Abstract
Large RNAs often utilize GNRA tetraloops as structural elements to stabilize the overall tertiary fold. These tetraloop-receptor (TR) interactions have a conserved geometry in which the tetraloop docks into the receptor at an angle of ~15 degrees from the helix containing the receptor. Here we show that the conserved GUAAY pentaloop found in domain III of group IIB1 introns participates in a novel class of RNA tertiary interaction with a geometry and mode of binding that is significantly different from that found in GNRA TR interactions. This pentaloop is highly conserved within the IIB1 class and interacts with the minor groove of the catalytic DV. The base planes of the loop and receptor nucleotides are not coplanar and greatly deviate from standard A-minor motifs. The helical axis of the GUAAY stem loop diverges ~70° from the angle of insertion found in a typical GNRA TR interaction. Therefore, the loop architecture and insertion orientation is distinctive, with in vitro splicing data indicating that a GNRA tetraloop is incompatible at this position. The GUAAY pentaloop-receptor motif is also found in the structure of the eukaryotic thiamine pyrophosphate riboswitch in the context of a hexanucleotide loop sequence. We therefore propose, based on phylogenetic, structural, and biochemical data, that the GUAAY pentaloop-receptor interaction represents a novel structural motif that is present in multiple structured RNAs.
Graphical abstract
Introduction
One of the most ubiquitous RNA structures is the tetraloop [1, 2], a four nucleotide sequence that caps an A-form double helix. While numerous tetraloop motifs have been identified, three are the most prevalent: CUUG [3], UNCG [4], and GNRA [5], with the latter demonstrating a robust ability to form RNA tertiary structures. GNRA tetraloops can engage in tetraloop-receptor interactions, where the loop nucleotides insert into the minor groove of an RNA receptor helix. In addition, different GNRA tetraloop sequences have varying affinities for receptor sequences, allowing this tertiary structure motif to be sequence specific [6, 7]. GNRA tetraloop-receptor interactions have a defined geometry between the two interacting motifs, with the tetraloop docking into the minor groove of the receptor helix at an angle of ~15°. Furthermore, the nucleobases of the tetraloop and the receptor nucleotides are coplanar. Despite many large RNA structures being determined over the past two decades, there are only two identified classes of tetraloop-receptor interactions (along with the GANC tetraloop [8, 9]).
Previously, we determined the crystal structure of a eukaryotic group II intron from the brown algae Pylaiella littoralis (P.li.LSUI2) in which we described the conformation of the μ-μ′ interaction (PDB 4R0D) [10]. This interaction was first biochemically identified through nucleotide analogue interference mapping (NAIM) and involves a GUAAY pentaloop docking into the base of the catalytic domain V [11] (Figure 1a). This μ-μ′ interaction occurs directly adjacent to the κ-κ′ interaction in the group IIB intron crystal structure [10] and results in a quintuple adenosine base stack inserting into the minor groove (Figure 1b). Here we present phylogenetic, structural, and biochemical evidence that distinguishes the GUAAY pentaloop from the ubiquitous GNRA tetraloop and reveals a new mode of interaction between an RNA loop and a receptor helix.
Figure 1. DV acts as a receptor to two tertiary interactions.
A) Secondary structure shows the three tertiary interactions involving DV. μ and κ insert five adenosines into the minor groove of DV. B) The tertiary structure of the minor groove of DV. The five adenosines form an extended base stack, which is perpendicular to the direction of the minor groove. The μ-μ′ interaction positions three nucleotides into the minor groove of DV. The base planes of the μ loop are approximately perpendicular to that of the receptor helix.
Results
The consensus secondary structure of the group IIB1 phylogenetic subclass reveals that nucleotides 1 to 4 of the pentaloop sequence are highly conserved as GUAA [12] (Table 1). At the fifth position, a pyrimidine residue is always observed, with a U and C representing 71% and 19% of the sequences, thus forming the GUAAY consensus sequence. Additionally, the stem containing the loop contains a closing conserved U-G wobble pair (85%), but there are no observed occurrences of the G-U wobble pair. This differs from GNRA tetraloops, which tend to have a preference for C-G base pairs at this position, but can also contain a closing G-C pair [13, 14]. In addition to the sequence conservation, the loop is attached to the domain IIIa helix, which has a highly conserved length of seven base-pairs in group IIB1 introns (Figure 1a).
