Abstract
Background
We reported on a pilot study of minor histocompatibility antigen vaccination using constructs expressing male-specific gene disparities of selected mouse CDNA on Y and sex determining region Y in the canine model. We performed reduced-intensity hematopoietic cell transplantation with female donors and male recipients, producing stable mixed donor-recipient hematopoietic chimeras. We then performed a vaccine series in three female transplant donors followed by donor lymphocyte infusion (DLI) into their respective mixed chimeras. One mixed chimera experienced a significant shift in the percentage of donor chimerism, but no response occurred in the other 2 recipients. We then hypothesized that inadequate donor sensitization was responsible for these results.
Methods
To test this hypothesis, we added 4 monthly booster vaccinations to 2 of the original hematopoietic cell transplantation donors, including the donor that drove the partial response, followed by a second DLI.
Results
Strong T cell responses were shown by ELISpot and confirmed by intracellular cytokine staining in both donors. A second DLI resulted in a further increase in donor chimerism in the same mixed chimera that experienced the previous increase, but no change in donor chimerism was again seen in the other recipient. Evaluation of RNA expression of the target antigens demonstrated that conversion occurred in the recipient that expressed both selected mouse CDNA on Y and sex determining region Y.
Conclusions
T cell responses against Y chromosome-encoded disparities were not necessarily sufficient to drive in vivo female antimale responses. Other factors including the presence of specific haplotypes or the heterogeneous expression of the target antigen may affect T cell responses against minor histocompatibility antigens. These results warrant future vaccine studies in a larger transplant cohort using epigenetic modulation of the recipient to promote target gene expression.
In the major histocompatibility complex (MHC)-matched hematopoietic cell transplantation (HCT) setting, minor histocompatibility antigens (miHAs) are implicated in curative graft-versus-tumor (GVT) responses for patients with hematologic malignancies, as well as the morbidity of graft rejection and graft-versus-host disease (GVHD). Minor histocompatibility antigens are MHC class I- and class II-presented endogenous peptides derived from nonsynonymous disparities within coding regions between the donor and recipient. These include unique Y chromosome disparities (H-Y) in female into male HCT. Genetic disparities that give rise to miHAs including H-Y are only antigenic when presented in the context of specific MHC molecules, a requirement termed HLA-restricted and dog leukocyte antigen (DLA)-restricted, in humans and dogs, respectively. Tissue-selective expression of miHAs suggests that it may be possible to augment and separate GVT responses from GVHD using a miHA vaccine.1 Although some miHAs are known in humans, formidable obstacles of efficacy, safety, and feasibility currently prevent the translation of our knowledge of miHAs into an established immunotherapy.2
We seek to establish a recombinant miHA vaccine in the canine model of allogeneic HCT to provide a large outbred animal model capable of addressing the challenges faced in implementing a miHA vaccine in human allogeneic HCT. With minimum-intensity conditioning, DLA-identical marrow infusion, and a short course of postgrafting immunosuppression, the canine model produces stable mixed donor-recipient hematopoietic chimeras.3 This mixed chimerism is a state of tolerance between donor and recipient cells and is not affected by “unsensitized” donor lymphocyte infusions (DLIs).4,5 However, if the donor is first sensitized to miHAs via recipient-derived skin implants, organ transplantation, or injections of allogeneic peripheral blood mononuclear cells (PBMCs), then a sensitized DLI breaks tolerance resulting in full donor chimerism that is often accompanied by GVHD.4-7 Thus, stable mixed chimerism provides a reproducible in vivo model to test donor T cell sensitization against recipient miHAs. A graphic reproduction of published results on chimerism analyses after unsensitized and miHA-sensitized DLI into DLA-identical mixed chimeras is provided in Figure 1 as a reference to interpret the results of this pilot study.4
FIGURE 1.
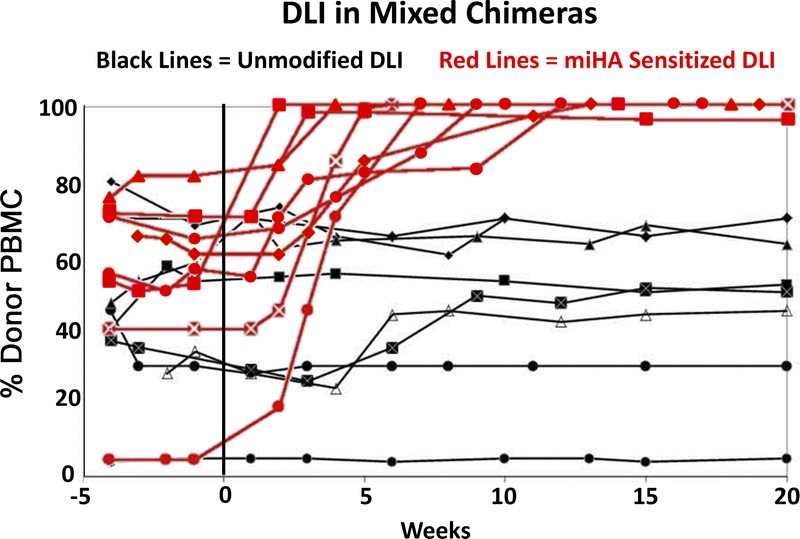
Reproduction of published results following unsensitized DLI and miHA-sensitized DLI into eight stable mixed chimeric recipients.4 The chimerism results were shown as percent donor PBMC on the y axis, with weeks after the DLI shown on the x axis. Eight recipients were infused with unsensitized donor lymphocytes and followed by chimerism analysis (black lines). Eight donors were then sensitized to a miHA via 4 weekly skin grafts from their respective recipients. Eight recipients, 6 of whom had first received an unsensitized DLI, then received a miHA-sensitized DLI 1 week after their respective donor's last skin graft, followed by chimerism analysis (red lines).
