Abstract
The Membranome database was developed to assist analysis and computational modeling of single-pass (bitopic) transmembrane (TM) proteins and their complexes by providing structural information about these proteins on a genomic scale. The database currently collects data on >6000 bitopic proteins from Homo sapiens, Arabidopsis thaliana, Dictyostelium discoideum, Saccharomyces cerevisiae, Escherichia coli and Methanocaldococcus jannaschii. It presents the following data: (i) hierarchical classification of bitopic proteins into 15 functional classes, 689 structural superfamilies and 1404 families; (ii) 446 complexes of bitopic proteins with known three-dimensional (3D) structures classified into 129 families; (iii) computationally generated three-dimensional models of TM α-helices positioned in membranes; (iv) amino acid sequences, domain architecture, functional annotation and available experimental structures of bitopic proteins; (v) TM topology and intracellular localization, (vi) physical interactions between proteins from the database along with links to other resources. The database is freely accessible at http://membranome.org. There is a variety of options for browsing, sorting, searching and retrieval of the content, including downloadable coordinate files of TM domains with calculated membrane boundaries.
INTRODUCTION
Proteins with a single transmembrane (TM) α-helix (i.e. bitopic) represent the most abundant and functionally diverse category of membrane proteins (1). These proteins participate in cell adhesion and communication, regulation of signaling, transport and metabolism, immune response, cell death and complex developmental processes (1,2).
Most bitopic proteins are composed of one or several water-soluble domains and a TM α-helix that frequently forms dimers or larger complexes with other membrane proteins. Water-soluble domains are usually crystallized and studied after removal of TM helices. Structural studies of TM α-helices and their oligomers are seriously impeded by their instability, flexibility and high heterogeneity that depend on surrounding lipids or detergents. Therefore, experimental three-dimensional (3D) structures of bitopic proteins from the protein data bank (PDB) (3) are mostly represented by water-soluble domains, and to a much lesser extent by their TM α-helices in monomeric or oligomeric forms or as a part of large multiprotein complexes.
The limited structural information on bitopic proteins can be significantly expanded by computational modeling of their hydrophobic TM α-helices, which are structurally simple and can be reliably predicted. Locations and topologies of TM segments in protein sequences can be determined using more than 20 computational methods (4–7). 3D models of predicted TM α-helices can be generated and optimized in membrane using FMAP (8), Ez-3D (9) servers or Coarse-Grained Molecular Dynamic simulations (10). Structural modeling of TM α-helical dimers can be performed by PREDDIMER (11), CATM (12) or EFDOCK-TM (13) methods.
Nevertheless, our understanding of structure-function relationships of bitopic proteins, including their ability to form functional oligomers in membranes is still very limited. Even the exact number and topology of bitopic proteins from different proteomes remains unknown due to their frequent misprediction, which usually happens due to confusion between functional TM α-helices and cleavable hydrophobic N-terminal signal sequences (5,7,14,15) or C-terminal helices that are substituted by glycosyl-phosphatidylinositol (GPI) anchors during protein maturation (16). Hence, UniProt (17) and HTP (Human Transmembrane Proteome) (18) databases provide partly different sets of human bitopic proteins (2377 and 2507, respectively).
The lack of resources that combine and classify all available experimental and structural information on bitopic proteins at the genomic scale hampers their systematic analysis, which is essential to understand functional dynamics and evolution of this most abundant class of membrane proteins. Creation of a comprehensive database for bitopic proteins is a starting point for comparative structural analysis and modeling of individual TM helices and functionally relevant dimeric complexes of bitopic proteins.
DATABASE CONTENT
The Membranome database (http://membranome.org) was created to facilitate the large-scale modeling and analysis of individual TM α-helices that serve as structural and evolutionary units of membrane proteins. The database compiles and organizes diverse data on bitopic proteins with links to a variety of external resources. This information includes: amino acid sequence, domain architecture, protein topology in membrane, intracellular localizations, interaction partners, available 3D structures of TM and water-soluble domains and their complexes, oligomeric states, protein functions and involvement in metabolic pathways. More importantly, it provides the following data not found in other databases: (i) a browsable hierarchical classification of whole proteins, rather than of protein domains; and (ii) 3D models of TM α-helical domains in membranes, generated by our FMAP method (8). The database also includes a set of homo- and hetero-oligomeric bitopic protein complexes whose 3D structures (usually for the water-soluble domains) are available in the PDB. The complexes are classified into families, similar to that for individual proteins.
