Abstract
Objectives
Circadian rhythm (CR) was identified by RNA sequencing as the most dysregulated pathway in human osteoarthritis (OA) in articular cartilage. This study examined circadian rhythmicity in cultured chondrocytes and the role of the CR genes NR1D1 and BMAL1 in regulating chondrocyte functions.
Methods
RNA was extracted from normal and OA-affected human knee cartilage (n=14 each). Expression levels of NR1D1 and BMAL1 mRNA and protein were assessed by quantitative PCR and immunohistochemistry. Human chondrocytes were synchronized and harvested at regular intervals to examine circadian rhythmicity in RNA and protein expression. Chondrocytes were treated with small interfering RNA (siRNA) for NR1D1 or BMAL1, followed by RNA sequencing and analysis of the effects on the TGF-β pathway.
Results
NR1D1 and BMAL1 mRNA and protein levels were significantly reduced in OA compared to normal cartilage. In cultured human chondrocytes, a clear circadian rhythmicity was observed for NR1D1 and BMAL1. Increased BMAL1 expression was observed after knocking down NR1D1, and decreased NR1D1 levels were observed after knocking down BMAL1. Sequencing of RNA from chondrocytes treated with NR1D1 or BMAL1 siRNA identified 330 and 68 significantly different genes, respectively, and this predominantly affected the TGF-β signaling pathway.
Conclusions
The circadian rhythm pathway is dysregulated in OA cartilage. Interference with circadian rhythmicity in cultured chondrocytes affects TGF-β signaling, which is a central pathway in cartilage homeostasis.
Keywords: Circadian rhythm, NR1D1, BMAL1, Osteoarthritis, TGF-β
INTRODUCTION
Osteoarthritis (OA) involves destruction of articular cartilage and remodeling of other joint tissues 1. The main OA pathogenesis pathways in cartilage include destruction of the extracellular matrix, cell death, abnormal cell differentiation and production of inflammatory mediators 2. A large number of signaling mechanisms are abnormally activated in OA and contribute to cartilage damage. These include inflammation-related pathways such as NFκB and MAP kinases, Wnt, hypoxia, PI3K and TGF-β pathways 1,3, 4.
Global transcriptome analysis by RNA sequencing provides an unbiased approach to obtain vast amounts of information on genes and pathways that are abnormally activated or inhibited in disease 5. We completed an RNA sequencing study on normal and OA human knee cartilage that led to the discovery that the circadian rhythm (CR) pathway was inhibited and the most significantly dysregulated pathway in OA. Among the differentially expressed genes in this pathway, NR1D1 and BMAL1 showed the largest degree of suppression in OA cartilage.
CR is critical in coordinating cell functions throughout all tissues 6,7. In mammals, circadian rhythm is a fundamental regulatory factor for many aspects of behavior and physiology, including sleep/wake cycles, blood pressure, body temperature and metabolism 6. Disruption of CR leads to increased incidence of many diseases, such as cancer, metabolic disease, and mental illness 6. CR is regulated by the central oscillator in the hypothalamic suprachiasmatic nucleus (SCN), and local oscillators throughout the body coordinate daily cycles by integrating signals from the SCN with other internal and external time cues 8. The pacemaker consists of a core group of genes with transcriptional-translational feedback loops that involve multiple clock genes such as CLOCK, BMAL1, NPAS2, PER1, 2 and 3, CRY1 and 2, and NR1D1 9, 10. These clock genes and their protein products function in a feedback loop resulting in a nearly 24-hour cycle. The transcription factor BMAL1 is the core driver of the molecular clock. Positive regulators (BMAL1, CLOCK, NPAS2) drive the expression of negative feedback regulators (PER, CRY, NR1D1), which in turn inhibit the expression and activity of the positive regulators 11.
NR1D1 encodes a member of the nuclear receptor family, and is expressed in liver, adipose tissue, and skeletal muscle 12–14. It is a transcriptional repressor that is activated by heme 15, 16, and recruits nuclear receptor co-repressor (NCoR)-Histone Deacetylase (HDAC) 3 complexes to Rev-Erb response elements in enhancers and promoters of target genes 17–19. BMAL1 encodes a main positive transcriptional regulator of the circadian oscillation, which functions as a heterodimeric complex with CLOCK 20. NR1D1 transcription is activated by BMAL1/CLOCK through its binding to E-box in NR1D1 promoter 21. In turn NR1D1 acts as the major regulator of BMAL1 by repressing its transcription 19, 22, thus forming a negative feedback loop to maintain circadian rhythmicity. Aside from its function in circadian rhythm control, these genes are also involved in the control of metabolism, autophagy, and inflammatory responses 9,12, 19, 23–27. Interestingly, even cells isolated from peripheral tissues generate a CR in culture using the same clock factor network 7, 28. These rhythmically expressed genes control the expression of many other genes (clock controlled genes), which in turn drive cascades of rhythmic gene expression. At least 4–10% of total cellular transcripts in any given organ are thought to oscillate in a circadian manner and this set of oscillating genes has a tissue specific pattern 29, 30.
The objectives of this study were to study CR in cultured chondrocytes and determine the consequences of CR gene dysregulation in chondrocyte function. This study is the first to determine in a systematic approach (i) which CR genes are expressed in normal and OA cartilage, (ii) how expression of these genes is regulated, (iii) whether cartilage cells display endogenous CR and (iv) whether clock genes are involved in regulating expression of genes that are associated with OA pathogenesis.
MATERIALS AND METHODS
Cartilage donors
Normal human knee cartilage tissues were procured by tissue banks from 5 female (age 26–57 years, mean 39 years) and 18 male (age 18–44 years, mean 30 years) donors (approved by Scripps Institutional Review Board) and processed within 24–72 hours post mortem. Full thickness cartilage was harvested for RNA isolation from identical locations on the medial femoral condyles. OA-affected cartilage was harvested from the tissue removed during knee replacement surgery from 10 female (age 61–82 years, mean 69 years) and 6 male (age 66–84 years, mean 71 years) donors.