A major challenge facing the development of a recombinant miHA vaccine in the canine model is the lack of characterized miHAs. T cell cloning reagents used to characterize miHAs in humans are not yet available in the canine model. Instead, we postulated that making a vaccine encoding large sections of Y chromosome gene disparities may overcome the lack of peptide-level characterization of miHAs in the canine model and allow us to further develop this model through the use of female transplant donors and male transplant recipients.
At the time of vaccine development, the canine genome had only 3 Y chromosome gene sequences available including ubiquitously transcribed tetratricopeptide repeat containing, Y-linked (UTY), selected mouse CDNA on Y (SMCY), and sex determining region Y (SRY). Attempts were made to clone the most disparate sections with respect to their X homologues of the large genes UTY and SMCY, as well as the entire small SRY gene. After 2 cloning attempts, 3 domains of canine SMCY encoding approximately 60% of the total disparities with the X-homologue SMCX were cloned, as well as the entire SRY gene. These clones were shuttled into DNA expression plasmids and replication-deficient human adenovirus type 5 (rAd5) vectors. Three female HCT donors then received 2 doses of the expression plasmids delivered by particle-mediated epidermal delivery (PMED) 4 weeks apart, and then 4 weeks later received an intramuscular boost of rAd5. Four weeks after the rAd5 boost injection, a DLI into their respective male mixed hematopoietic chimeric recipients resulted in a significant increase in donor chimerism in 1 of 3 hosts, representing the first functional miHA response to a recombinant miHA vaccine in a large animal model.8
In our initial vaccine series, we observed relatively weak antigen-specific T cell responses at the time of the first DLI. We hypothesized that “inadequate donor sensitization” may explain why the observed increase in donor chimerism did not reach full donor chimerism and explain the lack of change in donor chimerism in the other 2 recipients. Because 2 of the original donor-recipient transplant pairs were still available, including the pair whose recipient demonstrated an increase in donor chimerism after the first DLI, we were able to address this hypothesis with 4 additional PMED boost injections in the 2 female donors followed by a second DLI into their respective male mixed hematopoietic chimeric recipients.
MATERIALS AND METHODS
Experimental Animals, DLA Typing, DLA-Identical HCT, and Chimerism Analysis
The donor-recipient HCT pairs in this study were H353-H519 and H592-H597. These dogs were mini-mongrel cross breeds that were raised at the Fred Hutchinson Cancer Research Center, Seattle, WA, and housed in kennels certified by the American Association for Accreditation of Laboratory Animal Care. All study designs were approved by Institutional Animal Care and Use Committee. The DLA typing method was previously described9-11 and reported.8 The DLA typing for H353-H519 included DRB1 9/22 and DLA-88 03801/50101, whereas typing for H592-H597 was DRB1 9/15 and DLA-88 01101/01201. Minimal-intensity DLA-identical HCT was previously described.3 Chimerism analyses were previously described.12
Vaccine Preparation, Administration, and DLI
The amino acid sequence of the 4 expression plasmids was previously reported.8 The 4 plasmids were mixed at equal concentrations, and 2 μg of plasmid DNA was coated onto 0.5 mg of 1 micron gold particles per cartridge, as described.13 The dogs underwent general anesthesia. Eight cartridges were then delivered to each side of the abdomen lateral to the mammary gland using the PMED device, Powderject XR1, set between 400 to 500 psi, as previously reported.8 The protocol for DLI was previously described.14,15 Cell counts with differentials were obtained using an ADVIA 2120i (Siemens, Deerfield, IL). T cell counts were determined using canine-specific antibody for CD3 as described.4
Interferon-γ ELISpot, Interferon-γ Intracellular Cytokine Staining, and Peptide Nomenclature
Enzyme-linked immunospot (ELISpots) and intracellular cytokine staining (ICS) were performed as described.8 The same amount of dimethyl sulfoxide (DMSO) used to dissolve the peptide or peptides was used as a negative control. Phytohemagglutinin at a final concentration of 25 μg/mL was used as a positive control for ELISpots, and phorbol myristate acetate at 100 ng/mL and ionomycin at 1 μg/mL was used as a positive control for ICS. Positive ELISpot responses were defined as greater than 50 spot-forming cells per 106 PBMCs on average, with nonoverlapping standard deviations with the DMSO-negative control. Positive ICS responses were based on the percentage of CD3+ CD4+ or CD3+ CD8+ T cells expressing interferon-γ that was determined by placing a gate with an adequate proximity to the DMSO-negative control and maintaining that gate for all samples tested. A positive response was defined as a 4-fold greater response than the DMSO-negative control as long as the DMSO-negative control remained below 0.05%, and the response was greater than or equal to 0.1% interferon-γ–expressing CD3+ CD4+ or CD3+ CD8+ T cells.16
The 168 overlapping 15-mer peptide sequences covering the coding sequence of SMCY domain 1 (D1), SMCY domain 2 (D2), SMCY domain 3 (D3), and SRY expression plasmids used in these studies were previously provided.8 Six overlapping peptides covered SMCY D1 peptide (P) 1 to 6, 40 peptides covered SMCY D2 P1-40, 69 peptides covered SMCY D3 P1-69, and 53 peptides covered SRY P1-53. For the pooled peptide ELISpots and ICS, the 168 peptides were separated into 26 pools that allowed for a 13 × 13 peptide pool matrix as shown in Figures 3A and B.
FIGURE 3.