PREDICTION AND MODELING OF TM α-HELICES
Over 30 computational methods can be used to predict TM segments in protein sequences (4–7). However, none of them focuses on correct determination of TM helix ends that usually lay outside membrane boundaries and are important for helix–helix interactions. Besides, these methods were not designed to evaluate the free energies of helix folding and transfer to the membrane environment, which are important physico-chemical characteristics of TM α-helices. To address these issues and perform a large-scale 3D modeling of TM α-helices of bitopic proteins, we used our FMAP (Folding of Membrane-Associated Peptides) method (19). FMAP was applied for the following purposes: (i) to identify individually stable TM α-helices in protein sequences by calculating the free energy of folding of an α-helix in membrane relative to coil in water (ΔGfold); (ii) to define helix hydrophobicity by calculating the transfer energy of an α-helix from water to the lipid environment (ΔGtransf); (iii) to generate all-atom 3D models of TM helices; and (iv) to define spatial positions of TM helices in the lipid bilayer along with corresponding parameters (hydrophobic thicknesses and tilt angles).
FMAP was developed as a thermodynamics-based approach to predict stability and lengths of α-helices in different environments (water, micelles, membrane and protein interior). It combines a thermodynamic model of helix-coil transition implemented in Framework (20) and the PPM2.0 method for Positioning of Proteins in Membranes (21). Both methods have been extensively tested for peptides and proteins in micelles and membranes (20–23). Generation of all-atom 3D models (including the N-cap residue) of predicted α-helical segments, is based on structural templates that represent straight or Pro-kinked α-helices. The side chain rotamers are automatically selected to represent energetically preferred conformers from the library (24) and optimize transfer energy of side-chains from water to the lipid bilayer. In calculating ΔGtransf of modeled TM α-helices, PPM2.0 approximates solvent properties of the anisotropic membrane environment by transbilayer profiles of polarity parameters of the fluid dioleoyl-phosphatidylcholine bilayer (21). Details of the method are described in the Membranome website. (http://membranome.org/about.php?subject=methods). The method is available through the FMAP web server (http://membranome.org/server.php).
Testing FMAP on a set of 139 bitopic proteins with known structures from 83 PDB entries (Supplementary Table S1) showed its ability to correctly identify all TM α-helices in these proteins with average errors in N- and C-termini prediction of 2.2 and 3.0 residues per helix, respectively (Supplementary Figures S1A and B, S2). The average accuracy in prediction of hydrophobic thicknesses for single-pass TM-helices was 2.4 ± 2.0 Å (Supplementary Figure S1C). The latter is similar to the accuracy of the underlying PPM method that was shown to successfully reproduce hydrophobic thicknesses and tilt angles of experimentally studied peptides and proteins in the lipid bilayer (21,22). Tilt angles calculated by FMAP for individual TM helices were found to deviate from those in experimental structures, especially for protein oligomers and proteins with loops or water-soluble domains. In these cases, changes in orientations of TM α-helices are caused by helix–helix association or by interactions of peripheral domains and loops with the membrane interface. In addition, the calculations with PPM and FMAP demonstrated that single TM α-helices can experience much greater fluctuations of tilt angles (up to 10° within the energy gap of 1 kcal/mol) than multi-helical TM proteins (22). Our testing also demonstrated that FMAP performed similarly to Phobius in prediction of location of TM helices in amino acid sequences: 86.1% of TM helices of 4831 proteins identified as bitopic by FMAP (similarly annotated in UniProt) showed 80–100% sequential overlap with Phobious predictions, 10.4% of helices predicted by FMAP had 1–79% overlap with Phobius predictions, and only 3.5% of helices were not predicted by both methods simultaneously (Supplementary Table S2). However, FMAP should not be used as a general method of TM helix prediction in membrane proteins, as its performance has not yet been sufficiently tested on polytopic proteins.
