Abstract
PTEN is a negative regulator of PI3K-AKT signaling and a potent tumor suppressor in many types of cancer. To test a tumor suppressive role of PTEN in pre-B acute lymphoblastic leukemia (ALL), we induced Cre-mediated deletion of Pten in mouse models of pre-B ALL. In contrast to its role as a tumor suppressor in other cancers, loss of one or both alleles of Pten caused rapid cell death of pre-B ALL cells and was sufficient to clear transplant recipient mice of leukemia. Small molecule inhibition of PTEN in human pre-B ALL cells resulted in AKT hyperactivation, p53 checkpoint activation and cell death. Loss of PTEN function in pre-B ALL cells was functionally equivalent to acute activation of autoreactive pre-BCR signaling, which engaged a deletional checkpoint for removal of autoreactive B cells. We propose that targeted inhibition of PTEN and hyperactivation of AKT triggers a checkpoint for elimination of autoreactive B cells and represents a new strategy to overcome drug-resistance in human ALL.
Introduction
The majority of newly generated pre-B cells in the bone marrow are eliminated at the pre-B cell receptor (pre-BCR) checkpoint1. Critical survival and proliferation signals come from the pre-BCR: If pre-B cell clones fail to express a functional pre-BCR, signaling output is too weak. If the pre-BCR binds to ubiquitous self-antigen (autoreactive immunoglobulin μ heavy chain; μ–HC), pre-BCR signals are strong. Both attenuation below minimum (e.g. non-functional pre-BCR) and hyperactivation above maximum (e.g. autoreactive pre-BCR) thresholds of signaling strength trigger negative selection and cell death. Approximately 75% of newly generated pre-B cells express an autoreactive μ–HC2-3, highlighting the importance of stringent negative selection of autoreactive clones at the pre-BCR checkpoint. While autoreactive pre-B cell clones are eliminated owing to the toxicity of strong pre-BCR signaling1-3, sustained activation of Phosphoinositide 3-kinase-AKT (PI3K-AKT) signaling is sufficient to rescue B cell survival in the absence of a functional BCR4 and required for pre-B cell survival5. Likewise, germline mutations in humans that result in either loss or hyperactivation of PI3K-AKT signaling have equally deleterious effects on human early B cell development6, suggesting that early B cells are selected for an intermediate level of PI3K signaling.
Phosphatase and tensin homolog (PTEN) is a key negative regulator of the PI3K-AKT pathway and functions as a dual protein and lipid phosphatase, which dephosphorylates PtdIns(3,4,5)P3 (PIP3). PTEN counteracts PI3K, which phosphorylates PtdIns(4,5)P2 to generate PIP3, the membrane anchor and ligand of the AKT-PH domain7. Deletions or inactivating mutations of PTEN are frequently observed in all main types of human cancer (on average 8.3% among 37,898 samples studied)8. The common outcome of these lesions is increased membrane levels of PIP3 and AKT-hyperactivation. Genetic lesions of PTEN mutations also play a major role in hematopoietic malignancies. For instance, lesions in PTEN, PI3K and AKT pathway component genes are present in up to 50% of T cell lineage ALL cases9.
Results
Pten is required for initiation and maintenance of pre-B ALL in vivo
To study a potential role of PTEN and negative regulation of PI3K-AKT signaling, we developed BCR-ABL1- and NRASG12D-driven models of pre-B ALL (Fig. 1). To this end, we transformed IL7-dependent pre-B cells from bone marrow of Ptenfl/fl and Pten+/fl mice10 with oncogenic BCR-ABL1 or NRASG12D. BCR-ABL1 represents the driver oncogene in Ph+ ALL, the most common subtype of pre-B ALL in adults (~30%). RAS pathway lesions affect NRAS, KRAS, PTPN11 and NF1 and occur in ~50% of both adult and pediatric ALL11. Together, BCR-ABL1 and NRASG12D driven pre-B ALL reflect two major non-overlapping types of human ALL (~80%). For inducible deletion of Pten, pre-B ALL cells were transduced with tamoxifen (Tam)-inducible Cre-ERT2 (Cre) or ERT2 (estrogen receptor) empty vector (EV) controls. After selection of the transduced cells with puromycin, Cre was activated by Tam (1 μmol/l), which induced excision of LoxP-flanked Pten alleles and depletion of PTEN protein within two days (Fig. 1a). Notably, inducible Cre-mediated deletion of Pten in pre-B ALL cells resulted in rapid cell death of leukemia cells (Fig. 1b, Supplementary Fig. 1a). To address whether loss of PTEN not only affected survival of established leukemia but also leukemia-initiation, we reversed the order and first induced deletion of Pten in IL7-dependent Ptenfl/fl pre-B cells and subsequently induced BCR-ABL1-mediated transformation. Two days after induction of Cre, Pten+/+ and Pten−/− pre-B cells were transduced with GFP-tagged BCR-ABL1. Pten+/+ pre-B cells showed rapid outgrowth of BCR-ABL1GFP-expressing clones, indicating leukemic transformation. In contrast, Pten−/− pre-B cells carrying BCR-ABL1GFP were not susceptible to malignant transformation by BCR-ABL1 (Fig. 1c; Supplementary Fig. 1b). These findings were recapitulated in an in vivo transplant setting. BCR-ABL1-transformed Ptenfl/fl pre-B ALL cells caused fatal leukemia in transplant recipient mice within 18 days. Luciferase bioimaging revealed that Cre-mediated deletion of Pten did not interfere with engraftment of pre-B ALL cells. However, pre-B ALL cells failed to initiate fatal disease in the absence of PTEN and transplant recipients survived for indefinite periods of time (Fig. 1d). Minimal residual disease (MRD) analysis by genomic PCR revealed no trace of covert leukemia clones (Supplementary Fig. 1c).
Figure 1. Pten is required for leukemic transformation of pre-B cells.
(a) Deletion of Pten was confirmed after induction of Cre by tamoxifen in BCR-ABL1 transformed Ptenfl/fl pre-B cells at the genomic level by PCR (left) and by Western blot (right). β-actin was used as loading control. Representative blot, n = 3 (b) Viability of BCR-ABL1-transformed pre-B ALL cells, as measured by flow cytometry, after tamoxifen dependent induction of Pten deletion. P < 0.0001 was calculated by contingency table. (c) The fraction of BCR-ABL1GFP+ cells was measured over time by flow cytometry in pre-B cells. P value was calculated by contingency table (P < 0.0001). (d) Ptenfl/fl BCR-ABL1-transformed pre-B ALL cells transduced with tamoxifen-inducible Cre or an empty vector (EV) control were injected into NOD/SCID recipient mice (n = 7 per group). Pten deletion was induced 24 h before injection. P = 0.0002 was calculated by Mantel-Cox log-rank test. (e) Microarray gene expression analysis after 48 hours of induction of Pten deletion in BCR-ABL1-transformed pre-B ALL cells. (f) Confirming gene expression results for eight surface molecules by flow cytometry against forward scatters (FSC). (g) Expression of B cell markers were measured in vitro (left, n = 3) and in vivo (right, n = 4). Representative flow cytometry plots are shown. (h)Western blot measurement in Ptenfl/fl BCR-ABL1-transformed pre-B ALL cells using β-actin as loading control. Representative blot, n = 4. Error bars (b, c) represent S.D.
Pten mediates feedback regulation of pre-BCR and its co-receptor CD19
To elucidate the mechanism of how the tumor suppressor PTEN, seemingly paradoxically, enables oncogenic transformation of pre-B cells, we studied gene expression changes upon inducible Pten-deletion (Fig. 1e). Loss of Pten induced expression of multiple markers of lymphocyte activation including Cd80, Cd86, Cd25 (Il2rα) and Ly6a (Sca-1). Pten-deletion resulted in downregulation of the IL7 receptor, both at the mRNA level and surface expression (Fig. 1e, f). In addition, multiple components of the pre-BCR (Ighm, Vpreb1, Syk) and its co-receptor CD19 were all downregulated upon deletion of Pten (Fig. 1e-f). In pre-B cells, PI3K-AKT signaling is initiated from the pre-BCR via Syk4-5,12-13 and CD19 via recruitment of PI3K14,15 to a YXXM motif in the cytoplasmic tail of CD19. For this reason, loss of pre-BCR and CD19 expression in response to deletion of Pten suggests PTEN-mediated feedback regulation of these PI3K-AKT activating receptors. Likewise, B cell precursors developing in Mb1-Cre Ptenfl/fl mice lost CD19 expression upon early deletion of Pten in the B cell lineage (Fig. 1g). Western blot analysis confirmed near complete loss of CD19 protein expression upon deletion of Pten, demonstrating loss of both surface and intracellular expression of CD19 (Fig. 1h).