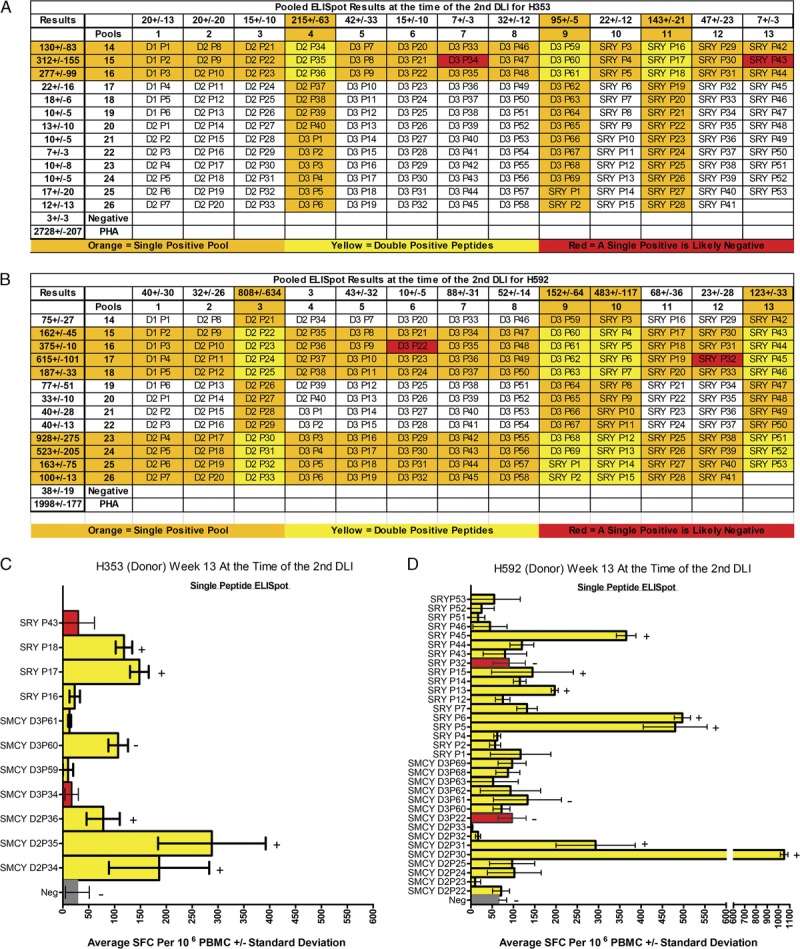
Determining single peptide responses from within the pooled ELISpot results in both female donors at the time of the second DLI. Pooled ELISpot results at the time of the second DLI for H353 (A) and H592 (B) were shown in the context of a 13 × 13 overlapping peptide pool matrix with the DMSO-negative and phytohemagglutinin-positive control included as a reference. Each column and each row display the peptides included in a pool. D1, D2, and D3 refer to the 3 domains of SMCY encoded within the vaccine; SMCY was dropped from the spreadsheet for space constraints. The P refers to peptide and the number after the P refers to the overlapping peptide, as discussed in the Materials and Methods section. Results were listed either horizontally or vertically with respect to each pool and expressed as average SFC per 106 PBMCs ± standard deviation. Strongly positive pools were highlighted in orange. Individual peptides found at the intersection of 2 strongly positive pools were highlighted in yellow and underwent single peptide ELISpot confirmation for H353 (C) and H592 (D). Two peptides that overlap a negative and positive pool, thus predicted not to elicit a response, were highlighted in red and included in the single peptide analysis. Specific single peptide responses underwent ICS confirmation. A plus sign (+) indicates a positive ICS response and a minus sign (−) indicates a negative ICS response shown in Table 1 and Figure 4. SFC, spot-forming cell.
Reverse Transcription-Polymerase Chain Reaction
RNA was isolated from frozen PBMCs using the RNeasy Plus kit; to maximize genomic DNA elimination, the samples went through the gDNA eliminator with on-column DNase digestion (Qiagen, Venlo, Netherlands). First-strand cDNA synthesis was performed with random hexamers using SuperScript III First-Strand Synthesis System (Thermo Fisher, Waltham, MA). Polymerase chain reaction (PCR) was performed using heat-activated immomix (Bioline, London, UK). To prevent carry over DNA contamination, 2'-deoxyuridine 5'triphosphate was spiked into each PCR reaction, and all subsequent PCR reactions underwent an initial uracil-DNA glycosylase incubation step (Amresco, Solon, OH). Gene-specific primers were created using sequences from Canis Familiaris GenBank reference sequence DQ156494.1 for SMCY and sequence AF107021.1 for SRY. All the primer sets spanned introns with the exception of the SRY gene, which has no introns.
RESULTS
Extra Rounds of Sensitization Produce Strong Pooled Peptide Responses in Both Donors
The entire vaccine schema for the 2 female donors H353 and H592 is shown vertically in Figures 2A and B. The initial vaccine series consisted of priming these donors with 2 monthly rounds of the expression plasmids delivered by PMED, which was followed by a boost from an injection of rAd5 viral vectors encoding the same disparities. Overlapping peptide pools that covered the entire coding sequence of the expression plasmids were used to evaluate responses by interferon-γ ELISpot and ICS.13 At the end of the initial vaccine series and at the time of the first DLI, only a few peptide pools were weakly positive by ELISpot and able to be confirmed by ICS. No positive responses were seen for individual peptides at the time of the first DLI for either donor (data not shown).
FIGURE 2.
