Abstract
This paper describes a new Cohort Selection application implemented to support streamlining the definition phase of multi-centric clinical research in oncology. Our approach aims at both ease of use and precision in defining the selection filters expressing the characteristics of the desired population. The application leverages our standards-based Semantic Interoperability Solution and a Groovy DSL to provide high expressiveness in the definition of filters and flexibility in their composition into complex selection graphs including splits and merges. Widely-adopted ontologies such as SNOMED-CT are used to represent the semantics of the data and to express concepts in the application filters, facilitating data sharing and collaboration on joint research questions in large communities of clinical users. The application supports patient data exploration and efficient collaboration in multi-site, heterogeneous and distributed data environments.
Introduction
Retrospective use of clinical data by clinical researchers covers a wide variety of research investigations including assessment of clinical trial feasibility, generation and retrospective validation of research hypotheses, data exploration, quality assessment, etc. While clinicians involved in research hold the clinical knowledge and expertise that would enable them to formulate and test hypotheses based on the exploration and analysis of the collected data, they are often limited by the unavailability of user-friendly and intuitive visual tools giving them easy access to the data. The filtering of datasets to select relevant cohorts of patients to be used for further analysis is often time-consuming and requires a lot of manual steps and/or advanced informatics skills (e.g. scripting, querying, etc.).
In this paper we describe a novel Cohort Selection application enabling the definition and management of cohorts of clinical data, i.e. sets of patients or cases with desired characteristics, out of large heterogeneous and distributed datasets. To support the needs of large clinical networks that focus on jointly carrying out collaborative research, we developed a highly expressive tool that leverages an underlying standards-based Semantic Interoperability Solution and a Groovy DSL for the flexible definition of complex filter stacks. To streamline the process of exploring and filtering retrospective datasets, the application enables clinical users to efficiently and intuitively build patient cohorts, perform basic analyses and counts, and export these cohorts to be further used in advanced analyses. The application provides two mechanisms of interaction between the clinical researcher and the underlying data repositories. Predefined templates/filters can be used for building complex cohorts by clinical users without any need for informatics knowledge. For the power-users (e.g. bioinformaticians, biostatisticians) we also provide the possibility to build cohorts by advanced scripting in a dedicated DSL and by directly sending queries to the data repositories through the Semantic Interoperability Layer (SIL). The solution implements reasoning with existing widely-used ontologies (e.g. SNOMED-CT, LOINC) in the process of filters execution and allows the clinical users to explore the concepts available in each dataset and their hierarchies, make use of synonyms, etc. To facilitate collaboration we provide functionality for sharing both the filter-stacks and the corresponding cohorts among specified teams of end-users who jointly carry out research. Functionality allowing for the splitting and merging of defined filter stacks is also implemented.
This work was carried out in the INTEGRATE1 project, which focused on supporting a novel research approach in oncology through the development of innovative biomedical infrastructures. The project implemented a semantic interoperability solution and reusable core infrastructure components to enable data sharing and collaboration within the clinical networks participating in INTEGRATE. Leveraging the underlying core components among which the SIL, we developed tools that facilitate efficient execution of post-genomic multi-centric clinical trials in breast cancer. Another application developed in the project on the same underlying infrastructure components as the Cohort Selection is Patient Screening for clinical trials (1). This application assesses the eligibility of patients for relevant clinical trials by automatically comparing the patient data available through the SIL with formalized clinical trials’ eligibility criteria represented with shared semantics.
The INTEGRATE project has also implemented a security framework that provides a technological solution covering the security requirements and guarantees compliance of the complete platform to the legal framework governing the project. It consists of modular components, respectively dealing with authentication, authorization, audit and privacy enhancing techniques.
The development of the cohort selection application has been driven by a clinical scenario focused on supporting clinical research in oncology, defined by end-users from organizations that are vanguards in oncology research and care. The users have provided feedback during the implementation of the application and participated in the evaluation sessions. Key use cases are to select cohorts with desired characteristics for data analysis, to facilitate feasibility analysis during trial design, and to help provide epidemiology reporting.
