Abstract
We describe binding free energy calculations in the D3R Grand Challenge 2015 for blind prediction of the binding affinities of 180 ligands to Hsp90. The present D3R challenge was built around experimental datasets involving Heat shock protein (Hsp) 90, an ATP-dependent molecular chaperone which is an important anticancer drug target. The Hsp90 ATP binding site is known to be a challenging target for accurate calculations of ligand binding affinities because of the ligand-dependent conformational changes in the binding site, the presence of ordered waters and the broad chemical diversity of ligands that can bind at this site. Our primary focus here is to distinguish binders from nonbinders. Large scale absolute binding free energy calculations that cover over 3000 protein–ligand complexes were performed using the BEDAM method starting from docked structures generated by Glide docking. Although the ligand dataset in this study resembles an intermediate to late stage lead optimization project while the BEDAM method is mainly developed for early stage virtual screening of hit molecules, the BEDAM binding free energy scoring has resulted in a moderate enrichment of ligand screening against this challenging drug target. Results show that, using a statistical mechanics based free energy method like BEDAM starting from docked poses offers better enrichment than classical docking scoring functions and rescoring methods like Prime MM-GBSA for the Hsp90 data set in this blind challenge. Importantly, among the three methods tested here, only the mean value of the BEDAM binding free energy scores is able to separate the large group of binders from the small group of nonbinders with a gap of 2.4 kcal/mol. None of the three methods that we have tested provided accurate ranking of the affinities of the 147 active compounds. We discuss the possible sources of errors in the binding free energy calculations. The study suggests that BEDAM can be used strategically to discriminate binders from nonbinders in virtual screening and to more accurately predict the ligand binding modes prior to the more computationally expensive FEP calculations of binding affinity.
Keywords: D3R, GC2015, Hsp90, Binding free energy, Docking, ROC
Introduction
Predicting prospectively binding affinities of receptor-ligand complexes with sufficient accuracy is of great importance to structure-based drug design and remains one of the most challenging problems in computational biophysics. A number of powerful binding free energy methods based on statistical mechanics and all-atom Molecular Dynamics simulation have been developed in the past decade [1–5]. A recent large-scale FEP study [6] has shown that relative binding free energies for congeneric series of ligands can be achieved to the order of 1.5 kcal/mol and correlation coefficient to ~0.7. However, accurate prediction of absolute binding free energies on a large scale has not been achieved. It is not uncommon that a method can work well in reproducing the experimental free energies for a small number of congeneric ligands, but fail to predict the activities for a different, more diverse set of compounds in prospective studies. The problem becomes even more challenging when large structural reorganization of the receptor occurs upon ligand binding.
The SAMPL series of blind challenges typically consists of a large number of chemically diverse compounds and therefore has provided a useful platform for testing the accuracy of different methods in a more realistic and statistically significant setting relevant to structure-based drug discovery projects [7]. We have participated in the SAMPL3, SAMPL4, SAMPL5 and D3R Grand Challenge 2015 challenges, using the in-house developed BEDAM method for absolute binding free energy calculations [8–10]. BEDAM is based on statistical mechanics of receptor-ligand binding and uses an implicit solvation model and Hamiltonian replica exchange MD to enhance the sampling of protein–ligand dynamic coupling [4, 11]. Compared with the popular MM-GB/SA protocol, which also uses implicit solvation, BEDAM can more effectively account for the entropic effects and reorganization upon binding. BEDAM’s ability to sample larger amounts of conformational space as its simulation method spans from the fully coupled receptor-ligand to fully decoupled alchemical states makes it a potentially powerful tool for predicting binding modes prior to the application of higher resolution explicit solvent FEP methods. Using BEDAM binding free energy scoring, we have achieved the second best enrichment among the participants in SAMPL4 [9].
