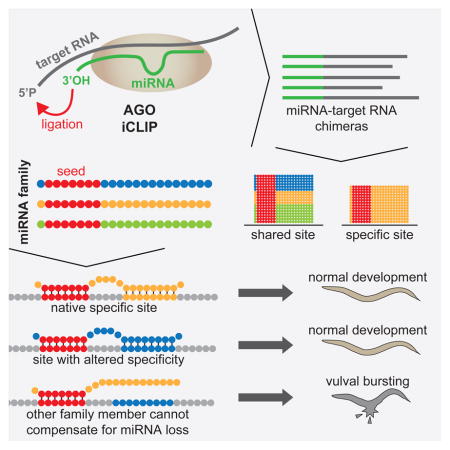
SUMMARY
To identify endogenous miRNA target sites, we isolated AGO-bound RNAs from Caenorhabditis elegans by individual-nucleotide resolution crosslinking-immunoprecipitation, which fortuitously also produced miRNA-target chimeric reads. Through the analysis of thousands of reproducible chimeras, pairing to the miRNA seed emerged as the predominant motif associated with functional interactions. Unexpectedly, we discovered that additional pairing to 3′ sequences is prevalent in the majority of target sites and leads to specific targeting by members of miRNA families. By editing an endogenous target site, we demonstrate that 3′ pairing determines targeting by specific miRNA family members and that seed pairing is not always sufficient for functional target interactions. Finally, we present a simplified method, Chimera PCR (ChimP), for the detection of specific miRNA-target interactions. Overall, our analysis revealed that sequences in the 5′ as well as the 3′ regions of a miRNA provide the information necessary for stable and specific miRNA target interactions in vivo.
Keywords: microRNA, iCLIP, chimeras, C. elegans, miRNA family, ALG-1
eTOC Blurb
Argonaute iCLIP produces miRNA-target chimeras that identify miRNA targets. Broughton et al. analyze these chimeras, reporting that 3′ regions of miRNAs confer specificity to miRNA target interactions in vivo. They develop a simple, versatile, and economic method called Chimera PCR (ChimP) for testing miRNA-target interactions.
INTRODUCTION
MicroRNAs (miRNAs) are small RNA molecules that are bound by Argonaute (AGO) proteins. The AGO-miRNA duplex forms the core of the miRNA-induced silencing complex (miRISC), which is directed by the bound miRNA to complementary sequences in the messenger RNA (mRNA) (Pasquinelli, 2012). The miRISC co-factors then promote translational inhibition and transcript destabilization of the target RNA. Canonical miRNA-target interactions featuring complementarity to the seed sequence, nucleotides (nts) 2–8 of the miRNA, have long been recognized as critical for miRNA targeting (Bartel, 2009). Recent structural and single molecule studies have emphasized the importance of seed pairing for stable AGO binding (Chandradoss et al., 2015; Elkayam et al., 2012; Jo et al., 2015; Nakanishi et al., 2012; Salomon et al., 2015; Schirle and MacRae, 2012).
However, there are examples of functional miRNA target interactions that occur without perfect seed pairing. For example, the well-established miRNA target lin-41 in Caenorhabditis elegans features two sites that are complementary to the let-7 miRNA (Slack et al., 2000; Vella et al., 2004). Neither site supports canonical seed pairing interactions; one of the sites forms a one-nucleotide bulge in the target and the other requires an unfavorable GU pair. Imperfect seed matches have been suggested to be compensated by more extensive pairing with the 3′ end of the miRNA (Brennecke et al., 2005; Grimson et al., 2007). However, examples of conserved sites with 3′ compensatory binding for weak seeds are relatively rare (Friedman et al., 2009). Although studies using 3′ untranslated region (UTR) reporters have suggested that non-canonical seed sites are functional (Helwak et al., 2013), a recent analysis of non-canonical target sites revealed that, even though these sites are bound by the miRNA complex, they do not appear to be broadly functional (Agarwal et al., 2015).
Many mature miRNAs can be classified based on the presence of identical seed sequences into groups called miRNA families (Lewis et al., 2003; Lim et al., 2003). Due to the shared seed sequence of miRNA family members, it is predicted that these miRNAs will regulate similar target RNAs. Phenotypic analyses of miRNA deletions in nematodes suggest that members of miRNA families have cooperative or redundant functions with each other (Alvarez-Saavedra and Horvitz, 2010). However, recent work suggests that individual family members may have independent targets, even when co-expressed (Moore et al., 2015).
Since miRNAs can regulate their targets by base pairing with as few as six nucleotides or through non-canonical interactions, the prediction of miRNA targets from sequence alone is difficult. Crosslinking immunoprecipitation and sequencing (CLIP-seq) and similar methods have been used to identify AGO binding sites on RNAs (Chi et al., 2009; Hafner et al., 2010; Zisoulis et al., 2010). However, CLIP-based approaches do not identify the miRNA that is responsible for a given interaction. Recently, methods have been developed to capture the miRNA associated with specific target sites bound by AGO (Grosswendt et al., 2014; Helwak et al., 2013; Moore et al., 2015). These methods (CLASH, modified iPAR-CLIP, CLEAR-CLIP) involve similar procedures to isolate AGO complexes, induce the ligation of miRNAs to nearby target RNA sequences, and then prepare sequencing libraries. The hybrid reads produced by these methods are known as miRNA-target chimeras.
Here we report that individual-nucleotide resolution crosslinking immunoprecipitation (iCLIP) of endogenous AGO in C. elegans produces miRNA-target chimeric reads at similar efficiencies as methods designed to yield chimeras. Our analysis of thousands of reproducible miRNA-target chimeric reads unambiguously reveals the identity of the miRNA at AGO mediated target sites and points to features that promote target mRNA regulation in the endogenous context. We demonstrate the importance of interactions beyond seed pairing in specifying miRNA target sites using an endogenous in vivo reporter. Furthermore, we present a method for the identification of miRNA-target chimeras that does not require the use of radioactivity or the analysis of sequencing datasets.
RESULTS
ALG-1 iCLIP generates miRNA-target chimeras
In C. elegans, the Argonaute-Like Gene 1 (ALG-1) protein is essential for normal miRNA expression and function. To generate a more complete map of ALG-1 target sites, we carried out ALG-1 iCLIP in wild-type (WT) C. elegans animals at the last larval stage of development, as iCLIP has been shown to recover more unique cDNAs than traditional CLIP-seq (Sugimoto et al., 2012). We analyzed ALG-1 binding sites using the CLIPper peak-finding algorithm (Lovci et al., 2013) and identified 5,006 ALG-1 binding sites that were reproducible in at least two biological replicates (Table S1). 79.9% of these binding sites occurred in 3′UTRs (Figure S1A).