Background
A cohort selection solution requires functionality for querying the underlying medical datasets. Querying of medical data is a complex matter; problem areas include the formalization of medical query statements, execution of complex context-related queries (e.g. “find if finding X was administered to a patient in the last 3 months before diagnosis Y was made”) nesting of queries and capturing of medical information semantics. The medical query language that we proposed, SNAggletooth Query Language (SNAQL), addresses all of the above topics (2) and was previously integrated in comparable medical applications based in the trial domain (3) (4). Contrary to other approaches focusing on developing a fully defined formal medical language (5) (6), the SNAQL language focusses on balancing the implementation effort with end-user usability.
The participation of multiple institutions in joint clinical research requires advanced methods to integrate data from heterogeneous sources. Semantic interoperability is essential for feasibility and sustainability of clinical research (7). In this context, several previous projects focused on related challenges: Observational Medical Outcomes Partnership (OMOP) (8), Informatics for Integrating Biology and the Bedside (i2b2) (9) or the Patient-Centered Outcomes Research Network (10) among others. As a foundation of the cohort selection application described in this paper, we have relied on our Semantic Interoperability Layer (SIL) (11) (12) based on widely adopted standards such as the HL7 Reference Information Model (13). In (11) we provide an evaluation of the SIL and a comprehensive comparison between the proposed HL7 RIM-based SIL and i2b2 and OMOP. In (14) we describe our data transformation pipeline and evaluate the effort of integrating a new clinical site in our semantic environment.
Existing cohort selection tools usually target biomedical experts with deep technology knowledge and a particular data source (15). We propose an effective tool that leverages our SIL for filtering cohorts in multi-source, heterogeneous datasets. Our tool is accessible to specialists and non-specialists alike through the DSL and the predefined visual and textual filters. Defined filter graphs can be shared among researchers even when the datasets are not shared (e.g. filters can be reused across organizations on ‘private’ datasets).
Methods
The cohort selection application:
In Figure 1 we introduce the components composing the Cohort Selection application. While several components (e.g. the Front-End Application) are specific to this application, most components (e.g. the Locker Service, the Query Builder Service) are generic and can provide relevant functionality to other applications as well.
Figure 1.
The main components of the cohort selection application and the relevant services used by the application
The Front-End Application implements GUI concepts enabling users to define, save and apply filters and to build filter stacks, to select and explore the datasets, to share filters with other users, and to save, share and export defined cohorts. The SNAQL service interprets the DSL statements in the filters and queries the underlying data sources through the SIL. The use of SNAQL facilitates very high expressiveness allowing users to build filters carrying out complex computations on the data. The Analytics Service provides functionality for displaying basic analytics and counts on the cohorts to support data exploration. The Locker Service provides the functionality that enables users to save cohorts and filter stacks in their own workspace and to share them with other users.
The Semantic Integration Services are part of the SIL and support the Front-End Application with retrieving metadata (Metadata Service) for data exploration and with providing autocomplete (Autocomplete Service) for ontology concepts in the definition of filters. The Query Normalization Service is used by the SNAQL Service to generate queries to and to retrieve data through the underlying Query Execution Service from the Common Data Model (CDM). In this phase query expansion based on the SNOMED-CT hierarchy of concepts is also provided.
In the next sections we describe the functionality of the main components of Cohort Selection: The Front-End Application, the Locker Service, the SNAQL Service and the DSL, and the key components of the SIL (Semantic Integration and Data Access Services).
The Front-End Application:
The role of the front-end application is to support the users to select and explore datasets, define and apply filters on selected datasets, and save, export and share results (filters, filter stacks, selected cohorts). The GUI implements several methods for specifying the filters. The application provides (i) pre-defined visual filters and (ii) generic textual filters that can be defined and extended by the users, supporting both ease of use and expressiveness. In Figure 2 we depict the GUI implementations of the filters specifying age ranges and gender respectively. In the UI, users can also define filters by implementing scripts of any degree of complexity with our DSL that reads close to natural language. The DSL is intuitive and can be used by non-programming experts. Defined textual filters can be used as templates and modified to suit the needs of the users for instance by replacing the ontology concepts or parts of the DSL as further explained in the Results section.
Figure 2.
Visual filters for intuitive selection of ranges of values and of discrete categories. Age range (top) and gender (bottom) of patients in the desired cohort.
