Abstract
FGF23, CYP24A1 and VDR altogether play a significant role in genetic susceptibility to chronic kidney disease (CKD). Identification of possible causative mutations may serve as therapeutic targets and diagnostic markers for CKD. Thus, we adopted both sequence and sequence-structure based SNP analysis algorithm in order to overcome the limitations of both methods. We explore the functional significance towards the prediction of risky SNPs associated with CKD. We assessed the performance of four widely used pathogenicity prediction methods. We compared the performances of the programs using Mathews correlation Coefficient ranged from poor (MCC = 0.39) to reasonably good (MCC = 0.42). However, we got the best results for the combined sequence and structure based analysis method (MCC = 0.45). 4 SNPs from FGF23 gene, 8 SNPs from VDR gene and 13 SNPs from CYP24A1 gene were predicted to be the causative agents for human diseases. This study will be helpful in selecting potential SNPs for experimental study from the SNP pool and also will reduce the cost for identification of potential SNPs as a genetic marker.
Keywords: Chronic kidney disease, SNP analysis, Combined sequence and sequence-structure based methods, FGF23, CYP24A1, VDR
1. Introduction
In the past few decades, enormous implementation has been made to complete human genome and high throughput genome analysis technologies. However, documentation of specific causative genetic markers could trigger common complex traits viz. diabetes, hypertension, CKD etc., which continue to pose a major challenge. Different human genome variations such as single nucleotide polymorphisms (SNPs), microsatellites and variable number of tandem repeats (VNTRs) are used as genetic markers for many diseases (Prasad and Thelma, 2007).
FGF23, CYP24A1 and VDR genes play an important role in the pathogenesis of CKD (Cozzolino and Malindretos, 2010, Petkovich and Jones, 2011, Wahl and Wolf, 2012), tumoral calcinosis (Farrow et al., 2011) and cancer (Slattery, 2007, Sakaki et al., 2014). FGF23 is the recently discovered regulator of phosphate and mineral metabolism. FGF23 mainly regulates the renal phosphate excretion. FGF23 levels are increased among CKD patients and many cross sectional studies demonstrated that an inverse relationship has been observed in glomerular filtration rate (GFR) with an inverse kidney function (Liu and Quarles, 2007, Damasiewicz et al., 2011, Wan et al., 2012). The increased level of FGF23 leads to the over expression of CYP24A1 mRNA in the kidney (Bai et al., 2003, Larsson et al., 2004, Shimada et al., 2005, Inoue et al., 2005, Perwad et al., 2007). The CYP24A1 enzyme is responsible for the catabolism of 25 hydroxyvitamin D3 (25–OHD3) and its hormonal form, 1,25-dihydroxyvitamin D3(1,25-(OH)2D3) into 24-hydroxylated products for excretion. The 1,25(OH)2D3 is the target hormone to induce the VDR expression (Petkovich and Jones, 2011). Further, the active form of the VDR mediates a wide variety of biological actions such as cell proliferation and differentiation, calcium homeostasis, immune modulation, neurological functions and bone mineralization (Norman, 2008). The over-expression of the CYP24A1 leads to VDR dysfunction as it over metabolized the 25OHD3 and 1,25(OH)2D3. Thus, CKD patients ought to experience vitamin D deficiency and subsequent osteoporosis (Loh et al., 2012). Fig. 1 shows the schematic representation of the disease mechanism.
Fig. 1.

The schematic representation of the disease mechanism.
Discrepancies are observed while establishing the treatment/diagnostic targets for complex multifactorial traits like CKD, hypertension by single locus analysis. This problem is mainly due to the small sample size, varying effects of several disease-predisposing variants, population structure, gene–environment interactions, poor study design or less number of polymorphisms selected for the analysis. These are some of the important factors which can hamper the detection of modest contribution of an individual locus to a trait such as hypertension and CKD. Haplotype based analysis explored different variants segregating at particular loci which will be helpful in studying complex disease. But still it is a daunting task to consider all genetic and non-genetic information in the analytic process. A single nucleotide polymorphism (SNP) is a nucleotide (A, T, G and C) change in the genome, which leads to genetic variation, occurring at each 100–300 bases along the 3-billion-base human genome, even though their density varies between regions (De Alencar and Lopes, 2010). A non-synonymous SNP (nsSNP) is known as a single base change in the coding region of a gene. This change leads to the amino acid substitutions (AAS) in the corresponding protein product. If SNP occurs in a primary amino acid sequence, the protein structure and function might be altered, which could lead to drastic phenotype and drug effect changes (Mah et al., 2011).
Experimental studies are crucial evidence to identify disease associated SNPs from a large number of reported SNPs and to study the functional role of SNPs. Although numerous studies have been carried out on how SNPs are associated with the diseases, it could not be confirmed by subsequent independent studies. In this case, computational analysis could help in saving the time, reducing costs and prioritize SNPs for analysis by quantitative ranking of functionally significant SNPs (De Alencar and Lopes, 2010). In this study, we implemented both sequence and sequence-structure-based computational approaches to analyze the SNPs in FGF23, VDR and CYP24A1 genes.
2. Materials and methods
Initially, the SNPs and their related sequences of FGF23, CYP24A1 and VDR genes were retrieved from the National Center for Biotechnology Information (NCBI) database of SNPs, dbSNP (http://www.ncbi.nlm.nih.gov./SNP/) for our computational analysis. We strict our list to missense mutation, that is mainly associated with the diseases (Boillee et al., 2006, Minde et al., 2011).
Sequence based and structure based methods are the two common approaches used in SNP prediction tools. Compared to the structure based predictions, sequence-structure based predictions are more precise one, since it includes all types of effect at the protein level, and can be applied to any human protein with known relatives (Yue et al., 2005, Mooney et al., 2010, Singh Kh and Karthikeyan, 2014). Sequence based predictions are failed to explain the underlying mechanism of how the single nucleotide polymorphism will alter the protein phenotype, whereas the structure based approaches may solve this limitation. Thus, we used the combination of structure based and sequence based approaches to validate the different aspects of SNP analysis (Yue and Moult, 2006, Singh Kh and Karthikeyan, 2014).
3. Sequence based tools
3.1. SIFT
The human nsSNP which is available in dbSNP was analyzed by sorting intolerance from tolerant (http://sift.jcvi.org/www/SIFT_dbSNP.html). The difference between functional and non-functional SNPs in coding regions was predicted by SIFT. The results from this software helped to predict the substitutions of an amino acid on phenotypic effect. SIFT predictions are mainly based on physiochemical properties of amino acid and sequence homology (Ng and Henikoff, 2002).
The SIFT algorithm uses a modified version of PSI Blast (Altschul et al., 1997) from NCBI (Wheeler et al., 2001) and Dirichlet mixture regulation (Sjolander et al., 1996) in order to construct multiple sequence alignment of protein sequences. It aligned the query sequences globally and all the sequences which are in same clad. The SIFT scores > 0.05 are considered by the algorithm to be tolerant (Sherry et al., 2001).
3.2. SNP & GO
The SNPs which are likely to be involved in the pathogenesis of human disease might be predicted by the SNP & GO server. It predicts the disease related mutations from a protein sequence and the functional annotation of the protein on the basis of support vector machines (SVMs).
The SNP & GO server collected the information from different sources such as protein sequence, the local sequence environment of the SNPs, the protein sequence profile, features generated from sequence alignment, and protein function. This server annotated the information from the gene ontology database (GO). This database included the gene products in terms of their associated biological processes, cellular components and molecular functions (Calabrese et al., 2009).
4. Combined sequence and structure based prediction tools
4.1. PolyPhen-2
The possible impact of an amino acid on the structure and function of a human protein was predicted by polymorphism phenotyping V2 (http://genetics.bwh.harvard.edu/pph2/) using physical and comparative considerations. The results from the PolyPhen-2 output encompass a score that ranges from 0 to a positive number. The zero indicates the neutral effect of SNP on protein structure whereas the large positive number indicates the substitution that may have severe effects (Ramensky et al., 2002, Xi et al., 2004, Ng and Henikoff, 2006).
4.2. I-Mutant
Protein stability changes upon single-site mutations were calculated by a neural-network-based web-server I-Mutant. The tool generated an output in connection with dataset derived from ProTherm (Bava et al., 2004). I-Mutant predicted the protein mutation which stabilizes or destabilizes the protein structure. The free energy value was also computed with the energy-based FOLD-X tool. The reliability index value was calculated by coupling the FOLD-X predictions with I-Mutant (Guerois et al., 2002).