Similar to other computational methods (4–7), FMAP occasionally produces false positive predictions by identifying cleavable N- and C-terminal hydrophobic fragments and long hydrophobic helices from water-soluble domains as potential TM helices. Therefore, it was important to filter out all false-positive predictions from the initial set of potential TM helices generated by FMAP. That was done by excluding all predicted TM helices that were annotated as cleavable signal or C-terminal segments in UniProt and Pfam (release 28) (25). All hydrophobic helices that overlap with water-soluble domains found in Pfam, InterPro (26) or PDB databases were also removed. Human intervention and analysis of related publications were required to resolve some ambiguous cases.
PROTEIN SET
A set of bitopic proteins was prepared using amino acid sequences from UniProt Swiss-Prot and TrEMBL (http://www.uniprot.org/help/2016/01/20/release) (17). Fragments and alternative isoforms of the same protein were not included. Identification of bitopic proteins in six proteomes was performed in two steps. First, all proteins with one or two potential TM α-helices were predicted by FMAP. Only helices of 15 to 40 residues with negative ΔGfold and ΔGtransf values and predicted TM orientation in the lipid bilayer were selected. Second, this initial protein set was filtered as described above to exclude secreted proteins (with N-terminal signal sequence), GPI-anchored proteins, water-soluble proteins with a hydrophobic helix within protein interior, as well as remaining two-helical TM proteins.
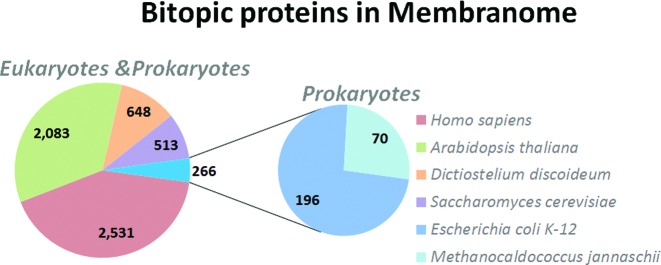
The database currently contains 6041 single-pass TM proteins from six organisms: 2531 proteins from Homo sapiens, 2083 proteins from Arabidopsis thaliana, 648 proteins from Dictyostelium discoideum, 513 proteins from Saccharomyces cerevisiae, 196 proteins from Escherichia coli K-12 and 70 proteins from Methanocaldococcus jannaschii (Figure 1).
Figure 1.
Numbers of bitopic proteins from different organisms in the Membranome database (2016-02).
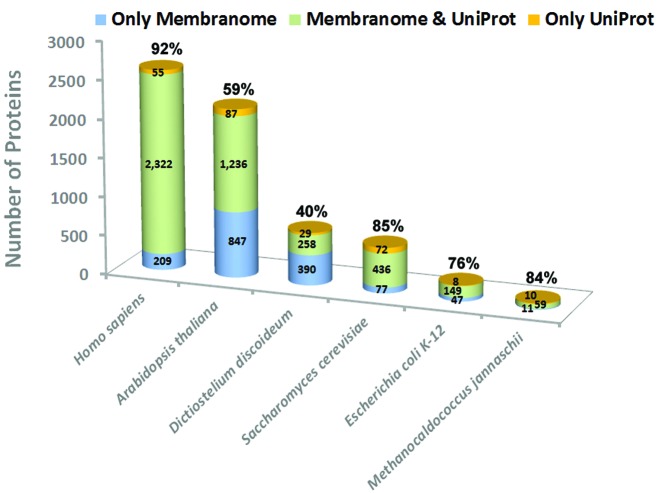
These protein sets show 76%, 85% and 92% overlap with bitopic protein sets from UniProt Swiss-Prot for well annotated proteomes (of E. coli, S.cerevisiae, H. sapiens, respectively) (Figure 2). A few proteins annotated as bitopic in UniProt were predicted as not having any TM helices by FMAP. All such proteins were not included in Membranome because we could not find any experimental evidence in publications that they actually have a single TM helix. On the other hand, we identified a significant number of single-pass TM proteins in sequences of unreviewed proteins from TrEMBL, mostly for the least annotated proteomes of A.thaliana and D. discoideum (847 and 390 additional proteins, respectively) (Figure 2). As a result, protein sets for these species were almost 2-fold expanded relative to the annotated UniProt sets. In most cases, same TM helices in TrEMBL entries were also predicted by Phobius and annotated as bitopic proteins in Pfam. Many newly added proteins of A.thaliana and D. discoideum are homologous to human and yeast bitopic proteins.
