Abstract
The Neurospora VS ribozyme is a catalytic RNA that has the unique ability to specifically recognize and cleave a stem-loop substrate through formation of a highly stable kissing-loop interaction (KLI). In order to explore the engineering potential of the VS ribozyme to cleave alternate substrates, we substituted the wild-type KLI by other known KLIs using an innovative engineering method that combines rational and combinatorial approaches. A bioinformatic search of the protein data bank was initially performed to identify KLIs that are structurally similar to the one found in the VS ribozyme. Next, substrate/ribozyme (S/R) pairs that incorporate these alternative KLIs were kinetically and structurally characterized. Interestingly, several of the resulting S/R pairs allowed substrate cleavage with substantial catalytic efficiency, although with reduced activity compared to the reference S/R pair. Overall, this study describes an innovative approach for RNA engineering and establishes that the KLI of the trans VS ribozyme can be adapted to cleave other folded RNA substrates.
INTRODUCTION
The Neurospora VS ribozyme belongs to the family of natural nucleolytic ribozymes, which are <200 nt in size and include the hairpin, HDV, hammerhead, glmS, twister, twister sister, pistol and hatchet ribozymes (1–5). Like many other ribozymes, the VS ribozyme provides a good model system for fundamental studies of RNA structure and function, which are of value for understanding more complex RNA-mediated processes (6). So far, a large body of in vitro work has led to a comprehensive functional and structural understanding of the VS ribozyme both in its cis and trans forms, which vary according to the covalent attachment of the stem-loop I (SLI) substrate domain [for reviews see (1,7–9)]. These studies have highlighted a unique mode of substrate recognition for the VS ribozyme, which involves specific recognition of the SLI domain in a folded hairpin state (10). Only a few studies have addressed the possibility of using the VS ribozyme to target and cleave RNA stem-loops that differ from the natural SLI substrate. In contrast, several other small nucleolytic ribozymes, particularly the hairpin, hammerhead and HDV ribozymes, have been modified to cleave numerous alternative single-stranded RNA substrates. For the VS ribozyme, the engineering principles that govern recognition of alternate substrates are likely more complex, but remain to be elucidated.
The catalytic domain or trans VS ribozyme (helical domains II–VI; Figure 1A) recognizes its hairpin substrate (SLI) via the formation of a kissing-loop interaction (KLI) required for efficient cleavage activity (10,11). In earlier work, mutagenesis and chemical modification studies provided evidence that this KLI contains three WC/WC (12) base pairs (bp) between loop I (G630, U631 and C632) and loop V (G697, A698 and C699), and that both loops likely form U-turn motifs (11). More recently, the nuclear magnetic resonance (NMR) structure of an SLI/SLV complex (13) and the crystal structure of the complete VS ribozyme (14) confirmed formation of the U-turn motifs and three cis WC/WC bp, but they also revealed the presence of a cis WC/sugar edge bp between C629 and A701 (between C10 of SLI and A14 of SLV in the SLI/SLV complex; Figure 1B). In vitro selection and biochemical studies of the VS ribozyme (15,16) as well as NMR studies of several SLI/SLV complexes (13) have provided evidence that formation of the I/V KLI is associated with a helix shift in the SLI substrate that is essential for catalysis. SLI variants that adopt a pre-shifted conformation bind with higher affinity to both the trans ribozyme (17) and an isolated SLV (18). Moreover, isothermal titration calorimetry (ITC) studies of SLI/SLV complexes conducted with a pre-shifted SLI RNA demonstrated that the I/V KLI is remarkably stable (Kd = 0.24 ± 0.02 μM) and constitutes the predominant energetic contribution for substrate binding by the trans VS ribozyme (18). The I/V KLI is also important for substrate docking with the catalytic domain, which allows for the formation of the active site (19). In the proposed general acid-base mechanism of the VS ribozyme cleavage reaction, G638 from the internal loop of SLI (G638 loop) acts as the general base and A756 from the internal loop of SLVI (A756 loop) acts as the general acid (20–27). The recent crystal structure of the VS ribozyme confirms that the active site is formed through the association of these two internal loops, in which G638 and A756 are properly positioned with respect to the scissile phosphate to fulfill their proposed roles in catalysis (14).
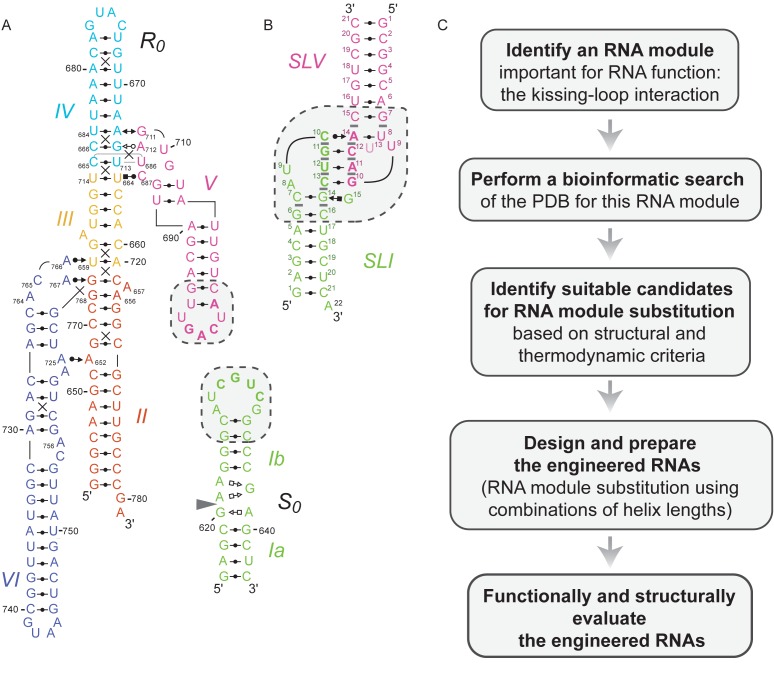
Figure 1.
Rational engineering of KLIs for substrate recognition by the Neurospora VS ribozyme. (A) Structural schematic of a trans VS ribozyme system formed by the S0 substrate and the R0 ribozyme (32). The schematic is derived from NMR and X-ray structures (14,43). The cleavage site is located between G620 and A621 as indicated by the gray arrowhead. (B) Structural characteristics of the VS ribozyme I/V KLI derived from the NMR structure of the SLI/SLV complex [PDB ID: 2MI0; (13)]. (C) Flowchart of the engineering strategy used in this study. In (A and B), the dashed boxes enclose residues that contribute to the KLI and the closing base pairs of the two stems, whereas base-pairing interactions are represented using the Leontis-Westhof notation (12). In addition, base-pairing residues at the KLI are in bold. In (B), gray bars represent base stacking at the KLI.
Only a limited number of studies have investigated the possibility of using the VS ribozyme to selectively cleave RNA stem-loops other than its natural SLI substrate. Although minor changes to the sequence of the natural SLI substrate are compatible with VS ribozyme cleavage (11,15,28–31), larger sequence changes in the substrate may only be possible by incorporating compensating changes in the catalytic domain. To test this possibility, we recently investigated the cleavage of variant SLI substrates, in which the number of base pairs in stem Ib was either increased or decreased, using variant trans ribozymes containing a variable number of base pairs in stem V (32). We observed helix-length compensation between stem Ib and stem V, since the highest activity for each ribozyme variant was observed with a substrate that contains the compensatory number of base pairs in stem Ib (or one less). Moreover, this study establishes that VS ribozyme derivatives efficiently cleave SLI substrates that contain an alternate number of base pairs in stem Ib.