Deletion of Pten compromises BCR-ABL1 and NRAS-driven leukemogenesis
BCR-ABL1 and NRASG12D-driven ALL subtypes were dependent on PTEN function to a similar degree as inducible, Cre-mediated deletion of Pten induced cell death and reduced colony forming ability (Fig. 2a-b). Genotyping of colonies revealed that Ptenfl/fl pre-B ALL clones that were able to form colonies had retained Ptenflox alleles despite activation of Cre. Hence, these few colonies arose from pre-B ALL clones that had evaded Cre-mediated deletion of Pten (Supplementary Fig. 2).
Figure 2. Deletion of Pten compromises BCR-ABL1 and NRAS-driven leukemogenesis.
(a) The percentage of GFP+ cells were measured in BCR-ABL1 (left) or NRASG12D (right) transformed Ptenfl/fl pre-B cells transduced with inducible Cre-ERT2-GFP or GFP empty vector (EV) control after induction of Cre with tamoxifen by flow cytometry. For each plot, P < 0.0001 was obtained by contingency table. (b) Comparison of colony forming ability of BCR-ABL1 (left) or NRASG12D (right) transformed Ptenfl/fl pre-B cells after Pten deletion. (c) Measuring the induction of cell cycle arrest following 24 h of tamoxifen induced Pten deletion in BCR-ABL1 (left) or NRASG12D (right) transformed Ptenfl/fl pre-B cells. The percentages of each phase of the cell cycle are indicated. (d) The prevalence of cellular senescence was measure using β-galactosidase assay after deletion of Pten in BCR-ABL1 (left) or NRASG12D (right) transformed Ptenfl/fl pre-B cells. The percentages of cells stained with blue dots are depicted on the panels. Results in (a–d) are representative of two independent experiments. Scale bars in (b) and (d) represent 1 mm and 15 μm respectively. P values (b–d) were calculated by Student’s t test. (e) NOD/SCID recipient mice injected with 105 Ptenfl/fl BCR-ABL1-transformed pre-B ALL transduced with Cre-ERT2 (left) were treated with tamoxifen (0.4 mg/mouse) or corn oil (vehicle) (n = 7 per group). In a parallel experiment, NOD/SCID recipient mice were injected with 105 Ptenfl/fl NRASG12D transformed pre-B ALL cells transduced with Cre-ERT2 or empty vector (EV) control (right). In the latter experiment all mice were treated with Tamoxifen (0.4 mg/mouse; n = 7 per group). P values were calculated by Mantel-Cox log-rank test. All cells were labeled with luciferase before injection. Treatments were started on day 5 after transplantation and continued for ten consecutive days. Error bars (a–b, d) represent S.D.
Pten-deletion also induced G0/G1 cell cycle arrest and senescence in pre-B ALL cells (Fig. 2c-d). To assess the effect of acute ablation of Pten in fully established BCR-ABL1 and NRASG12D-driven pre-B ALL cells in vivo, 100,000 Ptenfl/fl pre-B ALL cells carrying tamoxifen-inducible Cre were injected into sublethally irradiated NOD/SCID recipient female mice. 5 days after transplantation of Ptenfl/fl pre-B ALL cells, leukemia-initiation and engraftment was confirmed by bioluminescence imaging and Cre was induced by 10 consecutive daily injections of tamoxifen. Consistent with in vitro results, Pten-deletion caused leukemia regression in vivo (Fig. 2e) and prolonged overall survival in the recipient mice. We conclude that Pten is required for both initiation and maintenance of pre-B ALL in vivo.
Pre-B ALL cells are exempt from genetic lesions of PTEN
Given the unexpected sensitivity of pre-B ALL cells to even a moderate dose-reduction of Pten, we reassessed the concept of PTEN as a tumor suppressor in human cancer and leukemias and lymphomas. A reanalysis of genetic lesions of PTEN in human cancer revealed a high frequency of mutations in solid cancer (8.3% in 37,898 samples) and hematological malignancies (8.4% in 2,548 samples)8. In addition to point mutations, deletions of PTEN at chromosome 10q23 are frequent in cancer (5.3% in 8,071 samples studied; Fig. 3a). These findings are in agreement with previous mechanistic studies demonstrating a tumor suppressor role of PTEN in acute myeloid16 and chronic myeloid leukemia (CML)17, in T cell lineage acute lymphoblastic leukemia9,18-19, and in mature B cell lymphoma20-22. However, point mutations in PTEN were not detected in a single case of 694 pre-B ALL patient samples. Likewise, no PTEN deletions were found in 231 pre-B ALL cases (Fig. 3a). Similarly, while oncogenic activation of the PI3K-AKT pathway in leukemia and lymphoma also occurs through activating mutations of agonists of the PI3K-AKT pathway, pre-B ALL cases do not harbor such mutations (Supplementary Fig. 3). Together, these genetic data suggest that PTEN lesions and other mutations that lead to oncogenic activation of the PI3K-AKT pathway are not favorable in pre-B ALL and that PTEN may have a fundamentally different role in pre-B ALL compared to other hematopoietic malignancies.
Figure 3. Pre-B ALL cells do not harbor genetic lesions in PTEN and do not tolerate hyperactivation of PI3K-AKT signaling.
(a) Frequencies of genetic lesions of PTEN including mutations and deletions from the COSMIC data base8 for 16 types of human cancer. (b) Comparing the level of CpG methylation for PTEN promoter region in pre-B cells from healthy donors (n = 12), bone marrow biopsies from patients with pre-B ALL (n = 83) and patient-derived B cell Non-Hodgkin’s lymphoma cells (n = 68). (c) Comparing PTEN expression using RPPA assay for cases with newly diagnosed adult ALL (n = 155), T cell lineage ALL (n = 22) and mature B cell lymphoma (n = 11). P values in (b–c) were calculated by two-sided Mann–Whitney Wilcoxon test. (d) PTEN expression in a panel of normal human CD19+ sorted pre-B cells (n = 3), patient-derived pre-B ALL (n = 8) and B cell Non-Hodgkin lymphoma samples (n = 4). β-actin was used as loading control. P values were calculated by Student’s t test. (e) Measuring the activity of Akt after deletion of Pten in BCR-ABL1- and NRASG12D- transformed pre-B ALL cells. β-actin was used as loading control. Representative blot, n = 3 (f) Schematic of pre-B cell receptor signaling and its interaction with PI3K/AKT pathway. (g) Rescuing cell death after Pten deletion in BCR-ABL1 (left) or NRASG12D (right) transformed Ptenfl/fl pre-B cells by using AKT inhibitor (AZD, 3 μmol/L). The percentage of GFP+ cells were measured by flow cytometry. (h) measuring the percentages of GFP+ cells after Pten deletion was induced in BCR-ABL1-transformed pre-B ALL cells in the presence or absence of AKT inhibitor (AZD, 3 μmol/L), PI3K inhibitor (BKM, 1 μmol/L) and Syk inhibitor (PRT; 3 μmol/L). For each EV and Cre transduced cells, P values were calculated by contingency table to compare treatment group with untreated control group (Ctrl). All P values for EV transduced cells are not significant (P>0.5, not shown). Error bars represent S.D. Results are representative of three independent experiments.