5. Computational site directed mutagenesis
The human CYP24A1 protein crystal structure was not solved, but the rat CYP24A1 crystal structure was available in the protein data bank (PDB) (Berman et al., 2000) (PDB id: 3K9V) (Annalora et al., 2010). The sequence similarity between both the sequences was 85%. Thus we modeled the human CYP24A1 protein using rat CYP24A1 in Prime module of Schrodinger software (Prime, version 3.9, Schrödinger, LLC, New York, NY, 2015). The FGF23 (PDB id: 2p39) (Goetz et al., 2007) and VDR (PDB id: 3B0T) (Kakuda et al., 2010) crystal structures were downloaded from the PDB. Computational mutagenesis was performed using Maestro, version 9.10, Schrodinger, LLC, New York, 2015. After mutagenesis, each protein was optimized and energy minimized using OPLS_2005 force field in the protein preparation wizard of Schrodinger, LLC. After energy minimization, the mutant structure was superimposed with the corresponding native structure and the root mean square deviation (RMSD) was calculated. The RMSD is the square root of the mean of the square of the distance between the matched atoms.
| (1) |
where dii is the distance between the ith atom of structure l and ith atom of structure 2 and N is the number of atoms matched in each structure.
6. Analysis of effect of mutation on protein solvent assessable area and secondary structure
The accessible surface area (ASA) was calculated by rolling a sphere size of a water molecule over the protein space which was accessible to a solvent (Chothia and Finkelstein, 1990). The ASA was mostly transformed to the relative surface area (RSA) for the comparative and predictive purpose. It was calculated to the given amino acid residue in the polypeptide chain, relative to the maximum possible exposure of the residue in the center of a tri-peptide flanked with either glycine (Connolly, 1983) or alanine (Chothia, 1976). Understanding the degree of surface exposure of an amino acid was valuable since it was used to enhance the understanding of a variety of biological problems such as protein–ligand interactions (Ahmad et al., 2003) and protein–protein interactions (Jones and Thornton, 1997a, Jones and Thornton, 1997b), active sites (Haste Andersen et al., 2006), and structural epitopes (Jones and Thornton, 1997a, Jones and Thornton, 1997b) and the prediction of disease related SNPs (Panchenko et al., 2004). The RSA can be calculated as follows,
| (2) |
where ASAmax is the maximum obtained solvent exposed area (Petersen et al., 2009).
In order to compare the surface accessibility, from exposed to buried regions were calculated. Geneious Pro (Kearse et al., 2012) software (Aukland, New Zealand) was used to compare the secondary structure of the wild and mutant type of the protein. The pI for protein folding and unfolding free energy, optimum pH for protein stability was further calculated using PROKA 3.0 (Copenhagen, Denmark) (Li et al., 2005, Olsson et al., 2011).
7. Statistical analyses
In statistical prediction the following three cross-validation methods are often used to evaluate the anticipated success rate of a predictor: independent dataset test, sub-sampling (or K-fold cross-validation) test, and jackknife test (Chou and Zhang, 1995). Among the three, however, the jackknife test is deemed the least arbitrary and most objective as elucidated by Eqs. 28–32 of Chou, 2011. Therefore, the jackknife test has been widely recognized and increasingly used to test the quality for various predictors (Chen et al., 2012, Chen et al., 2013, Chen et al., 2014, Chen et al., 2016a, Chen et al., 2016b, Lin et al., 2014, Liu et al., 2015a, Liu et al., 2015b, Liu et al., 2015c, Liu et al., 2016a, Liu et al., 2016b, Qiu et al., 2015, Jia et al., 2016a, Jia et al., 2016b).
Six different parameters were widely used to describe the predictions quality viz. accuracy, precision, sensitivity, specificity, negative predictive value (NPV) and Matthews correlation coefficient (MCC). In the following equations true positives, true negatives, false positives and false negatives are represented as tp, tn, fp and fn respectively.
| (3) |
| (4) |
| (5) |
| (6) |
Unfortunately, the four metrics formulated in Eqs. (3), (4), (5), (6), are not intuitive and easy-to-understand to most biologists especially the equation for MCC. Hence, we adopted the formulation proposed by Chou et al. (2012). According to the formulation, the same four metrics can be expressed as
| (7) |
| (8) |
| (9) |
| (10) |
where N+ is the total number of SNPs investigated, whereas N − + is the number of the disease caused by SNPs which were incorrectly predicted as neutral; N− is the total number of non-synonymous SNPs investigated, and N + − is the number of the non-synonymous SNPs wrongly predicted as deleterious.
The MCC (Matthews, 1975) is a good evaluation statistics, because it was unaffected by the different proportions of neutral and pathogenic datasets predicted by different programs. Overall the MCC was insensitive to different test set sizes and thus it gives a more balanced assessment of performance than the other performance measures (Baldi et al., 2000). The use of these metrics and their merits has been concurred by a series of recent studies (Chen et al., 2016a, Chen et al., 2016b, Jia et al., 2016a, Jia et al., 2016b, Liu et al., 2016a, Liu et al., 2016b). The set of metrics is valid only for the single-label systems. For the multi-label systems whose existence has become more frequent in system biology (Chou et al., 2012) and system medicine (Xiao et al., 2013), a completely different set of metrics as defined by Chou, 2013 is needed.
8. Results
The main objective of the present study is to identify the pathogenic SNPs from the pool of SNPs reported in NCBI using the web based analysis tools. We have used both the combined sequence and sequence-structure-based tools in order to overcome the limitations of both the methods towards the prediction of risky SNPs associated with CKD. The workflow followed in this study is shown in Fig. 2.
Fig. 2.
The workflow followed in the study.
Thusberg et al. (2011) had reported the accuracy of SNP & GO (0.82) and that it is comparably good with PolyPhen 2 (0.69) and SIFT (0.65). The SNP & GO software predicted a high precision value (0.90) in comparison to PolyPhen-2 (0.71), SIFT (0.64). SNP & GO, SIFT, PolyPhen-2, and I-Mutant software were used to analyze all our dataset including SNPs from the Uniprot disease database (664 SNPs) and 287 non-sense mutations.
9. Statistical analysis of the performance from in silico prediction methods
We used six different statistical measures, namely accuracy, precision, specificity, sensitivity, negative predictive value (NPV), and Matthews correlation coefficient (MCC) to evaluate the performance of the tools. Initially a dataset comprising of deleterious SNPs from Uniprot disease database and nsSNPs was formed and we predicted the performance of the tools. Based on the computational method predictions, the dataset was evaluated to obtain tp (true positive), tn (true negative), fp (false positive) and fn (false negative) values in order to calculate the statistics measures (Table 1). Based on the statistical analyses, I-Mutant (0.89) and SNP & GO (0.72) performed well in terms of accuracy, I-Mutant (0.91) and SIFT (0.85) performed well in terms of precision, SIFT (0.72) and PolyPhen-2 (0.61) performed well in terms of specificity and I-Mutant (0.97) and SNP & GO (0.89) performed well in terms of sensitivity and SNP & GO (0.75) and PolyPhen-2 (0.75) performed good in terms of NPV and PolyPhen-2 (0.42) performed well in terms of MCC. Overall the accuracy predictions were worst in the case of SIFT tool (0.71) and PolyPhen-2 (0.71), PolyPhen-2 performed worst in terms of precision (0.67), I-Mutant and SIFT performed worst in terms of specificity (0.32) and sensitivity (0.71) respectively. Further, we performed the statistical analysis for the combined sequence based and sequence-structure based prediction methods. Interestingly, our findings clearly exhibit that the predictions based on both sequence and sequence-structure based method produced good statistical method (MCC = 0.45) rather than single individual method.
Table 1.
Statistical evaluation of various computational methods.
| SIFT | SNP & GO | PolyPhen-2 | I-Mutant | Combined sequence and sequence-structure based method | |
|---|---|---|---|---|---|
| Tp | 270 | 286 | 164 | 553 | 1273 |
| Tn | 123 | 110 | 126 | 25 | 384 |
| Fp | 48 | 117 | 80 | 52 | 297 |
| Fn | 110 | 36 | 40 | 16 | 202 |
| Cases + | 380 | 322 | 204 | 569 | 1475 |
| Cases − | 171 | 227 | 206 | 77 | 681 |
| Accuracy | 0.71 | 0.72 | 0.71 | 0.89 | 0.77 |
| Specificity | 0.72 | 0.48 | 0.61 | 0.32 | 0.56 |
| Sensitivity | 0.71 | 0.89 | 0.80 | 0.97 | 0.86 |
| MCC | 0.40 | 0.41 | 0.42 | 0.39 | 0.45 |
10. SNP dataset
FGF23, CYP24A1 and VDR genes play a very important role in the CKD pathogenesis, which were selected for computational analysis of deleterious SNPs. We have selected SNPs only from the coding regions, since coding regions are critical for the determination of protein tertiary structure and function.
