Abstract
Plasma membrane NADPH oxidases (NOXs) are key producers of reactive oxygen species under both normal and stress conditions in plants and they form functional subfamilies. Studies of these subfamilies indicated that they show considerable evolutionary selection. We performed a comparative genomic analysis that identified 50 ferric reduction oxidases (FRO) and 77 NOX gene homologs from 20 species representing the eight major plant lineages within the supergroup Plantae: glaucophytes, rhodophytes, chlorophytes, bryophytes, lycophytes, gymnosperms, monocots, and eudicots. Phylogenetic and structural analysis classified these FRO and NOX genes into four well-conserved groups represented as NOX, FRO I, FRO II, and FRO III. Further analysis of NOXs of phylogenetic and exon/intron structures showed that single intron loss and gain had occurred, yielding the diversified gene structures during the evolution of NOXs family genes and which were classified into four conserved subfamilies which are represented as Sub.I, Sub.II, Sub.III, and Sub.IV. Additionally, both available global microarray data analysis and quantitative real-time PCR experiments revealed that the NOX genes in Arabidopsis and rice (Oryza sativa) have different expression patterns in different developmental stages, various abiotic stresses and hormone treatments. Finally, coexpression network analysis of NOX genes in Arabidopsis and rice revealed that NOXs have significantly correlated expression profiles with genes which are involved in plants metabolic and resistance progresses. All these results suggest that NOX family underscores the functional diversity and divergence in plants. This finding will facilitate further studies of the NOX family and provide valuable information for functional validation of this family in plants.
Keywords: NADPH oxidase (NOX), phylogenetic analysis, abiotic stress, hormone, coexpression
Introduction
Reactive oxygen species (ROS) have been shown to be toxic but also function as signaling molecules. This biological paradox of ROS and their underlie mechanism confer them vital roles in the integrity, fitness and aging of living organisms (D’Autréaux and Toledano 2007). NADPH oxidases (NOXs) are key enzymes of ROS generation and thus play crucial roles in a variety of biological processes in different kingdoms of life (Torres and Dang 2005; Bedard and Krause 2007; Bedard et al. 2007). While they are not found in prokaryotes and most unicellular eukaryotes, they are universally present in fungi, animals, and plants (Bedard et al. 2007). In fungi, only ancestral NOXs including one ferric reductase (FRE) and three fungal NOXs were identified (Aguirre et al. 2005). In animals, total seven types of NOXs, namely NOX1, NOX2, NOX3, NOX4, NOX5, DUOX1, and DUOX2, were reported (Bedard and Krause 2007; Bedard et al. 2007). However, in plants, only NOX5-like type NOXs were found even although multiple members exist in different species (Sagi and Fluhr 2006; Bedard et al. 2007; Wang et al. 2013).
The plant NOXs, also named as respiratory burst oxidase homologs (RBOHs), are proposed to be major ROS producers of plants under normal and stress conditions (Foreman et al. 2003; Sagi and Fluhr 2006). The first plant NOX was isolated from rice as a gp91phox homolog of mammalian NOX (Groom et al. 1996). Thereafter, plant NOXs were identified in Lycopersicon esculentum (Amicucci et al. 1999), Nicotiana tabacum (Yoshioka et al. 2001), Solanum tuberosum (Yoshioka et al. 2003), Arabidopsis thaliana (Sagi and Fluhr 2006), Medicago truncatula (Marino et al. 2011), Phaseolus vulgaris (Arthikala et al. 2014), and Zea mays (Nestler et al. 2014). In Arabidopsis, ten members of typical NOXs were reported with assigned to be AtRBOHA–J, respectively (Sagi and Fluhr 2006). In rice, at least nine typical NOXs were identified (Wong et al. 2007; Wang et al. 2013). Those rice NOXs were named as OsRBOHA–I or OsNOX1–9, respectively, and found to be diverse in protein domain composition and functioning (Wang et al. 2013). Besides the typical NOXs, some ferric reduction oxidases (FROs), which were considered as the isoforms of yeast ferric-chelate reductase (FRE), were also found in high plants (Sagi and Fluhr 2006). In Arabidopsis total eight FROs (AtFRO1–8) were identified (Wu et al. 2005) whereas in rice only two FROs were reported (OsFRO1 and OsFRO7) (Wang et al. 2013). These proteins are closely related but still different from the typical plant NOX. They contain six membrane-spanning domains, two hemes, and conserved motifs involved in NADPH and FAD binding but lack NADPH_Ox domain and several calcium-binding EF-hand motifs that the typical NOX proteins have (Bedard et al. 2007; Wang et al. 2013).
Different NOX might have different functions both in animals and plants (Bedard and Krause 2007; Kaye et al. 2011). In Arabidopsis, two NOXs, AtRBOHD and AtRBOHF, were found to be essential for jasmonic acid-induced expression of genes regulated by the MYC2 transcription factor (Maruta et al. 2011) while AtRBOHD be participating in ABA-mediated ROS production and stomatal closure (Zhang et al. 2009). Although the activation mechanisms of AtRBOHD and AtRBOHF are similar in stress responses, AtRBOHD has a significantly greater ROS-producing activity than does AtRBOHF (Kimura et al. 2012). AtRBOHD and AtRBOHF also showed to be functioning in disease resistance (Chaouch et al. 2012) and salt stress tolerance (Xie et al. 2011). In addition, it was reported that AtRBOHD participates in endosperm development (Penfield et al. 2006), and AtRBOHB involves in seed after-ripening (Müller et al. 2009) whereas AtRBOHC functions in root-hair-tip growth (Takeda et al. 2008). Moreover, Evans et al. (2005) found that AtRBOHJ functions in salt stress tolerance and, more recently, it was reported that AtRBOHH and AtRBOHJ play essential roles in pollen tube tip growth via Ca2+-activated ROS production (Kaya et al. 2014, 2015). In maize, the expression of the four NOX genes could be induced by ABA treatment, implying that they also function in plant stress tolerance (Lin et al. 2009). In rice, the expression profiles of NOX-encoding genes showed unique stress-response characteristics (Wang et al. 2013). In wheat roots, NOXs ameliorate the oxidative stress induced by nickel (Hao et al. 2006). As a whole, NOXs participate in the plant immune response (Yoshioka et al. 2011), polar growth of root hairs (Nestler et al. 2014) and pollen tubes (Wudick and Feijó 2014; Kaya et al. 2014, 2015), abscisic acid (ABA)-mediated stomatal closure (Zhang et al. 2009; Shi et al. 2012), apoptosis (Tewari et al. 2012), tapetal programmed cell death (PCD) and pollen development (Xie et al. 2014), control of cell differentiation and proliferation (Cano-Domínguez et al. 2008), and seed after ripening (Müller et al. 2009) in higher plants.
The ancient forms of NOXs also exhibit important roles. These ancient forms including fungal NOXs and yeast FREs are responsible either for ROS production or for iron uptake (Bedard et al. 2007). Fungal NOXs were found playing key roles in fungal cellular differentiation, pathogenicity, and fungal-plant symbiosis (Scott and Eaton 2008; Segmüller et al. 2008). Their counterparts in higher plants such as AtRFOs and OsRFOs, however, mainly function in iron acquisition, metabolism, and/or homeostasis (Wu et al. 2005; Sperotto et al. 2010).
As the major producer of ROS during cell growth, plant development, and stress responses, the increasing evidence also show that plant NOXs participate in a number of signaling pathways involving mitogen-activated protein kinases (Zhang et al. 2014), Ca2+-dependent protein kinases (Asano et al. 2012), receptor activated C-kinases (Nakashima et al. 2008), phosphatidylinositol (Kaye et al. 2011), phospholipase Dα1 and phosphatidic acid (Zhang et al. 2009), Ca2+ (Wudick and Feijó 2014), NO (Delledonne et al. 2002), cGMP (Li et al. 2011), extracellular ATP (Song et al. 2006), and hormone-signaling transduction cascades (Zhang et al. 2014). Recently, it was suggested that a cyclin-dependent protein kinase/NOX activation circuit is required for rapid defense signal propagation in Arabidopsis (Dubiella et al. 2013). In addition, the clathrin- and microdomain-dependent endocytic pathways were found to cooperatively regulate AtRBOHD dynamics in Arabidopsis and the regulation of AtRBOHD activity by clustering and endocytosis could facilitate the activation of redox signaling pathways under salt stress (Hao et al. 2014). Thus, NOXs may serve as molecular ‘hubs’ during ROS-mediated signaling in plants (Marino et al. 2012), therefore, crucially involve in plant stress response and normal growth and development.
Although these studies in fungi, yeast, human, and Arabidopsis have led to an understanding of the biochemical properties and physiological functions of NOXs, there has been no systematic study of the evolution and functional divergence of the NOX gene family, especially in Plantae. Here, we performed a comprehensive analysis of the NOX gene family in 20 species, representing the eight major plant lineages within the supergroup Plantae. Phylogenetic analysis was performed to delineate the evolutionary history of the NOX family in Plantae, and exon/intron structure analysis was performed to gain insight into the possible mechanisms of the structural diversity of NOX gene family. Finally, the tissue-specificity, inducibility, and coexpression networks of NOX gene expression in Arabidopsis and rice (Oryza sativa) were characterized by examining publicly available microarray data sets and quantitative real-time PCR (qRT-PCR) experiments. The results obtained here will broaden our understanding of the roles of plant NOXs and provide a framework for further functional investigations of these genes in plants.