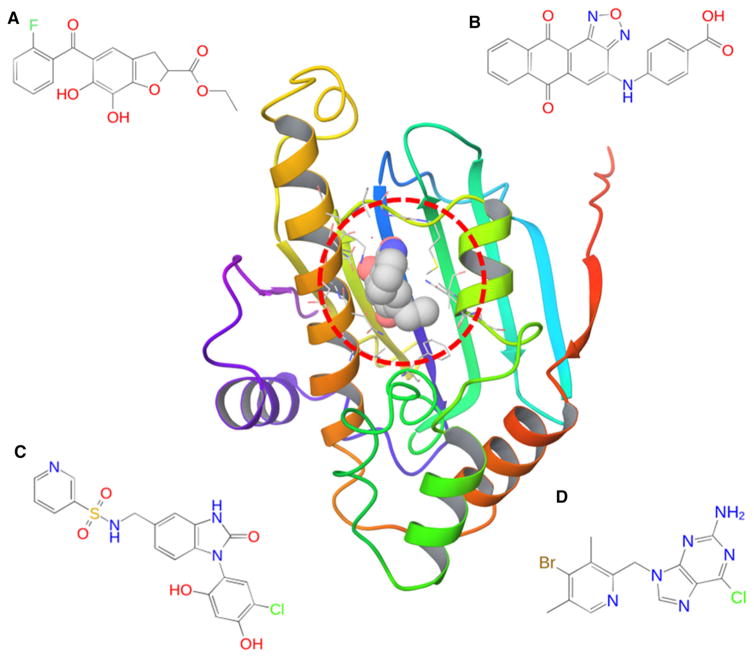
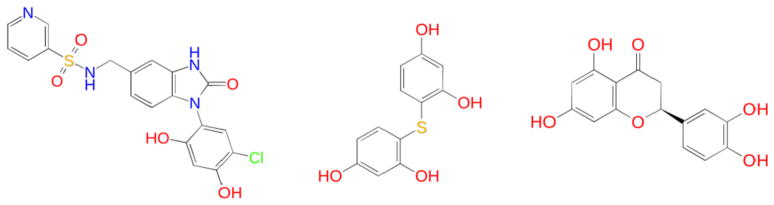
The D3R Grand Challenge 2015 (GC2015) consists of 180 ligands (147 actives, 33 inactives) targeting the Hsp90 ATP binding site [12]. Figure 1 shows the binding site of the Hsp90 and several representative ligands with different chemical scaffolds. The size and chemical diversity of the ligand data set makes it less practical to apply FEP methods to cover all the ligands. The ATP binding site in Hsp90 is known to be particularly difficult for binding free energy calculations [13] as ligand binding involves binding site conformational changes, burial of charged side chains inside the binding site and varying numbers of enclosed water molecules in the binding pocket [14]. Therefore, rather than aiming to predict the rank ordering of ligand binding affinities, our primary goal in this challenge is to test how well the different methods, such as BEDAM, MM-GB/SA and docking, can discriminate binders from nonbinders in this large scale blind prediction [15]. Statistical analysis of our results show that a moderate enrichment has been achieved using BEDAM binding free energy scoring, even though the ratio of actives to inactives in this data set is the opposite of that in typical virtual screening experiments where the goal is to identify the few active molecules in the compound library. Compared to scoring using Glide SP and single-snapshot MM-GB/SA using Prime, the BEDAM method which requires docked poses as a starting point, is significantly better in distinguishing binders from nonbinders, a result largely consistent with that in the SAMPL4 blind challenge against a different protein target, the LEDGF/p75-binding pocket of HIV-1 integrase [9]. The study suggests that BEDAM can be strategically employed to (1) more reliably distinguish binders from nonbinders in order to obtain enhanced enrichment compared to docking and MM-GB/SA; and (2) for accurate pose predictions to prepare for subsequent, more accurate FEP calculations of binding affinity.
Fig. 1.
Hsp90 ATP binding site and representative ligands with different chemical scaffolds. a Benzofuran; b anthraquinone; c benzoimidazolone with resorcinol substitution; d aminopurine derivative
Methods and materials
The Hsp90 dataset contains 180 compounds in which 147 are actives and 33 are inactives. The IC50 of the actives ranges from 5.2 nM to 50 μM. The 3D structures of the 180 ligands were created using the LigandPrep protocol from Schrodinger Inc. to generate 311 protonated states. To account for the effect of protein conformational change upon binding, both an open structure PDB ID 2XDX and a closed structure PDB ID 4YKR of Hsp90 are used as receptor structures for docking. Glide docking was performed on the 311 protonated ligands against the open and closed forms of the receptor. The ionization penalties for each protonation state were added to the final docking scores and BEDAM scores. Two highly conserved bound waters near the buried Asp93 side chain [14] were retained during docking but are removed during subsequent BEDAM simulations which use an implicit solvation model to mimic the solvent effect. For each of the open and closed receptor conformations, five top docked poses per ligand were retained for BEDAM calculations. These yield 311 × 5 × 2 ≈ 3110 BEDAM binding free energy calculations, making this study the largest scale absolute binding free calculations reported to date. For a given ligand, the best BEDAM score among all the combinations of protonation states × docked poses for this ligand is taken as the unique free energy score of the ligand for submission.
In the BEDAM (Binding Energy Distribution Analysis Method) approach [4], the protein–ligand system is described by the OPLS2005 force field [16, 17] and an implicit solvation model AGBNP2 [18, 19]. The standard binding free energy is computed using a hybrid effective potential connecting the unbound state (λ = 0) and the bound state (λ = 1), without going through the gas phase ligand state as in the case of explicit solvent double decoupling method. The methodology of BEDAM has been described in previous papers [4, 9]. The setup of the BEDAM simulations in the present study is the same as that in the SAMPL4 challenge, which has been described in the SAMPL4 paper [9].
