ABSTRACT
The organization of intracellular transport processes is adapted specifically to different cell types, developmental stages, and physiologic requirements. Some protein traffic routes are universal to all cells and constitutively active, while other routes are cell-type specific, transient, and induced under particular conditions only. Small GTPases of the Rab (Ras related in brain) subfamily are conserved across eukaryotes and regulate most intracellular transit pathways. The complete sets of Rab proteins have been identified in model organisms, and molecular principles underlying Rab functions have been uncovered. Rabs provide intracellular landmarks that define intracellular transport sequences. Nevertheless, it remains a challenge to systematically map the subcellular distribution of all Rabs and their functional interrelations. This task requires novel tools to precisely describe and manipulate the Rab machinery in vivo. Here we discuss recent findings about Rab roles during development and we consider novel approaches to investigate Rab functions in vivo.
KEYWORDS: cell polarity, lysosome related organelles (LROs), Rab GTPases, Staccato/Munc13-4
Introduction
Intracellular transport is adjusted to the demands and functions of diverse cell types. Differentiating and mature cells within tissues have distinct requirements for membrane and protein transport. However, independent of cell type and organismal developmental stage, cells must maintain Essential Permanent protein/membrane Traffic (EPT) routes. Well-known EPTs are the endocytic pathway or the transport route between the Endoplasmic reticulum (ER) and the Golgi apparatus. Rab proteins that control EPTs are among the first transport regulators to be identified.1 However, since these Rab proteins regulate early trafficking steps at the basis of the cellular transport machineries, the loss-of-function phenotypes of these EPT regulators are often the sum of perturbations of multiple intracellular pathways, making it difficult to pinpoint the mechanistic role of individual Rabs or transport pathways.2 Therefore, novel approaches are required to study individual pathways and to perturb traffic regulators in a temporally and spatially controlled manner.
While EPTs are commonly required in most cell types, specialized cellular functions and developmental processes require Dedicated Transient Traffic (DTT) routes. DTTs usually do not control essential cellular functions, but are induced on demand in response to external signals, such as mechanical or immunological stimuli.3,4 These signals often trigger intracellular events, such as the release of second messengers or transient increases in intracellular Ca2+ levels.5 Well-studied examples are neurotransmitter release in neurons6 and pulsed secretion by salivary gland cells.7 Genetic disruption of DTT transport often results in morphological or functional defects, although cell viability is usually not affected upon loss of Rabs controlling DTTs.
Here we discuss recent work on Rab-regulated transport routes in Drosophila melanogaster. Small GTPases are master regulators of intracellular membrane and protein transport and control the spatio-temporally targeted delivery of membrane and protein payloads.8 In the past decade all Rab family members in Drosophila were identified9 and their regulative elements characterized.10 In addition, comprehensive collections of inducible Rab transgenes were generated that allow targeted misexpression of tagged Rab proteins.9,10 While these genetic resources have provided many important insights into the organization of the Rab machinery, they are based on overexpression of Rab proteins. However, excess of certain Rab proteins has been shown to change transport and to generate mutant phenotypes.11-13 Therefore, maintaining endogenous expression levels of Rab proteins is a key prerequisite for faithfully charting intracellular traffic. Dunst et al. reported the generation of endogenous YFP-tagged rab alleles (YRabs) and characterized them in 6 different organs consisting of 23 different cell types.14 This resource is suitable to predict and compare traffic in different cell types, distinct cell differentiation stages or allows screening Rabs involved in cargo-specific transport. Caviglia et al. used the YRab collection to investigate the identity of the membrane material that mediates the formation of tubular connections during tracheal morphogenesis in Drosophila.15 They identified Rab7 and Rab39, which, together with Munc13–4/Staccato, act as regulators of lysosome-related organelles (LROs) in tracheal tip cells.
The Rab activity cycle: Hard facts and open questions
Rabs are switchable protein-binding platforms. Active Rabs are GTP-bound and anchored to membranes, where they recruit effector proteins involved in specific traffic steps16,17 or signaling cascades.18,19 Rab proteins are directly controlled by GDP/GTP exchange factors (GEFs) and GTPase activating proteins (GAPs). Once Rab-containing vesicles reach their target (delivery) sites, local GAPs trigger hydrolysis of Rab-bound GTP, thus rendering Rabs inactive. Deactivated GDP-bound Rabs are then extracted from membranes through mechanisms that are not yet fully understood. It has been suggested that lipid translocation proteins are involved. Recently, one study using attenuated total reflection-Fourier transform infrared (ATR-FTIR) spectroscopy proposed that GDP-dissociation factors (GDI)20-22 directly mediate membrane extraction.23 ATR-FTIR involves the collection of infrared radiation reflected from the interphase between an aqueous phase and a prism, in which the evanescent wave penetrated from the prism into the aqueous solution is absorbed by substances solved in the aqueous phase.24
The subsequent formation of cytosolic Rab-GDP/GDI complexes enables inactive Rab-GDPs to recycle and reintegrate into their original membranes.25 How are Rab-GDP/GDI assemblies trafficked? One possibility is that cytosolic Rab-GDP/GDI complexes are present in excess over local membrane-associated Rab-GTP pools. In this scenario no directed transport is required between different Rab pools. Alternatively, Rab-GDP/GDIs bind Rab-GDP effectors that move the complex along retrograde transport routes. Although neither speculation is easy to validate, recent technological advances will aid in resolving Rab-GDP/GDI traffic. Especially the availability of endogenously tagged Rabs provides an opportunity to study and manipulate transport with unprecedented resolution (Fig. 4). Once Rab-GDPs reach the membrane, GDI-displacement factors26 or GEFs27 promote their membrane integration and conversion into GTP-bound Rabs, and the traffic cycle starts over.28
Figure 2.
