Abstract
When isolated squid giant axons are incubated in radioactive amino acids, abundant newly synthesized proteins are found in the axoplasm. These proteins are translated in the adaxonal Schwann cells and subsequently transferred into the giant axon. The question as to whether any de novo protein synthesis occurs in the giant axon itself is difficult to resolve because the small contribution of the proteins possibly synthesized intra-axonally is not easily distinguished from the large amounts of the proteins being supplied from the Schwann cells. In this paper, we reexamine this issue by studying the synthesis of endogenous neurofilament (NF) proteins in the axon. Our laboratory previously showed that NF mRNA and protein are present in the squid giant axon, but not in the surrounding adaxonal glia. Therefore, if the isolated squid axon could be shown to contain newly synthesized NF protein de novo, it could not arise from the adaxonal glia. The results of experiments in this paper show that abundant 3H-labeled NF protein is synthesized in the squid giant fiber lobe containing the giant axon’s neuronal cell bodies, but despite the presence of NF mRNA in the giant axon no labeled NF protein is detected in the giant axon. This lends support to the glia–axon protein transfer hypothesis which posits that the squid giant axon obtains newly synthesized protein by Schwann cell transfer and not through intra-axonal protein synthesis, and further suggests that the NF mRNA in the axon is in a translationally repressed state.
Keywords: Squid axon, Neurofilament protein, Local protein synthesis, mRNA, Ribonuclease protection, Schwann cell
Introduction
It was shown almost 50 years ago that when isolated squid giant axons are incubated in radioactive amino acids there are abundant newly synthesized proteins found in the axoplasm (Giuditta et al. 1968). There were two possible interpretations of this finding: (1) that this was due to de novo protein biosynthesis in the axoplasm, and (2) that radioactive proteins were synthesized in the adaxonal Schwann cells and then transported into the giant axon. Subsequent work showed that the principal source of the newly synthesized proteins found in the axoplasm of the isolated squid giant axon was from the adaxonal Schwann cells, and this finding was put forth as the Glia-Neuron Protein Transfer Hypothesis (Gainer et al. 1977b; Lasek et al. 1977). While the concept that glia provide proteins to axons has received wide acceptance [see recent review by Tytell et al. (2016)], the idea that axons do not synthesize proteins de novo continues to be controversial (Alvarez et al. 2000; Giuditta et al. 2008, 2002). Detailed considerations of this issue in both invertebrate and vertebrate axons can be found in several reviews (Holt and Schuman 2013; Jung et al. 2012; Perry and Fainzilber 2014; Piper and Holt 2004).
Much of the current evidence for axonal protein synthesis in mature axons comes from reports of protein synthetic machinery (i.e., mRNAs, t-RNAs, and other translational components) being present in axons and the assumption that the presence of this machinery implies de novo protein synthesis (Alvarez et al. 2000; Kalinski et al. 2015; Koenig and Giuditta 1999; Piper and Holt 2004; Van Minnen 1994). In this regard, it is notable that canonical ribosomes are rarely, if ever, found in uninjured, mature axons. In comparison, the presence of canonical ribosomes and the existence of protein synthesis in dendrites has been accepted for some time (Steward 1983a, b; Steward and Levy 1982; Steward and Schuman 2003). Many components of translational machinery have also been reported as being present in the squid giant axon (Alvarez et al. 2000; Giuditta et al. 2008, 2002; Koenig and Giuditta 1999), and a unique feature of this experimental preparation is that it is possible to directly assess de novo protein synthesis in the axoplasm. However, one problem for studies on the giant axon is how to discriminate the possibly small contribution of the radioactive proteins synthesized in the axon from the enormous amount of the radioactive protein supplied from the Schwann cells (Gainer et al. 1977b; Lasek et al. 1977).
In view of the continuing interest in axonal protein synthesis, and the increasing tendency of investigators to accept the presence of machinery in axons to synthesize proteins as evidence that they in fact do, we reexamined whether the squid axon can synthesize neuron-specific proteins from endogenous neuron-specific mRNAs found in the axoplasm. Our laboratory previously showed that neither NF mRNA (Way et al. 1992) nor NF protein(Cohen et al. 1987) was present in the squid giant axon but not in the surrounding adaxonal glia. Therefore, if the isolated squid axon could be shown to contain newly synthesized NF protein de novo, it could not arise from the adaxonal glia. Our ability to address this issue was also greatly facilitated by knowing the predicted NF amino acid sequences (Szaro et al. 1991; Way et al. 1992), and with this information, we could generate highly specific riboprobes to assess NF mRNAs in axons by RNase protection assay methods and polyclonal antibodies for NF proteins using immunoprecipitation methods. The results of experiments in this paper show that abundant 3H-labeled and immunoprecipitated NF protein was synthesized from radioactive amino acid substrates in the squid giant fiber lobe containing the neuron cell bodies, but despite the presence of detectable NF mRNA in the giant axon, no labeled immunoprecipitated NF protein could be detected in the giant axon. This lends additional support to the proposal that the squid giant axon does not synthesize proteins de novo in significant amounts (Gainer et al. 1977b; Lasek et al. 1977), and further challenges the view that the demonstration of mRNA in axons is a sufficient evidence for its translation.
Materials and Methods
Animals
Live squid (Loligo pealii) were obtained during the months of June–August of the years 1992–1994, at the Marine Biological Laboratory (MBL, Woods Hole, MA). Animals were kept in tanks with running seawater at ambient temperature (18–21 °C) and were used within 24 h of their delivery. All invertebrates including cephalopods are not covered under Animal Welfare Act (AWA) regulations, and thus procedures are not formally reviewed and approved. Nevertheless, these experiments done at the MBL used research procedures that follow the basic tenants prescribed by the AWA for all animal research.
Dissection of Squid Tissues and Extraction of Their Proteins
Several neural tissues were dissected for RNA and protein analyses. These include the squid giant axons, the giant fiber lobe (GFL) in the stellate ganglion that contains the cell bodies giving rise to the giant axon, and the optic lobes in the squid central nervous system. The gills were also dissected as a non-neural tissue control. Giant axons were carefully dissected from squid that had been killed by decapitation using previously described methods (Cohen et al. 1987; Lasek et al. 1977; Tytell and Lasek 1984). Each end of the axon was ligated with thread prior to excision and a dissecting microscope with dark-field illumination was used to clean the giant axon of adhering small axons and other extraneous tissues from its surface. The giant fiber lobe (GFL), containing the cell bodies that give rise to the giant axon, was dissected from the stellate ganglion by previously described methods (Cohen et al. 1987; Lasek et al. 1977; Tytell et al. 1990). The optic lobes of the squid were dissected as described elsewhere (Cohen et al. 1987; Szaro et al. 1991).
To extract the proteins from the above tissues, the dissected tissues were rinsed briefly in ice-cold isosmolar sucrose to remove the external seawater, blotted and immediately homogenized in SDS-gel homogenizing buffer (2 % SDS, 5 % 2-beta-mercaptoethanol), and stored frozen at −20 °C for subsequent SDS-PAGE. Alternatively, the tissues were rinsed briefly in ice-cold isosmolar sucrose and placed into Eppendorf tubes on ice containing-either a 5 mM EDTA, 40 mM Tris, 1 mg/ml leupeptin. DH 7.0 solution or a 1 mM EGTA, 100 mM NaCl: 20’m ~ HEPES, 50 % glycerol, pH 7.0 solution, and frozen for further use.
Preparation of Total RNA from Squid Tissues and RNase Protection Assays of Neurofilament RNAs
Total RNAs were prepared from freshly dissected and rapidly frozen squid tissues (optic lobe, GFL, giant axon, and gills) using a guanidium thiocyanate–phenol–chloroform extraction method (Chomczynski and Sacchi 1987).