11. Prediction of deleterious nsSNPs using sequence based prediction tools
In the initial process, we analyzed all the SNPs with sequence based prediction tools. SIFT algorithm was used for the protein conversion and predicted whether an amino acid substitution had an impact on protein function by aligning similar proteins. Further, a score was generated to determine the evolutionary conversion status of the amino acid of interest. The retrieved 739 SNPs were submitted to the SIFT program to check its tolerance and 454 SNPs have found to be having missense mutation in the coding region.
The output scores for the SIFT analysis ranges from 0 to 1, while 0 represents damaging whereas 1 denotes neutral. If the SIFT cutoff score is lower than the 0.05, the amino acid change at a particular position is tolerated (no effect). Further, the repetitive amino acid substitutions would be predicted as deleterious. The SIFT algorithm predicted 4 SNPs from FGF23 gene, 15 SNPs from VDR gene and 13 SNPs from CYP24A1 gene which were found to be having a critical deleterious role (Table 2).
Table 2.
Analysis of SNPs detected in the coding region of FGF23, VDR and CYP24A1 genes.
| SIFT |
I-Mutant |
SNP & GO |
PolyPhen-2 |
RMSD (Å) | ||||
|---|---|---|---|---|---|---|---|---|
| GENE | Uniprot ID | SNP id | Amino acid change | Prediction | Prediction | Effect | Prediction | |
| FGF23 | Q9GZV9 | rs104894342 | S71G | Damaging | Decrease | Disease | Probably damaging | 5.72 |
| Q9GZV9 | rs104894343 | M96T | Damaging | Decrease | Disease | Probably damaging | 6.66 | |
| Q9GZV9 | rs104894344 | S129F | Damaging | Increase | Disease | Probably damaging | 5.77 | |
| Q9GZV9 | rs575204793 | R160Q | Damaging | Decrease | Disease | Possibly damaging | 5.77 | |
| VDR | P11473 | rs121909796 | R274L | Damaging | Decrease | Disease | Possibly damaging | 7.85 |
| P11473 | rs121909799 | I314S | Damaging | Decrease | Disease | Benign | 6.87 | |
| P11473 | rs121909800 | R391C | Damaging | Decrease | Disease | Probably damaging | 6.83 | |
| P11473 | rs121909802 | E329K | Damaging | Decrease | Disease | Probably damaging | 7.70 | |
| P11473 | rs11574090 | L230V | Damaging | Decrease | Disease | Possibly damaging | 8.13 | |
| P11473 | rs75590999 | I367M | Damaging | Decrease | Disease | Probably damaging | 7.19 | |
| P11473 | rs114678556 | R358H | Tolerated | Decrease | Disease | Possibly damaging | 7.14 | |
| P11473 | rs199705103 | R154W | Damaging | Decrease | Disease | Probably damaging | 8.63 | |
| CYP24A1 | Q07973 | rs6068812 | L409S | Damaging | Decrease | Disease | Probably damaging | 3.51 |
| Q07973 | rs114368325 | R396W | Damaging | Decrease | Disease | Probably damaging | 3.08 | |
| Q07973 | rs387907322 | R159Q | Damaging | Decrease | Disease | Probably damaging | 4.92 | |
| Q07973 | rs387907324 | E322K | Damaging | Decrease | Disease | Probably damaging | 3.61 | |
| Q07973 | rs58713852 | T248K | Damaging | Decrease | Disease | Probably damaging | 4.04 | |
| Q07973 | rs114476330 | R120H | Damaging | Decrease | Disease | Probably damaging | 5.14 | |
| Q07973 | rs114579367 | D202H | Damaging | Decrease | Neutral | Probably damaging | 3.47 | |
| Q07973 | rs116548533 | R344H | Damaging | Decrease | Neutral | Probably damaging | 3.88 | |
| Q07973 | rs139763321 | L148P | Damaging | Decrease | Disease | Probably damaging | 4.67 | |
| Q07973 | rs140189382 | Y407N | Damaging | Decrease | Disease | Probably damaging | 3.46 | |
| Q07973 | rs141152573 | R439H | Damaging | Decrease | Disease | Probably damaging | 3.26 | |
| Q07973 | rs143934667 | R396Q | Damaging | Decrease | Disease | Probably damaging | 3.53 | |
| Q07973 | rs146980218 | R439Q | Damaging | Decrease | Disease | Probably damaging | 3.24 | |
The SNP & GO tool is a collection of unique framework, and includes information derived from protein sequence, and evolutionary information and function as encoded in the Gene Ontology terms. The software predicts the human disease related SNPs in proteins with functional annotations. 12 SNPs from FGF23 gene, 60 SNPs from VDR gene and 22 SNPs from CYP24A1 gene were predicted to be associated with human diseases (Table 2).
12. Prediction of deleterious nsSNPs using sequence-structure based prediction tool
The PolyPhen-2 program was used to determine the structural level alterations. Various parameters such as evolutionary conservation, physicochemical differences and the proximity of the substitution were considered in order to predict functional domains, and structural features and functional effects of amino acid changes. PolyPhen-2 score in the dataset ranges from 0 to 1. If the PolyPhen-2 score is < 0.5 then the mutation is a benign one. The changes are possibly damaging if the score is > 0.5 and > 0.9 are probably damaging. 13 SNPs from FGF23 gene, 45 SNPs from VDR gene and 62 SNPs from CYP24A1 gene were predicted to be probably/possibly damaging and these SNPs may affect the structural stability and the phenotype of the protein (Table 2).
I-Mutant program was used to check the stability of the protein caused by nsSNPs. This program calculated the energy difference between native and variant proteins based on Gibbs free energy values. I-Mutant predictions were classified into three different classes viz. neutral mutations (− 0.5 ≤ kcal/mol), mutations which decreased the Gibbs free energy (− 0.5 < kcal/mol), and mutations which produce a larger increased energy (0.5 > kcal/mol). 21 SNPs from FGF23 gene, 174 SNPs from VDR gene and 83 SNPs from CYP24A1 gene might decrease the protein stability (Table 2).
The wild type protein was mutated using Maestro, Schrodinger, LLC, New York, 2015. Further, the mutated protein was optimized and energy minimized using protein preparation wizard, Schrodinger, LLC, New York, 2015. The RMSD between the wild type and mutant type was calculated and reported in the Table 2.
We adopted four online SNP prediction tools (two sequence based and two sequence-structure based) to reduce the false positive errors. These online servers were used for different parameters such as sequence, evolutionary approach, physicochemical, secondary structure, solvent accessibility, and free energy calculations for analysis. After analysis, all the results predicted by four different SNP prediction servers, we anticipated that those SNPs which were predicted to be disease/disorder/damaging etc., by at least three different algorithms, had high RMSD and may show functional significance and it may be the reason behind the cause of disorder to the human body (Table 1). Such SNPs are listed below: FGF23 rs104894342 (S71G), rs104894343 (M96T), rs104894344 (S129F), rs575204793 (R160Q); SNP id's of VDR rs121909796 (R274L), rs121909799 (I314S), rs121909800 (R391C), rs121909802 (E329K), rs11574090 (L230V), rs75590999 (I367M), rs114678556 (R358H), rs199705103 (R154W); SNP id's of CYP24A1 rs6068812 (L409S), rs114368325 (R396W), rs387907322 (R159Q), rs387907324 (E322K), rs58713852 (T248K), rs114476330 (R120H), rs114579367 (D202H), rs116548533 (R344H), rs139763321 (L148P), rs140189382 (Y407N), rs141152573 (R439H), rs143934667 (R396Q), rs146980218 (R439Q). Fig. 3 shows the superimposed structure of wild and mutant proteins.
Fig. 3.
Superimposed structure of wild type FGF23 and S71G mutant (A), wild type of VDR and R274L mutant (B), wild type CYP24A1 and L409S mutant. The SNPs in this figure are randomly selected from each gene for the easy interpretation of the result.
Additionally, the solvent accessible areas of 25 deleterious SNPs were analyzed to better understand the relationship between sequence and structure. Thus the solvent accessible area was calculated by NetsurfP server (Kongens Lyngby, Denmark). NetSurfP predicted the amino acids, whether in exposed region or buried region at 25% threshold (residues may be predicted to be exposed/buried based on a 25% threshold). The changes in exposed to buried or buried to expose regions due to mutation were shown in Table 3. Mutations in the buried sites are more prone to disrupt the protein structure compared to mutations introduced in the solvent exposed structures. Thus, the latter tend to destabilize proteins, through steric hindrance and the introduction of strained conformations. Mutation in FGF23 and VDR genes shows the number of changes from buried to expose and exposed to buried when compared to CYP24A1 gene.
Table 3.