Materials and Methods
Data Retrieval and Identification of FRO and NOX Genes
The protein sequences of 20 completely or partially sequenced plant genomes representing the eight major plant lineages (i.e., the glaucophyte Cyanophora paradoxa, the rhodophytes Cyanidioschyzon merolae and Galdieria sulphuraria, the chlorophytes Ostreococcus tauri, Ostreococcus lucimarinus, Micromonas pusilla strain RCC299 and strain CCMP1545, Chlorella variabilis NC64A, Coccomyxa subellipsoidea C-169, Chlamydomonas reinhardtii and Volvox carteri, the bryophyte Physcomitrella patens, the lycophyte Selaginella moellendorffii, the gymnosperm Picea abies, the monocots Brachypodium distachyon, O. sativa and Z. mays and the eudicots A. thaliana, Populus trichocarpa, and Vitis vinifera) were retrieved from public databases. All of the protein sequences were the most current nonredundant sequences and all data sources and versions were the same as in our previous study (Li et al. 2014), except for the gymnosperm P. abies version from ConGenIE 1.0 (http://congenie.org/start, last accessed February 29, 2016; Nystedt et al. 2013). Additionally, the most current and nonredundant protein sequences of Homo sapiens (e!Ensembl release 78, http://asia.ensembl.org/Homo_sapiens/Info/Index, last accessed February 29, 2016), Saccharomyces cerevisiae (SGD release R64.2, http://www.yeastgenome.org/, last accessed February 29, 2016) and Escherichia coli K-12 (EcoGene v3.0, http://ecogene.org/, last accessed February 29, 2016; Zhou et al. 2013) were also collected from public databases. All of the above protein sequences were integrated into a local protein database and the same methods in our previous study (Li et al. 2014) were used for the subsequent identification of the FRO and NOX homologs. The only difference was that the three family specific domain viz., Ferric_reduct (PF01794), FAD_binding_8 (PF08022), NAD_binding_6 (PF08030) and four family specific domain viz., NADPH_Ox (PF08414), Ferric_reduct (PF01794), FAD_binding_8 (PF08022), NAD_binding_6 (PF08030) HMM profiles were used in HMM search against the local protein database for the identification of FRO and NOX homologous genes, respectively.
Phylogenetic Relationships and Exon/Intron Structure Analysis
The amino acid sequences alignment of NADPH_Ox, Ferric_reduct, FAD_binding_8 and NAD_binding_6 domains of the candidate FRO and NOX proteins were used for the construction of phylogenetic trees, and the detailed methods were referenced to our previous study (Li et al. 2014). The exon/intron structures of individual FRO and NOX genes were obtained through the online Gene Structure Display Server (GSDS, http://gsds.cbi.pku.edu.cn, last accessed February 29, 2016; Hu et al. 2015) by aligning the coding or cDNA sequences with their corresponding genomic DNA sequences from Phytozome v9.1 (http://www.phytozome.net/, last accessed February 29, 2016; Goodstein et al. 2012). To illustrate the evolution of introns and the predictions pertaining to the types of introns, we constructed a gene model for annotation of introns and the detailed methods were referenced to our previous study (Li et al. 2014).
Cis-Regulatory Elements, In Silico Expression Profiles
The 1,500 bp upstream of the transcription start site of all NOX genes in Arabidopsis and rice were obtained from Phytozome v9.1 (http://www.phytozome.net/), and the cis-regulatory elements were identified using the PlantCARE program (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/, last accessed February 29, 2016; Lescot et al. 2002).
In Silico Expression Profiles and Coexpression Network Analysis
For the expression profile analysis of NOX genes in Arabidopsis and rice, ATH1 22k and Os 51k microarray data in the Genevestigator V3 database were used, and then the heat maps were constructed using the obtained gene expression data sets, respectively (Hruz et al. 2008).
Coexpression network analysis of NOX genes in Arabidopsis and rice were performed using Mutual Rank (MR) values calculated by using the online tools ATTED-II (version 7.1, http://atted.jp/, last accessed February 29, 2016; Obayashi et al. 2007) and RiceFREND (version 2.0, http://ricefrend.dna.affrc.go.jp/, last accessed February 29, 2016; Sato 2012) coexpression databases, respectively. The following criteria for interactome frameworks were performed at the cut-off MR value ≤ 20 between two genes and hierarchy =2 with the NOX gene as the center. The Cytoscape v3.2.1 software (http://www.cytoscape.org/, last accessed February 29, 2016; Shannon et al. 2003) was used to drawn the coexpression networks.
Plant Materials, Treatments, and qRT-PCR Analysis
Preparation of the Arabidopsis (Col-0) and rice (O. sativa ssp. japonica cv. Dongjin) seedlings were referenced to our previous study (Li et al. 2014). Meanwhile, the detailed methods for seedling under various abiotic stresses, that is, cold (4 °C), heat (30 °C for Arabidopsis/40 °C for rice), salt (200 mM NaCl), drought (20% PEG6000), and oxidation (30 μM MV) and hormones, that is, ABA (100 μM) and MeJA (100 μM) were the same as in our previous study as well (Li et al. 2014). All the MV, MeJA, and ABA used for treatments were purchased from Sigma-Aldrich. All samples were collected following treatment and immediately frozen at −80 °C.
Total RNA was extracted by using Trizol reagent (Takara, Japan) according to the manufacturer’s instructions and the subsequent qRT-PCR analyses were referenced to our previous study (Li et al. 2014). All gene-specific primers were designed to avoid the conserved region and span introns or cross an exon-exon junction. The detailed primer sequences are shown in supplementary table S3, Supplementary Material online. The AtTub6 (AT5G12250) and OsActin1 (Gene ID: KC140126) genes were chosen as the internal control in the analysis of Arabidopsis and rice, respectively.
Results
Identification of FRO and NOX Family Members in Plants
To comprehensively investigate and characterize the FRO and NOX gene families in plants, 20 species representing the eight major plant lineages within the supergroup Plantae, were selected for analysis (fig. 1). A Hidden Markov Model (HMM) search was performed with the obtained sequences and total 50 FRO and 77 NOX homologs were identified in Plantae (fig. 1; supplementary table S1, Supplementary Material online). One or more FRO genes and/or four or more NOX genes were identified in each genome of the selected species (fig. 1). Most of the aquatic algae, including glaucophytes, rhodophytes, and chlorophytes, contained none NOX genes per genome and only one FRO genes except one species in the chlorophytes, C. reinhardtii, carried three FRO genes per genome (fig. 1). In contrast, two or more FRO genes and four or more NOX genes were found in all land plants, including bryophytes, lycophytes, gymnosperms, monocots, and eudicots (fig. 1). In addition, none plant-type FRO gene was identified per genome in bacteria (E. coli K-12) and none plant-type NOX gene was identified per genome in animal (H. sapiens), fungi (S. cerevisiae), and bacteria (E. coli K-12; fig. 1).
Fig. 1.—
The FRO and NOX gene family in Plantae. (A) Systematic evolutionary relationships of 20 species among eight lineages within the supergroup Plantae. (B) The numbers of FRO and NOX homologs in each species. (C) The typical domain organization of FRO and NOX proteins. The transmembrane regions are shown with dotted pink box.
The Pfam and SMART databases were used to analyze the functional domains of the identified FRO and NOX candidates. All of the putative FROs possess a transmembrane region with Ferric_reduction domain (Pfam accession number PF01794), and two cytosol regions, FAD_binding_8 domain (PF08022) and NAD_binding_6 domain (PF08030). However, besides the domains as FROs have, all NOXs also posses a typical NADPH_Ox domain (PF08414).
Phylogenetic Analysis and Classification of the FRO and NOX Family
To explore the phylogenetic relationships among FRO and NOX family members in plants, we first generated an unrooted maximum-likelihood phylogenetic tree with the 50 FROs and 77 NOXs (fig. 2A), which were inferred from the amino acid sequences of the Ferric_reduct, FAD_binding_8 and NAD_binding_6 domains (supplementary fig. S1, Supplementary Material online). FRO and NOX homologs in plants can be classified into four well-conserved subfamilies (NOX, FRO I, FRO II, FRO III; fig. 2) with high statistical support, according to the topology and the deep duplication nodes of FRO and NOX paralogues in the maximum-likelihood tree. Interestingly, the topological relationship of members within subfamilies was highly consistent with the evolutionary relationships between species in Plantae (figs. 1B and 2A). The majority of aquatic algae contained none FRO and NOX genes per genome, except for chlorophytes C. reinhardtii carried three FROs, V. carteri, Coccomyxa subeuipsoidea c-169, and Chlorella variabius nc64A carried one FRO and grouped into FRO II, whereas the majority of land plants, except for the gymnosperm Picea sitchensis, carried ≥2 FRO and 4 NOX genes per genome and were clustered into NOX and FRO II (fig. 2). Further analysis of the functional domains showed that domain organization of the proteins between FRO and NOX subfamilies varied considerably (fig. 2B). All of the subfamilies contained a transmembrane domain Ferric_reduct, FRO I was considered as ancestor NOX, FRO II and FRO III contained FAD_binding_8 and NAD_binding_6 domains, but NOX also contained a typical NADPH_Ox domain (fig. 2B).
Fig. 2.—
Phylogenetic relationships and domain organization of FRO and NOX genes in plants. (A) The unrooted maximum-likelihood phylogenetic tree of FRO and NOX family members was inferred from the amino acid sequence alignment of the Ferric_reduct, FAD_binding_8 and NAD_binding_6 domains. The four conserved groups are marked by different colors and represented as NOX, FRO I, FRO II and FRO III. Scale bar represents 0.2 amino acid substitution per site. (B) Domain organizations of the four conserved groups of FRO and NOX proteins. The transmembrane regions are shown with dotted pink box.