Figure 2.
Comparison of bitopic protein sets in Membranome and UniProt databases. Percent of overlap between protein sets is indicated above each column.
Comparison of human bitopic protein sets from Membranome (2531 proteins) and HTP (2507 proteins in d.1.4.) (18) show significant overlap (80% overlap with d.1.3. released on 9.15.2015). The main differences appeared because of inclusion in Membranome of many proteins from TrEMBL. Further, our protein set was curated to remove all false-positive predictions and to comply with published data. In contrast, the HTP database, which is based on fully-automated methods, has a number of obvious mispredictions. For example, HTP considers some water-soluble or peripheral proteins with hydrophobic segments as TM proteins (e.g. PPL13_HUMAN, SPB4_HUMAN, SNX1_HUMAN), because TM helices were consistently predicted in these proteins by different computational methods. In some cases, HTP incorrectly defines the type of TM proteins, e.g it assigns the well-studied single-pass TM protein TYOBP_HUMAN (27) to two-helical TM proteins.
PROTEIN CLASSIFICATION, LOCALIZATION AND TOPOLOGY
Whole proteins, rather than individual domains, were classified into 15 functional classes, 689 superfamilies and 1404 families. The assignment to functional classes (receptors, ligands/regulators, structural/adhesion, proteins involved in biogenesis, membrane remodeling and vesicular trafficking, molecular transport, gene regulation, 6 classes of enzymes and proteins with undefined function) was based on UniProt, Pfam, InterPro and other published annotations (2).
Proteins with similar domain architecture were grouped into superfamilies, which frequently correspond to clans or families in Pfam, MEROPS (28), CAZy (29) or superfamilies in Uniprot or other classifications (2). Allocation of proteins to families followed UniProt and InterPro classifications, if available. Otherwise, the proteins were classified into families using Panther (30) or by sequence analysis of paralogues and orthologues from KEGG (31). The links to the corresponding Pfam or InterPro families were provided, whenever possible. Different subunits of multiprotein complexes were included as families of the same superfamily. A similar classification to families and classes was created for bitopic protein complexes with experimentally determined structures.
Assignment of eukaryotic proteins to 22 types of cellular and intracellular membranes was based on information from UniProt and COMPARTMENTS (32) databases. Protein topology (membrane side associated with N-terminus, ‘In’ or ‘Out’) was assigned using information from UniProt, TOPDB (33), PDB and publications, including direct experimental studies of TM protein topology, locations of water-soluble domains in specific intracellular compartments and post-translational modifications.
DATABASE ORGANIZATION
Access to the database is provided through a menu on the website that allows browsing of the content for individual proteins or their complexes by protein classes, superfamilies, families, localizations in different membrane types and species. Users can also access each of the six proteomes separately by using ‘Select Species’ button. Then the picture of the selected organism appears below the button. The database can be searched using text (protein names, keywords), UniProt ID and PDB IDs.
Pages are dynamically generated for every level of hierarchical classification including superfamilies, families, individual protein and protein complexes. To facilitate retrieval and analysis of data, these pages were organized as sortable lists and tables supplemented by images, internal and external links. For example, one can compare membrane interaction modes of evolutionarily related proteins from the database by navigating to a protein superfamily page that simultaneously displays images of TM domains of all proteins from the superfamily with calculated membrane boundaries, along with the table presenting all calculated parameters. Sorting in the tables can be done based on the content of different fields, such as protein family code, protein name, UniProtKB ID, localization (membrane type) and parameters of TM segments (position in sequence, hydrophobic thickness, tilt angle, ΔGfold and ΔGtransf).
