLICATIONS OF ARSENIC BIOTRANSFORMATION IN ENVIRONMENTAL CONTAMINATION AND REMEDIATION
Arsenic Biotransformation and Rice Uptake of Arsenic
Rice is particularly efficient in accumulating arsenic into its grain (Williams et al. 2007, Zhao et al. 2010) (Figures 4 and 5). Therefore, in addition to arsenic-tainted groundwater, rice is the major route of arsenic exposure for populations subsisting on rice (Meharg & Zhao 2012). Rice paddies irrigated with arsenic-contaminated groundwater represent a showcase for arsenic biogeochemistry. In this system, arsenic biotransformation, particularly reduction, is the key process in transporting arsenic from groundwater to the rice on dining tables of millions of people. Similar to sediment arsenic mobilization, iron/arsenate reduction under flooded conditions causes the release of arsenic to soil pore water, with arsenite as the dominant species, contributing to high arsenic accumulation in rice (Li et al. 2009b, Xu et al. 2008). Moreover, arsenite behaves like a silicic acid analog and is taken up adventitiously via the highly expressed silicon transporters in rice roots, further exacerbating the problem (Ma et al. 2008). Application of organic matter can enhance arsenic release by promoting iron and arsenate reduction, as indicated by enhanced microbial activities and increased copy number of arsC genes in soil, and arsenic speciation in soil solution is further modulated by other microbial processes, such as oxidation and methylation (Huang et al. 2012b). Soils with similar levels of total arsenic can show different patterns of arsenic release upon flooding (Stroud et al. 2011), and the differences may result from soil chemical properties and/or microbial communities.
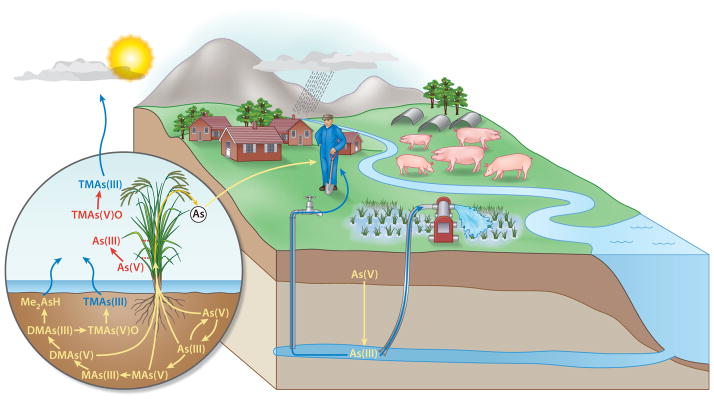
Figure 5.
Human exposure to groundwater arsenic through irrigation of rice paddy and drinking water. Arsenic from groundwater affects human health both directly (through drinking) and indirectly (through consuming rice irrigated with As-tainted groundwater). The pathway through rice was underestimated until recently (Zhu et al. 2008). Abbreviations: As(III), arsenite; As(V), arsenate; DMAs(III), dimethylarsenite; DMAs(V), dimethylarsenate; Me2AsH, dimethylarsine; MAs(III), methylarsenite; MAs(V), methylarsenate; TMAs(III), trimethylarsine; TMAs(V)O, trimethylarsine oxide.
Flooding conditions in paddy soil make methylation an important microbial process affecting arsenic speciation and mobility. Despite the wide detection of methylated arsenic species in rice, the rice plant is unlikely to methylate arsenic unless the plant expresses a bacterial arsM (Meng et al. 2011). Soil microbes are likely responsible for methylated arsenic species in soil-rice systems (Lomax et al. 2011, Zhao et al. 2013) (Figure 5). Addition of organic matter to paddy soil can increase arsenic methylation and volatilization (Huang et al. 2012a). Amendment with organic matter significantly increases both the amount of arsenic in rice grain and the proportion of methylated species ( Jia et al. 2012). Degenerate arsM primers can be used to monitor the diversity and abundance of microbes harboring this functional gene ( Jia et al. 2013). The arsM gene is present in diverse soils sampled from different parts of China, and both organic matter amendment and rhizospheric effects enhance the abundance of arsM in soil, implicating microbial arsM as an important player in rice uptake of methylated arsenic species.
Overall, our understanding of microbial transformation of arsenic in paddy soils is meager. We know little about the abundance and diversity of microbes harboring ars operons and how environmental conditions drive their dynamics. As paddy soils contain diverse and complex microbial assemblages, net biogeochemical cycling is controlled by the interplay of different guilds of contributing microbes. Detailed analysis of the composition and patterns of arsenic-related functional genes in soils will be required in the future to construct predictive models to guide the exploration of the potential of microbial arsenic transformation in mitigating health risks associated with arsenic in rice.
Arsenic Biomethylation and Bioremediation
Both chemical and biological approaches to remediation of arsenic-contaminated environments have been utilized. Chemical remediation can induce secondary pollution and is often costly. Bioremediation, if efficiently designed, can be advantageous. Biomethylation, because its end product TMAs(III) is volatile, can remove arsenic from contaminated water or soil (Ye et al. 2012). The natural process of methylation and volatilization is rather slow and is not practical on the industrial scale of bioremediation. Although amendment with specific organic matter (such as dried distillers grain) can significantly improve this process, it is still not efficient enough (Huang et al. 2011, Jia et al. 2012). Introduction and overexpression of arsM genes in soil organisms may accelerate the production of volatile arsenicals (Chen et al. 2013).
CURRENT CHALLENGES AND CONCLUDING REMARKS
Arsenic from both natural and anthropogenic sources contaminates the environment. Knowledge about how Earth abides arsenic will provide insights and incentives to mitigate risks associated with arsenic pollution of water, soil, and food. As an element, arsenic cannot be destroyed and will always be present in the environment, but managing its biotransformation can modify its physical location and chemical speciation. Arsenic biotransformations are complex, and different pathways predominate in different environments. To understand these pathways and how they affect life on Earth, future research endeavors will need to involve
identification of molecular details of the pathways of arsenosugar and arsenobetaine biosynthesis, particularly in marine organisms;
understanding the linkage between microbial diversity and the kinetics of arsenic biotrans-formation in order to predict which pathways will predominate and hence the fate of arsenic in specific environments;
developing and deploying bioremediation and biomitigation strategies based on rational design of pathways for arsenic biotransformation in microbial communities; and
systems approaches to modeling and manipulating the fate of arsenic in various ecosystems at various temporal and spatial scales to harness the toxicity of arsenic.
SUMMARY POINTS.
Arsenic is a ubiquitous environmental toxin that enters the human biosphere through microbial transformations.
Microbial enzymes catalyze cycles of arsenic oxidation and reduction that mobilize or immobilize arsenic.