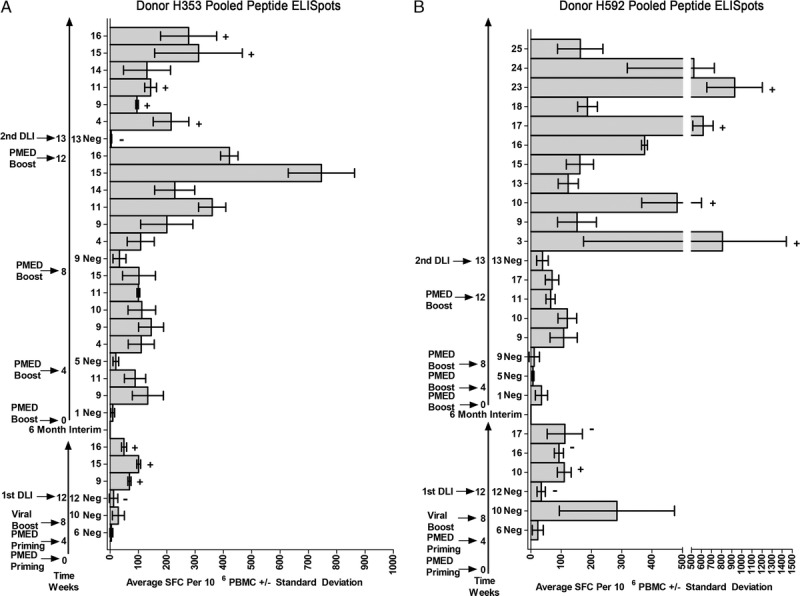
Summary of the pooled ELISpot results in the two donors. The entire treatment schema is shown vertically for donors H353 (A) and H592 (B). The 2 long, vertical arrows plot out time in weeks; the numbers to the left indicate the timing of the various rounds of sensitization and subsequent DLIs; the numbers to the right refer to the time at which the ELISpots were performed. The first long vertical arrow represents the first vaccine regimen and first DLI from the previous study, provided as a reference. The second long vertical arrow, following the six-month interim, depicts the four PMED boost sensitizations and the second DLI. The numbers to the right of the 2 long vertical arrows refer to the time at which the ELISpots were performed and were placed next to the DMSO negative (Neg) control for the ELISpot performed at that time point. Only positive pool responses were shown after the DMSO negative control. Specific pooled peptide responses underwent ICS confirmation. A plus sign (+) indicates a positive ICS response and a minus sign (−) indicates a negative ICS response shown in Table 1 and Figure 4.
Based on the review of a vaccine series used to validate the expression plasmids by PMED in 2 test females,8 it was thought that strong ELISpot responses would be detected with 2 extra PMED boosts because these donors were already previously sensitized. Two additional PMED boosts for a total of four injections would then allow an assessment of the number of rounds of sensitizations necessary to reach maximal in vitro responses. Additionally, based on preliminary validation studies, the maximum ELISpot response after PMED boosting was observed 1 week after the injection, and began to decrease after that time (unpublished results). Therefore, ELISpot responses were assessed 1 week after each PMED boost.
At the time of the second DLI into their respective mixed chimeric recipients, multiple strong pooled peptide responses were observed for both female donors. For H353, the in vitro responses peaked after the third PMED boost, but were still increasing after the fourth PMED boost for H592. Sufficient samples were available to confirm a variety of CD4+ and CD8+ T cell responses to many of these pooled peptides in both female donors (Table 1 and Figure 4).
TABLE 1.
Summary of the intracellular cytokine staining resultsa

FIGURE 4.
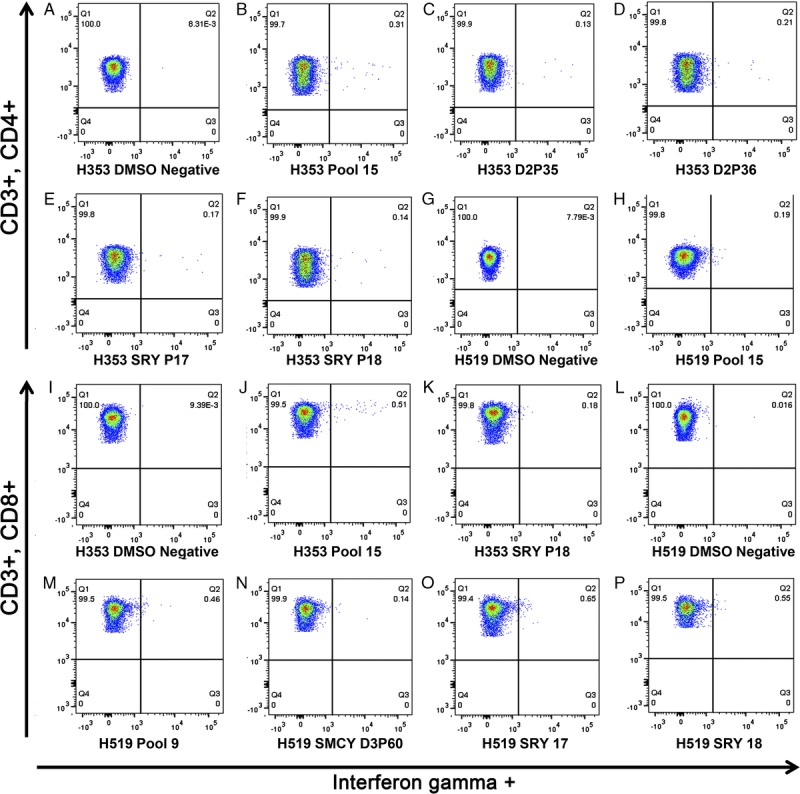
Three-color intracellular cytokine staining for interferon-γ. The percentage of interferon-γ CD3 + CD4+ T cells of H353 responding to (A) the DMSO-negative control, (B) pool 15, (C) SMCY D2P35, (D) SMCY D2P36, (E) SRY P17, and (F) SRY P18. The percentage of interferon-γ CD3 + CD4+ T cells of H519 responding to (G) DMSO-negative control, and (H) pool 15. The percentage of interferon-γ CD3 + CD8+ T cells of H353 responding to (I) DMSO-negative control, (J) pool 15, and (K) SRY P18. The percentage of interferon-γ CD3 + CD8+ T cells of H519 responding to (L) DMSO-negative control, (M) pool 9, (N) SMCY D3P60, (O) SRY P17, and (P) SRY P18. The remainder of the data is listed in table format (Table 1).
Antigen-Specific Responses to Individual Peptides in Both Donors
The strong pooled peptide results suggested that it would be possible to detect in vitro responses to individual peptides. From the onset of these studies, the 26 peptide pools were arranged into a 13 × 13 matrix, shown in a spreadsheet format for the pooled peptide ELISpot performed at the time of the second DLI for both female donors in Figures 3A and B. The matrix allows for a quick determination of candidate peptides that were contributing to the responses seen in the pooled ELISpot and ICS assays. Strongly positive pools were highlighted in orange. Individual peptides at the intersection of 2 positive pools were highlighted in yellow and underwent a single peptide ELISpot confirmation (Figures 3C and D). As further validation of the matrix, 2 peptides at the intersection of a positive and negative pool, thus predicted to produce a negative response, were highlighted in red and used as negative controls in the single peptide ELISpot. Many of these ELISpot responses were confirmed by ICS (Table 1 and Figure 4).