Tissue processing and RNA isolation
Cartilage was stored at −20°C in Allprotect Tissue Reagent (Qiagen, Valencia, CA) immediately after resection from the subchondral bone. For RNA isolation, cartilage was pulverized in a 6770 Freezer/Mill Cryogenic Grinder (SPEX SamplePrep, Metuchen, NJ), and homogenized in Qiazol Lysis Reagent (Qiagen) using 25mg tissue per 700μl Qiazol. RNA was isolated using the miRNeasy Mini kit (Qiagen) with on-column DNAse digestion, followed by removal of proteoglycans using RNAmate (BioChain Institute, Newark, CA). RNA from cultured chondrocytes was extracted using Direct-zol RNA MiniPrep Kit (Zymo Research, Irvine, CA).
RNA sequencing and data analysis
RNA from 8 normal (2 female, 6 male) and 10 OA (5 female, 5 male) cartilage donors was sequenced using 125–150ng of total RNA as input. mRNA libraries were prepared using the Encore Complete RNA-Seq DR Multiplex System 1–8 and 9–16 (NuGen, San Carlos, CA) with 16 unique indexed adapters (L2V6DR-BC2-L2V6DR-BC16). Two lanes of an Illumina HiSeq 2000 instrument were used to generate a total of 8–30 million single-end 100bp reads.
Raw data were checked for quality with the software FastQC (v0.10.1) (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). We mapped the RNA-seq reads for each library to the UCSC human hg19 reference genome using TopHat2 (v2.0.9) 31. Read abundances were estimated using Cufflinks (v2.1.1) as Fragments Per Kilobase of exon per Million fragments mapped (FPKM) 32. Cuffdiff2 was used to calculate differentially expressed genes between normal and OA samples 33. Genes with a q-value < 0.05 were considered significantly differentially expressed and were included in the downstream pathway analysis. Signaling Pathway Impact Analysis was conducted using the Bioconductor packages SPIA and Graphite, using the differentially expressed gene list and their log2 fold changes as input 34, 35. Pathway databases included in the analysis include KEGG, Biocarta, NCI and Reactome. Pathways were considered significantly differentially expressed if the pGFWER were < 0.05.
Quantitative polymerase chain reaction (qPCR)
RNA was extracted from normal and OA human cartilage samples as well as from cultured chondrocytes and gene expression levels were analyzed by qPCR. The following pre-designed TaqMan gene expression assays (Life Technologies) were used: NR1D1 (Hs00253876_m1), BMAL1 (Hs00154147_m1), TGFBR1 (Hs00610320_m1), TGFBR2 (Hs00234253_m1), TGFBR3 (Hs01114253_m1), TGFB1 (Hs00998133_m1), TGFB2 (Hs00234244_m1), TGFB3 (Hs01086000_m1), ID2 (Hs041787239_m1), TNC (Hs01115665_m1), and ELN (Hs00355783_m1).
Immunohistochemistry
Immunohistochemistry was performed to assess protein expression patterns in human and mouse cartilage using anti-NR1D1 antibody (Abcam ab174309, Cambridge, MA) and anti-BMAL1 antibody (Thermo Scientific PA1-523, Waltham, MA). Rabbit IgG (1 μg/ml) was used as a negative control in all experiments. For human cartilage, expression patterns were compared between normal and OA samples. In C57 BL/6 mice, we analyzed young normal and aged knees as a model of aging-related OA. We also analyzed knees from mice with surgically induced OA by destabilization of medial meniscus and medial collateral ligament resection 36. The methods for tissue processing and immunohistochemistry were described earlier 36.
Western blotting
At indicated time points, cultured human chondrocytes were lysed in RIPA buffer supplemented with Halt protease inhibitor cocktail and phosphatase inhibitor cocktail (Thermo Scientific) and samples were analyzed by western blotting as previously described 37. The following antibodies were used: anti-GAPDH (Life Technologies AM4300), anti-Rev-Erbα (Cell Signaling 13418), anti-BMAL1 (Bethyl Laboratories A302-616A, Montgomery, TX).
Circadian rhythmicity in cultured chondrocytes
Cultured chondrocytes were maintained in DMEM with 10% calf serum supplemented with penicillin and streptomycin. Medium was changed to 0.5% serum 16 hours before synchronization with 100nM dexamethasone. T0 was defined as 24 hours after dexamethasone application to the medium. Dexamethasone-containing media was removed and replaced with serum-free media at the time points indicated in figure legends. Cells were collected every four hours for the time course analysis of gene and protein expression. For subsequent experiments with synchronization, cells were collected at T0, T8, T16, T24, T36, and T48 according to the highest and lowest expression time points of NR1D1 and BMAL1.
siRNA knockdown of NR1D1 and BMAL1 in chondrocytes
Small interfering RNAs (siRNAs) for NR1D1 (s18386), BMAL1 (s1616) and negative control (AM4635) were purchased from Life Technologies. Human normal chondrocytes were transfected with siRNA using Lipofectamine RNAiMAX transfection reagent (Life Technologies) at a concentration of 12.5 pmol/ml. Three siRNAs targeting different regions were tested in advance to knock down NR1D1 (s18386, 5940, s18387), and the single most effective siRNA (s18387) was used in subsequent experiments. Since we were able to achieve sufficient knock down by the first siRNA tested, we did not further explore other siRNAs for BMAL1. Effects of NR1D1 and BMAL1 knock down on circadian rhythm genes were analyzed by qPCR, and relative gene expressions were compared to a control group treated with negative control siRNA. We also performed a global analysis of gene expression using next generation sequencing of RNA from chondrocytes that had been transfected with NR1D1 and BMAL1 siRNA (harvested at T24 and T12, respectively to account for their peak expression) with corresponding samples treated with control siRNA (n=2 for each time point). NEBNext Ultra Directional RNA Library Prep Kit for Illumina (New England Biolabs, Ipswich, MA) was used for library preparation. RNA sequencing and data analysis was performed as described above for the RNA samples from normal and OA human cartilage using miRNeasy Mini kit (Qiagen). Quality control for the raw data was performed as described above. The raw RNA-seq reads were aligned to the UCSC human hg19 genome using the STAR aligner (v2.3.0) 38. Read counts were quantified using the Python package HTSeq-Count (v0.6.1) with UCSC RefSeq hg19 annotation (Release 57). Differential expression analysis of the knockdown versus control was conducted on raw count data normalized by the Bioconductor Limma package using the Bioconductor package DESeq2 39. Genes were considered significantly differentially expressed if they had an adjusted p value of < 0.05. Signaling Pathway Impact Analysis was performed on the differentially expressed genes as described above.
