Abstract
Background
CD200 is reported to be involved in tumor progression and can serve as a prognostic marker in several cancers. The purpose of this study was to evaluate the prognostic significance of CD200 in cutaneous squamous cell carcinoma (CSCC).
Material/Methods
The relative mRNA and protein expression of CD200 in the tumor tissues and corresponding normal tissues of 102 CSCC patients were detected by quantitative real-time polymerase chain reaction (qRT-PCR) and Western blot analysis, respectively. The chi-square test was used to analyze the association between CD200 expression and clinical features of CSCC patients. In addition, the overall survival of the patients according to the expression level of CD200 was estimated by Kaplan-Meier analysis and the prognostic significance of the gene was analyzed by Cox regression analysis.
Results
Increased expression of CD200 was detected in the tumor tissues compared with the corresponding normal tissues both at mRNA and protein level. And CD200 expression level was associated with tumor differentiation grade (P=0.041) and clinical stage (P=0.004). Patients with high expression level of CD200 had a shorter overall survival than those with low expression (31.3 months vs. 41.9 months) and there was a significant difference between them (log-rank test, P<0.001). Cox regression analysis indicated that CD200 could be an independent marker for the prognosis of CSCC.
Conclusions
CD200 is up-regulated and may be a novel biomarker for the prognosis in CSCC, and it may be a potential therapeutic target for CSCC.
MeSH Keywords: Antigens, CD; Prognosis; Skin Neoplasms
Background
Cutaneous squamous cell carcinoma (CSCC) is one of the most common skin cancers in the world, accounting for approximately 20% of all cutaneous malignancies [1]. The cancer often occurs on the highly visible areas of the skin, such as the head, neck, and face [2]. CSCC in most patients can be completely eradicated with surgery or other dermatological treatments, but some cases suffer with a substantial risk of metastasis and high mortality [3–5]. Therefore, a number of studies have concentrated on exploring of the molecular markers, which would provide responsibly prognostic information for tumor treatments. However, the specific biomarkers for CSCC prognosis, which could provide significantly clinical information, remain substantially limited [2].
CD200, also known as OX-2, is a member of the Ig superfamily (IgSF) of proteins and is important in the regulation and fine control of immune responses, as well as in helping to maintain immune homeostasis [6,7]. It is a membrane glycoprotein and plays a role in inducing signaling in cells expressing its cognate cell surface receptor, the CD200 receptor (CD200R) [7]. In CD200-deficient mouse models, hyper-activation of macrophages was observed and the immune response to autoimmune disease was also enhanced [8]. Recently, the abnormal expression of CD200 has been detected in several types of cancer, such as plasma cell myeloma, head and neck squamous cell carcinoma, and melanoma cells [9–11]. Daniel et al. reported that the expression level of CD200 in tumor tissues was higher than that in normal tissues in CSCC patients and that the gene might accelerate tumor progression [12]. However, the clinical significance of CD200 in CSCC has rarely been reported.
In this study we aimed to investigate the expression level of CD200 in clinical CSCC specimens and normal tissues. Moreover, the relationship between CD200 expression and clinical factors of patients with CSCC was analyzed. The clinical significance of CD200 in CSCC patients was estimated via Kaplan-Meier and Cox regression analysis.
Material and Methods
Patients and tissue specimens
We collected 102 CSCC patients who were confirmed by pathological and clinical diagnoses in General Hospital of Beijing Military Region from October 2009 to February 2015. The study was approved by the Ethics Committee of the hospital. None of the patients had received any chemotherapy or radiotherapy before the surgery. Written informed consents were signed by all participants in advance.
Tumor tissues and corresponding normal tissues of the patients were obtained and frozen in liquid nitrogen immediately, then stored at −80°C until use. The clinicopathologic characteristics of patients with CSCC, including age, sex, tumor size, tumor differentiation grade, and clinical stage, were recorded in a database. All of the patients were arranged for 5-year follow-up and the information about the clinical features and survival were collected for analyzing the clinical significance of CD200.