High expression levels of PTEN in patient-derived pre-B ALL cells
Consistent with these findings, our analysis of patient-derived samples showed that PTEN promoter regions are hypermethylated in B cell lymphomas (n = 68) but not in pre-B ALL samples (n = 83; Fig. 3b). Reverse Phase Protein Array (RPPA) measurements for 155 newly diagnosed cases of adult ALL, 22 cases of T cell lineage ALL and 11 cases of mature B cell lymphoma (MDACC, 1983-2007)23 revealed higher PTEN protein levels in pre-B ALL samples compared to T cell lineage ALL and mature B cell lymphoma (Fig. 3c). Studying a panel of normal human CD19+ sorted pre-B cells (n = 3), patient-derived pre-B ALL (n = 8) and B cell Non-Hodgkin lymphoma samples (n = 4), we confirmed these findings by Western blot (Fig. 3d). The MDACC (1983-2007) RPPA data set replicated results from previous work24 that identified high expression levels of PTEN as predictor of favorable outcome in patients with T cell lineage ALL (Supplementary Fig. 4). In contrast, RPPA data for diagnostic samples from patients with pre-B ALL showed the opposite trend and higher than median expression levels of PTEN in diagnostic pre-B ALL samples predicted shorter relapse-free survival (Supplementary Fig. 4).
Pten regulates AKT activity downstream of the pre-B cell receptor
To elucidate the mechanistic basis of Pten-dependency in pre-B ALL, we first tested the hypothesis that loss of Pten results in hyperactivation of PI3K-AKT signaling and subsequent cell death. In agreement with this hypothesis, we found that inducible deletion of Pten in both BCR-ABL1 and NRASG12D-driven pre-B ALL cells resulted in increased phosphorylation of AKT at both S473 and T308 (Fig. 3e). Consistent with PTEN-dependent feedback regulation, deletion of Pten caused hyperactivation of PI3K-AKT and was followed by downregulation of pre-BCR, IL7R and CD19 (Fig. 1f-h; Fig. 3f), all of which mediate PI3K-AKT activation in pre-B cells. Syk is a B cell-specific tyrosine kinase that links pre-BCR signaling to PI3K activation12-15. To measure the contribution of individual components of the PI3K-AKT pathway to induce toxicity, we used selective small molecule inhibitors of the Syk (PRT06207), PI3K (BKM120) and AKT (AZD5363) kinases (Fig. 3f). Consistent with hyperactivation of AKT in pre-B ALL cells, pharmacological inhibition of AKT (AZD) largely reduced toxicity of Pten-deletion (Fig. 3g, Supplementary Fig. 5a). Also inhibition of Syk (PRT) and PI3K (BKM) mitigated the toxic effects of Pten-deletion. Given that BCR-ABL1 and NRASG12D can activate AKT independently from Syk and PI3K, the AKT inhibitor (AZD) had the strongest protective effect in reducing cell death induced by Pten-deletion (Fig. 3h, Supplementary Fig. 5b).
Both alleles of Pten are required for survival and proliferation of pre-B ALL cells
Deletion of PTEN in human cancer typically affects one allele while the other copy is retained25, suggesting that cancer cells are selected for deletion of one but not both alleles of PTEN. Indeed, deletion of both alleles of Pten induces p53-dependent senescence in a model of prostate cancer, unless Tp53 is also deleted25. For this reason, we studied the consequences of both heterozygous and homozygous deletion of Pten in mouse pre-B ALL cells (Supplementary Fig. 6). Comparing deletion of both alleles (Ptenfl/fl) or one (Pten+/fl) allele of Pten, we found that—in contrast to solid tumor cells—both homozygous and heterozygous deletion of Pten affected colony formation and proliferation of pre-B ALL cells and induced cellular senescence. The deleterious effects of mono- and bi-allelic Pten-ablation in pre-B ALL cells were indistinguishable (Supplementary Fig. 6a-c). Likewise, deletion of either both or only one allele of Pten compromised leukemogenesis in an in vivo transplant experiment and no difference was noted between pre-B ALL cells carrying deletion of one or both alleles of Pten (Supplementary Fig. 6d). Western blot analysis revealed that deletion of one allele of Pten resulted in increased phosphorylation of AKT (Supplementary Fig. 6e). However, this increase was smaller compared to AKT-phosphorylation in response to biallelic Pten-deletion.
AKT-hyperactivation eliminates autoreactive B cells
Unlike other cell types, pre-B cells are selected for an intermediate level of PI3K-AKT signaling strength4-6. Both loss- and gain-of-function mutations of PI3K trigger negative selection of pre-B cells and cell death4-6. PI3K-AKT signaling is activated downstream of the pre-BCR and its coreceptor CD194-5. Previous work had demonstrated that tonic activation of PI3K-AKT signaling can rescue early B cell development in the absence of a functional pre-BCR4-5. Here, we determined whether pharmacological inhibition of PI3K-AKT signaling can rescue negative selection and cell death in response to acute activation of an autoreactive pre-BCR. To test this hypothesis, we employed a reconstitution system26 for inducible activation of pre-BCR signaling downstream of non-autoreactive (μ–HCNA) or autoreactive μ heavy chains (μ–HCAuto) or an empty vector control (EV; Fig. 4a). Tamoxifen (Tam)-induced assembly of the proximal BCR signalosome in the presence of EV or μ–HCNA had no effect on the viability of BCR-ABL1 pre-B ALL cells (Fig. 4b, Supplementary Fig. 7a). However, inducible activation of autoreactive pre-BCR signaling (μ–HCAuto) caused rapid cell death, which could be rescued by small molecule inhibition of Syk (PRT) and AKT (AZD) kinase activity (Fig. 4b, Supplementary Fig. 7a). Activation of signaling from an autoreactive μ–HC (μ–HCAuto) and induction of cell death involved phosphorylation of Syk and AKT, whereas activation of pre-BCR signaling downstream of μ–HCNA and EV had no effect on phosphorylation levels of Syk and AKT (Fig. 4c). Treatment with Syk (PRT) and AKT (AZD) kinase inhibitors rescued toxicity and demonstrated that hyperactivation of Syk and AKT downstream of autoreactive pre-BCR signaling was required to engage a deletional checkpoint (Fig. 4b). We conclude that hyperactivation of PI3K-AKT signaling upon deletion of Pten is functionally equivalent to hyperactivation of PI3K-AKT downstream of an autoreactive pre-BCR via Syk. Both events trigger a deletional checkpoint for removal of autoreactive B cell clones.
Figure 4. Hyperactivation of AKT is a defining feature of autoreactive pre-BCR signaling and triggers a checkpoint for removal of autoreactive pre-B cells.
(a) Schematic of reconstitution system for inducible activation of pre-BCR signaling. (b) Relative changes of GFP+ cells relative to EV-transduced cells are shown after activation of pre-B cell signaling in BCR-ABL1 transformed pre-B ALL cells with two-day pre-treatment with inhibitors of Syk (PRT, 3 μmol/L) or AKT (AZD, 3 μmol/L). For each curve, P value was calculated by contingency table comparing treatment groups with untreated control group (Ctrl). P values for μHCNA-transduced cells were not significant (>0.5, not shown). (c) Comparing the phosphorylation of Syk, Akt and Rps6 by Western blot 5 minutes after addition of tamoxifen. Representative blot, n = 2. (d) Relative changes in the percentages of GFP+ cells after transduction of pre-B ALL cells with a GFP-tagged constitutive active Syk (SykCA; SykY348E/Y352E) or empty vector (EV) control in the presence of vehicle or inhibitors of AKT (AZD, 3 μmol/L), PI3K (BKM, 1 μmol/L) and Syk (PRT, 3 μmol/L). (e) Relative changes in the percentages of GFP+ cells after transduction of pre-B ALL cells with a GFP-tagged retroviral vector encoding myristoylated AKT (AKTCA) or GFP empty vector (EV) control in the presence or absence of AKT inhibitor (AZD, 3 μmol/L). (f) B→myeloid reprogramming of BCR-ABL1 transformed Ptenfl/fl pre-B ALL (CD19+ Mac1−) to myeloid lineage (CD19− Mac1+) cells after induction of C/EBPα. (g) Expression of C/EBPα, the B cell lineage-specific transcription factor PAX5, total and phospho-AKTS473 and PTEN in B-lineage and B→myeloid reprogrammed cells. Representative blot, n = 2. (h) Changes in the frequency of GFP+ cells in B→myeloid (CD19− Mac1+) and pre-B cells (CD19+ Mac1−) after induction of Cre-GFP by tamoxifen. P < 0.0001 was calculated using contingency table. All results presented in this figure are representative of two independent experiments with triplicates. Error bars (b, d–e, h) represent S.D. of the mean. For curves in (b, d and e), P values were calculated by contingency table comparing treatment groups with untreated control group (Ctrl).