Effect of TGF-β1 on circadian rhythm gene expression
To test the effect of knocking down NR1D1 and BMAL1 on the chondrocyte response to TGF-β, recombinant human TGF-β1 (PeproTech, Rocky Hill, NJ) was added to the culture medium of chondrocytes from four independent donors with normal cartilage at a concentration of 10ng/ml 48 hours after siRNA transfection. Chondrocytes were collected 6 hours later for RNA extraction and qPCR as described above. The time point was selected according to a preliminary experiment comparing 6 hours and 24 hours after TGF-β1 stimulation, resulting in maximum effect at the early time point.
Statistical analysis
The qPCR values and positively stained cell counts in immunohistochemistry were normally distributed and equally variable. The mRNA expression values for NR1D1 and BMAL1 between normal and OA human cartilage, and between cultured chondrocytes with or without TGF-β1 treatment were analyzed by Student’s t-test. The comparison between groups in rates of NR1D1 and BMAL1 positive cells in immunohistochemistry slides were performed by multiple comparison using ANOVA with Bonferroni adjustment. Ratios of positively stained cells in each group (18–24 months old, 30–36 months old, and surgically induced OA) were compared against 6 months old samples, therefore 3 hypotheses with a desired α=0.05 were tested. For experiments with cultured chondrocytes treated with siRNA, statistically significant differences in qPCR values were determined with multiple comparison using Dunnett’s test. Samples treated with negative control siRNA were used as control condition. When TGF-β1 was added to the culture medium, samples with both TGF-β1 and negative control siRNA treatment, were considered as the control condition. The results are reported as mean ± S.E. P values less than 0.05 were considered significant.
RESULTS
Expression patterns of clock genes and proteins in normal and OA human cartilage
RNA sequencing data from 8 normal and 10 OA human cartilage samples were analyzed for dysregulated pathways and this showed that the circadian rhythm was the most significantly dysregulated pathway in OA cartilage. Among genes listed in the circadian rhythm pathway in KEGG data base, the mRNA levels of NR1D1, BHLHE40, BMAL1 (ARNTL), PER1, PRKAG2, PER2, RORA, NPAS2, and CRY2 were significantly lower in OA while RBX1 was significantly increased in OA cartilage (Table 1). Among these genes, NR1D1, BHLHE40 and BMAL1 showed the largest differences between normal and OA, with a log2 fold change >2.4. Since there is a known close interaction between NR1D1 and BMAL1 as core clock genes, we chose to further focus on the role of these two genes in cartilage homeostasis.
Table 1.
Differentially expressed circadian rhythm pathway genes in normal and OA human cartilage.
| Gene | Normal | OA | log2FC | q value |
|---|---|---|---|---|
| NR1D1 | 384.80 | 56.79 | −2.76 | 0.0030 |
| BHLHE40 | 405.97 | 65.06 | −2.64 | 0.0030 |
| BMAL1 | 8.03 | 1.51 | −2.41 | 0.0088 |
| PER1 | 131.68 | 28.29 | −2.22 | 0.0053 |
| PRKAG2 | 23.04 | 6.63 | −1.80 | 0.0030 |
| PER2 | 17.59 | 5.68 | −1.63 | 0.0088 |
| RORA | 22.94 | 10.19 | −1.17 | 0.0161 |
| NPAS2 | 8.82 | 4.08 | −1.11 | 0.0280 |
| CRY2 | 16.57 | 7.89 | −1.07 | 0.0332 |
| RBX1 | 46.48 | 108.46 | 1.22 | 0.0104 |
Among genes listed in KEGG database for circadian rhythm pathway, NR1D1, BHLHE40, and BMAL1 showed the largest differences between normal and OA cartilage.
Quantitative PCR was performed to validate RNA sequencing results on human normal and OA cartilage (n=14 each). The expression levels of NR1D1 and BMAL1 were confirmed to be significantly lower in OA cartilage (Figure 1A). Protein expression of NR1D1 and BMAL1 in cartilage was assessed by immunohistochemistry in both human and mice cartilage. NR1D1 protein was expressed throughout the human cartilage tissue, most strongly in the superficial and mid zones (Figure 1B). In OA cartilage, NR1D1 positive cells were diminished even in areas of preserved full thickness cartilage, with few positive cells remaining in the superficial zone. In lesions with fibrillations, strong staining was observed in chondrocyte clusters. In young normal mice, NR1D1 protein distribution was similar as in normal human tissue, with positive cells mainly in the superficial and upper mid zone. Statistically significant differences were detected between 6 months old normal knees and all other groups in NR1D1 expression, while BMAL1 expression was significantly different in 6 months old normal knees and 30–36 months old aged knees, as well as knees with surgically induced OA. Both NR1D1 and BMAL1 presented reduced expression with aging and in surgically induced OA, compared to young normal knees (Figure 1C).
Figure 1. Expression of NR1D1 and BMAL1 mRNA and protein in cartilage.

(A) qPCR was performed on 14 normal and 14 OA human articular cartilage samples (* p<0.05, ** p<0.01). (B) NR1D1 and BMAL1 Protein expression was analyzed by immunohistochemistry. Representative images are shown for young normal, aged normal and OA normal appearing and OA lesion area. Images are representative of 5–8 samples for each tissue type. NR1D1 and BMAL1 positive cells were observed predominantly in the superficial and upper-mid zone of normal cartilage. Protein expression was reduced in OA cartilage even in the normal areas, although strong expression was observed in the cluster cells. (C) Immunohistochemistry of mouse cartilage. 6 mouse joints for each sample type were analyzed. Bar graphs indicate percentages of positive cells. NR1D1 and BMAL1 positive cells were distributed mainly in the superficial to upper-mid zone. Expression levels were significantly reduced with aging and with induction of surgical OA.