Table 1. Sequence alignment of the GUAAY pentaloop in group IIB1 introns.
P.li.I2 (P.li.LSUI2) sequence is bolded. Sequence positions that are 100% conserved are denoted with a black background, while positions that are >75% conserved are denoted with a light grey background. Positions that were >95% pyrimidine and purine are denoted in light blue and light green, respectively.
| Intron name | N | Y | U | G | U | A | A | Y | G | R | N |
|---|---|---|---|---|---|---|---|---|---|---|---|
| P.li.I2 | G | U | U | C | G | A | U | ||||
| Sh.fr.I1 | A | U | U | G | U | A | A | C | G | A | U |
| Vi.an.I1 | A | U | U | G | U | A | A | C | G | A | U |
| Vi.ha.I1 | A | U | U | G | U | A | A | C | G | A | U |
| Wo.sp.I1 | U | U | U | G | U | A | A | C | G | A | A |
| Wo.sp.I3 | U | U | U | G | U | A | A | C | G | A | A |
| A.v.I3 | C | C | U | G | U | A | A | C | G | G | G |
| A.v.I1 | C | C | U | G | U | A | A | C | G | G | G |
| Th.e.I1 | U | C | U | G | U | A | A | C | G | G | A |
| Al.sh.I1 | C | C | U | G | U | A | A | C | G | G | G |
|
| |||||||||||
| Cu.me.I1 | G | C | U | G | U | A | A | U | G | G | U |
| Cu.me.I2 | G | C | U | G | U | A | A | U | G | G | U |
| Ag.r.I1 | G | C | U | G | U | A | A | U | G | G | U |
| Al.bo.I1 | C | C | U | G | U | A | A | U | G | G | G |
| Al.sh.I2 | C | C | U | G | U | A | A | U | G | G | G |
| B.t.I1 | U | C | U | G | U | A | A | U | G | G | G |
| B.vu.I1 | C | C | U | G | U | A | A | U | G | G | G |
| B.vu.I2 | U | C | U | G | U | A | A | U | G | G | G |
| Ci.ro.I1 | G | C | U | G | U | A | A | U | G | G | U |
| E.c.I5 | U | C | U | G | U | A | A | U | G | G | A |
| P.st.I1 | C | C | U | G | U | A | A | U | G | G | G |
| H.s.I1 | A | C | U | G | U | A | A | U | G | G | U |
| Hm.ar.I1 | G | C | U | G | U | A | A | U | G | G | U |
| Kl.pn.I2 | U | C | U | G | U | A | A | U | G | G | A |
| Le.pn.I1 | U | C | U | G | U | A | A | U | G | G | A |
| Le.pn.I2 | G | C | U | G | U | A | A | U | G | G | C |
| P.st.I2 | C | C | U | G | U | A | A | U | G | G | G |
| P.p.I2 | C | C | U | G | U | A | A | U | G | G | G |
| P.p.I4 | G | C | U | G | U | A | A | U | G | G | C |
| Po.sp.I3 | U | C | U | G | U | A | A | U | G | G | A |
| Po.sp.I4 | U | C | U | G | U | A | A | U | G | G | A |
| Po.sp.I1 | U | C | U | G | U | A | A | U | G | G | A |
| Po.sp.I2 | U | C | U | G | U | A | A | U | G | G | A |
| Re.sp.I1 | G | C | U | G | U | A | A | U | G | G | C |
| Sh.sp.I2 | U | C | U | G | U | A | A | U | G | G | A |
| Th.e.I7 | U | C | U | G | U | A | A | U | G | G | A |
| X.f.I1 | C | C | U | G | U | A | A | U | G | G | G |
| Th.e.I3 | U | C | U | G | U | A | A | U | G | G | A |
| Eu.re.I1 | C | U | U | G | U | A | A | U | G | A | G |
| An.pr.I1 | C | U | U | G | U | A | A | U | G | A | G |
| Eu.si.I2 | C | U | U | G | U | A | A | U | G | A | G |
| Fa.pr.I1 | C | U | U | G | U | A | A | U | G | A | G |
| S.ce.I5 | A | U | U | G | U | A | A | U | G | A | U |
| Sh.ba.I1 | A | U | U | G | U | A | A | U | G | A | U |