Structure Analysis of FRO and NOX Family Genes
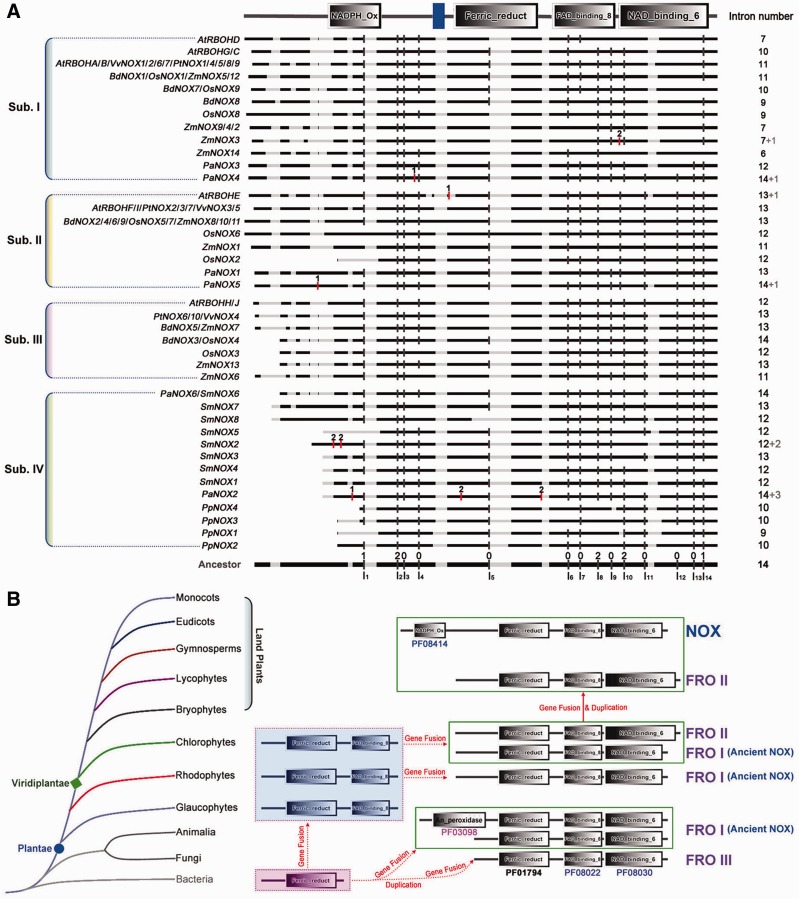
To better understand the evolution of the FRO and NOX gene families, we generated unrooted maximum-likelihood phylogenetic trees of the FRO and NOX family members (fig. 3A; supplementary fig. S4, Supplementary Material online), which were inferred from the amino acid sequences of the Ferric_reduct, FAD_binding_8, NAD_binding_6, and/or NADPH_Ox domains (supplementary figs. S2 and S3, Supplementary Material online). Numbers above the nodes represent bootstrap values from 1,000 replications. Intron position is generally very well-conserved in orthologous genes over long evolutionary time intervals, whereas exon/intron structure is slightly less, but sufficiently conserved in paralogous genes to reveal evolutionary relationships between introns (Rogozin et al. 2003; Li et al. 2009). To investigate the gene structural diversity and possible mechanisms for the structural evolution of FRO and NOX homologs in land plants, the rhodophytes (Galdiria ssulphuraria), chlorophytes (V. carteri, C. reinhardtii, Co. subellipsoidea C-169, Chlorella variabilis NC64A), bryophytes (Ph. patens), lycophytes (Se. moellendorffii), gymnosperms (Picea abies), eudicots (A. thaliana, Po. trichocarpa, and Vi. vinifera), and monocots (B. distachyon, O. sativa, and Z. mays), were considered to analyze the exon/intron organization in the coding sequence (fig. 3; supplementary fig. S4, Supplementary Material online). According to the exon/intron organization, the plant NOXs examined here can be divided into four conserved subfamilies which were represented as Sub. I, Sub. II, Sub. III, and Sub. IV, respectively (fig. 3); whereas, FRO II proteins can be grouped into three subfamilies named as FRO IIa, FRO IIb, and FRO IIc, respectively (supplementary fig. S4, Supplementary Material online). Overall, there was considerable diversity in the number and the length of introns in the FRO and NOX family genes, and NOX family members shared similar gene structure in terms of intron numbers and intron phases within the same subfamily except Sub. IV (fig. 3; supplementary fig. S4, Supplementary Material online). For instance, NOX genes in Sub. I have 6–14 introns, 44.8% (13/29) genes have 11 introns, 48.3% (15/29) genes have 6–10 introns, and two genes have 12 and 14 introns, respectively. Whereas NOX genes in Sub. II, III, and IV have 11–13, 11–14, and 7–17 introns, respectively. We also investigated intron phases with respect to codons in the NOX and FRO genes. The intron phases were remarkably well conserved among subfamily members, whereas the intron arrangement and phases were strikingly distinct between subfamilies (fig. 3B; supplementary fig. S4, Supplementary Material online).
Fig. 3.—
Phylogenetic relationships and exon/intron structure of NOX family genes in land plants. (A) The unrooted maximum-likelihood phylogenetic tree of FRO and NOX family members was inferred from the amino acid sequence alignment of the NADPH_Ox, Ferric_reduct, FAD_binding_8 and NAD_binding_6 domains. Numbers above the nodes represent bootstrap values from 1,000 replications. (B) Exon/intron structure of NOX family genes in land plants. Black boxes represent exons; black lines represent introns; The length of the boxes and lines are scaled relative to the length of the gene, and longer introns are denoted by a double slash; numbers 0, 1, and 2 are intron phases, which indicates the positions of introns between or within codons. Phase 0 introns are located between two consecutive codons, phase 1 introns are located between the first and second nucleotides of a codon, and phase 2 introns are located between the second and third nucleotides of a codon. The introns labeled with red mumber, that is, “1” or “2” were gained introns. The four conserved subfamilies of NOXs are represented as Sub. I, Sub. II, Sub.III and Sub.IV at the left side of the gene structures.
To further explore intron loss or gain within the NOX family genes, we examined the exon/intron organization of paralogous and orthologs in the land plants (fig. 4A). This analysis revealed that single intron loss and gain likely occurred during the structural evolution of NOX family genes in land plants. For example, the paralogous genes AtRBOHC/G and PtNOX8/9 showed conserved exon/intron structure in terms of the number of introns and exon length, whereas a single intron appears to have been lost during the evolution of the SmNOX6/7/8 paralogs (figs. 3B and 4A). By contrast, a single intron gain occurred between the paralogs ZmNOX3 and ZmNOX2 (figs. 3B and 4A). Similar intron loss were seen with BdNOX7/8 (subfamily I), OsNOX5/6 (subfamily II), BdNOX3/5 (subfamily III), and PpNOX3/4 (subfamily IV; fig 4A). Single intron gain was also observed in the NOX genes of land plants. For instance, ZmNOX3 and PaNOX4 in subfamily I and AtRBOHE and PaNOX5 in subfamily II appear to have gained an intron, whereas SmNOX2 and PaNOX2 gain 2 and 3 introns, respectively (fig. 4A). So the intron gain or loss may play a very important role in the structure evolution of NOX family.
Fig. 4.—
The expansion and evolution of the FRO and NOX gene family in plants. (A) Schematic comparison of intron distribution in NOX orthologs of land plants generated with the CIWOG software. Black horizontal lines are aligned sequences; gray horizontal lines are gaps in the alignment; black vertical bars are conserved common introns and represented as I1~I14; red vertical bars are gained introns. The numbers 0, 1, and 2 are intron phases. The intron numbers of each NOX genes were showed on the right side and the gray numbers, that is, “+1,” “+2,” or “+3” are represented the number of gained introns. (B) A model for the expansion and evolution of the NOX gene family in Plantae. The evolutionary relationships among eight lineages (left) and a model for the expansion and evolution of the FRO and NOX gene family (right) within the supergroup Plantae.
Examination of the chromosomal locations of the NOX family genes in the genomes of bryophytes (Ph. patens), lycophytes (Se. moellendorffii), monocots (B. distachyon, O. sativa, and Z. mays), and eudicots (Arabidopsis, Po. trichocarpa, and Vi. vinifera) showed that NOX genes are randomly distributed (supplementary fig. S5, Supplementary Material online). Moreover, a search for NOX paralogs using the Plant Genome Duplication Database (PGDD; http://chibba.agtec.uga.edu/duplication/, last accessed February 29, 2016) (Lee et al. 2013) revealed twelve paralogous gene pairs in A. thaliana, Po. trichocarpa, B. distachyon, and O. sativa (supplementary fig. S6A, Supplementary Material online), but none in the other species.
To further explore association of positive selection with duplication and divergence of NOX family genes, the rate of nonsynonymous substitution (Ka), synonymous substitution (Ks), and the Ka/Ks ratios were calculated for the 12 paralogous gene pairs and used to estimate duplication and divergence times. The Ka/Ks ratios varied from 0.831 to 1.197 among the four different species (supplementary fig. S6B, Supplementary Material online). The fact that the Ka/Ks ratios were less than 1 suggests that the NOX family genes have undergone strong negative selection pressure and the duplication event was estimated to have occurred 6.217–39.499 Ma.
Based on all of these information, we construct a model of evolution of FRO and NOX gene families in plants (fig. 4B). In this model, it seems that all FRO family members originated from a common ancestor which contained only Ferric_reduct domain. Ferric_reduct domain then obtained FAD_binding_8 and NAD_binding_6 domains by first gene fusion and duplication and clustered into FRO I, FRO II, and FRO III subfamilies. FRO III mainly existed in fungi, FRO I perceived as ancient NOX mainly existed in animal which also gained an_peroxidase domain (Pfam accession number PF03098) and in two kinds of algae, rhodophytes and chlorophytes. FRO I obtained another NADPH_Ox domain (PF08414) which is represented as NOX family. For the evolutionary way in plants, FRO IIs existed both in chlorophytes and land plants but NOXs only in land plants (figs. 2 and 4B), implying their functional divergence.
Tissue-Specific Expression Patterns of NOX Genes in Arabidopsis and Rice
To investigate differences in the expression of NOX genes in Arabidopsis and rice during plant development, we analyzed the expression profiles of AtNOXs (i.e., AtRBOHA–J) and OsNOXs (i.e., OsNOX1–9) in ten and nine developmental stages/tissues, respectively, based on the microarray data reported in Genevestigator (fig. 5). Essentially identical developmental expression profiles for the AtNOX and OsNOX genes were also obtained with data from the Arabidopsis and rice eFP browsers in the Bio-Analytic Resource (http://bar.utoronto.ca/welcome.htm, last accessed February 29, 2016) database (Winter et al. 2007). The expression patterns of AtNOXs and OsNOXs from the microarray data analyses are shown as heat maps in white/gray/red (low to high) that reflect the percent of expression (fig. 5).
Fig. 5.—
Developmental expression patterns of NOX family genes in Arabidopsis and rice. Expression profiles of (A) AtNOXs (i.e., AtRBOHA–J) and (B) OsNOXs (i.e., OsNOX1–9) in different developmental stages obtained from microarray data reported in Genevestigator. Results are shown as heat maps in white/gray/red (low to high) that reflect the percent of expression.