DNA fragments corresponding to neurofilament (NF) and beta-actin mRNA sequences were generated on GFL cDNA templates by PCR and subcloned into a pCRII vector (Invitrogen, San Diego, CA, USA). Four pairs of the primers for neurofilament transcripts and a primer pair for beta-actin that were used for amplification reactions and primer sequences are listed in Table 1. The forward primers used for amplification reactions for NF 60 corresponded to nucleotide residue numbers 1383-1400 and the reverse primers corresponded to 1605-1622 as in GenBank sequence with accession number M64717 (Szaro et al. 1991). Forward primers used for amplification reactions for NF 70 corresponded to nucleotide residue numbers 1469-1486 and the reverse primers corresponded to 1676-1693 as in GenBank sequence with accession number M64718 (Szaro et al. 1991). Forward primers used for amplification reactions for NF 220L corresponded to nucleotide residue numbers 3464-3482 and the reverse primers corresponded to 3673-3690 as in GenBank sequence with accession number M64717 (Way et al. 1992). Forward primers used for amplification reactions for NF 220L corresponded to nucleotide residue numbers 3464-3482 and the reverse primers corresponded to 3673-3690 as in GenBank sequence with accession number M94389 (Way et al. 1992). Forward primers used for amplification reactions for NF 220S corresponded to nucleotide residue numbers 3464-3482 and the reverse primers corresponded to 3581-3598 as in GenBank sequence with accession number M94389 (Way et al. 1992). For beta-actin, the forward primers corresponded to nucleotide residue numbers 1119-1136 and the reverse primers corresponded to 1233-1252 as in GenBank sequence with accession number AY701849 (Kaplan et al. 1992).
Table 1.
Primer set for RT-PCR for neurofilaments and beta-actin
| Genes | Forward primer (5′–3′) | Reverse primer (5′–3′) | GenBank accession no. | Protected fragment size (bp) |
|---|---|---|---|---|
| NF60 | TGGAATTGGAAATTGCTGC | TCCGATGAACCCATGGAC | M64717 | 240 |
| NF70 | GGAGCCAGACCTAACGAG | TCTTGACAACCGTTCCGGT’ | M64718 | 315 |
| NF220L | AGTCCAGTAGCCTCCGAGA | TCCGATGAACCCATGGAC | M94389 | 226 |
| NF220S | AGTCCAGTAGCCTCCGAGA’ | ATCCGGGTGATCCTGCAGT’ | M94389 | 135 |
| Beta-actin | GCTCCACCAGAAAGGAAA | AAGCACTTCCTATGGACGAT | AY701849 | 133 |
Cycling conditions consisted of an initial denaturation step at 94 °C for 5 min, followed by 28 cycles of 1 min at 94 °C, 2 min at 55 °C, and 2 min at 72 °C. At the end of cycling, the final extension was carried out at 72 °C for 10 min. 32P-labeled antisense cRNAs were synthesized in vitro transcription with T7 RNA polymerase using each linearized plasmid DNA.
Nine micrograms of total RNA from each tissue sample were hybridized overnight to each of the 32P-labeled riboprobes at 42 °C and RNase digestion was performed by using RPAIl kit (Ambion). Hybridized mixtures were treated with RNase A/T1 (2.5 U/ml RNase A and 100 U/ml RNase T1) for 30 min at 37 °C. After RNase inactivation and precipitation of protected fragments, pellets were dissolved in loading buffer and electrophoresed on 6 % polyacrylamide-8 M urea gels. The gel was dried and subjected to autoradiographic exposure on KODAK BioMaxMR film. The labeled probes were also hybridized to 10 μg of yeast tRNA as a negative control.
For quantification, the dried gel was exposed to a Phosphor screen and analyzed using a Phosphoimager (Model No. 455A, Molecular Dynamics). Integrated values expressed as a number of pixels were converted into radioactivity by using a standard curve that was generated from values obtained from the known amount of cpm applied to the denaturing gel. The cpm associated with the gill sample was subtracted from the radioactivity associated with each sample to correct for background and the final value was normalized to the beta-actin loading control in each sample. The protected fragment sizes were 240-bp for NF60, 315-bp for NF70, 226-bp for NF220L, 135-bp for NF220S, and 133-bp for beta-actin (Table 1).
Incorporation of Radioactive Amino Acids into Proteins in Squid Giant Axons and GFLs
Cleanly dissected giant axons and GFLs were incubated in radioactive amino acids as previously described (Lasek et al. 1977; Tytell and Lasek 1984). In one experiment, the incubation media contained 300 Ci of L- (4-5-3H-leucine (specific activity 120–190 Ci/ml, Amersham, TRK636) which was rapidly dried in a test tube under vacuum and then dissolved in artificial seawater (ASW) composed of 425 mM NaCI, 9 mM KCL, 10 mM CaCI2, 50 mM MgCI2, 5 mM glucose, and 10 mM Tris-HC1, pH 7.6) at a final concentration of 1 mCi/ml. In a second experiment, the incubation media contained a cocktail of three radioactive amino acids (at 100 mCi each) that were designed to maximize the radioactivity incorporated into the squid NFs that were rapidly dried together in a test tube under vacuum and then dissolved in ASW. These three amino acids were L-(3-3H) serine (specific activity 10–20 Ci/ml, Amersham TRK 636), 3H L-(4,5-(N)-3H Lysine monohydrochloride (specific activity 75–100 Ci/ml, Amersham, TRK520), and L-(G–3H) glutamic acid (specific activity 20–40 Ci/ml, Amersham TRK445) at an average final concentration of 1 mCi/ml. The latter amino acid cocktail was designated the 3H–SKE medium.
The axons and GFLs were incubated in 1 ml volumes of the above media on cleaned microscope slides on which wells were formed with petroleum jelly to retain the solutions. Evaporation was minimized by enclosing the slides in a petri dish that contained a piece of moistened tissue paper. The incubations of the squid tissues in the above media were for 4 h at the ambient, seawater temperature (18–21 °C), and were terminated by rapidly washing the axons and GFLs in several volumes of ASW to remove the unincorporated amino acids and the washed tissues were transferred to a glass–glass microhomogenizers (Micromedic, Cleveland) containing extraction buffers for SDS-PAGE and/or immunoprecipitation. The extraction buffer that was routinely used for of SDS-PAGE analysis contained 2 % beta-mercaptoethanol, 8 M urea, 1 % sodium dodecyl sulfate (SDS) and 100 mM Tris–HCl, pH 7.2 (referred to as BUST in (Cohen et al. 1987; Tytell and Lasek 1984). The extraction buffer used in immunoprecipitation experiments was a standard RIPA buffer with a composition of 150 mM NaCl, 1 % NP–40,0.5 % sodium deoxycholate, 0.1 % SDS, and 50 mM Tris HCl, pH 8.0. The homogenates were centrifuged at 10,000g for 20 min at 20 °C to remove any insoluble debris and the supernatants were recovered & frozen until further use. The SDS-PAGE and Western blot procedures were done as previously described (Cohen et al. 1987).
Immunoprecipitation of Squid NF Proteins
Rabbit antibodies were generated against specific subunits of the squid neurofilament proteins based on their specific amino acid sequences (see (Szaro et al. 1991; Way et al. 1992)). Since all the NF subunits contain identical 5′ N-terminal and rod sequences, an antibody directed at this conserved NF N-terminal domain (termed NF–NT) was made in rabbits (Rabbits 821 and 822) to a common N-terminal sequence (amino acids 9–24) EISTTTTYEGESRPSS by the Immunodynamics Corp. The NF subunits do differ significantly in the 3′ C-terminal regions, except for a short region shared by the NF 60 and NF 70 proteins. Therefore, another rabbit antibody directed at a domain shared by both the NF 60 and NF 70 proteins (termed NF 60/70) was made against the peptide EAEVLSTILTRSEGG (amino acids 465–479) by Peptide Technologies, Inc. A third rabbit antibody (termed NF 70) was made against a peptide KGEDKANYTQNTVYQ (amino acids 601–615) that was specific for the NF 70 subunit by Peptide Technologies, Inc. Attempts to make an antibody specific for the NF 220 subunit were unsuccessful. Prebleed sera were also taken from donor rabbits for controls. A commercial Anti-beta-actin monoclonal antibody (Amersham) was used in some experiments. All of these antibodies were previously validated for their subunit specificities and specific neuronal localizations by Western blots and immunohistochemistry, respectively (Grant et al. 1995).
The immunoprecipitation procedure used was based on a published Santa Cruz Biotechnology protocol (http://www.scbt.com/fr/protocols.html?protocol=immunoprecipitation).
After harvesting the immunoprecipitates, the immunoreactive labeled NF proteins in the samples were subjected to SDS-PAGE and visualized by autoradiography/fluorography using the Entensify–Amplify product, NAMP100 from Amersham (GE Healthcare Life Sciences, Inc).