| Sh.pi.I1 | G | U | U | G | U | A | A | U | G | A | U |
| Sb.mo.I1 | U | U | U | G | U | A | A | U | G | A | A |
| Sh.sp.I1 | A | U | U | G | U | A | A | U | G | A | U |
| Vi.ha.I2 | A | U | U | G | U | A | A | U | G | A | U |
| Sh.se.I1 | A | U | U | G | U | A | A | U | G | A | U |
| E.c.I8 | G | C | U | G | U | A | A | U | A | A | C |
| T.e.I8 | U | G | U | G | U | A | A | U | A | C | A |
| R.pi.I1 | G | C | U | G | U | A | A | U | C | G | U |
| P.ae.I2 | G | C | U | G | U | A | A | U | C | G | U |
| Ch.lu.I1 | C | C | U | G | U | A | A | U | C | G | G |
| Ch.ph.I1 | C | C | U | G | U | A | A | U | C | G | G |
| Pr.ae.I4 | C | C | G | G | U | A | A | U | G | G | G |
| S.eq.I1 | G | C | A | G | U | A | A | U | G | G | C |
| Hp.au.I1 | U | C | C | G | U | A | A | U | C | G | A |
The first nucleotide of the pentaloop (G448) stacks directly on top of the capping U-G wobble pair (Figure 2a). Interestingly, instead of stacking on the 5′ adjacent base U447, G448 instead stacks almost entirely on G453, which is paired with U447. This may explain the preference for a U-G over G-U wobble pair, as the latter would orient the loop further from its receptor interaction. The second pentaloop nucleotide (U449) continues the base stack above the first nucleotide. The Watson-Crick edges of these two nucleotides are oriented in a similar direction, which keeps the phosphate backbone in an A-form helix-like conformation. U449 stacks on G448 and forms a base-phosphate interaction with C452 using its Watson-Crick edge. In this orientation, U449 (N3) hydrogen bonds with the non-bridging oxygen of the phosphate of C452. This type of base-phosphate interaction is typical of a U-turn, which was first observed in tRNA anti-codon loops [15]. Substitution of cytosine for U449 could only be maintained if N3 were protonated, while a purine substitution would result in a steric clash or force the backbone into a sub-optimal configuration. Altogether, the configuration of the second nucleotide in context within the overall loop structure provides a rationale for its conservation. The third loop nucleotide (A450) initiates the U-turn [15] and causes its Watson-Crick edge to deviate from an A-form helix rotation. This contrasts with the GNRA tetraloop, where the U-turn is initiated between nucleotides one and two (G|NRA) [16]. The fourth pentaloop nucleotide (A451) stacks beneath the previous nucleotide and also inserts its nucleobase into the minor groove of DV. Finally, the last nucleotide of the pentaloop (C542) finishes the base stack below A451. It makes a single hydrogen bond between O2 and N2 of G555 from the receptor. Because O2 is present in both uracil and cytosine, this provides a rationale for the conservation of a pyrimidine at this position.
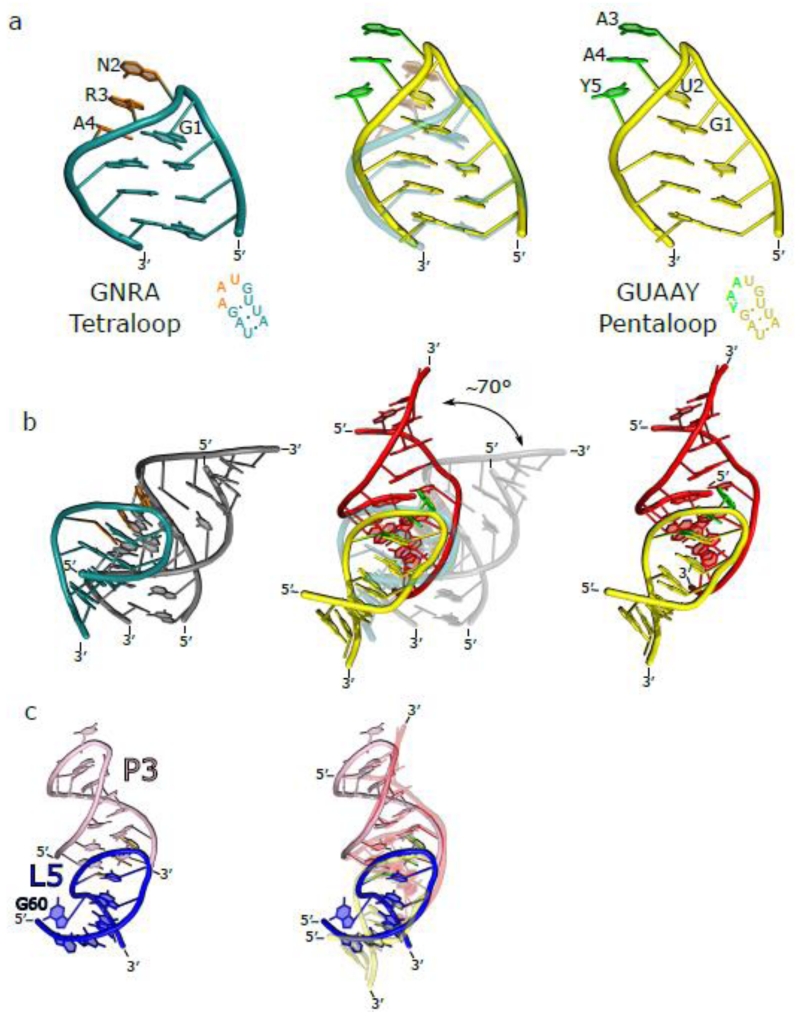
Figure 2. Comparison of the GNRA tetraloop versus the GUAAY pentaloop.