Other microbial enzymes catalyze cycles of arsenic methylation and demethylation that alter the toxicity and availability of arsenic in our food supply.
Through a combination of genomics, biochemistry, and plant and food science, we are beginning to understand the effects of arsenic biogeochemistry on life and how life remodels the environment.
Acknowledgments
The research described in this study was supported by US Public Health Service Grant GM55425 to B.P.R. Y.G.Z. was supported by the Natural Science Foundation of China (41090282) and the Ministry of Science & Technology of China (2011DFB91710). Research in F.J.Z.’s lab was supported by the Natural Science Foundation of China (41330853) and the Program for Changjiang Scholars and Innovative Research Team in University (IRT1256). We also thank Dr. Jun Ye, Ms. Yalan Shi, Mr. San’an Nie, and Dr. Zheng Chen for their help in preparing this manuscript.
Glossary
- GSH
reduced glutathione
- MAs(V)
methylarsenate
- DMAs(V)
dimethylarsenate
- TMAs(V)O
trimethylarsine oxide
- SAM
S-adenosylmethionine
- TMAs(III)
trimethylarsine
- MAs(III)
methylarsenite
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- Abernathy CO, Thomas DJ, Calderon RL. Health effects and risk assessment of arsenic. J Nutr. 2003;133:S1536–38. doi: 10.1093/jn/133.5.1536S. [DOI] [PubMed] [Google Scholar]
- Afkar E, Lisak J, Saltikov C, Basu P, Oremland RS, Stolz JF. The respiratory arsenate reductase from Bacillus selenitireducens strain MLS10. FEMS Microbiol Lett. 2003;226:107–12. doi: 10.1016/S0378-1097(03)00609-8. [DOI] [PubMed] [Google Scholar]
- Ahmann D, Roberts AL, Krumholz LR, Morel FM. Microbe grows by reducing arsenic. Nature. 1994;371:750. doi: 10.1038/371750a0. [DOI] [PubMed] [Google Scholar]
- Ajees AA, Marapakala K, Packianathan C, Sankaran B, Rosen BP. Structure of an As(III) S-adenosylmethionine methyltransferase: insights into the mechanism of arsenic biotransformation. Biochemistry. 2012;51:5476–85. doi: 10.1021/bi3004632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akkari KH, Frans RE, Lavy TL. Factors affecting degradation of MSMA in soil. Weed Sci. 1986;34:781–88. [Google Scholar]
- Anderson GL, Williams J, Hille R. The purification and characterization of arsenite oxidase from Alcaligenes faecalis, a molybdenum-containing hydroxylase. J Biol Chem. 1992;267:23674–82. [PubMed] [Google Scholar]
- Andreae MO. Arsenic speciation in seawater and interstitial waters: the influence of biological-chemical interactions on the chemistry of a trace element. Limnol Oceanogr. 1979;24:440–52. [Google Scholar]
- Bhattacharjee H, Mukhopadhyay R, Thiyagarajan S, Rosen BP. Aquaglyceroporins: ancient channels for metalloids. J Biol. 2008;7:33. doi: 10.1186/jbiol91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharjee H, Rosen BP. Arsenic metabolism in prokaryotic and eukaryotic microbes. In: Nies DH, Silver S, editors. Molecular Microbiology of Heavy Metals. Berlin: Springer; 2007. pp. 371–406. [Google Scholar]
- Bobrowicz P, Wysocki R, Owsianik G, Goffeau A, Ulaszewski S. Isolation of three contiguous genes, ACR1, ACR2 and ACR3, involved in resistance to arsenic compounds in the yeast Saccharomyces cerevisiae. Yeast. 1997;13:819–28. doi: 10.1002/(SICI)1097-0061(199707)13:9<819::AID-YEA142>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Brammer H, Ravenscroft P. Arsenic in groundwater: a threat to sustainable agriculture in South and South-East Asia. Environ Int. 2009;35:647–54. doi: 10.1016/j.envint.2008.10.004. [DOI] [PubMed] [Google Scholar]
- Burló F, Guijarro I, Carbonell-Barrachina AA, Valero D, Martínez-Sánchez F. Arsenic species: effects on and accumulation by tomato plants. J Agric Food Chem. 1999;47:1247–53. doi: 10.1021/jf9806560. [DOI] [PubMed] [Google Scholar]
- Canfield DE, Rosing MT, Bjerrum C. Early anaerobic metabolisms. Philos Trans R Soc B. 2006;361:1819–34. doi: 10.1098/rstb.2006.1906. discussion 35–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlin A, Shi W, Dey S, Rosen BP. The ars operon of Escherichia coli confers arsenical and antimonial resistance. J Bacteriol. 1995;177:981–86. doi: 10.1128/jb.177.4.981-986.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo R, Saier MH. Functional promiscuity of homologues of the bacterial ArsA ATPases. Int J Microbiol. 2010;2010:187373. doi: 10.1155/2010/187373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castlehouse H, Smith C, Raab A, Deacon C, Meharg AA, Feldmann J. Biotransformation and accumulation of arsenic in soil amended with seaweed. Environ Sci Technol. 2003;37:951–57. doi: 10.1021/es026110i. [DOI] [PubMed] [Google Scholar]
- Chen CJ, Chen CW, Wu MM, Kuo TL. Cancer potential in liver, lung, bladder and kidney due to ingested inorganic arsenic in drinking water. Br J Cancer. 1992;66:888–92. doi: 10.1038/bjc.1992.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Qin J, Zhu YG, de Lorenzo V, Rosen BP. Engineering the soil bacterium Pseudomonas putida for arsenic methylation. Appl Environ Microbiol. 2013;79:4493–95. doi: 10.1128/AEM.01133-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YC, Su HJ, Guo YL, Hsueh YM, Smith TJ, et al. Arsenic methylation and bladder cancer risk in Taiwan. Cancer Causes Control. 2003;14:303–10. doi: 10.1023/a:1023905900171. [DOI] [PubMed] [Google Scholar]
- Cortinas I, Field JA, Kopplin M, Garbarino JR, Gandolfi AJ, Sierra-Alvarez R. Anaerobic biotransformation of roxarsone and related N-substituted phenylarsonic acids. Environ Sci Technol. 2006;40:2951–57. doi: 10.1021/es051981o. [DOI] [PubMed] [Google Scholar]