Overall, all of the AtNOX and OsNOX genes are expressed during the vegetative and reproductive development stages, and they display strong tissue specificity. In Arabidopsis, AtRBOHD shows higher expression in most tissues and developmental stages than other AtNOXs (fig. 5A). The AtRBOHD expression is almost seen in all the tissues except germinate seed and mature siliques, whereas AtRBOHG and AtRBOHF are the highest expressed in the seedling and young rosette, AtRBOHA is mainly expressed in the young rosette and mature siliques (fig. 5A), the AtRBOHE expression is the highest in the mature siliques, whereas AtRBOHH and AtRBOHJ are mainly expressed in mature siliques and senescence leaves, respectively. In rice, the OsNOX1, OsNOX2, OsNOX3, OsNOX5, OsNOX6, OsNOX7, and OsNOX8 transcripts tend to accumulate to higher levels than the OsNOX4 and OsNOX9 transcripts in most tissues/developmental stages, but their individual expression patterns differ (fig. 5B). Apparently, most OsNOX genes have higher expression at the vegetative development stages, but OsNOX2, OsNOX5, OsNOX7, and OsNOX8 genes are also higher expressed at the reproductive development stages. It seems that OsNOX4 is only highly expressed at the reproductive stages (fig. 5B).
Response Profiles of NOX Genes under Abiotic Stresses and Hormone Treatments in Arabidopsis and Rice
To understand the molecular mechanism of AtNOXs and OsNOXs transcriptional regulation under abiotic stresses and hormone treatments, we first identified potential cis-elements in the promoter regions of each NOX gene using the PlantCARE program. The results obtained show that the promoter regions of AtNOX and OsNOX genes contain many response elements for several abiotic/biotic stresses, such as low temperature, heat, drought (MYB binding sites), anaerobic conditions, and pathogens (TC-rich repeats, W box, GCC box, Box S, Box-W1, and EIRE) (supplementary tables S4 and S5, Supplementary Material online). In addition, the AtNOX and OsNOX gene promoters contain cis-elements for responding to several hormones, such as auxin, gibberellin, ABA, ethylene, salicylic acid (SA), and methyl jasmonic acid (MeJA) (supplementary tables S4 and S5, Supplementary Material online). Further analysis showed that the majority of the AtNOX and OsNOX promoters contain anaerobic-, ABA-responsive elements, heat stress, drought (MYB binding sites), and MeJA response elements (supplementary tables S4 and S5, Supplementary Material online).
To further demonstrate the expression profiles of AtNOX and OsNOX genes induced by abiotic stresses, we again examined Arabidopsis and rice microarray data in the Genevestigator database. As shown in supplementary figures S7 and S8, Supplementary Material online, the expression of AtNOX and OsNOX genes is induced to varying degrees by cold, heat, drought (PEG), salt (NaCl), and oxidative (methyl viologen, MV) stresses. As can be seen, AtRBOHB and AtRBOHD, both belong to Sub. I, exhibit strongly upregulations to hypoxia treatments both in seedlings and roots (supplementary fig. S7A, Supplementary Material online). AtRBOHD is also upregulated by drought, osmotic stress and salt treatments (supplementary fig. S7A, Supplementary Material online). In contrast, the two Sub. III genes (AtRBOHH and AtRBOHJ) have no much changes in expression under most abiotic stresses (supplementary fig. S7, Supplementary Material online). The OsNOX genes exhibit more complicated expression profiles in response to environmental stress treatments and no marked distinct orderliness in expression was observed (supplementary fig. S8, Supplementary Material online). It should be noticed that OsNOX2 and OsNOX9 are greatly upregulated during seed germination of rice under either anaerobic or aerobic conditions, whereas OsNOX1, OsNOX3, OsNOX4, and OsNOX8 are downregulated at early stages during the seed germination process (supplementary fig. S8A, Supplementary Material online).
The expression profiles of AtNOXs and OsNOXs under different abiotic stresses were also examined by qRT-PCR experiments. In brief, cold greatly upregulates the transcripts of AtRBOHA but downregulates those of AtRBOHD, E, I, and H; heat markedly upregulates the transcripts of AtRBOHA, B, C, and F but downregulates those of AtRBOHE and I; dehydrate stress (PEG treatment) significantly upregulates the transcripts of AtRBOHB but downregulates those of AtRBOHE and I; in contrast, salt stress (NaCl treatment) greatly downregulates almost all the AtNOX genes, especially AtRBOHE and I; for oxidative stress (MV treatment), except AtRBOHH, all the other eight genes are significantly upregulated (fig. 6). More complicated expression profiles of NOX genes are presented in rice. As can be seen, the expression profiles of OsNOX genes exhibit obvious temporal and spatial variations (fig. 7). Under cold stress, almost all the OsNOX genes are upregulated in both shoots and roots at 12 h timepoint except OsNOX9 is slightly downregulated at this timepoint. However, the transcripts of most OsNOX genes are fallen back at 24 h timepoint by cold stress but some are still upregulated at this timepoint, for example OsNOX3 is upregulated both in shoots and roots whereas OsNOX2, 4, 5, 7, and 9 are upregulated only in roots (fig. 7A). For heat treatment, almost all of the OsNOX genes are upregulated at 12 h timepoint in shoots and roots except OsNOX1 and 3 are downregulated at this timepoint in roots, however, almost all the genes are markedly downregulated at 24 h timepoint in roots by this stress treatment (fig. 7B). In dehydrate stress experiment (PEG treatment), all OsNOXs are significantly upregulated at both 12 and 24 h timepints in shoots but significant upregulations in roots were only observed from OsNOX1, 3, and 5 genes at these two timepoints (fig. 7C). For NaCl treatment, great upregulation of gene transcripts were found from OsNOX2, 4, 5, 6, and 9 at both 12 and 24 h timepoints in shoots whereas most of the genes keep no big changes in roots at these timepoints (fig. 7D). In contrast, oxidative stress (MV treatment) significantly upregulates the transcripts of all the genes detected in both shoots and roots, especially OsNOX2, 4, 6, and 9 in shoots (fig. 7E).
Fig. 6.—
Inducible expression patterns of NOX family genes in Arabidopsis under abiotic-stresses and hormone treatments. Expression levels of AtRBOHA–I assayed by qRT-PCR under (A) cold (4 °C), (B) heat (30 °C), (C) drought (20% PEG6000), (D) salt (200 mM NaCl), (E) oxidative (30 µM MV) stresses and (F) ABA (100 µM), (G) MeJA (100 µM) hormone treatments. Data are means ±SD (n = 3) and are representative of similar results from three independent experiments.
Fig. 7.—
Inducible expression patterns of NOX family genes in rice under abiotic-stresses and hormone treatments. Expression levels of OsNOX1–9 assayed by qRT-PCR under (A) cold (4 °C), (B) heat (40 °C), (C) drought (20% PEG6000), (D) salt (200 mM NaCl), (E) oxidative (30 µM MV) stresses and (F) ABA (100 µM), (G) MeJA (100 µM) hormone treatments. Data are means ± SD (n = 3) and are representative of similar results from three independent experiments.
As for several hormone response elements were found in the promoter regions of AtNOX and OsNOX genes, we also examined Arabidopsis and rice microarray data in the Genevestigator database (supplementary figs. S7 and S8, Supplementary Material online), as well as by qRT-PCR experiments. The expression of AtNOX and OsNOX genes is induced by hormones such as ABA, SA, MEJA, Zeatin, GA, IAA, KT, 6-BA, JA, and ACC. As can be seen, AtRBOHB and AtRBOHD are markedly downregulated by ABA, Zaetin, and IAA treatments but upregulated by MeJA (supplementary fig. S7B, Supplementary Material online). Three others in this subfamily genes (AtRBOHA, AtRBOHC, and AtRBOHG) show similar expression profiles under these hormone treatments except they are not obviously induced by MeJA (supplementary fig. S7B, Supplementary Material online). Interestingly, the genes in Sub. II (AtRBOHE, AtRBOHF, and AtRBOHI) exhibit almost totally opposite expression profiles as the Sub. I genes have under ABA and IAA treatments (supplementary fig. S7B, Supplementary Material online). The OsNOX genes exhibit more complicated and irregular expression profiles (supplementary fig. S8, Supplementary Material online). As for the results of qRT-PCR, ABA treatment pronounced upregulates the transcripts of most AtNOX genes including AtRBOHA, C, D, E, F, and I, but downregulates those of AtRBOHB and H; similar expression profiles of the AtNOX genes were also found by treatment with MeJA (fig. 6). In rice, ABA treatment markedly upregulates the transcripts of OsNOX3 and 7 at 12 h timepoint and the transcripts of OsNOX2 and 4 at 24 h timepoint in shoots, but only the transcripts of OsNOX1 are obviously upregulated in roots at 12 h timepoint. However, ABA treatment markedly downregulates the transcripts of OsNOX5, 6, 7, and 8 in roots at the two timepoints (fig. 7F). MeJA treatment significantly upregulates the transcripts of OsNOX1, 2, 3, 5, 6, 7, 8, and 9 at both 12 and 24 h timepoints in shoots. In roots, however, MeJA treatment only greatly upregulates the transcripts of OsNOX3, 4, and 7 at both 12 and 24 h timepoints (fig. 7G). It should be noticed that the expression profiles of the members of both AtNOX and OsNOX genes have no distinct subfamily characteristics in response to these environmental stresses and hormone treatments.
Coexpression Analysis of NOX Genes in Arabidopsis and Rice
To further explore the system-level functionality of OsNOXs and AtNOXs, we constructed the framework for coexpression analysis of NOX genes using the Cytoscapev3.2.1 software (supplementary fig. S9, Supplementary Material online). MR values were labeled on the connecting lines between the two coexpression genes. The coexpression of directly linked genes with OsNOXs and AtNOXs were predicted by RiceFREND version 2.0 and ATTED-II version 7.1, respectively (supplementary table. S2, Supplementary Material online). In the frameworks of the coexpression networks, we observed that the patterns of correlation of expression profiles among NOXs and related genes which are involved in plant resistance and many metabolism progresses. For AtNOXs, AtRBOHA/C/D/F are mainly involved in plant-pathogen interaction and they are related to genes which are involved in biosynthesis of secondary metabolites, plant hormone signal transduction, phenylalanine metabolism, alpha-linolonic acid metabolism, amino sugar, and nucleotide sugar metabolism, phenylpropanoid biosynthesis (supplementary fig. S9A, Supplementary Material online). For example, AtRBOHA is coexpressed with a bZIP transcription factor family protein TGA10 (At5g06839), AtRBOHB is related with C2H2 like zinc finger protein (At5g44160), whereas AtRBOHC has relatedness between At3g01190 which belongs to peroxidase superfamily protein, and AtRBOHD shows a correlation with lipoxygenase protein LOX4 (At1g72520). Additionally, AtRBOHE is coexpressed with pollen Ole e 1 allergen and extensin family protein (At5g47635), AtRBOHG/H/I shows a correlation with Leucine-rich repeat protein kinase family protein, and AtRBOHH/J are involved in FRE-like transmembrane component family protein (supplementary fig. S9A and table S2, Supplementary Material online).