A) The GNRA tetraloop (left) and the GUAAY pentaloop (right) are connected to a three base-pair helix and aligned to the bottom of the stem. Central panel shows the alignment of the two loops. This alignment shows that the the GUAAY pentaloop extends approximately one base pair higher and at a different orientation compared to the GNRA tetraloop. Nucleotides of the GNRA tetraloop and GUAAY pentaloop that interact with their cognate receptor are colored in orange and green, respectively. B) The GNRA tetraloop (left) nucleotides insert parallel to the base planes of the receptor, while the GUAAY pentaloop (right) nucleotides are nearly perpendicular to the receptor base planes. This allows a greater number of nucleotides to insert into the minor groove. The alignment of the two structures (central panel) shows a ~70° deviation in terms of the angle of attack of the loop for the receptor. C) The structure of the equivalent GUAAY-containing hexanucleotide loop-receptor (left) from the thiamine pyrophosphate riboswitch (PDB 2CKY). The alignment with the GUAAY pentaloop-receptor from the group IIB intron (right) shows an almost identical mode of recognition between the two components of the interaction.
There is some commonality between the GNRA tetraloop and the GUAAY pentaloop. Both loop structures allow the last three nucleobases to splay out in a manner that allows them to interact with a receptor (Figure 2b). However, the base planes of the loops represent a significant difference between the two systems. In the GNRA tetraloop, the first and last nucleotide are on the same plane and form a trans-Watson-Crick/Hoogsteen pair between the guanosine sugar edge and the Hoogsteen edge of the adenosine. In contrast, the guanosine of the pentaloop is the only base in its plane. Rather, the second nucleotide (U449) is located on the same base plane as the final nucleotide (GUAAY). Furthermore, the second and last nucleobase are not interacting with each other. Instead, the second nucleobase interacts with the phosphate backbone and Watson-Crick edge of the last nucleotide is oriented in the opposite direction. Because the first and second nucleotide stack with one another, this allows the pentaloop to extend one base plane higher, with respect to the GNRA tetraloop (Figure 2a).
The last three nucleotides of the GNRA and GUAAY loop are positioned so that their nucleobases are ordered and oriented outward from the helix. This allows the nucleobases to insert into their respective receptors, but the orientation of insertion differs greatly. The inserting nucleotides of the GNRA tetraloop are approximately planar with respect to the receptor base planes (Figure 2b). In contrast, the GUAAY nucleotides insert into the minor groove of DV ~70° relative to the receptor nucleotides. While this orientation precludes the formation of canonical A-minor motifs, it allows the possibility of more nucleotides to stack above and below without clashing with the phosphate backbone.