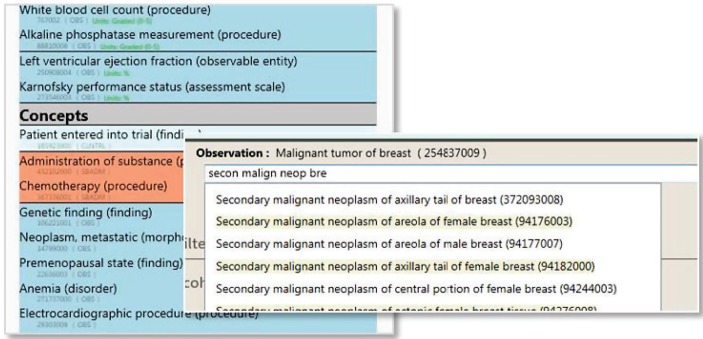
Leveraging the SIL, the GUI enables the exploration of the datasets and the selection of ontology concepts to be used in the filters, including autocomplete functionality (Figure 3). In Figure 4 we show the capabilities of the GUI for exploring the datasets and for saving, sharing with other users and exporting cohorts. The users can select the colleagues with whom they want to share the defined cohorts and can specify the desired data categories to be exported for further analysis.
Figure 3.
GUI capabilities for exploring the available datasets and for selecting ontology concepts in the filter templates with the use of autocomplete.
Figure 4.

GUI capabilities for saving, sharing with other users and exporting cohorts, and for exploring the datasets.
The SNAQL Service and the Domain Specific Language (DSL):
The cohort selection application makes use of the medical query language SNAQL for querying the underlying datasets. SNAQL is designed as an abstraction layer that hides the technical specifics of underlying semantic data models to the end-user when creating new queries during the cohort selection process. The main objective of the language was to allow the definition of queries in a language close to natural language, while keeping the implementation effort to a minimum.
The SNAQL syntax enables the definition of medical concepts, the specification contextual (like temporary) constraints on these concepts and the combination of concepts using unary and binary operators (and/or/not logic). In the table below we provide an example of a SNAQL script.
| Karnofsky performance status evaluation greater than or equals to 60% and confirmed carcinoma of breast occurred within 2 years |
|---|
| //SNOMED Codes of medical concepts def karnofsky_performance_status = ‘273546003’ def carcinoma_of_breast = ‘254838004’ //SNAQL Script result = measurement of: karnofsky_performance_status, gteq: 60.’%’ and {observation of: carcinoma_of_breast occurred before:’now’, within: 2.years} |
SNAQL is a Domain Specific Language (DSL) leveraging on the DSL-facilitating features of the Groovy scripting language for offering advanced functionality (e.g. defining temporal constraints, logical operators) at minimal effort.
These include operator overloading, closures, metaprogramming (Meta Object Protocol (MOP)) support and named parameter support through argument maps. SNAQL queries are executed by the SNAQE (SNAggletooth Query Engine) engine which is not an interpreter, but rather the definition of a context in which the SNAQL code makes sense.
The locker service:
The cohort selection application revolves around creating patient cohorts (consisting of lists of patient identifiers) by selecting data sources (e.g. databases containing medical information of patients), and including patients by expressing filter criteria on each patient’s data.
Naturally, the result of the cohort selection application is not the end goal but enables a user to use a defined patient cohort as a base for his subsequent work. This subsequent work might involve tooling that is not connected to the cohort selection infrastructure (e.g. takes as input an exported list of patient identifiers and patient information) but it could also involve tooling that is integrated with the cohort selection infrastructure. When the tooling is integrated, application and services can connect to a service to store and retrieve cohorts. In order to provide this service, a generic document sharing service (named “Locker service”) has been developed. The Locker service manages json documents and blobs for a user (using user identity information from the security framework – WS-Trust in this case) given a namespace and a key.
The cohort selection application uses the Locker service to manage cohorts and share them between applications and services within the infrastructure. Additionally, the locker service allows for sharing documents (cohorts) with other users (given a key and namespace of the cohort and a user id). In the application, the user can share a cohort by selecting a username and the key and namespace of a cohort and subsequently the cohort will appear in the cohort list of the recipient.