- Covey AK, Furbish DJ, Savage KS. Earthworms as agents for arsenic transport and transformation in roxarsone-impacted soil mesocosms: a μXANES and modeling study. Geoderma. 2010;156:99–111. [Google Scholar]
- Crecelius EA. Changes in the chemical speciation of arsenic following ingestion by man. Environ Health Perspect. 1977;19:147–50. doi: 10.1289/ehp.7719147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen WR, McBride BC, Pickett AW. The transformation of arsenicals by Candida humicola. Can J Microbiol. 1979;25:1201–5. doi: 10.1139/m79-187. [DOI] [PubMed] [Google Scholar]
- Cullen WR, Reimer KJ. Environmental arsenic chemistry. Chem Rev. 1989;89:713–64. [Google Scholar]
- Davies PC, Benner SA, Cleland CE, Lineweaver CH, McKay CP, Wolfe-Simon F. Signatures of a shadow biosphere. Astrobiology. 2009;9:241–49. doi: 10.1089/ast.2008.0251. [DOI] [PubMed] [Google Scholar]
- Delnomdedieu M, Basti MM, Otvos JD, Thomas DJ. Reduction and binding of arsenate and dimethylarsinate by glutathione: a magnetic resonance study. Chem Biol Interact. 1994;90:139–55. doi: 10.1016/0009-2797(94)90099-x. [DOI] [PubMed] [Google Scholar]
- Dhar RK, Zheng Y, Saltikov CW, Radloff KA, Mailloux BJ, et al. Microbes enhance mobility of arsenic in Pleistocene aquifer sand from Bangladesh. Environ Sci Technol. 2011;45:2648–54. doi: 10.1021/es1022015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drobná Z, Del Razo LM, García-Vargas GG, Sánchez-Peña LC, Barrera-Hernández A, et al. Environ-mental exposure to arsenic, AS3MT polymorphism and prevalence of diabetes in Mexico. J Expo Sci Environ Epidemiol. 2012;23:151–55. doi: 10.1038/jes.2012.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drobná Z, Xing W, Thomas DJ, Stýblo M. shRNA silencing of AS3MT expression minimizes arsenic methylation capacity of HepG2 cells. Chem Res Toxicol. 2006;19:894–98. doi: 10.1021/tx060076u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duval S, Ducluzeau AL, Nitschke W, Schoepp-Cothenet B. Enzyme phylogenies as markers for the oxidation state of the environment: the case of respiratory arsenate reductase and related enzymes. BMC Evol Biol. 2008;8:206. doi: 10.1186/1471-2148-8-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmonds JS, Francesconi KA. Arseno-sugars from brown kelp (Ecklonia radiata) as intermediates in cycling of arsenic in a marine ecosystem. Nature. 1981;289:602–4. [Google Scholar]
- Edmonds JS, Francesconi KA. Transformations of arsenic in the marine environment. Experientia. 1987;43:553–57. doi: 10.1007/BF02143584. [DOI] [PubMed] [Google Scholar]
- Edmonds JS, Francesconi KA, Stick RV. Arsenic compounds from marine organisms. Nat Prod Rep. 1993;10:421–28. [Google Scholar]
- Ellis PJ, Conrads T, Hille R, Kuhn P. Crystal structure of the 100 kDa arsenite oxidase from Alcaligenes faecalis in two crystal forms at 1.64 Å and 2.03 Å. Structure. 2001;9:125–32. doi: 10.1016/s0969-2126(01)00566-4. [DOI] [PubMed] [Google Scholar]
- Fendorf S, Herbel MJ, Tufano KJ, Kocar BD. Biogeochemical processes controlling the cycling of arsenic in soils and sediments. In: Violante A, Huang PM, Gadd GM, editors. Biophysico-Chemical Processes of Heavy Metals and Metalloids in Soil Environments. Hoboken, NJ: Wiley; 2008. pp. 313–38. [Google Scholar]
- Fendorf S, Michael HA, van Geen A. Spatial and temporal variations of groundwater arsenic in South and Southeast Asia. Science. 2010;328:1123–27. doi: 10.1126/science.1172974. [DOI] [PubMed] [Google Scholar]
- Feng M, Schrlau JE, Snyder R, Snyder GH, Chen M, et al. Arsenic transport and transformation associated with MSMA application on a golf course green. J Agric Food Chem. 2005;53:3556–62. doi: 10.1021/jf047908j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher JC, Hollibaugh JT. Selenate-dependent anaerobic arsenite oxidation by a bacterium from Mono Lake, California. Appl Environ Microbiol. 2008;74:2588–94. doi: 10.1128/AEM.01995-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francesconi KA, Kuehnelt D. Arsenic compounds in the environment. In: Frankenberger JWT, editor. Environmental Chemistry of Arsenic. New York: Dekker; 2002. pp. 51–94. [Google Scholar]
- Gao S, Buran RG. Environmental factors affecting rates of arsine evolution from mineralization of arsenicals in soil. J Environ Qual. 1997;26:753–63. [Google Scholar]
- Garbarino JR, Bednar AJ, Rutherford DW, Beyer RS, Wershaw RL. Environmental fate of roxarsone in poultry litter. I Degradation of roxarsone during composting. Environ Sci Technol. 2003;37:1509–14. doi: 10.1021/es026219q. [DOI] [PubMed] [Google Scholar]
- Ghosh M, Shen J, Rosen BP. Pathways of As(III) detoxification in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1999;96:5001–6. doi: 10.1073/pnas.96.9.5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladysheva TB, Oden KL, Rosen BP. Properties of the arsenate reductase of plasmid R773. Biochemistry. 1994;33:7288–93. doi: 10.1021/bi00189a033. [DOI] [PubMed] [Google Scholar]
- Grube A, Donaldson D, Kiely T, Wu L. Pesticides Industry Sales and Usage: 2006 and 2007 Market Estimates. Washington, DC: EPA; 2011. http://www.epa.gov/pesticides/pestsales/07pestsales/market_estimates2007.pdf. [Google Scholar]
- Hanaoka K, Hasegawa S, Kawabe N, Tagawa S, Kaise T. Aerobic and anaerobic degradation of several arsenicals by sedimentary micro-organisms. Appl Organometal Chem. 1990;4:239–43. [Google Scholar]
- Hansen HR, Raab A, Price AH, Duan GL, Zhu YG, et al. Identification of tetramethylarsonium in rice grains with elevated arsenic content. J Environ Monit. 2011;13:32–34. doi: 10.1039/c0em00460j. [DOI] [PubMed] [Google Scholar]
- Harada N, Takagi K, Baba K, Fujii K, Iwasaki A. Biodegradation of diphenylarsinic acid to arsenic acid by novel soil bacteria isolated from contaminated soil. Biodegradation. 2009;21:491–99. doi: 10.1007/s10532-009-9318-3. [DOI] [PubMed] [Google Scholar]