Before we started to perform the binding free energy calculations for the 180 Hsp90 ligands, we computed binding free energies for the known Hsp90 ligands reported in a study by Astex Therapeutics [12], which included 14 aminopyrimidines and 18 resorcinol compounds. The calculated BEDAM binding free energies for the non-aminopyrimidines match well the experimental values (estimated as −kT log IC50). However the computed values of for the aminopyrimidines were less favorable than the experiments by a constant factor of about 8 kcal/mol. We have tried to identify the source of the error by testing various possibilities (hydration, tautomerization, protonation, force field parameterization). For example, we find that the hydration free energies of aminopyrimidines are well reproduced by the AGBNP2 implicit solvent model, yet the solvent mediated effective interaction energy between the aminopyrimidines and the Hsp90 binding site is underestimated by the implicit solvent model. It is still unclear whether the problem is in the AGBNP2 implicit solvation parameters or in the OPLS2005 force field specific for the amoinopyrimidine functional group. We comment on this later in the manuscript. To remedy this energy function deficiency for the aminopyrimidines and the closely related aminopurines, we took an ad hoc approach by adding uniformly −8 kcal/mol to the ΔGbind(calc) for the 62 aminopyrimidine/aminopurine compounds in the D3R GC2015 challenge.
The Prime MM-GBSA protocol built into the Schrodinger Maestro package was used to compute the MM-GB/SA binding scores using the Glide docked structures of the 180 Hsp90 Ligand–protein complexes. In this protocol, each of the Glide docked complexes was subject to a short energy minimization prior to the MM-GB/SA rescoring.
For the explicit solvent double decoupling calculations (DDM) [1, 3, 20–22] of binding free energy performed in this study, the protein receptor is modeled with the Amber ff99sb-ILDN force field [23], and the ligands are described by the Amber GAFF parameters set [24] and the AM1-BCC charge model [25]. A DDM calculation involves two legs of simulation, in which a restrained ligand is gradually decoupled from the receptor binding pocket or from the aqueous solution. In each leg of the decoupling simulations, the Coulomb interaction is turned off first using 11 λ-windows, and the Lennard-Jones interactions are then turned off in 17 λ-windows. (Coulomb decoupling: λ = 0.0, 0.1, 0.2, 0.3, 0.4, 0.5, 0.6, 0.7, 0.8, 0.9, 1.0; Lennard-Jones decoupling: λ = 0.0, 0.1, 0.2, 0.3, 0.4, 0.5, 0.55, 0.6, 0.65, 0.7, 0.75, 0.8, 0.85, 0.9, 0.94, 0.985, 1.0). The DDM simulations were performed using the Gromacs4.6.4 package for 3 ns in each of the alchemical λ windows.
Results
Using the BEDAM technology on a distributed computing network at Temple University and CUNY Brooklyn College, we are able to perform binding free energy calculations for a total of 3110 protein–ligand complexes for the 180 Hsp90 ligands in a time frame of 1 week. Because of the complexity of the receptor binding site which includes receptor flexibility and structured waters, our focus is on evaluating how the fast BEDAM free energy method distinguishes binders from nonbinders, rather than the more challenging task to rank order the binding affinities. We note that there are more actives than inactives in this data set, which is unlike typical virtual screening experiments where the task is to identify the few active molecules distributed among a large number of inactive compounds.
We first examine how well BEDAM calculations identify true binders among the top ligands ranked by BEDAM. Among the top 10 % or 18 of the predicted binders, only one is nonbinder. This yields an enrichment at 10 % of the data set as: . Compared with the maximum achievable enrichment , BEDAM performed well in identifying the true binders among the top ranked ligands.
Next we look at the BEDAM’s performance in picking out the nonbinders: in the bottom 10 % ranked ligands, 5 out of the 18 ligands or 28 % are nonbinders. This translates into an enrichment of nonbinders at bottom 10 % of the database as: . Here, the maximum achievable enrichment picking nonbinders is . The comparison of the BEDAM generated EF with the maximum achievable value shows that a significant number of true binders were incorrectly classified as nonbinders (false negatives).