To demonstrate the toxic effects of Syk- and AKT-hyperactivation in a genetic experiment, we transduced pre-B ALL cells with constitutively active (CA) forms of Syk and AKT and measured pre-B ALL cell viability in the presence and absence of Syk (PRT), PI3K (BKM) and AKT (AZD) kinase inhibitors. As predicted, SykCA rapidly induced cell death in pre-B ALL cells, which could be nearly completely rescued by kinase inhibition of Syk, PI3K and AKT (Fig. 4d, Supplementary Fig. 7b). Likewise, hyperactivation of AKT (AKTCA) was toxic in pre-B ALL cells and toxicity was reduced by AKT kinase inhibition (AZD; Fig. 4e, Supplementary Fig. 7c). Collectively, these findings suggest that hyperactivation of PI3K-AKT signaling engages a deletional checkpoint for removal of autoreactive B cells.
B cell-specific expression and activity of PTEN
To test the basic premise of a checkpoint for elimination of autoreactive pre-B cells, we reprogrammed pre-B ALL cells into myeloid lineage cells using an inducible C/EBPα vector system27. To this end, we engineered Ptenfl/fl pre-B ALL cells with a doxycycline inducible vector system for expression of the myeloid transcription factor C/EBPα, which results in B→myeloid lineage conversion (Fig. 4f-h). Inducible expression of C/EBPα in pre-B ALL cells not only downregulated Pax5, a transcription factor that determines B cell lineage identity but also resulted in downregulation of PTEN levels, demonstrating that PTEN expression levels depend on B cell lineage identity (Fig. 4g). C/EBPα-mediated myeloid lineage conversion also resulted in a 45-fold increase in AKT-S473 phosphorylation compared to the EV transduced cells, which mirrors transcriptional repression of PTEN in myeloid cells (Fig. 4g). These findings are consistent with the notion that myeloid cells, unlike B lineage cells, are permissive to loss of PTEN function and hyperactivation of AKT signaling.
To test whether B→myeloid lineage conversion erases dependency on PTEN-mediated negative regulation of AKT activity, Ptenfl/fl pre-B ALL cells carrying inducible C/EBPα or empty vector controls (EV), were also induced to express Cre (Fig. 5g-h). Inducible deletion of Pten resulted in a rapid elimination of B cell lineage ALL cells (CD19+, Mac1−) as in previous experiments (Fig. 1c-f, Supplementary Fig. 1). By contrast, deletion of Pten in B→myeloid reprogrammed (CD19−, Mac1+) resulted in a slight increase in the frequency of Cre-GFP+ cells, reflecting a relative survival advantage of Pten-deletion in myeloid cells (Fig. 4h). These findings are consistent with a previous study17, which identified PTEN as a tumor suppressor in BCR-ABL1-transformed myeloid leukemia, and reinforce the notion that normal pre-B and pre-B ALL cells are uniquely dependent on negative regulation of AKT by PTEN.
Figure 5. pre-B cell-specific functions of PTEN in normal progenitor cells and leukemia.
(a) PTEN protein levels in mouse cytokine-dependent myeloid progenitor cells (Sca1+, c-kit+, Lin−), IL-7 dependent pre-B cells, BCR-ABL1-transformed myeloid and pre-B leukemia. PTEN/β-actin densitometry ratios were normalized to ratios calculated for myeloid progenitor cells. P-values were calculated by Student’s t test. Representative blot, n = 3. (b) Western blot measurement for PTEN in patient-derived CML samples (n = 5) and patient derived B-lineage ALL (n = 5). PTEN/β-actin densitometry ratios were calculated and the P = 0.00001 was calculated by Student’s t test. (c, d) The percentages of viable cells were measured after Cre–induced Pten deletion in myeloid progenitor cells (c, left) and pre-B cells (c, right) and their BCR-ABL1 transformed counterparts CML-like (d, left) and pre-B ALL (d, right) cells. P values were calculated using contingency tables. (e) The protein expression of cell cycle checkpoint molecules Arf, p53 and p21 after deletion of Pten in BCR-ABL1-transformed myeloid leukemia and pre-B ALL. Representative blot, n = 2. (f) Measuring total and phosphorylated form of p53 after 48 hours of Cre-induced Pten deletion in BCR-ABL1-transformed pre-B ALL in the presence of AKT inhibitor (AZD, 3 μmol/L) or vehicle. Representative blot, n = 3. (g) Glucose consumption, lactate production, and ATP levels were measured on day two following induction of Cre in Ptenfl/fl BCR-ABL1-transformed pre-B ALL or myeloid reprogrammed cells. Values obtained were normalized to number of viable cells and are shown as average relative levels. P values were calculated by Student’s t test. β-actin was used as loading control (a–b, e–f). Error bars (c–d, g) represent SD of the mean.
The deleterious effects of Pten-deletion in pre-B ALL are linked to B cell lineage identity
Pre-B (Ph+ ALL) and chronic myeloid leukemia (CML) are both driven by the oncogenic BCR-ABL1 tyrosine kinase but markedly differ with respect to PTEN expression levels (Fig. 5a-b). PTEN protein levels were high in BCR-ABL1-driven mouse pre-B ALL and in patient-derived Ph+ ALL (n = 5) but barely detectable in mouse CML-like and patient-derived CML cells (n = 5; Fig. 5a-b). This difference is consistent with regulation of Pten levels during B→myeloid reprogramming (Fig. 4g). While pre-B cells are subject to a deletional checkpoint for removal of autoreactive clones, this is not the case for other cell types (including myeloid cells). Likewise, deletion of Pten in normal myeloid progenitor cells had no effects but caused toxicity in normal pre-B cells (Fig. 5c). Moreover, Cre-mediated Pten-deletion had no deleterious effects in BCR-ABL1 myeloid lineage CML but caused cell death in BCR-ABL1-transformed pre-B ALL cells (Fig. 5d). Induction of cell death upon Pten-deletion in pre-B ALL cells was paralleled by accumulation of Arf, p53 and p21 checkpoint molecules. In contrast, Pten-deletion in myeloid leukemia cells neither induced activation of checkpoint molecules nor cell death (Fig. 5d-e). Consistent with its function as a regulator of an autoimmunity checkpoint, these findings suggest that PTEN is specifically required for the survival of normal and transformed pre-B cells and dispensable in myeloid cells. Activation of p53 upon acute deletion of Pten involved increased p53-phosphorylation at S15, which increases p53 stability and its half-life (Fig. 5f). Interestingly, both S15-phosphorylation and accumulation of p53 were reversed by AKT-inhibition (AZD5363), indicating that activation of p53 in response to Pten-deletion is caused by AKT-hyperactivation (Fig. 5f).
AKT hyperactivity exacerbates glucose and energy depletion
AKT promotes glycolysis and the use of cellular ATP towards translation, cell growth and proliferation through activation of mTORC128-29. We studied the metabolic consequences of Pten-deletion on glycolysis and energy supply in the context of mTORC1 inhibition by rapamycin. As expected, inducible deletion of Pten resulted in an increase in glycolysis, as measured by increased glucose consumption and increased production of lactate (Supplementary Fig. 8). These effects were reversed by rapamycin, indicating that increased levels of glycolysis upon Pten-deletion were caused by mTORC1 activation. Consistent with increased energy-inefficient glycolysis, inducible deletion of Pten in pre-B ALL cells reduced ATP levels, which were restored by rapamycin (Supplementary Fig. 8). These metabolic effects of Pten-deletion were specific for B-lymphoid cells. B→myeloid reprogramming of pre-B ALL cells using inducible expression of CEBPα (Fig. 4f-h) revealed that ablation of Pten had divergent outcomes in B lymphoid and B→myeloid reprogrammed leukemia cells. While loss of Pten moderately increased glycolysis in myeloid cells, resulting in a substantial net gain of cellular ATP levels, glycolytic activity was excessively increased in B lymphoid cells resulting in depletion of glucose reserves and in near exhaustion of cellular ATP (Fig. 5g). Taken together, these findings indicate that deletion of Pten in pre-B ALL cells caused profound metabolic distress in conjunction with strong activation of p53 checkpoint activation in pre-B ALL cells. It should be noted that we tested alternative mechanisms of toxicity related to Pten deletion, including dephosphorylation of STAT5 and accumulation of cellular reactive oxygen species (ROS). Unlike metabolic stress and p53-mediated cell death, loss of STAT5-phosphorylation and increased ROS levels were only observed in BCR-ABL1-transformed ALL cells and could not be mechanistically validated in other ALL subtypes (data not shown).