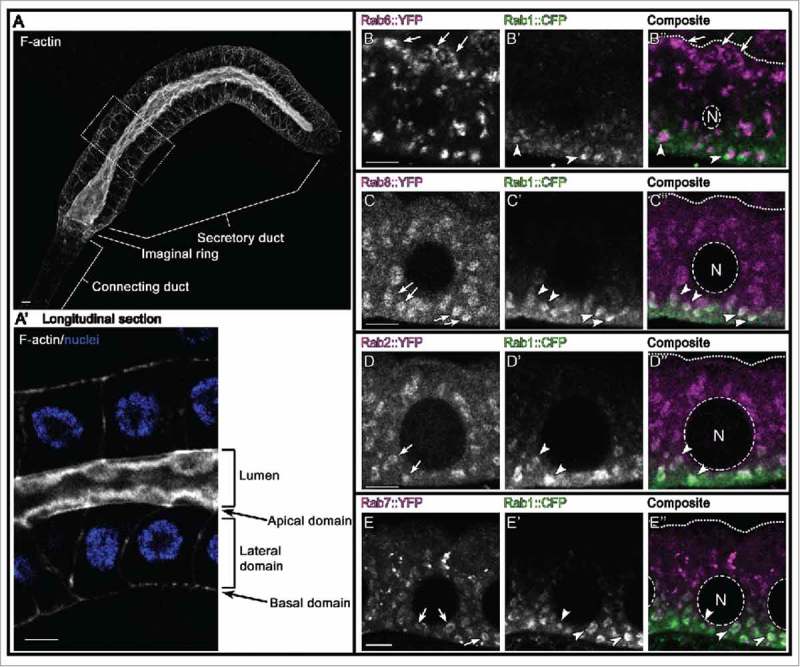
ER associated Rab proteins in salivary gland cells. Rab1 and Rab2 are canonical Rab proteins that regulate traffic between ER and the Golgi apparatus. We show here that Rab7 and 8, but not Rab6 (trans-Golgi Rab protein) are present on Rab1 membranes in salivary gland cells. It remains to be shown what role Rab7 and Rab8 may play in ER/Golgi related transport. Of note, all used Yrab and Crab alleles reflect the endogenous expression of respective Rab proteins. A,A’ Confocal micrograph shows wild-type salivary gland (A) stained with phalloidin (F-actin, white) and DAPI (nuclei, blue). Cells in boxed region are magnified in (A’). Scale bars indicate 5µm. B-E” Confocal cross-sections of salivary gland cells from YFPrab6;CFPrab1 (B-B″), YFPrab8;CFPrab1 (C-C″), YFPrab2;CFPrab1 (D-D″) and YFPrab7;CFPrab1 (E-E″) larvae probed for direct YFP fluorescence (B-E, in gray and B”-E” in magenta) or stained against CFP-HA (B’-E’, in gray and B″-E″ in green). Arrows and arrowheads point to individual Rab domains; in B″-E″ dashed lines indicate cell nucleus (N) and dotted lines mark the apical domain; scale bars indicate 5µm. Note that Rab6 accumulates apically and in a domain adjacent to Rab1 basally, whereas Rab2, Rab7 and Rab8 partially overlap with Rab1.
How is Rab traffic spatially and temporally organized? A sequence of Rab proteins regulates the directed transport of specific cargo species and the Rab target-membrane specificity is controlled by GAPs and GEFs.29,30 However, how GEF/GAP specificity and regulatory activity is orchestrated in time and space remains unclear.28 Yeast Rabs can recruit GEFs that activate the next Rab in the traffic sequence, as well as GAPs that deactivate the predecessor.31,32 Similar regulatory mechanisms have also been reported for Rabs in higher eukaryotes.33,34 Other studies show that the hypervariable C-terminal domain of each Rab is instructive for insertion into specific membranes,35 and that active Rabs are able to induce self-amplifying loops by generating functional lipid micro-domains.16,36 Interestingly, on the same membrane entity multiple different Rab micro-domains can coexist.37 Therefore, specific combinations of Rabs may constitute a “Rab code,” defining the identities of intracellular transport vehicles.38
Rab proteins in drosophila melanogaster
Genetic resources that allow systematic analyses of protein localization in vivo are established for baker's yeast,39 C. elegans40 and fruit flies.41 However, comprehensive collections of tagged rab alleles suitable to study endogenous Rab dependent transport in a complex model organism emerged only recently for Drosophila. In Drosophila melanogaster, 31 members of the Rab-GTPase family were identified based on sequence comparisons.9 Many Rabs are expressed in specific patterns during embryogenesis,9 and transcription profiles indicate dynamic expression levels of many Rab transcripts throughout development and FlyBase.org. However, it is important to explore how mRNA expression patterns correlate with the localization, levels and activities of the corresponding proteins. To this aim, Dunst et al. generated a comprehensive set of endogenously modified rab alleles (Yrabs). The YRab-collection comprises 27 strains; each different Rab is N-terminally tagged with YFP14 (rablibrary.mpi-cbg.de). The YFP-tag allows comparing the distributions of different Rab proteins directly and provides a common target for RNAi or nanobody-induced protein depletion approaches. Dunst and colleagues systematically analyzed the expression, localization and abundance of Yrabs in 6 different organs, which harbor a total of 23 different cell types. Each cell type is characterized by a unique composition of the Rab machinery. For example, all cell types sampled have high levels of Rab1 mRNA and protein. In contrast, Rab10 mRNA levels are generally high but only imaginal disk cells show correspondingly high protein levels, whereas all other cell types tested have low Rab10 protein levels. Therefore, some Rabs may have cell type specific mRNA translation or protein turnover rates. In addition, the organization of Rab compartments is dependent on the state of cellular differentiation. For example, Rab3 and Rab27 regulate synaptic activity in a redundant fashion, and their expression is limited to very few cell types.14,42 However, their expression and localization differ strikingly. Rab3 is expressed during early axonogenesis and the protein is distributed throughout the cytosol (Fig. 1). Only upon initiation of synaptogenesis does Rab3 become restricted to presynaptic contact sites. In contrast, Rab27 protein is not detectable in embryonic neurons and only appears in interconnected neurons at mature synaptic contacts (rablibrary.mpi-cbg.de).
Figure 1.
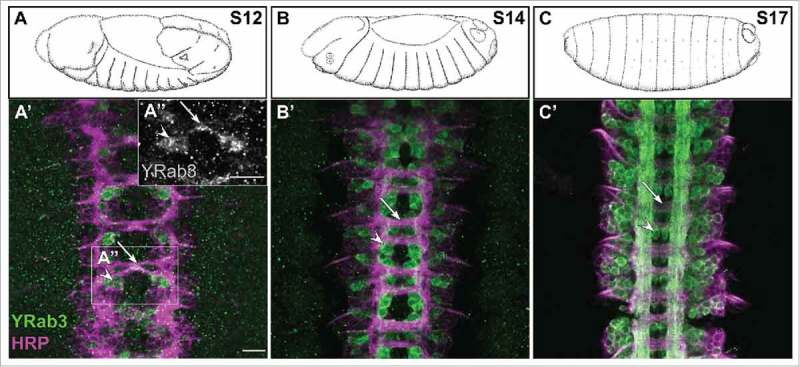
Rab3 expression in embryonic neurons. Rab3 and Rab27 are pan-neuronal presynaptic vesicle identifiers. In contrast to Rab27 (no detectable YRab27 in embryonic neurons, data not shown), Rab3 is expressed in a subset of wiring neurons (before synapse formation). Interestingly, after axogenesis all interconnected neurons show Rab3 and Rab27 restricted to presynaptic membranes. A-C Cartoons (adapted from Fly atlas (Hartenstein, V. (1993) Atlas of Drosophila development and flybase.org) depicting different developmental stages of Drosophila embryos. (A’-C’) Images from corresponding Yrab3 embryonic ventral nerve cords. Of note, YRab3 (green) is detectable in neural cell bodies (A’-C’, white arrowheads) and projections (A’-C’, white arrows). HRP (magenta) labels all neuronal membranes, developmental stages are indicated (A-C, upper left corner), scale bars indicate 20µm.