A relatively few peptides were responsible for the majority of the peptide pool responses in both donors at the time of the second DLI, and demonstrated that a detectable immune response was generated to a relatively small amount of the encoded disparities. For example, 6 peptides accounted for the majority of the pooled peptide responses for H353. These 6 peptides implicated three small sections within 3 of the 4 expression plasmids used in the vaccine including SMCY domain 2 overlapping peptides 34 to 36, SMCY domain 3 peptide 60, and SRY overlapping peptides 17 and 18. The ICS showed a predominantly CD4+ response to SMCY domain 2 overlapping peptides 35 to 36, but a detectable CD8+ T cell response was also shown for SMCY domain 2 peptides 34 and 36. The ICS response for SMCY domain 3 peptide 60 did not reach significance. Finally, SRY peptides 17 and 18 both demonstrated a CD4+ response, whereas a CD8+ response was also shown for SRY peptide 18. In a similar fashion, the majority of peptide pool responses for H592 were attributable to small sections of the encoded disparity. Using the 13 × 13 matrix, 31 yellow highlighted peptides were tested. Single peptide ELISpot implicated SMCY domain 2 overlapping peptides 30 to 31, and several SRY covering peptides. The ICS confirmed that these peptides induced various CD4+ and CD8+ T cell responses. In summary, CD4+ and CD8+ T cell responses were shown against sections of SMCY and SRY at the time of the second DLI for both donors.
Chimerism and Evaluation of GVHD After Vaccine-Sensitized DLI
After the first DLI, H519 experienced an increase in donor chimerism, donor PBMC increased from 50% to 70%, and donor granulocytes increased from 30% to 90% (red lines, Figures 5A and B); no change in the percent donor chimerism occurred in the other 2 recipients H597 and H382. In the second DLI, H519 received 0.4 × 108 CD3+ T cells/kg and H597 received 0.34 × 108 CD3+ T cells/kg. H519 experienced a further increase in donor chimerism with donor PBMC increasing from 70% to 90%, and donor granulocytes increased from 90% to 100% (green lines, Figures 5A and B). No change in donor chimerism was again observed for H597. No clinical or pathologic evidence of GVHD was seen in either H519 or H597 at necropsy.
FIGURE 5.
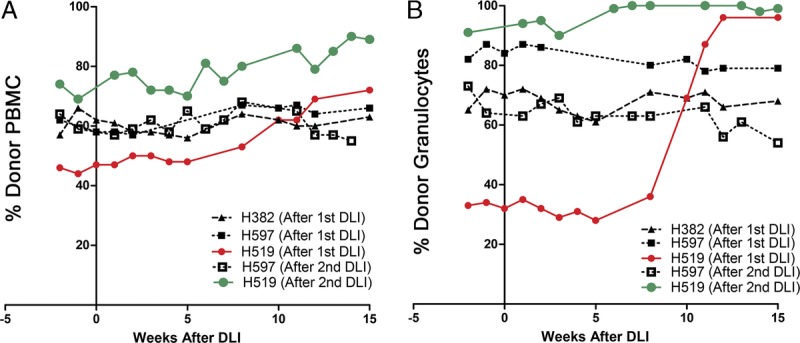
The chimerism results were shown as percent donor PBMC (A) and percent donor granulocytes (B) on the y axis, with weeks after the DLI shown on the x axis. The results for the PMED boosting study were labeled “(After second DLI)”, and the chimerism results following the first vaccine study were labeled “(After first DLI).” The chimerism results are shown for recipient H519 after the first DLI (red) and after the second DLI (green).
Assessing the Immune Responses in the 2 Mixed Chimeric Recipients
A major goal of this study was to isolate a T cell response to an individual peptide in a recipient that underwent an increase in donor chimerism after a vaccine-sensitized DLI as evidence of peptide-level characterization of miHA. No positive peptide pool responses were seen in any of the 3 recipients 1 week and 5 weeks after the first DLI. However, after H519 experienced an increase in donor chimerism, an ELISpot performed 13 weeks after the DLI was positive for pools 15 and 16 (Figure 6). These responses were confirmed by ICS with pool 15 producing a CD4+ T cell response and pool 16 producing a CD8+ response (Table 1). No positive responses were seen for single peptides from within pools 15 and 16 by ELISpot after 2 attempts (data not shown).
FIGURE 6.
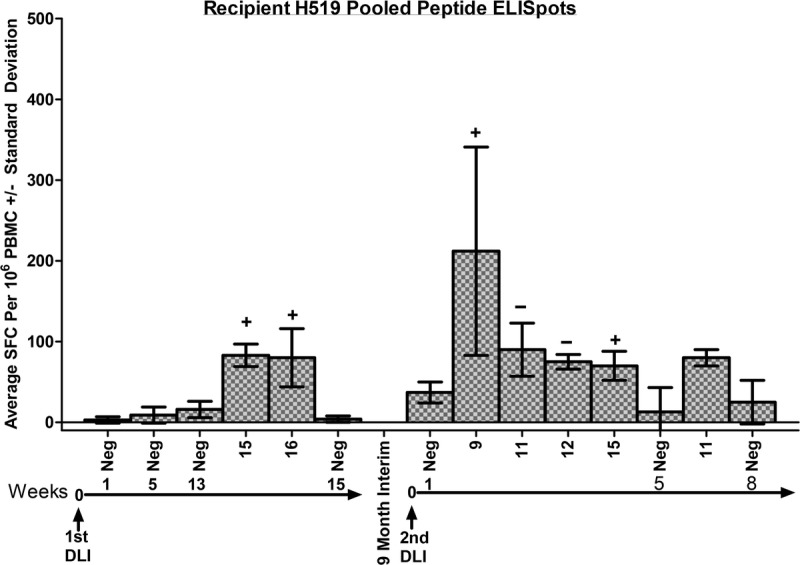
Summary of the pooled ELISpot results for recipient H519. The 2 long horizontal arrows represent the time after each DLI, separated by a 9-month interval. The numbers above the arrows refer to the time after the DLI at which the ELISpots were performed and are placed below the DMSO negative (neg) controls for each time point tested. Only positive pools were shown after the DMSO-negative control from that time. Specific pooled peptide responses underwent ICS confirmation. A plus sign (+) indicates a positive ICS response, and a minus sign (−) indicates a negative ICS response shown in Table 1 and Figure 4.