Rhythmicity of gene expression and regulation of clock genes in cultured human chondrocytes
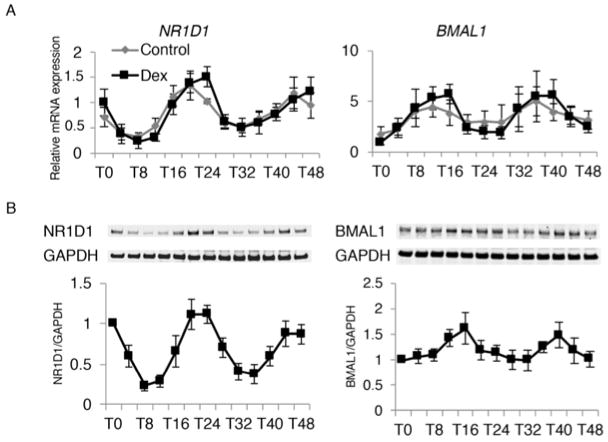
A spontaneous robust circadian rhythmicity was observed in cultured human chondrocytes where NR1D1 had peak expression at T24 and T48 after synchronization, whereas BMAL1 fluctuated in a slightly earlier opposite phase with highest expression at T16 and T40 (Figure 2A). The rhythmicity was confirmed for both NR1D1 and BMAL1 protein expression by western blotting (Figure 2B).
Figure 2. Rhythmicity of NR1D1 and BMAL1 expression in cultured chondrocytes.
(A) (B) Chondrocytes isolated from 5 individual donors were analyzed with and without dexamethasone (DEX) synchronization. qPCR and western blotting were performed on samples collected at 4 hour intervals. T0 indicates 24 hours after DEX was added to the culture medium. (A) NR1D1 mRNA expression peaked at T24 and T48, with lowest expression observed at T8 and T32. BMAL1 expression followed a reversed phase. (B) NR1D1 protein fluctuated in an identical pattern with the mRNA expression. BMAL1 protein expression had less fluctuation, with approximately 4 hours delayed phase from mRNA expression.
Knock down of NR1D1 and BMAL1 was used to examine the interaction of clock genes in cultured chondrocytes. Effective siRNA-mediated knock down in protein levels was confirmed by western blotting for NR1D1 and BMAL1 (Figure 3A). When transfected with specific siRNA 48 hours before synchronization, the expression levels of NR1D1 and BMAL1 were repressed throughout the time course up to T48. NR1D1 still displayed rhythmic fluctuation with reduced peak expression at T24 and T48. BMAL1 siRNA completely disrupted the rhythmic expression pattern of BMAL1 (Figure 3B). BMAL1 knock down reduced the level of NR1D1 mRNA and protein, and NR1D1 knock down increased BMAL1 transcription (Figure 3B). These changes were mainly observed at T24 and 48, when the expression level of BMAL1 was low. In relation to other genes in the circadian rhythm pathway, NR1D1 knock down increased BHLHE40 from T16 to T48, while having subtle effect on PER2, RORA, RORC, and RBX1. In contrast, BMAL1 knock down increased BHLHE40 at T8 and decreased its expression at T24, while the expression level of PER2 was reduced at T0, T24 and T36. RORA, RORC, and RBX1 were not significantly affected by BMAL1 knock down. (Supp.1) These results establish that a cell autonomous circadian rhythmicity exists in cultured chondrocytes and demonstrate the interdependence of CR genes in their temporal expression patterns.
Figure 3. Effect of NR1D1 and BMAL1 knock down on expression of NR1D1 and BMAL1.

(A) Western blotting was performed on cultured chondrocytes from 7 individual donors after siRNA transfection, collected at T12 and T24 after DEX synchronization. Effective knock down of both NR1D1 and BMAL1 was confirmed at both time points. Knocking down BMAL1 decreased the expression level of NR1D1, while knocking down NR1D1 had less effect on BMAL1 protein level. (B) qPCR was performed on samples after siRNA transfection, and RNA collected at time points as indicated. NR1D1 expression was reduced at all time points by knocking down NR1D1 or BMAL1, while still showing rhythmicity. BM AL1 expression was reduced and dysregulated by knocking down BMAL1, whereas knocking down NR1D1 resulted in increased expression levels of BMAL1 after T24.
Genome-wide impact of circadian rhythm gene knock down in cultured human chondrocytes
To determine the role of CR genes in regulating global gene expression patterns, we used next generation RNA sequencing on normal human articular chondrocytes that were transfected with siRNA against NR1D1 and BMAL1. This unbiased approach revealed 330 and 68 differentially expressed genes, respectively, as compared to control siRNA (Table 2). Importantly, a large number of these differentially expressed genes (n=50) were altered by both siRNAs treatments (Supp. Table S2). Most (48 of these 50) genes were up regulated and only 2 genes were down regulated compared to treatment with control siRNAs. The global probability value from the signaling pathway impact analysis (SPIA) was significant for 4 pathways in NR1D1 knock down, and for 5 pathways in BMAL1 knock down (Table 2). Among the differentially expressed pathways, the TGF-β signaling pathway was significantly altered by both siRNA treatments. Altered expression levels of TGF-β signaling pathway genes were validated by qPCR analysis of RNA from additional chondrocyte cultures that were treated with the siRNAs (Figure 4A). While expression levels of TGFBR1, TGFB1, TGFB2 and TGFB3 were increased, levels of TGFBR2 and TGFBR3 were reduced.
Table 2.
Pathways affected by NR1D1 or BMAL1 knock down
| NR1D1 knock down (330 differentially expressed genes) | |
|---|---|
| Pathway name | pG |
| Protein processing in endoplasmic reticulum | 1.89E-06 |
| Viral carcinogenesis | 1.03E-05 |
| Oocyte meiosis | 0.0016 |
| TGF-beta signaling pathway | 0.0017 |
| BMAL1 knock down (68 differentially expressed genes) | |
|---|---|
| Pathway name | pG |
| TGF-beta signaling pathway | 0.0088 |
| p53 signaling pathway | 0.0093 |
| Focal adhesion | 0.0103 |
| Retrograde endocannabinoid signaling | 0.0103 |
| Oocyte meiosis | 0.0131 |
RNA sequencing was performed on samples after siRNA transfection for NR1D1 and BMAL1. Among the differentially expressed pathways between NR1D1 or BMAL1 knock down and control, the TGF-β signaling pathway was significantly altered by both siRNA treatments.
