Abstract
Background
Cell-free DNA has been proposed as a means of predicting complications among severely injured patients. The purpose of this systematic review was to assess whether cell-free DNA was useful as a prognostic biomarker for outcomes in trauma patients in the intensive care unit.
Methods
We searched Pubmed, Embase, Scopus and the Cochrane Central Register for Controlled Trials and reference lists of relevant articles for studies that assessed the prognostic value of cell-free DNA detection in trauma patients in the intensive care unit. Outcomes of interest included survival, posttraumatic complications and severity of trauma. Due to considerable heterogeneity between the included studies, a checklist was formed to assess quality of cell-free DNA measurement.
Results
A total of 14 observational studies, including 904 patients, were eligible for analysis. Ten studies were designed as prospective cohort studies; three studies included selected patients from a cohort while one study was of a retrospective design. We found a significant correlation between higher values of cell-free DNA and higher mortality. This significant correlation was evident as early as on intensive care unit admission. Likewise, cell-free DNA predicted the severity of trauma and posttraumatic complications in a majority of patients.
Conclusion
The amount of cell-free DNA can function as a prognostic tool for mortality and to a lesser extent severity of trauma and posttraumatic complications. Standardizing cell-free DNA measurement is paramount to ensure further research in cell-free DNA as a prognostic tool.
Electronic supplementary material
The online version of this article (doi:10.1186/s13054-016-1578-9) contains supplementary material, which is available to authorized users.
Keywords: cfDNA, mtDNA, nDNA, Trauma, Intensive care unit
Background
In recent years cell-free DNA (cfDNA) has become increasingly used as a clinical and noninvasive biomarker in the fields of cancer [1–3], pre-natal diagnostics [4], organ transplantation [5], and in several emergency conditions [6–8]. cfDNA, defined as extracellular DNA circulating freely in the blood, can be further subcategorized to circulating mitochondrial DNA (mtDNA) and circulating nuclear DNA (nDNA). Within cancer research, cfDNA has been proposed to have the ability to act as a noninvasive biopsy of the tumor (i.e., liquid-biopsy) [9] and as a prognostic marker for clinical outcomes such as disease burden [3], progression-free survival [10], and overall survival [11].
In patients admitted to an intensive care unit (ICU) as a result of trauma, cfDNA has received increasing attention under the hypothesis that cfDNA originates from cell death [12, 13] and could correlate with the severity of trauma with prognostic and predictive abilities. Preliminary reports have confirmed that the amount of cfDNA correlates inversely to mortality [14], trauma severity [15], and post-traumatic complications [16]. Due to the short half-life of cfDNA [17], it is suitable as a marker of the patient’s condition in the immediate emergency phase. mtDNA has also been increasingly investigated in trauma patients in recent years and it has been argued that mtDNA could be considered a damage-associated molecular pattern (DAMP). It is well established that circulating DAMPs lead to an increase in the innate immune response [18], possible leading to a systematic inflammatory response syndrome (SIRS) [19].
The use of cfDNA as a predictive marker of clinical outcome have not been systematically analyzed. The aim of this study was to review the literature on cfDNA as a predictive marker of clinical outcomes as measured in trauma patients in ICUs.
Methods
This systematic review was conducted according to the Preferred Reporting Items for Systematic reviews and Meta-Analyses (PRISMA) statement [20].
Eligibility criteria
The inclusion criteria for studies in this review were cohort human studies that investigated levels of cfDNA in plasma or serum in trauma patients aged ≥18 years in the ICU. Trauma was defined as all grades of trauma ranging from minor to severe trauma, including isolated traumatic brain injury (TBI) that resulted in ICU admission. Studies exclusively evaluating circulating RNA as well as studies conducted outside the ICUs were excluded. A thorough assessment of the quality of DNA sampling and processing was conducted for all included studies using previous definitions [21] (see Additional file 1). We included studies that analyzed DNA by specific sequencing (beta-globin, GAPDH, NADH dehydrogenase) or fluorescent methods. We only included published studies and only studies published in the English language.