Solvent accessibility of the wild type and mutant type of FGF23, VDR, CYP24A1 proteins.
| Gene | Mutation | Exposed to buried | Buried to exposed |
|---|---|---|---|
| FGF23 | M96T | 36W, 40I, 50S, 108F, 122N, 166L, 167I, 168H | 48R, 68T, 170N, 171T |
| R160Q | 36W, 49N, 81G, 167I | 154Y, 160R, 170N, | |
| S71G | 33G, 81G, 166L, 167I, 168H | 48R, 68T, 170N, 171T | |
| S129F | 36W, 40I, 50S, 108F, 122N, 130P, 131Q, 133H, 143R, 166L, 167I, 168H | 30P, 48R, 68T, 154Y, 169F, 170N | |
| VDR | E329K | 142T, 239Q, 284M, 300V, 341P | 312P, 376S |
| I314S | 142T, 239Q, 280T, 300V, 385Q, 389D | 145P | |
| I367M | 142T, 239Q, 385Q, 389D | 145P, 290N, 306S | |
| L230V | 142T, 143Y, 389D | 290N, 303A | |
| R154K | 389D, 142T, 341P, 389D | 415T | |
| R274L | 239Q, 264K, 284M, 341P, 389D | 145P, 290N, 303A, 314I, 376S | |
| R358H | 142T, 239Q, 389D | 145P, 285S, 290N, 295Y, 306S | |
| R391C | 239Q, 385Q, 389D | 145P, 295Y, 376S, 410C, 419L | |
| CYP24A1 | D202H | 264N | 143E |
| E322K | 232G | 87V, 353L | |
| L148P | 140Y, 143E, 353L | 136A, 264N | |
| L409S | 262S, 264N | – | |
| R120H | 264N | 353L | |
| R159Q | 176M, 232G, 264N | 140Y | |
| R344H | 193L, 232G, 264N | 140Y, 353L | |
| R396K | 264N, 300D | 140Y, 353L | |
| R396Q | 129L, 300D | 353L | |
| R439H | 136A, 232G | 140Y, 353L |
13. Relative surface area
The analysis of the RSA and ASA of the wild type and mutant type for all the residues is shown in Fig. 4, Fig. 5. After analyzing the graph it was found that the FGF23 and VDR have changes in their RSA and ASA value of the wild type except CYP24A1. In FGF23, Q54K SNP produced slight different RSA and ASA when compared to the wild type. The same type of slight difference was observed in the CYP24A1:I367M SNP. The effect of SNP in the formation of secondary structure was analyzed and displayed in the Fig. 6.
Fig. 4.
The relative surface area (RSA) of wild type and selected mutant type of FGF23 gene (A), CYP24A1 gene (B) and VDR gene (C).
Fig. 5.
The accessible surface area (ASA) of wild type and selected mutant type of FGF23 gene (A), CYP24A1 gene (B) and VDR gene (C).
Fig. 6.
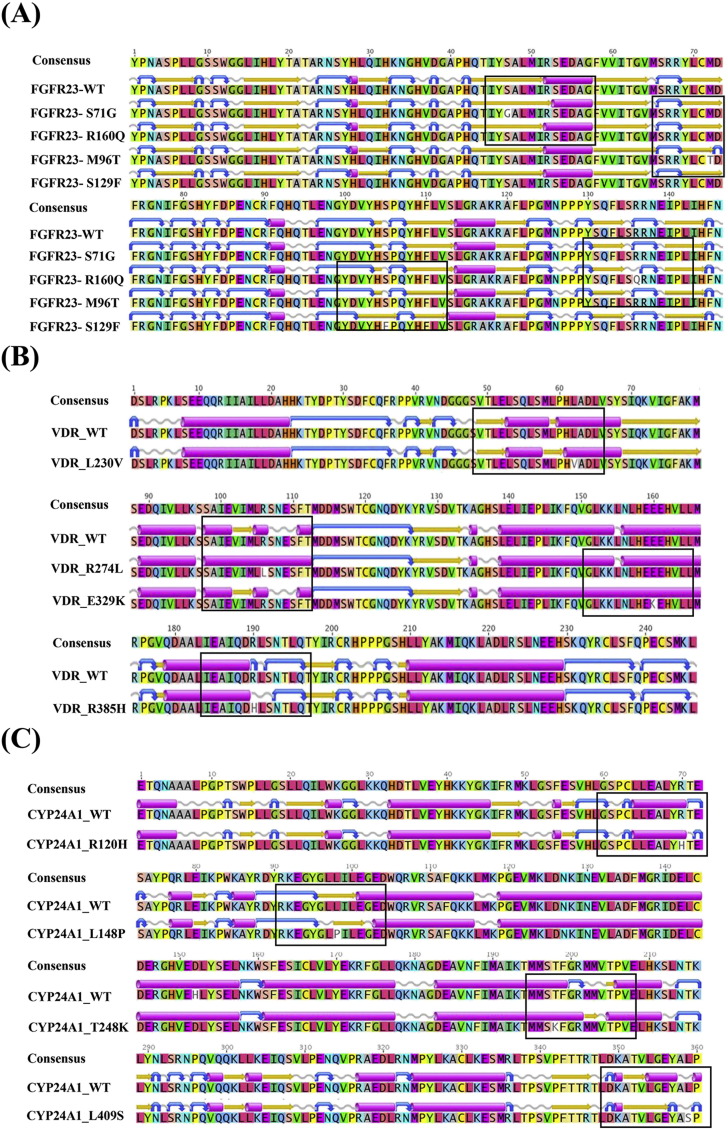
Multiple sequence alignment and secondary structure prediction of FGF23, CYP24A1 and VDR genes. Alignment of secondary structure identified the β–strand to alpha change in S71G mutant, β–strand to turn change in M96T mutant, addition of β–strand in S129F mutant, addition of coil in R160Q mutant (A), addition of β–strand in L230V mutant, addition of alpha helix in R274L and E329K mutants, turn to coil change in R385H mutant (B), Turn to coil change in R120H mutant, β–strand to coil change in L148P mutant, coil to β–strand mutant in T248K mutant and alpha helix to β–strand and turn change in L409S mutant.
In FGF23, a small deletion of alpha helix was observed in S71G mutation. In M96T, one extra turn was noticed near the helical region. Moreover, the addition of an extend beta strand was observed in FGF23–S129F SNP. Finally, in R160Q the coil was extended towards its right side.
In VDR, L230V SNP led to small changes in the beta strand. In R274L mutation, the small beta strand coil changed and instead a long alpha helix was formed. The E329K formed a linear alpha helix. In R358H, the small turn was changed into a coil. The remaining SNPs could not cause significant changes in the secondary structure.
In CYP24A1, R120H led to a change into turn. L148P mutation brought about change in the formation of small coil structure. An extension of alpha helix was observed in the T248K mutation. Further, in L409S mutation a coil is presented instead of alpha helix. The remaining mutations could not cause significant change in the protein secondary structure.
Further, we analyzed the pH for optimum stability, pI for folding and unfolding are free energy of the wild and mutant protein and found that all the three proteins were stable at different pH. FGF23 was stable at 9.6 pH, VDR was stable at 7.8 to 8.5 pH and CYP24A1 was stable at 7.9 to 8.6 pH. There were no vigorous changes observed in the optimum pH of wild and mutant proteins. The CYP24A1 enzyme had more binding energy when compared to the remaining two proteins. The predicted pH value is shown in Table 4.
Table 4.
Predicted value of pH of optimum stability, pI of folding and unfolding and free energy for wild type and selected mutant type genes.
| Protein | Amino acid change | pH of optimum stability | pI value folded | pI value unfolded | Free energy (kcal/mol) |
|---|---|---|---|---|---|
| FGF23 | WT | 9.6 | 9.32 | 9.42 | 0.7 |
| M96T | 9.6 | 9.32 | 9.42 | 0.6 | |
| 160Q | 9.6 | 8.79 | 9.11 | 0.2 | |
| S71G | 9.6 | 9.32 | 9.42 | 0.6 | |
| S129F | 9.6 | 9.32 | 9.42 | 0.8 | |
| VDR | WT | 7.9 | 6.20 | 6.61 | 14.8 |
| E329K | 8.5 | 6.50 | 7.03 | 19 | |
| I314S | 7.9 | 6.22 | 6.31 | 18 | |
| I367M | 7.9 | 6.23 | 6.61 | 18.2 | |
| L230V | 7.9 | 6.23 | 6.61 | 18.1 | |
| R154K | 7.9 | 6.23 | 6.61 | 21.6 | |
| R274L | 7.8 | 5.98 | 6.44 | 13.4 | |
| R358H | 7.9 | 6.07 | 6.52 | 17.6 | |
| R391C | 7.8 | 5.98 | 6.44 | 17.8 | |
| CYP24A1 | WT | 8.3 | 9.01 | 8.86 | 52.3 |
| D202H | 9.3 | 9.13 | 8.97 | 51.8 | |
| E322K | 9.0 | 9.08 | 9.06 | 54.3 | |
| L148P | 8.3 | 9.02 | 8.86 | 54.5 | |
| L409S | 8.3 | 9.01 | 8.86 | 52.2 | |
| R120H | 8.3 | 8.88 | 8.74 | 51.2 | |
| R159Q | 8.2 | 8.91 | 8.74 | 47.4 | |
| R344H | 8.3 | 8.88 | 8.74 | 52.0 | |
| R396K | 8.6 | 8.94 | 8.86 | 50.5 | |
| R396Q | 7.9 | 8.94 | 8.74 | 47.4 | |
| R439H | 8.3 | 8.87 | 8.74 | 51.4 |
WT—Wild type.