Results
Previous SDS-PAGE and Western blot studies identified two major NF proteins in the squid giant axon, a high molecular weight phosphorylated protein, NF 220, a low molecular protein, NF 60 (Brown and Lasek 1990; Cohen et al. 1987; Pant et al. 1978), and a dephosphorylated form of NF220, approximately 190 kD, found together with NF60 in the cell bodies in the GFL (Tytell et al. 1990). Subsequent molecular cloning studies in our laboratory revealed that there were two low molecular weight NF mRNAs in the GFL, an abundant NF60 mRNA and a small amount of a NF70 mRNA, in addition to a higher molecular weight mRNA corresponding to NF 190, all of which were formed from a single gene by alternative splicing (Szaro et al. 1991; Way et al. 1992). Given the availability of the nucleotide sequences for the NF proteins, we could generate probes for use in quantitative RNase protection assays (Table 1) in order to determine the relative amounts of these NF subtype mRNAs in the squid giant axon and the GFL.
The Presence of NF Subtype mRNAs in the Squid GFL and the Giant Axon
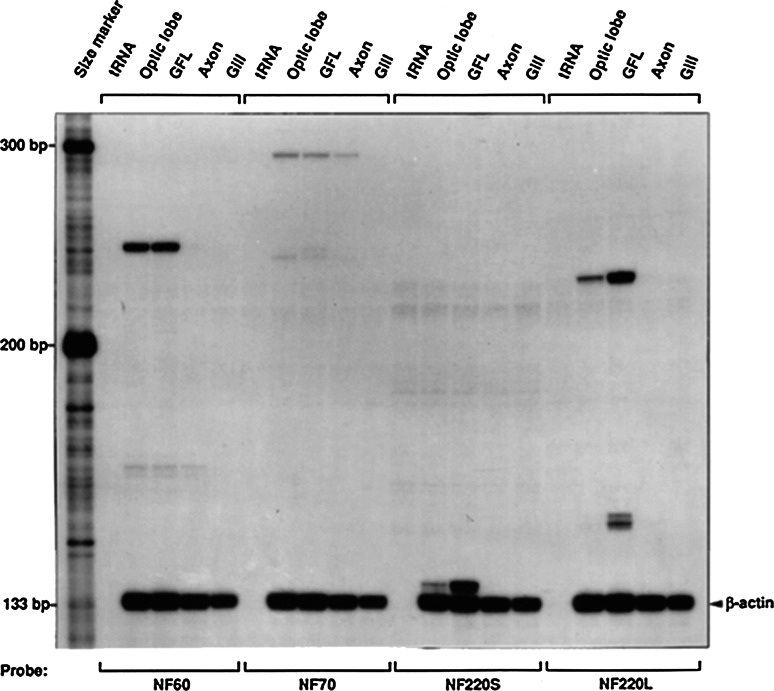
In order to determine the relative expression of the NF subtype mRNAs, total RNA was obtained from various neural and non-neural tissues in the squid, and analyzed for the amounts of the three NF subtype mRNAs by RNase protection assays. Figure 1 shows that the protected fragments of mRNAs for each subtype (NF60, NF70, and NF220) were detected in neural tissues (e.g., optic lobe and GFL) but not in non-neural tissues (e.g., as shown in Fig. 1 for gill RNAs which also served as the background control value in the quantitative analysis shown in Fig. 2). The presence of NF220 was detected by using two independent riboprobes directed at different nucleotide sequences in its C-terminal tail, notated as NF220s and NF220L (see “Methods” section, and Fig. 1). Consistent with previous reports (Szaro et al. 1991; Way et al. 1992), the most abundant NF-protected fragments (mRNAs) were found in the optic lobe and GFL. The NF60 and NF220 mRNAs were the most abundant subtype mRNAs, and very little NF 70 mRNA was expressed in these tissues. The NF220 and NF60 mRNA expression were much greater in the GFL than in the optic lobe (Fig. 1). Notably, the expression of the NF mRNAs in the giant axon was very much lower than in either the optic lobe or the GFL and appears undetectable in Fig. 1. The quantitative analyses of these data are illustrated in Fig. 2, in which the gill values were subtracted as background controls. It is clear that there were small but detectable amounts of all the subtypes of NF mRNA present in the giant axon.
Fig. 1.
Ribonuclease protection assay (RPA) analysis of neurofilament (NF) mRNAs in squid tissues. 9 µg of total RNA prepared from either squid optic lobe, Giant fiber lobe (GFL) containing the neuronal perikarya in the stellate ganglion which give rise to the giant axons, the giant axon (AXON), or the gill (non-neuronal control tissue) were hybridized with specific probes to the NF protein subunits (NF 60, 70, 220S, 220L) or beta-actin (1× 10E5 CPM each) and then treated with RNase A/T1 as described in “Methods” section. The protected fragment sizes for the transcripts are 133 bp for beta-actin, 136 bp for NF220S, 228 bp for NF220L, 250 bp for NF60, and 316 bp for NF70. Quantitative analyses of these data are shown in Fig. 2
Fig. 2.
Relative abundance of NF subunit mRNAs in squid neural tissues. After RPA was performed, the radioactive content of each band (see Fig. 1) was measured using a phosphoimager as described in “Methods” section. The data are expressed as relative (percentage) of the radioactivity in the NF220S protected band in the GFL (100 %) after normalization of each band to beta-actin mRNA, and the subtraction of background radioactivity in Gill mRNA. The data shown represent Means ± SEMs of three independent measurements. While there is detectable NF mRNA in axons, it is expressed at considerably lower levels than in the GFL (perikarya) samples
Figure 2 shows the relative amounts of the NF mRNAs of the various NF subunits in each tissue relative to the level of the NF220s in the GFL (taken as 100 %). The NF60 and NF220 mRNAs in the OL and the GFL that contain the neuron somata that give rise to the giant axon (Brown and Lasek 1990) are both many fold greater than in the giant axon. Thus, we conclude that some NF mRNA is present in the giant axon, although it is very low compared to the neuron cell bodies in the GFL. This conclusion is similar to that determined by an independent in situ hybridization study of the presence of NF220 in the giant axons and GFL (Way et al. 1992). Most importantly, our previous studies showed that there is neither NF mRNA (Way et al. 1992) nor NF protein (Cohen et al. 1987) expressed n the adaxonal glia surrounding he giant axon.
Does the Squid Giant Axon Synthesize NF Proteins?