Total RNA extraction and qRT-PCR analysis
Total RNA was extracted from all the specimens by Trizol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. DNAse was used for the residuary DNA in the extracted RNA. The concentration of the RNA was detected by UV absorbance (A260/A280), and 1% agarose gel electrophoresis was used for evaluating its quality. Reverse transcription was conducted to synthesize the first chain of cDNA using Prime Scrip RT reagent kit (Takara, Dalian, China). Then RT-PCR reaction was performed with SYBR Green assay (Takara, Dalian, China). GAPDH acted as internal control and 2−ΔΔCt method was used to calculate the relative mRNA expression of CD200 data analyses. The sequences of the primers used in this study are listed in Table 1.
Table 1.
The primer sequences of CD200 and GAPDH in RT-PCR reaction.
| Genes | Sequences | |
|---|---|---|
| CD200 | Forward | GAGCTCCAGGCGCACATCCGC |
| Reverse | TGCGGATGTGCGCCTGGAGCTC | |
| GAPDH | Forward | GAAGGTGAAGCTCGCAGTC |
| Reverse | GAAGATGGTGATGGGATTTC | |
Western blot analysis
Total protein was extracted from all samples and separated by 10% SDS-PAGE gels. Then the brands were transferred to PVDF membrane (Bio-Rad, CA, USA) after blocking for 1 h with 5% non-fat milk. Subsequently, the membranes were incubated with primary monoclonal antibodies against CD200 or GAPDH (at a 1:1000 dilution) for 1 h and washed 6 times over a period of 1 h in TBS/Tween-20. Blots were then incubated at 1:10000 with anti-goat, 500 μg/0.5ml IgG HRP (Jackson ImmunoResearch) in blocking buffer for 1 h and washed 6 times over a period of 1 h in TBS/Tween-20. Finally, the membranes were developed using ECL and exposed to X-ray film for 30 s.
Statistical analysis
All data are showed as mean ± standard deviation (SD). The difference between the 2 groups was analyzed by Students’ t test. Association between CD200 expression and the clinical features were estimated by chi-square test. Kaplan-Meier analysis was used for evaluating the overall survival of the patients according to the CD200 expression. The prognostic significance of the gene was analyzed by Cox regression analysis. All of the statistical analyses were performed in SPSS 18.0 software (SPSS Inc, IL, USA), and Sigma Plot software (Systat Software Inc., CA, USA) was used for drawing. P<0.05 was considered to be statistically significant.
Results
The expression level of CD200 was increased in tumor tissues
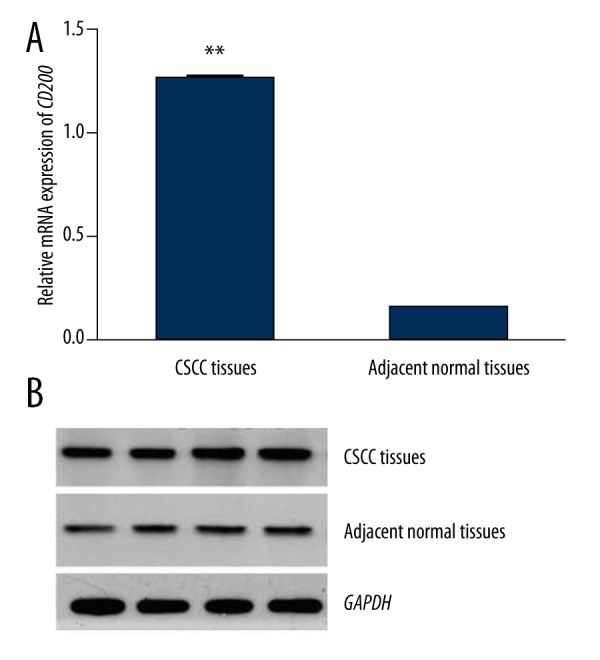
Relative mRNA and protein expression of CD200 was detected by qRT-PCR and Western blot analysis, respectively. As shown in Figure 1, relative mRNA expression of CD200 was significantly higher in tumor tissues than in adjacent normal tissues (P<0.001). The protein expression of CD200 was also increased in tumor tissues compared to adjacent normal tissues (P<0.001, Figure 2)
Figure 1.