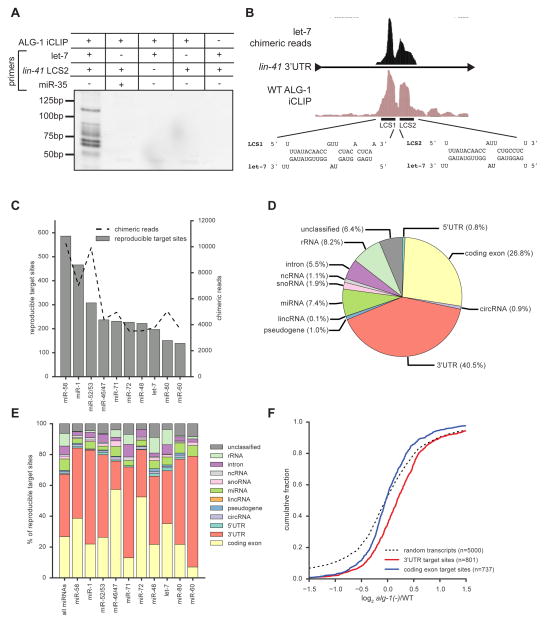
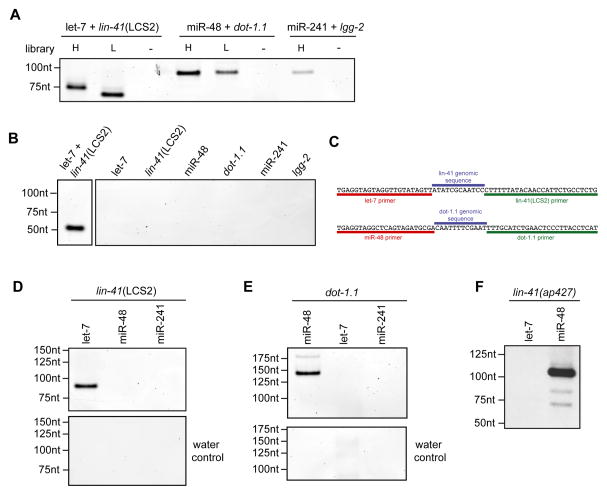
Chimera-generating methods have provided unambiguous miRNA targeting data in a variety of organisms and cell types (Grosswendt et al., 2014; Helwak et al., 2013; Moore et al., 2015). Interestingly, these miRNA-target chimeric reads have been reported to also occur in CLIP-seq and PAR-CLIP libraries, even without the addition of the biochemical steps intended to increase their frequency (Grosswendt et al., 2014). We tested for the presence of miRNA-target chimeras in our ALG-1 iCLIP libraries by PCR using primers for mature let-7 and the second let-7 complementary site (LCS2) in the 3′UTR of lin-41. PCR products were detected only for this well-established target interaction, and not for another miRNA and the same target site, or when using single primers (Figure 1A). This result shows that chimeric reads for authentic miRNA target sites occur in ALG-1 iCLIP and that these reads occur at a high enough frequency for PCR to detect.
Figure 1. ALG-1 iCLIP produces miRNA-target chimeric reads.
(A) The presence of miRNA-target chimeras in ALG-1 iCLIP libraries were tested by PCR using the indicated primers. (B) let-7 chimeric reads map to LCS1 and LCS2 in the lin-41 3′UTR. (C) The number of target sites and chimeric reads detected for the ten miRNAs with most target sites. (D) Distribution of target sites among transcript types. (E) Genic locations of target sites for the indicated miRNAs. (F) Up-regulation in alg-1(−) of transcripts with target sites only in 3′UTR (red) (Mann-Whitney U test, P<2×10–19) but not in coding exons (blue) (Mann-Whitney U test, P<0.35) in comparison to randomly selected transcripts. See also Figure S1 and Table S2.
To identify all of the miRNA-target chimeric reads in our ALG-1 iCLIP data, we developed a computational pipeline for their detection and mapping. We anticipated that low numbers of chimeric reads (<5000) might be recovered, as has been described for CLIP libraries that lack the intermolecular ligation step (Grosswendt et al., 2014; Moore et al., 2015). Remarkably, our analysis revealed 153,684 non-redundant chimeric reads that mapped to the C. elegans genome at 46,910 sites for 112 guide and 47 passenger strand miRNAs. Sites with at least two overlapping reads represented 20.6% of total sites. Non-redundant chimeric reads ranged from 1.3% to 5.1% of all reads from five independent ALG-1 iCLIP libraries. For comparison, ~2% of CLASH, ~0.2% of modified iPAR-CLIP, and ~1.5–5% of CLEAR-CLIP libraries consisted of chimeric reads (Grosswendt et al., 2014; Helwak et al., 2013; Moore et al., 2015). Initial analysis of the chimeric reads from ALG-1 iCLIP revealed that they map to known miRNA target sites. For example, let-7 chimeric reads map specifically to the two let-7 complement sites in the lin-41 3′UTR (Figure 1B). To assess the frequency of non-specific chimera formation in ALG-1 iCLIP, we mapped our chimeric reads to the E. coli genome. E. coli bacteria are the food source for C. elegans, and reads that map to the E. coli genome are commonly recovered in CLIP-based experiments (Grosswendt et al., 2014). <7% of total non-redundant miRNA-containing reads were ligated to E. coli sequences, indicating that nonspecific ligation events were rare. These analyses show that ALG-1 iCLIP produces miRNA-target chimeras with similar efficiency to methods specifically designed to generate chimeric reads. Moreover, these chimeric reads correctly match specific miRNAs to previously characterized miRNA target sites.
To explore the mechanism by which chimeric reads might have been generated, we examined the prevalence of full-length and 3′ truncated miRNAs in our chimeric read data. Previously, it was suggested that trimming of the miRNA 3′ end by RNase treatment may allow endogenous enzymes present in the lysate to ligate the miRNA to proximal target RNA sequence (Grosswendt et al., 2014). However, the majority of miRNA-target chimeras produced by ALG-1 iCLIP were composed of full-length miRNAs. The inclusion of truncated miRNAs increased chimera identification by ~20% (Figure S1B), whereas >90% of the chimeras were formed by shortened miRNAs in unmodified iPAR-CLIP. Since the majority of miRNA-target chimeric reads were composed of intact miRNAs, it is likely that most iCLIP chimeras form during the biochemistry used to produce chemical moieties compatible with linker ligation steps. However, since the inclusion of 3′ truncated miRNAs increased the identification of miRNA-target chimeras, it remains possible that the action of an endogenous ligase is responsible for a subset of chimeric reads. During our analysis of ALG-1 iCLIP chimeric reads, we noticed that many reads contained an untemplated nucleotide on the 5′ end of the miRNA. The inclusion of the untemplated nucleotide when searching for chimeric reads increased read identification by ~30% (Figure S1B). This nucleotide is primarily an adenosine or thymidine and is likely added during reverse transcription (Figure S1C). For our computational identification of miRNA-target chimeric reads, we included both 3′ truncated and 5′ untemplated nucleotide addition miRNA variations.
For all subsequent analyses we considered only the 4,920 chimera-producing sites that were reproducible in at least two biological replicates (Table S2), hereafter referred to as target sites. The miRNAs with the greatest number of target sites tended to be those that were highly expressed, as determined by the number of chimeric reads (Figure 1C). In addition, of the 20 miRNAs with the greatest number of target sites, 80% were identified as the top 20 highest expressed miRNAs at mid-L4 (Kato et al., 2009). Similarly, 83% of the guide strand miRNAs with reproducible target sites were previously shown to be associated with ALG-1 (Zisoulis et al., 2010) (Figure S1D). These experiments show that ALG-1 iCLIP generates reproducible miRNA-target chimeric reads that reveal the miRNA targeting landscape in C. elegans.
Targets identified by chimeras are misregulated in alg-1(−) animals
AGO proteins are generally guided by miRNAs to the 3′UTR of mRNAs (Pasquinelli, 2012). The majority of target sites identified in this study occurred in mRNAs and 40.5% of all target sites mapped to 3′UTRs (Figure 1D). Of these 3′UTR target sites, 87% overlapped an ALG-1 binding site in at least one ALG-1 iCLIP library (Figure S1E). We found relatively few target sites in introns (5.5%) compared to miRNA-target chimeras from human CLASH (15%) and mouse CLEAR-CLIP (36%) (Moore et al., 2015). However, both CLASH and CLEAR-CLIP considered clustered sites, whereas we considered only reproducible target sites. When we analyzed all chimeric reads (including non-reproducible) found in our ALG-1 iCLIP libraries, 16.4% of the reads mapped to introns (Figure S1F). The fewer miRNA intronic target sites observed when considering only reproducible sites suggests that these interactions are either unstable or occur infrequently.
Chimeras also formed with other mature miRNAs and snoRNAs. In addition, we observed target sites that mapped to published circRNAs (Ivanov et al., 2015; Memczak et al., 2013). Within mRNA sequences, individual miRNAs exhibited distinct binding patterns with some miRNAs having primarily 3′UTR (e.g., miR-71 and miR-60) or coding exon target sites (e.g., miR-46/47 and miR-72) (Figure 1E and Figure S1G).