Validation of PTEN as a therapeutic target in patient-derived pre-B ALL cells
Despite the well-characterized function of PTEN as a tumor suppressor, to test whether PTEN represents a therapeutic target in human pre-B ALL, we used two distinct shRNAs against PTEN and a scrambled control for lentiviral delivery in patient-derived pre-B ALL cells. The two shRNA hairpins reduced PTEN protein levels by 2- to 4-fold compared to the non-targeting (scrambled) control as determined by Western blot (Fig. 6a). Lentiviral hairpin-mediated knockdown of PTEN in three human myeloid lineage CML cell lines did not affect viability compared to scrambled controls (Fig. 6b, Supplementary Fig. 9). In contrast, PTEN knockdown induced cell death in cells from four individuals with pre-B ALL bearing BCR-ABL1 (LAX2, ICN1), TCF3-PBX1 (ICN12) or KRASG12V (LAX7R) oncogenes (Fig. 6b; Supplementary Fig. 9). We conclude that PTEN is essential for the survival of human pre-B ALL, validating PTEN as a potential therapeutic target in human pre-B ALL.
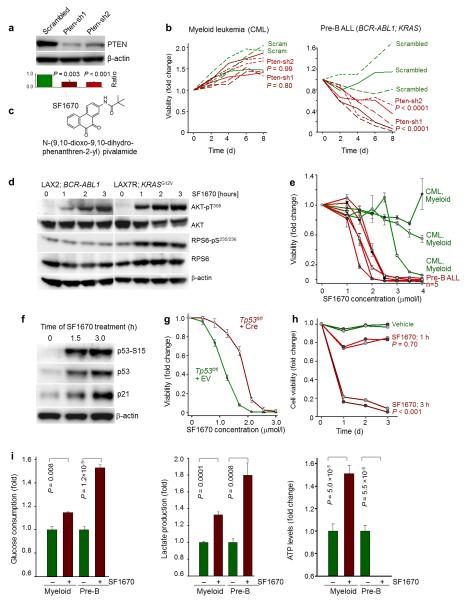
Figure 6. Small molecule inhibitor of PTEN is specifically toxic in pre-B ALL.
(a) Validating PTEN knockdown by two shRNA against PTEN using β-actin as loading control. Representative blot, n = 3. P values were calculated by Student’s t test comparing each PTEN shRNA to the scrambled shRNA. (b) Measuring the viability of three CML cell lines (KYO-1, KU812, JURL-MK1) and three patient derived pre-B ALL xenografts (LAX7R, LAX2 and ICN12) after transduction with two shRNA against PTEN and a scrambled sequence. P values were calculated using contingency tables comparing average viability of cells for each shRNA vs scrambled control. (c) Chemical structures of PTEN inhibitor SF1670. (d) Western blot measuring the activity AKT signaling pathway after treatment of two pre-B ALL cases with SF1670 (10 μmol/L). β-actin was used as loading control. Representative blot, n = 4. (e) Cell viability assay after SF1670 treatment of human pre-B ALL (n = 5, red) and CML (n = 3, green) cases. P < 0.0001 was calculated using contingency table comparing Pre-B ALL vs CML cells. Error bars represent SD. (f) Western blot measurement of p21, p53 and p53 (Ser15) on a pre-B ALL cells treated with SF1670 (10 μmol/L). β-actin was used as loading control. Representative blot, n = 3. (g) Cell viability assay after SF1670 treatment of BCR-ABL1- transformed pre-B ALL cells with or without Tp53 deletion. Error bars represent SD. P < 0.001 was calculated by contingency table. (h) Measuring viability of two pre-B ALL cases treated for only 1 or 3 h with SF1670 (10 μmol/L) for three consecutive days. P values were calculated by contingency table comparing average cell viability after each treatment with vehicle treated cells. The results are representative of three independent experiments. (i) Measuring Glucose consumption, lactate production, and ATP levels were after 24 hours treatment of CML cell line (KYO-1) and patient derived pre-B ALL case (LAX7R) with SF1670 (2.5 μmol/L). Values obtained were normalized to number of viable cells and are shown as average relative levels. P values were calculated by Student’s t test. Error bars represent SD.
Preclinical evaluation of PTEN inhibitor in pre-B ALL treatment
To test potential therapeutic usefulness of pharmacological PTEN inhibition, we studied the effects of a small molecule inhibitors of PTEN, SF167030-31 in human pre-B ALL cells (Fig. 6c). Biochemical validation of SF1670 activity revealed that the PTEN small molecule inhibitor induced activation of AKT signaling in pre-B ALL cases carrying BCR-ABL1 or KRASG12V oncogenes within one hour of treatment (Fig. 6d). SF1670 strongly and selectively induced cell death in pre-B ALL cells at similar concentrations that were previously used to stimulate neutrophils in vivo31 (IC50, 1.2 μmol/L; Fig. 6e, Supplementary Fig. 10a). In addition, SF1670 exhibits a similar B cell lineage-specific effect as in genetic Pten-deletion and shRNA-mediated knockdown of PTEN, since myeloid CML cells were either not affected by SF1670 or only at higher concentrations (Fig. 6e). In addition, treatment of human pre-B ALL cells with SF1670 phenocopied the effects of genetic deletion of Ptenfl/fl in pre-B ALL cells. Importantly, acute deletion of Pten in Ptenfl/fl in pre-B ALL cells resulted in phosphorylation (S15) and activation of p53 (Fig. 5f). Likewise, small molecule inhibition of PTEN resulted in rapid activation of p53 and p21 checkpoint molecules and p53-phosphorylation at S15 (Fig. 6f). To test whether p53-checkpoint activation represents an important aspect of toxicity in pre-B ALL cells resulting from acute PTEN-inhibition, we transduced Tp53fl/fl BCR-ABL1 transformed pre-B cells with Cre or empty vector (EV). Interestingly, Tp53 deletion caused a substantial shift in dose response curves of cells treated with SF1670. (Fig. 6g). While Cre-mediated deletion of Pten resulted in rapid loss of CD19 and pre-BCR (VpreB) surface expression (Fig. 1f, Supplementary Fig. 10b), we observed the same effects in human pre-B ALL cells that were treated for two days with SF1670 (Supplementary Fig. 10b). Because prolonged systemic inhibition of PTEN would likely raise safety concerns based on its role as tumor suppressor in a wide range of cell types8,18, we tested the effects of short, transient inhibition of PTEN using SF1670. One single exposure to SF1670 for three hours (and subsequent washout of SF1670) induced AKT hyperactivation and caused significant cell death in pre-B ALL cells (P < 0.001; Fig. 6h).
Small molecule inhibition of PTEN (SF1670) also recapitulated metabolic features of genetic deletion of Pten (Fig. 6i). While SF1670-treatment resulted in moderate stimulation of glycolysis (glucose consumption and lactate production) in CML cells, stimulation of glycolytic responses were much stronger in B lymphoid ALL cells (Fig. 5g). However, SF1670-treatment resulted in a net increase in cellular ATP in myeloid CML cells but in near-complete depletion of energy reserves in patient-derived pre-B ALL cells. Recapitulating our measurements in response to Cre-mediated deletion of Pten in myeloid leukemia and pre-B ALL (Fig. 5g), SF1670-treatment induced pre-B cell-specific depletion of ATP and energy crisis.
Discussion
Unlike other cell types, pre-B cells are under intense selective pressure and have to pass a central B cell tolerance checkpoint for removal of autoreactive clones2. The vast majority of newly generated pre-B cells express an autoreactive immunoglobulin μ-heavy chain resulting in strong pre-B cell receptor signaling and are, hence, eliminated at this checkpoint 2-3. Humans carrying germline mutations resulting in hyperactive PI3K-AKT signaling suffer from profound B cell lymphopenia6, presumably because virtually all newly generated pre-B cells are subject to negative selection due to hyperactive PI3K-AKT signaling. Recent work from our group demonstrated that central B cell tolerance checkpoints are fully functional in human pre-B ALL cells, despite malignant transformation31,32. Here we demonstrated that PTEN represents a critical gatekeeper of this checkpoint. Despite its tumor suppressor function in all major subtypes of cancer, PTEN is required to prevent indiscriminate checkpoint activation. Reflecting the unique function of PTEN in central B cell tolerance, pre-B ALL was the only subtype of human cancer that we found exempt from PTEN lesions (Fig. 3a). Previous work demonstrated that pre-B cells need a minimum level of PI3K-AKT signaling to survive5 and targeting of this pathway is potentially useful in the treatment of human pre-B ALL33. However, here we propose that acute inhibition of PTEN or direct pharmacological hyperactivation of PI3K-AKT signaling may represent a strategy to trigger central B cell tolerance checkpoints and thereby overcome conventional drug resistance in human pre-B ALL.