Relative mRNA expression of CD200 in tumor tissues and corresponding normal tissues of CSCC patients (GAPDH as normalized control). CD200 expression was higher in tumor tissues than in adjacent normal tissues (P<0.001).
Figure 2.
Relative protein expression of CD200 in tumor tissues and corresponding normal tissues of CSCC patients. CD200 protein expression was higher in tumor tissues than in adjacent normal tissues (P<0.001).
Correlation between CD200 expression and clinicopathologic characteristics of CSCC patients
All clinical data are listed in Table 2. The 102 CSCC patients included 39 males and 63 females, with average age of 57.4 years. The patients were divided into a high expression group and a low expression group according to the median expression level of CD200. The results indicated that CD200 expression was significantly associated with tumor grade of differentiation (P=0.041) and clinical stage (P=0.004). However, no distinct differences between sex, age, and CD200 expression were observed (Table 2).
Table 2.
Association between CD200 expression and the clinical features of CSCC patients.
| Characteristics | Cases (n) | CD200 expression | χ2 | P | |
|---|---|---|---|---|---|
| High (n, %) | Low (n, %) | ||||
| Gender | 1.533 | 0.216 | |||
| Male | 39 | 18 (46.2%) | 21 (53.8%) | ||
| Female | 63 | 37 (58.7%) | 26 (41.3%) | ||
| Age | 0.532 | 0.466 | |||
| ≥55 | 59 | 30 (50.8%) | 29 (49.2%) | ||
| <55 | 43 | 25 (58.1%) | 18 (41.9%) | ||
| Tumor size | 0.710 | 0.399 | |||
| ≥5 cm | 54 | 37 (68.5%) | 17 (31.5%) | ||
| <5 cm | 48 | 18 (37.5%) | 30 (62.5%) | ||
| Tumor differentiation grade | 4.156 | 0.041 | |||
| Well to moderate | 67 | 41 (61.2%) | 26 (38.8%) | ||
| Poor | 35 | 14 (40.0%) | 21 (60.0%) | ||
| Clinical stage | 8.447 | 0.004 | |||
| I+II | 45 | 17 (37.8%) | 28 (62.2%) | ||
| III+IV | 57 | 38 (66.7%) | 19 (33.3%) | ||
Association between CD200 expression and overall survival of patients with CSCC
To explore the prognostic value of CD200, we conducted 5-year follow-up with all patients. According to Kaplan-Meier analysis, patients with high expression level of CD200 had a shorter overall survival than those with low expression (31.3 months vs. 41.9 months, log-rank test, P<0.001, Figure 3). Then a univariate and multivariate analysis were performed to estimate the prognostic value of CD200 using Cox regression analysis. The result demonstrated that high expression of CD200 (HR=4.558, 95%CI=2.397–8.666, P=0.000), tumor differentiation grade (HR=1.833, 95%CI=1.036–3.242, P=0.037), and clinical stage (HR=2.374, 95%CI=1.369–4.118, P=0.002) were tightly related to the prognosis of CSCC and they could act as independent prognostic biomarkers in CSCC.
Figure 3.

Overall survival of CSCC patients according to the expression of CD200. Patients with high level of CD200 had a shorter overall survival time than those with low-expression. There was a significant difference between the groups (log-rank test, P<0.001).
Discussion
To improve the clinical outcomes of CSCC patients, a growing number of studies have focused on looking for the molecular markers associated with cancer prognosis. LRIG-1 was proved to be an excellent prognostic indicator in CSCC [13]. P300, a member of the histone acetyltransferase family of transcriptional coactivators, was reported to be an independent biomarker in the prognosis of CSCC by Chen et al. [2]. Some members of the HER family might be potential therapeutic and prognostic markers in CSCC due to its association with tumor progression and lymph node metastasis [14]. In this study, we engaged in discovering more molecular markers to better predict the prognosis of patients with CSCC.