What is the biologic relevance of the intracellular compartmentalization of Rab proteins? Total Rab protein levels do not reflect the proportion of active GTP-bound Rabs, and visualizing the localization of YRabs does not allow discriminating between different Rab activity states. However, active GTP-Rabs are generally integrated into membranes, whereas inactive GDP-Rabs are GDI-bound and distributed in the cytosol. Biochemical approaches can be used to separate these Rab pools. Isopycnic density gradients can be used to sediment cytosolic Rabs.43 Consequently, the non-sedimenting Rab protein fraction represents GTP-Rabs engaged in membrane traffic. However, an in-depth understanding of traffic routes requires systematic mapping of the subcellular localization of all Rabs and characterizing their functional interconnections. The most common strategy is to use immunohistochemical approaches. The detection of Rab proteins with fluorescent probes often reveals prominent puncta (rablibrary.mpi-cbg.de). Notably, however, the subcellular distribution of membrane proteins in fixed samples is prone to artifacts,44 and only live imaging can reveal the dynamic morphologies of intracellular membrane compartments. For instance, compartments that move from the trans-Golgi network toward the plasma membrane often resemble tubular shapes, distinct from spherical vesicles.45 Thus, fixation and staining methods need to be carefully validated to minimize experimental artifacts. Dunst and colleagues used fixed samples and annotated systematically the localization of each Rab in many different cell types using defined and unbiased terminology for grouping all Rabs into similarity-clusters. For instance, Rab dependent ER traffic is well studied, and Rab1, 2, 10 and 18 are known to regulate traffic between ER and other organelles46,47 or maintain ER structure.48 In Drosophila salivary gland cells these ER-Rabs show similar distribution and the authors grouped these Rabs into one annotation cluster (Fig. 2, D-D″). Rab6 is linked to Golgi and post-Golgi transport.49 Although Rab6 is localized in puncta, the Rab6 annotation tree is different from the ER-Rab cluster (Fig. 2, B-B″). Surprisingly, also Rab7 and 8 clustered together with ER-Rabs. Only Rab7 was found to localize at ER contact sites.50 So far, Rab851 is not reported to organize ER transport. Are Rab7 and 8 associated with ER membranes in Drosophila cells? In salivary gland cells Rab7 and Rab8 partially co-localize with Rab1, thus confirming that the annotation correctly grouped Rab7 and Rab8 with Rab1 (Fig. 2, C-C″ and E-E″).
The collection of tagged rab alleles in Drosophila represents a unique resource to systematically chart intracellular transport in vivo. The findings indicate a very complex interplay between protein expression, localization and activity. Thus, membrane and protein traffic pathways are likely to depend to a large extent on the particular cell types, developmental stages and physiologic conditions analyzed in a given study, suggesting that existing models of cellular traffic may in some cases be too superficial.
Essential permanent transport and dedicated transient traffic crossroads
Differentiation and tissue morphogenesis are interconnected processes that follow a hard-wired sequence of events. How do cells organize their protein and membrane traffic according to morphogenetic programs, and what kinds of signals control the spatial and temporal constitution of the intracellular transport machinery?
DTT pathways are induced locally, and ensure spatially and temporally targeted delivery of membrane material and cargo. However, the initiation, regulation and composition of DTTs are not well understood. Recently, Caviglia and colleagues reported new insights into one particular DTT route. They studied the formation of tubular connections in the tracheal system in Drosophila, which consists of a network of epithelial tubes.52,53 The tracheal network transports oxygen to tissues. It originates from segmentally repeated invaginations of the embryonic ectoderm that extend and undergo a series of branching events.54,55 Tracheal tubes consist of an epithelial monolayer that surrounds a lumen lined by apical extracellular matrix (cuticle) that prevents tracheal tubes from collapsing.
In the tracheal system, tubular connections (anastomoses) are built between pre-existing lumenized branches. These connections are made by specialized tip-cells called fusion cells (FCs; Fig. 3). FCs migrate toward each other, and, upon contact, form a new adherens junction at the contact site. The fusion cells then repolarize and form a secondary apical domain at the cell-cell contact site. The original primary apical side of each tip cell faces the invading stalk cell lumen, where the 2 cell types are connected via adherens junctions. The stalk cell ‘invades’ the tip cell (resembling a finger poking into a balloon), bringing the primary FC apical membrane very close to the secondary apical membrane domain at the contact between the 2 tip cells.52 The subsequent intracellular fusion of the 2 apical membrane domains leads to the formation of a luminal connection between the adjacent branches (Fig. 3).
Figure 3.
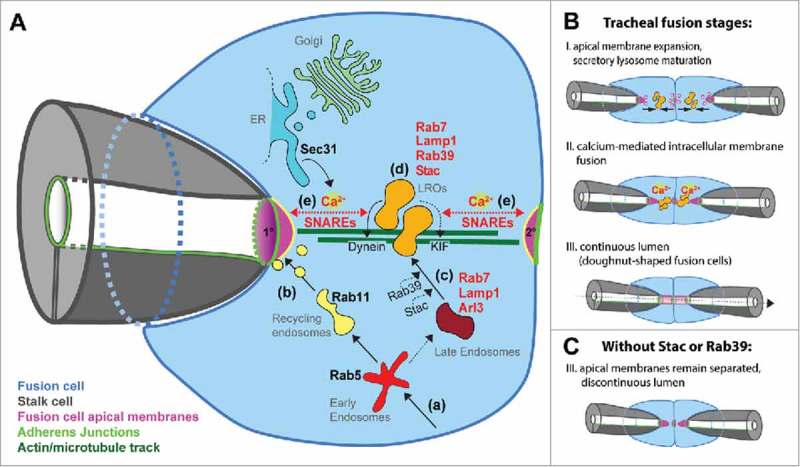
Overview of membrane trafficking steps during tracheal tube fusion. (A) Scheme depicting a fusion tip cell (blue) invaded by a stalk cell (gray). The primary (1°) and secondary (2°) apical membranes are in magenta, adherens junctions in bright green, actin/microtubule track in dark green, ER in blue, Golgi apparatus in light green, LROs in orange, late endosomes in dark red, early endosomes in light red and recycling endosomes in yellow. Broken lines represent hypothetical trafficking steps; continuous lines represent experimentally validated steps. Endocytic compartments (a) donate membrane material to apical membranes (b) or mature to give rise to late endosomes (c). Arl3 is recruited to Rab7- and Lamp1- positive compartments, and Arl3 effectors recruit Munc13–4/Staccato and Rab39. Dynein and kinesin (KIF) motor proteins are effectors of Rab7 and/or Rab39, and link LROs to the actin/microtubule track (d). Once the apical plasma membrane domains are reached, Ca2+-bound Munc13–4/Staccato promotes SNARE-dependent membrane fusion (e). Ca2+-enriched microdomains are established by localized release from Sec 31-positive ER portions close to the fusing membranes. (B) Stages of lumen fusion in the wild type (I-III). (C) In the absence of Stac or Rab39, LROs fail to form. As a consequence the 2 apposed apical membranes fail to fuse, although fusion cells are still able to expand their apical domains. (Modified after Caviglia et al. NCB, 2016).