More complicated coexpression networks were found in rice NOXs. In brief, the OsNOXs are mainly coexpressed with the genes functioning in plant-pathogen interaction, metabolic pathways, N-glycan biosythesis, endocytosis, and fatty acid and unsaturated fatty acid biosynthesis (supplementary fig. S9B and table S2, Supplementary Material online). For example, OsNOX1 shows relatedness with RabGAP/TBC domain containing protein and SBP-domain protein 4 (SBP, SPL2), OsNOX2 is coexpressed with serine/threonine-protein kinase PBS1, and OsNOX3 has correlation with proteins which are similar to quinone-oxidoreductase QR1, HvPIP2;1 protein (PIP2;2) and a Nodulin-like domain containing protein. OsNOX4, OsNOX5, OsNOX6, OsNOX7, OsNOX8, and OsNOX9 were found having relatedness with genes which encode an flavin-containing monooxygenase FMO family protein, cellulose synthase-8 (CESA6), metacaspase1, an protein kinase domain containing protein, TRANSPARENT TESTA GLABRA 1 protein (TTG1 protein), and an AP2 domain containing protein RAP2.2 (AP2-EREBP) and cinnamate 4-hydroxylase CYP73, respectively (supplementary fig. S9B and table S2, Supplementary Material online).
For verifying the results of coexpression analysis, we choose some interesting genes to examine their expression profiles under MeJA treatment by qRT-PCR method. As can be seen, the selected genes exhibit very well coexpression patterns with their linked OsNOX genes (fig. 8). Among the examined eight genes, the transcripts of seven are up-regulated by MeJA treatment and also for their predicted coexpression genes. Although the transcripts of OsNOX4 shows no match with those of its two predicted coexpression genes in this experiment (fig. 8), the coexpression could also be observed since its transcription is greatly upregulated in roots under MeJA treatment (fig. 7H).
Fig. 8.—
Expression profiles analysis of coexpression genes with OsNOXs by qRT-PCR under MeJA hormone treatment on the shoot of rice seedling. Data are means ± SD (n = 3) and are representative of similar results from three independent experiments. Abbreviations and locus names of genes used are available in supplementary table S2, Supplementary Material online.
Discussion
Expansion, Diversity, and Evolution of the FROs and NOXs in Plants
Gene duplication is a common phenomenon in eukaryotes and contributes to biological diversity during evolution (Van de Peer et al. 2009; Magadum et al. 2013). The FROs and NOXs are represented by at least one gene in land plants. Although some previous studies suggested that FRO proteins are the ancestral NOXs (Bedard et al. 2007; Wong et al. 2007), but the evolution history did not well examined, especially that in Plantae. In this study, we found that the three ancestors (cyanophytes, phodophytes, and chlorophytes) of Plantae, contain very few FRO homologs and many species have zero or only one FROs per genome and no NOX homologs were found in these species (fig. 1). Based on their domain composition and gene structure characters of the two family proteins (figs. 1 and 2; supplementary fig. S1, Supplementary Material online), FROs in Plantae should be truly considered to be ancient forms of NOXs. A single gene duplication might lead to the expansion of the FRO and NOX gene families in plants, which occurred during the divergence of ancestral cyanophytes from Plantae. The scattered distribution of NOX family genes on chromosomes (supplementary fig. S5, Supplementary Material online) and the 12 paralogous NOX gene pairs found in seven land plants (supplementary fig. S6, Supplementary Material online) suggest that segmental duplications may have been involved in the expansion of the NOX gene family and caused differences in the number of NOX genes within subfamilies and species of land plants.
Gene fusion and exon shuffling after gene duplication are mechanisms that can enhance the functional divergence of duplicated genes by creating additional domains or rearranging the original functional domains (Morgante et al. 2005; Kaessmann 2010). In this study, phylogenetic analysis, together with the domain organizations and gene structures of FRO and NOX family genes, showed distinct evolutionary differences among subfamilies (figs. 2 and 4; supplementary figs. S2 and S3, Supplementary Material online). As suggested in our proposed model for FRO and NOX gene family evolution in plants (fig. 4B), there are two distinct evolutionary ways for the families. One leads to the emergence of fungi and animal FROs and another leads to evolution of plant FROs and NOXs. FRO III subfamily proteins, also named as FREs, are more ancient forms of FROs, which were only identified in fungi. FRO II subfamily proteins are dominantly identified in algae and most land plants. Based on domain characteristics, FRO I subfamily proteins might be considered to be ancient NOXs, which were mainly identified in C. reinhardtii and H. sapiens (fig. 2). Apparently, the ancestral FRO I protein gained a NADPH_Ox domain is an important event for the evolution of both plant and animal NOX proteins. Therefore, it is reasonable to conclude that the gene fusion and duplication might contribute to the expansion and evolution of the two family genes in plants. All FRO family members originated from a common ancestor, which contained only Ferric_reduct domain and existed in all living organisms from prokaryotic bacteria to eukaryotic angiosperms. Then at least two gene fusion events occurred to yield the FROs of rhodophytes and green algae by obtained the FAD_binding_8 and a NAD_binding_6 domains. And then at least one gene duplication occurred to yield three FRO genes (one FRO II and two FRO I) in green algae and one FRO II gene duplication bring 2–9 FRO II proteins in land plants (figs. 1, 2, and 4B). Finally, another gene fusion event occurred in red algae (contains one FRO I protein) or green algae (contains two FRO I proteins) to yield plant NOXs and then one or two gene duplication bring four NOX genes in bryophytes (figs. 1 and 4B).
Gene structural diversity within gene families is another evolutionary mechanism that promotes variability, and intron loss or gain is important in generating structural diversity and complexity (Zhang and Kishino 2004; Li et al. 2009). Analyzing the exon/intron structures of FRO and NOX genes (fig. 3; supplementary fig. S4, Supplementary Material online), we found that the numbers of introns and intron phases among subfamily members are remarkably conserved, whereas the intron arrangement and intron phases are strikingly distinct between subfamilies. Further analysis of the exon/intron organization of orthologous and paralogous genes in land plants within the NOX family (fig. 4A) suggests that single intron loss and gain likely occurred and led to the diversification of gene structure. This consequently contributes to the functional diversity and divergence of the FRO and NOX family proteins during the evolution process in plants. This evolutionary mechanism was also found in our previous study (Li et al. 2014).
The NADPH_Ox domain only occurred in land plants, implying this domain might specifically contribute to adaptation of the plants to terrestrial habitats. The increasing literatures have showed that NOXs are mainly responsible for the stress tolerance and development by producing ROS (Kaya et al. 2014; Wang et al. 2015) whereas FROs are mainly responsible for iron uptake and iron homeostasis in higher plants (Gross et al. 2003; Sperotto et al. 2010). However, the most homologs of FROs in fungi show dual functions of both ROS-generating and metal ion-reducing (Aguirre et al. 2005). Previously, several NOX homologs were identified from green algae and red algae but considering their lack of NADPH_Ox domain in amino acid sequences (Hervé et al. 2006; Anderson et al. 2011), they therefore, should be assigned to FROs as suggested in our current study (figs. 1, 2, and 4). These algae FROs seem only functioning in ROS production but not iron homeostasis (Hervé et al. 2006; Anderson et al. 2011). These results clearly show the functional divergence of FROs and NOXs in Plantae during their long-term evolution history.
Plant NOXs Respond to Abiotic Stresses and Hormone Treatments
Many studies have shown that ROS production and NOX activity were stimulated in plants under various environmental stress conditions, such as cold, heat, drought, salt, oxidative stress, and hormone treatments. In Arabidopsis, AtRBOHD and AtRBOHF were found participating in the ROS-involved response of the plants to pathogen attacks (Torres et al. 2002) and the ABA-mediated regulation of stomatal closure (Kwak et al. 2003). In rice, OsRbohB (OsNOX1) was reported to have a ROS-producing activity (TakahaShi et al. 2012) and using suspension cells, Yoshie et al. (2005) found that the two others, OsRbohA (OsNOX2) and OsRbohE (OsNOX3), participated in ROS-dependent immune responses. Recently, we examined the expression profiles of nine NOX and two FRO genes in rice under various environmental conditions and found unique stress response characteristics, indicating their related but distinct functions in response to different environmental stresses including drought, heat and salt (Wang et al. 2013). Among these NOX genes, OsRbohA (OsNOX2) was proved to be essential to rice normal developmental regulation and drought stress tolerance (Wang et al. 2015).