Given these encouraging results with the VS ribozyme, we were interested to gain further insights into the adaptability of this unique ribozyme for specific cleavage of folded RNA stem-loops. Previous studies have led to the idea that the I/V KLI of the VS ribozyme is an independent RNA module (13,16,18,31), and we hypothesized that this KLI can be substituted by other KLIs to create variant ribozymes that efficiently cleave stem-loop substrates with a modified terminal loop. In the present study, we developed a rational engineering approach to replace the I/V KLI of the Neurospora VS ribozyme by other KLIs and achieve efficient substrate cleavage (Figure 1C). This rational approach is supported by the recently available NMR structure of an SLI/SLV complex (13) and the crystal structure of the complete VS ribozyme (14). We first performed a bioinformatic search for KLIs in the protein data bank (PDB), which allowed us to identify two KLIs that share structural characteristics with the natural KLI of the VS ribozyme. Next, VS substrate/ribozyme (S/R) pairs, in which the I/V KLI was substituted by each of these two KLI candidates, were prepared using several combinations of helix length for stems Ib and V (32) in order to fine-tune the structural context of the surrogate KLIs necessary to obtain efficient cleavage of the modified substrates. The engineered S/R pairs were then kinetically characterized to identify the most efficient ones and structurally evaluated to validate the procedure. Interestingly, several novel S/R pairs were identified that display substantial catalytic activity, further illustrating the adaptability of the VS ribozyme architecture for cleavage of alternate stem-loop substrates.
MATERIALS AND METHODS
Search for KLIs with WebFR3D
A bioinformatic search was performed with WebFR3D (33) to find KLIs in the PDB. The search query was defined as an interaction matrix between a set of 6 nt with no specific nucleotide identity (NT1 to NT6; Supplementary Figure S1B). Nucleotides NT1 and NT3 form a cis WC/WC base pair that closes the first loop, and this loop contains a non-self-pairing strand of 4–10 nt. Nucleotides NT4 and NT6 form a cis WC/WC base pair that closes the second loop, and this loop also contains a non-self-pairing strand of 4–10 nt. Nucleotides NT2 and NT5 are part of the first and second loops, respectively, and must form a cis WC/WC base pair. Finally, residues NT1-NT3 must be either sequentially located 5′ respectively to residues NT4-NT6 or part of a different chain. For this search, we used the WebFR3D list of non-redundant RNA-containing structures determined by X-ray crystallography, Cryo-EM or NMR spectroscopy.
Clustering of WebFR3D results
A custom Python script was used to parse the HTML output file of WebFR3D. The script extracts two key features about each entry: the PDB ID of the associated structure and the type and numbering of the nucleotides (NT1-NT6) matching the query. It then uses this information to cluster all fragments associated with a single KLI and quantifies the number of cis WC/WC base pairs associated with this interaction.
Structural data retrieval and result listing
A custom Python script was used to automatically gather information about each KLI. The size of each loop and the total number of cis WC/WC base pairs in the KLI were retrieved directly from the cluster of WebFR3D results and used to define the secondary structure of each KLI. Additional information was retrieved from the RCSB repository via the PDB ID of the KLI. The information gathered about each KLI structure was listed as an HTML page enriched with JavaScript (Supplementary Figure S1D and Table S1).
Plasmids
The Avapl ribozyme (R0, Figure 1A) is a derivative of the AvaI ribozyme and its parental plasmid (pAvapl) was constructed by polymerase chain reaction from the A-3 clone (10). The plasmids for transcription of the RTAR*, RL22 and RL88 ribozyme variants (Figure 3A and Supplementary Figures S3A and 4A) were prepared by QuikChange II site-directed mutagenesis (Stratagene, CA, USA). All plasmids were validated by DNA sequencing.
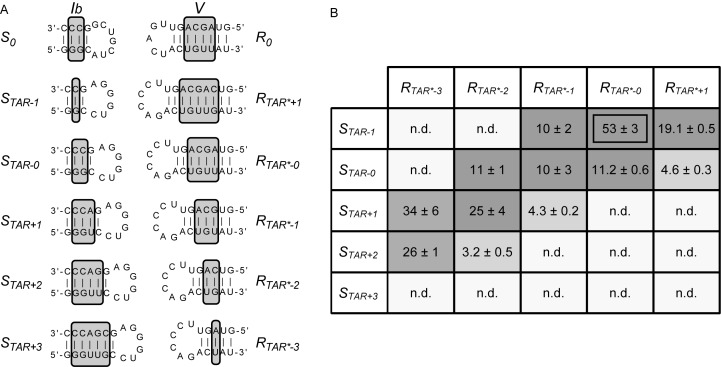
Figure 3.
Sequences and kinetic data of the STAR/RTAR*variants. (A) Sequences of the STAR/RTAR* variants. Only the regions of the S0 substrate and R0 ribozyme (stem-loops Ib and V shown above; Figure 1A) that were substituted in the STAR and RTAR* variants are shown. The number of base pairs varies in stems Ib and V, as indicated by the gray boxes on the putative secondary structures. (B) Grayscale heat map of the kcat/KM values (10−2 min−1μM−1) for the STAR/RTAR* variants. Dark gray shading corresponds to kcat/KM values within 5-fold of the most active STAR/RTAR* pair (STAR-1/RTAR*-0); medium gray shading corresponds to lower kcat/KM values (5–17-fold lower than the most active STAR/RTAR* pair), and light gray shading reflects the absence of detectable cleavage activity (n.d.: kobs ≤ 0.002 min−1) after 24 min at [R] ≤ 15 μM.
ITC studies
Preparation of RNA and ITC studies were performed as previously described (18). Relevant experimental details are provided in the legend of Supplementary Figure S2.
Preparation of RNAs for kinetic studies
The RTAR*, RL22 and RL88 ribozymes were prepared by in vitro T7 RNA polymerase transcription from plasmid templates linearized at the AvaI site. All ribozymes were purified by gel electrophoresis and ion-exchange HPLC, as previously described (32). The STAR, SL88 and SL22 substrates were synthesized in vitro using the T7 RNA polymerase and partially single-stranded synthetic oligonucleotide templates (Integrated DNA Technologies, IA, USA). These substrates were purified by gel electrophoresis, dephosphorylated with calf alkaline phosphatase and 5′-end-labeled with γ-(32P) adenosine triphosphate (PerkinElmer, Ont.) using established procedures (32).
Kinetic studies
Mutiple-substrate and single-substrate kinetic studies were carried out as single-turnover reactions at 37°C in 1× Kinetic Reaction buffer (50 mM Tris at pH 7.6, 25 mM KCl, 2 mM spermidine and 25 mM MgCl2), as previously described (32). Briefly, multiple-substrate kinetic studies were typically performed with a mixture of 5′-32P-labeled nucleic acids containing all purified substrates for a given S/R pair (35 pM of each) and excess R (0.005–25 μM). These multiple-substrate kinetic studies were used to identify quickly the S/R pairs that lead to detectable substrate cleavage. All multiple-substrate kinetic experiments were performed at least three times.