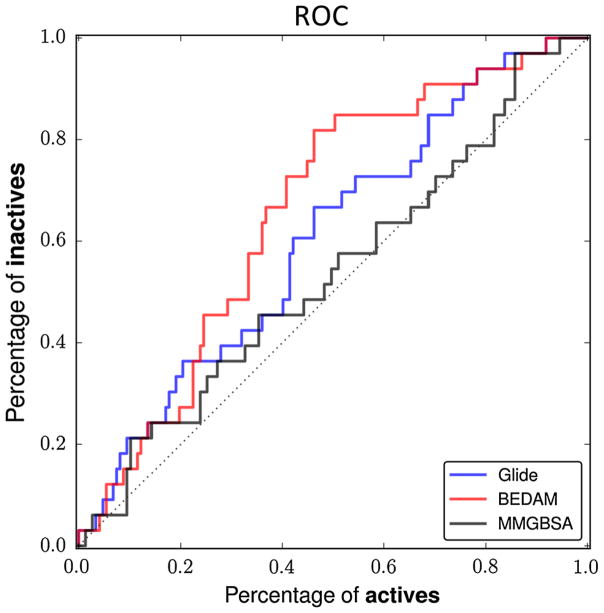
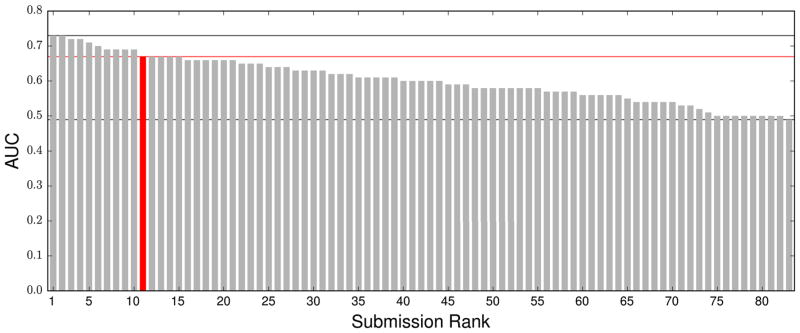
The overall results of BEDAM in separating nonbinders from binders are summarized by the Receiver Operating Characteristic (ROC) curve: see Fig. 2. The BEDAM free energy scoring preformed better than both the Prime MM-GB/SA protocol and Glide SP scoring function in our hands in identifying the few nonbinders from the large number of binders. The distribution of AUC among the 83 submissions in the scoring stage of the Hsp90 section of the D3R GC2015 challenge is shown in Fig. 3. It is to be noted that the differences in the AUC within the upper half of the submissions are small.
Fig. 2.
ROC curves obtained with BEDAM, glide SP and prime MM-GB/SA
Fig. 3.
The distribution of AUC among 83 submissions in the D3R GC2015 challenge. The BEDAM result is colored red
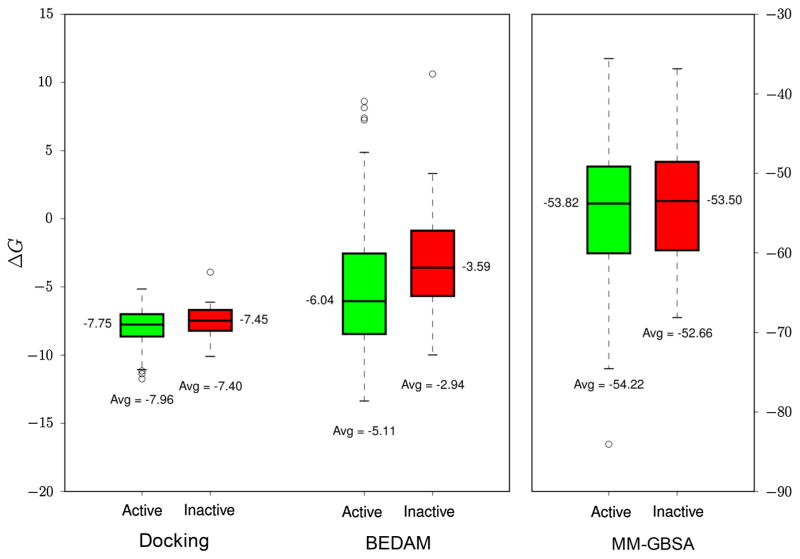
The ability of the different methods to separate binders from nonbinders can be analyzed in more detail by examining the distributions of binding scores among binders and nonbinders: as seen from Fig. 4, among the three methods, only the mean value of BEDAM binding free energy scores is able to separate binders from nonbinders with a gap of 2.4 kcal/mol. However, in the BEDAM result, there is still significant overlap in ΔG values for binders and nonbinders, which is consistent with the fact that the enrichment factor is far from the maximum value achievable. Overall, the BEDAM results shown in Figs. 2, 3, 4 are promising and suggest that further improvement in its energy function and sampling algorithm [11] could enable it to play a useful role for virtual screening in early stage structure-based drug discovery projects.
Fig. 4.