- Hasegawa H, Rahman MA, Kitahara K, Itaya Y, Maki T, Ueda K. Seasonal changes of arsenic speciation in lake waters in relation to eutrophication. Sci Total Environ. 2010;408:1684–90. doi: 10.1016/j.scitotenv.2009.11.062. [DOI] [PubMed] [Google Scholar]
- Hohmann-Marriott MF, Blankenship RE. Evolution of photosynthesis. Annu Rev Plant Biol. 2011;62:515–48. doi: 10.1146/annurev-arplant-042110-103811. [DOI] [PubMed] [Google Scholar]
- Huang H, Jia Y, Sun GX, Zhu YG. Arsenic speciation and volatilization from flooded paddy soils amended with different organic matters. Environ Sci Technol. 2012a;46:2163–68. doi: 10.1021/es203635s. [DOI] [PubMed] [Google Scholar]
- Huang H, Zhu YG, Chen Z, Yin XX, Sun G. Arsenic mobilization and speciation during iron plaque decomposition in a paddy soil. J Soils Sediments. 2012b;12:402–10. [Google Scholar]
- Huang JH, Hu KN, Decker B. Organic arsenic in the soil environment: speciation, occurrence, transformation, and adsorption behavior. Water Air Soil Pollut. 2011;219:401–15. [Google Scholar]
- Huang JH, Scherr F, Matzner E. Demethylation of dimethylarsinic acid and arsenobetaine in different organic soils. Water Air Soil Pollut. 2007;182:31–41. [Google Scholar]
- Hughes MF. Arsenic toxicity and potential mechanisms of action. Toxicol Lett. 2002;133:1–16. doi: 10.1016/s0378-4274(02)00084-x. [DOI] [PubMed] [Google Scholar]
- Indriolo E, Na G, Ellis D, Salt DE, Banks JA. A vacuolar arsenite transporter necessary for arsenic tolerance in the arsenic hyperaccumulating fern Pteris vittata is missing in flowering plants. Plant Cell. 2010;22:2045–57. doi: 10.1105/tpc.109.069773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam FS, Gault AG, Boothman C, Polya DA, Charnock JM, et al. Role of metal-reducing bacteria in arsenic release from Bengal delta sediments. Nature. 2004;430:68–71. doi: 10.1038/nature02638. [DOI] [PubMed] [Google Scholar]
- Ji G, Garber EAE, Armes LG, Chen CM, Fuchs JA, Silver S. Arsenate reductase of Staphylococcus aureus plasmid pI258. Biochemistry. 1994;33:7294–99. doi: 10.1021/bi00189a034. [DOI] [PubMed] [Google Scholar]
- Jia Y, Huang H, Sun GX, Zhao FJ, Zhu YG. Pathways and relative contributions to arsenic volatilization from rice plants and paddy soil. Environ Sci Technol. 2012;46:8090–96. doi: 10.1021/es300499a. [DOI] [PubMed] [Google Scholar]
- Jia Y, Huang H, Zhong M, Wang FH, Zhang LM, Zhu YG. Microbial arsenic methylation in soil and rice rhizosphere. Environ Sci Technol. 2013;47:3141–48. doi: 10.1021/es303649v. [DOI] [PubMed] [Google Scholar]
- Kaufman AJ, Johnston DT, Farquhar J, Masterson AL, Lyons TW, et al. Late Archean biospheric oxygenation and atmospheric evolution. Science. 2007;317:1900–3. doi: 10.1126/science.1138700. [DOI] [PubMed] [Google Scholar]
- Kile ML, Houseman EA, Breton CV, Smith T, Quamruzzaman O, et al. Dietary arsenic exposure in Bangladesh. Environ Health Perspect. 2007;115:889–93. doi: 10.1289/ehp.9462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch I, Feldmann J, Wang LX, Andrewes P, Reimer KJ, Cullen WR. Arsenic in the Meager Creek hot springs environment, British Columbia, Canada. Sci Total Environ. 1999;236:101–17. doi: 10.1016/s0048-9697(99)00273-9. [DOI] [PubMed] [Google Scholar]
- Köhler M, Hofmann K, Völsgen F, Thurow K, Koch A. Bacterial release of arsenic ions and organoarsenic compounds from soil contaminated by chemical warfare agents. Chemosphere. 2001;42:425–29. doi: 10.1016/s0045-6535(00)00060-6. [DOI] [PubMed] [Google Scholar]
- Krafft T, Macy JM. Purification and characterization of the respiratory arsenate reductase of Chrysiogenes arsenatis. Eur J Biochem. 1998;255:647–53. doi: 10.1046/j.1432-1327.1998.2550647.x. [DOI] [PubMed] [Google Scholar]
- Kulp TR, Hoeft SE, Asao M, Madigan MT, Hollibaugh JT, et al. Arsenic(III) fuels anoxygenic photosynthesis in hot spring biofilms from Mono Lake, California. Science. 2008;321:967–70. doi: 10.1126/science.1160799. [DOI] [PubMed] [Google Scholar]
- Kulp TR, Hoeft SE, Miller LG, Saltikov C, Murphy JN, et al. Dissimilatory arsenate and sulfate reduction in sediments of two hypersaline, arsenic-rich soda lakes: Mono and Searles Lakes, California. Appl Environ Microbiol. 2006;72:6514–26. doi: 10.1128/AEM.01066-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langner HW, Jackson CR, McDermott TR, Inskeep WP. Rapid oxidation of arsenite in a hot spring ecosystem, Yellowstone National Park. Environ Sci Technol. 2001;35:3302–9. doi: 10.1021/es0105562. [DOI] [PubMed] [Google Scholar]
- Le XC, Cullen WR, Reimer KJ. Human urinary arsenic excretion after one-time ingestion of seaweed, crab, and shrimp. Clin Chem. 1994;40:617–24. [PubMed] [Google Scholar]
- Lebrun E, Brugna M, Baymann F, Muller D, Lièvremont D, et al. Arsenite oxidase, an ancient bioenergetic enzyme. Mol Biol Evol. 2003;20:686–93. doi: 10.1093/molbev/msg071. [DOI] [PubMed] [Google Scholar]
- Lehr CR, Polishchuk E, Delisle MC, Franz C, Cullen WR. Arsenic methylation by micro-organisms isolated from sheepskin bedding materials. Hum Exp Toxicol. 2003;22:325–34. doi: 10.1191/0960327103ht353oa. [DOI] [PubMed] [Google Scholar]
- Leslie EM. Arsenic-glutathione conjugate transport by the human multidrug resistance proteins (MRPs/ABCCs) J Inorg Biochem. 2011;108:141–49. doi: 10.1016/j.jinorgbio.2011.11.009. [DOI] [PubMed] [Google Scholar]