Figure 4. Correlation of NR1D1 and BMAL1 knock down and TGF-β signaling.

(A) qPCR for TGF-β signaling pathway genes was performed on cultured chondrocytes isolated from 3 individual donors after siRNA transfection for NR1D1 and BMAL1. (B) qPCR for NR1D1 and BMAL1 were performed on cultured chondrocytes isolated from 4 individual donors after TGF-β1 stimulation. TGF-β1 treatment significantly reduced NR1D1 expression and significantly increased BMAL1 expression. (C) qPCR was performed for ELN and TNC after siRNA transfection with and without TGF-β1 stimulation. (* p<0.05, ** p<0.01).
Interactions between CR genes and TGF signaling in cultured human chondrocytes
TGF-β1 treatment significantly reduced NR1D1 mRNA expression, while increasing BMAL1 mRNA expression level (Figure 4B). Knocking down NR1D1 or BMAL1 before TGF-β1 treatment affected expression levels of TGF-β pathway genes (Supp. Table S3, Supp. 4) in the same manner as the previously mentioned experiment without TGF-β stimulation. The expression levels of some TGF-β induced genes, such as ELN and TNC were increased by knocking down NR1D1 or BMAL1 (Figure 4C).
Discussion
Since the report of circadian rhythmicity in the mitosis of epiphyseal cartilage by Simmons 40, several authors have described daily variation in endochondral ossification, extracellular matrix synthesis, chondrocyte proliferation and cartilage growth 41. Attempts to seek diurnal variations in OA markers have also been made 42, but the correlation between CR and cartilage homeostasis in mature articular cartilage has remained largely unclear. In a recent study, Gossan et al. reported that 619 genes (3.9% of the expressed genes) displayed circadian pattern of expression in cartilage. This included genes involved in cartilage homeostasis and survival, as well as genes with potential importance in the pathogenesis of OA. Several clock genes were disrupted in the early stages of cartilage degeneration in a mouse model of OA 30, thus suggesting the role of circadian rhythm in maintaining cartilage homeostasis. Furthermore, Kc et al. reported that environmental disruption of circadian rhythms by altered light:dark cycle promoted osteoarthritic changes in mouse knee joint 43.
Our results indicate a time-dependent variability of mRNA expression in cultured chondrocytes with a clear circadian rhythmicity up to 48 hours with nearly opposite phase in NR1D1 and BMAL1. This observation is in agreement with previous reports using different cell types, as well as mice cartilage 8, 22, 30, 44–47. We identified lower expression levels of multiple circadian rhythm genes by genome wide RNA sequencing in OA, and bioinformatics analysis revealed that circadian rhythm is the most dysregulated pathway in human OA cartilage. Down regulation of NR1D1 and BMAL1 mRNA expression levels in OA cartilage was confirmed by qPCR, and expression of corresponding proteins was reduced with age and with the induction of surgical OA in a mouse model, before severe cartilage degradation occurred. This seems to be inconsistent from the report of Chaturvedi et al., proposing that NR1D1 (Rev-ERbAα) expression was highest among all the nuclear receptors expressed in OA cartilage, thus assuming over expression of this gene to be involved in OA pathogenesis 48. Our RNA sequencing data also revealed that NR1D1 expression was highest among the 48 nuclear receptors in both normal and OA cartilage (supp.5). However, the expression level in OA cartilage was significantly lower than the expression level in normal cartilage (Figure 1), which the previous report was not able to identify, as it did not include analysis of normal cartilage. Recently, Dudek et al. performed a series of experiments on BMAL1, reporting the rhythmicity of this gene in chondrocytes and showing the effect of knock down and knock out of this gene on cartilage degeneration49. Our results are in agreement with their report, further adding data on NR1D1.
Results from previous reports show altered expression levels of circadian rhythm genes by aging, induction of mechanical stress, TNF-α, and development of OA 30, 50, 51. Our results indicate that the circadian rhythm pathway is overall down regulated in OA. Although each circadian rhythm gene is potentially affected by external stimuli such as inflammatory mediators, it is surprising that both the positive and negative limbs of the feedback loop are down regulated. Interestingly, nearly 70% of gene expression changes induced by BMAL1 knock down were common to those influenced by NR1D1 knock down, despite its opposing function. The most likely interpretation seems to be that the major impact caused by down regulation of the main positive stimulator BMAL1, due to whatever external cause (e.g. aging, mechanical stress, cytokines) outside the circadian feedback loop, subsequently induces the down regulation of the whole circadian rhythm pathway. Indeed, down regulation of RORC and RORA, the only positive regulators of BMAL1 52, may contribute to forming a feedback loop leading to further down regulation of BMAL1. Knock down of the negative regulator including NR1D1 would theoretically counteract BMAL1 knock down by stimulating BMAL1 transcription, but the effect seems to be masked by a larger effect of BMAL1 down regulating stimuli. However, our results suggest that most of the final output of BMAL1 knock down is mediated via NR1D1 and other downstream genes, rather than a direct effect of BMAL1.
TGF-β is a potent multi-functional regulator of cell growth and differentiation abundantly expressed in cartilage. The TGF-β pathway has been identified as a key signaling pathway in osteoarthritis, but evidence for both protective and catabolic roles of TGF-β has been reported 3, 53. TGF-β activation in subchondral bone is suggested to contribute to the development of osteoarthritis 54, but conversely, loss of TGF-β signaling in cartilage induces chondrocyte hypertrophy and ultimately results in cartilage degeneration 55. TGF-β is implicated in all stages of chondrogenesis, including mesenchymal condensation, proliferation of chondroblasts and the deposition of cartilage-specific ECM molecules 4.