Online methods
Patient samples, human cells and cell lines
We obtained primary patient samples (Supplementary Table 1) in compliance with the Institutional Review Board of the University of California San Francisco. Human leukemia cells were maintained in Minimum Essential Medium α (MEM α, Invitrogen, Carlsbad, CA) supplemented with GlutaMAX, 20% fetal bovine serum, 100 IU/ml penicillin and 100 μg/ml streptomycin and incubated at 37°C in a humidified incubator with 5% CO2. We obtained informed consent from all participants. Primary human ALL cells (patient samples) were cultured on irradiated OP9 stromal cells (ATCC; Manassas, Virginia) in same media as described above.
Retroviral transduction
Complete list of vectors in this study is provided in Supplementary Table 2. Transfections of the MSCV-based retroviral constructs were performed using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) with Opti-MEM media (Invitrogen). Retroviral supernatant was produced by co-transfecting 293FT cells with the plasmids pHIT60 (gag-pol) and pHIT123 (ecotropic env). Cultivation was performed in high glucose Dulbecco’s modified Eagle’s medium (DMEM, Invitrogen) with GlutaMAX containing 10% fetal bovine serum, 100 IU/mL penicillin, 100 μg/mL streptomycin, 25 mM HEPES, 1 mM sodium pyruvate and 0.1 mM non-essential amino acids. Regular media were replaced after 16 hours by growth media containing 10 mM sodium butyrate. After 8 h incubation, the media was changed back to regular growth media. 24 hours later, the virus supernatant were harvested, filtered through a 0.45 μm filter and loaded by centrifugation (2000 × g, 90 min at 32°C) two times on 50 μg/mL RetroNectin (Takara, Madison, WI) coated non-tissue 6-well plates. Three million cells were transduced per well by centrifugation at 600 × g for 30 minutes and maintained for 72 h at 37°C with 5% CO2 before transferring into culture flasks.
Preparation of bone marrow cells from mice
Complete list of mice in this study is provided in Supplementary Table 3. Bone marrow cells were extracted from mice younger than 6 weeks of age. Bone marrow cells were obtained by flushing cavities of femur and tibia with PBS. After filtration through a 70 μm filter and depletion of erythrocytes using a lysis buffer (BD PharmLyse, BD Biosciences), washed cells were either frozen for storage (in 10% DMSO, 90% FBS) or subjected to further experiments.
BCR-ABL1 –driven mouse model of pre-B ALL and myeloid leukemia
Bone marrow cells from Ptenfl/fl (n = 3, female) and Ptenfl/+ (n = 2, female) mice (Supplementary Table 3) were collected and retrovirally transformed with BCR-ABL1. For B cell lineage leukemia, transduction was performed after culturing of bone marrow cells with 10 ng IL-7/ml for one week. To generate myeloid lineage leukemia bone marrow cells were cultured with 10 ng/ml recombinant mouse IL-3, 25 ng/ml recombinant mouse IL-6, and 50 ng/ml recombinant mouse SCF for one week (PeproTech). This treatment leads to selection for Lin− Sca-1+ c-Kit+ (LSK) cells. Transformation of precursor B cells and LSK cells with BCR-ABL1 generate pre-B ALL and CML-like models respectively. After BCR-ABL1 transduction all cytokines were removed from the culture to negatively select the non-transduced cells. Both ALL and CML cells were maintained in Iscove’s modified Dulbecco’s medium (IMDM, Invitrogen) supplemented with GlutaMAX, 20% fetal bovine serum, 100 IU ml−1 penicillin, 100 μg ml−1 streptomycin and 50 μM 2-mercaptoethanol. BCR–ABL1-transformed ALL cells were propagated only for short periods of time and usually not longer than 2 months to avoid acquisition of additional genetic lesions during long-term cell culture.
In vivo transplantation of leukemia cells
For in vivo experiments, mouse ALL cells were labeled with retroviral firefly luciferase, selected based on antibiotic resistance (Blasticidin), and injected via tail into sublethally irradiated (300 cGy) NOD/SCID recipient mice. Engraftment was monitored using luciferase bioimaging (VIS 100 bioluminescence/optical imaging system; Xenogen). D-Luciferin (Xenogen) dissolved in PBS was injected intraperitoneally at a dose of 2.5 mg per mouse 15 min before measuring the light emission. All mouse experiments were subject to institutional approval by the University of California San Francisco Institutional Animal Care and Use Committee. 6-8 week old female NOD-SCID mice (Jackson Laboratories, ME) were randomly allocated into each treatment group. The minimal number of mice in each group was calculated by using the ‘cpower’ function in R/Hmisc package. No blinding was used.
C/EBPα-mediated reprogramming of pre-B cells into myeloid cells
The TetO-C/EBPα-GFP-construct was generated as a modification of the previously published TetO-CEBPα-tdTomato lentivirus27. Ptenfl/fl pre-B ALL cells were first transduced with rtTA, selected for puromycin resistance and subsequently infected with the TetO-C/EBPα-GFP-construct. C/EBPα was induced in the Ptenfl/fl pre-B ALL cells by addition of 2 μg/ml doxycycline (Sigma-Aldrich). Staining for cell surface antigens was done with directly conjugated antibodies against CD11b (PE; Mac1) and CD19 (PerCP-Cy5.5) EBioscience. To measure the effects of Pten-deletion in C/EBPα-reprogrammed pre-B ALL cells, deletion was induced with a tamoxifen-inducible Cre or an empty vector control (0.5 μmol/L). DAPI and 7AAD at 1 μg/ml were used as viability markers.
Array-based promoter methylation analysis using HELP
The HELP (HpaII tiny fragment enrichment by ligation mediated PCR) assay was performed as previously described. One microgram of high molecular weight DNA was digested overnight with isoschizomer enzymes HpaII and MspI respectively (NEB, Ipswich, MA). DNA fragments were purified using phenol/chloroform, resuspended in 10mM Tris-HCl pH 8.0, and used immediately to set up the ligation reaction with MspI/HpaII-compatible adapters and T4 DNA ligase. Ligation-mediated PCR was performed with enrichment for the 200 to 2,000 bp products and then submitted for labeling and hybridization onto a human HG_17 promoter custom-designed oligonucleotide array covering 25,626 HpaII amplifiable fragments within the promoters of the genes. To analysis, raw data (.pair) files were generated using NimbleScan software. Signal intensities at each HpaII amplifiable fragment were calculated as a robust (25% trimmed) mean of their component probe-level signal intensities. Any fragments found within the level of background MspI signal intensity, measured as 2.5 mean-absolute- deviation (MAD) above the median of random probe signals, were considered as “failed” probes and removed. A median normalization was performed on each array by subtracting the median log-ratio (HpaII/MspI) of that array (resulting in median log-ratio of 0 for each array).
Western blotting
Cells were lysed in CelLytic buffer (Sigma, St. Louis, MO) supplemented with PMSF, phosphatase and protease inhibitor cocktail (Pierce, Rockford, IL). 15 μg of protein mixture per sample were separated on NuPAGE (Invitrogen, Carlsbad, CA) 4-12% Bis-Tris gradient gels and transferred on PVDF membranes (Immobilion, Millipore, Temecula, CA). For the detection of mouse and human proteins by Western blot, primary antibodies were used together with the WesternBreeze immunodetection system (Invitrogen). Details of antibodies used are in Supplementary Table 4.
Flow cytometry
Antibodies used for flow cytometry experiments are listed in Supplementary Table 4. For apoptosis analyses, Annexin V, propidium iodide and 7-AAD (BD Biosciences) were used. Fortessa scanner was used for flow cytometry and FlowJo-V10 was used for data analysis.