- Li RY, Ago Y, Liu WJ, Mitani N, Feldmann J, et al. The rice aquaporin Lsi1 mediates uptake of methylated arsenic species. Plant Physiol. 2009a;150:2071–80. doi: 10.1104/pp.109.140350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li RY, Stroud JL, Ma JF, McGrath SP, Zhao FJ. Mitigation of arsenic accumulation in rice with water management and silicon fertilization. Environ Sci Technol. 2009b;43:3778–83. doi: 10.1021/es803643v. [DOI] [PubMed] [Google Scholar]
- Liscombe DK, Louie GV, Noel JP. Architectures, mechanisms and molecular evolution of natural product methyltransferases. Nat Prod Rep. 2012;29:1238–50. doi: 10.1039/c2np20029e. [DOI] [PubMed] [Google Scholar]
- Lomax C, Liu WJ, Wu L, Xue K, Xiong J, et al. Methylated arsenic species in plants originate from soil microorganisms. New Phytol. 2011;193:665–72. doi: 10.1111/j.1469-8137.2011.03956.x. [DOI] [PubMed] [Google Scholar]
- Lopez DL, Bundschuh J, Birkle P, Armienta MA, Cumbal L, et al. Arsenic in volcanic geothermal fluids of Latin America. Sci Total Environ. 2012;429:57–75. doi: 10.1016/j.scitotenv.2011.08.043. [DOI] [PubMed] [Google Scholar]
- Ma JF, Yamaji N, Mitani N, Xu XY, Su YH, et al. Transporters of arsenite in rice and their role in arsenic accumulation in rice grain. Proc Natl Acad Sci USA. 2008;105:9931–35. doi: 10.1073/pnas.0802361105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma LQ, Komar KM, Tu C, Zhang W, Cai Y, Kennelley ED. A fern that hyperaccumulates arsenic. Nature. 2001;409:579. doi: 10.1038/35054664. [DOI] [PubMed] [Google Scholar]
- Maejima Y, Arao T, Baba K. Transformation of diphenylarsinic acid in agricultural soils. J Environ Qual. 2011;40:76–82. doi: 10.2134/jeq2009.0496. [DOI] [PubMed] [Google Scholar]
- Maki T, Hasegawa H, Watarai H, Ueda K. Classification for dimethylarsenate-decomposing bacteria using a restrict fragment length polymorphism analysis of 16S rRNA genes. Anal Sci. 2004;20:61–68. doi: 10.2116/analsci.20.61. [DOI] [PubMed] [Google Scholar]
- Maki T, Hirota W, Ueda K, Hasegawa H, Rahman MA. Seasonal dynamics of biodegradation activities for dimethylarsinic acid (DMA) in Lake Kahokugata. Chemosphere. 2009;77:36–42. doi: 10.1016/j.chemosphere.2009.06.016. [DOI] [PubMed] [Google Scholar]
- Maki T, Takeda N, Hasegawa H, Ueda K. Isolation of monomethylarsonic acid–mineralizing bacteria from arsenic contaminated soils of Ohkunoshima Island. Appl Organometal Chem. 2006a;20:538–44. [Google Scholar]
- Maki T, Watarai H, Kakimoto T, Takahashi M, Hasegawa H, Ueda K. Seasonal dynamics of dimethyl-arsenic acid degrading bacteria dominated in Lake Kibagata. Geomicrobiol J. 2006b;23:311–18. [Google Scholar]
- Makris KC, Quazi S, Punamiya P, Sarkar D, Datta R. Fate of arsenic in swine waste from concentrated animal feeding operations. J Environ Qual. 2008;37:1626–33. doi: 10.2134/jeq2007.0479. [DOI] [PubMed] [Google Scholar]
- Malasarn D, Saltikov W, Campbell KM, Santini JM, Hering JG, Newman DK. arrA is a reliable marker for As(V) respiration. Science. 2004;306:455. doi: 10.1126/science.1102374. [DOI] [PubMed] [Google Scholar]
- Marapakala K, Qin J, Rosen BP. Identification of catalytic residues in the As(III) S-adenosylmethionine methyltransferase. Biochemistry. 2012;51:944–51. doi: 10.1021/bi201500c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin P, DeMel S, Shi J, Gladysheva T, Gatti DL, et al. Insights into the structure, solvation, and mechanism of ArsC arsenate reductase, a novel arsenic detoxification enzyme. Structure. 2001;9:1071–81. doi: 10.1016/s0969-2126(01)00672-4. [DOI] [PubMed] [Google Scholar]
- Meharg AA, Zhao FJ. Arsenic and Rice. Dordrecht: Springer; 2012. [Google Scholar]
- Meng XY, Qin J, Wang LH, Duan GL, Sun GX, et al. Arsenic biotransformation and volatilization in transgenic rice. New Phytol. 2011;191:49–56. doi: 10.1111/j.1469-8137.2011.03743.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messens J, Silver S. Arsenate reduction: thiol cascade chemistry with convergent evolution. J Mol Biol. 2006;362:1–17. doi: 10.1016/j.jmb.2006.07.002. [DOI] [PubMed] [Google Scholar]
- Mestrot A, Feldmann J, Krupp EM, Hossain MS, Roman-Ross G, Meharg AA. Field fluxes and speciation of arsines emanating from soils. Environ Sci Technol. 2011a;45:1798–804. doi: 10.1021/es103463d. [DOI] [PubMed] [Google Scholar]
- Mestrot A, Merle JK, Broglia A, Feldmann J, Krupp EM. Atmospheric stability of arsine and methyl-arsines. Environ Sci Technol. 2011b;45:4010–15. doi: 10.1021/es2004649. [DOI] [PubMed] [Google Scholar]
- Miyashita S, Fujiwara S, Tsuzuki M, Kaise T. Rapid biotransformation of arsenate into oxo-arsenosugars by a freshwater unicellular green alga, Chlamydomonas reinhardtii. Biosci Biotechnol Biochem. 2011;75:522–30. doi: 10.1271/bbb.100751. [DOI] [PubMed] [Google Scholar]
- Miyashita S, Shimoya M, Kamidate Y, Kuroiwa T, Shikino O, et al. Rapid determination of arsenic species in freshwater organisms from the arsenic-rich Hayakawa River in Japan using HPLC-ICP-MS. Chemosphere. 2009;75:1065–73. doi: 10.1016/j.chemosphere.2009.01.029. [DOI] [PubMed] [Google Scholar]
- Morrison J. Distribution of arsenic from poultry litter in broiler chickens, soil, and crops. J Agric Food Chem. 1969;17:1288–90. [Google Scholar]