In most cases, miRISC promotes transcript destabilization of bound targets (Eichhorn et al., 2014). Hence, loss of AGO proteins or deletion of specific miRNAs results in increased target mRNA abundance. To examine if the mRNAs containing target sites identified by ALG-1 iCLIP are up-regulated in alg-1 mutants, we performed RNA-seq on WT and alg-1(gk214) animals, referred to hereafter as alg-1(−). Compared to randomly selected genes, the mean change in expression was higher for genes with target sites in 3′UTRs but not coding exons (Figure 1F). Similarly in alg-1(−), only transcripts with ALG-1 binding sites in 3′UTRs were up-regulated (Figure S1H). This observation is consistent with previous microarray analysis of alg-1(−) animals that showed up-regulation primarily of transcripts with ALG-1 binding in 3′UTRs (Zisoulis et al., 2010).
Seed-pairing is enriched in miRNA target sites
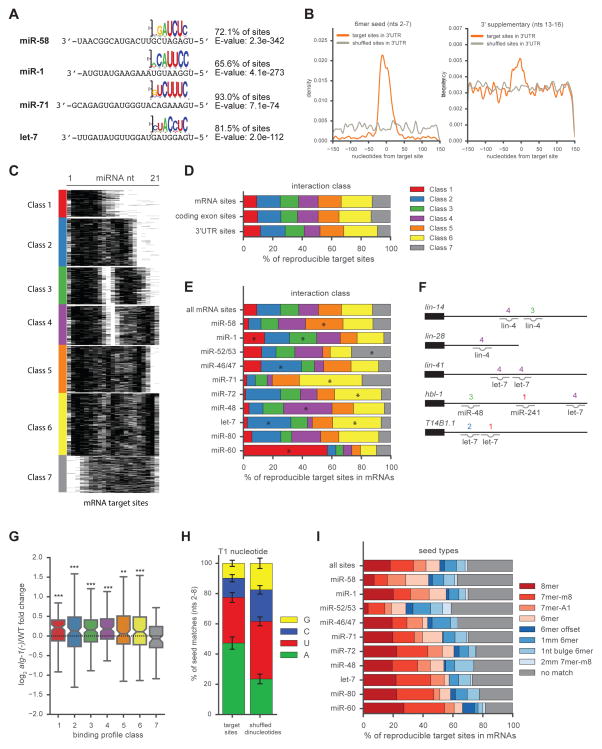
The miRNA sequence can be separated into five functional domains that affect miRNA target recognition: 5′ anchor (nt 1), seed sequence (nts 2–8), central region (nts 9–12), 3′ supplementary region (nts 13–16), and 3′ tail (nts 17–22) (Wee et al., 2012). We anticipated that complementarity to the seed sequence of the cognate miRNA would be a prominent feature in our target sites. However, it was also possible that the target sites identified by ALG-1 iCLIP would share a common sequence motif unrelated to the identity of the cognate miRNA. Using MEME motif analysis (Bailey and Elkan, 1994), the primary motif we identified was the seed complement for the cognate miRNA that was ligated to target sites of highly expressed miRNAs (Figure 2A). This demonstrates that the chimeras produced by ALG-1 iCLIP are dependent on the identity of the ligated miRNA. Complementarity to the 3′ supplementary region of the miRNA, in addition to the seed sequence, has been suggested to moderately enhance miRNA targeting (Grimson et al., 2007). To assess the prevalence of seed and 3′ supplementary pairing, we looked for enrichment of the complementary nucleotides to these miRNA domains in and around the target sites. For both seed and 3′ supplementary pairing, the presence of the complement to these sites is enriched at the target site compared to surrounding sequences (Figure 2B).
Figure 2. miRNA-target chimeric reads are enriched for seed pairing.
(A) The seed complement is the primary motif identified by MEME of targets for the indicated miRNAs. (B) Density plot showing enrichment of 6mer seed and 3′ supplementary complementarity to cognate miRNAs at 3′UTR target sites. (C) miRNA-target duplex structure predictions calculated by RNAhybrid and partitioned into seven classes by k-means clustering. Black pixels represent pairing. (D) Distribution of classes among all mRNA, coding exon or 3′UTR target sites. (E) Distribution of classes for the indicated miRNAs. Significantly enriched classes (one-sided Fisher’s exact test, P<0.001) are indicated (*). (F) Class interactions of established miRNA target sites. For sites with multiple miRNAs bound, the miRNA with the greatest number of chimeric reads at that site was chosen. (G) Up-regulation of transcripts in alg-1(−) for each interaction class (Mann-Whitney U test, *** P<0.001, ** P<0.005). (H) T1A is enriched after the seed complement for the ten miRNAs with the greatest number of target sites compared to sites with shuffled dinucleotides (Fisher’s exact test, P<0.0001). Error bars represent ±SEM. (I) Distribution of seed complements for all mRNA target sites and the indicated miRNAs. See also Figure S1.
To examine globally how miRNAs interact with their target sites in mRNAs, we paired each miRNA with its target site using RNAhybrid (Rehmsmeier et al., 2004) and grouped similar interactions by k-means clustering on the predicted miRNA-target duplexes. When k=7, six groups feature seed interactions and six feature 3′ interactions. (Figure 2C). Unlike chimeras from human miRNAs identified by CLASH, we did not detect a class of distributed interactions (Helwak et al., 2013). Our classes exhibit distinct pairing among the different miRNA functional domains. For example, Class 1 features seed-only pairing, whereas Class 3 and Class 4 exhibited seed pairing with 3′ supplemental pairing but no central pairing. Class 7 interactions did not exhibit seed pairing interactions, but instead contained complementarity to interactions throughout the other miRNA domains.
Categorization of the target sites found in coding exons and 3′UTRs did not reveal enrichment for a specific class in either genic region (Figure 2D). However, individual miRNAs exhibited enrichment for specific classes of interactions (Figures 2E and S1I). We detected differential enrichment for each of the seven classes among the miRNAs that produce the greatest number of target sites, including differences between members of the same miRNA family, such as let-7 and miR-48. Well-established miRNA targets sites, such as those in lin-14, lin-28, lin-41, and hbl-1, generally featured Class 3 and Class 4 interactions (Figure 2F).
To examine if there might be functional differences among the classes of pairing interactions, we examined the expression of transcripts with each class of interaction in their 3′UTRs in alg-1(−) animals compared to WT (Figure 2G). Among the seven classes, only the seedless Class 7 was not significantly up-regulated in comparison to randomly selected transcripts. Although it is possible that the Class 7 target sites primarily direct translational repression, it is striking that seed pairing seems to be broadly important for the regulation of target mRNA stability.
Another feature that has been associated with functional miRNA target sites is the presence of an adenosine immediately 3′ of the seed complement in the target RNA, known as T1A (Lewis et al., 2005). Human AGO contains a binding pocket that recognizes T1A, which likely functions to anchor the AGO protein at the target site (Schirle et al., 2015, 2014). T1A is over-represented in our chimera derived target sites compared to sites with shuffled dinucleotides (Figure 2H). This analysis lends genome-wide support for previous computational and structural work pointing to the importance of the identity of the nucleotide after seed pairing in the target sequence.
Although all but one class of miRNA-target interactions exhibited general seed pairing, many sites within these classes appeared to have imperfect seed matches. We examined the complementarity of target sites to their cognate miRNAs for various classes of seed matching. Among all mRNA target sites, ~50% of interactions included 6mer (nts 2–7), 7mer-m8 (nts 2–8), 7mer-A1 (nts 2–7 with T1A), or a 8mer (nts 2–8 with T1A) seed interaction with the cognate miRNA, whereas ~20% of sites included a 6mer-offset (nts 3–8), mismatch, or bulge seed interaction and ~30% of sites featured no match to an established seed type (Figure 2I). Taken together, these findings support the importance of seed pairing in miRNA function and reveal that the majority of miRNA target sites support additional 3′ end complementary interactions.