Study search
A computerized comprehensive search strategy was conducted using four databases (PubMed, EMBASE, SCOPUS, and the Cochrane Central Register for Controlled Trials) from January 1974 to January 2016. The search was performed on 20 January 2016. The following literature search was used in PubMed: “(circulating cell free dna) OR cfdna) OR circulating nucleic acids) OR cell free mitochondrial DNA) OR nDNA) AND (injury) OR trauma) OR stress) OR surgery) OR intensive care unit) OR perioperative) OR postoperative) OR intraoperative) OR preoperative)”.
The literature search was adapted from the PubMed literature search to EMBASE, SCOPUS, and the Cochrane Central Register for Controlled Trials. We supplemented the structured literature search with searching of the reference lists from the included articles in order to find additional eligible studies.
Study selection
The Cochrane systematic review tool Covidence.org was used in the screening process. Two reviewers (MG and JB) independently screened titles and abstracts until the full-text articles were found. Two authors (MG and JB) independently assessed the full-text articles. Whenever different opinions emerged a third author (IG) was included in the discussion until consensus was reached.
Data collection and data items
All included articles were assessed for the following information: publication details, study method, patient details, specific time points of cfDNA measurements, mortality, post-traumatic complications, and trauma severity. This data was collected using a data sheet.
Risk of bias of individual studies
Risk of bias assessment was performed by two authors (MG and JB) using an adapted version of the Scottish Intercollegiate Guidelines Network (SIGN) (http://www.sign.ac.uk/methodology/checklists.html) checklist for cohort studies [22]. The SIGN checklist evaluates selection bias, performance bias, attrition bias, detection bias, and statistical analysis. All studies were assigned up to 8 points if they met the SIGN checklist criteria. Studies with 8 points were considered to be high quality (HQ) studies with little or no risk of bias, while studies between 6 and 7 and 0 and 5 were considered acceptable quality (AQ) with an associated risk of bias and unacceptable quality with a high risk of bias, respectively.
One point was given for every checkpoint met, while studies with insufficient information regarding a checkpoint were given 0 points. Checkpoints not applicable for certain studies were given 1 point as it was not considered a shortcoming of the quality of the study.
Different techniques and methods have been developed over the past years to process and analyze cfDNA. This was taken into consideration by a thorough assessment of DNA sampling and processing in all included studies using previously described methods [21] on the quality of cfDNA processing and analyses (see Additional file 1). We adopted these recommendations of cfDNA processing into a list of checkpoints for each study. Checkpoints considered the type of medium for analysis (plasma or serum), the amount and speed of centrifugation, type of sample tube, freezing of samples and under which conditions (–20 or –80 °C), and finally if blood had been processed before 4 hours. The results of these assessments were collected on the same data sheet as outcomes. Greater than 5 points were deemed as high-quality circulating DNA processing, 3–4 points were deemed as acceptable quality, while 0–2 points were considered unacceptable quality. The quality of the DNA processing was taken into account in evaluating the robustness of study conclusions.
Results
Study selection
The search identified 2728 potential studies (see Fig. 1), where 2710 studies by title and abstract screening were deemed irrelevant. Of the remaining 18 studies, six studies were excluded for the following reason: four studies were based on nontrauma patients [23–26] and two studies included both ICU and nonICU patients [27, 28]. A total of 15 studies from 14 references were included in this systematic review of which two studies were added after examining the included studies’ references lists (see Fig. 1). Ten studies were designed as prospective cohort studies, three studies included selected patients from a cohort, while one study included retrospective patients.
Fig. 1.