- Mukherjee SC, Rahman MM, Chowdhury UK, Sengupta MK, Lodh D, et al. Neuropathy in arsenic toxicity from groundwater arsenic contamination in West Bengal, India. J Environ Sci Health A. 2003;38:165–83. doi: 10.1081/ese-120016887. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay R, Dey S, Xu N, Gage D, Lightbody J, et al. Trypanothione overproduction and resistance to antimonials and arsenicals in Leishmania. Proc Natl Acad Sci USA. 1996;93:10383–87. doi: 10.1073/pnas.93.19.10383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay R, Rosen BP. The Saccharomyces cerevisiae ACR2 gene encodes an arsenate reductase. FEMS Microbiol Lett. 1998;168:127–36. doi: 10.1111/j.1574-6968.1998.tb13265.x. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay R, Rosen BP. Arsenate reductases in prokaryotes and eukaryotes. Environ Health Perspect. 2002;110(Suppl 5):745–48. doi: 10.1289/ehp.02110s5745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay R, Rosen BP, Phung LT, Silver S. Microbial arsenic: from geocycles to genes and enzymes. FEMS Microbiol Rev. 2002;26:311–25. doi: 10.1111/j.1574-6976.2002.tb00617.x. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay R, Zhou Y, Rosen BP. Directed evolution of a yeast arsenate reductase into a protein tyrosine phosphatase. J Biol Chem. 2003;278:24476–80. doi: 10.1074/jbc.M302610200. [DOI] [PubMed] [Google Scholar]
- Mushak P, Crocetti AF. Risk and revisionism in arsenic cancer risk assessment. Environ Health Perspect. 1995;103:684–89. doi: 10.1289/ehp.95103684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamiya K, Nakayama T, Ito H, Edmonds JS, Shibata Y, Morita M. Degradation of arylarsenic compounds by microorganisms. FEMS Microbiol Lett. 2007;274:184–88. doi: 10.1111/j.1574-6968.2007.00835.x. [DOI] [PubMed] [Google Scholar]
- Natl. Res. Counc. (NRC) Arsenic in Drinking Water. Washington, DC: Natl. Acad. Press; 1999. http://www.nap.edu/openbook.php?isbn=0309063337. [Google Scholar]
- Natl. Res. Counc. (NRC) Arsenic in Drinking Water: 2001 Update. Washington, DC: Natl. Acad. Press; 2001. http://www.nap.edu/books/0309076293/html/ [Google Scholar]
- Naujokas MF, Anderson B, Ahsan H, Aposhian HV, Graziano JH, et al. The broad scope of health effects from chronic arsenic exposure: update on a worldwide public health problem. Environ Health Perspect. 2013;121:295–302. doi: 10.1289/ehp.1205875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman DK, Ahmann D, Morel FMM. A brief review of microbial arsenate respiration. Geomicrobiol J. 1998;15:255–68. [Google Scholar]
- Oden KL, Gladysheva TB, Rosen BP. Arsenate reduction mediated by the plasmid-encoded ArsC protein is coupled to glutathione. Mol Microbiol. 1994;12:301–6. doi: 10.1111/j.1365-2958.1994.tb01018.x. [DOI] [PubMed] [Google Scholar]
- Oremland RS, Hoeft SE, Santini JM, Bano N, Hollibaugh RA, Hollibaugh JT. Anaerobic oxidation of arsenite in Mono Lake water and by a facultative, arsenite-oxidizing chemoautotroph, strain MLHE-1. Appl Environ Microbiol. 2002;68:4795–802. doi: 10.1128/AEM.68.10.4795-4802.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oremland RS, Saltikov CW, Wolfe-Simon F, Stolz JF. Arsenic in the evolution of Earth and extraterrestrial ecosystems. Geomicrobiol J. 2009;26:522–36. [Google Scholar]
- Oremland RS, Stolz JF. The ecology of arsenic. Science. 2003;300:939–44. doi: 10.1126/science.1081903. [DOI] [PubMed] [Google Scholar]
- Oremland RS, Stolz JF. Arsenic, microbes and contaminated aquifers. Trends Microbiol. 2005;13:45–49. doi: 10.1016/j.tim.2004.12.002. [DOI] [PubMed] [Google Scholar]
- Petrick JS, Ayala-Fierro F, Cullen WR, Carter DE, Aposhian HV. Monomethylarsonous acid (MMA(III)) is more toxic than arsenite in Chang human hepatocytes. Toxicol Appl Pharmacol. 2000;163:203–7. doi: 10.1006/taap.1999.8872. [DOI] [PubMed] [Google Scholar]
- Petrick JS, Jagadish B, Mash EA, Aposhian HV. Monomethylarsonous acid (MMAIII) and arsenite: LD50 in hamsters and in vitro inhibition of pyruvate dehydrogenase. Chem Res Toxicol. 2001;14:651–56. doi: 10.1021/tx000264z. [DOI] [PubMed] [Google Scholar]
- Planer-Friedrich B, Lehr C, Matschullat J, Merkel BJ, Nordstrom DK, Sandstrom MW. Speciation of volatile arsenic at geothermal features in Yellowstone National Park. Geochim Cosmochim Acta. 2006;70:2480–91. [Google Scholar]
- Planer-Friedrich B, London J, McCleskey RB, Nordstrom DK, Wallschlaeger D. Thioarsenates in geothermal waters of Yellowstone National Park: determination, preservation, and geochemical importance. Environ Sci Technol. 2007;41:5245–51. doi: 10.1021/es070273v. [DOI] [PubMed] [Google Scholar]
- Planer-Friedrich B, Suess E, Scheinost AC, Wallschlager D. Arsenic speciation in sulfidic waters: reconciling contradictory spectroscopic and chromatographic evidence. Anal Chem. 2010;82:10228–35. doi: 10.1021/ac1024717. [DOI] [PubMed] [Google Scholar]
- Price RE, Amend JP, Pichler T. Enhanced geochemical gradients in a marine shallow-water hydrothermal system: unusual arsenic speciation in horizontal and vertical pore water profiles. Appl Geochem. 2007;22:2595–605. [Google Scholar]
- Price RE, Pichler T. Distribution, speciation and bioavailability of arsenic in a shallow-water submarine hydrothermal system, Tutum Bay, Ambitle Island, PNG. Chem Geol. 2005;224:122–35. [Google Scholar]
- Putila JJ, Guo NL. Association of arsenic exposure with lung cancer incidence rates in the United States. PLoS ONE. 2011;6:e25886. doi: 10.1371/journal.pone.0025886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Lehr CR, Yuan C, Le XC, McDermott TR, Rosen BP. Biotransformation of arsenic by a Yellowstone thermoacidophilic eukaryotic alga. Proc Natl Acad Sci USA. 2009;106:5213–17. doi: 10.1073/pnas.0900238106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Rosen BP, Zhang Y, Wang G, Franke S, Rensing C. Arsenic detoxification and evolution of trimethylarsine gas by a microbial arsenite S-adenosylmethionine methyltransferase. Proc Natl Acad Sci USA. 2006;103:2075–80. doi: 10.1073/pnas.0506836103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman MA, Hasegawa H, Lim RP. Bioaccumulation, biotransformation and trophic transfer of arsenic in the aquatic food chain. Environ Res. 2012;116:118–35. doi: 10.1016/j.envres.2012.03.014. [DOI] [PubMed] [Google Scholar]