The semantic interoperability layer (SIL):
Services exploiting patient data from different sources require a solution to solve semantic heterogeneities. The cohort application is based on a SIL to homogenize access among clinical systems. Based on widely adopted standards, the SIL facilitates uniform access to legacy systems. To provide a common data representation for storing and querying clinical data used within clinical trials, three main components are required for the SIL (Figure 5): (i) Common Data Model (CDM), (ii) Core Dataset and (iii) Query and Data Normalization.
Figure 5.

Semantic Interoperability Layer (SIL) components for the cohort selection application
The CDM is based on the HL7 RIM to define the schema of the data warehouse. The Core Dataset (mainly based on SNOMED-CT and is extended with LOINC and HGNC for laboratory tests and gene names) contains medical concepts and relations describing the semantics of the data sources. To provide a semantically uniform access to the data stored in the data warehouse, a two-phase normalization process has been proposed: (i) Data normalization (Normalization Pipeline) and (ii) Query normalization. Both phases use the semantic knowledge of the Core Dataset to normalize data in the CDM and to subsequently retrieve data through the query execution service. Such common set up should be deployed at each institution to execute a common cohort selection application.
Expert-driven clinical scenario definition, requirements elicitation and evaluation:
The application has been implemented based on a scenario developed within the INTEGRATE collaborative project with clinical research organizations representing a large community of clinical users focused on research and care in oncology. The datasets used in the development of the prototypes and for validation and evaluation originate from completed clinical trials in breast cancer and were provided by the clinical collaborators in the project. In the underlying clinical scenario, a researcher would like to define a patient cohort with specific characteristics. The researcher has access to our application and to the INTEGRATE platform for facilitating the tasks. The tool supports the researcher with formulating the selection filters and applying them on relevant data sources. The cohort selection is carried out on multisource distributed heterogeneous datasets. The tool keeps track of all filtering results and allows exporting the selected cohort to continue with detailed analysis. Teams of researchers can collaborate on defining cohorts, either in a single- or multi-site context. They can share both the defined cohorts and the sets of selection filters applied (for instance to apply them on other datasets in local environments). The tool should address both the needs of expert bioinformaticians (providing flexibility and expressiveness in defining the filters), and of the non-informatics experts for instance to allow clinical experts to efficiently define their research questions. The clinical users involved in the definition of the scenario have driven the development process, evaluating the prototypes at multiple time points during implementation. Next to that, we have set up evaluation sessions with external clinical users for feedback, additional requirements and to identify new contexts of use for the application.
Results
In this section we first present main capabilities of the application with examples relevant in the clinical domain in which it was defined, then we discuss the evaluation with clinical users and the identified contexts of use.
Introducing the capabilities of the cohort selection application:
With the use of widely-adopted ontologies (e.g. SNOMED-CT) we provide reasoning during execution of filters and enable users to explore the concepts available in each dataset and their hierarchies, make use of synonyms, etc. Functionality allowing the splitting and merging of defined filter stacks is also implemented. To facilitate collaboration we provide functionality for sharing both the filter-stacks and the corresponding cohorts among teams of clinical users. Figure 6 demonstrates the use of SNOMED-CT to support reasoning: First filter selects all patients that have received a treatment with any Anthracycline (which is a class of drugs while the patient data only includes the actual drugs in that class that were prescribed) and the second filter selects out of those the patients that have received Epirubicin (a particular drug in that class). Figure 7 depicts an example of a filter that evaluates a temporal condition taking as reference a relative time point – another data element in the dataset which in this example is the time of diagnosis. The DSL fragment in the filter uses SNOMED-CT codes and normal form representations (according to SNOMEDT guidelines) for the concepts.
Figure 6.
Anthracyclines versus Epirubicin – reasoning based on SNOMED-CT.
Figure 7.
Breast cancer patients with no relapse 5 years after diagnosis – temporal conditions. SNOMED-CT codes of concepts represented in SNOMED-CT normal form are used in the DSL fragment.
In Figure 8 we show a more complex filter graph that demonstrates how the application can be used to split and merge cohorts. To define this cohort, we first build for a selected dataset a cohort of ER+ breast cancer patients and look at the age distribution. Then we apply filters to select metastatic patients per target site (liver, lung, bone and CNS). Finally we merge the four cohorts and visualize again the age distribution.