MiRNA family members bind specific sets of target sites
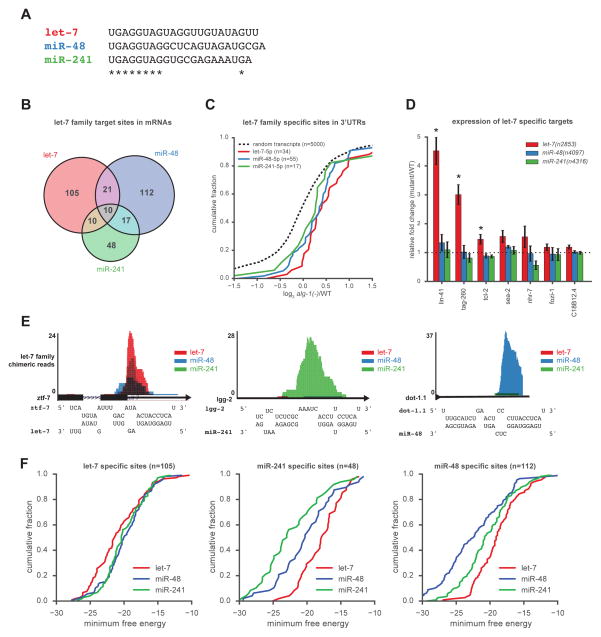
Considering that seed pairing has been proposed to be not only necessary but also sufficient for miRNA targeting (Doench and Sharp, 2004; Lewis et al., 2005, 2003) and that miRNA families in C. elegans may act redundantly (Alvarez-Saavedra and Horvitz, 2010), we predicted that miRNA family members would bind largely overlapping sets of targets. To investigate this possibility, we examined target sites for the let-7 family of miRNAs. Three of the let-7 family of miRNAs, let-7, miR-48, and miR-241, are expressed during the last larval stage in overlapping sets of tissues (Abbott et al., 2005; Kato et al., 2009; Martinez et al., 2008). Additionally, miR-48 and miR-241 are processed from the same primary transcript. Whereas the first eight nucleotides of let-7, miR-48, and miR-241 are identical, the rest of their sequences diverge (Figure 3A). Surprisingly, the chimeras formed by let-7, miR-48, and miR-241 revealed that the majority of their target sites were non-overlapping (Figure 3B).
Figure 3. The let-7 family of miRNAs binds divergent sets of target sites.
(A) Mature sequences of the three most abundant let-7 family miRNAs. (B) Overlap of target sites for let-7 family miRNAs. (C) Transcripts specifically bound by single let-7 family miRNAs in 3′UTRs are up-regulated in alg-1(−) compared to random transcripts (Mann-Whitney U test: let-7 targets P<1.3×10- 5, miR-48 targets P<4.4×10-5, miR-241 targets P<0.18). (D) qRT-PCR of let-7 specific target candidates in the indicated strains (* P<0.05). Error bars represent ±SEM. (E) Examples of shared and specific let-7 family targets with predicted pairing interactions. (F) RNAhybrid analysis of minimum free energy of miRNA-target duplex for each let-7 family member to its specific mRNA target sites. See also Figure S2 and S3.
Target sites that were shared by multiple let-7 family members included the established let-7 family targets, daf-12 and hbl-1 (Abbott et al., 2005; Grosshans et al., 2005) (Figure S2A and B). Along the 3′UTRs of both daf-12 and hbl-1, multiple let-7 family members share some target sites, but other target sites are specific or highly biased for binding to a single family member. The general binding preferences of these sites agree with early observations that some let-7 family members are predicted to pair more favorably with specific complementary sites in the hbl-1 3′UTR (Lin et al., 2003).
In general, transcripts bound by a single let-7 family member tended to be regulated by the miRNA pathway. We found that transcripts targeted by an individual let-7 family miRNA in their 3′UTR were significantly up-regulated in alg-1(−) animals (Figure 3C). Although the majority of let-7 target sites did not produce chimeras with other let-7 family members (Figure 3B), most of the transcripts containing these let-7 specific sites produced chimeras or strong peaks representing ALG-1 binding at additional locations. While these observations suggest combinatorial regulation, we were still able to detect specific misregulation of some of these targets in animals deficient for let-7 activity; whereas these same targets were not up-regulated in miR-48 or miR-241 null strains (Figure 3D). We were unable to detect targets for miR-48 and miR-241 that appeared to be specifically misregulated, likely because both miR-48 and miR-241 are significantly up-regulated in miR-241 and miR-48 deletion strains, respectively (Figure S3A and B), unlike in let-7 mutant animals (Figure S3C and D).
Some let-7 target sites, such as the established site in ztf-7 (Jovanovic et al., 2010), are shared by multiple let-7 family members, whereas others produced chimeric reads almost exclusively with a specific member (Figure 3E). Since nucleotides 1–8 of let-7, miR-48, and miR-241 are identical, other sequences in these miRNAs must dictate specific target interactions. With the limited number of specific sites for each let-7 family miRNA, we did not detect a miRNA region common to all let-7 family members that would be responsible for exclusive interactions. Instead, the overall binding energy is more favorable for each miRNA and its cognate sites than for other family members paired to those sites. We hybridized each let-7 family member to specific target sites using RNAhybrid to determine the minimum free energy of the miRNA-target duplex. In general, pairing was most favorable for the let-7 family member with its experimentally defined set of specific target sites (Figure 3F).
Specific binding by miRNA family members appears to be common for other miRNAs. The miR-58 and miR-238 families of miRNAs are expressed at mid-L4 (Kato et al., 2009) and also have divergent 3′ ends (Figure S3E and F). For both of these families, we found primarily specific target sites with similar patterns of favorable pairing as those observed for the let-7 family (Figure S3G–J). We found that shared sites were more likely to contain perfect seed matches than specific sites (Figure S3K), but a strong bias for T1A in shared sites versus specific sites was not detected (Figure S3L). These analyses reveal that miRNA family members can exhibit divergent target interactions and that these preferences likely arise from 3′ end pairing of the miRNA to its target site.
MiRNA 3′ end pairing directs specific target interactions
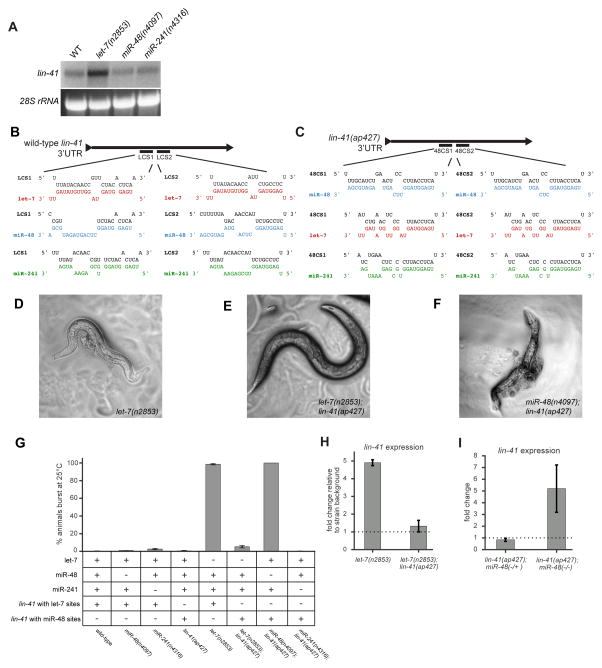
Our detection of chimeras specific for let-7 in the lin-41 3′UTR is consistent with the requirement for let-7-mediated regulation of lin-41. Loss of let-7 results in a lethal phenotype where animals burst through their vulvas (Reinhart et al., 2000; Slack et al., 2000), and this phenotype can be suppressed by restoring regulation of just lin-41 (Ecsedi et al., 2015). Additionally, versions of let-7 that contain mutations in 3′ end sequences do not fully rescue bursting of let-7 mutants, pointing to the importance of the 3′ end of this miRNA (Zhang et al., 2015). To further test if regulation of lin-41 is entirely dependent on let-7, we analyzed the expression of lin-41 in let-7(n2853) animals and in null mutants of miR-48 and miR-241, at mid-L4 stage. Compared to WT, the levels of lin-41 were misregulated only in let-7(n2853) (Figure 4A).
Figure 4. Seed pairing is not sufficient for target regulation in vivo.