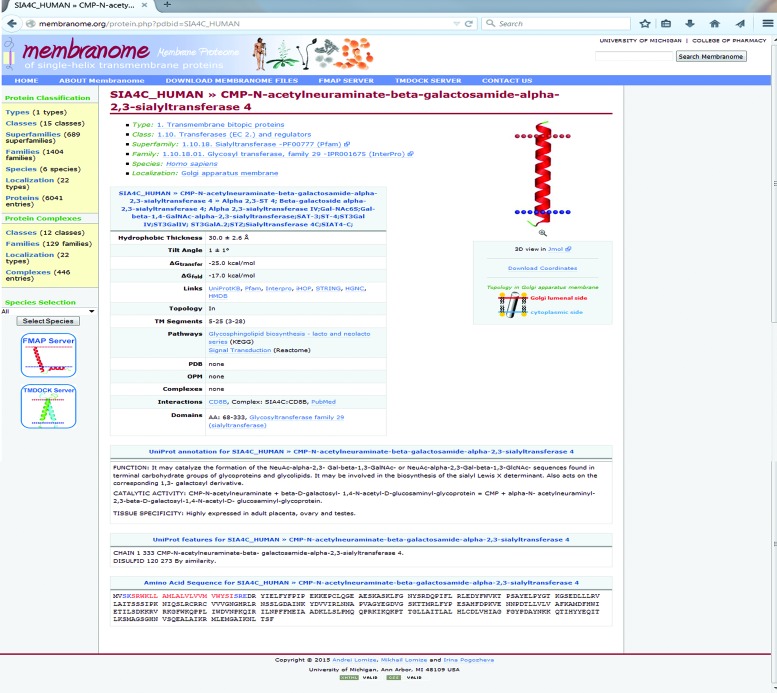
Each ‘protein page’ (Figure 3) provides protein names, classification, source organism, intracellular localization, structural and functional information, association with metabolic pathways and interactions with other bitopic proteins. Structural information includes amino acid sequence with TM segment indicated in red (whole TM helices marked by blue color), domain architecture, topology in membrane and links to experimental structures of water-soluble and TM domains and their complexes available in PDB and OPM (34). Protein sequences and annotations were taken from UniProt. Further, for each protein, the database provides an FMAP-generated 3D model of a TM α-helical domain as a coordinate file supplemented by the static and dynamic images and the following calculated parameters: position in the amino acid sequence, topology in membrane, hydrophobic thickness, tilt angle, stability of the helix relative to membrane-bound coil (ΔGfold) and transfer free energy of the helix from water to membrane (ΔGtransf).
Figure 3.
Example of page for an individual bitopic protein from the Membranome database.
Each ‘protein complex page’ is constructed similarly and provides intracellular localization and classification of the complex. Currently these pages were created only for complexes with experimentally determined 3D structures. Information on other direct interactions between bitopic proteins from Membranome was included on ‘protein pages’. This information was compiled from Arabidopsis Interactome (35), ConsensusPathDB (36), IntAct (37) and BioGRID data (38). Potential interactions with other proteins not included in the database can be found by following links to STRING (39). The ‘protein complex pages’ and ‘protein pages’ are linked to each other.
To expand information on each protein, we included relevant links to gene repository HGNC (40), to metabolic pathway databases, KEGG, Reactome (41) and HMDB (42), and to information-citation databases, iHOP (43) and PubMed.
The visualization of 3D models is provided by static images generated using scripts for PyMOL (44) and dynamic images that can be interactively displayed in Jmol. Coordinate files of 3D models of TM domains with hydrocarbon core boundaries marked by dummy atoms can be downloaded in PDB format either individually for each protein or as a single file for all proteins.
To help the first-time user, the database has an ‘ABOUT Membranome’ section that introduces general database features, such as protein sets and classification, and describes the underlying FMAP method and its verification (http://membranome.org/about.php). The ‘Analysis of Bitopic Proteins’ subsection shows differences in lengths of TM α-helices, their hydrophobic thicknesses, helix stabilities and transfer energies between proteins associated with different types of biological membranes. For example, this analysis shows that TM α-helices of bitopic proteins from plasma membranes have significantly higher stability and hydrophobicity (ΔGfold∼−20 kcal/mol, ΔGtransf∼−26 kca/mol) than TM α-helices of bitopic proteins from inner mitochondrial membranes (ΔGfold∼−17 kcal/mol, ΔGtransf∼−8 kca/mol) (Figure 7 in website).
MAINTENANCE AND UPDATE
Membranome is a relational database of 18 tables. It was developed with PHP, MySQL and the Smarty engine, which separates the program logic (PHP, MySQL) and presentation (XHTML, CSS, JavaScript), and enables caching. The database content is partially curated and will be annually updated using SQL queries. The curation includes protein filtering and selection, as well as classification of individual proteins and their complexes into families and superfamilies and cross-verification of protein topology and intracellular localization obtained from different sources. The database content is provided as downloadable files (sql and text).