Although many studies have found that NOX family genes functionally in variety abiotic stresses, there has been no systematic studies of the functions of NOX genes response to various abiotic stresses. According to identify potential cis- elements in the promoter regions of Arabdopsis and rice NOXs, we found that the promoter regions of NOXs contain multiple response elements for various abiotic stresses (supplementary tables S4 and S5, Supplementary Material online). Further exploration of the expression profiles of NOXs in Arabidopsis using silico data and qRT-PCR results demonstrated that AtNOXs belong to different subfamilies might possess different expression profiles under abiotic stresses, although the results obtained from the two experiment ways were somewhat inconsistent (fig. 6; supplementary fig. S7, Supplementary Material online). For example, the two Sub I genes, AtRBOHB and AtRBOHD, are strongly upregulated by hypoxia, MV, and AtRBOHD is also upregulated by drought and osmotic stresses (supplementary fig. S7A, Supplementary Material online). Considering its strongly expression in various tissues and organs (fig. 5A; Suzuki et al. 2011), these results suggest a putative housekeeping role of this gene in various plant developmental progresses. However, the genes in Sub. II (AtRBOHE, AtRBOHF, and AtRBOHI) exhibit almost totally opposite expression profiles as the Sub. I genes have according to the silico analysis (supplementary fig. S7B, Supplementary Material online). The qRT-PCR data also show somewhat differences in expression profiles between the two subfamily genes, especially under PEG (fig. 6). It was reported that AtRBOHF plays a crucial role in the interplay between intracellular oxidative stress and pathogenesis responses (Chaouch et al. 2012). In contrast, the Sub. III genes (AtRBOHH and AtRBOHJ) have no much changes in expression under most abiotic stresses and hormone treatments (fig. 6; supplementary fig. S7, Supplementary Material online) but they were found to be specifically expressed in stamens and pollen (Sagi and Fluhr 2006) and play crucial roles in pollen tube tip growth via Ca2+_activated ROS production (Kaya et al. 2014, 2015). The rice OsNOX genes also show great differential expression under various stresses and hormone treatments but their expression profiles exhibit no markedly distinct orderliness between the subfamilies (fig. 7; supplementary fig. S8, Supplementary Material online), implying their more complicated regulatory mechanisms and functions during the response to environmental stresses. Apparently, each NOX gene in rice has its unique response characteristics to different stresses but complicated cross-talks existing among the members of the gene family. OsNOX1 (OsRbohB) was found to modulate cytosolic Ca2+ concentration by directly interaction with OsRac1, whereas OsNOX2 (OsRbohA) and OsNOX3 (OsRbohE) were proved to participate in the plant immune response and/or drought tolerance (Yoshie et al. 2005; Wong et al. 2007; Wang et al. 2015). All the functions of these NOXs were found to be related their activity in ROS production (Yoshie et al. 2005; Wong et al. 2007; TakahaShi et al. 2012; Wang et al. 2015).
Hormone response elements were also found in the promoter regions of Arabdopsis and rice NOXs (supplementary tables S4 and S5, Supplementary Material online). AtRBOHB and AtRBOHD, are strongly upregulated by MeJA treatments but markedly downregulated by ABA, Zaetin, and IAA treatments(supplementary fig. S7A, Supplementary Material online), AtRBOHE, AtRBOHF, and AtRBOHI exhibit different expression profiles under ABA and IAA treatments (fig. 6 and supplementary fig. S7B, Supplementary Material online). In rice, almost all the rice NOX genes exhibit great upregulation under oxidative stress (MV) and the hormone (ABA, SA, and MeJA) treatments (fig. 7), indicating that their functions in stress responses might be involved in hormone signaling since ROS are important components of the cross-talk among ABA, JA, and SA signaling pathways during plant stress responses (Atkinson and Urwin 2012). It has been reported that the cross-talk between NOXs (mainly OsNOX1and OsNOX3), H2O2 and MAPK in ABA signaling plays crucial roles in the functioning of a novel C2H2-type zinc finger protein, ZFP36, in rice stress tolerance (Zhang et al. 2014). However, as reported recently, OsNOX2 might function with an ABA-independent defense pathway and mediate very complicated signaling pathways might be involved in rice normal growth processes and drought stress response (Wang et al. 2015). All these results suggest the crucial roles of NOXs in the plant defense system.
Coexpression Analysis of NOXs in Arabidopsis and Rice Reflects Their Multiple Functions Both in Normal Growth Regulation and Stress Response
As the key producers of ROS, NOXs play a crucial role in the development of plants, particularly in tip growing systems (Kaur et al. 2014). It was reported that loss function of AtRBOHF or AtRBOHD/F reduced the plant size and root length whereas knockout of AtRBOHC suppressed the root hair elongation (Torres et al. 2002; Foreman et al. 2003; Kwak et al. 2003). In addition, AtRBOHD was found participating in endosperm development (Penfield et al. 2006) and be closely related with vesicle cycle (Hao et al. 2014), AtRBOHB involving in seed after-ripening (Müller et al. 2009) whereas AtRBOHH and AtRBOHJ functioning in pollen tube tip growth (Kaya et al. 2014). In N. tabacum, Potocký et al. (2007) found ROS-generated from NOX in polarized growth of pollen tubes and lipid microdomain polarization was found playing a role in pollen tube tip growth in Picea meyeri (Liu et al. 2009). Kaur et al. (2014) conclude that the NOX may associate with polarized lipid microdomain in targeting apical plasma membrane region. MtRBOHA from M. truncatula and PvRBOHB from P. vulgaris were reported to be involved in plant-rhizobial symbiosis (Marino et al. 2011; Montiel et al. 2012). ROS production is also involved in various stages of seed, seedling development, and blue light responses of seedlings in plants (Kaur et al. 2014). Our coexpression analysis shows that both AtNOXs and OsNOXs are significantly correlated with many genes which are involved in metabolism and development progresses in plants (supplementary table S2 and fig. S9, Supplementary Material online). Such like, AtRBOHC was observed relatedness to peroxidase superfamily protein (supplementary table S2, Supplementary Material online), and which are involved in biosynthesis of secondary metabolism, phenylpropanoid biosynthesis, and plant-pathogen interaction (supplementary table S2, Supplementary Material online). In rice, OsNOX1 shows a significantly correlated expression profiles with RabGAP/TBC domain containing protein and a protein similar to SBP-domain protein 4, implying it might participate in endocytic trafficking process and floral development since the RabGAP/TBC Armus integrates autophagy with signaling and endocytic trafficking by coordinates Rac1 and Rab7 functions (Carroll et al. 2013) and many SBP-box genes highly expressed in inflorescence apex, floral meristem, leaf, and floral primordial (Birkenbihl et al. 2005). In addition, one of the largest SBP-box genes, SPL14, was characterized as conferring resistance to the PCD-inducing fungal toxin fumonisin B1 (FB1) (Stone et al. 2005). OsNOX2 was found to be coexpressed with a serine/threonine-protein kinase PBS1 (supplementary table S2, Supplementary Material online). PBS1 and Pto were found to fulfill different functions in the recognition of pathogen avirulence proteins in Arabidopsis (Swiderski and Innes 2001). OsNOX8 has a correlation with TTG1 protein (supplementary table S2, Supplementary Material online) which was found to regulate several developmental and biochemical pathways such as trichome differentiation and anthocyanin biosynthesis in Arabidopsis (Zhang et al. 2003).
As mentioned above, plant NOXs are mainly ROS generated enzymes, and ROS in turn mediate various abiotic responses and biotic interactions (Kaur et al. 2014). In early reports, various abiotic stresses and the role of phytohormones such as ABA, JA, brassinosteroid (BR), auxins, and SA in plants have achieved important progress. AtRBOHD and AtRBOHF are involved in pathogenic response (Torres et al. 2002) and AtRBOHD is upregulated by auxin (Peer et al. 2013) and responsible for negative regulation of cell death by triggering SA accumulation under Botrytis cinerea treatment when it was overexpressed (Torres and Dang 2005). A recent study found that AtSRC2 can regulate the activation of AtRBOHF under cold treatment (Kawarazaki et al. 2013). Furthermore, ROS production was decreased in maize apoplast under salinity stress, suggesting that NaCl play a negative effect on NOX activity (Rodriguez et al. 2007). Comparing the role of NOXs (RbohD and RbohF) and peroxidases (Prx103, Prx107, Prx112, and Prx118) in Triticum aestivum which were infected by Puccinia triticina, Dmochowska-Boguta et al. (2012) concluded that NOXs are more complex and might be playing diverse roles in different kinds of interactions in dicots and monocots. In our result, AtRBOHA is coexpressed with a bZIP transcription factor family protein TGA10 (supplementary table S2, Supplementary Material online), TGA transcription factors have been identified overlapping roles in plant disease resistance and stress responses (Despres et al. 2003; Mueller et al. 2008) . AtRBOHB shows a relatedness with C2H2-type zinc finger protein which has found playing a role in rice stress tolerance (Zhang et al. 2014). AtRBOHD shows a correlation with lipoxygenase protein LOX4, which play a role in plant-pathogen reaction, while researches have shown that OsRBOHE (OsNOX3) is involved in the regulation of defense-related genes EI2 and LOX (Yoshie et al. 2005). It was also reported that LOX3 and LOX4 interfere with distinct metabolic and that LOX4 plays a major role in controlling plant defence against nematode infection (Ozalvo et al. 2014). OsNOX3 also shows a correlation with a protein similar to Quinone-oxidoreductase QR1 (supplementary table S2, Supplementary Material online). QR1 is one of the earliest genes on the haustorium signal transduction pathway and is necessary for the redox bioactivation of haustorial inducing factors (Bandaranayake et al. 2010). OsNOX6 exhibits to be highly coexpressed with a protein similar to Metacaspase 1 (supplementary table S2, Supplementary Material online), implying it might be involved in plant hypersensitive response and cell death. Metacaspase proteins belong to caspase-like proteins which may contain zinc finger motifs resembling those in the plant hypersensitive response and cell death protein Isd-1 (Uren et al. 2000). OsNOX9 has a correlation with proteins similar to AP2 domain containing protein RAP2.2 (AP2-EREBP) and two other AP2-EREBP proteins (supplementary table S2, Supplementary Material online), indicating its potential functions in redox balance and stress tolerance of the plant since AP2/EREBP transcription factors actively participate in hormone and redox signalings and gene regulatory networks under abiotic stresses (Shaikhali et al. 2008; Karim et al. 2009). OsNOX9 is also highly coexpressed with cinnamate 4-hydroxylase CYP73. In tobacoo, a class II cinnamate 4-hydroxylase, CYP73A15, was found participating in the production of lignin (Blee et al. 2000.
All these results suggest that NOXs in plants play a complicated role in many metabolic and resistance progresses. Different NOXs can be involved in similar stimuli but also show a complex and cross-talk signaling networks in response to a number of environmental stresses. However, the function of each NOX and the molecular mechanism to resistance are still unclear, and the interactions among NOXs are still barely reported. Further studies will be committed to explore the functions of single or multiple NOX genes in plants.
Supplementary Material
Supplementary figures S1–S9 and tables S1–S5 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
This work was supported by the Natural Science Foundation of China (No. 31270299 and 31401300), the Natural Science Foundation of Guangdong Province (No. 2015A030313572), and the Program for New Century Excellent Talents in University of China (NCET-11-0440) and the Foundation of President of Guangdong Academy of Agricultural Sciences (No. 201614).