A structural motif search was done to determine if similar loop structures existed in the PDB that were not yet annotated. Using the AMIGOS II program [17], we initiated an (η,θ) search, with the pentaloop values corresponding to (182.1, 218.3), (175.2, 256.4), (18.7, 231.2), (162.9, 223.6), and (167.8, 273.0). Based on this search, no pentaloop match was identified. However, the RNA 3D Motif Atlas [18, 19] revealed a hexanucleotide loop containing the sequence GUAAU from the eukaryotic thiamine pyrophosphate riboswitch (PDB 3D2V and 2CKY) that docks into a receptor helix in a similar manner as seen in the group IIB1 intron (Figure 2c). In this case, there is no analogous κ-κ′ interaction found near the inserting base stack of the GUAAY sequence, therefore suggesting that this novel loop-receptor combination can exist as an independent functional unit. In addition, the presence of the GUAAY sequence in a hexaloop indicates that it can function in a variety of sequence contexts in a manner similar to that seen for GNRA tetraloops [20].
The initial discovery of μ-μ′ indicated a strict requirement for the loop to be a pentaloop [11]. Due to the lack of structural information at the time, only two tetraloop substitutions were conducted: mutating the entire loop to UUCG (Δμ), and deletion of the last loop nucleotide to a GUAA sequence. With the information obtained from the crystal structure of the group IIB1 intron [10], the latter loop mutation actually results in a significant change of sequence register of the loop structure. Specifically, shortening the loop from five to four nucleotides results in the substitution of a uracil at a position that usually involves an adenosine inserting into the receptor. In the same study, a point mutation at that position from an adenosine to uracil resulted in defects in splicing activity. Utilizing the structural data as a guide, we aimed to characterize the sequence and spatial requirements that would promote the formation of μ-μ′ through mutagenesis (Figure 3). While the in vitro splicing assay is an indirect measure for the formation of μ-μ′, the only known function of this tertiary interaction is to stabilize the active conformation of the active site. We are therefore using splicing as an indication for the formation of the μ-μ′ interaction. Magnesium is an essential cation for ribozymes, both as a structural element [21-23] to stabilize the negative phosphate backbone as well as for catalytic function [24, 25]. In the case of the group II intron, two catalytic metals are bound in the active site [26, 27]. One serves to activate the nucleophile for attack at the scissile phosphate, while the other stabilizes the transition state of the leaving group. In standard splicing conditions for P.li.LSUI2 [28], 10 mM Mg2+ is used to stimulate splicing in vitro. However, at this concentration we observed no significant difference in splicing activity for any of the D3a mutants (Figure 4). This led us to believe that this magnesium concentration is capable of overcoming structural defects caused by mutagenesis, as seen in Figure 4 (orange bars). This is especially apparent in the Δμ mutant, which should be incapable of forming the loop-receptor interaction (Figure 4b). To address this, the Mg2+ concentration was lowered to 2.5mM. However, it is still useful to test the splicing at 10 mM Mg2+ in order to validate the retention of activity in the mutants assayed in this study.
Figure 3. Secondary structure of P.li.LSUI2 D3a mutants.
Mutants were grouped into four categories: a) loop point mutants, b) loop mutants, c) helical length mutants, and d) helical length/tetraloop mutants. Red letters indicate changes to the wild-type sequence. Red lines indicate where helical deletions took place. Values below the mutants correspond to Figure 4 for in vitro splicing assays at 2.5mM (blue) and 10mM (orange) MgCl2.
Figure 4. In vitro splicing assay of P.li.LSUI2 D3a mutants.
a) Point mutants made either in the closing base pair of the stem or in the pentaloop. The single point mutants retained activity comprable to wild-type. However, mutating the loop entirely to adenosines resulted in a drastic reduction in activity. b) Mutagenesis of the loop to any of the tetraloop sequences results in severe defects in splicing activity. c) Changing the helical length from the native seven base-pairs perturbs activity. d) The defects caused by the GAAA-tetraloop cannot be rescued with different helical length mutations. Thirty minute time points taken for all splicing assays. *<0.05 by two-tailed Student’s t-test. **< 0.001 by two-tailed Student’s t-test.
The high level of conservation of the GUAAY pentaloop is remarkable for group IIB1 introns, with the first four positions being completely conserved across all the sequences. Despite this conservation, mutants with single base substitutions seemed to retain splicing activity comparable to wild type (Figure 4a). By comparison, mutating the loop entirely to adenosines leads to ~3-fold difference. This suggests that there is some level of redundancy with related sequences having a capacity to form a similar three-dimensional structure as the wild-type loop.