- Richey C, Chovanec P, Hoeft SE, Oremland RS, Basu P, Stolz JF. Respiratory arsenate reductase as a bidirectional enzyme. Biochem Biophys Res Commun. 2009;382:298–302. doi: 10.1016/j.bbrc.2009.03.045. [DOI] [PubMed] [Google Scholar]
- Rosen BP, Ajees AA, McDermott TR. Life and death with arsenic. BioEssays. 2011;33:350–57. doi: 10.1002/bies.201100012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossman TG, Uddin AN, Burns FJ, Bosland MC. Arsenite is a cocarcinogen with solar ultraviolet radiation for mouse skin: an animal model for arsenic carcinogenesis. Toxicol Appl Pharmacol. 2001;176:64–71. doi: 10.1006/taap.2001.9277. [DOI] [PubMed] [Google Scholar]
- Rutherford DW, Bednar AJ, Garbarino JR, Needham R, Staver KW, Wershaw RL. Environmental fate of roxarsone in poultry litter. Part II Mobility of arsenic in soils amended with poultry litter. Environ Sci Technol. 2003;37:1515–20. doi: 10.1021/es026222+. [DOI] [PubMed] [Google Scholar]
- Saltikov CW, Newman DK. Genetic identification of a respiratory arsenate reductase. Proc Natl Acad Sci USA. 2003;100:10983–88. doi: 10.1073/pnas.1834303100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders JG. Microbial role in the demethylation and oxidation of methylated arsenicals in seawater. Chemosphere. 1979;8:135–37. [Google Scholar]
- Shibata Y, Morita M, Fuwa K. Selenium and arsenic in biology: their chemical forms and biological functions. Adv Biophys. 1992;28:31–80. doi: 10.1016/0065-227x(92)90022-j. [DOI] [PubMed] [Google Scholar]
- Sierra-Alvarez R, Yenal U, Field JA, Kopplin M, Gandolfi AJ, Garbarino JR. Anaerobic biotransformation of organo-arsenical pesticides monomethylarsonic acid and dimethylarsinic acid. J Agric Food Chem. 2006;54:3959–66. doi: 10.1021/jf053223n. [DOI] [PubMed] [Google Scholar]
- Smedley PL, Kinniburgh DG. A review of the source, behaviour and distribution of arsenic in natural waters. Appl Geochem. 2002;17:517–68. [Google Scholar]
- Smith AH, Lingas EO, Rahman M. Contamination of drinking-water by arsenic in Bangladesh: a public health emergency. Bull World Health Organ. 2000;78:1093–103. [PMC free article] [PubMed] [Google Scholar]
- Song WY, Park J, Mendoza-Cózatl DG, Suter-Grotemeyer M, Shim D, et al. Arsenic tolerance in Ara-bidopsis is mediated by two ABCC-type phytochelatin transporters. Proc Natl Acad Sci USA. 2010;107:21187–92. doi: 10.1073/pnas.1013964107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- States JC, Srivastava S, Chen Y, Barchowsky A. Arsenic and cardiovascular disease. Toxicol Sci. 2009;107:312–23. doi: 10.1093/toxsci/kfn236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stellman JM, Stellman SD, Christinan R, Weber T, Tomasallo C. The extent and patterns of usage of Agent Orange and other herbicides in Vietnam. Nature. 2003;422:681–87. doi: 10.1038/nature01537. [DOI] [PubMed] [Google Scholar]
- Stolz JF, Basu P, Santini JM, Oremland RS. Arsenic and selenium in microbial metabolism. Annu Rev Microbiol. 2006;60:107–30. doi: 10.1146/annurev.micro.60.080805.142053. [DOI] [PubMed] [Google Scholar]
- Stolz JF, Perera E, Kilonzo B, Kail B, Crable B, et al. Biotransformation of 3-nitro-4-hydroxybenzene arsonic acid (roxarsone) and release of inorganic arsenic by Clostridium species. Environ Sci Technol. 2007;41:818–23. doi: 10.1021/es061802i. [DOI] [PubMed] [Google Scholar]
- Stone R. Arsenic and paddy rice: a neglected cancer risk? Science. 2008;321:184–85. doi: 10.1126/science.321.5886.184. [DOI] [PubMed] [Google Scholar]
- Stroud JL, Khan MA, Norton GJ, Islam MR, Dasgupta T, et al. Assessing the labile arsenic pool in contaminated paddy soils by isotopic dilution techniques and simple extractions. Environ Sci Technol. 2011;45:4262–69. doi: 10.1021/es104080s. [DOI] [PubMed] [Google Scholar]
- Sun W, Sierra-Alvarez R, Hsu I, Rowlette P, Field JA. Anoxic oxidation of arsenite linked to chemolithotrophic denitrification in continuous bioreactors. Biotechnol Bioeng. 2010a;105:909–17. doi: 10.1002/bit.22611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W, Sierra-Alvarez R, Milner L, Field JA. Anaerobic oxidation of arsenite linked to chlorate reduction. Appl Environ Microbiol. 2010b;76:6804–11. doi: 10.1128/AEM.00734-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor VF, Jackson BP, Siegfried M, Navratilova J, Francesconi KA, et al. Arsenic speciation in food chains from mid-Atlantic hydrothermal vents. Environ Chem. 2012;9:130–38. doi: 10.1071/EN11134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchounwou PB, Patlolla AK, Centeno JA. Carcinogenic and systemic health effects associated with arsenic exposure—a critical review. Toxicol Pathol. 2003;31:575–88. doi: 10.1080/01926230390242007. [DOI] [PubMed] [Google Scholar]
- Tofail F, Vahter M, Hamadani JD, Nermell B, Huda SN, et al. Effect of arsenic exposure during pregnancy on infant development at 7 months in rural Matlab, Bangladesh. Environ Health Perspect. 2009;117:288–93. doi: 10.1289/ehp.11670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng CH, Chong CK, Chen CJ, Lin BJ, Tai TY. Abnormal peripheral microcirculation in seemingly normal subjects living in blackfoot-disease-hyperendemic villages in Taiwan. Int J Microcirc Clin Exp. 1995;15:21–27. doi: 10.1159/000178945. [DOI] [PubMed] [Google Scholar]
- Tseng CH, Chong CK, Tseng CP, Hsueh YM, Chiou HY, et al. Long-term arsenic exposure and ischemic heart disease in arseniasis-hyperendemic villages in Taiwan. Toxicol Lett. 2003;137:15–21. doi: 10.1016/s0378-4274(02)00377-6. [DOI] [PubMed] [Google Scholar]
- Tseng CH, Tseng CP, Chiou HY, Hsueh YM, Chong CK, Chen CJ. Epidemiologic evidence of diabetogenic effect of arsenic. Toxicol Lett. 2002;133:69–76. doi: 10.1016/s0378-4274(02)00085-1. [DOI] [PubMed] [Google Scholar]