Caviglia et al. investigated the mechanism of intracellular membrane fusion in the tip cells and identified lysosome-related secretory organelles (LROs) essential for lumen fusion. LROs are pleomorphic organelles in several specialized cell types (including, among others cytotoxic T-cells, endothelial cells and pigment cells56) and have cell specific origin membranes. These compartments store specific cargo and acquire specific structural proteins/luminal environments. In response to external stimuli LROs release their cargo by fusing with the plasma membrane in a spatiotemporally, tightly regulated manner.15,57 Interestingly, early findings identified Rab geranylgeranyl transferases important for successful LRO maturation and secretion.58 Later, individual Rab proteins were associated with specific LRO maturation and traffic steps.56,59,60 However, only recently Azous et al. published a comprehensive screen aimed to identify all Rabs that regulate granule secretion in mast cells. The authors identified 30 Rabs involved in exocytosis, including Rabs that affect selectively secretion triggered by a Ca2+ ionophore. Among these Rabs responsible to control Ca2+-induced release is Rab39A.61 Interestingly, in tracheal fusion cells Caviglia and colleagues found Arl3-dependent recruitment of Rab39 and Munc13–4/Staccato, a Ca2+-binding protein,62 onto Rab7- and LAMP1-positive LROs. Active Rab7 and Rab39 are known to bind effectors, including Dynein63 and kinesin-3 KIF1A64 motor proteins, which link LROs to microtubules65 and can initiate transport along microtubules toward the apical plasma-membrane. In addition, the authors describe the appearance of local transient intracellular Ca2+ sparks specific to the tip cell type that mediates lumen fusion, by triggering the fusion of LROs to each apical plasma membrane domain, thus creating a connection between pre-existing lumina. Thus, in tip cells the Rab machinery may act to position the membrane fusion machinery in the right place to mediate the connection of growing lumina.
Experimental challenges
Intracellular transport is tightly regulated. However, the plasticity of such regulative networks is hardly assessed and comprehensive expression studies of endogenous Rabs show large spatial, temporal and quantitative differences. Therefore experimental setups need to be evaluated and standardised. Available genetic tools yielded many important findings. However, misexpression and perturbation of multiple pathways may blur results and make interpretations difficult. Therefore, here we propose novel strategies to manipulate Rab dependent transport with a high spatio-temporal resolution.
Closing remarks
What are the next aims? Although some principles apply to all Rabs, such as GDP/GTP-switch regulation by GEFs and GAPs, other paradigms may not apply generally (e.g., mechanisms of Rab membrane insertion and extraction). A challenging task is to validate existing molecular and mechanistic models for all members of the Rab family. A second major challenge is to systematically chart the Rab machinery in each cell type. Many Rabs are allocated, based on published findings in different cell types, to specific organelles or traffic routes. However, recent findings show cell-type specific complements of Rab proteins, consistent with the idea of a “Rab code” that is characteristic of each cell type. Comprehensive libraries of endogenously tagged rab alleles provide a powerful tool to faithfully characterize cell type specific transport routes and Rab codes.
Rab protein localization and loss of function rescue strategies
Many Rabs are small proteins and their surface shows only limited immunogenic activity. Therefore, antibodies that specifically detect the intracellular localization of endogenous Rab proteins are rare.66 As an alternative to antibodies, expression of tagged transgenes (for instance YFP-tagged wild-type rab alleles) can be used to study Rab protein localization. However, experiments using over- or misexpression of Rab transgenes need to be interpreted with great care. First, increased Rab levels or ectopic Rab activity can lead to mutant phenotypes. Second, large amounts of inactive cytosolic transgenic protein can mask the localization of the limited pool of active Rab proteins.11-14,67 Genomic rescue constructs41 in mutant backgrounds or manipulation of endogenous rab loci by homologous recombination14 are powerful approaches that allow targeting the introduced tag for controlled Rab RNA and/or protein knock-down experiments.42,68
Rab protein depletion
Mutagenesis, RNAi knock-down or nanobody approaches reduce the entire cellular Rab protein pool. The expression of dominant-negative alleles is another way to impede cellular Rab function.9 Dominant negative Rabs preferentially bind GDP and outcompete the endogenous Rab pool for the binding of regulatory elements. Therefore, the GTP-bound Rab pool is depleted and all traffic controlled by the affected Rab is jammed. However, overexpressed dominant-negative Rabs may interfere not only with the targeted Rab protein, but also with other Rabs, and therefore the effects may be to some extent ambiguous. Moreover, a given Rab can be involved in multiple traffic steps and the complete Rab depletion can result in phenotypes that may reflect the perturbation of multiple traffic routes. In addition, the plasticity of the transport machinery may be able to compensate for the affected traffic step.16,38
Genetic tweezers to dissect Rab traffic steps
To manipulate individual traffic steps we propose a new approach for local deactivation of Rab proteins. Particular cellular membrane compartments and organelles are characterized by distinct Rabs, and membrane maturation/differentiation processes are characterized by a change in the set of Rabs that decorate the corresponding membranes (“Rab identity”). In consequence, for a period of time different Rabs share the very same membrane compartment and therefore come in close spatial proximity. The idea is to use that time frame and local proteolytic activity to perturb the membrane maturation sequence. Our experimental approach uses a 2-component system with one Rab protein tagged with the Tobacco-etch virus Protease (TevP, Fig. 4A) and a second Rab tagged with a TevP cleavage site (TEV, Fig. 4B) inserted between the GTPase domain and the membrane anchor. The correct genetic combination of TevP-Rab and TEV-Rab allows blocking one specific membrane traffic step without impeding Rab functionality in other trafficking steps.
Figure 4.