The distribution of binding score values from BEDAM, Glide SP and MM-GB/SA for the 180 Hsp90 ligands
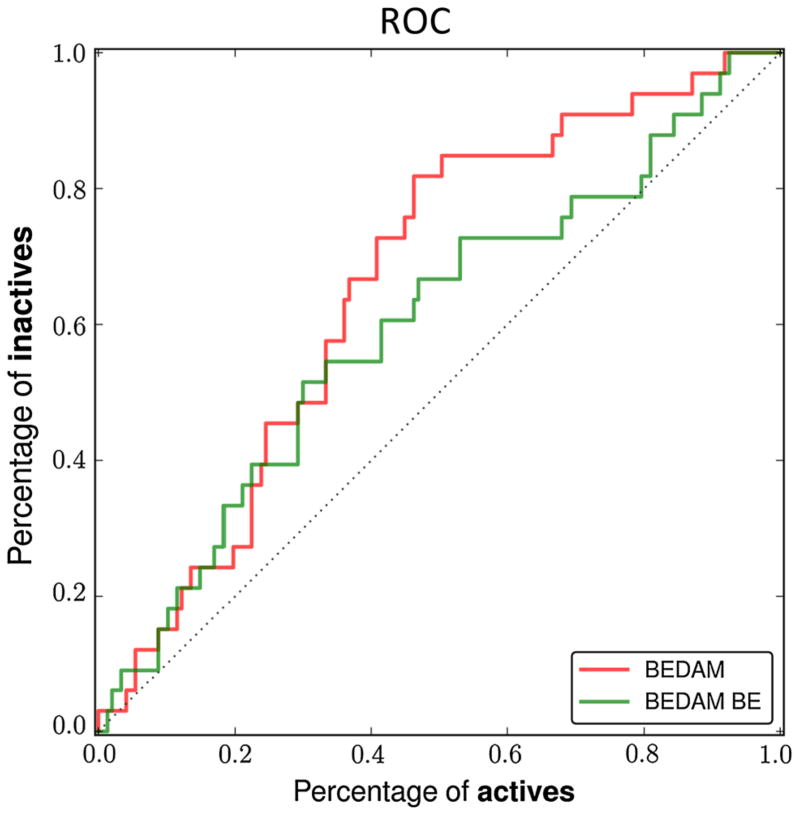
The results of BEDAM calculations also demonstrate the importance of including conformational reorganization in scoring protein–ligand complexes: as seen from Fig. 5, the ROC curve obtained using binding free energy is superior compared with that obtained using BEDAM binding energy alone. This result is consistent with our prior findings in the SAMPL4 dataset for HIV-1 Integrase ligands [9].
Fig. 5.
ROC curves for BEDAM binding free energy scoring and binding energy scoring
We also examined the BEDAM results for the 102 phenolic compounds (Fig. 6) among the 180 Hsp90 ligands. This group of ligands which include 87 binders and 15 nonbinders, is not hampered by the energy function error related to the aminopyrimidine/aminopurine functional groups. In the bottom 10 % predicted phenols or 10 ligands, 3 are true nonbinders, which yields an enrichment factor . This result is better than the enrichment factor at 10 % of 1.52 for the full ligand set. In addition, all top 10 % predicted ligands are true binders, reaching the maximum achievable ER of 1.17.
Fig. 6.
Representative phenol compounds
While the BEDAM was developed for early stage drug discovery where the emphasis is on enrichment of focused ligand libraries and not for rank ordering the binding affinities of active compounds, we found that the Spearman correlation coefficient between the BEDAM predicted ranking and experimental results for the 102 phenolic compounds is 0.45. This value is identical to the highest Spearman value reported by the D3R GC2015 participants for the full ligand set.
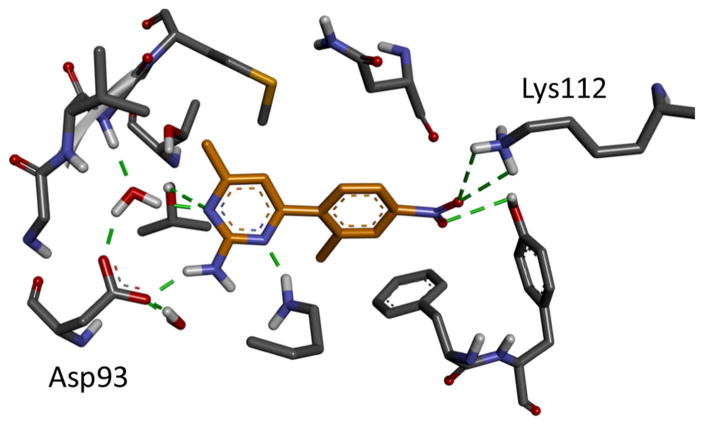
We have also performed explicit solvent double decoupling calculations (DDM) on two aminopyrimidines, 73 and 179, and one resorcinol compound, 40, partly to investigate the error of the overly positive ΔG we have seen with BEDAM simulations for aminopyrimidines and also to explore at a higher level of detail the thermodynamics of binding of different ligands. These calculations were also performed blindly, prior to the release of experimental data by the D3R GC2015 challenge organizers: see Table 1. The top docked structures of the complexes were used as the starting point of the calculations. The results show that the problem of underestimated binding free energy observed for aminopyrimidines/aminopurines are not seen in the explicit solvent DDM calculations. In fact, the computed ΔG for one of the aminopyrimidines 73 is overestimated by −4.8 kcal/mol compared with the experiment (Table 1). This suggests that the error observed earlier for aminopyrimidines may be due to the inability of the implicit solvent model to correctly capture the specific hydrogen bonding patterns involving explicit waters around the Asp93 side chain in the Hsp90 binding site (Fig. 7). Table 1 also revealed the important Coulomb electrostatic contribution to the total ΔG in these ligands, which is consistent with the observation of extensive, correlated hydrogen bonding network involving aminopyrimidine or resorcinol moieties, ordered waters and protein side chains in the polar Hsp90 binding pocket: see Fig. 7 for an example.