(A) Northern blot for lin-41 in the indicated strains. (B–C) Diagram with binding profiles for let-7, miR-48, and miR-241 pairing to sites in WT lin-41 and lin-41(ap427) where the let-7 complementary sites (LCS) have been switched miR-48 complementary sites (48CS). (D) let-7(n2853) animals burst through the vulva at 25°C. (E) Suppr ession of let- 7(n2853) bursting by lin-41(ap427). (F) lin-41(ap427);miR-48(n4097) double mutants phenocopy let-7(n2853) vulval bursting. (G) Quantification of bursting in the presence (+) and absence (−) of the indicated gene products. Error bars represent ±SEM (H) qRT-PCR of lin-41 in let-7(n2853) and lin-41(ap427);let-7(n2853), normalized to WT and lin-41(ap427) strains, respectively. Error bars represent ±SEM. (I) Single worm qRT-PCR of lin-41 in lin-41(ap427);miR-48(n4097) strains heterozygous or homozygous for the miR-48 deletion, normalized to lin-41(ap427);miR-48(+/+). Error bars represent ±SEM.
The imperfect seed pairing of LCS1 and LCS2 in lin-41 likely necessitates strong miRNA 3′ end interactions, which are much more favorable for let-7 than for miR-48 or miR-241. However, our chimera data indicated that some target sites are capable of perfect seed pairing (up to 8mer) to any of the let-7 family members yet appear to be exclusively bound by a single member (Figure S3K). To test the functional importance of specific miRNA targeting in vivo, we decided to replace LCS1 and LCS2 in the lin-41 3′UTR (Figure 4B) with a target site that was specific for another let-7 family member. We reasoned that the altered specificity of this site might switch the identity of the miRNA required to regulate lin-41. To minimally perturb the endogenous context, we used CRISPR/Cas9 genome editing and homologous recombination methods to introduce two copies of the miR-48 specific target site from the dot-1.1 3′UTR (Figure 3E) at the same positions as LCS1 and LCS2 in the lin-41 3′UTR. The dot-1.1 miR-48 site has 7mer-A1 seed pairing with a GU-wobble at the nucleotide 8 position and more extensive 3′ end complementarity to miR-48 than to let-7 or miR-241 (Figure 4C). Importantly, all let-7 family members are expected to be capable of interacting with the dot-1.1 site through seed mediated interactions. Thus, if seed pairing is sufficient for regulation, this version of lin-41 should no longer be dependent on any one let-7 family miRNA. Animals harboring the mutant lin-41(ap427) allele, which has the miR-48 specific sites in the lin-41 3′UTR, display no observable phenotypes, suggesting that lin-41 is sufficiently regulated in these animals.
The let-7(n2853) mutation results in a single nucleotide change to the seed sequence of mature let-7 and decreased levels of mature let-7 (Figure S3C and D). At 25°C, let-7(n2853) animals burst through the vulva and die due to the specific misregulation of lin-41 (Figure 4A and D). Remarkably, when we combined the let-7(n2853) mutation with lin-41(ap427), vulval bursting was suppressed, indicating that regulation of this edited version of lin-41 is no longer dependent on let-7 (Figure 4E). We next attempted to generate a double mutant with lin-41(ap427) and miR-48(n4097), a deletion allele of the miR-48 miRNA. However, we were unable to generate a strain of homozygous double mutants because all animals burst through the vulva and died once they reached the L4 stage, regardless of culture temperature (Figure 4F). Finally, we generated lin-41(ap427);miR-241(n4316) double mutants, which had no observable phenotypes.
To quantify the dependence of lin-41(ap427) regulation on each let-7 family member, we conducted bursting assays at 25°C ( Figure 4G). WT, miR-48 null, miR-241 null, and lin-41(ap427) animals did not burst at 25°C, whereas ~98% of let-7(n2853) animals burst and died. However, only ~7% of these let-7 mutants burst in the lin-41(ap427) background. As described above, a lin-41(ap427);miR-48(n4097) strain could not be isolated due to bursting and lethality of all double mutants, as confirmed by genotyping of the corpses. For lin-41(ap427);miR-241 null animals, there was no detectable bursting. The apparent miRNA specific regulation of WT lin-41 by let-7 and lin-41(ap427) by miR-48 is not due to down-regulation of other let-7 family members in the miRNA mutant backgrounds (Figure S3A and B).
Consistent with the phenotypes, the misregulation of lin-41 mRNA levels in let-7(n2853) was prevented in the let-7(n2853);lin-41(ap427) strain (Figure 4H). Instead, expression of lin-41 in the edited strain was found to be strongly misregulated in the absence of miR-48, as detected by single worm qRT-PCR (Figure 4I). Thus, the potential for pairing to miRNA 3′ end sequences drives miRNA specific regulation at the molecular and phenotypic levels, which in the case of miR-48 regulation of lin-41(ap427) is not compensated by the presence of let-7 or miR-241. Taken together, these experiments reveal the importance of miRNA 3′ end interactions in dictating target specificity among miRNA family members, even when targets share a canonical seed complement.
Simplified detection of endogenous miRNA target interactions by Chimera PCR (ChimP)
Although several groups have developed chimera-generating methods that can be applied in a variety of model systems, each of these protocols requires the use of radioactivity and complex sequencing data analysis. As a consequence, these methods are impractical for many research groups, which may be interested in a single miRNA or target. Considering that miRNA-target chimeras were detectable in standard iCLIP libraries using PCR (Figure 1A), we developed a method to facilitate the detection of miRNA-target chimeras by PCR. This method, called Chimera PCR (ChimP), allows the detection of miRNA-target chimeras without the use of radioactivity or the need for complex sequencing analyses (Figure S4A).
In brief, the ChimP protocol is similar to other chimera-generating methods that include an intermolecular ligation step to ligate miRNAs to their target RNAs (Grosswendt et al., 2014; Helwak et al., 2013; Moore et al., 2015). However, ChimP does not require radioactive tagging and isolation of RNA from a membrane. Instead, AGO-miRNA-target RNA tertiary complexes are treated with Proteinase K to isolate the RNAs. Libraries are then generated in a similar manner to standard iCLIP. The resulting library can then be used as the template in a PCR reaction using an oligonucleotide with the miRNA sequence as the forward primer and an oligonucleotide complementary to the target RNA as the reverse primer.
We used ChimP to confirm the miRNA-target chimeras for let-7 and lin-41, miR-48 and dot-1.1, and miR-241 and lgg-2, which were originally detected in our ALG-1 iCLIP reads (Figure 5A). To control for the possibility that the PCR products produced by ChimP were the result of amplification from a single primer, we also performed single primer controls to demonstrate that primers for both the miRNA and target sequence are required (Figure 5B). Furthermore, we cloned and sequenced the PCR products for let-7 and lin-41 and miR-48 and dot-1.1 (Figure 5C). In both cases, the miRNA-target chimera contained the sequence of the two primers used for amplification separated by a small sequence that originated from the target RNA. This demonstrates that ChimP is capable of producing bona fide miRNA-target chimeras.
Figure 5. Chimera PCR (ChimP) enables the identification of miRNA-target chimeras by PCR.
(A) Detection of let-7 family chimeras using ChimP. Libraries were generated using higher (H) and lower (L) molecular weight cDNAs. (B) Single primer negative controls along with a let-7 + lin-41(LCS2) positive control from another part of the gel. (C) Examples of sequenced ChimP products for let-7 and lin-41 LCS2, and miR-48 and dot-1.1. (D) Detection of the lin-41 LCS2 with let-7 but not with other let-7 family member primers by ChimP. (E) Specific detection of dot-1.1 with a primer for miR-48. (F) The miR-48 complement sites in the lin-41(ap427) 3′UTR interact with miR-48 and not let-7. See also Figure S4.
To test the fidelity of ChimP, we asked whether it could distinguish between miRNA family specific target sites. For both, lin-41 and dot-1.1, we were only able to detect miRNA-target chimeras for let-7 and miR-48, respectively (Figure 5D and E). To further demonstrate the utility of ChimP, we also applied our method to detect miR-71 target sites to the alg-1 and C44F1.1 3′UTRs that had been identified from our chimeric data (Figure S4B and C) and include examples of the biological reproducibility of ChimP (Figure S4D and E).
In some cases, we noted that primers generated products in the minus template control PCR reactions. Sequencing of these products showed that they were primer-dimers that amplified due to overlapping end complementarity. Furthermore, on occasion we found that low annealing temperatures, particularly for miRNAs with low GC content, or when library amplification primers were not fully removed from the library, led to the amplification of non-specific products, such as miRNA-rRNA reads. As a consequence, we recommend including no template control reactions as well as cloning and Sanger sequencing of fragments in pilot experiments employing ChimP.