Our RNA sequencing results on NR1D1 and BMAL1 siRNA treated chondrocytes identified TGF-β signaling pathway as a common differentially expressed pathway. In particular, ELN and TNC, previously reported to be induced by TGF-β 56, 57, were among the most overexpressed genes after NR1D1 and BMAL knock down. Our invitro experiments show that (1) TGF-β signaling genes are affected by NR1D1 and BMAL1 alteration, (2) TGF-β itself alters NR1D1 and BMAL1 expression, (3) alteration in NR1D1 and BMAL1 expression would affect TGF-β downstream gene expressions, such as ELN and TNC. We investigated whether there are potential effects on SMAD signaling but did not observe any difference in the levels of SMAD2/3, Smad1,5,8 or in TGF-β induced SMAD phosphorylation after NR1D1 or BMAL1 knock down.
This link between circadian rhythm genes and TGF- β signaling is in agreement with previous reports 26, 58, and suggest that alterations in circadian rhythmicity may result in abnormal ECM protein synthesis, potentially contributing to OA pathophysiology. However, at this point we were not able to further investigate the direct mechanism of interaction between TGF-β and clock genes, and further research is necessary.
Biomarkers of OA show diurnal variation 42, 59–61. While some of this variation may be attributed to physical activity or food consumption, the present findings that cartilage has an intrinsic CR suggests that it also may be a determinant of OA biomarker fluctuation.
This study has several limitations. First, we were unable to control the time of human sample collection. Since NR1D1 and BMAL1 expression level present large variation across the day, it is possible that the comparison between normal and OA would have been influenced by the time of death and/or sample collection. However, the difference of expression level between normal and OA was significantly large, thus suggesting that the baseline expression level is reduced in OA and this reduction has a bigger impact on cartilage homeostasis than the daily rhythmic change. Secondly, aging is a considerable factor that may have influenced our gene expression level differences between normal and OA human cartilage samples. Gossan et al. presented that the circadian clock becomes less robust during aging and the oscillation amplitude is reduced in aged mice 30. However, our observation of reduced NR1D1 and BMAL1 protein in mice surgical OA model indicates that the reduction of NR1D1 and BMAL1 occurs by OA progression, independent from aging. Finally, we do not know whether circadian rhythm gene expression patterns in articular cartilage change after death. However, our samples were collected at an early time point within 72 hours after death or resection during knee replacement surgery, and we consider only minimum changes would occur.
In conclusion, we show that expression of NR1D1 and BMAL1 is reduced in OA cartilage. NR1D1 and BMAL1 present circadian rhythmicity in cultured chondrocytes with an opposite phase. Reduced expression of NR1D1 and BMAL1 in chondrocytes affects TGF-β signaling.
Supplementary Material
Acknowledgments
Role of Funding
This study was supported by National Institutes of Health grants AG007996 and AG049617, and the Sam and Rose Stein Endowment Fund.
Footnotes
Author contributions
ML, RA, TS, AS conceived of the study, and participated in its design and coordination.
RA, MS, KF, AS performed gene and bioinformatics expression analyses.
YA, YM, OA, TT, YT carried out histology, immunohistochemistry and cell culture experiments and performed quantitative analysis.
All authors read and approved the final manuscript.
Dr. Lotz had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Confict of interest
The authors have no conflicts of interest.
Ethics approval
This study was conducted with the approval of the Human Subjects Committee and the Institutional Animal Care and Use Committee at The Scripps Research Institute.
References
- 1.Loeser RF, Goldring SR, Scanzello CR, et al. Osteoarthritis: a disease of the joint as an organ. Arthritis and rheumatism. 2012;64(6):1697–707. doi: 10.1002/art.34453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Loeser RF. Osteoarthritis year in review 2013: biology. Osteoarthritis Cartilage. 2013;21(10):1436–42. doi: 10.1016/j.joca.2013.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bush JR, Beier F. TGF-beta and osteoarthritis--the good and the bad. Nature medicine. 2013;19(6):667–9. doi: 10.1038/nm.3228. [DOI] [PubMed] [Google Scholar]
- 4.Wu L, Huang X, Li L, et al. Insights on biology and pathology of HIF-1alpha/-2alpha, TGFbeta/BMP, Wnt/beta-catenin, and NF-kappaB pathways in osteoarthritis. Current pharmaceutical design. 2012;18(22):3293–312. doi: 10.2174/1381612811209023293. [DOI] [PubMed] [Google Scholar]
- 5.Mortazavi A, Williams BA, McCue K, et al. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nature methods. 2008;5(7):621–8. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 6.Gachon F, Nagoshi E, Brown SA, et al. The mammalian circadian timing system: from gene expression to physiology. Chromosoma. 2004;113(3):103–12. doi: 10.1007/s00412-004-0296-2. [DOI] [PubMed] [Google Scholar]
- 7.Dibner C, Schibler U, Albrecht U. The mammalian circadian timing system: organization and coordination of central and peripheral clocks. Annu Rev Physiol. 2010;72:517–49. doi: 10.1146/annurev-physiol-021909-135821. [DOI] [PubMed] [Google Scholar]
- 8.Balsalobre A, Brown SA, Marcacci L, et al. Resetting of circadian time in peripheral tissues by glucocorticoid signaling. Science. 2000;289(5488):2344–7. doi: 10.1126/science.289.5488.2344. [DOI] [PubMed] [Google Scholar]