- Tseng WP, Chu HM, How SW, Fong JM, Lin CS, Yeh S. Prevalence of skin cancer in an endemic area of chronic arsenicism in Taiwan. J Natl Cancer Inst. 1968;40:453–63. [PubMed] [Google Scholar]
- Tufano KJ, Reyes C, Saltikov CW, Fendorf S. Reductive processes controlling arsenic retention: revealing the relative importance of iron and arsenic reduction. Environ Sci Technol. 2008;42:8283–89. doi: 10.1021/es801059s. [DOI] [PubMed] [Google Scholar]
- van Lis R, Nitschke W, Duval S, Schoepp-Cothenet B. Arsenics as bioenergetic substrates. Biochim Biophys Acta. 2013;1827:176–88. doi: 10.1016/j.bbabio.2012.08.007. [DOI] [PubMed] [Google Scholar]
- Von Endt DW, Kearney PC, Kafman DD. Degradation of monosodium methanearsonic acid by soil microorganisms. J Agric Food Chem. 1968;16:17–20. [Google Scholar]
- Warelow TP, Oke M, Schoepp-Cothenet B, Dahl JU, Bruselat N, et al. The respiratory arsenite oxidase: structure and the role of residues surrounding the Rieske cluster. PLoS ONE. 2013;8:e72535. doi: 10.1371/journal.pone.0072535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzel WW. Arsenic. In: Alloway BJ, editor. Heavy Metals in Soils: Trace Metals and Metalloids in Soils and Their Bioavailability. Dordrecht, Neth: Springer; 2013. pp. 241–81. [Google Scholar]
- Wilkie JA, Hering JG. Rapid oxidation of geothermal arsenic(III) in streamwaters of the eastern Sierra Nevada. Environ Sci Technol. 1998;32:657–62. [Google Scholar]
- Williams PN, Raab A, Feldmann J, Meharg AA. Market basket survey shows elevated levels of As in South Central U.S. processed rice compared to California: consequences for human dietary exposure. Environ Sci Technol. 2007;41:2178–83. doi: 10.1021/es061489k. [DOI] [PubMed] [Google Scholar]
- Wolfe-Simon F, Blum JS, Kulp TR, Gordon GW, Hoeft SE, et al. A bacterium that can grow by using arsenic instead of phosphorus. Science. 2010;332:1163–66. doi: 10.1126/science.1197258. [DOI] [PubMed] [Google Scholar]
- Woolson EA, Aharonson N, Iadevaia R. Application of the high-performance liquid chromatography–flameless atomic absorption method to the study of alkyl arsenical herbicide metabolism in soil. J Agric Food Chem. 1982;30:580–84. [Google Scholar]
- Xu XY, McGrath SP, Meharg AA, Zhao FJ. Growing rice aerobically markedly decreases arsenic accumulation. Environ Sci Technol. 2008;42:5574–79. doi: 10.1021/es800324u. [DOI] [PubMed] [Google Scholar]
- Xue J, Zartarian V, Wang SW, Liu SV, Georgopoulos P. Probabilistic modeling of dietary arsenic exposure and dose and evaluation with 2003–2004 NHANES data. Environ Health Perspect. 2010;118:345–50. doi: 10.1289/ehp.0901205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang HC, Fu HL, Lin YF, Rosen BP. Pathways of arsenic uptake and efflux. Curr Top Membr. 2012;69:325–58. doi: 10.1016/B978-0-12-394390-3.00012-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Rensing C, Rosen BP, Zhu YG. Arsenic biomethylation by photosynthetic organisms. Trends Plant Sci. 2012;17:155–62. doi: 10.1016/j.tplants.2011.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye WL, Wood BA, Stroud JL, Andralojc PJ, Raab A, et al. Arsenic speciation in phloem and xylem exudates of castor bean. Plant Physiol. 2010;154:1505–13. doi: 10.1104/pp.110.163261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshinaga M, Cai Y, Rosen BP. Demethylation of methylarsonic acid by a microbial community. Environ Microbiol. 2011;13:1205–15. doi: 10.1111/j.1462-2920.2010.02420.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakharyan RA, Sampayo-Reyes A, Healy SM, Tsaprailis G, Board PG, et al. Human monomethylarsonic acid (MMAV) reductase is a member of the glutathione-S-transferase superfamily. Chem Res Toxicol. 2001;14:1051–57. doi: 10.1021/tx010052h. [DOI] [PubMed] [Google Scholar]
- Zargar K, Conrad A, Bernick DL, Lowe TM, Stolc V, et al. ArxA, a new clade of arsenite oxidase within the DMSO reductase family of molybdenum oxidoreductases. Environ Microbiol. 2012;14:1635–45. doi: 10.1111/j.1462-2920.2012.02722.x. [DOI] [PubMed] [Google Scholar]
- Zargar K, Hoeft S, Oremland R, Saltikov CW. Identification of a novel arsenite oxidase gene, arxA, in the haloalkaliphilic, arsenite-oxidizing bacterium Alkalilimnicola ehrlichii strain MLHE-1. J Bacteriol. 2010;192:3755–62. doi: 10.1128/JB.00244-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Liu C-Q, Liu H, Jin Z, Han G, Li L. Geochemistry of the Rehai and Ruidian geothermal waters, Yunnan Province, China. Geothermics. 2008;37:73–83. [Google Scholar]
- Zhao FJ, Dunham SJ, McGrath SP. Arsenic hyperaccumulation by different fern species. New Phytol. 2002;156:27–31. [Google Scholar]
- Zhao FJ, McGrath SP, Meharg AA. Arsenic as a food chain contaminant: mechanisms of plant uptake and metabolism and mitigation strategies. Annu Rev Plant Biol. 2010;61:535–59. doi: 10.1146/annurev-arplant-042809-112152. [DOI] [PubMed] [Google Scholar]
- Zhao FJ, Zhu YG, Meharg AA. Methylated arsenic species in rice: geographical variation, origin, and uptake mechanisms. Environ Sci Technol. 2013;47:3957–66. doi: 10.1021/es304295n. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Messier N, Ouellette M, Rosen BP, Mukhopadhyay R. Leishmania major LmACR2 is a pentavalent antimony reductase that confers sensitivity to the drug Pentostam. J Biol Chem. 2004;279:37445–51. doi: 10.1074/jbc.M404383200. [DOI] [PubMed] [Google Scholar]
- Zhu YG, Williams PN, Meharg AA. Exposure to inorganic arsenic from rice: a global health issue? Environ Pollut. 2008;154:169–71. doi: 10.1016/j.envpol.2008.03.015. [DOI] [PubMed] [Google Scholar]