Using ChimP, we were able to demonstrate that the let-7 specific binding of lin-41 switches to miR-48 specific binding in the edited lin-41(ap427) strain (Figure 5D and F). These results further illustrate the utility of ChimP and show that specific binding of a miRNA target can be determined by miRNA 3′ end sequences. Overall, these analyses demonstrate that ChimP is a versatile method to rapidly test for the presence of endogenous miRNA-target interactions. With this assay, researchers can avoid laborious and computationally intensive CLIP-based chimera-producing experiments when specific miRNAs and potential target sites are of interest.
DISCUSSION
The initial aim of this study was to refine the catalog of AGO binding sites using iCLIP. Our serendipitous discovery that ALG-1 iCLIP produces miRNA-target chimeras led to the most comprehensive map of unambiguous miRNA target sites in C. elegans to date. Investigation of this reproducible dataset of endogenous miRNA target sites provided insights into functional target interactions in a living animal. On a genome-wide scale, we found that miRNA-target interactions associated with regulatory outcomes generally involve seed pairing, an adenosine at the T1 position, and binding sites in mRNA 3′UTRs. Moreover, we were surprised to observe that miRNA families with divergent 3′ ends target largely distinct sets of sites. We demonstrated that pairing to the miRNA 3′ end not only provides specificity but can also be essential for target regulation fidelity in vivo. Finally, we developed ChimP to allow for the detection of chimeric reads by PCR, and anticipate that this will be a widely accessible method for interrogating potential miRNA-target site interactions.
Identification and validation of endogenous miRNA binding sites
The target sites identified by ALG-1 iCLIP chimeras represent endogenous miRNA-target interactions that occur at mid-L4. Presently, it is unclear why we recovered a larger fraction of chimeric reads in our iCLIP libraries (~1.3–5.1%) than did a previous directed attempt to form chimeras in C. elegans (~0.2%) (Grosswendt et al., 2014). Of the >150,000 miRNA-target chimeric reads generated by ALG-1 iCLIP, we conservatively used only those that were reproducible in at least two biological replicates. This requirement focused our studies on high-confidence sites and helped to potentially eliminate off-target or transient binding events.
Although ALG-1 iCLIP produces miRNA-target chimeric reads with similar efficiencies as CLEAR-CLIP (Moore et al., 2015) and CLASH (Helwak et al., 2013), and more efficiently than modified iPAR-CLIP (Grosswendt et al., 2014), the biochemical steps were not specifically designed to promote the optimal 5′- and 3′-end chemistry that is required for chimera formation. It has been proposed that chimeric reads that form in CLIP-seq and PAR-CLIP libraries occur due to the action of an endogenous ligase present in the lysates (Grosswendt et al., 2014). In the CLIP-seq and PAR-CLIP libraries analyzed by Grosswendt et al. 2014, the majority of the miRNA sequences in chimeras were truncated at the 3′-end, likely by the RNase used to trim unprotected RNA fragments. However, full-length miRNA sequences account for the majority of miRNA-target chimeric reads in ALG-1 iCLIP data. Furthermore, our analysis of AGO-2 iCLIP data, where the 3′-linker was ligated after RNA isolation (Bosson et al., 2014), found almost no miRNA-target chimeric sequences (data not shown). This suggests that the T4 RNA ligase used to ligate the 3′-linker is responsible for the efficiency seen in ALG-1 iCLIP.
We have also demonstrated that ChimP can be used to identify miRNA-target sites for specific miRNA-target interactions of interest. An advantage of ChimP is the ability to detect chimeric events without the use of radioactivity or the bioinformatics expertise required to identify chimeras from CLIP-based methods. ChimP is sensitive enough to reproduce the specificity seen for the let-7 family miRNA targets observed in this study. Although ChimP does not identify the miRNA-target landscape across the transcriptome, it allows for the investigation of specific interactions that may be of interest to laboratories focused on certain miRNAs and targets.
Features of endogenous miRNA targeting
By capturing the endogenous miRISC, we were able to examine miRNA-target interactions in vivo without changing the stoichiometry between ALG-1, the miRNAs, and target RNAs. Perfect complementarity to at least a 6mer seed sequence was found in the majority of target sites. Interestingly, we did notice that the frequency of different types of seed interactions depended on the identity of the miRNA. For example, miR-60 is highly enriched for Class 1 pairing interactions, which involve complementarity to only the 5′ end of the miRNA, and has greater potential for perfect seed pairing than any of the other most abundant miRNAs (Figure 2E and I). This pattern could be related to the relatively low GC content of the miR-60 5′ region, which might then be compensated for by strong seed pairing interactions to stabilize miRISC binding.
Similar to the mammalian target sites identified by CLEAR-CLIP and CLASH (Helwak et al., 2013; Moore et al., 2015), we also observed extensive predicted pairing to the 3′ end of the miRNA, in addition to seed pairing. We identified seven classes of base pairing interactions between miRNAs and mRNAs, six of which featured various degrees of miRNA 3′ end base pairing. Additionally, target sites were enriched for T1A, consistent with predicted features of miRNA target sites (Grimson et al., 2007; Lewis et al., 2005) and structural evidence that AGO contains a binding pocket for adenosine in the T1 position (Schirle et al., 2015, 2014).
In addition to distinct classes of pairing interactions being associated with different degrees of target regulation, the location of the target site also seems to be important for functional targeting. Similar to previous observations (Zisoulis et al., 2010), we noted that transcripts with target sites in coding exons were less up-regulated in alg-1(−) animals than those with target sites in 3′UTRs. It is possible that some of these are regulated primarily at the level of translational inhibition with no detectable mRNA destabilization. Alternatively, competition between miRISC and translating ribosomes may reduce the residence time of AGO at exonic sites, thwarting effective regulation of the mRNA (Gu et al., 2009). However, not only were many target sites in coding exons reproducible but some gave rise to abundant chimeras, suggesting that these interactions are unusually stable. We noticed that some of these target sites overlap with published circRNAs for C. elegans (Ivanov et al., 2015; Memczak et al., 2013). Thus, it is possible that some of these stable, chimera generating target sites are derived from miRISC interactions with circRNAs and not the mRNA. In addition, circRNAs in Drosophila have been reported to be enriched for conserved miRNA seed complement sites, which further suggests that some circRNAs are bound by miRNAs (Westholm et al., 2014). One hypothesis is that these sites act as sponges to absorb excess miRNA load (Tay et al., 2014); however, other work has demonstrated that many circRNAs and other potential competing endogenous RNAs, due to their low expression, are not capable of functioning as miRNA sponges that titrate miRISC from mRNA targets (Bosson et al., 2014; Denzler et al., 2014). Thus, it remains to be determined what role coding-exon sites play or whether the substrates for AGO binding of this class are linear or circular RNA.
Target sites identified by ALG-1 iCLIP and other chimera-generating methods have also mapped to ncRNAs (Grosswendt et al., 2014; Helwak et al., 2013; Moore et al., 2015). Some of these interactions may represent novel functions for AGO, whereas others may arise from background ligation to highly expressed cellular RNAs. We anticipate that interactions between miRNAs and ncRNAs that are identified by chimera formation with a single miRNA are more likely to represent functional interactions (Figure S5A) than sites that are bound by many unrelated miRNAs (Figure S5B). It remains unclear, outside of base pairing specificity, why some miRNAs seem to have preferences for particular types of ncRNA interactions. For example, miR-46/47 has twice as many snoRNA target sites, compared to all other expressed miRNAs, whereas miR-71 is devoid of snoRNA interactions (Figure 1E).
Specificity role for miRNA 3′ ends
Since members of a miRNA family typically have identical 5′ sequences but divergent 3′ ends, they provide an ideal source for assessing contributions of the 3′ supplementary region to specificity and function. Unexpectedly, specific targeting by miRNA family members seems to be much more common than anticipated and exists even in the presence of strong seed complementarity. For specific target sites, the miRNA that is bound to those sites is predicted to be more thermodynamically stable than other miRNAs of the same family. This suggests that base pairing interactions, beyond the seed sequence, are responsible for miRNA targeting specificity.