The second vaccine-sensitized DLI provided another opportunity to demonstrate an immune response in a recipient that underwent an increase in donor chimerism. In contrast to the first DLI, responses were evident within 1 week after the second DLI into recipient H519 with positive peptide pools by ELISpot to 9, 11, 12, and 15 (Figure 6). The ICS analysis showed that pool 9 produced a CD8+ response, whereas pool 15 demonstrated a CD4+ response, and pools 11 and 12 were negative (Table 1, Figure 4). Two single peptide ELISpots failed to show a positive response to peptides shown to produce a response in the vaccinated donor H353. An attempt was then made to isolate a response to a single peptide using ICS, because differences exist with respect to response detection between ELISPOT and ICS.17,18 Assaying these same peptides by ICS showed a CD8+ T cell response for SMCY domain 3 peptide 60, SRY peptide 17, and SRY peptide 18 (Table 1 and Figure 4). Further characterization of the memory or naive T cell subset analysis of these responses awaits the development of that capability in the canine model. By 5 weeks after the second DLI, only pool 11 remained weakly positive by ELISpot, and no positive responses were seen after that time (Figure 6). No ELISpot responses were observed 1 week and 5 weeks after the second DLI for recipient H597 (not shown). In summary, T cells were shown to respond to peptides encoded from within SMCY and SRY in the recipient that experienced an increase in donor chimerism.
RNA Expression of SMCY and SRY
We then asked the question whether the transplant recipients transcribed the target genes in PBMC at the time of the DLI (Figure 7). RNA was isolated from frozen PBMC samples on all the male mixed chimeric recipients involved in the vaccine studies including H382, H597, and H519. The SMCY gene expression was ubiquitously detected, but SRY was only detected in H519, and this expression remained after the second DLI. We sequenced recipient genomic DNA through the regions corresponding to each primer to eliminate the possibility that an unknown polymorphism inhibited amplification of SRY in some animals. The genomic sequences were perfectly homologous at all primer positions in all animals, arguing that the observed difference in reverse transcription-PCR between the 2 recipients was due to a difference in expression.
FIGURE 7.
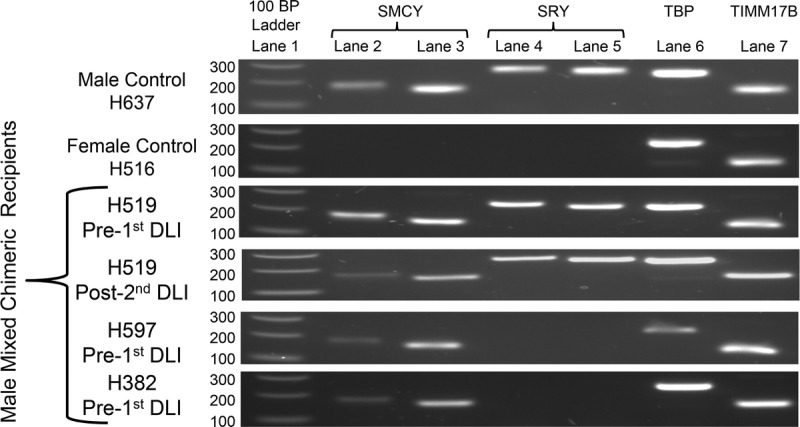
Recipient RNA expression of SMCY and SRY by RT-PCR. Lane 1 was loaded with 100 BP DNA ladder. Lanes 2–5 were loaded with reverse transcribed-PCR products from different primer pairs for SMCY and SRY. Lanes 6 and 7 were loaded with RT-PCR products from primer pairs for the housekeeping genes TBP and TIMM17B. Shown were the RT plus (+) samples for a male control, a female control, and the mixed chimeric recipients that underwent DLI. RT minus controls was performed for each sample (not shown). Positive SRY and SMCY bands were verified by sequencing. The following forward and reverse primers were used for SMCY lane 2 (accaactgcaggctgaaacc/agatctgagccctgcacact), SMCY lane 3 (ctgcaggagccctatcattc/agatctgagccctgcacact), SRY lane 4 (agcctcctcctccatgctat/ttagagtaaaaagaagcgtcagcg), SRY lane 5 (ggtaccagtggaaaatgcttacag/cagctgtccgtgtaggtgaa), TBP lane 6 (gaaatgctgaatataatcccaagc/cagctccccaccatgttct), and TIMM17B lane 7 (atcaagggcttccgcaatg/cacagtcgatggtggagaacag).TBP, TATA-box binding protein; TIMM17B, translocase of inner mitochondrial membrane 17 homology B (yeast).
DISCUSSION
This is a second report on a pilot study designed to develop a recombinant miHA vaccine in the canine model of allogeneic HCT. The overall numbers of transplant pairs used in these studies are small, which prevent definitive conclusions. However, important insights into developing immunotherapies targeting genetic disparities can be gleaned from these studies to address the still unmet clinical goal of boosting GVT responses by targeting specific miHAs.