While the previous study deleted the terminal pyrimidine to test the effects of a GUAA-tetraloop sequence, a GAAA sequence was also tested. The latter allows for a canonical GNRA-tetraloop fold to still maintain an adenosine at the very tip of the loop to potentially interact with the DV minor groove. Both of these mutants result in ~3-fold reduction in splicing activity (Figure 4b), consistent with the findings of Fedorova et al. (2005). This still does not rule out the possibility that a tetraloop is incompatible at this position, because a tetraloop substitution would effectively lower the loop by one base plane (vide supra). Different helical lengths were tested (−2 to +2) to favor docking of this tetraloop and affect splicing activity. Increasing the helix by 1 base pair had a modest reduction on the effect of splicing, however the other helical changes (−2, −1, +2) resulted in significant defects in splicing (Figure 4c). Therefore, the addition of a GAAA-loop with varying helical lengths (−2 to +3) does not rescue splicing activity, rather it exacerbated the defect further to levels comparable to Δμ (Figure 4d).
Discussion
The GUAAY pentaloop-receptor interaction found in group IIB1 introns and the eukaryotic thiamine pyrophosphate riboswitch represents a new class of loop-receptor interactions in addition to the known GNRA and GANC [8, 9] tetraloop-receptors. Consistent with previous studies, the GUAAY loop cannot be replaced with a GNRA tetraloop. In addition, the defect caused by a tetraloop substitution cannot be rescued by changing the length of the attached helix. The helix attached to the μ-loop emanates from a highly conserved four-way junction, with two other tertiary interactions in close proximity to this junction. It could be possible to adapt a GNRA-tetraloop at this position, but would likely require the remodeling of the aforementioned junction. It is probable, however, that a GNRA tetraloop is completely incompatible in this position. Due to the close proximity of κ-κ′, the receptor must be able to accommodate five adenosines in the minor groove. Rather than inserting planar with respect to the receptor nucleotides, the loop nucleotides insert at an unusual angle that allows the base stacking vector to be parallel to the direction of the minor groove. This orientation allows five, and potentially more, nucleotides to base stack effectively into the minor groove without clashing with the surrounding phosphate backbone. Additionally, there may be other stable variants of the pentaloop, similar to the GUAAY, but unable to accommodate splicing activity in the specific context of the μ-μ′ interaction. It should be added that this new class of loop-helix interaction seems to tolerate insertions as seen in the hexanucleotide sequence of the loop from the thiamine pyrophosphate riboswitch. GNRA tetraloops behave in a similar manner with additional inserted nucleotides in the form GNR[Xn]A[7, 29].
Materials and Methods
IIB1 intron sequences were obtained from the Group II Intron Database [30]. Alignment was done using Bioedit [31]. Sequence logos were generated using the WebLogo [32]. AMIGOS II [17] motif search was done on the following data set: PDBs accessed on September 2015, with a minimum of 30 RNA nucleotides, and a resolution greater than 3.5Å.
The constructs used for the in vitro self-splicing assays contained wild-type P.li.LSUI2 sequence with a 250-nt 5′ exon and 75-nt 3′ exon. The domain IV sequence in this construct was deleted and replaced with a UUCG stem loop. This construct was inserted into the pUC57 plasmid at the EcoRV site. Overlapping PCR was used to generate the mutant constructs and inserted into pUC57 with EcoRV and T4 DNA ligase. Plasmid was linearized using HindIII and used for in vitro transcription with T7 RNA polymerase. Radiolabeled transcripts were prepared using 10 μCi [α-32P] UTP (3000 Ci/mmole), 0.5 mM UTP, 1 mM other NTPs, and 2.5mM MgCl2. Transcripts were gel purified on a 4% polyacrylamide (19:1)/8 M urea gel, RNA was recovered by diffusion into 300 mM NaCl, 0.01% SDS, 1 mM EDTA. Self-splicing experiments were performed for 30 min at 45°C in a splicing buffer containing, 1 M NH4Cl, 40 mM Tris-HCl (pH 7.5), and 0.02% SDS, and either 2.5 mM or 10 mM MgCl2. Reactions were stopped by addition of 2.5 volumes of 95% EtOH and 0.5 volume of 3M sodium acetate pH 5.2. This mixture was then incubated at −80° C for 30 mins to precipitate the RNA followed centrifugation. Supernatant was removed and pellets resuspended in 5 μl of 0.5× TE, 50% formamide. Splicing products were resolved using a denaturing 4% polyacrylamide (19:1)/8 M urea gels. The Bio-Rad PMI Imager was used to image and quantitate gels. All splicing assays were done in triplicate.