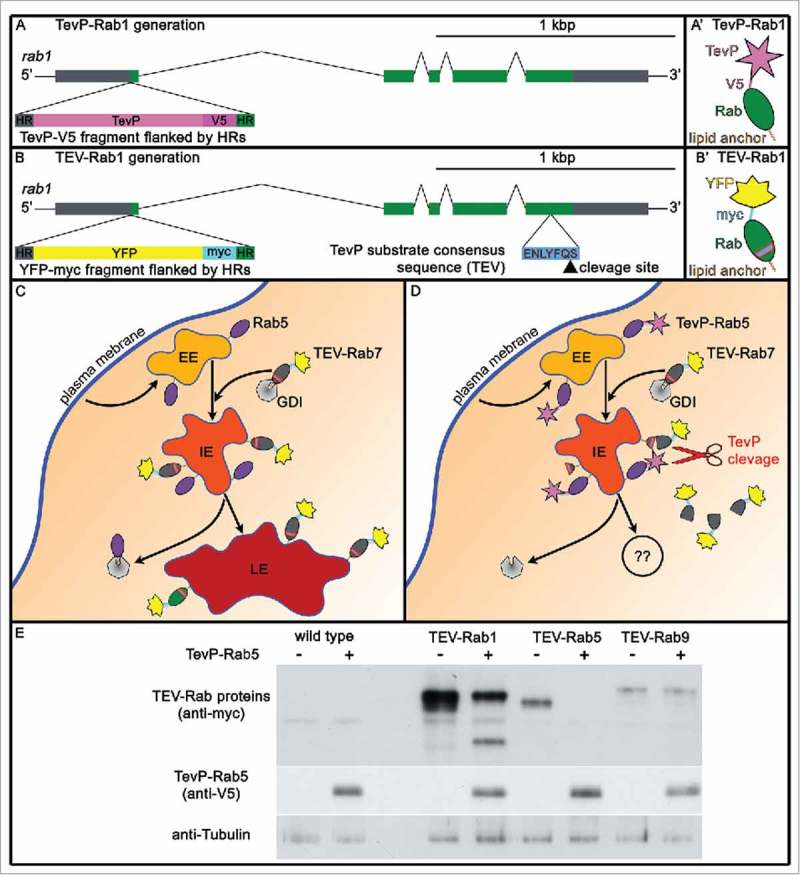
(A) novel strategy to impede intracellular protein/membrane traffic. A,B Homologous recombination strategy for generating TevP-rab and TEV-rab alleles. A and B are the genomic regions with the recombination points and the exogenous DNA inserted, which is indicated in each case. A' and B' are the proteins generated. TevP-rab1 (A,A’) and TEV-rab1 (B,B’) are shown as an example. 5′ and 3′ UTRs, gray; rab1 coding regions, green; TevP, pink; YFP, yellow; V5, magenta; MYC, cyan; HR stands for ‘Homology Region’. C,D Sketch depicts endosomal maturation without processing (C) and upon TEV-Rab7 cleavage by TevP-Rab5 (D). TevP cleavage of TEV-Rabs disconnects the Rab-GTPase domain from membranes. Early endosomes (EE) are in light-orange, intermediate endosomes (IE) in orange and late endosomes (LE) in red. TevP (pink star) tagged Rab5, and YFP (yellow) tagged TEV-Rab7 are depicted. E Larval fat body protein lysates from wild type (lanes 1 and 2), TEV-Rab1 (lane 3), TEV-Rab1;TevP-Rab5 (lane 4), TEV-Rab5 (lane 5), TEV-Rab5;TevP-Rab5 (lane 6), TEV-Rab9 (lane 7), TEV-Rab9;TevP-Rab5 (lane 8) probed for MYC (top), V5 (middle) and Tubulin (bottom). Note, TEV-Rab1 and TEV-Rab5 are cleaved by TevP-Rab5 whereas TEV-Rab9 is not. This result indicates that one traffic step requires Rab1 and Rab5 in close vicinity. Note that expression of TevP-Rab5 in a TEV-Rab5 background leads to complete degradation of TEV-Rab5 (lane 6).
For instance, flies that express TevP-Rab5 and TEV-Rab7 may show hampered endosomal maturation69,70 while Lysosome-ER transport, which depends on Rab9 and Rab7,71 should remain unperturbed (Fig. 4C-E). Systematically probing the functional relationships of Rabs using TevP-Rab/TEV-Rab combinations may prove useful to chart the sequence of Rabs required to shuttle particular protein cargo. In an analogous approach, a protein of interest is tagged with TevP (TevP-cargo) and its intracellular transport can be mapped using TEV-Rabs.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We apologize to the authors whose work is not referenced in this review due to space limitations. We thank Natalie Dye for critical reading of the manuscript, Marino Zerial for his constructive comments on TevP/TEV Rabs, and Eli Knust and Suzanne Eaton for providing resources to generate presented original research.
Funding
Work was supported by grants from the Swiss National Science Foundation (early Postdoc Mobility Fellowship) to S.C., the Max-Planck Society (MPG) to D.B-F. and J.L., the German Research Foundation (DFG, BR5490) to M.B., the Swiss National Science Foundation (SNF_31003A_141093/1, the “Cells-in-Motion” Cluster of Excellence (EXC 1003-CiM), and the DFG Collaborative Research Center SFB 1009 “Breaking barriers” to S.L.