Reactive S/R combinations were further investigated by single-substrate kinetic studies to derive the values of kobs and kcat/KM. These studies were typically performed with [S] = 250 pM and excess R (0.005–15 μM). The percentage of remaining substrate [F = St/(St + Pt) * 100%; where St and Pt are the amount of substrate and product at time t] was plotted against time. The resulting time courses were fit to the equation F = Ae−(kobs * t) + F∞, where A is the amplitude, kobs is the rate of the reaction and F∞ is a variable that represents the unreactive fraction of the substrate. The value of kobs, the standard error for kobs and the F∞ parameter were estimated by non-linear regression analysis within the OriginPro 8.1 software (OriginLab, MA, USA). The F∞ values are below 10% for all S/R combinations. A linear dependence of kobs on [R] was observed for each S/R combination at the lowest values of [R] and analyzed by linear regression to derive the second order rate constant of the reaction, kcat/KM. The quality of the fit for the linear dependence of the kobs on [R] was obtained from the square of the correlation coefficient (R2), and in all cases R2 was ≥ 0.99. For the single-substrate kinetic experiments, three independent experiments were performed, and reported kcat/KM and their errors are average values and standard deviations, respectively, from these multiple measurements.
Native gel studies of RNA substrates
Non-denaturing polyacrylamide gel electrophoresis was carried out on a 7.5% polyacrylamide bisacrylamide (37.5:1) gel in Tris-glycine buffer (25 mM Tris pH 8.0 and 200 mM glycine). Prior to loading, the RNA substrates (1 and 4 μg) were heated at 95°C for 2 min in storage buffer [10 mM Tris pH 7.6 and 1 mM ethylenediaminetetraacetic acid (EDTA)] and transferred on ice for 5 min. Then, 4 μl of 2× Kinetic Reaction buffer was added and the samples were incubated at 37°C for 5 min. The gel was pre-run for 30 min at 100 V, run for 2 h at 250 V and 4°C and then stained with Stains All (Sigma-Aldrich, Ont.).
SHAPE studies of ribozymes
For SHAPE (Selective 2′-hydroxyl Acylation analyzed by Primer Extension) experiments, variant ribozymes (20 pmol) in 4 μl of H2O were first incubated at 37°C for 5 min with 5 μl of 2× Kinetic Reaction buffer and then treated with 1 μl of 130 mM N-methylisatoic anhydride (NMIA; dissolved in anhydrous dimethyl sulfoxide). The resulting mix was incubated for 54 min at 37°C and then transferred on ice. Reverse transcription reactions were performed on mutant ribozymes with 5′-32P-labeled primers and SuperScript III reverse transcriptase (ThermoFisher Scientific Inc.), as previously described (34). The primer extension products were separated on an 8% denaturing polyacrylamide gel (7 M urea, 90 mM Tris/Borate, 2 mM EDTA). The gel was visualized by phosphorimager (Personal Molecular Imager™ System, Bio-Rad) and quantified using Image Lab Software 4.1.
Three-dimensional modeling of S/R complexes with a substituted KLI
Models of the S0/R0, SL88-0/RL22-0 and STAR-0/RTAR*-0 complexes were constructed by fragment assembly in PyMOL (The PyMOL Molecular Graphics System, Version 1.7.4 Schrödinger, LLC.) based on heavy-atom superpositions with the SX/RX complex obtained from the X-ray structure of the VS ribozyme [PDB ID: 4R4P; (14)] by taking the SLI substrate of one protomer (SX) and the trans VS ribozyme (helical domains II–VI) of the other protomer (RX). Starting with representative high-resolution structures of the studied KLIs [PDB ID: 2MI0—model 2 (13)], 1KIS—model 2 (35) and 4IOA (36)], the hairpin stems within these complexes were modified by stems generated with MC-Sym (37) that match those of SLI (residues 623–626 and 634–637) and SLV (residues 688–694 and 702–708) from the S0/R0 complex (Figure 1A), and the resulting hybrid structures retained the hairpin loop residues of the original high-resolution structures (2MI0: residues 4–17 and 104–118; 1KIS: residues 5–12 and 21–28; and 4IOA: residues 423–430 and 2383–2392). The hairpin stem corresponding to stem V within these hybrid structures was superposed to stem V of RX. Subsequently, SX was copied one time for each hybrid structure and its stem Ib was superposed to stem Ib of each hybrid structure. The final S/R complexes retain all residues from the hybrid structures, all residues of RX except for residues 688–708 of SLV and only residues 616–622 and 638–643 of SX (according to residue numbering in Figure 1).
RESULTS
Search for KLIs in the PDB
We searched for three-dimensional structures of KLIs in the PDB using a three-step computational workflow (Supplementary Figure S1A) that is based on the motif-search tool WebFR3D (33). This online version of the FR3D program was chosen because it has the capacity to perform searches of RNA motifs composed of non-contiguous nucleotides in a curated ensemble of non-redundant structures. In the first step, a search query was launched with WebFR3D to identify single cis Watson–Crick (WC) base pairings between any two residues belonging to two different terminal loops of 4–10 residues in length (Supplementary Figure S1B). This search used the WebFR3D list of non-redundant RNA-containing structures determined by X-ray crystallography, Cryo-EM or NMR spectroscopy. In the second step, the individual entries of the WebFR3D output were clustered to identify all existing WC pairings for a specific pair of terminal loops, and thereby provide a list of KLIs. In the third and final step, additional structural data and related information were retrieved for each KLI, and these data were listed in an interactive HTML/JavaScript table (Supplementary Figure S1D and Table S1). From this motif search of the PDB, 113 KLIs were identified and these form between one and ten cis WC/WC base pairs at the kissing-loop interface.
Selection of KLIs that could substitute for the VS ribozyme I/V interaction
To identify KLIs that could substitute for the VS ribozyme I/V KLI, we automatically filtered the list of 113 structures to eliminate all entries originating from X-ray and Cryo-EM structures with resolution ≥3.6 Å and only selected those that form heterodimeric KLI interactions with 3–10 cis WC/WC base pairs. Heterodimeric KLIs were selected because substitutions of SLI and SLV in the VS ribozyme S/R system (Figure 1A) by stem-loops that form an homodimeric KLI might lead to self-association of substrates and ribozymes, which could interfere with trans cleavage. In addition, KLIs with a minimum of three cis WC/WC base pairs were selected to prevent formation of an unstable KLI that could lead to inefficient trans cleavage (11,31). This filtering step produced a list of 17 structures that represent 6 types of KLIs (Table 1): the TAR/TAR* interaction derived from HIV-1; the L6/L7 and L22/L88 interactions from the large subunit of rRNAs; the L5/L13 interaction from the cobalamin riboswitch; the L3/L4 interaction from the human SRP Alu domain; and the ColE1 KLI. To identify suitable KLIs for our engineering studies, we selected those that share structural similarities with the VS ribozyme I/V KLI. More specifically, a suitable KLI must fit certain structural characteristics: (i) form consecutive cis WC/WC base pair with non-modified nucleotides, (ii) the adjoining stems must contain at least three consecutive cis WC/WC base pairs and (iii) the interhelical angle must fall between 125° and 175° (13,38). Such structural criteria limited our choice to two classes of KLIs: the HIV-1 TAR/TAR* interaction and the L22/L88 (loop 22/loop 88) interaction from the large subunit of rRNAs, and those with PDB ID codes 1KIS (35) and 4IOA (36) were selected for our engineering studies.
Table 1. Filtered results of the bioinformatics search to identify KLIs from the PDB.

|
Thermodynamic stability of the selected KLIs
To ascertain that the two selected KLIs are good candidates for our engineering studies, we verified their thermodynamic stability. Although the TAR/TAR* and L22/L88 KLIs incorporate a larger number of WC base pairs than the VS ribozyme I/V KLI (Figures 1B and 2A and B), the latter is known to be exceptionally stable (18). Based on ITC studies, the Kd of the TAR/TAR* and L22/L88 complexes are 40-fold lower and 16-fold higher, respectively, than that of the SLI/SLV complex (Figure 2C and Supplementary Figure S2). These results confirm that the thermodynamic stability of the two selected KLIs is not considerably different from that of the I/V KLI. Thus, these two KLIs were considered suitable for our engineering studies.