Research Highlights.
We have discovered a novel class of interacting loops present in a group IIB intron and a previous structure of the thiamine pyrophosphate riboswitch
This pentaloop sequence interacts with a receptor helix at a 70° angle deviation from the more common GNRA tetraloop-receptor interaction
This work highlights the fact that there may be additional classes of RNA tertiary interactions to be discovered in the future
Acknowledgements
We thank Timothy Wiryaman for comments on the manuscript. R.T.C. was supported by the Cell, Molecular, and Genetics (CMG) Training Program funded by NIH predoctoral training grant 5T32GM007240. This work was supported by NIH grant 5R01GM102216 awarded to N.T.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Cate JH, Gooding AR, Podell E, Zhou K, Golden BL, Kundrot CE, et al. Crystal structure of a group I ribozyme domain: principles of RNA packing. Science. 1996;273:1678–85. doi: 10.1126/science.273.5282.1678. [DOI] [PubMed] [Google Scholar]
- [2].Woese CR, Winker S, Gutell RR. Architecture of ribosomal RNA: constraints on the sequence of “tetra-loops”. Proc Natl Acad Sci U S A. 1990;87:8467–71. doi: 10.1073/pnas.87.21.8467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Jucker FM, Pardi A. Solution structure of the CUUG hairpin loop: a novel RNA tetraloop motif. Biochemistry. 1995;34:14416–27. doi: 10.1021/bi00044a019. [DOI] [PubMed] [Google Scholar]
- [4].Molinaro M, Tinoco I. Use of ultra stable UNCG tetraloop hairpins to fold RNA structures: thermodynamic and spectroscopic applications. Nucleic Acids Res. 1995;23:3056–63. doi: 10.1093/nar/23.15.3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Jaeger L, Michel F, Westhof E. Involvement of a GNRA tetraloop in long-range RNA tertiary interactions. J Mol Biol. 1994;236:1271–6. doi: 10.1016/0022-2836(94)90055-8. [DOI] [PubMed] [Google Scholar]
- [6].Fiore JL, Nesbitt DJ. An RNA folding motif: GNRA tetraloop-receptor interactions. Q Rev Biophys. 2013;46:223–64. doi: 10.1017/S0033583513000048. [DOI] [PubMed] [Google Scholar]
- [7].Wu L, Chai D, Fraser ME, Zimmerly S. Structural variation and uniformity among tetraloop-receptor interactions and other loop-helix interactions in RNA crystal structures. PLoS One. 2012;7:e49225. doi: 10.1371/journal.pone.0049225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Keating KS, Toor N, Pyle AM. The GANC tetraloop: a novel motif in the group IIC intron structure. J Mol Biol. England. 2008:475–81. doi: 10.1016/j.jmb.2008.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Ishikawa J, Furuta H, Ikawa Y. An in vitro-selected RNA receptor for the GAAC loop: modular receptor for non-GNRA-type tetraloop. Nucleic Acids Res. 2013;41:3748–59. doi: 10.1093/nar/gkt040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Robart AR, Chan RT, Peters JK, Rajashankar KR, Toor N. Crystal structure of a eukaryotic group II intron lariat. Nature. 2014;514:193–7. doi: 10.1038/nature13790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Fedorova O, Pyle AM. Linking the group II intron catalytic domains: tertiary contacts and structural features of domain 3. EMBO J. 2005;24:3906–16. doi: 10.1038/sj.emboj.7600852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Toor N, Hausner G, Zimmerly S. Coevolution of group II intron RNA structures with their intron-encoded reverse transcriptases. RNA. 2001;7:1142–52. doi: 10.1017/s1355838201010251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Moody EM, Feerrar JC, Bevilacqua PC. Evidence that folding of an RNA tetraloop hairpin is less cooperative than its DNA counterpart. Biochemistry. 2004;43:7992–8. doi: 10.1021/bi049350e. [DOI] [PubMed] [Google Scholar]