References
- [1].Bourne HR, Sanders DA, McCormick F. The GTPase superfamily: a conserved switch for diverse cell functions. Nature 1990; 348:125-132; PMID:2122258; http://dx.doi.org/ 10.1038/348125a0 [DOI] [PubMed] [Google Scholar]
- [2].Zeigerer A, Bogorad RL, Sharma K, Gilleron J, Seifert S, Sales S, Berndt N, Bulik S, Marsico G, D'Souza RC, et al.. Regulation of liver metabolism by the endosomal GTPase Rab5. Cell Reports 2015; 11:884-892; PMID:25937276; http://dx.doi.org/ 10.1016/j.celrep.2015.04.018 [DOI] [PubMed] [Google Scholar]
- [3].Aderem A, Underhill DM. Mechanisms of phagocytosis in macrophages. Annual Rev Immunol 1999; 17:593-623; PMID:10358769; http://dx.doi.org/ 10.1146/annurev.immunol.17.1.593 [DOI] [PubMed] [Google Scholar]
- [4].Beningo KA, Wang YL. Fc-receptor-mediated phagocytosis is regulated by mechanical properties of the target. J Cell Sci 2002; 115:849-856; PMID:11865040 [DOI] [PubMed] [Google Scholar]
- [5].Jaconi ME, Lew DP, Carpentier JL, Magnusson KE, Sjogren M, Stendahl O. Cytosolic free calcium elevation mediates the phagosome-lysosome fusion during phagocytosis in human neutrophils. J Cell Biol 1990; 110:1555-1564; PMID:2110568; http://dx.doi.org/ 10.1083/jcb.110.5.1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Zucker RS. Calcium- and activity-dependent synaptic plasticity. Curr Opin Neurobiol 1999; 9:305-313; PMID:10395573; http://dx.doi.org/ 10.1016/S0959-4388(99)80045-2 [DOI] [PubMed] [Google Scholar]
- [7].Talbot WS, Swyryd EA, Hogness DS. Drosophila tissues with different metamorphic responses to ecdysone express different ecdysone receptor isoforms. Cell 1993; 73:1323-1337; PMID:8324824; http://dx.doi.org/ 10.1016/0092-8674(93)90359-X [DOI] [PubMed] [Google Scholar]
- [8].Zerial M, McBride H. Rab proteins as membrane organizers. Nat Rev Mol Cell Biol 2001; 2:107-117; PMID:11252952; http://dx.doi.org/ 10.1038/35052055 [DOI] [PubMed] [Google Scholar]
- [9].Zhang J, Schulze KL, Hiesinger PR, Suyama K, Wang S, Fish M, Acar M, Hoskins RA, Bellen HJ, Scott MP. Thirty-one flavors of Drosophila rab proteins. Genetics 2007; 176:1307-1322; PMID:17409086; http://dx.doi.org/ 10.1534/genetics.106.066761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Chan CC, Scoggin S, Wang D, Cherry S, Dembo T, Greenberg B, Jin EJ, Kuey C, Lopez A, Mehta SQ, et al.. Systematic discovery of Rab GTPases with synaptic functions in Drosophila. Curr Biol 2011; 21:1704-1715; PMID:22000105; http://dx.doi.org/ 10.1016/j.cub.2011.08.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Bucci C, Parton RG, Mather IH, Stunnenberg H, Simons K, Hoflack B, Zerial M. The small GTPase rab5 functions as a regulatory factor in the early endocytic pathway. Cell 1992; 70:715-728; PMID:1516130; http://dx.doi.org/ 10.1016/0092-8674(92)90306-W [DOI] [PubMed] [Google Scholar]
- [12].van der Sluijs P, Hull M, Webster P, Male P, Goud B, Mellman I. The small GTP-binding protein rab4 controls an early sorting event on the endocytic pathway. Cell 1992; 70:729-740; PMID:1516131; http://dx.doi.org/ 10.1016/0092-8674(92)90307-X [DOI] [PubMed] [Google Scholar]
- [13].Mottola G, Classen AK, Gonzalez-Gaitan M, Eaton S, Zerial M. A novel function for the Rab5 effector Rabenosyn-5 in planar cell polarity. Development 2010; 137:2353-2364; PMID:20534670; http://dx.doi.org/ 10.1242/dev.048413 [DOI] [PubMed] [Google Scholar]
- [14].Dunst S, Kazimiers T, von Zadow F, Jambor H, Sagner A, Brankatschk B, Mahmoud A, Spannl S, Tomancak P, Eaton S, et al.. Endogenously tagged rab proteins: a resource to study membrane trafficking in Drosophila. Dev Cell 2015; 33:351-365; PMID:25942626; http://dx.doi.org/ 10.1016/j.devcel.2015.03.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Caviglia S, Brankatschk M, Fischer EJ, Eaton S, Luschnig S. Staccato/Unc-13-4 controls secretory lysosome-mediated lumen fusion during epithelial tube anastomosis. Nat Cell Biol 2016; 18:727-739; PMID:27323327 [DOI] [PubMed] [Google Scholar]
- [16].Grosshans BL, Ortiz D, Novick P. Rabs and their effectors: achieving specificity in membrane traffic. Proc Natl Acad Sci U S A 2006; 103:11821-11827; PMID:16882731; http://dx.doi.org/ 10.1073/pnas.0601617103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Jean S, Kiger AA. Coordination between RAB GTPase and phosphoinositide regulation and functions. Nat Rev Mol Cell Biol 2012; 13:463-470; PMID:22722608; http://dx.doi.org/ 10.1038/nrm3379 [DOI] [PubMed] [Google Scholar]
- [18].Platta HW, Stenmark H. Endocytosis and signaling. Curr Opin Cell Biol 2011; 23:393-403; PMID:21474295; http://dx.doi.org/ 10.1016/j.ceb.2011.03.008 [DOI] [PubMed] [Google Scholar]
- [19].Sadowski L, Pilecka I, Miaczynska M. Signaling from endosomes: location makes a difference. Exp Cell Res 2009; 315:1601-1609; PMID:18930045; http://dx.doi.org/ 10.1016/j.yexcr.2008.09.021 [DOI] [PubMed] [Google Scholar]
- [20].Pfeffer S, Aivazian D. Targeting Rab GTPases to distinct membrane compartments. Nat Rev Mol Cell Biol 2004; 5:886-896; PMID:15520808; http://dx.doi.org/ 10.1038/nrm1500 [DOI] [PubMed] [Google Scholar]
- [21].Ullrich O, Horiuchi H, Alexandrov K, Zerial M. Use of Rab-Gdp dissociation inhibitor for solubilization and delivery of Rab Proteins to Biological-Membranes in Streptolysin O-Permeabilized cells. Method Enzymol 1995; 257:243-253 [DOI] [PubMed] [Google Scholar]
- [22].Ullrich O, Stenmark H, Alexandrov K, Huber LA, Kaibuchi K, Sasaki T, Takai Y, Zerial M. Rab Gdp dissociation inhibitor as a general regulator for the membrane association of Rab Proteins. J Biol Chem 1993; 268:18143-18150; PMID:8349690 [PubMed] [Google Scholar]
- [23].Gavriljuk K, Itzen A, Goody RS, Gerwert K, Kotting C. Membrane extraction of Rab proteins by GDP dissociation inhibitor characterized using attenuated total reflection infrared spectroscopy. Proc Natl Acad Sci U S A 2013; 110:13380-13385; PMID:23898197; http://dx.doi.org/ 10.1073/pnas.1307655110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Wrobel TP, Marzec KM, Majzner K, Kochan K, Bartus M, Chlopicki S, Baranska M. Attenuated total reflection Fourier transform infrared (ATR-FTIR) spectroscopy of a single endothelial cell. Analyst 2012; 137:4135-4139; PMID:22854681; http://dx.doi.org/ 10.1039/c2an35331h [DOI] [PubMed] [Google Scholar]