14. Discussion
Identification of the disease causing mutations from those which are functionally neutral is very essential to understand the molecular pathophysiology of the diseases. In recent days, amino acid substitutions account for approximately half of the known gene lesions responsible for human inherited disease (Cooper et al., 1998). Thus, identification of nsSNPs which affect protein functions and causing disease is crucial. In natural selection, many of the nsSNPs effects are neutral since mutations are removed in essential positions. Therefore, researchers have the ability to discriminate accurately significant, protein function altering SNPs from those that are functionally neutral (Boillee et al., 2006). However, there is increasing evidence of availability for the role of coding or non-coding mutations in protein regulatory functions and subsequent diseases (Yan et al., 2002, Hudson, 2003). Analyzing the vast number of SNPs might not be reasonable for researchers to carry out in vitro experiments on each and every SNP to infer from their biological significance. Thus, the vast number of SNPs causes challenge to biologists as well as bioinformaticians. Apart from these, numerous studies are in progress to study the effect of SNPs in genetic profiles and alteration pharmacogenomic drug profiles using a molecular epidemiological approach.
In this paper, we attempted to predict the SNPs which can alter the protein expression and function in three interlinked genes (FGF23, VDR and CYP24A1). The mutations among these genes have associated with several diseases (Bai et al., 2003, Shimada et al., 2005, Liu and Quarles, 2007, Perwad et al., 2007, Damasiewicz et al., 2011).
Thus, the changes of amino acids in particular region might be associated with several diseases. Therefore, our study would pave way in selecting SNPs that were likely to have potential complexity to refine SNP prediction. GO based score was incorporated in the SNP & GO prediction algorithm which enables correlation between given SNP and its corresponding gene product function. PANTHER predicted classification data that is also included in the SNP & GO prediction. SNP & GO tool was more advanced than PANTHER. As PANTHER requires Gene or dbSNP IDs which cannot be entered directly as search inputs, limiting the scope of searches to the protein sequence level and require information on protein alignment for search input. PolyPhen-2 ranking of the SNPs on the basis of protein phenotype changes which caused by severe SNP effects. I-Mutant server uses a neural network based web server for the analysis of the protein stability upon the single mutation.
Out of 740 missense SNPs reported in dbSNP, we found 25 missense SNPs in the coding region which may affect the normal gene regulation or protein stability. Mutation in FGF23 gene was associated with hyper and hypo phosphatemia (Gupta et al., 2004, Saito and Fukumoto, 2009), familial tumoral calcinosis (Farrow et al., 2011) and autosomal dominant hyophosphatemic rickets (ADHR Consortium, 2000) etc. Five mutations (H41Q, S71G, M96T, S129F, and Q54K) in the coding region were already reported (Garringer et al., 2008). Interestingly, in our in silico findings, we also found that these five mutations have a significant effect on protein structure and stability. Consistent with in vitro findings, we hypothesized that the mutations in these regions lead to alter in peptide folding and decreased in FGF23 secretion. Moreover, we predicted that the protein stability was decreased with respect to these mutations.
CYP24A1 gene mutations were known to cause hypercalcemia, nephrocalcinosis and nephrolithiasis etc. R159Q mutation in the coding region disrupts the hydrogen bond interaction in the CYP24A1 active site (Ji and Shen, 2011). Thus, this SNP analysis also revealed that mutation decreased the protein structure stability. L409S mutation affected the enzyme activity since it leads to weakening the binding with 1,25(OH)2D3 (Nesterova et al., 2013). In secondary structure analysis, we found that the alpha helix was changed into beta turn and might be a structural change to cause this effect. Moreover, the enzyme activity decreased in L148P mutation because of the direct interaction with enzyme substrate (Nesterova et al., 2013), therefore, this mutation leads to decrease in enzyme activity. Small coil was changed in this particular region and may cause the protein stability and activity.
VDR has long been known for its important role in regulating body levels of calcium (Ca) and phosphorous (P) and in mineralization of bone (Holick, 2010). In VDR gene, we found eight polymorphisms as more deleterious. VDR mutations were associated with rickets, cancer, osteoporosis etc. VDR activation is essential for different types of cellular processes. R274L mutation in the active site region causes changes in VDR structure between helices H1 & H2 (Nakabayashi et al., 2013). Secondary structure analysis predicted the deletion of beta strand and coil formation of alpha helix. Moreover, I314S (Whitfield et al., 1996) and R391C (Nguyen et al., 2006) mutation was found to have changed the conformations and leads to changes in hormonal binding domain. Among these, R391C mutation was well known for its ability to reduce the binding with steroid receptor co-activator 1 (SRC-1). Interestingly, our in silico findings elucidate the deleterious nature of these polymorphisms. Therefore, our findings conceal that these mutations may affect the gene expression and the protein structure.
To the best of our knowledge, no comprehensive evaluation of the performance of missense variant pathogenicity predictors has been made outside the performance studies of individual methods in the context of identification of SNPs associated with risk. We selected test sets which have not been used in the training set of all methods, but the pathogenic subset was comprised of dataset from Uniprot disease database mutations. Testing the performance of a method with the same cases when it was trained would lead to biased results, thus data set from Uniprot disease database mutations would have an advantage over the other methods. The performance decreased in all methods regardless whether trained on Uniprot data or not. But, if we combined the sequence based and sequence-structure based results it outperforms than the individual methods.
The neutral dataset was generated from dbSNP entries that had > 1% frequency when there was data at least for 25 individuals (50 chromosomes). By this way we minimized the number of false negatives in the test set.
Out of 25 deleterious SNP reports from our study, 8 SNPs were already reported in the Uniprot disease database. Different parameters such as sequence, evolutionary approach, physiochemical, secondary structure, solvent accessibility, and free energy calculations were used for the analysis of SNPs.
As demonstrated in a series of recent publications (Chen et al., 2016a, Jia et al., 2016a, Jia et al., 2016b, Jia et al., 2016c, Liu et al., 2016a, Liu et al., 2016b, Liu et al., 2016c) in developing new prediction methods, user-friendly and publicly accessible web-servers will significantly enhance their impacts (Chou, 2015, Chen et al., 2015), we shall make efforts in our future work to provide a web-server for the prediction methods presented in this paper.
15. Conclusion
In the present study, we investigated the functional and structural impact of SNPs caused by the CKD associated genes (FGF23, CYP24A1 and VDR) using different computational prediction tools. The approach can also be applied to study the relationship between SNP conservation levels and epidemiological studies among these studied genes. 25 SNPs were predicted to be disorder/diseases/damaging etc., by three or four different algorithms and high RMSD will show functional significance and it may cause disorder in the human body. Out of which four SNPs (S71G, M96T, S129F, R160Q) of FGF23 gene, eight SNPs (R274L, I314S, R391C, E329K, L230V, I367M, R358H, R154W) of VDR gene and thirteen SNPs (L409S, R396W, R159Q, E322K, T248K, R120H, D202H, R344H, L148P, Y407N, R439H, R396Q, R439Q) of CYP24A1 gene were found to have a possible functional effect in the coding region of our comparative sequence and structure–SNP based analysis tools with low RMSD value. Further, experimental study needs to be carried out for further validation to analyze the functional effect of the mutations reported in the Table 1. As we mentioned earlier, our combined sequence and sequence-structure based methods outperformed than the available methods. Thus, our method is the best one for prioritizing nsSNPs out of SNP pool.
The in silico data presented here demonstrate the comparative computational approach for classification of three difference gene variants which is a powerful and fast technique and can be used for large scale analyses. The present study will also be helpful to understand the functional variation from the perspective of structure, expression, evolution, physiochemical property, and phenotypes and can help the experimental geneticists to carry out their large scale SNP analysis.