CD200 is a cell surface glycoprotein that can inhibit alloimmune and autoimmune responses via its receptor CD200R [15–17]. The immune suppression capacity of CD200 could maintain homeostasis in various tissues, such as lymphoid cells, the nervous system, and skin [16,18,19]. It might also help protect migratory neoplastic cells [15]. In previous studies, CD200 was reported to take part in several cancers. For instance, CD200 was over-expressed and correlated with progression of metastatic melanoma as well as acting as a potential therapeutic target [20]. Siva et al. reported that CD200 was expressed in ovarian cancer, melanoma, neuroblastoma, and renal carcinoma, thereby potentially suppressing anti-tumor immune responses, but it was absent in prostate, lung, breast, astrocytoma, or glioblastoma [21]. CD200 co-expression with stem cell markers was found in prostate, breast, brain, and colon cancers [22]. According to Podnos et al., CD200 expression could prevent the delivery of an immunosuppressive signal and influence metastatic growth, suggesting it as a potential therapy for breast cancer [23]. Zhang et al. detected the expression of CD200 in acute myeloid leukemia and proved that its antigen expression was related to the poor prognosis of this disease [24]. The over-expression of CD200 is linked with expansion of suppressive Treg cells and elevation of cytokines, and plays a vital role in the progression of multiple myeloma [25]. The diagnostic and prognostic value of CD200 was also confirmed in plasma cell myeloma [26]. CD200 protein was verified to be a prognostic indictor for the poor prognosis of multiple myeloma [24]. Using gene expression data from the databases, Jerome et al. concluded that CD200 expression is associated with the progression of various cancers, such as bladder cancer, lung cancer, chronic myelogenous leukemia, breast cancer, primary melanoma, metastatic melanoma, prostatic intraepithelial neoplasia, and prostate carcinoma [28]. However, the clinical significance of CD200 in CSCC is still unclear.
In the present study, we detected the expression of CD200 both at mRNA and protein level in CSCC tissues and adjacent normal tissues. The over-expression of CD200 at 2 levels was found, which indicated CD200 might be an oncogene in CSCC. Based on this result, we explored wether it was involved in the progression of CSCC. Finally, tumor differentiation grade and clinical stage were verified to tightly influence the expression of CD200. All of these results suggest that CD200 is associated with tumor progression in CSCC.
Because the prognostic value of CD200 was also verified in many malignancies, we investigated the prognostic value of CD200 in CSCC. After 5-year follow-up, Kaplan-Meier analysis of the survival rate of patients with CSCC in our study indicated that patients with low CD200 expression had a longer survival compared to those with high expression (log-rank test, P<0.001). A multivariate analysis based on univariate analysis validated the prognostic value of CD200 expression adjusted for the clinicopathologic characteristics, and indicated that CD200, as well as differentiation grade and clinical stage, could serve as independent predictors for the prognosis of CSCC.
Conclusions
The expression level of CD200 in tumor tissues is up-regulated compared to that in adjacent normal tissues. Its expression is significantly affected by tumor differentiation grade and clinical stage. Furthermore, the over-expression of CD200 is related to shorter survival time of patients with CSCC. It seems that CD200 is a potential prognostic predictor in CSCC.
Table 3.
The multivariate analysis adjusted for clinical factors for estimating the prognostic value of CD200 in CSCC.