- [25].Goody RS, Rak A, Alexandrov K. The structural and mechanistic basis for recycling of Rab proteins between membrane compartments. Cell Mol Life Sci 2005; 62:1657-1670; PMID:15924270; http://dx.doi.org/ 10.1007/s00018-005-4486-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Suh HY, Lee DW, Lee KH, Ku B, Choi SJ, Woo JS, Kim YG, Oh BH. Structural insights into the dual nucleotide exchange and GDI displacement activity of SidM/DrrA. EMBO J 2010; 29:496-504; PMID:19942850; http://dx.doi.org/ 10.1038/emboj.2009.347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Wu YW, Oesterlin LK, Tan KT, Waldmann H, Alexandrov K, Goody RS. Membrane targeting mechanism of Rab GTPases elucidated by semisynthetic protein probes. Nat Chem Biol 2010; 6:534-540; PMID:20512138; http://dx.doi.org/ 10.1038/nchembio.386 [DOI] [PubMed] [Google Scholar]
- [28].Barr FA. Review series: Rab GTPases and membrane identity: causal or inconsequential? J Cell Biol 2013; 202:191-199; PMID:23878272; http://dx.doi.org/ 10.1083/jcb.201306010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Blumer J, Rey J, Dehmelt L, Mazel T, Wu YW, Bastiaens P, Goody RS, Itzen A. RabGEFs are a major determinant for specific Rab membrane targeting. J Cell Biol 2013; 200:287-300; PMID:23382462; http://dx.doi.org/ 10.1083/jcb.201209113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Cherfils J, Zeghouf M. Regulation of small GTPases by GEFs, GAPs, and GDIs. Physiol Rev 2013; 93:269-309; PMID:23303910; http://dx.doi.org/ 10.1152/physrev.00003.2012 [DOI] [PubMed] [Google Scholar]
- [31].Rivera-Molina FE, Novick PJ. A Rab GAP cascade defines the boundary between two Rab GTPases on the secretory pathway. Proc Natl Acad Sci U S A 2009; 106:14408-14413; PMID:19666511; http://dx.doi.org/ 10.1073/pnas.0906536106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Nottingham RM, Pfeffer SR. Defining the boundaries: Rab GEFs and GAPs. Proc Natl Acad Sci U S A 2009; 106:14185-14186; PMID:19706500; http://dx.doi.org/ 10.1073/pnas.0907725106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Bryant DM, Datta A, Rodriguez-Fraticelli AE, Peranen J, Martin-Belmonte F, Mostov KE. A molecular network for de novo generation of the apical surface and lumen. Nat Cell Biol 2010; 12:1035-1045; PMID:20890297; http://dx.doi.org/ 10.1038/ncb2106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Langemeyer L, Barr FA. Analysis of rab GTPases. Curr Protocols Cell Biol 2012; 57:15.18:15.18.1–15.18.17; PMID:23208545 [DOI] [PubMed] [Google Scholar]
- [35].Li F, Yi L, Zhao L, Itzen A, Goody RS, Wu YW(. The role of the hypervariable C-terminal domain in Rab GTPases membrane targeting. Proc Natl Acad Sci U S A 2014; 111:2572-2577; PMID:24550285; http://dx.doi.org/ 10.1073/pnas.1313655111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Simonsen A, Lippe R, Christoforidis S, Gaullier JM, Brech A, Callaghan J, Toh BH, Murphy C, Zerial M, Stenmark H. EEA1 links PI(3)K function to Rab5 regulation of endosome fusion. Nature 1998; 394:494-498; PMID:9697774; http://dx.doi.org/ 10.1038/28879 [DOI] [PubMed] [Google Scholar]
- [37].Sonnichsen B, De Renzis S, Nielsen E, Rietdorf J, Zerial M. Distinct membrane domains on endosomes in the recycling pathway visualized by multicolor imaging of Rab4, Rab5, and Rab11. J Cell Biol 2000; 149:901-914; PMID:10811830; http://dx.doi.org/ 10.1083/jcb.149.4.901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Fukuda M. Regulation of secretory vesicle traffic by Rab small GTPases. Cell Mol Life Sci 2008; 65:2801-2813; PMID:18726178; http://dx.doi.org/ 10.1007/s00018-008-8351-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Kumar A, Agarwal S, Heyman JA, Matson S, Heidtman M, Piccirillo S, Umansky L, Drawid A, Jansen R, Liu Y, et al.. Subcellular localization of the yeast proteome. Gene Dev 2002; 16:707-719; PMID:11914276; http://dx.doi.org/ 10.1101/gad.970902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Sarov M, Schneider S, Pozniakovski A, Roguev A, Ernst S, Zhang YM, Hyman AA, Stewart AF. A recombineering pipeline for functional genomics applied to Caenorhabditis elegans. Nat Methods 2006; 3:839-844; PMID:16990816; http://dx.doi.org/ 10.1038/nmeth933 [DOI] [PubMed] [Google Scholar]
- [41].Sarov M, Barz C, Jambor H, Hein MY, Schmied C, Suchold D, Stender B, Janosch S, K JV, Krishnan RT, et al.. A genome-wide resource for the analysis of protein localisation in Drosophila. eLife 2016; 5:e12068; PMID:26896675; http://dx.doi.org/ 10.7554/eLife.12068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Brankatschk M, Dunst S, Nemetschke L, Eaton S. Delivery of circulating lipoproteins to specific neurons in the Drosophila brain regulates systemic insulin signaling. eLife 2014; 3:e02862; PMID:25275323; http://dx.doi.org/ 10.7554/eLife.02862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Sivars U, Aivazian D, Pfeffer SR. Yip3 catalyses the dissociation of endosomal Rab-GDI complexes. Nature 2003; 425:856-859; PMID:14574414; http://dx.doi.org/ 10.1038/nature02057 [DOI] [PubMed] [Google Scholar]
- [44].Schnell U, Dijk F, Sjollema KA, Giepmans BN. Immunolabeling artifacts and the need for live-cell imaging. Nat Methods 2012; 9:152-158; PMID:22290187; http://dx.doi.org/ 10.1038/nmeth.1855 [DOI] [PubMed] [Google Scholar]
- [45].Keller P, Toomre D, Diaz E, White J, Simons K. Multicolour imaging of post-Golgi sorting and trafficking in live cells. Nat Cell Biol 2001; 3:140-149; PMID:11175746; http://dx.doi.org/ 10.1038/35055042 [DOI] [PubMed] [Google Scholar]
- [46].Tisdale EJ, Bourne JR, Khosravi-Far R, Der CJ, Balch WE. GTP-binding mutants of rab1 and rab2 are potent inhibitors of vesicular transport from the endoplasmic reticulum to the Golgi complex. J Cell Biol 1992; 119:749-761; PMID:1429835; http://dx.doi.org/ 10.1083/jcb.119.4.749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Dejgaard SY, Murshid A, Erman A, Kizilay O, Verbich D, Lodge R, Dejgaard K, Ly-Hartig TB, Pepperkok R, Simpson JC, et al.. Rab18 and Rab43 have key roles in ER-Golgi trafficking. J Cell Sci 2008; 121:2768-2781; PMID:18664496; http://dx.doi.org/ 10.1242/jcs.021808 [DOI] [PubMed] [Google Scholar]