- [14].Antao VP, Lai SY, Tinoco I. A thermodynamic study of unusually stable RNA and DNA hairpins. Nucleic Acids Res. 1991;19:5901–5. doi: 10.1093/nar/19.21.5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Quigley GJ, Rich A. Structural domains of transfer RNA molecules. Science. 1976;194:796–806. doi: 10.1126/science.790568. [DOI] [PubMed] [Google Scholar]
- [16].Jucker FM, Pardi A. GNRA tetraloops make a U-turn. RNA. 1995;1:219–22. [PMC free article] [PubMed] [Google Scholar]
- [17].Wadley LM, Keating KS, Duarte CM, Pyle AM. Evaluating and learning from RNA pseudotorsional space: quantitative validation of a reduced representation for RNA structure. J Mol Biol. 2007;372:942–57. doi: 10.1016/j.jmb.2007.06.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Parlea LG, Sweeney BA, Hosseini-Asanjan M, Zirbel CL, Leontis NB. The RNA 3D Motif Atlas: Computational methods for extraction, organization and evaluation of RNA motifs. Methods. 2016;103:99–119. doi: 10.1016/j.ymeth.2016.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Petrov AI, Zirbel CL, Leontis NB. Automated classification of RNA 3D motifs and the RNA 3D Motif Atlas. RNA. 2013;19:1327–40. doi: 10.1261/rna.039438.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Abramovitz DL, Pyle AM. Remarkable morphological variability of a common RNA folding motif: the GNRA tetraloop-receptor interaction. J Mol Biol. 1997;266:493–506. doi: 10.1006/jmbi.1996.0810. [DOI] [PubMed] [Google Scholar]
- [21].Cate JH, Hanna RL, Doudna JA. A magnesium ion core at the heart of a ribozyme domain. Nat Struct Biol. 1997;4:553–8. doi: 10.1038/nsb0797-553. [DOI] [PubMed] [Google Scholar]
- [22].Misra VK, Draper DE. On the role of magnesium ions in RNA stability. Biopolymers. 1998;48:113–35. doi: 10.1002/(SICI)1097-0282(1998)48:2<113::AID-BIP3>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- [23].Draper DE. A guide to ions and RNA structure. RNA. 2004;10:335–43. doi: 10.1261/rna.5205404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Steitz TA, Steitz JA. A general two-metal-ion mechanism for catalytic RNA. Proc Natl Acad Sci U S A. 1993;90:6498–502. doi: 10.1073/pnas.90.14.6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Bowman JC, Lenz TK, Hud NV, Williams LD. Cations in charge: magnesium ions in RNA folding and catalysis. Curr Opin Struct Biol. 2012;22:262–72. doi: 10.1016/j.sbi.2012.04.006. [DOI] [PubMed] [Google Scholar]
- [26].Toor N, Keating KS, Taylor SD, Pyle AM. Crystal structure of a self-spliced group II intron. Science. United States. 2008:77–82. doi: 10.1126/science.1153803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Toor N, Rajashankar K, Keating KS, Pyle AM. Structural basis for exon recognition by a group II intron. Nat Struct Mol Biol. United States. 2008:1221–2. doi: 10.1038/nsmb.1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Costa M, Fontaine JM, Loiseaux-de Goër S, Michel F. A group II self-splicing intron from the brown alga Pylaiella littoralis is active at unusually low magnesium concentrations and forms populations of molecules with a uniform conformation. J Mol Biol. 1997;274:353–64. doi: 10.1006/jmbi.1997.1416. [DOI] [PubMed] [Google Scholar]
- [29].Lemieux S, Major F. Automated extraction and classification of RNA tertiary structure cyclic motifs. Nucleic Acids Res. 2006;34:2340–6. doi: 10.1093/nar/gkl120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Dai L, Toor N, Olson R, Keeping A, Zimmerly S. Database for mobile group II introns. Nucleic Acids Res. 2003;31:424–6. doi: 10.1093/nar/gkg049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Tippmann HF. Analysis for free: comparing programs for sequence analysis. Brief Bioinform. 2004;5:82–7. doi: 10.1093/bib/5.1.82. [DOI] [PubMed] [Google Scholar]
- [32].Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–90. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]