| Characteristics | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|
| HR | 95%CI | P | HR | 95%CI | P | |
| CD200 (high vs. low) | 5.512 | 3.035–10.011 | 0.000 | 4.558 | 2.397–8.666 | 0.000 |
| Gender (male vs. female) | 1.474 | 0908–2.395 | 0.117 | – | – | – |
| Age (≥55 vs. <55) | 0.932 | 0.579–1.500 | 0.773 | – | – | – |
| Tumor size (>5 cm vs. <5 cm) | 1.312 | 0.822–2.094 | 0.256 | – | – | – |
| Tumor differentiation grade (well vs. poor) | 2.441 | 1.456–4.091 | 0.001 | 1.833 | 1.036–3.242 | 0.037 |
| Clinical stage (I+II vs. III+IV) | 3.239 | 1.962–5.347 | 0.000 | 2.374 | 1.369–4.118 | 0.002 |
Footnotes
Source of support: Departmental sources
References
- 1.Kwa RE, Campana K, Moy RL. Biology of cutaneous squamous cell carcinoma. J Am Acad Dermatol. 1992;26(1):1–26. doi: 10.1016/0190-9622(92)70001-v. [DOI] [PubMed] [Google Scholar]
- 2.Chen MK, Cai MY, Luo RZ, et al. Overexpression of p300 correlates with poor prognosis in patients with cutaneous squamous cell carcinoma. Br J Dermatol. 2015;172(1):111–19. doi: 10.1111/bjd.13226. [DOI] [PubMed] [Google Scholar]
- 3.Martorell-Calatayud A, Sanmartin Jimenez O, Cruz Mojarrieta J, Guillen Barona C. Cutaneous squamous cell carcinoma: defining the high-risk variant. Actas Dermosifiliogr. 2013;104(5):367–79. doi: 10.1016/j.adengl.2011.12.012. [DOI] [PubMed] [Google Scholar]
- 4.Cerroni L. Cutaneous squamous-cell carcinoma. New Engl J Med. 2001;345(4):296. author reply 296–97. [PubMed] [Google Scholar]
- 5.Yanofsky VR, Mercer SE, Phelps RG. Histopathological variants of cutaneous squamous cell carcinoma: a review. J Skin Cancer. 2011;2011:210813. doi: 10.1155/2011/210813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Talebian F, Bai XF. The role of tumor expression of CD200 in tumor formation, metastasis and susceptibility to T lymphocyte adoptive transfer therapy. Oncoimmunology. 2012;1(6):971–73. doi: 10.4161/onci.20034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Estep RD, Rawlings SD, Li H, et al. The rhesus rhadinovirus CD200 homologue affects immune responses and viral loads during in vivo infection. J Virol. 2014;88(18):10635–54. doi: 10.1128/JVI.01276-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoek RM, Ruuls SR, Murphy CA, et al. Down-regulation of the macrophage lineage through interaction with OX2 (CD200) Science. 2000;290(5497):1768–71. doi: 10.1126/science.290.5497.1768. [DOI] [PubMed] [Google Scholar]
- 9.Douds JJ, Long DJ, Kim AS, Li S. Diagnostic and prognostic significance of CD200 expression and its stability in plasma cell myeloma. J Clin Pathol. 2014;67(9):792–96. doi: 10.1136/jclinpath-2014-202421. [DOI] [PubMed] [Google Scholar]
- 10.Jung YS, Vermeer PD, Vermeer DW, et al. CD200: association with cancer stem cell features and response to chemoradiation in head and neck squamous cell carcinoma. Head Neck. 2015;37(3):327–35. doi: 10.1002/hed.23608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Talebian F, Liu JQ, Liu Z, et al. Melanoma cell expression of CD200 inhibits tumor formation and lung metastasis via inhibition of myeloid cell functions. PloS One. 2012;7(2):e31442. doi: 10.1371/journal.pone.0031442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Belkin DA, Mitsui H, Wang CQ, et al. CD200 upregulation in vascular endothelium surrounding cutaneous squamous cell carcinoma. JAMA Dermatol. 2013;149(2):178–86. doi: 10.1001/jamadermatol.2013.1609. [DOI] [PubMed] [Google Scholar]
- 13.Tanemura A, Nagasawa T, Inui S, Itami S. LRIG-1 provides a novel prognostic predictor in squamous cell carcinoma of the skin: immunohistochemical analysis for 38 cases. Dermatol Surg. 2005;31(4):423–30. doi: 10.1111/j.1524-4725.2005.31108. [DOI] [PubMed] [Google Scholar]