Table 1.
Binding free energies of two aminopyrimidines and one resorcinol calculated from DDM (Double Decoupling in explicit solvent)
| ΔG°(calc) = ΔG(Coulomb) + ΔG (VDW) + ΔG(restraint) | ||||||
|---|---|---|---|---|---|---|
|
| ||||||
| Ligand | 2D-structure | ΔG (Coulomb) | ΔG (VDW) | ΔG (restraint) | ΔG° (calc) | ΔG° (experiment) |
| 179 |

|
−2.0 ± 0.2 | −8.2 ± 0.2 | 3.22 | −7.0 ± 0.02 | −7.0 |
| 73 |

|
−6.4 ± 0.01 | −11.8 ± 0.6 | 3.22 | −14.4 ± 0.6 | −9.6 |
| 40 |

|
−8.4 ± 0.08 | −9.8 ± 0.2 | 3.22 | −15.0 ± 0.1 | −10.44 |
Fig. 7.
The final frame of the complex of Hsp90 and ligand 73 in the λ = 1 DDM trajectory. The carbon atoms of the ligand are shown in yellow. The intermolecular hydrogen bonds are shown as green dashed lines
Finally, we note that while the absolute binding free energies computed from BEDAM are systematically too positive for aminopyrimidines/aminopurines, both BEDAM and DDM simulations correctly identified the crystallographic binding modes for the two aminopyrimidines examined here.
Discussion
We analyzed possible sources of errors in the BEDAM binding free energy calculations. One obvious source of error is the energy function error specific for the amoinopyrimidine/aminopurine functional groups described earlier in Methods. This problem in the energy function affects the calculated ΔGbind for the 62 aminopyrimidines/aminopurines or 1/3 of the compounds. As shown earlier, focusing on the subset of 102 phenolic compounds which are not affected by the same energy function error yields significantly improved enrichment factors.
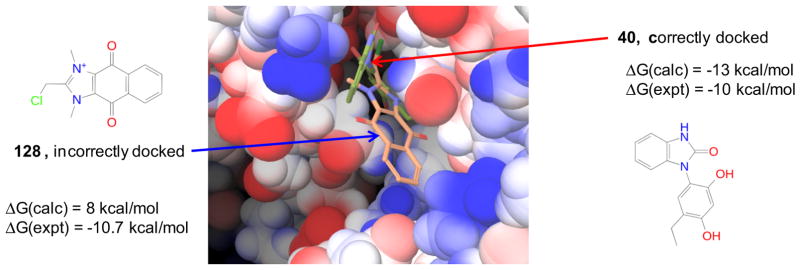
We also find that several of the strongest binders were incorrectly predicted to be nonbinders because of the failure in obtaining good initial docked poses. Among this group of false negatives are the anthraquinone and naphthoquinone scaffold containing compounds 128, 129, 130, 131 and 161. Figure 8 compares the docked structures of the active compound 40 and 128, and their experimental and calculated binding free energies. While 40 is docked correctly into the receptor binding pocket (by comparing with the released crystal structure), the main functional moiety of 128 is left solvent exposed in the docked structure. As a result, for 128, the ΔGbind from subsequent BEDAM calculation is grossly underestimated. Since the sampling of the internal degrees of freedom of the receptor and ligand is not specifically enhanced in the standard BEDAM protocol used here, the pose prediction errors in the docked structures can be left uncorrected during the BEDAM simulations because of large internal energy barriers for the ligands to adopt the correct conformations. While we have included both open and closed conformations of Hsp90 receptor during docking, the failure to dock compounds containing the bulky scaffolds such as anthraquinone and naphthoquinone indicates that the protein conformational changes induced by such rigid ligands are not properly accounted for by the two conformations used in docking. One possible solution to such failures is to include a significantly larger number of diverse receptor structures in docking and to specifically accelerate intramolecular conformational sampling in BEDAM in addition to the accelerated sampling of the external degrees of freedom that BEDAM accomplishes [11].
Fig. 8.