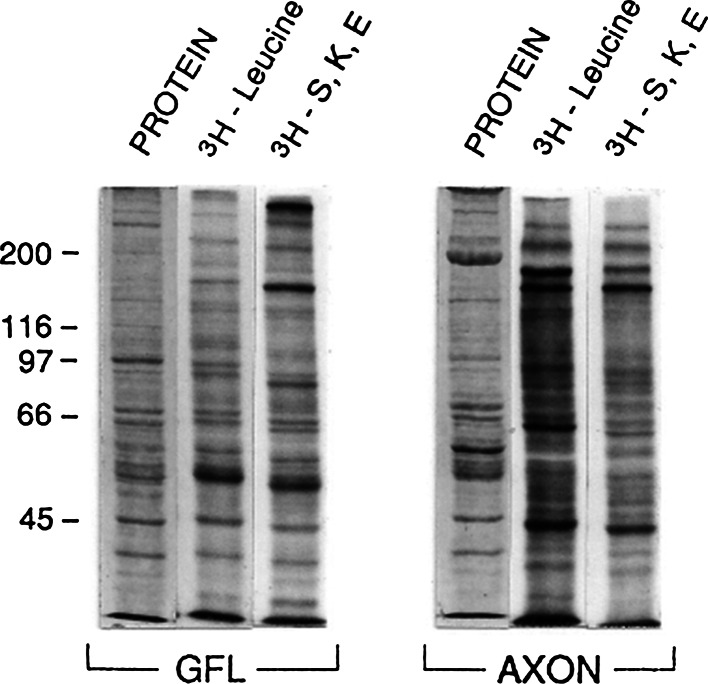
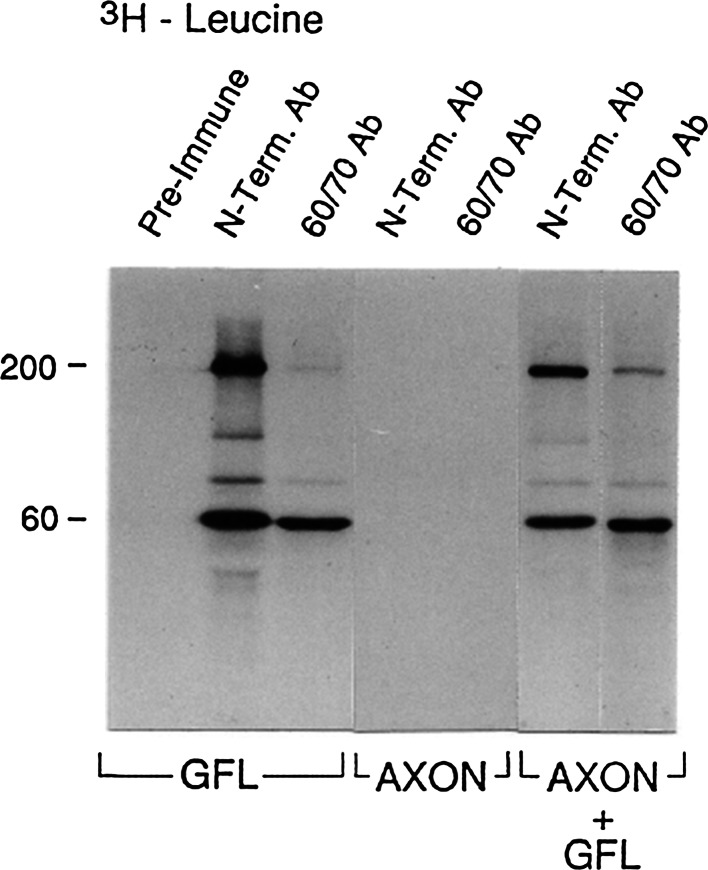
When isolated squid giant axons are incubated in radioactive amino acids, there are substantial amounts of newly synthesized radioactive protein found in the axoplasm of the giant axon (Giuditta et al. 1968; Lasek et al. 1977, 1974). This phenomenon is illustrated in Fig. 3 where SDS-PAGE patterns of such newly synthesized, radioactive proteins in axons are illustrated after incubation in either 3H-leucine or in a mixture of three 3H-labeled amino acids known to be enriched in NFs (3H-serine, 3H-lysine, and 3H-glutamate; 3H–S, K, E). These labeled proteins have been shown to be derived from translation occurring in the adaxonal Schwann cells and subsequent transfer of the newly synthesized, radioactive proteins into the giant axon (Gainer et al. 1977b; Lasek et al. 1977, 1974; Tytell and Lasek 1984; Tytell et al. 2016). Whether any of these radioactive proteins were intra-axonally synthesized was evaluated by the experiments in Figs. 4 and 5. Immunoprecipitations of the newly synthesized, labeled proteins in the axon and GFL (shown in Fig. 3) were done using a N-terminally directed (pan-specific) NF antibody that cross-reacts with all three NF subunits, and an NF60/70 antibody that specifically cross-reacts with NF60 and NF70. The data are shown in Fig. 4 for the 3H-leucine labeled proteins and in Fig. 5 for the 3H–S, K, E proteins that are shown in Fig. 3. Figure 4 shows that there was robust immunoprecipitation of the 3H-leucine-labeled NF subunit proteins by the N-terminally directed (pan-specific) NF antibody and by the NF60/70-specific antibody in the GFL sample, but no immunoprecipitable 3H-leucine-labeled NF proteins in the giant axon sample. An immunoprecipitation control experiment is illustrated in the last two columns in Fig. 4, where 1:1 mixtures of the GFL and axon samples were subjected to identical immunoprecipitation procedures, and showed that the immunoprecipitation of the 3H-labeled NF proteins from the GFL sample was not prevented by the presence of the large amounts of endogenous, nonradioactive NF protein in the axon sample. The lower intensity of the immunoprecipitated bands in this control experiment is due to the fact that half of the GFL sample’s radioactive protein was used in the mixture. Similar results were obtained with the 3H–S, K, E amino acids (Fig. 5) which appeared to preferentially label the NF220 protein in the GFL but much less robustly than the 3H–leucine did. From these experiments, we conclude that in contrast to the robust translation of NF mRNAs in the GFL, we can find no evidence at all for the translation of NF mRNA in the squid giant axon.
Fig. 3.
SDS-PAGE separations of radioactive proteins incorporated into and extracted from GFL and ensheathed giant axons after incubation of tissues in either 3H-leucine or 3H-serine, 3H-lysine, and 3H-glutamate (3H–S, K, E) mixtures. The lanes labeled PROTEIN illustrate the Commassie blue staining patterns of proteins in the GFL and giant axon extracts, and the numbers to the left represent the migration positions the proteins of known molecular weights (kD × 10E−3). The radioactive proteins in these extracts were also used in the immunoprecipitation experiments illustrated in Figs. 4 and 5
Fig. 4.
Autoradiographs of SDS-PAGE of immunoprecipitates of 3H-leucine-labeled squid neurofilament proteins, NF 60 and NF220, from extracts of GFL, giant axons, or GFL and giant axon (50:50) mixtures. The antibodies employed were reactive either with all of the NF subunits in squid (referred to as N-term Ab) or only with the low molecular weight forms (NF60/70 Ab). Data shown are representative of three independent immunoprecipitation experiments
Fig. 5.
Autoradiographs of SDS-PAGE of immunoprecipitates of 3H-SKE-labeled squid neurofilament proteins, NF 60 and NF220, from extracts of GFL, giant axons, or GFL and giant axon (50:50) mixtures. The antibodies employed were reactive either with all of the NF subunits in squid (referred to as N-term Ab) or only with the low molecular weight forms (NF60/70 Ab). Data shown are representative of three independent immunoprecipitation experiments
Discussion
The squid axon has served as a successful experimental model for the study of many fundamental cell biological and physiological processes (Gilbert et al. 1990). In particular it is a very favorable system to study whether axons can synthesize proteins de novo, mainly because it is possible to obtain large quantities of pure axoplasm by simple extrusion methods (Brown and Lasek 1990; Steinbach 1941). The earliest experiments using this model showed that after incubation of the isolated giant axon in radioactive amino acid precursors, substantial amounts of newly synthesized radioactive proteins were found in the axoplasm (Fischer and Litvak 1967; Giuditta et al. 1968). These findings raised the question of the origin of these radioactive proteins. Were they derived from protein biosynthesis in the axoplasm or by translation in the adaxonal Schwann cells with subsequent transfer of those newly synthesized proteins into the axoplasm? Subsequent experiments about a decade later showed that most, if not all, of these radioactive proteins found in the axoplasm were synthesized in the adaxonal Schwann cells and then transferred into the giant axon (Gainer et al. 1977b; Lasek et al. 1977, 1974).
Although it is generally accepted that adaxonal glia are the principal source of newly synthesized proteins in the squid giant axon, whether any intra-axonal protein synthesis occurs in highly differentiated, mature axons, including the squid giant axon, remains controversial. The proponents of intra-axonal protein synthesis contend that the axon provides an example of what is termed “local protein synthesis,” a term commonly used to refer to all locations of protein synthesis in neurons outside of the cell body (Alvarez et al. 2000; Giuditta et al. 2008; Jung et al. 2012; Martin 2004). These authors present a theoretical argument that the maintenance of cytoskeletal proteins in distal regions of long axons cannot be solely dependent on the process of slow axonal transport and requires supplementation by local intra-axonal translation (Alvarez et al. 2000; Giuditta et al. 1986). In addition, they challenge the classical radioactive pulse-labeling paradigm used to demonstrate the slow axonal transport of various cytoskeletal proteins (Black and Lasek 1980; Hoffman and Lasek 1975) and state there is no direct experimental evidence for such slow transport of cytoskeleton proteins (Alvarez et al. 2000). However, recent studies, using contemporary imaging methods directly visualized the fast and slow axonal transport of membranes and cytoskeleton proteins, respectively, in living axons (Brown 2000, 2003; Brown and Jung 2013; Li et al. 2014). The second argument is that many components of translation machinery are found in axons, including in squid giant axoplasm, and therefore this serves as prima facie evidence that translation must be occurring in this cellular compartment (Alvarez et al. 2000; Giuditta et al. 2008). These data include reports of abundant tRNAs (Black and Lasek 1977; Ingoglia et al. 1983), mRNAs (Capano et al. 1987; Gioio et al. 1994; Kaplan et al. 1992; Rapallino et al. 1988), various elongation and initiation factors (Giuditta et al. 1980, 1986; Giuditta et al. 1991; Kar et al. 2013), rRNA (Giuditta et al. 1980; Perrone-Capano et al. 1999), and even polyribosomes that can be translated in vitro to produce various proteins (Giuditta et al. 1991; Sotelo et al. 1999). Given this apparent translation capacity of squid axoplasm it should then be possible for isolated axoplasm to synthesize proteins directly from radioactive amino acids. However, whenever radioactive amino acids are incubated together with isolated squid axoplasm there are absolutely no newly synthesized radioactive proteins that can be detected in these experiments (Lasek et al. 1977). Nevertheless other energy-intensive processes such as axonal transport by kinesin motors are readily observed in the extruded axoplasm (Kang et al. 2016; Prahlad et al. 2000; Vale et al. 1985). Another key observation against the idea of intra-axonal protein synthesis in the squid giant axon is the absence of morphological evidence for canonical ribosomes in the giant axon, and in vertebrate axons in general (Conradi 1966; Palay and Palade 1955; Palay et al. 1968; Peters et al. 1968, 1970; Zelena 1972). This is in contrast to well-documented sites of “local protein synthesis” such as in dendrites where canonical ribosomes and polyribosomes have clearly been observed (Steward and Levy 1982; Steward and Schuman 2003).