Our initial vaccine trial demonstrated that adoptive transfer of female donor T cells sensitized with a recombinant vaccine encoding Y chromosome disparities can induce in vivo responses even when the responses are barely detectable by ELISpot and ICS. In this report, we add to our understanding by showing that strong SMCY-specific and SRY-specific CD4+ and CD8+ donor T cell responses did not affect which recipient responded to the vaccine-sensitized DLI. These results are not consistent with an inadequate donor sensitization hypothesis. Instead, future vaccine trials with a larger cohort are necessary to determine whether the observed in vivo response occurred exclusively due to the presence of a specific DLA haplotype or in the setting where recipient cells expressed both SMCY and SRY.
Historical experiments that used skin transplant rejection in congenic mice to map the locus of miHAs provide several insights applicable to the interpretation of our results.19 First, it is known that not all congenic strains of female mice are able to reject skin grafts from male donors, that is, possess the ability to strongly respond to the genetic disparity encoded within the Y chromosome.20-23 Instead, the strength, in terms of the rapidity of tissue rejection by H-Y antigens, depends on the presence of specific MHC alleles.24 Second, tissue rejection or the equivalent in our model of increasing donor chimerism after a vaccine-sensitized DLI likely requires a combination of CD8+ cytotoxic T lymphocytes as well as CD4+ helper T lymphocytes responding to different MHC class I- and class II-restricted epitopes, respectively, in the responding haplotype.25 Third, it is known that SMCY encodes a miHA in mice and men when presented by specific MHC molecules,26 hence the justification for cloning sections of SMCY into our vaccine. However, it remains unclear if SMCY encodes a miHA within all of the different MHC haplotypes in outbred populations. Additionally, our vaccine only encodes for 60% of the disparity between SMCX and SMCY; thus a potential miHA encoded within SMCY capable of driving in vivo responses may have been missed. Therefore, it is not surprising that our vaccine-sensitized DLI did not induce an anti-SMCY response capable of inducing an in vivo response in every male recipient, because they all possessed different haplotypes.
What about the anti-SRY immune response? Did it play a role in the increase in donor chimerism after DLI? Several lines of evidence suggest a working hypothesis that the increase in donor chimerism after the vaccine-sensitized DLI required SRY expression in the recipient's hematopoietic cells. First, both transplant donors showed strong antigen-specific responses to SRY. Second, the transplant recipient that experienced an increase in donor chimerism demonstrated anti-SRY responses and was the only recipient that had SRY expression in their PBMCs. Third, the heterogenous SRY expression in the hematopoietic cells of male dogs provides a plausible explanation for why the responding mixed chimeric recipient did not reach full donor chimerism, that is, a residual population of male cells did not express SRY and were not eliminated. The role that SRY played in these studies cannot be extrapolated from murine studies. The SRY gene does not encode a miHA in mice27 and, to our knowledge, has not yet been shown to encode a miHA in humans. In mice, SRY expression is tightly regulated by DNA methylation and limited to gonadal tissues with expression that begins to wane by 12.5 days postcoitum, providing a plausible explanation for why SRY does not encode a miHA in mice.28,29 However, in humans and canines, SRY expression is not limited to the gonads, and the role of SRY beyond testis development has not been determined.30 These findings require confirmation in a larger cohort and then validation in future vaccine trials by treating the male recipients with a DNA methyltransferase inhibitor, such as 5-azacytidine (Vidaza), to restore SRY expression before vaccine-sensitized DLI.31
In conclusion, we developed a recombinant miHA vaccine in the canine model of allogeneic HCT to address the barriers of safety and efficacy currently preventing miHA vaccination in human allogeneic HCT. Future vaccine trials are needed to clarify the role of different DLA alleles and to determine if SRY encodes a miHA in dogs. Once accomplished, this vaccine can then be combined with adjuvants to optimize donor T cell sensitization in the donor, and ultimately devise strategies to directly vaccinate the transplant recipient.
ACKNOWLEDGMENT
The authors thank Michele Spector, DVM, and the technicians at the Fred Hutchinson Cancer Research Center canine facility for caring for the dogs used in our experiments. The authors also thank Helen Crawford and Bonnie Larson for their help with article preparation.
Footnotes
Published online 7 April 2016.
The authors are grateful for research funding from the National Institutes of Health, Bethesda, MD, grants P01 CA078902, P30 CA015704, K12 CA76930, P30 DK56465, support from Gabrielle's Angel Foundation for Cancer Research (SLR), and by awards from the Joseph Steiner Krebsstiftung, Bern, Switzerland and Lupin Foundation, Metairie, LA (RS).
The authors declare no conflicts of interest.
S.L.R., B.S., and S.S.G. participated in research design, performance of research, data analysis, and writing of the article. D.H.F. and J.T.F. contributed new reagents, performance of research, and participated in research design. R.S. participated in research design and writing of the article.