Target sites with weak seed sequence complementarity, such as those with bulged or mismatched nucleotides, are thought to be compensated by more extensive interactions with the 3′ end of the miRNA (Brennecke et al., 2005; Grimson et al., 2007). As a consequence, sites with a weak seed may be more likely to be regulated by specific miRNA family members. However, single cell reporters have demonstrated that some sites with 6mer seed or 7mer-m8 seed with 3′ supplementary complementarity can direct miRNA family specific regulation (Moore et al., 2015). These experiments suggested the possibility that these interactions alone could be functionally relevant.
To test whether miRNA 3′ end interactions can direct functional specificity when multiple miRNA family members are present at endogenous levels, we engineered an in vivo reporter based on the regulation of lin-41. Normally, lin-41 is repressed solely by let-7 via two sites in its 3′UTR (Ecsedi et al., 2015; Slack et al., 2000; Vella et al., 2004). By editing those sites to become miR-48 target sites, we were able to transfer regulation of endogenous lin-41 to miR-48. In the miR-48 null background, animals with the edited lin-41 3′UTR phenocopy let-7 null strains with completely penetrant busting and lethality (Reinhart et al., 2000). Importantly, the new miR-48 sites retain perfect seed pairing to any of the let-7 family members, yet only miR-48 appears capable of binding, as demonstrated by ChimP, and regulating this version of lin-41. Altogether, this work shows that miRNA family members have many distinct targets and demonstrates that targeting specificity among miRNA family members can have functional consequences in vivo. However, the functional importance of miRNA family member-specific interactions, outside of the context of the let-7 family and the lin-41 3′UTR, remains an important question.
Crystal structures of AGO have revealed that when bound to the miRNA alone, only nucleotides 2–6 of the miRNA seed sequence are positioned to interact with target RNAs (Elkayam et al., 2012; Nakanishi et al., 2012; Schirle and MacRae, 2012). However, when the complex includes a complement RNA, AGO undergoes a conformational change that allows for full seed pairing (nts 2–8) and potentially exposes the miRNA 3′ supplementary region (nts 13–16) for additional pairing interactions (Schirle et al., 2014). Our data suggest that these suspected conformational changes allow miRNAs capable of 3′ end pairing interactions to outcompete miRNAs that support only seed pairing for a given site. Although in vitro studies have concluded that base pairing beyond the seed does not increase the affinity of AGO for a target site (Chandradoss et al., 2015; Jo et al., 2015; Salomon et al., 2015), it seems reasonable that target recognition in the more complex endogenous context could utilize additional base pairing interactions for specific and functional interactions in vivo.
Here we have reported the identification of thousands of examples of endogenous miRNA target sites in an intact organism. This work expands the dataset of experimentally captured miRNA-target interactions, providing a rich resource for improving target predictions and our understanding of miRNA targeting in vivo. For laboratories interested in select miRNA target sites, ChimP can be used to screen potential target interactions without having to analyze complex sequencing data. Our genome-wide analysis of chimeras formed by endogenous miRNA-target interactions revealed that pairing to the miRNA 3′ end provides a degree of specificity not previously considered in most target prediction methods. As we have shown, the ability of a miRNA to recognize more than just seed sequences shared by all members of a family can have important consequences in vivo. The specificity provided by the miRNA 3′ end may be especially relevant in humans where ~60% of miRNAs are part of a miRNA family (Kozomara and Griffiths-Jones, 2011). Specific miRNA family members are reportedly involved in numerous pathologies (Boyerinas et al., 2010), potentially because they have distinct targets that are not sufficiently regulated by the other family members. Overall, our results support the importance of seed pairing for functional miRNA-target interactions, but also reveal that this core motif might not always be sufficient. Instead, additional interactions with the miRNA 3′ end may be necessary for specific targeting in the endogenous context.
EXPERIMENTAL PROCEDURES
Nematode strains
Strains used and generation of lin-41(ap427) are described in the Supplemental Experimental Procedures.
ALG-1 iCLIP
ALG-1 iCLIP was performed as previously described (Broughton and Pasquinelli, 2013), using mid-L4 WT (N2) C. elegans animals grown at 25°C for 29 hours after L1 synchronization. Computational identification of ALG-1 binding sites and identification of chimeric reads is described in the Supplemental Experimental Procedures.
Supplementary Material
Highlights.
AGO iCLIP miRNA-target chimeras reveal miRNA targeting landscape in C. elegans
miRNA families target non-overlapping sets of target sites
miRNA 3′ end interactions contribute to target site specificity in vivo
Chimera PCR (ChimP) identifies specific miRNA target sites of interest
Acknowledgments
We thank G. Pratt for assistance with software development and members of the Pasquinelli Lab for critical reading of the manuscript. Some strains were provided by the C. elegans Genetics Center (CGC), which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). Support for this study was provided by a NSF-GRFP fellowship (DGE-1144086) (J.B.) and a UCSD Eureka Scholarship (J.H.). This work was supported by grants from NIH to G.Y. (HG004659, NS075449) and A.P. (GM071654).
Footnotes
ACCESSION NUMBERS
The sequencing data have been deposited in the SRA database under the accession numbers: SRP078361 (ALG-1 iCLIP) and SRP078368 (RNA-seq).
AUTHOR CONTRIBUTIONS
J.B. designed and carried out all experiments with assistance from J.H. Computational experiments were carried out by J.B. and M.L. G.Y. provided advice in experimental design and analyses. A.P. conceived and supervised the project. J.B. wrote the paper and all authors contributed to the final version.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abbott AL, Alvarez-Saavedra E, Miska Ea, Lau NC, Bartel DP, Horvitz HR, Ambros V. The let-7 MicroRNA family members mir-48, mir-84, and mir-241 function together to regulate developmental timing in Caenorhabditis elegans. Dev Cell. 2005;9:403–14. doi: 10.1016/j.devcel.2005.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4:1–38. doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Saavedra E, Horvitz HR. Many Families of C. elegans MicroRNAs Are Not Essential for Development or Viability. Curr Biol. 2010;20:367–373. doi: 10.1016/j.cub.2009.12.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosson AD, Zamudio JR, Sharp PA. Endogenous miRNA and Target Concentrations Determine Susceptibility to Potential ceRNA Competition. Mol Cell. 2014;56:347–359. doi: 10.1016/j.molcel.2014.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyerinas B, Park SM, Hau A, Murmann AE, Peter ME. The role of let-7 in cell differentiation and cancer. Endocr Relat Cancer. 2010;17:F19–36. doi: 10.1677/ERC-09-0184. [DOI] [PubMed] [Google Scholar]
- Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-target recognition. PLoS Biol. 2005;3:0404–0418. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broughton JP, Pasquinelli AE. Identifying Argonaute binding sites in Caenorhabditis elegans using iCLIP. Methods. 2013;63:119–25. doi: 10.1016/j.ymeth.2013.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandradoss SD, Schirle NT, Szczepaniak M, MacRae IJ, Joo C. A Dynamic Search Process Underlies MicroRNA Targeting. Cell. 2015;162:96–107. doi: 10.1016/j.cell.2015.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460:479–86. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denzler R, Agarwal V, Stefano J, Bartel DP, Stoffel M. Assessing the ceRNA hypothesis with quantitative measurements of miRNA and target abundance. Mol Cell. 2014;54:766–76. doi: 10.1016/j.molcel.2014.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench JG, Sharp PA. Specificity of microRNA target selection in translational repression. Genes (Basel) 2004;504:504–511. doi: 10.1101/gad.1184404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecsedi M, Rausch M, Großhans H. The let-7 microRNA directs vulval development through a single target. Dev Cell. 2015;32:335–44. doi: 10.1016/j.devcel.2014.12.018. [DOI] [PubMed] [Google Scholar]