The proponents of de novo protein synthesis in the squid giant axon posit that the absence of canonical ribosomes in the giant axon and in other axons is due to the unconventional location of ribosomes in axons, i.e., beneath the plasma membrane, and refer to these sites as “periaxoplasmic plaques” (Alvarez et al. 2000; Giuditta et al. 2008; Koenig and Martin 1996). These putative subaxolemmal ribosomes are detected by various indirect assays which include staining with a sensitive RNA biding dye, YOYO-1 (Koenig and Martin 1996), electron microscopic imaging (ESI) that detects ribosomal phosphate signals (Bleher and Martin 2001; Martin et al. 1989), and immunohistochemistry using the Y-10B antibody specific or rRNA (Sotelo et al. 1999). If such subaxolemmal ribosomes are present in the squid giant axon, it is then argued that they might not be included in the axoplasm following extrusion (Alvarez et al. 2000), and that this could explain why isolated axoplasm could not be shown to synthesize proteins. It is specifically to address this proposal of subaxolemmal ribosomes that we did the experiments in this paper, in which the intact axon containing an unperturbed subaxolemmal domain was used and the translation of an endogenous, neuron-specific mRNA in the axon was studied. We previously showed by in situ hybridization that the axon contained neurofilament mRNAs, albeit at low levels compared to the cell bodies in the GFL (Way et al. 1992). Most importantly, there is no neurofilament mRNA or protein present in the adaxonal Schwann cells (Way et al. 1992; Cohen et al. 1987); therefore, if any radioactive neurofilament protein had been found in the axon after incubation in radioactive amino acids, it could not have been derived from translation in the Schwann cells. The presence of low levels of neurofilament subunit mRNAs in the axon as compared to the cell bodies in the GFL was confirmed in this paper by RNase protection assays (Figs. 1, 2). Evaluation of the radioactive proteins synthesized by the intact squid axon preparation to detect newly synthesized neurofilament proteins showed none at all (Figs. 4, 5). In contrast, there was abundant radioactive neurofilament protein newly synthesized in the cell bodies in the GFL, consistent with the proposition that the squid axon does not significantly synthesize detectable amounts of NF proteins from its endogenous NF mRNA.
We revisited this issue of denovo protein synthesis in the squid giant axon in this paper because there is increasing interest in local protein synthesis in neurons (Holt and Schuman 2013; Jung et al. 2012; Perry and Fainzilber 2014; Piper and Holt 2004) and many investigators cite the squid giant axon as an example of intra-axonal protein synthesis. There is increasingly convincing evidence emerging for local protein synthesis not only in dendrites (Steward 1983a, b; Steward and Levy 1982; Steward and Schuman 2003), but also in immature axons and growth cones during development, in axons after injury and regeneration, and in immature axons in tissue culture (Doron-Mandel et al. 2015; Holt and Schuman 2013; Jung et al. 2012; Kalinski et al. 2015; Rishal and Fainzilber 2014; Van Minnen 1994; Yoon et al. 2012). There have also been elegant studies on neurons in gastropod molluscs, e.g., Aplysia californica and Lymnaea stagnalis, that indicate that de novo protein synthesis can occur in their axons (Spencer et al. 2000; Van Minnen et al. 1997). However, it has been pointed out by many authors (Sanchez-Soriano et al. 2005; Steward and Schuman 2003; Van Minnen and Syed 2001) that the axons in these species are very short and tapered and dendrites in these invertebrates extend from the axons as opposed to cell bodies. Therefore, in these species, the distinction between axons and dendrites is unclear, and hence, it has been proposed that these structures should more properly be called “neurites”(Sanchez-Soriano et al. 2005; Van Minnen and Syed 2001). The squid is a cephalopod mollusk and its giant axon is a long cylinder of uniform diameter as is typical of vertebrate long axons, and giant axons in crustacea (Bittner 1981, 1991). Therefore, it would seem that these are more appropriate invertebrate models of mature axons. We hypothesize that uninjured, mature axons do not normally synthesize proteins de novo. Various studies on long axons in invertebrates and vertebrates support this view. In their review on RNA translation in axons, Piper and Holt (2004) point out that the evidence for protein synthesis in mature axons is difficult to interpret.
In addition to newly synthesized proteins in adaxonal glia that can be transferred into the squid giant axon (Tytell et al. 2016), it has been reported that newly transcribed RNAs in Schwann cells can also be transferred to axoplasm (Eyman et al. 2007), a phenomenon that has also been found to occur in vertebrate axons. Court et al. (2008) showed that when axons in the sciatic nerves of the Wallerian degeneration slow (Wlds) mutant mouse were severed from their cell somata, their adaxonal Schwann cells were able to transfer polysomes to the “desomatized” axons which resisted Wallerian degeneration for several weeks following the injury in this mutant mouse (Freeman 2014). Whereas in control axons canonical ribosomes are scarce, under these conditions of injury canonical ribosomes in the axons of the Wld s mouse dramatically increased compared to axons in control mice. It is not clear whether low levels of glia to axon transfer of ribosomes occur in uninjured axons, but these findings are consistent with the general view that following injury vertebrate axons can activate their translational machinery (Court et al. 2008; Holt and Schuman 2013; Piper and Holt 2004; Twiss and Fainzilber 2009; Zheng et al. 2001) as well as their glia to neuron macromolecular transfer mechanisms (Tytell et al. 2016). Several speculations about the possible mechanisms for glia to neuron transfer include the invagination of glial membranes and cytoplasm into the axons (Court et al. 2008), transfer by nanotubes (Twiss and Fainzilber 2009), and via exosomes (Holt and Schuman 2013; Twiss and Fainzilber 2009; Tytell et al. 2016).
What mechanisms could underlie the fact that mature axons that have the capacity to synthesize proteins but do not, and yet can be activated by injury to do so? It is possible that the RNA species in the normal, mature axons are translationally repressed until they are activated by the injury. Adaxonal glia may play an important role in this activation of the intrinsic translational machinery in the axon in addition to supplying functional ribosomes. There is ample evidence for such a regulatory mechanism in nature, e.g., in the oocyte where mRNAs are packaged with ribonucleoprotein particles (RNPs) and are translationally repressed by multiple mechanisms until they are activated in later stages of oogenesis (Hubstenberger et al. 2015). Many other examples exist in which mRNA binding proteins (RBPs) store mRNAs in inactive forms until regulatory mechanisms are invoked to activate translation (Anderson and Kedersha 2008; Kiebler and Bassell 2006; Thomas et al. 2011; Wells 2006). Interestingly, Kosik and Kritchevsky (2002) previously proposed that mRNAs are normally axonally transported in storage granules in a translationally repressed state. Lui-Yesucevitz et al. (2011) discuss the roles of RBPs in regulating the times and places of local protein synthesis in the context of different storage structures, RNPs, stress granules (SGs), and processing bodies (P-bodies), and various known regulating agents such as miRNAs (Schratt 2009), FMRP (Ashley et al. 1993; Narayanan et al. 2008), and the cytoplasmic polyadenylation element binding protein, CPEB (Richter and Klann 2009).