Acknowledgment
Authors gratefully acknowledge the financial support provided by Indian Council of Medical Research (ICMR), Govt. of. India to S. N (F. No.: BIC/11(09)/14) in the form of Senior Research Fellowship.
References
- ADHR Consortium Autosomal dominant hypophosphataemic rickets is associated with mutations in FGF23. Nat. Genet. 2000;26:345–348. doi: 10.1038/81664. [DOI] [PubMed] [Google Scholar]
- Ahmad S., Gromiha M.M., Sarai A. Real value prediction of solvent accessibility from amino acid sequence. Proteins. 2003;50:629–635. doi: 10.1002/prot.10328. [DOI] [PubMed] [Google Scholar]
- Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annalora A.J., Goodin D.B., Hong W.X., Zhang Q., Johnson E.F., Stout C.D. Crystal structure of CYP24A1, a mitochondrial cytochrome P450 involved in vitamin D metabolism. J. Mol. Biol. 2010;396:441–451. doi: 10.1016/j.jmb.2009.11.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai X.Y., Miao D., Goltzman D., Karaplis A.C. The autosomal dominant hypophosphatemic rickets R176Q mutation in fibroblast growth factor 23 resists proteolytic cleavage and enhances in vivo biological potency. J. Biol. Chem. 2003;278:9843–9849. doi: 10.1074/jbc.M210490200. [DOI] [PubMed] [Google Scholar]
- Baldi P., Brunak S., Chauvin Y., Andersen C.A., Nielsen H. Assessing the accuracy of prediction algorithms for classification: an overview. Bioinformatics. 2000;16:412–424. doi: 10.1093/bioinformatics/16.5.412. [DOI] [PubMed] [Google Scholar]
- Bava K.A., Gromiha M.M., Uedaira H., Kitajima K., Sarai A. ProTherm, version 4.0: thermodynamic database for proteins and mutants. Nucleic Acids Res. 2004;32:D120–D121. doi: 10.1093/nar/gkh082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The protein data bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boillee S., Vande Velde C., Cleveland D.W. ALS: a disease of motor neurons and their nonneuronal neighbors. Neuron. 2006;52:39–59. doi: 10.1016/j.neuron.2006.09.018. [DOI] [PubMed] [Google Scholar]
- Calabrese R., Capriotti E., Fariselli P., Martelli P.L., Casadio R. Functional annotations improve the predictive score of human disease-related mutations in proteins. Hum. Mutat. 2009;30:1237–1244. doi: 10.1002/humu.21047. [DOI] [PubMed] [Google Scholar]
- Chen W., Ding H., Feng P., Lin H., Chou K.C. iACP: a sequence-based tool for identifying anticancer peptides. Oncotarget. 2016 doi: 10.18632/oncotarget.7815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Feng P., Ding H., Lin H., Chou K.C. Using deformation energy to analyze nucleosome positioning in genomes. Genomics. 2016;107:69–75. doi: 10.1016/j.ygeno.2015.12.005. [DOI] [PubMed] [Google Scholar]
- Chen W., Feng P.M., Lin H., Chou K.C. iRSpot-PseDNC: identify recombination spots with pseudo dinucleotide composition. Nucleic Acids Res. 2013;41:e68. doi: 10.1093/nar/gks1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Feng P.M., Lin H., Chou K.C. iSS-PseDNC: identifying splicing sites using pseudo dinucleotide composition. Biomed. Res. Int. 2014:623149. doi: 10.1155/2014/623149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Lin H., Chou K.C. Pseudo nucleotide composition or PseKNC: an effective formulation for analyzing genomic sequences. Mol. BioSyst. 2015;11:2620–2634. doi: 10.1039/c5mb00155b. [DOI] [PubMed] [Google Scholar]
- Chen W., Lin H., Feng P.M., Ding C., Zuo Y.C., Chou K.C. iNuc-PhysChem: a sequence-based predictor for identifying nucleosomes via physicochemical properties. PLoS One. 2012;7 doi: 10.1371/journal.pone.0047843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chothia C., Finkelstein A.V. The classification and origins of protein folding patterns. Annu. Rev. Biochem. 1990;59:1007–1039. doi: 10.1146/annurev.bi.59.070190.005043. [DOI] [PubMed] [Google Scholar]
- Chothia C. The nature of the accessible and buried surfaces in proteins. J. Mol. Biol. 1976;105:1–12. doi: 10.1016/0022-2836(76)90191-1. [DOI] [PubMed] [Google Scholar]
- Chou K.C. Some remarks on protein attribute prediction and pseudo amino acid composition. J. Theor. Biol. 2011;273:236–247. doi: 10.1016/j.jtbi.2010.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou K.C. Some remarks on predicting multi-label attributes in molecular biosystems. Mol. BioSyst. 2013;9:1092–1100. doi: 10.1039/c3mb25555g. [DOI] [PubMed] [Google Scholar]
- Chou K.C. Impacts of bioinformatics to medicinal chemistry. Med. Chem. 2015;11:218–234. doi: 10.2174/1573406411666141229162834. [DOI] [PubMed] [Google Scholar]
- Chou K.C., Wu Z.C., Xiao X. iLoc-Hum: using the accumulation-label scale to predict subcellular locations of human proteins with both single and multiple sites. Mol. BioSyst. 2012;8:629–641. doi: 10.1039/c1mb05420a. [DOI] [PubMed] [Google Scholar]
- Chou K.C., Zhang C.T. Prediction of protein structural classes. Crit. Rev. Biochem. Mol. Biol. 1995;30:275–349. doi: 10.3109/10409239509083488. [DOI] [PubMed] [Google Scholar]
- Connolly M.L. Analytical molecular surface calculation. J. Appl. Crystallogr. 1983;16:548–558. [Google Scholar]
- Cooper D.N., Ball E.V., Krawczak M. The human gene mutation database. Nucleic Acids Res. 1998;26:285–287. doi: 10.1093/nar/26.1.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cozzolino M., Malindretos P. The role of vitamin D receptor activation in chronic kidney disease. Hippokratia. 2010;14:7–9. [PMC free article] [PubMed] [Google Scholar]
- Damasiewicz M.J., Toussaint N.D., Polkinghorne K.R. Fibroblast growth factor 23 in chronic kidney disease: new insights and clinical implications. Nephrology (Carlton) 2011;16:261–268. doi: 10.1111/j.1440-1797.2011.01443.x. [DOI] [PubMed] [Google Scholar]
- De Alencar S.A., Lopes J.C. A comprehensive in silico analysis of the functional and structural impact of SNPs in the IGF1R gene. J. Biomed. Biotechnol. 2010 doi: 10.1155/2010/715139. (Airtcle ID: 715139) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrow E.G., Imel E.A., White K.E. Miscellaneous non-inflammatory musculoskeletal conditions. Hyperphosphatemic familial tumoral calcinosis (FGF23, GALNT3 and alphaKlotho) Best. Pract. Res. Clin. Rheumatol. 2011;25:735–747. doi: 10.1016/j.berh.2011.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garringer H.J., Malekpour M., Esteghamat F., Mortazavi S.M., Davis S.I., Farrow E.G., Yu X., Arking D.E., Dietz H.C., White K.E. Molecular genetic and biochemical analyses of FGF23 mutations in familial tumoral calcinosis. Am. J. Physiol. Endocrinol. Metab. 2008;295:E929–E937. doi: 10.1152/ajpendo.90456.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz R., Beenken A., Ibrahimi O.A., Kalinina J., Olsen S.K., Eliseenkova A.V., Xu C., Neubert T.A., Zhang F., Linhardt R.J., Yu X., White K.E., Inagaki T., Kliewer S.A., Yamamoto M., Kurosu H., Ogawa Y., Kuro-O M., Lanske B., Razzaque M.S., Mohammadi M. Molecular insights into the klotho-dependent, endocrine mode of action of fibroblast growth factor 19 subfamily members. Mol. Cell. Biol. 2007;27:3417–3428. doi: 10.1128/MCB.02249-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerois R., Nielsen J.E., Serrano L. Predicting changes in the stability of proteins and protein complexes: a study of more than 1000 mutations. J. Mol. Biol. 2002;320:369–387. doi: 10.1016/S0022-2836(02)00442-4. [DOI] [PubMed] [Google Scholar]
- Gupta A., Winer K., Econs M.J., Marx S.J., Collins M.T. FGF-23 is elevated by chronic hyperphosphatemia. J. Clin. Endocrinol. Metab. 2004;89:4489–4492. doi: 10.1210/jc.2004-0724. [DOI] [PubMed] [Google Scholar]