Senescence-associated β-galactosidase assay
Senescence-associated β-galactosidase activity was performed on cytospin preparations. Briefly, a fixative solution (0.25% glutaraldehyde, 2% paraformaldehyde in PBS pH 5.5 for mouse cells) was prepared fresh. 1 g paraformaldehyde was dissolved in 50 ml PBS at pH 5.5 by heating followed by addition of 250 μl of a 50% stock glutaraldehyde solution. 1× X-gal staining solution (10 ml) was prepared as follows: 9.3 ml PBS/MgCl2, 0.5 ml 20× KC solution (that is, 820 mg K3Fe(CN)6 and 1,050 mg K4Fe(CN)6 × 3H2O in 25 ml PBS) and 0.25 ml 40× X-gal (that is, 40 mg 5-bromo-4-chloro-3-indolyl β-D-galactoside per milliliter of N,N-dimethylformamide) solutions were mixed. 200,000 cells per cytospin were used (700 rpm, 8 min). The fixative solution was pipetted onto cytospins and incubated for 10 min at room temperature, then washed twice for 5 min in PBS/MgCl2. Cytospin preparations were submerged in 1× X-gal solution, incubated overnight at 37 °C in a humidified chamber and washed twice in PBS. Images were acquired with a regular microscopy and percentage of stained cells calculated by counting.
Colony forming assay
Methylcellulose colony-forming assays were performed with 10,000 cells after transformation with Cre-ERT2 or ERT2 and 2 days of treatment with 0.5 μmol/l tamoxifen. Cells were resuspended in murine MethoCult medium (StemCell Technologies, Vancouver, BC, Canada) and cultured on dishes (3 cm in diameter) with an extra water supply dish to prevent evaporation. After 7 to 14 days, total numbers of colonies per plate were counted by microscopy.
Cell-cycle analysis
For cell-cycle analysis, the BrdU flow cytometry kit for cell-cycle analysis (BD Biosciences) was used according to manufacturer’s instructions. BrdU incorporation (APC-labelled anti-BrdU antibodies) was measured along with DNA content (7-amino-actinomycin-D) in fixed and permeabilized cells. The analysis was gated on viable cells that were identified based on scatter morphology.
Cell viability assay
100,000 cells per well were seeded in a volume of 100 μl medium on Optilux 96-well plate (BD Biosciences, San Jose, CA). Inhibitors were diluted in medium and added at the indicated concentration in a total culture volume of 120 μl. After 3 days in culture, 12 μl of Resazurin (R&D, Minneapolis, MN) was added to each well and incubated for 4 hours at 37 °C. The fluorescent signal was monitored using 535 nm excitation wavelength and 590 nm emission wavelength. Fold changes were calculated using baseline values of untreated cells as a reference (set to 100 percent). Each sample was measured in triplicates.
RNA purification and Expression analysis
Total RNA was purified using the RNeasy kit (Qiagen, Valencia, CA). RNA quality was checked by using an Agilent Bioanalyzer (Agilent Technologies, Santa Clara, CA). cDNA was generated from 5 μg of total RNA using a poly(dT) oligonucleotide and the SuperScript III Reverse Transcriptase (Invitrogen, Carlsbad, CA). Biotinylated cRNA was generated and fragmented according to the Affymetrix protocol and hybridized to mouse Gene 1.0 ST (Affymetrix, High Wycombe, UK). After scanning (GeneChip Scanner 3000 7G; Affymetrix) of the GeneChip arrays, the generated CEL files were imported to BRB Array Tool (http://linus.nci.nih.gov/BRB-ArrayTools.html) and processed using the RMA algorithm (Robust Multi-array Average) for normalization and summarization. The GEO accession number for the arrays is GSE34829.
Proteomic profiling
Proteomic profiling was performed using Reverse Phase Protein Array (RPPA) analysis23 on peripheral blood and bone marrow specimens from 192 patients with acute lymphocytic leukemia (ALL) including 192 samples at diagnosis and 12 paired diagnosis-relapse samples evaluated at The University of Texas M.D. Anderson Cancer Center (MDACC) between 1983 and 2007 (two from the 1980s, 45 from the 1990s). Samples were acquired during routine diagnostic assessments in accordance with the regulations and protocols (Lab 01-473 after September 2001, prior protocols for samples before then) approved by the Investigational Review Board (IRB) of MDACC. Informed consent was obtained in accordance with Declaration of Helinski. Samples were analyzed under and IRB-approved laboratory protocol (Lab 05-0654). Samples were selected for inclusion based on availability in the Department of Leukemia Sample Bank. Patients aged 15 or over were included; another 8 pediatric ALL cases were on the array but are not included in this analysis. Samples underwent Ficoll separation to yield a mononuclear fraction. The samples were normalized to a concentration of 1 × 104 cells/μL and a whole cell lysate prepared as previously described28. All protein samples were prepared from fresh cells on the day of collection. Of the 192 newly diagnosed ALL cases most were treated with the hyper CVAD regimen either alone (n = 91) or in combination with rituximab (n = 31) or a tyrosine kinase inhibitor (n = 28) or nelarabine (n =3). A modification of the Berlin-Frankfurt-Mannheim regimen was used in 23 cases. The remaining 16 received miscellaneous combination regimens in use over this time period.
Glucose, Lactate and ATP Measurements
Glucose and lactate levels were measured using the Amplex Red Glucose/Glucose Oxidase Assay Kit (Invitrogen) and the L-Lactate Assay Kit (Cayman Chemical), respectively, according to the manufacturers’ protocols. Glucose and lactate concentrations were measured in fresh and spent medium. Total ATP levels were measured using the ATP Bioluminescence Assay Kit CLS II (Roche) according to the manufacturer’s protocol. 106 cells per mL were seeded in fresh medium and treated as indicated in figure legends. Relative levels of glucose consumed, lactate produced and total ATP are shown, and all values were normalized to number of viable cells. Inhibitors used in this study are listed in Supplementary Table 5.
Reconstitution model for pre-BCR signaling
Pre-B cells from Blnk−/− Rag2−/− Igll1−/− triple knockout transgenic mice26 were engineered to express either a non-auto-reactive (μ-HCNA), an autoreactive pre-BCR (μ-HCAuto) or an empty vector control (EV). The cells were transduced with a tamoxifen-inducible (Blnk-ER) form of the pre-BCR linker molecule Blnk (also known as SLP-65). Addition of tamoxifen (Tam) releases Blnk from its complex with HSP90 and allows for assembly of the pre-BCR signaling complex within minutes.
Statistical analysis
Unpaired, two-tailed Student’s t test was used to compare colony number, S phase percentage and frequency of cellular senescence between different groups. Two-sided Mann-Whitney Wilcoxon test was used to compare methylation values of ALL vs. normal pre-B or B-cell lymphoma groups and RPPA data of newly diagnosed adult ALL vs. T cell lineage ALL or mature B cell lymphoma , using R version 2.14.0. Mantel-Cox log rank test (two-sided) was used to compare survival data between different groups. R package “survival” version 2.35-8 was used for the survival analysis.
Supplementary Material
Acknowledgments
We would like to thank L. M. Staudt (Bethesda, MD), D. A. Fruman (Irvine, CA), T. Kurosaki (Osaka, Japan), S. Li (Boston, MA) and A. Weiss (San Francisco, CA) for comments and critical discussion of this study. This work is supported by grants from the National Institute of Health and National Cancer Institute through R01CA137060, R01CA139032, R01CA157644, R01CA169458, R01CA172558 (to M.M.), the William Lawrence and Blanche Hughes Foundation, the California Institute for Regenerative Medicine (CIRM; TR2-01816 to MM) and Leukaemia and Lymphoma Research (M.M.). T.G.G. is the recipient of a Research Scholar Award from the American Cancer Society (RSG-12-257-01-TBE) and an Established Investigator Award from the Melanoma Research Alliance (20120279), and is supported by NIH/National Center for Advancing Translational Science (NCATS) UCLA CTSI Grant Number UL1TR000124. M.M. is a Scholar of the Leukemia and Lymphoma Society and a Senior Investigator of the Wellcome Trust.
Footnotes
The authors have no conflicting financial interests.