Literature Cited
- Aguirre J, Rios-Momberg M, Hewitt D, Hansberg W. 2005. Reactive oxygen species and development in microbial eukaryotes. Trends Microbiol. 13:111–118. [DOI] [PubMed] [Google Scholar]
- Amicucci E, Gaschler K, Ward JM. 1999. NADPH oxidase genes from tomato (Lycopersicon esculentum) and curly-leaf pondweed (Potamogeton crispus). Plant Biol. 1:524–528. [Google Scholar]
- Anderson A, Bothwell JH, Laohavisit A, Smith AG, Davies JM. 2011. NOX or not? Evidence for algal NADPH oxidases. Trends Plant Sci. 16: 579–581. [DOI] [PubMed] [Google Scholar]
- Arthikala MK, et al. 2014. RbohB, a Phaseolus vulgaris NADPH oxidase gene, enhances symbiosome number, bacteroid size, and nitrogen fixation in nodules and impairs mycorrhizal colonization. New Phytol. 202:886–900. [DOI] [PubMed] [Google Scholar]
- Asano T, et al. 2012. A rice calcium-dependent protein kinase OsCPK12 oppositely modulates salt-stress tolerance and blast disease resistance. Plant J. 69:26–36. [DOI] [PubMed] [Google Scholar]
- Atkinson NJ, Urwin PE. 2012. The interaction of plant biotic and abiotic stresses: from genes to the field. J Exp Bot. 63:3523–3543. [DOI] [PubMed] [Google Scholar]
- Bandaranayake PCG, et al. 2010. A single-electron reducing quinine oxidoredutase is necessary to induce haustorium development in the root parasitic plant triphysaria. Plant Cell 22:1404–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedard K, Krause KH. 2007. The NOX family of ROS-generating NADPH oxidases: physiology and pathophysiology. Physiol Rev. 87:245–313. [DOI] [PubMed] [Google Scholar]
- Bedard K, Lardy B, Krause KH. 2007. NOX family NADPH oxidases: not just in mammals. Biochimie. 89:1107–1112. [DOI] [PubMed] [Google Scholar]
- Birkenbihl RP, Jach G, Saedler H, Huijser P. 2005. Functional dissection of the plant-specific SBP-domain, overlap of the DNA-binding and nuclear localization domains. J Mol Biol. 352:585–596. [DOI] [PubMed] [Google Scholar]
- Blee K, et al. 2000. Antisense and sense expression of cDNA coding for CYP73A15, a class II cinnamate 4-hydroxylase, leads to a delayed and reduced production of lignin in tobacco. Phytochemistry 57: 1159–1166. [DOI] [PubMed] [Google Scholar]
- Cano-Domínguez N, Alvarez-Delfín K, Hansberg W, Aguirre J. 2008. NADPH oxidases NOX-1 and NOX-2 require the regulatory subunit NOR-1 to control cell differentiation and growth in Neurospora crassa. Eukaryot Cell. 7:1352–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll B, et al. 2013. The TBC/RabGAP armus coodinates Rac1 and Rab7 functions during autophagy. Dev Cell. 25:15–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaouch S, Queval G, Noctor G. 2012. AtRbohF is a crucial modulator of defence-associated metabolism and a key actor in the interplay between intracellular oxidative stress and pathogenesis responses in Arabidopsis. Plant J. 69:613–627. [DOI] [PubMed] [Google Scholar]
- D’Autréaux B, Toledano MB. 2007. ROS as signalling molecules: mechanisms that generate specificity in ROS homeostasis. Nat Rev Mol Cell Biol. 8:813–824. [DOI] [PubMed] [Google Scholar]
- Delledonne M, et al. 2002. Reactive oxygen intermediates modulate nitric oxide signaling in the plant hypersensitive disease-resistance response. Plant Physiol Biochem. 40:605–610. [Google Scholar]
- Despres C, et al. 2003. The Arabidopsis NPR1 disease resistance protein is a novel cofactor that confers redox regulation of DNA binding activity to the basic domain/leucine zipper transcription factor TGA1. Plant Cell 15:2181–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dmochowska-Boguta M, Nadolska-Orczyk A, Orczyk W. 2012. Roles of peroxidases and NADPH oxidases in the oxidative response of wheat (Triticum aestivum) to brown rust (Puccinia triticina) infection. Plant Pathol. 62:993–1002. [Google Scholar]
- Dubiella U, et al. 2013. Calcium-dependent protein kinase/NADPH oxidase activation circuit is required for rapid defense signal propagation. Proc Natl Acad Sci U S A. 110:8744–8749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans NH, McAinsh MR, Hetherington AM, Knight MR. 2005. ROS perception in Arabidopsis thaliana: the ozone-induced calcium response. Plant J. 41:615–626. [DOI] [PubMed] [Google Scholar]
- Foreman J, et al. 2003. Reactive oxygen species produced by NADPH oxidase regulate plant cell growth. Nature 422:442–446. [DOI] [PubMed] [Google Scholar]
- Goodstein DM, et al. 2012. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40:D1178–D1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groom QJ, et al. 1996. RbohA, a rice homologue of the mammalian gp91phox respiratory burst oxidase gene. Plant J. 10:515–522. [DOI] [PubMed] [Google Scholar]
- Gross JS, Stein RJ, Fett-Neto AG, Fett JP. 2003. Iron homeostasis related genes in rice. Genet Mol Biol. 26:477–497. [Google Scholar]
- Hao FS, Wang XC, Chen J. 2006. Involvement of plasma-membrane NADPH oxidase in nickel-induced oxidative stress in roots of wheat seedlings. Plant Sci. 170:151–158. [Google Scholar]
- Hao HQ, et al. 2014. Clathrin and membrane microdomains cooperatively regulate RbohD dynamics and activity in Arabidopsis. Plant Cell 26:1729–1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hervé C, Tonon T, Collén J, Corre E, Boyen C. 2006. NADPH oxidases in Eukaryotes: red algae provide new hints!. Curr Genet. 49:190–204. [DOI] [PubMed] [Google Scholar]
- Hruz T, et al. 2008. Genevestigator v3: a reference expression database for the meta-analysis of transcriptomes. Adv Bioinformatics 2008:420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B, et al. 2015. GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31:1296–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaessmann H. 2010. Origins, evolution, and phenotypic impact of new genes. Genome Res. 20:1313–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karim MR, Hirota A, Kwiatkowska D, Tasaka M, Aida M. 2009. A role for Arabidopsis PUCHI in floral meristem identity and bract suppression. Plant Cell 21:1360–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur G, Sharma A, Guruprasad K, Pati PK. 2014. Versatile roles of plant NADPH oxidases and emerging concepts. Biotechnol Adv. 32: 551–563. [DOI] [PubMed] [Google Scholar]
- Kawarazaki T, et al. 2013. A low temperature-inducible protein AtSRC2 enhances the ROS-producing activity of NADPH oxidase AtRbohF. Biochim Biophys Acta. 1833:2775–2780. [DOI] [PubMed] [Google Scholar]
- Kaya H, et al. 2014. Ca2+-activated reactive oxygen species production by Arabidopsis RbohH and RbohJ is essential for proper pollen tube tip growth. Plant Cell 26:1069–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaya H, et al. 2015. Apoplastic ROS production upon pollination by RbohH and RbohJ in Arabidopsis. Plant Signal Behav. 10:e989050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaye Y, et al. 2011. Inositol polyphosphate 5-phosphatase7 regulates the production of reactive oxygen species and salt tolerance in Arabidopsis. Plant Physiol. 157:229–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura S, et al. 2012. Protein phosphorylation is a prerequisite for the Ca2+-dependent activation of Arabidopsis NADPH oxidases and may function as a trigger for the positive feedback regulation of Ca2+ and reactive oxygen species. Biochim Biophys Acta. 1823:398–405. [DOI] [PubMed] [Google Scholar]
- Kwak JM, et al. 2003. NADPH oxidase AtrbohD and AtrbohF genes function in ROS-dependent ABA signaling in Arabidopsis. EMBO J. 22:2623–2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TH, Tang H, Wang X, Paterson AH. 2013. PGDD: a database of gene and genome duplication in plants. Nucleic Acids Res. 41: D1152–D1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescot M, et al. 2002. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30:325–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JS, Wang XM, Zhang YL, Jia HL, Bi Y. 2011. cGMP regulates hydrogen peroxide accumulation in calcium-dependent salt resistance pathway in Arabidopsis thaliana roots. Planta 234:709–722. [DOI] [PubMed] [Google Scholar]
- Li W, et al. 2009. Phylogenetic analysis, structural evolution and functional divergence of the 12-oxo-phytodienoate acid reductase gene family in plants. BMC Evol Biol. 9:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li WY, Wang X, Li R, Li WQ, Chen KM. 2014. Genome-wide analysis of the NADK gene family in plants. PLoS One 9:e101051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin F, et al. 2009. Positive feedback regulation of maize NADPH oxidase by mitogen-activated protein kinase cascade in abscisic acid signaling. J Exp Bot. 60:3221–3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, et al. 2009. Lipid microdomain polarization is required for NADPH oxidase-dependent ROS signaling in Picea meyeri pollen tube tip growth. Plant J. 60:303–313. [DOI] [PubMed] [Google Scholar]
- Magadum S, Banerjee U, Murugan P, Gangapur D, Ravikesavan R. 2013. Gene duplication as a major force in evolution. J Genet. 92:155–161. [DOI] [PubMed] [Google Scholar]
- Marino D, Dunand C, Puppo A, Pauly N. 2012. A burst of plant NADPH oxidases. Trends Plant Sci. 17:9–15. [DOI] [PubMed] [Google Scholar]
- Marino D, et al. 2011. A Medicago truncatula NADPH oxidase is involved in symbiotic nodule functioning. New Phytol. 189:580–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruta T, et al. 2011. Arabidopsis NADPH oxidases, AtrbohD and AtrbohF, are essential for jasmonic acid-induced expression of genes regulated by MYC2 transcription factor. Plant Sci. 180:655–660. [DOI] [PubMed] [Google Scholar]
- Montiel J, et al. 2012. A Phaseolus vulgaris NADPH oxidase gene is required for root infection by rhizobia. Plant Cell Physiol. 53:1751–1767. [DOI] [PubMed] [Google Scholar]