Figure 2.
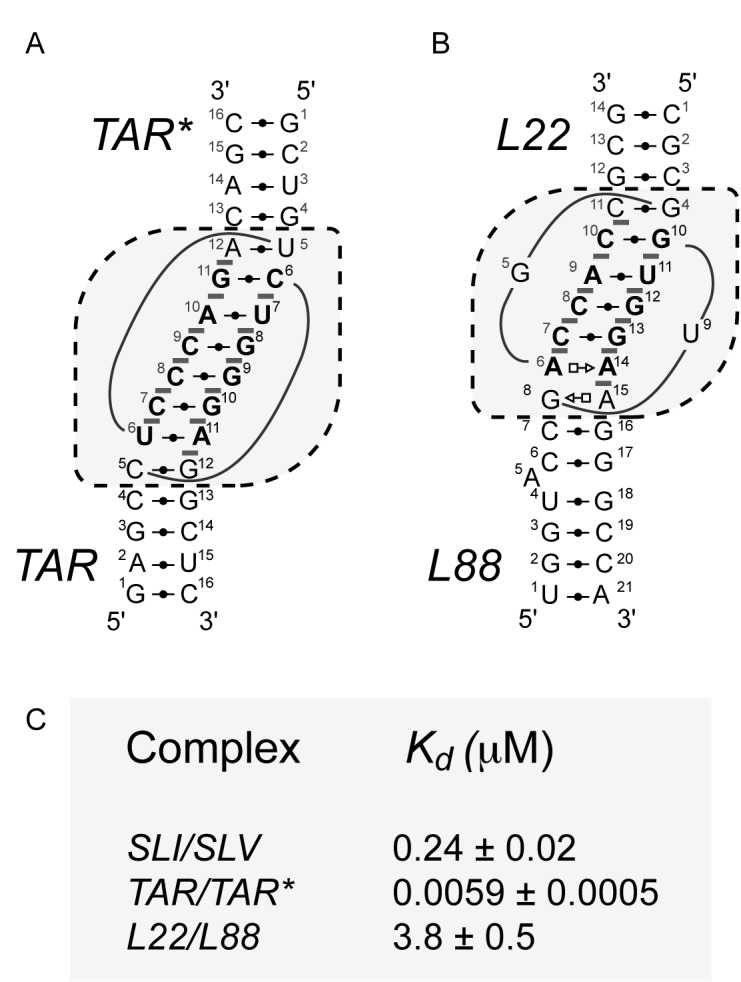
Schematics and thermodynamic stability of selected KLIs. Structural characteristics of the selected KLIs: (A) the HIV-1 TAR/TAR* KLI [PDB ID: 1KIS; (35)] and (B) the large ribosomal subunit L22/L88 KLI from Deinococcus radiodurans [PDB ID: 4IOA; (36)]. (C) Dissociation constants (Kd) of the selected KLIs determined in a previous study for the SLI/SLV and TAR/TAR* complexes (18) and here for the L22/L88 complex (Supplementary Figure S2). These Kd values were all determined at 25°C in 10 mM Tris pH 7.0 and 20 mM MgCl2. In (A and B), the dashed boxes enclose residues of the KLI and the closing base pairs of the two stems and base-pairing interactions are represented using the Leontis-Westhof notation (12). In addition, base-pairing residues at the KLI are in bold and gray bars highlight base stacking at the KLI.
Kinetic characterization of VS ribozyme S/R pairs with a modified I/V KLI
Next, kinetic studies were performed to test the hypothesis that the two selected KLIs can substitute for the natural I/V KLI interaction and yield active VS ribozyme variants. For these studies, we used the previously-characterized S0/R0 pair formed between a pre-shifted SLI substrate and a trans VS ribozyme (32). Since the selected KLIs may functionally replace the I/V KLI only when substituted in the proper orientation (e.g. either the L22 or the L88 loop replacing loop I) and with supporting helices of proper lengths, we prepared three combinatorial pools of S/R pairs that consist of 25 STAR/RTAR* pairs, 30 SL88/RL22 pairs and 36 SL22/RL88 pairs (Figure 3A and Supplementary Figures S3A and 4A). Within each pool, helix-length variations were introduced in both stem Ib of the variant substrates (STAR, SL88 and SL22) and stem V of the variant ribozymes (RTAR*, RL22 and RL88). In this context, compensatory S/R pairs are those for which the total number of base pairs in stems Ib and V is identical to that found for the S0/R0 reference pair (Figure 1). For all S/R combinations, the second-order rate constant kcat/KM was determined under single turnover conditions. It is important to note that the catalytic parameter kcat/KM is very useful when comparing the relative activity of a given enzyme for different competing substrates (e.g. RTAR*-0 acting on the different STAR substrates), but may be misleading when comparing the relative activity of different enzymes (39), particularly those of different S/R families. Thus, to compare the catalytic activity of different S/R families, we also relied on the values of the cleavage rate constant (kobs) measured under identical experimental conditions.
Kinetic studies of the VS ribozyme STAR/RTAR* pairs
Single turnover kinetic studies were performed to derive the second-order rate constant kcat/KM for all 25 STAR/RTAR* combinations. 32P-labeled STAR substrates were cleaved with an excess of RTAR* ribozymes, and the cleavage reaction monitored by denaturing gel electrophoresis. Initially, we used a multiple-substrate kinetic approach, where one enzyme is simultaneously incubated with a mixture of substrates (32). Given that the substrates are of different sizes (Figure 3A), the fate of any given substrate can be monitored by gel electrophoresis (32). For several of the STAR/RTAR* combinations, no cleavage activity was detected after 24 min at [R] ≤ 15 μM (kobs ≤ 0.002 min−1; light gray shading in Figure 3B). For the remaining STAR/RTAR* combinations that displayed detectable cleavage activity, a single-substrate kinetic approach was used to precisely derive kcat/KM values. The highest kcat/KM value (0.53 ± 0.03 min−1μM−1) was obtained for the STAR-1/RTAR*-0 combination (Figure 3B), which is 46-fold lower than that of S0/R0 (kcat/KM = 24.5 ± 0.9 min−1μM−1 at 37°C; data not shown). Several other STAR/RTAR* combinations have similar catalytic efficiency (within 5-fold; dark gray shading in Figure 3B), and these combinations involve all five RTAR* ribozymes and four of the five STAR substrates.
For RTAR*-0, efficient substrate cleavage is obtained with STAR-1 as well as with STAR-0, indicating limited substrate promiscuity. Other RTAR* ribozymes also display similar specificity, cleaving efficiently only two to three out of the five STAR substrates. Given that the highest kcat/KM values for each ribozyme is observed with the compensatory substrate or one that varies by one or two base pairs, it can be concluded that efficient substrate cleavage depends on helix-length compensation in stem V of the variant ribozyme. For example, the optimal ribozyme for STAR-1 is RTAR*-0, whereas the optimal one for STAR+ 2 is RTAR*-3. Thus, for the STAR/RTAR* family, there is limited substrate promiscuity for all ribozymes as well as helix-length compensation across the family, as previously observed for the reference S/R family (32).