Figure 8.
Distribution of ER+ BC patients with metastatic disease based on the target site.
Evaluation with clinical users:
The tool was evaluated and validated with clinical datasets provided by the clinical partners in the INTEGRATE project and by clinical users with several relevant areas of expertise. We first carried out an evaluation workshop in which the tool was evaluated by an oncologist, and three bioinformaticians on an anonymized dataset of 3 completed clinical trials of the German Breast Group(GBG) with a total of 4673 patients: TBP (Metastatic study), GAIN (Adjuvant study), GeparQuattro (NeoAdjuvant study) with ClinicalTrials.gov identifiers NCT00148876, NCT00196872 and NCT00288002. The evaluation sessions included semi-structured interviews and a discussions guided by scenarios in two contexts: (i) Trial feasibility analysis: Finding a triple negative cohort of sufficient size, (ii) Data analysis for hypothesis generation: Effect of ER status on metastatic relapse. The evaluation was positive both with respect to functionality and ease of use, and the feedback was used to refine and extend the tool for instance with functionality to support collaboration on definition of filter graphs and the sharing of defined cohorts among users within the tool.
The second evaluation took place at the sites of GBG and Institut Jules Bordet (IJB). The focus was on assessing the features and usability of the tool, and on identifying the most relevant contexts in which the application could be used. Participating users worked in clinical research with a wide range of areas of expertise: Bioinformatician, biostatistician, oncologist, epidemiologist, research nurse, and data manager. The evaluation yielded three contexts in which the tool can add value: (i) for data analysis to select cohorts with desired characteristics, (ii) during the clinical trial design phase for feasibility analysis, (iii) to help provide epidemiologic data to the researchers, the hospital, and the medical authorities (e.g. reporting to the cancer registry by filtering EHR data to identify patients with recurrent tumors). The application helps streamlining the definition of clinical trials by supporting (i) the definition of eligibility criteria and their evaluation on the available data, (ii) the assessment of feasibility of trial definitions with respect to criteria soundness and selectiveness, and estimation of enrollment power based on historical data, and (iii) the retrospective evaluation of hypotheses on combined datasets out of heterogeneous distributed sources.
Conclusions
In this paper we describe our cohort selection application that supports efficient data exploration, collaborative definition of research questions, and the design phase of clinical trials. We focused on flexibility and expressiveness in the definition of filters to support very complex (combinations of) criteria, for instance enabling complex computations and temporal comparisons among data.
The clinical researcher and clinician can either use existing (pre-defined) filters or define new filters using the DSL provided by the tool which emulates clinical logic (suited for non-programming experts). New filters can be added to the set of defined filters (i.e. saved) and they become available to all other users accessing the tool. With this approach the filter set is not fixed and grows with the tool use – new filters can be shared among users as soon as they are defined. The tool also provides templates: Existing filters can be modified to suit the needs of the user for instance by replacing the concepts in the filter with the relevant ontology concepts, which speeds up the definition of new filters.
We facilitate collaboration by providing mechanisms for sharing cohorts within the tool, for exporting cohorts to carry out analysis, and for sharing stacks of filters among users for instance to reapply them on other datasets. The filters can be reused and collaboratively extended by the users of the application. The SIL allows querying across heterogeneous distributed data sources enabling users to build cohorts across data sources. Users can also collaborate on jointly defining the research questions and the characteristics of the desired cohort (i.e. the filter stacks), but apply them locally on their own datasets.
Next to supporting clinical research, the application can be used to provide epidemiology data to the hospital, researchers and medical authorities (e.g. for reporting to the Cancer Registry). The application only offers basic analytics capabilities focused on data exploration, as the expert users already have very powerful tools they would like to use for advanced analysis. To support this we enable users to export and share defined cohorts and their data per categories (e.g. all data, clinical, genomic, etc.) and in formats that can be then taken as input in the analysis tools.
Footnotes
Driving excellence in Integrative Cancer Research through Innovative Biomedical Infrastructures. http://www.fp7-integrate.eu/. This work has been partially funded by the European Commission through this project (FP7-ICT-2009-6-270253).