- Haste Andersen P., Nielsen M., Lund O. Prediction of residues in discontinuous B-cell epitopes using protein 3D structures. Protein Sci. 2006;15:2558–2567. doi: 10.1110/ps.062405906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holick M. Physiology, Molecular Biology, and Clinical Applications. 2010. Vitamin D. (ISBN 978-1-60327-303-9) [Google Scholar]
- Hudson T.J. Wanted: regulatory SNPs. Nat. Genet. 2003;33:439–440. doi: 10.1038/ng0403-439. [DOI] [PubMed] [Google Scholar]
- Inoue Y., Segawa H., Kaneko I., Yamanaka S., Kusano K., Kawakami E., Furutani J., Ito M., Kuwahata M., Saito H., Fukushima N., Kato S., Kanayama H.O., Miyamoto K. Role of the vitamin D receptor in FGF23 action on phosphate metabolism. Biochem. J. 2005;390:325–331. doi: 10.1042/BJ20041799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H.F., Shen L. CYP24A1 mutations in idiopathic infantile hypercalcemia. N. Engl. J. Med. 2011;365:1741. doi: 10.1056/NEJMc1110226. author reply 1742-3. [DOI] [PubMed] [Google Scholar]
- Jia J., Liu Z., Xiao X., Liu B., Chou K.C. iSuc-PseOpt: identifying lysine succinylation sites in proteins by incorporating sequence-coupling effects into pseudo components and optimizing imbalanced training dataset. Anal. Biochem. 2016;497:48–56. doi: 10.1016/j.ab.2015.12.009. [DOI] [PubMed] [Google Scholar]
- Jia J., Liu Z., Xiao X., Liu B., Chou K.C. pSuc-Lys: predict lysine succinylation sites in proteins with PseAAC and ensemble random forest approach. J. Theor. Biol. 2016;394:223–230. doi: 10.1016/j.jtbi.2016.01.020. [DOI] [PubMed] [Google Scholar]
- Jia J., Liu Z., Xiao X., Liu B., Chou K.C. iPPBS-Opt: a sequence-based ensemble classifier for identifying protein–protein binding sites by optimizing imbalanced training datasets. Molecules. 2016;21 doi: 10.3390/molecules21010095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S., Thornton J.M. Analysis of protein–protein interaction sites using surface patches. J. Mol. Biol. 1997;272:121–132. doi: 10.1006/jmbi.1997.1234. [DOI] [PubMed] [Google Scholar]
- Jones S., Thornton J.M. Prediction of protein–protein interaction sites using patch analysis. J. Mol. Biol. 1997;272:133–143. doi: 10.1006/jmbi.1997.1233. [DOI] [PubMed] [Google Scholar]
- Kakuda S., Ishizuka S., Eguchi H., Mizwicki M.T., Norman A.W., Takimoto-Kamimura M. Structural basis of the histidine-mediated vitamin D receptor agonistic and antagonistic mechanisms of (23S)-25-dehydro-1alpha-hydroxyvitamin D3-26,23-lactone. Acta Crystallogr. D Biol. Crystallogr. 2010;66:918–926. doi: 10.1107/S0907444910020810. [DOI] [PubMed] [Google Scholar]
- Kearse M., Moir R., Wilson A., Stones-Havas S., Cheung M., Sturrock S., Buxton S., Cooper A., Markowitz S., Duran C., Thierer T., Ashton B., Meintjes P., Drummond A. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson T., Marsell R., Schipani E., Ohlsson C., Ljunggren O., Tenenhouse H.S., Juppner H., Jonsson K.B. Transgenic mice expressing fibroblast growth factor 23 under the control of the alpha1(I) collagen promoter exhibit growth retardation, osteomalacia, and disturbed phosphate homeostasis. Endocrinology. 2004;145:3087–3094. doi: 10.1210/en.2003-1768. [DOI] [PubMed] [Google Scholar]
- Li H., Robertson A.D., Jensen J.H. Very fast empirical prediction and rationalization of protein pKa values. Proteins. 2005;61:704–721. doi: 10.1002/prot.20660. [DOI] [PubMed] [Google Scholar]
- Lin H., Deng E.Z., Ding H., Chen W., Chou K.C. iPro54-PseKNC: a sequence-based predictor for identifying sigma-54 promoters in prokaryote with pseudo k-tuple nucleotide composition. Nucleic Acids Res. 2014;42:12961–12972. doi: 10.1093/nar/gku1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Fang L., Liu F., Wang X., Chen J., Chou K.C. Identification of real microRNA precursors with a pseudo structure status composition approach. PLoS One. 2015;10 doi: 10.1371/journal.pone.0121501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Fang L., Liu F., Wang X., Chou K.C. iMiRNA-PseDPC: microRNA precursor identification with a pseudo distance-pair composition approach. J. Biomol. Struct. Dyn. 2016;34:223–235. doi: 10.1080/07391102.2015.1014422. [DOI] [PubMed] [Google Scholar]
- Liu B., Fang L., Long R., Lan X., Chou K.C. iEnhancer-2L: a two-layer predictor for identifying enhancers and their strength by pseudo k-tuple nucleotide composition. Bioinformatics. 2016;32:362–369. doi: 10.1093/bioinformatics/btv604. [DOI] [PubMed] [Google Scholar]
- Liu B., Fang L., Wang S., Wang X., Li H., Chou K.C. Identification of microRNA precursor with the degenerate K-tuple or Kmer strategy. J. Theor. Biol. 2015;385:153–159. doi: 10.1016/j.jtbi.2015.08.025. [DOI] [PubMed] [Google Scholar]
- Liu S., Quarles L.D. How fibroblast growth factor 23 works. J. Am. Soc. Nephrol. 2007;18:1637–1647. doi: 10.1681/ASN.2007010068. [DOI] [PubMed] [Google Scholar]
- Liu Z., Xiao X., Qiu W.R., Chou K.C. iDNA-Methyl: identifying DNA methylation sites via pseudo trinucleotide composition. Anal. Biochem. 2015;474:69–77. doi: 10.1016/j.ab.2014.12.009. [DOI] [PubMed] [Google Scholar]
- Liu Z., Xiao X., Yu D.J., Jia J., Qiu W.R., Chou K.C. pRNAm-PC: predicting N(6)-methyladenosine sites in RNA sequences via physical–chemical properties. Anal. Biochem. 2016;497:60–67. doi: 10.1016/j.ab.2015.12.017. [DOI] [PubMed] [Google Scholar]
- Loh Z.Y., Yap C.W., Vathsala A., How P. Clinical and demographic predictors for vitamin D deficiency in multiethnic Asian patients with chronic kidney disease. Clin. Kidney J. 2012;5:303–308. doi: 10.1093/ckj/sfs074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maestro . Schrodinger, LLC; New York, NY: 2015. Version 9.10. [Google Scholar]
- Mah J.T., Low E.S., Lee E. In silico SNP analysis and bioinformatics tools: a review of the state of the art to aid drug discovery. Drug Discov. Today. 2011;16:800–809. doi: 10.1016/j.drudis.2011.07.005. [DOI] [PubMed] [Google Scholar]
- Matthews B.W. Comparison of the predicted and observed secondary structure of T4 phage lysozyme. Biochim. Biophys. Acta. 1975;405:442–451. doi: 10.1016/0005-2795(75)90109-9. [DOI] [PubMed] [Google Scholar]
- Minde D.P., Anvarian Z., Rudiger S.G., Maurice M.M. Messing up disorder: how do missense mutations in the tumor suppressor protein APC lead to cancer? Mol. Cancer. 2011;10:101. doi: 10.1186/1476-4598-10-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooney S.D., Krishnan V.G., Evani U.S. Bioinformatic tools for identifying disease gene and SNP candidates. Methods Mol. Biol. 2010;628:307–319. doi: 10.1007/978-1-60327-367-1_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakabayashi M., Tsukahara Y., Iwasaki-Miyamoto Y., Mihori-Shimazaki M., Yamada S., Inaba S., Oda M., Shimizu M., Makishima M., Tokiwa H., Ikura T., Ito N. Crystal structures of hereditary vitamin D-resistant rickets-associated vitamin D receptor mutants R270L and W282R bound to 1,25-dihydroxyvitamin D3 and synthetic ligands. J. Med. Chem. 2013;56:6745–6760. doi: 10.1021/jm400537h. [DOI] [PubMed] [Google Scholar]