- 9.Cho H, Zhao X, Hatori M, et al. Regulation of circadian behaviour and metabolism by REV-ERB-alpha and REV-ERB-beta. Nature. 2012;485(7396):123–7. doi: 10.1038/nature11048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takahashi JS, Hong HK, Ko CH, et al. The genetics of mammalian circadian order and disorder: implications for physiology and disease. Nature reviews Genetics. 2008;9(10):764–75. doi: 10.1038/nrg2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sato TK, Yamada RG, Ukai H, et al. Feedback repression is required for mammalian circadian clock function. Nature genetics. 2006;38(3):312–9. doi: 10.1038/ng1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Woldt E, Sebti Y, Solt LA, et al. Rev-erb-alpha modulates skeletal muscle oxidative capacity by regulating mitochondrial biogenesis and autophagy. Nature medicine. 2013;19(8):1039–46. doi: 10.1038/nm.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fontaine C, Dubois G, Duguay Y, et al. The orphan nuclear receptor Rev-Erbalpha is a peroxisome proliferator-activated receptor (PPAR) gamma target gene and promotes PPARgamma-induced adipocyte differentiation. The Journal of biological chemistry. 2003;278(39):37672–80. doi: 10.1074/jbc.M304664200. [DOI] [PubMed] [Google Scholar]
- 14.Lazar MA, Hodin RA, Darling DS, et al. A novel member of the thyroid/steroid hormone receptor family is encoded by the opposite strand of the rat c-erbA alpha transcriptional unit. Molecular and cellular biology. 1989;9(3):1128–36. doi: 10.1128/mcb.9.3.1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yin L, Wu N, Curtin JC, et al. Rev-erbalpha, a heme sensor that coordinates metabolic and circadian pathways. Science. 2007;318(5857):1786–9. doi: 10.1126/science.1150179. [DOI] [PubMed] [Google Scholar]
- 16.Raghuram S, Stayrook KR, Huang P, et al. Identification of heme as the ligand for the orphan nuclear receptors REV-ERBalpha and REV-ERBbeta. Nature structural & molecular biology. 2007;14(12):1207–13. doi: 10.1038/nsmb1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yin L, Lazar MA. The orphan nuclear receptor Rev-erbalpha recruits the N-CoR/histone deacetylase 3 corepressor to regulate the circadian Bmal1 gene. Molecular endocrinology. 2005;19(6):1452–9. doi: 10.1210/me.2005-0057. [DOI] [PubMed] [Google Scholar]
- 18.Feng D, Liu T, Sun Z, et al. A circadian rhythm orchestrated by histone deacetylase 3 controls hepatic lipid metabolism. Science. 2011;331(6022):1315–9. doi: 10.1126/science.1198125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yin L, Wu N, Lazar MA. Nuclear receptor Rev-erbalpha: a heme receptor that coordinates circadian rhythm and metabolism. Nucl Recept Signal. 2010;8:e001. doi: 10.1621/nrs.08001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bunger MK, Wilsbacher LD, Moran SM, et al. Mop3 is an essential component of the master circadian pacemaker in mammals. Cell. 2000;103(7):1009–17. doi: 10.1016/s0092-8674(00)00205-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Triqueneaux G, Thenot S, Kakizawa T, et al. The orphan receptor Rev-erbalpha gene is a target of the circadian clock pacemaker. Journal of molecular endocrinology. 2004;33(3):585–608. doi: 10.1677/jme.1.01554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Preitner N, Damiola F, Lopez-Molina L, et al. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110(2):251–60. doi: 10.1016/s0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- 23.Gibbs JE, Blaikley J, Beesley S, et al. The nuclear receptor REV-ERBalpha mediates circadian regulation of innate immunity through selective regulation of inflammatory cytokines. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(2):582–7. doi: 10.1073/pnas.1106750109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Migita H, Morser J, Kawai K. Rev-erbalpha upregulates NF-kappaB-responsive genes in vascular smooth muscle cells. FEBS letters. 2004;561(1–3):69–74. doi: 10.1016/S0014-5793(04)00118-8. [DOI] [PubMed] [Google Scholar]
- 25.Khapre RV, Kondratova AA, Patel S, et al. BMAL1-dependent regulation of the mTOR signaling pathway delays aging. Aging. 2014;6(1):48–57. doi: 10.18632/aging.100633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rey G, Cesbron F, Rougemont J, et al. Genome-wide and phase-specific DNA-binding rhythms of BMAL1 control circadian output functions in mouse liver. PLoS biology. 2011;9(2):e1000595. doi: 10.1371/journal.pbio.1000595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shimba S, Ishii N, Ohta Y, et al. Brain and muscle Arnt-like protein-1 (BMAL1), a component of the molecular clock, regulates adipogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(34):12071–6. doi: 10.1073/pnas.0502383102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Balsalobre A, Damiola F, Schibler U. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell. 1998;93(6):929–37. doi: 10.1016/s0092-8674(00)81199-x. [DOI] [PubMed] [Google Scholar]
- 29.Panda S, Antoch MP, Miller BH, et al. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109(3):307–20. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- 30.Gossan N, Zeef L, Hensman J, et al. The circadian clock in murine chondrocytes regulates genes controlling key aspects of cartilage homeostasis. Arthritis and rheumatism. 2013;65(9):2334–45. doi: 10.1002/art.38035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim D, Pertea G, Trapnell C, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome biology. 2013;14(4):R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trapnell C, Williams BA, Pertea G, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature biotechnology. 2010;28(5):511–5. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trapnell C, Hendrickson DG, Sauvageau M, et al. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nature biotechnology. 2013;31(1):46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tarca AL, Draghici S, Khatri P, et al. A novel signaling pathway impact analysis. Bioinformatics. 2009;25(1):75–82. doi: 10.1093/bioinformatics/btn577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sales G, Calura E, Cavalieri D, et al. graphite - a Bioconductor package to convert pathway topology to gene network. BMC bioinformatics. 2012;13:20. doi: 10.1186/1471-2105-13-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carames B, Hasegawa A, Taniguchi N, et al. Autophagy activation by rapamycin reduces severity of experimental osteoarthritis. Ann Rheum Dis. 2012;71(4):575–81. doi: 10.1136/annrheumdis-2011-200557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Akasaki Y, Hasegawa A, Saito M, et al. Dysregulated FOXO transcription factors in articular cartilage in aging and osteoarthritis. Osteoarthritis Cartilage. 2014;22(1):162–70. doi: 10.1016/j.joca.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dobin A, Davis CA, Schlesinger F, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Anders S, Huber W. Differential expression analysis for sequence count data. Genome biology. 2010;11(10):R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Simmons DJ. Circadian Mitotic Rhythm in Epiphyseal Cartilage. Nature. 1964;202:906–7. doi: 10.1038/202906a0. [DOI] [PubMed] [Google Scholar]