- Morgante M, et al. 2005. Gene duplication and exon shuffling by helitron-like transposons generate intraspecies diversity in maize. Nat Genet. 37:997–1002. [DOI] [PubMed] [Google Scholar]
- Mueller S, et al. 2008. General detoxification and stress responses are mediated by oxidized lipids through TGA transcription factors in Arabidopsis. Plant Cell 20:768–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller K, Carstens AC, Linkies A, Torres MA, Leubner-Metzger G. 2009. The NADPH-oxidase AtrbohB plays a role in Arabidopsis seed after-ripening. New Phytol. 184:885–897. [DOI] [PubMed] [Google Scholar]
- Nakashima A, et al. 2008. RACK1 functions in Rice innate immunity by interacting with the Rac1 immune complex. Plant Cell 20:2265–2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nestler J, et al. 2014. Roothairless5, which functions in maize (Zea mays L.) root hair initiation and elongation encodes a monocot-specific NADPH oxidase. Plant J. 79:729–740. [DOI] [PubMed] [Google Scholar]
- Nystedt B, et al. 2013. The Norway spruce genome sequence and conifer genome evolution. Nature 497:579–584. [DOI] [PubMed] [Google Scholar]
- Obayashi T, et al. 2007. ATTED-II: a database of co-expressed genes and cis elements for identifying co-regulated gene groups in Arabidopsis. Nucleic Acids Res. 35:D863–D869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozalvo R, et al. 2014. Two closely members of Arabidopsis 13-lipoxygenases(13-LOXs), LOX3 and LOX4, reveal distinct functions in response to plant-parasitic nematode infection. Mol Plant Pathol. 15:319–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peer WA, Cheng Y, Murphy AS. 2013. Evidence of oxidative attenuation of auxin signalling. J Exp Bot. 64:2629–2639. [DOI] [PubMed] [Google Scholar]
- Penfield S, Li Y, Gilday A, Graham S, Graham I. 2006. Arabidopsis ABA INSENSITIVE4 regulates lipid mobilization in the embryo and reveals repression of seed germination in the endosperm. Plant Cell 18: 1887–1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potocký M, Jones MA, Bezvoda R, Smirnoff N, Žárský V. 2007. Reactive oxygen species produced by NADPH oxidase are involved in pollen tube growth. New Phytol. 174:742–751. [DOI] [PubMed] [Google Scholar]
- Rodriguez AA, Ramiro Lascano H, Bustos D, Taleisnik E. 2007. Salinity-induced decrease in NADPH oxidase activity in the maize leaf blade elongation zone. J Plant Physiol. 164:223–230. [DOI] [PubMed] [Google Scholar]
- Rogozin IB, Wolf YI, Sorokin AV, Mirkin BG, Koonin EV. 2003. Remarkable interkingdom conservation of intron positions and massive, lineage-specific intron loss and gain in eukaryotic evolution. Curr Biol. 13:1512–1517. [DOI] [PubMed] [Google Scholar]
- Sagi M, Fluhr R. 2006. Production of reactive oxygen species by plant NADPH oxidases. Plant Physiol. 141:336–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y. 2012. RiceFREND: a platform for retrieving coexpressed gene networks in rice. Nucleic Acids Res. 41:D1214–D1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott B, Eaton CJ. 2008. Role of reactive oxygen species in fungal cellular differentiations. Curr Opin Microbiol. 11:488–493. [DOI] [PubMed] [Google Scholar]
- Segmüller N, et al. 2008. NADPH oxidases are involved in differentiation and pathogenicity in Botrytis cinerea. Mol Plant Microbe Interact. 21:808–819. [DOI] [PubMed] [Google Scholar]
- Shaikhali J, et al. 2008. The redox-sensitive transcription factor Rap2.4a controls nuclear expression of 2-Cys peroxire doxin A and other chloroplast antioxidant enzymes. BMC Plant Biol. 8(1):14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, et al. 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13:2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi YC, Fu YP, Liu WQ. 2012. NADPH oxidase in plasma membrane is involved in stomatal closure induced by dehydroascorbate. Plant Physiol Biochem. 51:26–30. [DOI] [PubMed] [Google Scholar]
- Song CJ, Steinebrunner I, Wang X, Stout SC, Roux SJ. 2006. Extracellular ATP induces the accumulation of superoxide via NADPH oxidases in Arabidopsis. Plant Physiol. 140:1222–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperotto RA, et al. 2010. Identification of putative target genes to manipulate Fe and Zn concentrations in rice grains. J Plant Physiol. 167:1500–1506. [DOI] [PubMed] [Google Scholar]
- Stone JM, Liang X, Nekl ER, Stiers JJ. 2005. Arabidopsis AtSPL14, a plant-specific SBP-domain transcription factor, participates in plant development and sensitivity to fumonisin B1. Plant J. 41:744–754. [DOI] [PubMed] [Google Scholar]
- Suzuki N, et al. 2011. Respiratory burst oxidases: the engines of ROS signaling. Curr Opin Plant Biol. 14:691–699. [DOI] [PubMed] [Google Scholar]
- Swiderski MR, Innes RW. 2001. The Arabidopsis PBS1 resistance gene encodes a member of a novel protein kinase subfamily. Plant J. 26:101–112. [DOI] [PubMed] [Google Scholar]
- Takahashi S, et al. 2012. Reactive oxygen species production and activation mechanism of the rice NADPH oxidase OsRbohB. J Biochem. 152:37–43. [DOI] [PubMed] [Google Scholar]
- Takeda S, Gapper C, Kaya H, Bell E, Kuchitsu K. 2008. Local positive feedback regulation determines cell shape in root hair cells. Science 319:1241–1244. [DOI] [PubMed] [Google Scholar]
- Tewari RK, Watanabe D, Watanabe M. 2012. Chloroplastic NADPH oxidase-like activity-mediated perpetual hydrogen peroxide generation in the chloroplast induces apoptotic-like death of Brassica napus leaf protoplasts. Planta 235:99–110. [DOI] [PubMed] [Google Scholar]
- Torres MA, Dangl JL. 2005. Functions of the respiratory burst oxidase in biotic interactions, abiotic stress and development. Curr Opin Plant Biol. 8:397–403. [DOI] [PubMed] [Google Scholar]
- Torres MA, Dangl JL, Jones JDG. 2002. Arabidopsis gp91phox homologues AtrbohD and AtrbohF are required for accumulation of reactive oxygen intermediates in the plant defense response. Proc Natl Acad Sci U S A. 99:517–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uren AG, et al. 2000. Identification of paracaspases and metacaspases: two ancient families of caspase-like proteins, one of which plays a key role in MALT lymphoma. Mol Cell. 6:961–967. [DOI] [PubMed] [Google Scholar]
- Van de Peer Y, Fawcett JA, Proost S, Sterck L, Vandepoele K. 2009. The flowering world: a tale of duplications. Trends Plant Sci. 14:680–688. [DOI] [PubMed] [Google Scholar]
- Wang GF, Li WQ, Wu GL, Zhou CY, Chen KM. 2013. Characterization of rice NADPH oxidase genes and their expression under various environmental conditions. Int J Mol Sci. 14:9440–9458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, et al. 2015. The plasma membrane NADPH oxidase OsRbohA plays a crucial role in developmental regulation and drought-stress response in rice. Physiol Plant. doi: 10.1111/ppl.12389. [DOI] [PubMed] [Google Scholar]
- Winter D, et al. 2007. An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS One 2:e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong HL, et al. 2007. Regulation of rice NADPH oxidase by binding of Rac GTPase to its N-terminal extension. Plant Cell 19:4022–4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, et al. 2005. Molecular and biochemical characterization of the Fe(III) chelate reductase gene family in Arabidopsis thaliana. Plant Cell Physiol. 46:1505–1514. [DOI] [PubMed] [Google Scholar]
- Wudick MM, Feijó JA. 2014. At the intersection: merging Ca2+ and ROS signaling pathways in pollen. Mol Plant. 7:1595–1597. [DOI] [PubMed] [Google Scholar]
- Xie HT, Wan ZY, Li S, Zhang Y. 2014. Spatiotemporal production of reactive oxygen species by NADPH oxidase is critical for tapetal programmed cell death and pollen development in Arabidopsis. Plant Cell 26:2007–2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie YJ, et al. 2011. Evidence of Arabidopsis salt acclimation induced by upregulation of HY1 and the regulatory role of RbohD-derived reactive oxygen species synthesis. Plant J. 66:280–292. [DOI] [PubMed] [Google Scholar]
- Yoshie Y, et al. 2005. Function of the rice gp91phox homologs OsrbohA and OsrbohE genes in ROS-dependent plant immune responses. Plant Biotechnol. 22:127–135. [Google Scholar]
- Yoshioka H, et al. 2001. Induction of plant gp91 phox homolog by fungal cell wall, arachidonic acid, and salicylic acid in potato. Mol Plant Microbe Interact. 14:725–736. [DOI] [PubMed] [Google Scholar]
- Yoshioka H, et al. 2003. Nicotiana benthamiama gp91phox homologues NbrbohA and NbrbohB participate in H2O2 accumulation and resistance to Phytophthora infestans. Plant Cell 15:706–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshioka H, Mase K, Yoshioka M, Kobayashi M, Asai S. 2011. Regulatory mechanisms of nitric oxide and reactive oxygen species generation and their role in plant immunity. Nitric Oxide 25:216–221. [DOI] [PubMed] [Google Scholar]
- Zhang F, Gonzalez A, Zhao M, Payne CT, Lloyd A. 2003. A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development 130:4859–4869. [DOI] [PubMed] [Google Scholar]
- Zhang H, et al. 2014. A novel rice C2H2-type zinc finger protein, ZFP36, is a key player involved in abscisic acid-induced antioxidant defence and oxidative stress tolerance in rice. J Exp Bot. 65:5795–5809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YY, et al. 2009. Phospholipase Dα1 and phosphatidic acid regulate NADPH oxidase activity and production of reactive oxygen species in ABA-mediated stomatal closure in Arabidopsis. Plant Cell 21: 2357–2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Kishino H. 2004. Genomic background predicts the fate of duplicated genes: evidence from the yeast genome. Genetics 166: 1995–1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou JR, Richardson AJ, Rudd KE. 2013. EcoGene-RefSeq: EcoGene tools applied to the RefSeq prokaryotic genomes. Bioinformatics 29:1917–1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.