- Nesterova G., Malicdan M.C., Yasuda K., Sakaki T., Vilboux T., Ciccone C., Horst R., Huang Y., Golas G., Introne W., Huizing M., Adams D., Boerkoel C.F., Collins M.T., Gahl W.A. 1,25-(OH)2D-24 hydroxylase (CYP24A1) deficiency as a cause of nephrolithiasis. Clin. J. Am. Soc. Nephrol. 2013;8:649–657. doi: 10.2215/CJN.05360512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng P.C., Henikoff S. Accounting for human polymorphisms predicted to affect protein function. Genome Res. 2002;12:436–446. doi: 10.1101/gr.212802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng P.C., Henikoff S. Predicting the effects of amino acid substitutions on protein function. Annu. Rev. Genomics Hum. Genet. 2006;7:61–80. doi: 10.1146/annurev.genom.7.080505.115630. [DOI] [PubMed] [Google Scholar]
- Nguyen M., D'alesio A., Pascussi J.M., Kumar R., Griffin M.D., Dong X., Guillozo H., Rizk-Rabin M., Sinding C., Bougneres P., Jehan F., Garabedian M. Vitamin D-resistant rickets and type 1 diabetes in a child with compound heterozygous mutations of the vitamin D receptor (L263R and R391S): dissociated responses of the CYP-24 and rel-B promoters to 1,25-dihydroxyvitamin D3. J. Bone Miner. Res. 2006;21:886–894. doi: 10.1359/jbmr.060307. [DOI] [PubMed] [Google Scholar]
- Norman A.W. From vitamin D to hormone D: fundamentals of the vitamin D endocrine system essential for good health. Am. J. Clin. Nutr. 2008;88:491S–499S. doi: 10.1093/ajcn/88.2.491S. [DOI] [PubMed] [Google Scholar]
- Olsson M.H.M., Sandergaard C.R., Rostkowski M., Jensen J.H. PROPKA3: consistent treatment of internal and surface residues in empirical pKa predictions. J. Chem. Theory Comput. 2011;7:525–537. doi: 10.1021/ct100578z. [DOI] [PubMed] [Google Scholar]
- Panchenko A.R., Kondrashov F., Bryant S. Prediction of functional sites by analysis of sequence and structure conservation. Protein Sci. 2004;13:884–892. doi: 10.1110/ps.03465504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perwad F., Zhang M.Y., Tenenhouse H.S., Portale A.A. Fibroblast growth factor 23 impairs phosphorus and vitamin D metabolism in vivo and suppresses 25-hydroxyvitamin D-1alpha-hydroxylase expression in vitro. Am. J. Physiol. Ren. Physiol. 2007;293:F1577–F1583. doi: 10.1152/ajprenal.00463.2006. [DOI] [PubMed] [Google Scholar]
- Petersen B., Petersen T.N., Andersen P., Nielsen M., Lundegaard C. A generic method for assignment of reliability scores applied to solvent accessibility predictions. BMC Struct. Biol. 2009;9:51. doi: 10.1186/1472-6807-9-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petkovich M., Jones G. CYP24A1 and kidney disease. Curr. Opin. Nephrol. Hypertens. 2011;20:337–344. doi: 10.1097/MNH.0b013e3283477a7b. [DOI] [PubMed] [Google Scholar]
- Prasad P., Thelma B.K. Normative genetic profiles of RAAS pathway gene polymorphisms in North Indian and South Indian populations. Hum. Biol. 2007;79:241–254. doi: 10.1353/hub.2007.0033. [DOI] [PubMed] [Google Scholar]
- Prime . Schrödinger, LLC; New York, NY: 2015. Version 3.9. [Google Scholar]
- Qiu W.R., Xiao X., Lin W.Z., Chou K.C. iUbiq-Lys: prediction of lysine ubiquitination sites in proteins by extracting sequence evolution information via a gray system model. J. Biomol. Struct. Dyn. 2015;33:1731–1742. doi: 10.1080/07391102.2014.968875. [DOI] [PubMed] [Google Scholar]
- Ramensky V., Bork P., Sunyaev S. Human non-synonymous SNPs: server and survey. Nucleic Acids Res. 2002;30:3894–3900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T., Fukumoto S. Fibroblast growth factor 23 (FGF23) and disorders of phosphate metabolism. Int. J. Pediatr. Endocrinol. 2009;2009:496514. doi: 10.1155/2009/496514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaki T., Yasuda K., Kittaka A., Yamamoto K., Chen T.C. CYP24A1 as a potential target for cancer therapy. Anti Cancer Agents Med. Chem. 2014;14:97–108. doi: 10.2174/18715206113139990307. [DOI] [PubMed] [Google Scholar]
- Sherry S.T., Ward M.H., Kholodov M., Baker J., Phan L., Smigielski E.M., Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada T., Yamazaki Y., Takahashi M., Hasegawa H., Urakawa I., Oshima T., Ono K., Kakitani M., Tomizuka K., Fujita T., Fukumoto S., Yamashita T. Vitamin D receptor-independent FGF23 actions in regulating phosphate and vitamin D metabolism. Am. J. Physiol. Ren. Physiol. 2005;289:F1088–F1095. doi: 10.1152/ajprenal.00474.2004. [DOI] [PubMed] [Google Scholar]
- Singh Kh D., Karthikeyan M. Combined sequence and sequence-structure-based methods for analyzing RAAS gene SNPs: a computational approach. J. Recept. Signal Transduct. Res. 2014;34:513–526. doi: 10.3109/10799893.2014.922575. [DOI] [PubMed] [Google Scholar]
- Sjolander K., Karplus K., Brown M., Hughey R., Krogh A., Mian I.S., Haussler D. Dirichlet mixtures: a method for improved detection of weak but significant protein sequence homology. Comput. Appl. Biosci. 1996;12:327–345. doi: 10.1093/bioinformatics/12.4.327. [DOI] [PubMed] [Google Scholar]
- Slattery M.L. Vitamin D receptor gene (VDR) associations with cancer. Nutr. Rev. 2007;65:S102–S104. doi: 10.1111/j.1753-4887.2007.tb00332.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thusberg J., Olatubosun A., Vihinen M.m. Performance of mutation pathogenicity prediction methods on missense variants. Hum. Mutat. 2011;32:358–368. doi: 10.1002/humu.21445. [DOI] [PubMed] [Google Scholar]
- Wahl P., Wolf M. FGF23 in chronic kidney disease. Adv. Exp. Med. Biol. 2012;728:107–125. doi: 10.1007/978-1-4614-0887-1_8. [DOI] [PubMed] [Google Scholar]
- Wan M., Smith C., Shah V., Gullet A., Wells D., Rees L., Shroff R. Fibroblast growth factor 23 and soluble klotho in children with chronic kidney disease. Nephrol. Dial. Transplant. 2012;28:153–161. doi: 10.1093/ndt/gfs411. [DOI] [PubMed] [Google Scholar]
- Wheeler D.L., Church D.M., Lash A.E., Leipe D.D., Madden T.L., Pontius J.U., Schuler G.D., Schriml L.M., Tatusova T.A., Wagner L., Rapp B.A. Database resources of the National Center for Biotechnology Information: 2002 update. Nucleic Acids Res. 2001;30:13–16. doi: 10.1093/nar/30.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield G.K., Selznick S.H., Haussler C.A., Hsieh J.C., Galligan M.A., Jurutka P.W., Thompson P.D., Lee S.M., Zerwekh J.E., Haussler M.R. Vitamin D receptors from patients with resistance to 1,25-dihydroxyvitamin D3: point mutations confer reduced transactivation in response to ligand and impaired interaction with the retinoid X receptor heterodimeric partner. Mol. Endocrinol. 1996;10:1617–1631. doi: 10.1210/mend.10.12.8961271. [DOI] [PubMed] [Google Scholar]
- Xi T., Jones I.M., Mohrenweiser H.W. Many amino acid substitution variants identified in DNA repair genes during human population screenings are predicted to impact protein function. Genomics. 2004;83:970–979. doi: 10.1016/j.ygeno.2003.12.016. [DOI] [PubMed] [Google Scholar]
- Xiao X., Wang P., Lin W.Z., Jia J.H., Chou K.C. iAMP-2L: a two-level multi-label classifier for identifying antimicrobial peptides and their functional types. Anal. Biochem. 2013;436:168–177. doi: 10.1016/j.ab.2013.01.019. [DOI] [PubMed] [Google Scholar]
- Yan H., Yuan W., Velculescu V.E., Vogelstein B., Kinzler K.W. Allelic variation in human gene expression. Science. 2002;297:1143. doi: 10.1126/science.1072545. [DOI] [PubMed] [Google Scholar]
- Yue P., Moult J. Identification and analysis of deleterious human SNPs. J. Mol. Biol. 2006;356:1263–1274. doi: 10.1016/j.jmb.2005.12.025. [DOI] [PubMed] [Google Scholar]
- Yue P., Li Z., Moult J. Loss of protein structure stability as a major causative factor in monogenic disease. J. Mol. Biol. 2005;353:459–473. doi: 10.1016/j.jmb.2005.08.020. [DOI] [PubMed] [Google Scholar]