Kinetic studies of the VS ribozyme SL88/RL22 and SL22/RL88 pairs
In a similar manner, multiple and single turnover kinetic studies were performed to evaluate the kcat/KM of the 30 SL88/RL22 and 36 SL22/RL88 combinations (Supplementary Figures S3 and 4). For the SL88/RL22 pairs, the most active combination is SL88 + 1/RL22-1 with a kcat/KM of 0.39 ± 0.03 min−1 μM−1, which is 63-fold lower than that of S0/R0. A few other SL88/RL22 combinations produce similar kcat/KM (within 5-fold; dark gray shading in Supplementary Figure S3B), and these combinations involve three of the five SL88substrates and three of the six RL22 ribozymes. Each of these three ribozymes displays similar cleavage activity for one or two SL88 substrates, and helix-length compensation is observed given that the most active SL88/RL22 combinations for these ribozymes are the SL88-0/RL22-0, SL88+1/RL22-1 and SL88+2/RL22-2 pairs. For the SL22/RL88 pairs, the highest kcat/KM obtained is 0.23 ± 0.02 min−1 μM−1 for SL22+1/RL88-1, which is 107-fold lower than that of S0/R0. In contrast to the SL88/RL22 combinations, several SL22/RL88 combinations display similar catalytic activity (within 5-fold; dark gray shading in Supplementary Figure S4B), and these combinations involve four of the six SL22 substrates and five of the six RL88 ribozymes. For each of the five active ribozymes, comparable kcat/KM values are obtained with one to four of the six SL88 substrates, indicating different degrees of substrate promiscuity. Furthermore, helix-length compensation is observed for all variant ribozymes given that the highest kcat/KM values for each ribozyme is observed with the compensatory substrate or one that varies by one base pair. For example, the optimal substrate for RL88+1 is SL22+0, whereas the optimal one for RL88-4 is SL22+4. Thus, for both the SL88/RL22 and SL22/RL88 families, there is limited substrate promiscuity for all ribozymes as well as helix-length compensation across the family, as previously observed for the reference S/R family (32).
Comparison of the catalytic activity of the different S/R families
Given that kcat/KM values can be misleading when comparing the relative activity of the various S/R families, we also compared the kobs values for the S/R pair of each family that displays the highest catalytic activity. These kobs values, determined under identical single turnover conditions, indicate that the activities of the STAR/RTAR*, SL88/RL22 and SL22/RL88 family representatives are reduced by 159-, 53- and 84-fold, respectively, compared to the parental S0/R0 pair (Supplementary Table S2). Thus, although both the kcat/KM and kobs values indicate that there is a significant level of catalytic activity for some variant S/R pairs, their activity is still lower than that of the parental S0/R0 pair.
Structural characterization of the variant substrates
To insure that the variant substrates examined in this study are properly folded, these substrates were analyzed by native gel electrophoresis. The migration of variant substrates on native gels was compared with that of two previously investigated VS ribozyme substrates (32), S-1 bp and S-2 bp, which respectively adopt a hairpin and a duplex conformation. Our results show that all variant substrates migrate on a native gel in a manner similar to the active hairpin conformation (Supplementary Figure S5).
Structural characterization of the variant ribozymes
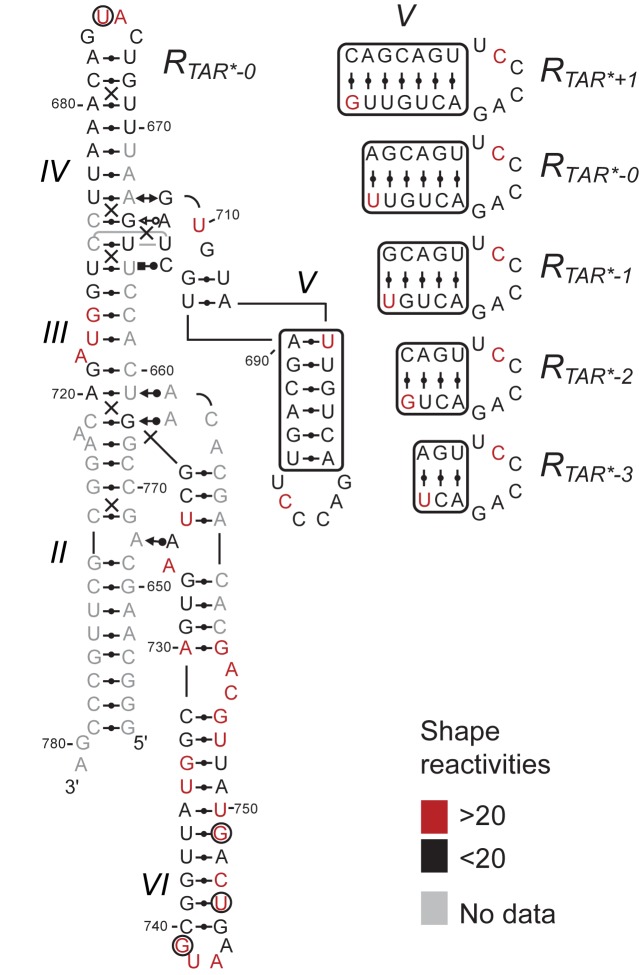
To verify the proper folding of variant ribozymes, the secondary and tertiary structures of these trans ribozymes were analyzed by SHAPE experiments. In these studies, the reactivity of each nucleotide to NMIA is quantified and interpreted as being directly linked to the local nucleotide flexibility (34). For the RTAR*-0 ribozyme (Figure 4), the nucleotides reactive to NMIA are found in non-helical regions that are likely to be dynamic, including the A718 and A725A726 bulges, the A756 loop and the terminal loops of stem-loops IV, V and VI (40–43). The reactivity patterns of the other RTAR* ribozymes are very similar to that of the RTAR*-0 ribozyme (Figure 4). In addition, the reactivity patterns of the R0, RL22 and RL88 ribozymes are similar to those of the RTAR* ribozymes in that known helical regions display lower reactivity to NMIA compared to non-helical regions (Supplementary Figure S6). As an exception, residues within and adjacent to G-U base pairs (in stem-loops V and VI) produce idiosyncratic patterns of NMIA reactivity, as previously demonstrated (44). Of note, the most NMIA-reactive nucleotide in all ribozymes is A726, an extruded residue from the II-VI bulge-bulge interaction and the most disordered residue in the NMR structures of the II-III-VI junction (14,43,45). Overall, the SHAPE results for all of the engineered ribozymes are in good agreement with the structural schematics of the R0 ribozyme derived from NMR and X-ray structures [Figure 1; (14,43)]. Thus, all of the variant VS ribozymes investigated here adopt a ground state conformation that is compatible with cleavage activity.
Figure 4.
SHAPE analysis of the RTAR* ribozyme variants. The normalized NMIA reactivity of each nucleotide within RTAR*-0 is color-coded on its structural schematic as per the SHAPE reactivities key. For the other RTAR*variants, the normalized NMIA reactivity is within the same category as for RTAR*-0, except for a few nucleotides in the variable SLV region (right panel) and outside this region (black circles indicate inverted reactivity compared to RTAR*-0; U675 for RTAR-1 and G741, U746 and G749 for RTAR-3).