Conclusions
The maintenance of axonal structure and function is clearly served by a variety of cell biological mechanisms including transport of newly translated proteins and transcribed mRNAs from the cell body to the axon (Brown 2003; Grafstein and Forman 1980; Hoffman and Lasek 1975), glia to axon transfer of RNAs and proteins (Tytell et al. 2016), and de novo protein synthesis in injured, regenerating and growing axons (Holt and Schuman 2013; Piper and Holt 2004; Rishal and Fainzilber 2014; Van Minnen 1994). It is also clear that the presence of mRNAs and other translational machinery in mature axons should not necessarily be taken as evidence of de novo protein synthesis in axons. For example, we show in this paper that squid giant axoplasm contains neurofilament protein mRNA, but does not synthesize neurofilament proteins. That the presence of mRNAs in axons does not necessarily indicate intra-axonal proteins synthesis also comes from studies in the hypothalamo-neurohypophysial system of posterior pituitary axons and terminals. Magnocellular neuron axons and terminals in the posterior pituitary contain mRNAs which code for oxytocin and vasopressin precursor proteins that are transported from the magnocellular neurons in the hypothalamus. These mRNAs increase in amount with functional activity (Mohr et al. 1991; Mohr and Richter 1992, 2003; Murphy et al. 1989; Trembleau et al. 1994, 1995, 1996). However, it is well established that these mRNAs are not translated in the axons and terminals in the posterior pituitary to the precursor proteins and peptides (Brownstein et al. 1980; Gainer et al. 1977a; Sachs et al. 1969). The synthetic pathway and post-translational processing for these precursor proteins to be converted into their peptide products require a rough endoplasmic reticulum (RER) and a Golgi apparatus, organelles that are present in the neuronal cell bodies, but are absent from the pituitary axons and nerve endings (Brownstein et al. 1980; Burbach et al. 2001). Studies in other mature axon systems with regard to the issue of whether the presence of their intra-axonal RNAs are used for intra-axonal protein synthesis will depend on the development of new methods to visualize protein synthesis in situ (Kim and Jung 2015; tom Dieck et al. 2015). In this regard, it is interesting that by using the elegant puromycylation method described by tom Dieck et al. (2015) protein synthesis can be detected in cell bodies and dendrites but not in axons. Finally, in view of the idea that mRNAs in mature axons may be translationally repressed (Kosik and Krichevsky 2002; Krichevsky and Kosik 2001) it would be interesting to determine whether any of the known repressive agents (Liu-Yesucevitz et al. 2011) are present and operative in squid axoplasm. Of particular interest is whether miRNAs involved in translational repression (Schratt 2009) may reside in the disproportionally large amount of 76nt RNAs that are present in squid axoplasm (Ingoglia and Jalloh 2016).
Acknowledgments
This research was supported by the Intramural Research Program of the NIH. We thank Dr.'s Michael Tytell, Raymond Lasek, and Phillip Grant for their useful comments about this manuscript.
References
- Alvarez J, Giuditta A, Koenig E (2000) Protein synthesis in axons and terminals: significance for maintenance, plasticity and regulation of phenotype. With a critique of slow transport theory. Prog Neurobiol 62:1–62 [DOI] [PubMed] [Google Scholar]
- Anderson P, Kedersha N (2008) Stress granules: the Tao of RNA triage. Trends Biochem Sci 33:141–150. doi:10.1016/j.tibs.2007.12.003 [DOI] [PubMed] [Google Scholar]
- Ashley CT Jr., Wilkinson KD, Reines D, Warren ST (1993) FMR1 protein: conserved RNP family domains and selective RNA binding. Science 262:563–566 [DOI] [PubMed] [Google Scholar]
- Bittner GD (1981) Trophic interactions of CNS giant axons in crayfish. Compd Biochem Physiol 68A:299–306 [Google Scholar]
- Bittner GD (1991) Long-term survival of anucleate axons and its implications for nerve regeneration. Trends Neurosci 14:188–193 [DOI] [PubMed] [Google Scholar]
- Black MM, Lasek RJ (1977) The presence of transfer RNA in the axoplasm of the squid giant axon. J Neurobiol 8:229–237. doi:10.1002/neu.480080306 [DOI] [PubMed] [Google Scholar]
- Black MM, Lasek RJ (1980) Slow components of axonal transport: two cytoskeletal networks. J Cell Biol 86:616–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleher R, Martin R (2001) Ribosomes in the squid giant axon. Neuroscience 107:527–534 [DOI] [PubMed] [Google Scholar]
- Brown A (2000) Slow axonal transport: stop and go traffic in the axon Nature reviews. Mol Cell Biol 1:153–156. doi:10.1038/35040102 [DOI] [PubMed] [Google Scholar]
- Brown A (2003) Axonal transport of membranous and nonmembranous cargoes: a unified perspective. J Cell Biol 160:817–821. doi:10.1083/jcb.200212017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown A, Jung P (2013) A critical reevaluation of the stationary axonal cytoskeleton hypothesis. Cytoskeleton 70:1–11. doi:10.1002/cm.21083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown A, Lasek RJ (1990) The cytoskeleton of the squid giant axon. In: Gilbert DM, Adelman WJ Jr., Arnold JM (eds) Squid as experimental animals. Plenum Press, New York, pp 235–302 [Google Scholar]
- Brownstein MJ, Russell JT, Gainer H (1980) Synthesis, transport, and release of posterior pituitary hormones. Science 207:373–378 [DOI] [PubMed] [Google Scholar]
- Burbach JP, Luckman SM, Murphy D, Gainer H (2001) Gene regulation in the magnocellular hypothalamo-neurohypophysial system. Physiol Rev 81:1197–1267 [DOI] [PubMed] [Google Scholar]
- Capano CP, Giuditta A, Castigli E, Kaplan BB (1987) Occurrence and sequence complexity of polyadenylated RNA in squid axoplasm. J Neurochem 49:698–704 [DOI] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N (1987) Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 162:156–159. doi:10.1006/abio.1987.9999 [DOI] [PubMed] [Google Scholar]
- Cohen RS, Pant HC, House S, Gainer H (1987) Biochemical and immunocytochemical characterization and distribution of phosphorylated and nonphosphorylated subunits of neurofilaments in squid giant axon and stellate ganglion. J Neurosci 7:2056–2074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conradi S (1966) Ultrastructural specialization of the initial axon segment of cat lumbar motoneurons. Preliminary observations. Acta Soc Med Ups 71:281–284 [PubMed] [Google Scholar]
- Court FA, Hendriks WT, MacGillavry HD, Alvarez J, van Minnen J (2008) Schwann cell to axon transfer of ribosomes: toward a novel understanding of the role of glia in the nervous system. J Neurosci 28:11024–11029. doi:10.1523/JNEUROSCI.2429-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doron-Mandel E, Fainzilber M, Terenzio M (2015) Growth control mechanisms in neuronal regeneration. FEBS Lett 589:1669–1677. doi:10.1016/j.febslet.2015.04.046 [DOI] [PubMed] [Google Scholar]
- Eyman M et al (2007) Local synthesis of axonal and presynaptic RNA in squid model systems. Eur J Neurosci 25:341–350. doi:10.1111/j.1460-9568.2007.05304.x [DOI] [PubMed] [Google Scholar]
- Fischer S, Litvak S (1967) The incorporation of microinjected 14C-amino acids into TCA insoluble fractions of the giant axon of the squid. J Cell Physiol 70:69–74. doi:10.1002/jcp.1040700110 [DOI] [PubMed] [Google Scholar]
- Freeman MR (2014) Signaling mechanisms regulating Wallerian degeneration. Curr Opin Neurobiol 27:224–231. doi:10.1016/j.conb.2014.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gainer H, Sarne Y, Brownstein MJ (1977a) Neurophysin biosynthesis: conversion of a putative precursor during axonal transport. Science 195:1354–1356 [DOI] [PubMed] [Google Scholar]
- Gainer H, Tasaki I, Lasek RJ (1977b) Evidence for the glia-neuron protein transfer hypothesis from intracellular perfusion studies of squid giant axons. J Cell Biol 74:524–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert DM, Adelman WJ Jr., Arnold JM (1990) Squid as experimental animals. Plenum Press, New York, p 516 [Google Scholar]