Importance of initial docked structure to binding free energy calculation
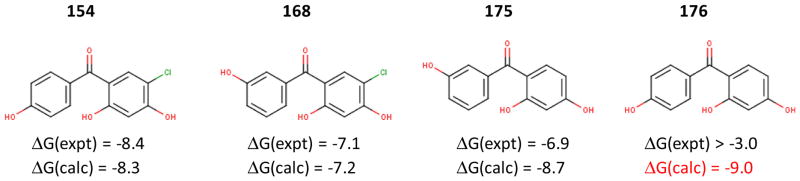
Lastly, the limited success achieved in distinguishing binders from nonbinders is simply attributable to the challenging nature of the problem. One example is given in Fig. 9, which shows the calculations for four chemically very similar benzophenone derivatives, which differ from one another by the presence of one hydroxyl or chlorine substituent. Here the BEDAM scoring correctly predicted the binding free energies for all but compound 176. Compounds 154 and 176 differ by just one chlorine substitution, yet their experimental −kT log IC50 differ by ≥5.4 kcal/mol. This is puzzling since in docking the two ligands adopt the identical binding mode and it is known that a chlorine substituent typically shifts the affinity by less than 1.5 kcal/mol. BEDAM predicted that the two ligands bind with similar affinities, in agreement with our chemical intuition yet in contradiction to experiments. Assuming the experiment is right, (note however the subtle possible difference between IC50 and Kd) this is an example of the difficulty of interpreting/predicting structure–activity trends in the Hsp90 ligands.
Fig. 9.
The experimental and calculated ΔG for the ligands containing benzophenone scaffold
Conclusion
We entered this blind challenge following our participation in the previous SAMPL challenges. The setting of the SAMPL and D3R series provides an excellent opportunity for more realistically testing different binding methods. The Hsp90 ATP binding site, compared to previous protein targets in the SAMPL series, is known to be among the most difficult ones, due to protein conformational changes, the presence of deeply buried charged side chain and several ordered waters in the binding pocket. In addition, the input ligands are chemically diverse; majority of the ligands are actives with a range of ΔGbind spanning a relatively narrow range of 5 kcal/mol. Our primary goal in this D3R GC2015 challenge was therefore to test how well BEDAM can distinguish actives from inactives, compared with other more rapid methods like docking and MM-GB/SA.
Using the BEDAM binding free energy scoring a moderate enrichment has been achieved for this challenging target. In our hands, compared to Glide SP and Prime MM-GB/SA, BEDAM performs better in discriminating inactives and actives, as indicated by various metrics such as the ROC curves, AUC values and enrichment factors. Among the three methods tested here, only BEDAM results show a gap averaging 2.4 kcal/mol separating the binders and nonbinders. However, the enrichment factor obtained from BEDAM is still relatively small. All methods we tested were unsuccessful in rank ordering the 147 active compounds for Hsp90.
The overall performance of BEDAM in the SAMPL series and D3R challenges show that good correlation and rank ordering can be achieved for relatively simpler host–guest systems that were the focus of SAMPL5. For the more complex protein–ligand systems, the results suggest that at this stage of the development, BEDAM is best used for enhanced enrichment in virtual screening of focused libraries of the kind that were studied in the SAMPL4 HIV Integrase challenge, and for predicting the binding pose prior to the higher resolution FEP calculations of binding affinity.
Acknowledgments
This work has been supported by the R01 GM30580 to RML, and by the NIH S10 OD020095 (cb2rr Shared Instrumentation Grant. Part of the free energy calculations were performed using NSF XSEDE computing resources. Parallel BEDAM calculations were also carried out on the BOINC distributed networks at Temple University and Brooklyn College of the City University of New York. The authors acknowledge invaluable technical support from Gene Mayro, Jaykeen Holt, Zachary Hanson-Hart from the IT department at Temple University, and James Roman, and John Stephen at Brooklyn College.