- Eichhorn SW, Guo H, McGeary SE, Rodriguez-Mias RA, Shin C, Baek D, Hsu SH, Ghoshal K, Villén J, Bartel DP. mRNA Destabilization Is the Dominant Effect of Mammalian MicroRNAs by the Time Substantial Repression Ensues. Mol Cell. 2014;56:104–115. doi: 10.1016/j.molcel.2014.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkayam E, Kuhn CD, Tocilj A, Haase AD, Greene EM, Hannon GJ, Joshua-Tor L. The structure of human argonaute-2 in complex with miR-20a. Cell. 2012;150:100–10. doi: 10.1016/j.cell.2012.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman RC, Farh KK-H, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimson A, Farh KKH, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosshans H, Johnson T, Reinert KL, Gerstein M, Slack FJ. The temporal patterning microRNA let-7 regulates several transcription factors at the larval to adult transition in C. elegans Dev Cell. 2005;8:321–30. doi: 10.1016/j.devcel.2004.12.019. [DOI] [PubMed] [Google Scholar]
- Grosswendt S, Filipchyk A, Manzano M, Klironomos F, Schilling M, Herzog M, Gottwein E, Rajewsky N. Unambiguous Identification of miRNA:Target Site Interactions by Different Types of Ligation Reactions. Mol Cell. 2014;54:1042–1054. doi: 10.1016/j.molcel.2014.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu S, Jin L, Zhang F, Sarnow P, Kay Ma. Biological basis for restriction of microRNA targets to the 3′ untranslated region in mammalian mRNAs. Nat Struct Mol Biol. 2009;16:144–50. doi: 10.1038/nsmb.1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jungkamp AC, Munschauer M, Ulrich A, Wardle GS, Dewell S, Zavolan M, Tuschl T. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–41. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helwak A, Kudla G, Dudnakova T, Tollervey D. Mapping the human miRNA interactome by CLASH reveals frequent noncanonical binding. Cell. 2013;153:654–65. doi: 10.1016/j.cell.2013.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov A, Memczak S, Wyler E, Torti F, Porath HT, Orejuela MR, Piechotta M, Levanon EY, Landthaler M, Dieterich C, Rajewsky N. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep. 2015;10:170–7. doi: 10.1016/j.celrep.2014.12.019. [DOI] [PubMed] [Google Scholar]
- Jo MH, Shin S, Jung SR, Kim E, Song JJ, Hohng S. Human Argonaute 2 Has Diverse Reaction Pathways on Target RNAs. Mol Cell. 2015;59:117–24. doi: 10.1016/j.molcel.2015.04.027. [DOI] [PubMed] [Google Scholar]
- Jovanovic M, Reiter L, Picotti P, Lange V, Bogan E, Hurschler Ba, Blenkiron C, Lehrbach NJ, Ding XC, Weiss M, Schrimpf SP, Miska Ea, Grosshans H, Aebersold R, Hengartner MO. A quantitative targeted proteomics approach to validate predicted microRNA targets in C. elegans. Nat Methods. 2010;7:837–42. doi: 10.1038/nmeth.1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, De Lencastre A, Pincus Z, Slack FJ. Dynamic expression of small non-coding RNAs, including novel microRNAs and piRNAs/21U-RNAs, during Caenorhabditis elegans development. Genome Biol. 2009;10:R54. doi: 10.1186/gb-2009-10-5-r54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozomara A, Griffiths-Jones S. MiRBase: Integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39:152–157. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Shih I, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–98. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Weinstein EG, Abdelhakim A, Yekta S, Rhoades MW, Burge CB, Bartel DP. The microRNAs of Caenorhabditis elegans. Genes Dev. 2003;17:991–1008. doi: 10.1101/gad.1074403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SY, Johnson SM, Abraham M, Vella MC, Pasquinelli A, Gamberi C, Gottlieb E, Slack FJ. The C. elegans hunchback homolog, hbl-1, controls temporal patterning and is a probable MicroRNA target. Dev Cell. 2003;4:639–650. doi: 10.1016/s1534-5807(03)00124-2. [DOI] [PubMed] [Google Scholar]
- Lovci MT, Ghanem D, Marr H, Arnold J, Gee S, Parra M, Liang TY, Stark TJ, Gehman LT, Hoon S, Massirer KB, Pratt Ga, Black DL, Gray JW, Conboy JG, Yeo GW. Rbfox proteins regulate alternative mRNA splicing through evolutionarily conserved RNA bridges. Nat Struct Mol Biol. 2013;20:1434–42. doi: 10.1038/nsmb.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez NJ, Ow MC, Reece-Hoyes JS, Barrasa MI, Ambros VR, Walhout AJM. Genome-scale spatiotemporal analysis of Caenorhabditis elegans microRNA promoter activity. Genome Res. 2008;18:2005–15. doi: 10.1101/gr.083055.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, Loewer A, Ziebold U, Landthaler M, Kocks C, le Noble F, Rajewsky N. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- Moore MJ, Scheel TKH, Luna JM, Park CY, Fak JJ, Nishiuchi E, Rice CM, Darnell RB. miRNA-target chimeras reveal miRNA 3′-end pairing as a major determinant of Argonaute target specificity. Nat Commun. 2015;6:8864. doi: 10.1038/ncomms9864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi K, Weinberg DE, Bartel DP, Patel DJ. Structure of yeast Argonaute with guide RNA. Nature. 2012;486:368–374. doi: 10.1038/nature11211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquinelli AE. MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship. Nat Rev Genet. 2012;13:271–82. doi: 10.1038/nrg3162. [DOI] [PubMed] [Google Scholar]
- Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10:1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- Salomon WE, Jolly SM, Moore MJ, Zamore PD, Serebrov V. Single-Molecule Imaging Reveals that Argonaute Reshapes the Binding Properties of Its Nucleic Acid Guides. Cell. 2015;162:84–95. doi: 10.1016/j.cell.2015.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirle NT, MacRae IJ. The crystal structure of human Argonaute2. Science. 2012;336:1037–40. doi: 10.1126/science.1221551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirle NT, Sheu-Gruttadauria J, Chandradoss SD, Joo C, MacRae IJ. Water-mediated recognition of t1-adenosine anchors Argonaute2 to microRNA targets. Elife. 2015;4:1–16. doi: 10.7554/eLife.07646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirle NT, Sheu-Gruttadauria J, MacRae IJ. Structural basis for microRNA targeting. Science (80- ) 2014;346:608–613. doi: 10.1126/science.1258040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slack FJ, Basson M, Liu Z, Ambros V, Horvitz HR, Ruvkun G. The lin-41 RBCC gene acts in the C. elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Mol Cell. 2000;5:659–69. doi: 10.1016/s1097-2765(00)80245-2. [DOI] [PubMed] [Google Scholar]
- Sugimoto Y, König J, Hussain S, Zupan B, Curk T, Frye M, Ule J. Analysis of CLIP and iCLIP methods for nucleotide-resolution studies of protein-RNA interactions. Genome Biol. 2012;13:R67. doi: 10.1186/gb-2012-13-8-r67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505:344–52. doi: 10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vella MC, Choi E, Lin S, Reinert K, Slack FJ. The C. elegans microRNA let-7 binds to imperfect let-7 complementary sites from the lin-41 3′UTR. Genes Dev. 2004;18:132–7. doi: 10.1101/gad.1165404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wee LM, Flores-Jasso CF, Salomon WE, Zamore PD. Argonaute divides Its RNA guide into domains with distinct functions and RNA-binding properties. Cell. 2012;151:1055–1067. doi: 10.1016/j.cell.2012.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westholm JO, Miura P, Olson S, Shenker S, Joseph B, Sanfilippo P, Celniker SE, Graveley BR, Lai EC. Genome-wide Analysis of Drosophila Circular RNAs Reveals Their Structural and Sequence Properties and Age-Dependent Neural Accumulation. Cell Rep. 2014;9:1966–1981. doi: 10.1016/j.celrep.2014.10.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Artiles KL, Fire AZ. Functional relevance of “seed” and “non-seed” sequences in microRNA-mediated promotion of C. elegans developmental progression. RNA. 2015:1–13. doi: 10.1261/rna.053793.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zisoulis DG, Lovci MT, Wilbert ML, Hutt KR, Liang TY, Pasquinelli AE, Yeo GW. Comprehensive discovery of endogenous Argonaute binding sites in Caenorhabditis elegans. Nat Struct Mol Biol. 2010;17:173–9. doi: 10.1038/nsmb.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.