CONCLUSIONS
The Membranome database is the first comprehensive resource on bitopic proteins from six complete genomes. This is a unique resource that provides all-atom 3D models of TM domains for more than 6000 bitopic proteins along with extensive structural and functional information on these proteins and their complexes. It also offers an original classification of whole proteins rather than their domains, which is different and more complete for bitopic proteins of selected species than classifications provided by other individual databases (UniProt, InterPro, Pfam). In the near future, the database will be populated by 3D models of TM α-helical dimers that can be either experimentally determined or generated using computational methods, such as (11–13). Integration of available information on bitopic proteins in a single place will be useful for the proteome-wide analysis of structure, function and evolution of this largest, but a poorly studied class of membrane proteins. The computational modeling and comparative analysis of structural features of single-pass membrane proteins and their complexes is important to understand energetics and structural features of TM helices that guide their insertion and oligomerization in membranes.
Acknowledgments
The authors are grateful to Keith Gingras, a student of the Department of Computational Science and Engineering of the University of Michigan, for his help in data analysis. The authors thank their reviewers for their constructive criticism.
Footnotes
Present address: Andrei Lomize, Department of Medicinal Chemistry, College of Pharmacy, University of Michigan, 428 Church St., Ann Arbor, MI 48109-1065, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
This work was supported by the Division of Biological Infrastructure of the National Science Foundation (NSF) [Award #1145367 to A.L., I.P.].
Conflict of interest statement. None declared.
REFERENCES
- 1.Hubert P., Sawma P., Duneau J.P., Khao J., Henin J., Bagnard D., Sturgis J. Single-spanning transmembrane domains in cell growth and cell-cell interactions: More than meets the eye? Cell Adhes. Migr. 2010;4:313–324. doi: 10.4161/cam.4.2.12430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Almen M.S., Nordstrom K.J., Fredriksson R., Schioth H.B. Mapping the human membrane proteome: A majority of the human membrane proteins can be classified according to function and evolutionary origin. BMC Biol. 2009;7 doi: 10.1186/1741-7007-7-50. doi:10.1186/1741-7007-7-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rose P.W., Prlic A., Bi C., Bluhm W.F., Christie C.H., Dutta S., Green R.K., Goodsell D.S., Westbrook J.D., Woo J., et al. The RCSB Protein Data Bank: views of structural biology for basic and applied research and education. Nucleic Acids Res. 2015;43:D345–D356. doi: 10.1093/nar/gku1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dobson L., Remenyi I., Tusnady G.E. CCTOP: A Consensus Constrained TOPology prediction web server. Nucleic Acids Res. 2015;43:W408–W412. doi: 10.1093/nar/gkv451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reeb J., Kloppmann E., Bernhofer M., Rost B. Evaluation of transmembrane helix predictions in 2014. Proteins. 2015;83:473–484. doi: 10.1002/prot.24749. [DOI] [PubMed] [Google Scholar]
- 6.Bernsel A., Viklund H., Falk J., Lindahl E., von Heijne G., Elofsson A. Prediction of membrane-protein topology from first principles. Proc. Natl. Acad. Sci. U.S.A. 2008;105:7177–7181. doi: 10.1073/pnas.0711151105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bernsel A., Von Heijne G. Improved membrane protein topology prediction by domain assignments. Protein Sci. 2005;14:1723–1728. doi: 10.1110/ps.051395305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lomize A.L., Pogozheva I. Membranome: A database of single-spanning transmembrane proteins. Biophys J. 2015;108:249a–250a. [Google Scholar]