References
- 1.Boresch S, Tettinger F, Leitgeb M, Karplus M. Absolute binding free energies: a quantitative approach for their calculation. J Phys Chem. 2003;107:9535–9551. [Google Scholar]
- 2.Chang C-E, Gilson MK. Free energy, entropy, and induced fit in host-guest recognition: calculations with the second-generation mining minima algorithm. J Am Chem Soc. 2004;126:13156–13164. doi: 10.1021/ja047115d. [DOI] [PubMed] [Google Scholar]
- 3.Deng Y, Roux B. Calculation of standard binding free energies: aromatic molecules in the T4 lysozyme L99A mutant. J Chem Theory Comput. 2006;2:1255–1273. doi: 10.1021/ct060037v. [DOI] [PubMed] [Google Scholar]
- 4.Gallicchio E, Lapelosa M, Levy RM. Binding energy distribution analysis method (BEDAM) for estimation of protein–ligand binding affinities. J Chem Theory Comput. 2010;6:2961–2977. doi: 10.1021/ct1002913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang L, Friesner RA, Berne BJ. Replica exchange with solute scaling: a more efficient version of replica exchange with solute tempering (REST2) J Phys Chem B. 2011;115:9431–9438. doi: 10.1021/jp204407d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang L, et al. Accurate and reliable prediction of relative ligand binding potency in prospective drug discovery by way of a modern free-energy calculation protocol and force field. J Am Chem Soc. 2015;137:2695–2703. doi: 10.1021/ja512751q. [DOI] [PubMed] [Google Scholar]
- 7.Mobley DL, et al. Blind prediction of HIV integrase binding from the SAMPL4 challenge. J Comput Aided Mol Des. 2014;28:327–345. doi: 10.1007/s10822-014-9723-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gallicchio E, Levy RM. Prediction of SAMPL3 host-guest affinities with the binding energy distribution analysis method (BEDAM) J Comput Aided Mol Des. 2012;26:505–516. doi: 10.1007/s10822-012-9552-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gallicchio E, et al. Virtual screening of integrase inhibitors by large scale binding free energy calculations: the SAMPL4 challenge. J Comput Aided Mol Des. 2014;28:475–490. doi: 10.1007/s10822-014-9711-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gallicchio E, et al. BEDAM binding free energy predictions for the SAMPL4 octa-acid host challenge. J Comput Aided Mol Des. 2015;29:315–325. doi: 10.1007/s10822-014-9795-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mentes A, et al. Binding energy distribution analysis method (BEDAM): Hamiltonian replica exchange with torsional flattening for binding mode prediction and binding free energy estimation. J Chem Theory Comput. 2016;12:2459–2470. doi: 10.1021/acs.jctc.6b00134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Murray CW, et al. Fragment-based drug discovery applied to Hsp90. discovery of two lead series with high ligand efficiency. J Med Chem. 2010;53:5942–5955. doi: 10.1021/jm100059d. [DOI] [PubMed] [Google Scholar]
- 13.Steinbrecher TB, et al. Accurate binding free energy predictions in fragment optimization. J Chem Inf Model. 2015;55:2411–2420. doi: 10.1021/acs.jcim.5b00538. [DOI] [PubMed] [Google Scholar]
- 14.Haider K, Huggins DJ. Combining solvent thermodynamic profiles with functionality maps of the Hsp90 binding site to predict the displacement of water molecules. J Chem Inf Model. 2013;53:2571–2586. doi: 10.1021/ci4003409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deng N, et al. Distinguishing binders from false positives by free energy calculations: fragment screening against the flap site of HIV protease. J Phys Chem B. 2014;119:976–988. doi: 10.1021/jp506376z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jorgensen WL, Maxwell DS, Tirado-Rives J. Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. J Am Chem Soc. 1996;118:11225–11236. [Google Scholar]
- 17.Kaminski GA, Friesner RA, Tirado-Rives J, Jorgensen WL. Evaluation and reparametrization of the OPLS-AA force field for proteins via comparison with accurate quantum chemical calculations on peptides. J Phys Chem B. 2001;105:6474–6487. [Google Scholar]
- 18.Gallicchio E, Levy RM. AGBNP: An analytic implicit solvent model suitable for molecular dynamics simulations and high-resolution modeling. J Comput Chem. 2004;25:479–499. doi: 10.1002/jcc.10400. [DOI] [PubMed] [Google Scholar]
- 19.Gallicchio E, Paris K, Levy RM. The AGBNP2 implicit solvation model. J Chem Theory Comput. 2009;5:2544–2564. doi: 10.1021/ct900234u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gilson M, Given J, Bush B, Mccammon J. The statistical-thermodynamic basis for computation of binding affinities: a critical review. Biophys J. 1997;72:1047–1069. doi: 10.1016/S0006-3495(97)78756-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deng Y, Roux B. Computations of standard binding free energies with molecular dynamics simulations. J Phys Chem B. 2009;113:2234–2246. doi: 10.1021/jp807701h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deng N, Zhang P, Cieplak P, Lai L. Elucidating the energetics of entropically driven protein–ligand association: calculations of absolute binding free energy and entropy. J Phys Chem B. 2011;115:11902–11910. doi: 10.1021/jp204047b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lindorff-Larsen K, et al. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins Struct Funct Bioinf. 2010;78:1950–1958. doi: 10.1002/prot.22711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA. Development and testing of a general amber force field. J Comput Chem. 2004;25:1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- 25.Jakalian A, Bush BL, Jack DB, Bayly CI. Fast, efficient generation of high-quality atomic charges. AM1-BCC model: I. Method. J Comput Chem. 2000;21:132–146. doi: 10.1002/jcc.10128. [DOI] [PubMed] [Google Scholar]