Structural modeling of the S0/R0, SL88-0/RL22-0 and STAR-0/RTAR*-0 pairs
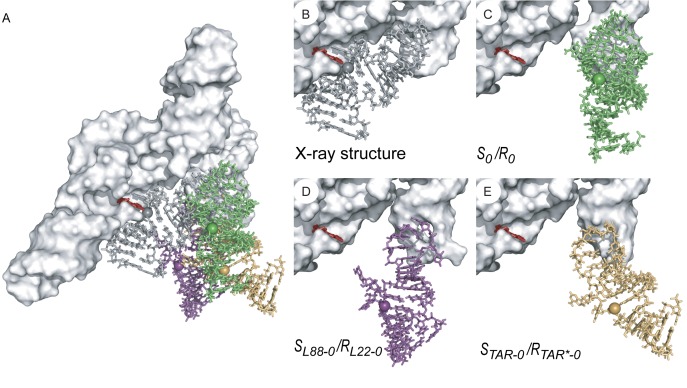
Once a stable KLI is formed between a well-folded VS ribozyme variant and its stem-loop substrate, the G638 internal loop of the substrate must properly interact with the A756 loop of the ribozyme to allow formation of a productive active site. In order to assess the effect of substituting the I/V KLIs on formation of the VS ribozyme active site, three-dimensional models of the SL88-0/RL22-0 and STAR-0/RTAR*-0 pairs were produced. These models were built by fragment assembly, combining either the X-ray structure of the L22/L88 KLI or the NMR structure of the TAR/TAR* KLI with the X-ray structure of the VS ribozyme [Figure 5; (14,35,36)]. As a control, the model of the S0/R0 pair was similarly built from the NMR structure of the SLI/SLV complex. In contrast with the close state observed in the X-ray structure (Figure 5B), the model of the S0/R0 pair (Figure 5C) adopts an open state with the G638 loop of SLI apart from the A756 loop of SLVI. This is not surprising, since the open state, previously described in the NMR-based model of the complete VS ribozyme, likely represents a ground-state conformation of the S/R complex (43). In the S0/R0 model, the scissile phosphate within the G638 loop is 26.6 Å from the corresponding phosphate in the crystal structure, but oriented directly toward A756, such that a simple repositioning of the helical domains containing the G638 and A756 loops would allow formation of the active site. The models of both the SL88-0/RL22-0 (Figure 5D) and STAR-0/RTAR*-0 pairs (Figure 5E) also represent an open state of the ribozyme with their scissile phosphates respectively 23.0 Å and 31.9 Å from the corresponding phosphate within the crystal structure. However, in both these models, the scissile phosphate is not oriented favorably with respect to the A756 loop to facilitate formation of the active site. Overall, these models suggest that substitutions of the KLI in the variant S/R pairs may reduce the probability of encounter between the G638 internal loop of the substrate and the A756 loop of the ribozyme and thus likely impair both the formation of the active site and the subsequent cleavage reaction.
Figure 5.
Three-dimensional models of substrate/ribozyme complexes. (A) Superposition of three-dimensional models of the variant S/R pairs on the X-ray structure of the VS ribozyme. Individual representations from this superposition are zoomed in and described in details for the subsequent panels. (B) X-ray structure of the VS ribozyme [PDB ID: 4R4P; (14)]. In this representation, the substrate of one protomer is shown in dark gray sticks with the phosphorus at the scissile phosphate as a sphere and the trans VS ribozyme (helical domains II–VI) of the other protomer is shown as a white surface, except for residue A756, which is shown as red sticks. (C) Model of the S0/R0 pair derived from the NMR structure of a VS ribozyme SLI/SLV complex [lime green; PDB ID: 2MI0; (13)]. (D) Model of the SL88-0/RL22-0 pair derived from the X-ray structure of the ribosomal L22/L88 KLI [purple; PDB ID: 4IOA; (36)]. (E) Model of the STAR-0/RTAR*-0 pair derived from the NMR structure of the TAR/TAR* complex [mustard yellow; PDB ID: 1KIS; (35)]. In (A–E), the trans VS ribozyme is shown as a white surface with A756 in red sticks and the individual substrates are shown as sticks with the phosphorus at the scissile phosphate as a sphere.
DISCUSSION
In this work, a comprehensive RNA engineering approach was developed, which demonstrates that the I/V KLI of the VS ribozyme is an RNA module that can be substituted by other KLIs while maintaining efficient substrate cleavage. Several KLIs were initially identified through a bioinformatics search of the PDB, of which two were selected as being structurally similar to the I/V KLI of the VS ribozyme: the HIV-1 TAR/TAR* KLI [PDB ID: 1KIS; (35)] and the L22/L88 KLI from the large subunit of rRNAs [PDB ID: 4IOA; (36)]. These two KLIs were substituted for the I/V KLI of the S0/R0 reference pair formed between a pre-shifted substrate and a trans version of the VS ribozyme. In addition, a combinatorial approach was used in which several variant S/R pairs were synthesized that contain different helix lengths in stems Ib and V. The resulting S/R pairs were kinetically evaluated, and several show substantial catalytic activity. Below, we compare the catalytic properties of the most active variant S/R pairs with that of the previously investigated S0/R0 pair in light of the thermodynamic and structural properties of the parental and surrogate KLIs.
Within the three families of S/R pairs investigated (STAR/RTAR*, SL88/RL22 and SL22/RL88), several S/R combinations show substantial catalytic efficiency (dark gray shading in Figure 3B and in Supplementary Figures S3B and 4B). For each family, helix-length compensation was observed for the most active S/R pairs, and this is consistent with what was observed in previous studies of the parental S0/R0 pair (32). For the STAR/RTAR* family, the trans VS ribozyme variants can efficiently cleave an SLI substrate with stem Ib lengths of 3–6 bp (Figure 3B). Both the SL88/RL22 and SL22/RL88 families also allow an appreciable level of catalytic efficiency for several SLI substrates, with a stem Ib length of 4–7 bp for SL22/RL88 and 4–6 bp for SL88/RL22 (Supplementary Figures S3 and 4). Within the three families of S/R pairs investigated, the STAR-1/RTAR*-0, SL88+1/RL22-1 and SL22+1/RL88-1 show the highest kcat/KM values, all within 2.5-fold of each other (0.2–0.5 min-1μM−1). Although these S/R pairs with surrogate KLIs behave like the reference S0/R0 pair in some respects, their kcat/KM values are reduced by 50- to 100-fold compared to S0/R0. Similarly, their kobs values determined under identical conditions are reduced 50- to 160- fold compared to S0/R0. The KLIs most likely form and assist catalysis for the highly-active variant S/R pairs, given that several other variant S/R pairs within the same family are inactive and that disruption of the KLI in the parental S0/R0 pair can lead to more than a 1,000-fold decrease in kcat/KM and more than a 6000-fold decrease in kobs (32). In addition, we verified by native gel electrophoresis and SHAPE analysis that all substrates and ribozymes of the variant S/R pairs adopt a stable fold equivalent to S0 and R0, respectively, and compatible with high catalytic efficiency. Thus, although surrogate KLIs allow high catalytic efficiency for some variant S/R pairs, it is puzzling that they do not allow the same high level of activity as that observed for the parental S0/R0 pair.
We have previously shown that mutations that decrease the affinity of the I/V KLI also lead to lower kcat/KM values when monitoring the trans cleavage reaction as done here (18,31). For example, deletion of U700 in SLV causes a 210-fold decrease in binding affinity for SLI and 140-fold decrease in kcat/KM (18,31). Thus, it is likely that the lower relative affinity of a surrogate KLI may affect the level of catalytic efficiency that could be attained in the variant S/R pairs. Given that the L22/L88 KLI is 16-fold less stable than the I/V KLI [Figure 2C; (18)], its higher Kd could explain in part the lower kcat/KM of the SL88/RL22 (>60-fold) and SL22/RL88 pairs (>100-fold) compared to S0/R0. However, given that the HIV-1 TAR/TAR* KLI is 40-fold more stable than the I/V KLI [Figure 2C; (18)], the difference in Kd of the KLI complex cannot explain the lower kcat/KM of the STAR/RTAR* pairs (>50-fold) compared to S0/R0.