- [48].English AR, Voeltz GK. Rab10 GTPase regulates ER dynamics and morphology. Nat Cell Biol 2013; 15:169-178; PMID:23263280; http://dx.doi.org/ 10.1038/ncb2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Kondylis V, Rabouille C. The Golgi apparatus: lessons from Drosophila. FEBS Letters 2009; 583:3827-3838; PMID:19800333; http://dx.doi.org/ 10.1016/j.febslet.2009.09.048 [DOI] [PubMed] [Google Scholar]
- [50].Rowland AA, Chitwood PJ, Phillips MJ, Voeltz GK. ER contact sites define the position and timing of endosome fission. Cell 2014; 159:1027-1041; PMID:25416943; http://dx.doi.org/ 10.1016/j.cell.2014.10.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Hsu VW, Prekeris R. Transport at the recycling endosome. Curr Opin Cell Biol 2010; 22:528-534; PMID:20541925; http://dx.doi.org/ 10.1016/j.ceb.2010.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Caviglia S, Luschnig S. Tube fusion: making connections in branched tubular networks. Seminars Cell Dev Biol 2014; 31:82-90; http://dx.doi.org/ 10.1016/j.semcdb.2014.03.018 [DOI] [PubMed] [Google Scholar]
- [53].Sigurbjornsdottir S, Mathew R, Leptin M. Molecular mechanisms of de novo lumen formation. Nat Rev Mol Cell Biol 2014; 15:665-676; PMID:25186133; http://dx.doi.org/ 10.1038/nrm3871 [DOI] [PubMed] [Google Scholar]
- [54].Ghabrial A, Luschnig S, Metzstein MM, Krasnow MA. Branching morphogenesis of the Drosophila tracheal system. Annual Rev Cell Dev Biol 2003; 19:623-647; PMID:14570584; http://dx.doi.org/ 10.1146/annurev.cellbio.19.031403.160043 [DOI] [PubMed] [Google Scholar]
- [55].Ochoa-Espinosa A, Affolter M. Branching morphogenesis: from cells to organs and back. Cold Spring Harbor Perspectives Biol 2012; 4; http://dx.doi.org/ 10.1101/cshperspect.a008243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Marks MS, Heijnen HF, Raposo G. Lysosome-related organelles: unusual compartments become mainstream. Curr Opin Cell Biol 2013; 25:495-505; PMID:23726022; http://dx.doi.org/ 10.1016/j.ceb.2013.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Pu J, Guardia CM, Keren-Kaplan T, Bonifacino JS. Mechanisms and functions of lysosome positioning. J Cell Sci 2016; 129:4329-4339; PMID:27799357; http://dx.doi.org/ 10.1242/jcs.196287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Detter JC, Zhang Q, Mules EH, Novak EK, Mishra VS, Li W, McMurtrie EB, Tchernev VT, Wallace MR, Seabra MC, et al.. Rab geranylgeranyl transferase alpha mutation in the gunmetal mouse reduces Rab prenylation and platelet sJynthesis. Proc Natl Acad Sci US A 2000; 97:4144-4149; PMID:10737774; http://dx.doi.org/ 10.1073/pnas.080517697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Stinchcombe J, Bossi G, Griffiths GM. Linking albinism and immunity: the secrets of secretory lysosomes. Science 2004; 305:55-59; PMID:15232098; http://dx.doi.org/ 10.1126/science.1095291 [DOI] [PubMed] [Google Scholar]
- [60].Raposo G, Marks MS, Cutler DF. Lysosome-related organelles: driving post-Golgi compartments into specialisation. Current Opin Cell Biol 2007; 19:394-401; PMID:17628466; http://dx.doi.org/ 10.1016/j.ceb.2007.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Azouz NP, Hammel I, Sagi-Eisenberg R. Characterization of mast cell secretory granules and their cell biology. DNA Cell Biol 2014; 33:647-651; PMID:24988214; http://dx.doi.org/ 10.1089/dna.2014.2543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Koch H, Hofmann K, Brose N. Definition of Munc13-homology-domains and characterization of a novel ubiquitously expressed Munc13 isoform. Biochem J 2000; 349:247-253; PMID:10861235; http://dx.doi.org/ 10.1042/bj3490247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Jordens I, Fernandez-Borja M, Marsman M, Dusseljee S, Janssen L, Calafat J, Janssen H, Wubbolts R, Neefjes J. The Rab7 effector protein RILP controls lysosomal transport by inducing the recruitment of dynein-dynactin motors. Curr Biol 2001; 11:1680-1685; PMID:11696325; http://dx.doi.org/ 10.1016/S0960-9822(01)00531-0 [DOI] [PubMed] [Google Scholar]
- [64].Gillingham AK, Sinka R, Torres IL, Lilley KS, Munro S. Toward a comprehensive map of the effectors of rab GTPases. Developmental Cell 2014; 31:358-373; PMID:25453831; http://dx.doi.org/ 10.1016/j.devcel.2014.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Kato K, Dong B, Wada H., Tanaka-Matakatsu M, Yagi Y, Hayashi S. Microtubule-dependent balanced cell contraction and luminal-matrix modification accelerate epithelial tube fusion. Nat Commun 2016; 7:11141; PMID:27067650; http://dx.doi.org/ 10.1038/ncomms11141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Riedel F, Gillingham AK, Rosa-Ferreira C, Galindo A, Munro S. An antibody toolkit for the study of membrane traffic in Drosophila melanogaster. Biol Open 2016; 5:987-992; PMID:27256406; http://dx.doi.org/ 10.1242/bio.018937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Pataki C, Matusek T, Kurucz E, Ando I, Jenny A, Mihaly J. Drosophila Rab23 is involved in the regulation of the number and planar polarization of the adult cuticular hairs. Genetics 2010; 184:1051-1065; PMID:20124028; http://dx.doi.org/ 10.1534/genetics.109.112060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Neumuller RA, Wirtz-Peitz F, Lee S, Kwon Y, Buckner M, Hoskins RA, Venken KJ, Bellen HJ, Mohr SE, Perrimon N. Stringent analysis of gene function and protein-protein interactions using fluorescently tagged genes. Genetics 2012; 190:931-940; PMID:22174071; http://dx.doi.org/ 10.1534/genetics.111.136465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Rink J, Ghigo E, Kalaidzidis Y, Zerial M. Rab conversion as a mechanism of progression from early to late endosomes. Cell 2005; 122:735-749; PMID:16143105; http://dx.doi.org/ 10.1016/j.cell.2005.06.043 [DOI] [PubMed] [Google Scholar]
- [70].Poteryaev D, Datta S, Ackema K, Zerial M, Spang A. Identification of the switch in early-to-late endosome transition. Cell 2010; 141:497-508; PMID:20434987; http://dx.doi.org/ 10.1016/j.cell.2010.03.011 [DOI] [PubMed] [Google Scholar]
- [71].Lombardi D, Soldati T, Riederer MA, Goda Y, Zerial M, Pfeffer SR. Rab9 functions in transport between late endosomes and the trans Golgi network. EMBO J 1993; 12:677-682; PMID:8440258 [DOI] [PMC free article] [PubMed] [Google Scholar]