References
- 1.Bucur A, van Leeuwen J, Chen NZ, Claerhout B, de Schepper K, Perez-Rey D, et al. Supporting Patient Screening to Identify Suitable Clinical Trials. In Stud Health Technol Inform. 2014;205:823–7. 2014. [PubMed] [Google Scholar]
- 2.Claerhout B, de Schepper K, Pérez-Rey D, Alonso-Calvo R, van Leeuwen J, Bucur A. Leveraging Dynamic Programming Languages for Efficient Implementation of a Patient Cohort Selection Engine. 2013.
- 3.van Leeuwen J, Bucur A, Keijser J, Claerhout B, de Schepper K, Pérez-Rey D, et al. Recruitment and feasibility tool: J. Clinical Bioinformatics. 2015 [Google Scholar]
- 4.Claerhout B, de Schepper K, Pérez-Rey D, Alonso-Calvo R, van Leeuwen J, Bucur A. Implementing patient recruitment on EURECA semantic integration platform through a Groovy query engine: BIBE. 2013.
- 5.Bache R, Taweel A, Miles S, Delaney B. An Eligibility Criteria Query Language for Heterogeneous Data Warehouses. Methods of Information in Medicine. 2014. p. to appear. [DOI] [PubMed]
- 6.Weng C, Tu S, Sim I, Richesson RL. Formal representation of eligibility criteria: a literature review. Journal of Biomedical Informatics. 2010:451–467. doi: 10.1016/j.jbi.2009.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hersh WR. Adding value to the electronic health record through secondary use of data for quality assurance, research, and surveillance. Am J Manag Care. 2007;81:126–128. [PubMed] [Google Scholar]
- 8.Stang PE, Ryan PB, Racoosin JA, Overhage JM, Hartzema AG, Reich C, et al. Advancing the science for active surveillance: rationale and design for the Observational Medical Outcomes Partnership. Annals of internal medicine. 2010;153(9):600–606. doi: 10.7326/0003-4819-153-9-201011020-00010. [DOI] [PubMed] [Google Scholar]
- 9.Murphy SN, Weber G, Mendis M, Gainer V, Chueh HC, Churchill S, et al. Serving the enterprise and beyond with informatics for integrating biology and the bedside (i2b2) Journal of the American Medical Informatics Association. 2010;17(2):124–130. doi: 10.1136/jamia.2009.000893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fleurence RL, Curtis LH, Califf RM, Platt R, Selby JV, Brown JS. Launching PCORnet, a national patient-centered clinical research network. Journal of the American Medical Informatics Association. 2014;21(4):578–582. doi: 10.1136/amiajnl-2014-002747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paraiso-Medina S, Perez-Rey D, Bucur A, Claerhout B, Alonso-Calvo R. Semantic normalization and query abstraction based on SNOMED-CT and HL7: Supporting multi-centric clinical trials. IEEE J Biomed Health Inform. 2015 May;19(3):1061–7. doi: 10.1109/JBHI.2014.2357025. 2015. [DOI] [PubMed] [Google Scholar]
- 12.Alonso-Calvo R., Perez-Rey D., Paraiso-Medina S, Claerhout B, Hennebert P, Bucur A. Enabling semantic interoperability in multi-centric clinical trials on breast cancer. Computer methods and programs in biomedicine. 2014;118(3):322–329. doi: 10.1016/j.cmpb.2015.01.003. [DOI] [PubMed] [Google Scholar]
- 13.Beeler G, Case J, Curry J, Hueber A, Mckenzie L, Schadow G, et al. HL7 Reference Information Model. 2003.
- 14.Ibrahim A, Bucur A, Perez-Rey D, Alonso E, de Hoog M, Dekker A, et al. Case Study for Integration of an Oncology Clinical Site in a Semantic Interoperability Solution based on HL7 v3 and SNOMED-CT: Data Transformation Needs. In AMIA Jt Summits Transl Sci Proc. 2015 Mar 25. 2015. [PMC free article] [PubMed]
- 15.Wilcox A, Vawdrey D, Chunhua Weng C, Velez M, Bakken S. Research Data Explorer: Lessons Learned in Design and Development of Context-based Cohort Definition and Selection; AMIA Jt Summits Transl Sci Proc. 2015; 2015. pp. 194–198. [PMC free article] [PubMed] [Google Scholar]