The VS ribozyme S/R pairs likely adopt both an open state and a close state that can be respectively represented by the recent NMR-based model and X-ray structure (14,43). The KLI is stably formed in both states, but only the close state captures the active site formed by the association of the G638 and A756 loops. In order to assess the effect of substituting the I/V KLI on formation of the VS ribozyme active site, the S0/R0, SL88-0/RL22-0 and STAR-0/RTAR*-0 pairs were modeled based on high-resolution structures of KLIs and the S/R complex derived from the X-ray structure of the VS ribozyme (Figure 5). Since all the modeled S/R pairs adopt an open state with the G638 loop apart from the A756 loop, it is difficult to precisely evaluate the compatibility of surrogate KLIs for formation of the active site. Nevertheless, we find that the distance between the scissile phosphates of the model and the crystal structure is longer for the STAR-0/RTAR*-0 pair than for the S0/R0 and SL88-0/RL22-0 pairs. In addition, it appears from these models that formation of the active site from the open state would involve the simple packing of two helical domains for the S0/R0 pair, but possibly be more restrained for the SL88-0/RL22-0 and STAR-0/RTAR*-0 pairs. Thus, the HIV-1 TAR/TAR* KLI and possibly the L22/L88 interaction, which both contain more base pairs than the I/V KLI, may partially restrict formation of the active site due to topological constraints. We envision that additional mutations of the VS ribozyme could be introduced to help compensate for such structural restriction and thereby increase the catalytic efficiency of the variant SL88/RL22 and STAR/RTAR* pairs.
Previous NMR studies of an SLI/SLV complex indicate that both the SLI and SLV loops contain several dynamic residues that could confer a certain degree of flexibility to the I/V KLI (13). Thus, the I/V KLI likely acts as a dynamic hinge that facilitates formation of the active site. It is not clear at this time if the surrogate KLIs allow the same degree of flexibility, and how their dynamics affect formation of the active site in the variant S/R pairs. In this regard, future studies investigating the dynamic behavior of these KLIs would provide valuable insights into how these KLIs can be further engineered to more efficiently assist formation of the active site in the VS ribozyme.
CONCLUDING REMARK
Despite the ability of the VS ribozyme to specifically cleave a stem-loop substrate, very few studies had previously explored the possibility of modifying the VS ribozyme for cleavage of alternate substrates. Using a novel RNA engineering approach, we demonstrated here that the KLI within the VS ribozyme could be substituted by at least two other known KLIs while maintaining substantial cleavage activity. Interestingly, the possibility of substituting the KLI broadens the substrate specificity of VS ribozyme derivatives, which can now target SLI variants in which the 7-nt terminal loop can be replaced by at least three other 6-nt loop sequences and the length of the adjoining stem can be varied to some extent. Future engineering efforts are now geared toward improving the cleavage efficiency of the VS ribozyme variants identified here as well as further broadening the substrate specificity of VS ribozyme derivatives.
Supplementary Material
Acknowledgments
The authors thank Dominique Chaussé for RNA preparation, Ryan Richter for computer support, as well as James G. Omichinski for discussions and critical reading of the manuscript. They also thank Sébastien Lemieux and Calcul Québec for providing computing facilities.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Canadian Institutes of Health Research [CIHR MOP-86502 to P.L.]; CIHR and Université de Montréal Ph.D. Scholarships (to J.L.-L.); FRQNT Ph.D. Scholarship (to P.D.). Funding for open access charge: CIHR.
Conflict of interest statement. None declared.
REFERENCES
- 1.Lilley D.M. Catalysis by the nucleolytic ribozymes. Biochem. Soc. Trans. 2011;39:641–646. doi: 10.1042/BST0390641. [DOI] [PubMed] [Google Scholar]
- 2.Jimenez R.M., Polanco J.A., Luptak A. Chemistry and biology of self-cleaving ribozymes. Trends Biochem. Sci. 2015;40:648–661. doi: 10.1016/j.tibs.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weinberg Z., Kim P.B., Chen T.H., Li S., Harris K.A., Lunse C.E., Breaker R.R. New classes of self-cleaving ribozymes revealed by comparative genomics analysis. Nat. Chem. Biol. 2015;11:606–610. doi: 10.1038/nchembio.1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harris K.A., Lunse C.E., Li S., Brewer K.I., Breaker R.R. Biochemical analysis of pistol self-cleaving ribozymes. RNA. 2015;21:1852–1858. doi: 10.1261/rna.052514.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li S., Lunse C.E., Harris K.A., Breaker R.R. Biochemical analysis of hatchet self-cleaving ribozymes. RNA. 2015;21:1845–1851. doi: 10.1261/rna.052522.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Herschlag D. Learning from ribozymes. RNA. 2015;21:527–528. doi: 10.1261/rna.050914.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collins R.A. The Neurospora Varkud satellite ribozyme. Biochem. Soc. Trans. Rev. 2002;30:1122–1126. doi: 10.1042/bst0301122. [DOI] [PubMed] [Google Scholar]
- 8.Lilley D.M. The Varkud satellite ribozyme. RNA. 2004;10:151–158. doi: 10.1261/rna.5217104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wilson T.J., Lilley D.M. A mechanistic comparison of the Varkud satellite and hairpin ribozymes. Prog. Mol. Biol. Transl. Sci. 2013;120:93–121. doi: 10.1016/B978-0-12-381286-5.00003-2. [DOI] [PubMed] [Google Scholar]
- 10.Guo H.C.T., Collins R.A. Efficient trans-cleavage of a stem-loop RNA substrate by a ribozyme derived from Neurospora VS RNA. EMBO J. 1995;14:368–376. doi: 10.1002/j.1460-2075.1995.tb07011.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rastogi T., Beattie T.L., Olive J.E., Collins R.A. A long-range pseudoknot is required for activity of the Neurospora VS ribozyme. EMBO J. 1996;15:2820–2825. [PMC free article] [PubMed] [Google Scholar]
- 12.Leontis N.B., Westhof E. Geometric nomenclature and classification of RNA base pairs. RNA. 2001;7:499–512. doi: 10.1017/s1355838201002515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bouchard P., Legault P. Structural insights into substrate recognition by the Neurospora Varkud satellite ribozyme: importance of U-turns at the kissing-loop junction. Biochemistry. 2014;53:258–269. doi: 10.1021/bi401491g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Suslov N.B., DasGupta S., Huang H., Fuller J.R., Lilley D.M., Rice P.A., Piccirilli J.A. Crystal structure of the Varkud satellite ribozyme. Nat. Chem. Biol. 2015;11:840–846. doi: 10.1038/nchembio.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Andersen A., Collins R.A. Rearrangement of a stable RNA secondary structure during VS ribozyme catalysis. Mol. Cell. 2000;5:469–478. doi: 10.1016/s1097-2765(00)80441-4. [DOI] [PubMed] [Google Scholar]
- 16.Andersen A.A., Collins R.A. Intramolecular secondary structure rearrangement by the kissing interaction of the Neurospora VS ribozyme. Proc. Natl. Acad. Sci. U.S.A. 2001;98:7730–7735. doi: 10.1073/pnas.141039198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zamel R., Collins R.A. Rearrangement of substrate secondary structure facilitates binding to the Neurospora VS ribozyme. J. Mol. Biol. 2002;324:903–915. doi: 10.1016/s0022-2836(02)01151-8. [DOI] [PubMed] [Google Scholar]