- 14.Vinicius de LV, Scapulatempo C, Perpetuo NM, et al. Prognostic and risk factors in patients with locally advanced cutaneous squamous cell carcinoma of the trunk and extremities. J Skin Cancer. 2011;2011:420796. doi: 10.1155/2011/420796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stumpfova M, Ratner D, Desciak EB, et al. The immunosuppressive surface ligand CD200 augments the metastatic capacity of squamous cell carcinoma. Cancer Res. 2010;70(7):2962–72. doi: 10.1158/0008-5472.CAN-09-4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wright GJ, Puklavec MJ, Willis AC, et al. Lymphoid/neuronal cell surface OX2 glycoprotein recognizes a novel receptor on macrophages implicated in the control of their function. Immunity. 2000;13(2):233–42. doi: 10.1016/s1074-7613(00)00023-6. [DOI] [PubMed] [Google Scholar]
- 17.Gorczynski R, Chen Z, Kai Y, et al. CD200 is a ligand for all members of the CD200R family of immunoregulatory molecules. J Immunol. 2004;172(12):7744–49. doi: 10.4049/jimmunol.172.12.7744. [DOI] [PubMed] [Google Scholar]
- 18.Meuth SG, Simon OJ, Grimm A, et al. CNS inflammation and neuronal degeneration is aggravated by impaired CD200-CD200R-mediated macrophage silencing. J Neuroimmunol. 2008;194(1–2):62–69. doi: 10.1016/j.jneuroim.2007.11.013. [DOI] [PubMed] [Google Scholar]
- 19.Akman-Karakas A, Yalcin AD, Koc S, et al. There might be a role for CD200 in the pathogenesis of autoimmune and inflammatory skin disorders. Med Sci Monit. 2013;19:888–91. doi: 10.12659/MSM.889624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Petermann KB, Rozenberg GI, Zedek D, et al. CD200 is induced by ERK and is a potential therapeutic target in melanoma. J Clin Invest. 2007;117(12):3922–29. doi: 10.1172/JCI32163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Siva A, Xin H, Qin F, et al. Immune modulation by melanoma and ovarian tumor cells through expression of the immunosuppressive molecule CD200. Cancer Immunol Immunother. 2008;57(7):987–96. doi: 10.1007/s00262-007-0429-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kawasaki BT, Mistree T, Hurt EM, et al. Co-expression of the toleragenic glycoprotein, CD200, with markers for cancer stem cells. Biochem Biophys Res Commun. 2007;364(4):778–82. doi: 10.1016/j.bbrc.2007.10.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Podnos A, Clark DA, Erin N, et al. Further evidence for a role of tumor CD200 expression in breast cancer metastasis: decreased metastasis in CD200R1KO mice or using CD200-silenced EMT6. Breast Cancer Res Treat. 2012;136(1):117–27. doi: 10.1007/s10549-012-2258-3. [DOI] [PubMed] [Google Scholar]
- 24.Zhang XL, Shen AL, Guo R, et al. [Expression Characteristics of CD200 in Acute Myeloid Leukemia and Its Clinical Significance]. Journal of experimental hematology/Chinese Association of Pathophysiology. 2014;22(6):1531–34. doi: 10.7534/j.issn.1009-2137.2014.06.006. [in Chinese] [DOI] [PubMed] [Google Scholar]
- 25.Aref S, Azmy E, El-Gilany AH. Upregulation of CD200 is associated with regulatory T cell expansion and disease progression in multiple myeloma. Hematol Oncol. 2015 doi: 10.1002/hon.2206. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 26.Douds JJ, Long DJ, Kim AS, et al. Diagnostic and prognostic significance of CD200 expression and its stability in plasma cell myeloma. J Clin Pathol. 2014;67(9):792–96. doi: 10.1136/jclinpath-2014-202421. [DOI] [PubMed] [Google Scholar]
- 27.Vela-Ojeda J, García-Ruiz EMA, Padilla-González Y, et al. [CD200 protein, bad prognostic in patients with multiple myeloma]. Rev Med Inst Mex Seguro Soc. 2015;53(4):438–43. [in Spanish] [PubMed] [Google Scholar]
- 28.Moreaux J, Veyrune JL, Reme T, et al. CD200: a putative therapeutic target in cancer. Biochem Biophys Res Commun. 2008;366(1):117–22. doi: 10.1016/j.bbrc.2007.11.103. [DOI] [PubMed] [Google Scholar]