- Gioio AE, Chun JT, Crispino M, Capano CP, Giuditta A, Kaplan BB (1994) Kinesin mRNA is present in the squid giant axon. J Neurochem 63:13–18 [DOI] [PubMed] [Google Scholar]
- Giuditta A, Dettbarn WD, Brzin M (1968) Protein synthesis in the isolated giant axon of the squid. Proc Natl Acad Sci USA 59:1284–1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuditta A, Cupello A, Lazzarini G (1980) Ribosomal RNA in the axoplasm of the squid giant axon. J Neurochem 34:1757–1760 [DOI] [PubMed] [Google Scholar]
- Giuditta A, Hunt T, Santella L (1986) Rapid important paper messenger RNA in squid axoplasm. Neurochem Int 8:435–442 [DOI] [PubMed] [Google Scholar]
- Giuditta A, Menichini E, Perrone Capano C, Langella M, Martin R, Castigli E, Kaplan BB (1991) Active polysomes in the axoplasm of the squid giant axon. J Neurosci Res 28:18–28. doi:10.1002/jnr.490280103 [DOI] [PubMed] [Google Scholar]
- Giuditta A, Kaplan BB, van Minnen J, Alvarez J, Koenig E (2002) Axonal and presynaptic protein synthesis: new insights into the biology of the neuron. Trends Neurosci 25:400–404 [DOI] [PubMed] [Google Scholar]
- Giuditta A, Chun JT, Eyman M, Cefaliello C, Bruno AP, Crispino M (2008) Local gene expression in axons and nerve endings: the glia-neuron unit. Physiol Rev 88:515–555. doi:10.1152/physrev.00051.2006 [DOI] [PubMed] [Google Scholar]
- Grafstein B, Forman DS (1980) Intracellular transport in neurons. Physiol Rev 60:1167–1283 [DOI] [PubMed] [Google Scholar]
- Grant P, Tseng D, Gould RM, Gainer H, Pant HC (1995) Expression of neurofilament proteins during development of the nervous system in the squid Loligo pealei. J Compd Neurol 356:311–326. doi:10.1002/cne.903560212 [DOI] [PubMed] [Google Scholar]
- Hoffman PN, Lasek RJ (1975) The slow component of axonal transport. Identification of major structural polypeptides of the axon and their generality among mammalian neurons. J Cell Biol 66:351–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt CE, Schuman EM (2013) The central dogma decentralized: new perspectives on RNA function and local translation in neurons. Neuron 80:648–657. doi:10.1016/j.neuron.2013.10.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubstenberger A, Cameron C, Noble SL, Keenan S, Evans TC (2015) Modifiers of solid RNP granules control normal RNP dynamics and mRNA activity in early development. J Cell Biol 211:703–716. doi:10.1083/jcb.201504044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingoglia NA, Jalloh B (2016) 76nt RNAsare transported axonally into regenerating axons and growth cones. What are they doing there? Neural Reg Res 11:390–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingoglia NA, Giuditta A, Zanakis MF, Babigian A, Tasaki I, Chakraborty G, Sturman JA (1983) Incorporation of 3H-amino acids into proteins in a partially purified fraction of axoplasm: evidence for transfer RNA-mediated, post-translational protein modification in squid giant axons. J Neurosci 3:2463–2473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung H, Yoon BC, Holt CE (2012) Axonal mRNA localization and local protein synthesis in nervous system assembly, maintenance and repair. Nat Rev Neurosci 13:308–324. doi:10.1038/nrn3210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinski AL, Sachdeva R, Gomes C, Lee SJ, Shah Z, Houle JD, Twiss JL (2015) mRNAs and protein synthetic machinery localize into regenerating spinal cord axons when they are provided a substrate that supports growth. J Neurosci 35:10357–10370. doi:10.1523/JNEUROSCI.1249-15.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang M, Baker L, Song Y, Brady ST, Morfini G (2016) Biochemical analysis of axon-specific phosphorylation events using isolated squid axoplasms. Methods Cell Biol 131:199–216. doi:10.1016/bs.mcb.2015.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan BB, Gioio AE, Capano CP, Crispino M, Giuditta A (1992) Beta-actin and beta-tubulin are components of a heterogeneous mRNA population present in the squid giant axon. Mol Cell Neurosci 3:133–144 [DOI] [PubMed] [Google Scholar]
- Kar AN, MacGibeny MA, Gervasi NM, Gioio AE, Kaplan BB (2013) Intra-axonal synthesis of eukaryotic translation initiation factors regulates local protein synthesis and axon growth in rat sympathetic neurons. J Neurosci 33:7165–7174. doi:10.1523/JNEUROSCI.2040-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiebler MA, Bassell GJ (2006) Neuronal RNA granules: movers and makers. Neuron 51:685–690. doi:10.1016/j.neuron.2006.08.021 [DOI] [PubMed] [Google Scholar]
- Kim E, Jung H (2015) Local protein synthesis in neuronal axons: why and how we study. BMB Rep 48:139–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenig E, Giuditta A (1999) Protein-synthesizing machinery in the axon compartment. Neuroscience 89:5–15 [DOI] [PubMed] [Google Scholar]
- Koenig E, Martin R (1996) Cortical plaque-like structures identify ribosome-containing domains in the Mauthner cell axon. J Neurosci 16:1400–1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosik KS, Krichevsky AM (2002) The message and the messenger: delivering RNA in neurons Science’s STKE: signal transduction knowledge environment. Sci Signal 2002:pe16. doi:10.1126/stke.2002.126.pe16 [DOI] [PubMed] [Google Scholar]
- Krichevsky AM, Kosik KS (2001) Neuronal RNA granules: a link between RNA localization and stimulation-dependent translation. Neuron 32:683–696 [DOI] [PubMed] [Google Scholar]
- Lasek RJ, Gainer H, Przybylski RJ (1974) Transfer of newly synthesized proteins from Schwann cells to the squid giant axon. Proc Natl Acad Sci USA 71:1188–1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasek RJ, Gainer H, Barker JL (1977) Cell-to-cell transfer of glial proteins to the squid giant axon. The glia-neuron protein trnasfer hypothesis. J Cell Biol 74:501–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Brown A, Jung P (2014) Deciphering the axonal transport kinetics of neurofilaments using the fluorescence photoactivation pulse-escape method. Phys Biol 11:026001. doi:10.1088/1478-3975/11/2/026001 [DOI] [PubMed] [Google Scholar]
- Liu-Yesucevitz L et al (2011) Local RNA translation at the synapse and in disease. J Neurosci 31:16086–16093. doi:10.1523/JNEUROSCI.4105-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin KC (2004) Local protein synthesis during axon guidance and synaptic plasticity. Curr Opin Neurobiol 14:305–310. doi:10.1016/j.conb.2004.05.009 [DOI] [PubMed] [Google Scholar]
- Martin R, Fritz W, Giuditta A (1989) Visualization of polyribosomes in the postsynaptic area of the squid giant synapse by electron spectroscopic imaging. J Neurocytol 18:11–18 [DOI] [PubMed] [Google Scholar]
- Mohr E, Richter D (1992) Diversity of mRNAs in the axonal compartment of peptidergic neurons in the rat. Eur J Neurosci 4:870–876 [DOI] [PubMed] [Google Scholar]
- Mohr E, Richter D (2003) Molecular determinants and physiological relevance of extrasomatic RNA localization in neurons. Front Neuroendocrinol 24:128–139 [DOI] [PubMed] [Google Scholar]
- Mohr E, Fehr S, Richter D (1991) Axonal transport of neuropeptide encoding mRNAs within the hypothalamo-hypophyseal tract of rats. EMBO J 10:2419–2424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy D, Levy A, Lightman S, Carter D (1989) Vasopressin RNA in the neural lobe of the pituitary: dramatic accumulation in response to salt loading. Proc Natl Acad Sci USA 86:9002–9005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan U, Nalavadi V, Nakamoto M, Thomas G, Ceman S, Bassell GJ, Warren ST (2008) S6K1 phosphorylates and regulates fragile X mental retardation protein (FMRP) with the neuronal protein synthesis-dependent mammalian target of rapamycin (mTOR) signaling cascade. J Biol Chem 283:18478–18482. doi:10.1074/jbc.C800055200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palay SL, Palade GE (1955) The fine structure of neurons. J Biophys Biochem Cytol 1:69–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palay SL, Sotelo C, Peters A, Orkand PM (1968) The axon hillock and the initial segment. J Cell Biol 38:193–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pant HC, Shecket G, Gainer H, Lasek RJ (1978) Neurofilament protein is phosphorylated in the squid giant axon. J Cell Biol 78:R23–R27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrone-Capano C, Crispino M, Menichini E, Kaplan BB, Giuditta A (1999) Ribosomal RNAs synthesized by isolated squid nerves and ganglia differ from native ribosomal RNAs. J Neurochem 72:910–918 [DOI] [PubMed] [Google Scholar]