- 9.Schramm C.A., Hannigan B.T., Donald J.E., Keasar C., Saven J.G., Degrado W.F., Samish I. Knowledge-based potential for positioning membrane-associated structures and assessing residue-specific energetic contributions. Structure. 2012;20:924–935. doi: 10.1016/j.str.2012.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stansfeld P.J., Goose J.E., Caffrey M., Carpenter E.P., Parker J.L., Newstead S., Sansom M.S. MemProtMD: Automated Insertion of Membrane Protein Structures into Explicit Lipid Membranes. Structure. 2015;23:1350–1361. doi: 10.1016/j.str.2015.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Polyansky A.A., Chugunov A.O., Volynsky P.E., Krylov N.A., Nolde D.E., Efremov R.G. PREDDIMER: A web server for prediction of transmembrane helical dimers. Bioinformatics. 2014;30:889–890. doi: 10.1093/bioinformatics/btt645. [DOI] [PubMed] [Google Scholar]
- 12.Mueller B.K., Subramaniam S., Senes A. A frequent, GxxxG-mediated, transmembrane association motif is optimized for the formation of interhelical Calpha-H hydrogen bonds. Proc. Natl. Acad. Sci. U.S.A. 2014;111:E888–E895. doi: 10.1073/pnas.1319944111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Y., Barth P. Evolutionary-guided de novo structure prediction of self-associated transmembrane helical proteins with near-atomic accuracy. Nat. Commun. 2015;6 doi: 10.1038/ncomms8196. doi:10.1038/ncomms8196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Petersen T.N., Brunak S., von Heijne G., Nielsen H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 15.Viklund H., Bernsel A., Skwark M., Elofsson A. SPOCTOPUS: A combined predictor of signal peptides and membrane protein topology. Bioinformatics. 2008;24:2928–2929. doi: 10.1093/bioinformatics/btn550. [DOI] [PubMed] [Google Scholar]
- 16.Galian C., Björkholm P., Bulleid N., von Heijne G. Efficient Glycosylphosphatidylinositol (GPI) modification of membrane proteins requires a C-terminal anchoring signal of marginal hydrophobicity. J. Biol. Chem. 2012;287:16399–16409. doi: 10.1074/jbc.M112.350009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.UniProt Consortium. UniProt: A hub for protein information. Nucleic Acids Res. 2015;43:D204–D212. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dobson L., Remenyi I., Tusnady G.E. The human transmembrane proteome. Biol. Direct. 2015;10 doi: 10.1186/s13062-015-0061-x. doi:10.1186/s13062-015-0061-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lomize A.L., Mosberg H.I., Pogozheva I.D. Thermodynamic approach to large-scale modeling of alpha-helices in membranes. Biophys. J. 2012;102:490a–491a. [Google Scholar]
- 20.Lomize A.L., Mosberg H.I. Thermodynamic model of secondary structure for alpha-helical peptides and proteins. Biopolymers. 1997;42:239–269. doi: 10.1002/(SICI)1097-0282(199708)42:2<239::AID-BIP12>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 21.Lomize A.L., Pogozheva I.D., Mosberg H.I. Anisotropic solvent model of the lipid bilayer. 2. Energetics of insertion of small molecules, peptides, and proteins in membranes. J. Chem. Inf. Model. 2011;51:930–946. doi: 10.1021/ci200020k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lomize A.L., Pogozheva I.D., Lomize M.A., Mosberg H.I. Positioning of proteins in membranes: a computational approach. Protein Sci. 2006;15:1318–1333. doi: 10.1110/ps.062126106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lomize A.L., Pogozheva I.D., Lomize M.A., Mosberg H.I. The role of hydrophobic interactions in positioning of peripheral proteins in membranes. BMC Struct. Biol. 2007;7 doi: 10.1186/1472-6807-7-44. doi:10.1186/1472-6807-7-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blaber M., Zhang X.J., Lindstrom J.D., Pepiot S.D., Baase W.A., Matthews B.W. Determination of alpha-helix propensity within the context of a folded protein. Sites 44 and 131 in bacteriophage T4 lysozyme. J. Mol. Biol. 1994;235:600–624. doi: 10.1006/jmbi.1994.1016. [DOI] [PubMed] [Google Scholar]