All mouse experiments were subject to institutional approval by the University of California San Francisco Institutional Animal Care and Use Committee.
Author Contributions
S.S., M.M., designed experiments and interpreted data. M.M. conceived the study, obtained funding, coordinated collaborations and wrote the paper. S.S., L.N.C., M.B., V.C., K.N.C. and H.G. performed experiments and analyzed data. Y.H. Q., A.M., and S.M.K. provided and characterized patient samples or cell lines and clinical outcome data. H. W. provided important reagents and mouse samples. M.Dv.M., T.E., A.H., G.C., S.M.K., T.G.G. and H.J. provided conceptual input to the design of the study.
References
- 1.Osmond DG. Proliferation kinetics and the lifespan of B cells in central and peripheral lymphoid organs. Curr Opin Immunol. 1991;3:179–185. doi: 10.1016/0952-7915(91)90047-5. [DOI] [PubMed] [Google Scholar]
- 2.Wardemann H, et al. Predominant autoantibody production by early human B cell precursors. Science.; 2003;301:1374–1377. doi: 10.1126/science.1086907. [DOI] [PubMed] [Google Scholar]
- 3.Keenan RA, et al. Censoring of autoreactive B cell development by the pre-B cell receptor. Science.; 2008;321:696–699. doi: 10.1126/science.1157533. [DOI] [PubMed] [Google Scholar]
- 4.Srinivasan L, et al. PI3 kinase signals BCR-dependent mature B cell survival. Cell. 2009;139:573–586. doi: 10.1016/j.cell.2009.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ramadani F, et al. The PI3K isoforms p110alpha and p110delta are essential for pre-B cell receptor signaling and B cell development. Sci Signal. 2010;3:ra60. doi: 10.1126/scisignal.2001104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deau MC, et al. A human immunodeficiency caused by mutations in the PIK3R1 gene. J Clin Invest.; 2014;124:3923–3928. doi: 10.1172/JCI75746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vivanco I, Sawyers CL. The phosphatidylinositol 3-Kinase AKT pathway in human cancer. Nat. Rev. Cancer. 2002;2:489–501. doi: 10.1038/nrc839. [DOI] [PubMed] [Google Scholar]
- 8.Futreal PA, et al. A census of human cancer genes. Nat Rev Cancer. 2004;4:177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gutierrez A, et al. High frequency of PTEN, PI3K, and AKT abnormalities in T-cell acute lymphoblastic leukemia. Blood. 2009;114:647–50. doi: 10.1182/blood-2009-02-206722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lesche R, et al. Cre/loxP-mediated inactivation of the murine Pten tumor suppressor gene. Genes. New York 2000. 2002;32:148–149. doi: 10.1002/gene.10036. [DOI] [PubMed] [Google Scholar]
- 11.Zhang J, et al. Key pathways are frequently mutated in high-risk childhood acute lymphoblastic leukemia: a report from the Children's Oncology Group. Blood. 2011;118:3080–7. doi: 10.1182/blood-2011-03-341412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guo B, Kato RM, Garcia-Lloret M, Wahl MI, Rawlings DJ. Engagement of the human pre-B cell receptor generates a lipid raft-dependent calcium signaling complex. Immunity. 2000;13:243–53. doi: 10.1016/s1074-7613(00)00024-8. [DOI] [PubMed] [Google Scholar]
- 13.Okada T, Maeda A, Iwamatsu A, Gotoh K, Kurosaki T. BCAP: the tyrosine kinase substrate that connects B cell receptor to phosphoinositide 3-kinase activation. Immunity. 2000;13:817–27. doi: 10.1016/s1074-7613(00)00079-0. [DOI] [PubMed] [Google Scholar]
- 14.Anzelon AN, Wu H, Rickert RC. Pten inactivation alters peripheral B lymphocyte fate and reconstitutes CD19 function. Nat Immunol. 2003;4:287–94. doi: 10.1038/ni892. [DOI] [PubMed] [Google Scholar]
- 15.Aiba Y, Kameyama M, Yamazaki T, Tedder TF, Kurosaki T. Regulation of B-cell development by BCAP and CD19 through their binding to phosphoinositide 3-kinase. Blood. 2008;111:1497–1503. doi: 10.1182/blood-2007-08-109769. [DOI] [PubMed] [Google Scholar]
- 16.Tesio M, et al. Pten loss in the bone marrow leads to G-CSF-mediated HSC mobilization. J Exp Med. 2013;210:2337–49. doi: 10.1084/jem.20122768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peng C, et al. PTEN is a tumor suppressor in CML stem cells and BCR-ABL-induced leukemias in mice. Blood. 2010;115:626–35. doi: 10.1182/blood-2009-06-228130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li J, et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science.; 1997;275:1943–1947. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- 19.Gutierrez A, et al. Pten mediates Myc oncogene dependence in a conditional zebrafish model of T cell acute lymphoblastic leukemia. J Exp Med. 2011;208:1595–603. doi: 10.1084/jem.20101691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lenz G, et al. Molecular subtypes of diffuse large B-cell lymphoma arise by distinct genetic pathways. Proc Natl Acad Sci U S A. 2008;105:13520–5. doi: 10.1073/pnas.0804295105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miletic AV, et al. Coordinate suppression of B cell lymphoma by PTEN and SHIP phosphatases. J Exp Med. 2010;207:2407–20. doi: 10.1084/jem.20091962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pfeifer M, et al. PTEN loss defines a PI3K-AKT pathway-dependent germinal center subtype of diffuse large B-cell lymphoma. Proc Natl Acad Sci U S A. 2013;110:12420–5. doi: 10.1073/pnas.1305656110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tibes R, et al. Reverse phase protein array: validation of a novel proteomic technology and utility for analysis of primary leukemia specimens and hematopoietic stem cells. Mol. Cancer Ther. 2006;5:2512–2521. doi: 10.1158/1535-7163.MCT-06-0334. [DOI] [PubMed] [Google Scholar]
- 24.Roman-Gomez J, et al. Lack of CpG island methylator phenotype defines a clinical subtype of T-cell acute lymphoblastic leukemia associated with good prognosis. J Clin Oncol. 2005;23:7043–9. doi: 10.1200/JCO.2005.01.4944. [DOI] [PubMed] [Google Scholar]
- 25.Chen Z, et al. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature. 2005;436:725–30. doi: 10.1038/nature03918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Köhler F, et al. Autoreactive B cell receptors mimic autonomous pre-B cell receptor signaling and induce proliferation of early B cells. Immunity. 2008;29:912–921. doi: 10.1016/j.immuni.2008.10.013. [DOI] [PubMed] [Google Scholar]
- 27.Xie H, Ye M, Feng R, Graf T. Stepwise reprogramming of B cells into macrophages. Cell. 2004;117:663–676. doi: 10.1016/s0092-8674(04)00419-2. [DOI] [PubMed] [Google Scholar]
- 28.Shackelford DB, et al. mTOR and HIF-1alpha-mediated tumor metabolism in an LKB1 mouse model of Peutz-Jeghers syndrome. Proc Natl Acad Sci U S A. 2009;106:11137–42. doi: 10.1073/pnas.0900465106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Düvel K, et al. Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol Cell. 2010;39:171–183. doi: 10.1016/j.molcel.2010.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rosivatz E, et al. A small molecule inhibitor for phosphatase and tensin homologue deleted on chromosome 10 (PTEN) ACS Chem. Biol. 2006;1:780–790. doi: 10.1021/cb600352f. [DOI] [PubMed] [Google Scholar]
- 31.Chen Z, et al. Signalling thresholds and negative B-cell selection in acute lymphoblastic leukaemia. Nature. 2015 May 21;521(7552):357–61. doi: 10.1038/nature14231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shojaee S, et al. Erk Negative Feedback Control Enables Pre-B Cell Transformation and Represents a Therapeutic Target in Acute Lymphoblastic Leukemia. Cancer Cell. 2015 Jul 13;28(1):114–28. doi: 10.1016/j.ccell.2015.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kharas MG, et al. Ablation of PI3K blocks BCR-ABL leukemogenesis in mice, and a dual PI3K/mTOR inhibitor prevents expansion of human BCR-ABL+ leukemia cells. J Clin Invest. 2008;118:3038–50. doi: 10.1172/JCI33337. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.