- Perry RB, Fainzilber M (2014) Local translation in neuronal processes—in vivo tests of a “heretical hypothesis”. Dev Neurobiol 74:210–217. doi:10.1002/dneu.22115 [DOI] [PubMed] [Google Scholar]
- Peters A, Proskauer CC, Kaiserman-Abramof IR (1968) The small pyramidal neuron of the rat cerebral cortex. The axon hillock and initial segment. J Cell Biol 39:604–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters S, Palay S, Webster HF (1970) The fine structure of the nervous system. Harper and Row, New York [Google Scholar]
- Piper M, Holt C (2004) RNA translation in axons. Annu Rev Cell Dev Biol 20:505–523. doi:10.1146/annurev.cellbio.20.010403.111746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prahlad V, Helfand BT, Langford GM, Vale RD, Goldman RD (2000) Fast transport of neurofilament protein along microtubules in squid axoplasm. J Cell Sci 113(Pt 22):3939–3946 [DOI] [PubMed] [Google Scholar]
- Rapallino MV, Cupello A, Giuditta A (1988) Axoplasmic RNA species synthesized in the isolated squid giant axon. Neurochem Res 13:625–631 [DOI] [PubMed] [Google Scholar]
- Richter JD, Klann E (2009) Making synaptic plasticity and memory last: mechanisms of translational regulation. Genes Dev 23:1–11. doi:10.1101/gad.1735809 [DOI] [PubMed] [Google Scholar]
- Rishal I, Fainzilber M (2014) Axon-soma communication in neuronal injury. Nat Rev Neurosci 15:32–42. doi:10.1038/nrn3609 [DOI] [PubMed] [Google Scholar]
- Sachs H, Fawcett P, Takabatake Y, Portanova R (1969) Biosynthesis and release of vasopressin and neurophysin. Recent Prog Horm Res 25:447–491 [DOI] [PubMed] [Google Scholar]
- Sanchez-Soriano N et al (2005) Are dendrites in Drosophila homologous to vertebrate dendrites? Dev Biol 288:126–138. doi:10.1016/j.ydbio.2005.09.026 [DOI] [PubMed] [Google Scholar]
- Schratt G (2009) microRNAs at the synapse. Nat Rev Neurosci 10:842–849. doi:10.1038/nrn2763 [DOI] [PubMed] [Google Scholar]
- Sotelo JR, Kun A, Benech JC, Giuditta A, Morillas J, Benech CR (1999) Ribosomes and polyribosomes are present in the squid giant axon: an immunocytochemical study. Neuroscience 90:705–715 [DOI] [PubMed] [Google Scholar]
- Spencer GE, Syed NI, van Kesteren E, Lukowiak K, Geraerts WP, van Minnen J (2000) Synthesis and functional integration of a neurotransmitter receptor in isolated invertebrate axons. J Neurobiol 44:72–81 [DOI] [PubMed] [Google Scholar]
- Steinbach HB (1941) Chloride in the giant axon of the squid. J Cell Compd Physiol 17:57–64 [Google Scholar]
- Steward O (1983a) Alterations in polyribosomes associated with dendritic spines during the reinnervation of the dentate gyrus of the adult rat. J Neurosci 3:177–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steward O (1983b) Polyribosomes at the base of dendritic spines of central nervous system neurons–their possible role in synapse construction and modification. Cold Spring Harb Symp Quant Biol 48(Pt 2):745–759 [DOI] [PubMed] [Google Scholar]
- Steward O, Levy WB (1982) Preferential localization of polyribosomes under the base of dendritic spines in granule cells of the dentate gyrus. J Neurosci 2:284–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steward O, Schuman EM (2003) Compartmentalized synthesis and degradation of proteins in neurons. Neuron 40:347–359 [DOI] [PubMed] [Google Scholar]
- Szaro BG, Pant HC, Way J, Battey J (1991) Squid low molecular weight neurofilament proteins are a novel class of neurofilament protein. A nuclear lamin-like core and multiple distinct proteins formed by alternative RNA processing. J Biol Chem 266:15035–15041 [PubMed] [Google Scholar]
- Thomas MG, Loschi M, Desbats MA, Boccaccio GL (2011) RNA granules: the good, the bad and the ugly. Cell Signal 23:324–334. doi:10.1016/j.cellsig.2010.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- tom Dieck S et al (2015) Direct visualization of newly synthesized target proteins in situ. Nat Methods 12:411–414. doi:10.1038/nmeth.3319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trembleau A, Morales M, Bloom FE (1994) Aggregation of vasopressin mRNA in a subset of axonal swellings of the median eminence and posterior pituitary: light and electron microscopic evidence. J Neurosci 14:39–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trembleau A, Melia KR, Bloom FE (1995) BC1 RNA and vasopressin mRNA in rat neurohypophysis: axonal compartmentalization and differential regulation during dehydration and rehydration. Eur J Neurosci 7:2249–2260 [DOI] [PubMed] [Google Scholar]
- Trembleau A, Morales M, Bloom FE (1996) Differential compartmentalization of vasopressin messenger RNA and neuropeptide within the rat hypothalamo-neurohypophysial axonal tracts: light and electron microscopic evidence. Neuroscience 70:113–125 [DOI] [PubMed] [Google Scholar]
- Twiss JL, Fainzilber M (2009) Ribosomes in axons–scrounging from the neighbors? Trends Cell Biol 19:236–243. doi:10.1016/j.tcb.2009.02.007 [DOI] [PubMed] [Google Scholar]
- Tytell M, Lasek RJ (1984) Glial polypeptides transferred into the squid giant axon. Brain Res 324:223–232 [DOI] [PubMed] [Google Scholar]
- Tytell M, Pant HC, Gainer H, Hill WD (1990) Characterization of the distinctive neurofilament subunits of the soma and axon initial segments in the squid stellate ganglion. J Neurosci Res 25:153–161. doi:10.1002/jnr.490250202 [DOI] [PubMed] [Google Scholar]
- Tytell M, Lasek RJ, Gainer H (2016) Axonal maintenance, glia, exosomes, and heat shock proteins. F1000Research 5(F1000 Faculty Rev). doi:10.12688/f1000research.7247.1 [DOI] [PMC free article] [PubMed]
- Vale RD, Schnapp BJ, Mitchison T, Steuer E, Reese TS, Sheetz MP (1985) Different axoplasmic proteins generate movement in opposite directions along microtubules in vitro. Cell 43:623–632 [DOI] [PubMed] [Google Scholar]
- Van Minnen J (1994) RNA in the axonal domain: a new dimension in neuronal functioning? Histochem J 26:377–391 [DOI] [PubMed] [Google Scholar]
- Van Minnen J, Syed NI (2001) Local protein synthesis. In Invertebrate axons: from dogma to dilemma. Results and Problems in Cell Differentiation, p 34 [DOI] [PubMed]
- Van Minnen J et al (1997) De novo protein synthesis in isolated axons of identified neurons. Neuroscience 80:1–7 [DOI] [PubMed] [Google Scholar]
- Way J, Hellmich MR, Jaffe H, Szaro B, Pant HC, Gainer H, Battey J (1992) A high-molecular-weight squid neurofilament protein contains a lamin-like rod domain and a tail domain with Lys-Ser-Pro repeats. Proc Natl Acad Sci USA 89:6963–6967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells DG (2006) RNA-binding proteins: a lesson in repression. J Neurosci 26:7135–7138. doi:10.1523/JNEUROSCI.1795-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon BC, Jung H, Dwivedy A, O’Hare CM, Zivraj KH, Holt CE (2012) Local translation of extranuclear lamin B promotes axon maintenance. Cell 148:752–764. doi:10.1016/j.cell.2011.11.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelena J (1972) Ribosomes in myelinated axons of dorsal root ganglia. Z fur Zellforsch Mikroskopische Anat 124:217–229 [DOI] [PubMed] [Google Scholar]
- Zheng JQ, Kelly TK, Chang B, Ryazantsev S, Rajasekaran AK, Martin KC, Twiss JL (2001) A functional role for intra-axonal protein synthesis during axonal regeneration from adult sensory neurons. J Neurosci 21:9291–9303 [DOI] [PMC free article] [PubMed] [Google Scholar]