- 25.Finn R.D., Coggill P., Eberhardt R.Y., Eddy S.R., Mistry J., Mitchell A.L., Potter S.C., Punta M., Qureshi M., Sangrador-Vegas A., et al. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 2016;44:D279–D285. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mitchell A., Chang H.Y., Daugherty L., Fraser M., Hunter S., Lopez R., McAnulla C., McMenamin C., Nuka G., Pesseat S., et al. The InterPro protein families database: the classification resource after 15 years. Nucleic Acids Res. 2015;43:D213–D221. doi: 10.1093/nar/gku1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Call M.E., Wucherpfennig K.W., Chou J.J. The structural basis for intramembrane assembly of an activating immunoreceptor complex. Nat. Immunol. 2010;11:1023–1029. doi: 10.1038/ni.1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rawlings N.D., Barrett A.J., Finn R. Twenty years of the MEROPS database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 2016;44:D343–D350. doi: 10.1093/nar/gkv1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lombard V., Golaconda Ramulu H., Drula E., Coutinho P.M., Henrissat B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014;42:D490–D495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mi H., Poudel S., Muruganujan A., Casagrande J.T., Thomas P.D. PANTHER version 10: Expanded protein families and functions, and analysis tools. Nucleic Acids Res. 2016;44:D336–D342. doi: 10.1093/nar/gkv1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kanehisa M., Sato Y., Kawashima M., Furumichi M., Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016;44:D457–D462. doi: 10.1093/nar/gkv1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Binder J.X., Pletscher-Frankild S., Tsafou K., Stolte C., O'Donoghue S.I., Schneider R., Jensen L.J. COMPARTMENTS: Unification and visualization of protein subcellular localization evidence. Database. 2014;2014:bau012. doi: 10.1093/database/bau012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dobson L., Lango T., Remenyi I., Tusnady G.E. Expediting topology data gathering for the TOPDB database. Nucleic Acids Res. 2015;43:D283–D289. doi: 10.1093/nar/gku1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lomize M.A., Pogozheva I.D., Joo H., Mosberg H.I., Lomize A.L. OPM database and PPM web server: resources for positioning of proteins in membranes. Nucleic Acids Res. 2012;40:D370–D376. doi: 10.1093/nar/gkr703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.A.I.M. Consortium. Evidence for network evolution in an arabidopsis interactome map. Science. 2011;333:601–607. doi: 10.1126/science.1203877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kamburov A., Stelzl U., Lehrach H., Herwig R. The ConsensusPathDB interaction database: 2013 update. Nucleic Acids Res. 2013;41:D793–D800. doi: 10.1093/nar/gks1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Orchard S., Ammari M., Aranda B., Breuza L., Briganti L., Broackes-Carter F., Campbell N.H., Chavali G., Chen C., del-Toro N., et al. The MIntAct project–IntAct as a common curation platform for 11 molecular interaction databases. Nucleic Acids Res. 2014;42:D358–D363. doi: 10.1093/nar/gkt1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chatr-Aryamontri A., Breitkreutz B.J., Oughtred R., Boucher L., Heinicke S., Chen D., Stark C., Breitkreutz A., Kolas N., O'Donnell L., et al. The BioGRID interaction database: 2015 update. Nucleic Acids Res. 2015;43:D470–D478. doi: 10.1093/nar/gku1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Szklarczyk D., Franceschini A., Wyder S., Forslund K., Heller D., Huerta-Cepas J., Simonovic M., Roth A., Santos A., Tsafou K.P., et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gray K.A., Yates B., Seal R.L., Wright M.W., Bruford E.A. Genenames.org: the HGNC resources in 2015. Nucleic Acids Res. 2015;43:D1079–D1085. doi: 10.1093/nar/gku1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fabregat A., Sidiropoulos K., Garapati P., Gillespie M., Hausmann K., Haw R., Jassal B., Jupe S., Korninger F., McKay S., et al. The Reactome pathway Knowledgebase. Nucleic Acids Res. 2016;44:D481–D487. doi: 10.1093/nar/gkv1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wishart D.S., Jewison T., Guo A.C., Wilson M., Knox C., Liu Y., Djoumbou Y., Mandal R., Aziat F., Dong E., et al. HMDB 3.0–The Human Metabolome Database in 2013. Nucleic Acids Res. 2013;41:D801–D807. doi: 10.1093/nar/gks1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fernandez J.M., Hoffmann R., Valencia A. iHOP web services. Nucleic Acids Res. 2007;35:W21–W26. doi: 10.1093/nar/gkm298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.DeLano W.L. The PyMOL molecular graphics system. San Carlos: DeLano Scientific LLC; 2003. [Google Scholar]