- 18.Bouchard P., Legault P. A remarkably stable kissing-loop interaction defines substrate recognition by the Neurospora VS Ribozyme. RNA. 2014;20:1451–1464. doi: 10.1261/rna.046144.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hiley S.L., Collins R.A. Rapid formation of a solvent-inaccessible core in the Neurospora Varkud satellite ribozyme. EMBO J. 2001;20:5461–5469. doi: 10.1093/emboj/20.19.5461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jones F.D., Strobel S.A. Ionization of a critical adenosine residue in the Neurospora Varkud satellite ribozyme active site. Biochemistry. 2003;42:4265–4276. doi: 10.1021/bi020707t. [DOI] [PubMed] [Google Scholar]
- 21.McLeod A.C., Lilley D.M. Efficient, pH-dependent RNA ligation by the VS ribozyme in trans. Biochemistry. 2004;43:1118–1125. doi: 10.1021/bi035790e. [DOI] [PubMed] [Google Scholar]
- 22.Zhao Z.Y., McLeod A., Harusawa S., Araki L., Yamaguchi M., Kurihara T., Lilley D.M. Nucleobase participation in ribozyme catalysis. J. Am. Chem. Soc. 2005;127:5026–5027. doi: 10.1021/ja0502775. [DOI] [PubMed] [Google Scholar]
- 23.Smith M.D., Collins R.A. Evidence for proton transfer in the rate-limiting step of a fast-cleaving Varkud satellite ribozyme. Proc. Natl. Acad. Sci. U.S.A. 2007;104:5818–5823. doi: 10.1073/pnas.0608864104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wilson T.J., McLeod A.C., Lilley D.M. A guanine nucleobase important for catalysis by the VS ribozyme. EMBO J. 2007;26:2489–2500. doi: 10.1038/sj.emboj.7601698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jaikaran D., Smith M.D., Mehdizadeh R., Olive J., Collins R.A. An important role of G638 in the cis-cleavage reaction of the Neurospora VS ribozyme revealed by a novel nucleotide analog incorporation method. RNA. 2008;14:938–949. doi: 10.1261/rna.936508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith M.D., Mehdizadeh R., Olive J.E., Collins R.A. The ionic environment determines ribozyme cleavage rate by modulation of nucleobase pKa. RNA. 2008;14:1942–1949. doi: 10.1261/rna.1102308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wilson T.J., Li N.S., Lu J., Frederiksen J.K., Piccirilli J.A., Lilley D.M. Nucleobase-mediated general acid-base catalysis in the Varkud satellite ribozyme. Proc. Natl. Acad. Sci. U.S.A. 2010;107:11751–11756. doi: 10.1073/pnas.1004255107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo H.C.T., De Abreu D.M., Tillier E.R.M., Saville B.J., Olive J.E., Collins R.A. Nucleotide sequence requirements for self-cleavage of Neurospora VS RNA. J. Mol. Biol. 1993;232:351–361. doi: 10.1006/jmbi.1993.1395. [DOI] [PubMed] [Google Scholar]
- 29.Beattie T.L., Olive J.E., Collins R.A. A secondary-structure model for the self-cleaving region of Neurospora VS RNA. Proc. Natl. Acad. Sci. U.S.A. 1995;92:4686–4690. doi: 10.1073/pnas.92.10.4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rastogi T., Collins R.A. Smaller, faster ribozymes reveal the catalytic core of Neurospora VS RNA. J. Mol. Biol. 1998;277:215–224. doi: 10.1006/jmbi.1997.1623. [DOI] [PubMed] [Google Scholar]
- 31.Bouchard P., Lacroix-Labonté J., Desjardins G., Lampron P., Lisi V., Lemieux S., Major F., Legault P. Role of SLV in SLI substrate recognition by the Neurospora VS ribozyme. RNA. 2008;14:736–748. doi: 10.1261/rna.824308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lacroix-Labonté J., Girard N., Lemieux S., Legault P. Helix-length compensation studies reveal the adaptability of the VS ribozyme architecture. Nucleic Acids Res. 2012;40:2284–2293. doi: 10.1093/nar/gkr1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Petrov A.I., Zirbel C.L., Leontis N.B. WebFR3D–a server for finding, aligning and analyzing recurrent RNA 3D motifs. Nucleic Acids Res. 2011;39:W50–W55. doi: 10.1093/nar/gkr249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Merino E.J., Wilkinson K.A., Coughlan J.L., Weeks K.M. RNA structure analysis at single nucleotide resolution by selective 2′-hydroxyl acylation and primer extension (SHAPE) J. Am. Chem. Soc. 2005;127:4223–4231. doi: 10.1021/ja043822v. [DOI] [PubMed] [Google Scholar]
- 35.Chang K.Y., Tinoco I., Jr The structure of an RNA “kissing" hairpin complex of the HIV TAR hairpin loop and its complement. J. Mol. Biol. 1997;269:52–66. doi: 10.1006/jmbi.1997.1021. [DOI] [PubMed] [Google Scholar]
- 36.Magee T.V., Han S., McCurdy S.P., Nguyen T.T., Granskog K., Marr E.S., Maguire B.A., Huband M.D., Chen J.M., Subashi T.A., et al. Novel 3-O-carbamoyl erythromycin A derivatives (carbamolides) with activity against resistant staphylococcal and streptococcal isolates. Bioorg. Med. Chem. Lett. 2013;23:1727–1731. doi: 10.1016/j.bmcl.2013.01.067. [DOI] [PubMed] [Google Scholar]
- 37.Major F., Turcotte M., Gautheret D., Lapalme G., Fillion E., Cedergren R. The combination of symbolic and numerical computation for three-dimensional modeling of RNA. Science. 1991;253:1255–1260. doi: 10.1126/science.1716375. [DOI] [PubMed] [Google Scholar]
- 38.Lavery R., Moakher M., Maddocks J.H., Petkeviciute D., Zakrzewska K. Conformational analysis of nucleic acids revisited: Curves+ Nucleic Acids Res. 2009;37:5917–5929. doi: 10.1093/nar/gkp608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eisenthal R., Danson M.J., Hough D.W. Catalytic efficiency and kcat/KM: a useful comparator? Trends Biotechnol. 2007;25:247–249. doi: 10.1016/j.tibtech.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 40.Campbell D.O., Bouchard P., Desjardins G., Legault P. NMR structure of Varkud satellite ribozyme stem-loop V in the presence of magnesium ions and localization of metal-binding sites. Biochemistry. 2006;45:10591–10605. doi: 10.1021/bi0607150. [DOI] [PubMed] [Google Scholar]
- 41.Desjardins G., Bonneau E., Girard N., Boisbouvier J., Legault P. NMR structure of the A730 loop of the Neurospora VS ribozyme: insights into the formation of the active site. Nucleic Acids Res. 2011;39:4427–4437. doi: 10.1093/nar/gkq1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bonneau E., Legault P. Nuclear magnetic resonance structure of the III-IV-V three-way junction from the Varkud satellite ribozyme and identification of magnesium-binding sites using paramagnetic relaxation enhancement. Biochemistry. 2014;53:6264–6275. doi: 10.1021/bi500826n. [DOI] [PubMed] [Google Scholar]
- 43.Bonneau E., Girard N., Lemieux S., Legault P. The NMR structure of the II-III-VI three-way junction from the Neurospora VS ribozyme reveals a critical tertiary interaction and provides new insights into the global ribozyme structure. RNA. 2015;21:1621–1632. doi: 10.1261/rna.052076.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wilkinson K.A., Vasa S.M., Deigan K.E., Mortimer S.A., Giddings M.C., Weeks K.M. Influence of nucleotide identity on ribose 2′-hydroxyl reactivity in RNA. RNA. 2009;15:1314–1321. doi: 10.1261/rna.1536209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McGinnis J.L., Dunkle J.A., Cate J.H., Weeks K.M. The mechanisms of RNA SHAPE chemistry. J. Am. Chem. Soc. 2012;134:6617–6624. doi: 10.1021/ja2104075. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.