- 41.Gossan N, Boot-Handford R, Meng QJ. Ageing and osteoarthritis: a circadian rhythm connection. Biogerontology. 2014 doi: 10.1007/s10522-014-9522-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kong SY, Stabler TV, Criscione LG, et al. Diurnal variation of serum and urine biomarkers in patients with radiographic knee osteoarthritis. Arthritis and rheumatism. 2006;54(8):2496–504. doi: 10.1002/art.21977. [DOI] [PubMed] [Google Scholar]
- 43.Kc R, Li X, Voigt RM, et al. Environmental Disruption of Circadian Rhythm Predisposes Mice to Osteoarthritis-Like Changes in Knee Joint. Journal of cellular physiology. 2015 doi: 10.1002/jcp.24946. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 44.Al-Nuaimi Y, Hardman JA, Biro T, et al. A Meeting of Two Chronobiological Systems: Circadian Proteins Period1 and BMAL1 Modulate the Human Hair Cycle Clock. The Journal of investigative dermatology. 2013 doi: 10.1038/jid.2013.366. [DOI] [PubMed] [Google Scholar]
- 45.Wongchitrat P, Mukda S, Phansuwan-Pujito P, et al. Effect of amphetamine on the clock gene expression in rat striatum. Neuroscience letters. 2013;542:126–30. doi: 10.1016/j.neulet.2013.03.009. [DOI] [PubMed] [Google Scholar]
- 46.Chalmers JA, Martino TA, Tata N, et al. Vascular circadian rhythms in a mouse vascular smooth muscle cell line (Movas-1) American journal of physiology Regulatory, integrative and comparative physiology. 2008;295(5):R1529–38. doi: 10.1152/ajpregu.90572.2008. [DOI] [PubMed] [Google Scholar]
- 47.Rath MF, Rohde K, Fahrenkrug J, et al. Circadian clock components in the rat neocortex: daily dynamics, localization and regulation. Brain structure & function. 2013;218(2):551–62. doi: 10.1007/s00429-012-0415-4. [DOI] [PubMed] [Google Scholar]
- 48.Chaturvedi P, Pratta M, Steplewski K, et al. Functional characterization of an orphan nuclear receptor, Rev-ErbAalpha, in chondrocytes and its potential role in osteoarthritis. Arthritis and rheumatism. 2006;54(11):3513–22. doi: 10.1002/art.22170. [DOI] [PubMed] [Google Scholar]
- 49.Dudek M, Gossan N, Yang N, et al. The chondrocyte clock gene Bmal1 controls cartilage homeostasis and integrity. The Journal of clinical investigation. 2016;126(1):365–76. doi: 10.1172/JCI82755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kanbe K, Inoue K, Xiang C, et al. Identification of clock as a mechanosensitive gene by large-scale DNA microarray analysis: downregulation in osteoarthritic cartilage. Modern rheumatology/the Japan Rheumatism Association. 2006;16(3):131–6. doi: 10.1007/s10165-006-0469-3. [DOI] [PubMed] [Google Scholar]
- 51.Haas S, Straub RH. Disruption of rhythms of molecular clocks in primary synovial fibroblasts of patients with osteoarthritis and rheumatoid arthritis, role of IL-1beta/TNF. Arthritis research & therapy. 2012;14(3):R122. doi: 10.1186/ar3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Takeda Y, Jothi R, Birault V, et al. RORgamma directly regulates the circadian expression of clock genes and downstream targets in vivo. Nucleic acids research. 2012;40(17):8519–35. doi: 10.1093/nar/gks630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang L, Almqvist KF, Veys EM, et al. Control of extracellular matrix homeostasis of normal cartilage by a TGFbeta autocrine pathway. Validation of flow cytometry as a tool to study chondrocyte metabolism in vitro. Osteoarthritis Cartilage. 2002;10(3):188–98. doi: 10.1053/joca.2001.0492. [DOI] [PubMed] [Google Scholar]
- 54.Zhen G, Wen C, Jia X, et al. Inhibition of TGF-beta signaling in mesenchymal stem cells of subchondral bone attenuates osteoarthritis. Nature medicine. 2013;19(6):704–12. doi: 10.1038/nm.3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang X, Chen L, Xu X, et al. TGF-beta/Smad3 signals repress chondrocyte hypertrophic differentiation and are required for maintaining articular cartilage. The Journal of cell biology. 2001;153(1):35–46. doi: 10.1083/jcb.153.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Alcazar MAA, Morty RE, Lendzian L, et al. Inhibition of TGF-beta Signaling and Decreased Apoptosis in IUGR-Associated Lung Disease in Rats. PloS one. 2011;6(10) doi: 10.1371/journal.pone.0026371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Estany S, Vicens-Zygmunt V, Llatjos R, et al. Lung fibrotic tenascin-C upregulation is associated with other extracellular matrix proteins and induced by TGFbeta1. BMC pulmonary medicine. 2014;14(1):120. doi: 10.1186/1471-2466-14-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gast H, Gordic S, Petrzilka S, et al. Transforming growth factor-beta inhibits the expression of clock genes. Annals of the New York Academy of Sciences. 2012;1261:79–87. doi: 10.1111/j.1749-6632.2012.06640.x. [DOI] [PubMed] [Google Scholar]
- 59.Karsdal MA, Byrjalsen I, Bay-Jensen AC, et al. Biochemical markers identify influences on bone and cartilage degradation in osteoarthritis--the effect of sex, Kellgren-Lawrence (KL) score, body mass index (BMI), oral salmon calcitonin (sCT) treatment and diurnal variation. BMC musculoskeletal disorders. 2010;11:125. doi: 10.1186/1471-2474-11-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Qvist P, Christgau S, Pedersen BJ, et al. Circadian variation in the serum concentration of C-terminal telopeptide of type I collagen (serum CTx): effects of gender, age, menopausal status, posture, daylight, serum cortisol, and fasting. Bone. 2002;31(1):57–61. doi: 10.1016/s8756-3282(02)00791-3. [DOI] [PubMed] [Google Scholar]
- 61.Gordon CD, Stabler TV, Kraus VB. Variation in Osteoarthritis Biomarkers from Activity not Food Consumption. Clinica chimica acta; international journal of clinical chemistry. 2008;398(1–2):21–6. doi: 10.1016/j.cca.2008.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.