REFERENCES
- 1. Goulmy E. Minor histocompatibility antigens: allo target molecules for tumor-specific immunotherapy (Review). Cancer J. 2004; 10: 1– 7. [DOI] [PubMed] [Google Scholar]
- 2. Spierings E. Minor histocompatibility antigens: past, present, and future (Review). Tissue Antigens. 2014; 84: 374– 60. [DOI] [PubMed] [Google Scholar]
- 3. Storb R, Yu C, Wagner JL, et al. Stable mixed hematopoietic chimerism in DLA-identical littermate dogs given sublethal total body irradiation before and pharmacological immunosuppression after marrow transplantation. Blood. 1997; 89: 3048– 3054. [PubMed] [Google Scholar]
- 4. Georges GE, Storb R, Thompson JD, et al. Adoptive immunotherapy in canine mixed chimeras after nonmyeloablative hematopoietic cell transplantation. Blood. 2000; 95: 3262– 3269. [PubMed] [Google Scholar]
- 5. Graves SS, Hogan WJ, Kuhr C, et al. Adoptive immunotherapy against allogeneic kidney grafts in dogs with stable hematopoietic trichimerism. Biol Blood Marrow Transplant. 2008; 14: 1201– 1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lesnikova M, Nikitine A, Mason N, et al. Ex vivo expanded T regulatory (Treg) cells block conversion of mixed chimeras to complete donor chimerism. Blood (ASH Annual Meeting Abstracts). 2006; 108: 5168 (abstract). [Google Scholar]
- 7. Lesnikova M, Nikitine A, Pogosov L, et al. Peripheral CD4+CD25+ regulatory T cells (Treg) block alloreactive host anti-donor T cells in canine mixed hematopoietic chimeras. Blood (ASH Annual Meeting Abstracts). 2005; 106: 3101 (abstract). [Google Scholar]
- 8. Rosinski SL, Stone B, Graves SS, et al. Development of a minor histocompatibility antigen vaccine regimen in the canine model of hematopoietic cell transplantation. Transplantation. 2015; 99: 2083– 2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Venkataraman GM, Stroup P, Graves SS, et al. An improved method for dog leukocyte antigen 88 typing and two new major histocompatibility complex class I alleles, DLA-88*01101 and DLA-88*01201. Tissue Antigens. 2007; 70: 53– 57. [DOI] [PubMed] [Google Scholar]
- 10. Wagner JL, Burnett RC, DeRose SA, et al. Histocompatibility testing of dog families with highly polymorphic microsatellite markers. Transplantation. 1996; 62: 876– 877. [DOI] [PubMed] [Google Scholar]
- 11. Wagner JL, Works JD, Storb R. DLA-DRB1 and DLA-DQB1 histocompatibility typing by PCR-SSCP and sequencing (brief communication). Tissue Antigens. 1998; 52: 397– 401. [DOI] [PubMed] [Google Scholar]
- 12. Yu C, Ostrander E, Bryant E, et al. Use of (CA)n polymorphisms to determine the origin of blood cells after allogeneic canine marrow grafting. Transplantation. 1994; 58: 701– 706. [PubMed] [Google Scholar]
- 13. Pertmer TM, Eisenbraun MD, McCabe D, et al. Gene gun-based nucleic acid immunization: elicitation of humoral and cytotoxic T lymphocyte responses following epidermal delivery of nanogram quantities of DNA. Vaccine. 1995; 13: 1427– 1430. [DOI] [PubMed] [Google Scholar]
- 14. Sandmaier BM, Storb R, Santos EB, et al. Allogeneic transplant of canine peripheral blood stem cells mobilized by recombinant canine hematopoietic growth factors. Blood. 1996; 87: 3508– 3513. [PubMed] [Google Scholar]
- 15. Lupu M, Gooley T, Zellmer E, et al. Principles of peripheral blood mononuclear cell apheresis in a preclinical canine model of hematopoietic cell transplantation. J Vet Intern Med. 2008; 22: 74– 82. [DOI] [PubMed] [Google Scholar]
- 16. De Rosa SC. Vaccine applications of flow cytometry. Methods. 2012; 57: 383– 391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Scheibenbogen C, Letsch A, Thiel E, et al. CD8 T cell responses to Wilms tumor gene product WT1 and proteinase 3 in patients with acute myeloid leukemia. Blood. 2002; 100: 2132– 2137. [DOI] [PubMed] [Google Scholar]
- 18. Karlsson AC, Martin JN, Younger SR, et al. Comparison of the ELISPOT and cytokine flow cytometry assays for the enumeration of antigen-specific T cells. J Immunol Methods. 2003; 283: 141– 153. [DOI] [PubMed] [Google Scholar]
- 19. Snell GD. Methods for the study of histocompatibility genes. J Genet. 1948; 49: 87– 108. [DOI] [PubMed] [Google Scholar]
- 20. Bailey DW. Allelic forms of a gene controlling the female immune response to the male antigen in mice. Transplantation. 1971; 11: 426– 428. [DOI] [PubMed] [Google Scholar]
- 21. Eichwald EJ, Silmser CR. Skin. Transplant Bull. 1955; 2: 148– 149. [PubMed] [Google Scholar]
- 22. Prehn RT, Main JM. The influence of sex on isologous skin grafting in the mouse. J Natl Cancer Inst. 1956; 17: 35– 36. [PubMed] [Google Scholar]
- 23. Scott DM, Ehrmann IE, Ellis PS, et al. Why do some females reject males? The molecular basis for male-specific graft rejection (Review). J Mol Med (Berl). 1997; 75: 103– 114. [DOI] [PubMed] [Google Scholar]
- 24. Silvers WK, Billingham RE. Genetic background and expressivity of histocompatibility genes. Science. 1967; 158: 118– 119. [DOI] [PubMed] [Google Scholar]
- 25. Roopenian DC. What are minor histocompatibility loci? A new look at an old question (Review). Immunol Today. 1992; 13: 7– 10. [DOI] [PubMed] [Google Scholar]
- 26. Simpson E, Roopenian D. Minor histocompatibility antigens (Review). Curr Opin Immunol. 1997; 9: 655– 661. [DOI] [PubMed] [Google Scholar]
- 27. Simpson E. Minor transplantation antigens: animal models for human host-versus-graft, graft-versus-host, and graft-versus-leukemia reactions (Review). Transplantation. 1998; 65: 611– 616. [DOI] [PubMed] [Google Scholar]
- 28. Nishino K, Hattori N, Tanaka S, et al. DNA methylation-mediated control of Sry gene expression in mouse gonadal development. J Biol Chem. 2004; 279: 22306– 22313. [DOI] [PubMed] [Google Scholar]
- 29. Koopman P, Münsterberg A, Capel B, et al. Expression of a candidate sex-determining gene during mouse testis differentiation. Nature. 1990; 348: 450– 452. [DOI] [PubMed] [Google Scholar]
- 30. Clépet C, Schafer AJ, Sinclair AH, et al. The human SRY transcript. Hum Mol Genet. 1993; 2: 2007– 2012. [DOI] [PubMed] [Google Scholar]
- 31. Dasari VK, Deng D, Perinchery G, et al. DNA methylation regulates the expression of Y chromosome specific genes in prostate cancer. J Urol. 2002; 167: 335– 358. [PubMed] [Google Scholar]