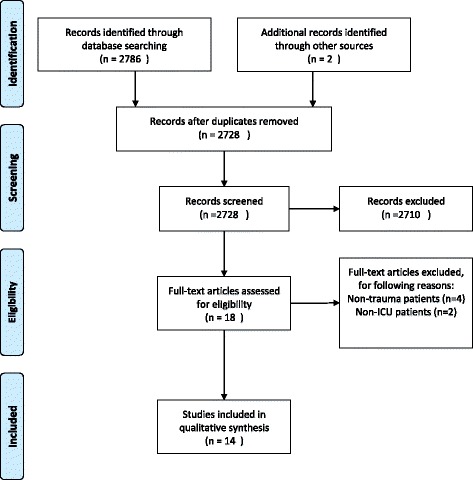
Flow diagram. PRISMA flow diagram of included studies in qualitative synthesis. ICU intensive care unit
Study characteristics
The main characteristics and cfDNA processing score (see Additional file 2 for information regarding specific points) of the included studies are summarized in Table 1. A total of 904 patients were included in the period 1996–2013 with a sample size range of 25–188. One reference presented results from two studies leading to two separate DNA processing and risk of bias assessments [29]. Ten studies included nonspecific trauma patients while five studies included patients with TBI with or without extracranial trauma [16, 30–36]. Nine of the nonspecific trauma studies based severity of trauma on the Injury Severity Score (ISS) [14, 15, 29, 34–38], while one used both the Acute Physiology and Chronic Health Evaluation (APACHE) II and Sepsis-related Organ Failure Assessment (SOFA) score [39]. All TBI studies used the Glasgow Coma Scale (GCS) to classify the effects of the trauma while using either the abbreviated injury scale (AIS), ISS, or APACHE II to describe the severity of trauma. The DNA processing score ranged from 2–5 (Table 1). All studies measured cfDNA on ICU admission, while eight studies also measured cfDNA at different time points during the admission period.
Table 1.
Study characteristics
| Author | Method | Patients, n | Age, years | Sex male, n (%) | Time of measurement | Severity score (n) [range] | DNA measurement scorea |
|---|---|---|---|---|---|---|---|
| Nonspecific trauma | |||||||
| Ren et al. (2013) [37] | Cohort study | 56 | 38 ± 14 | 47 (83.9%) | 1–6 h, 24–36 h, 60–90 h | ISS, minor: 4.8 ± 1.7 (16) ISS, moderate: 11.1 ± 1.9 (19) ISS, severe: 33.7 ± 20.0 (21) | 4 |
| Margraf et al. (2008) [15] | Prospective cohort | 37 | 45 ± 21.3 | 24 (64.9%) | ICU admission, 1–10 days | ISS, 31.6 ± 11 [16–50] | 5 |
| Lo et al. (2000) [14] | Retrospective study | 84 | ICU admission | ISS, minor/moderate: <16 (47) ISS, major: ≥16 (37) | 3 | ||
| Wijeratne et al. (2004) [39] | Cohort study | 96 | 63 (25–85) | 69 (73.4%) | ICU admission | ? | 5 |
| Lam et al. (2003) [29] | Observational study | 25 | 38 | 24 (63.2%) | Study 1: ICU admission, every 20 min for 180 min Study 2: ICU admission every day for 28 days | ISS, minor: <9 (4) ISS, moderate: 8–15 (10) ISS, severe:16–24 (7) ISS, very severe: >25 (4) | 3 |
| McIlroy et al. (2014) [43] | Prospective cohort and healthy controls | 35 | 38 (29–48) | 25 (71.4%) | Preoperative, immediately postoperative, 7 h postoperative, 24 h postoperative, 3 days postoperative, 5 days postoperative | Median ISS: 14 [9-22] | 4 |
| Yamanouchi et al. (2013) [34] | Prospective study | 37 | 56 (35–70) | 26 (70.2%) | ICU admission, days 1–2 and day 4 | AIS >3 SOFA score (day 1) 2 [2-4] APACHE II score (day 1) 11 [6-15] | 2 |
| Lam et al. (2004) [36] | Prospective cohort and healthy controls | 38 | NA | NA | ICU admission | ISS <16 (28) ISS >16 (10) | 4 |
| Gu et al. (2013) [35] | Prospective cohort and healthy controls | 86 | 45.5 (28.75–57.25) | 61 (70.1%) | ICU admission | SIRS absent ISS 14 [9.75-18.25] (50) SIRS present ISS 22 [18-29] (36) SIRS present APACHE II 11.5 [8-16] SIRS absent APACHE II 9 [6-11] | 4 |
| Traumatic brain injury | |||||||
| Macher et al. (2012) [30] | Prospective cohort and healthy controls | 65 | 38.18 ± 2.02 | 56 (86%) | ICU admission, 24 h, 48 h, 72 h and 96 h postoperative | GCS: 7 [3-9] ISS: 36 [9-75] APACHE II: 19 [5-34] | 2 |
| Shaked et al. (2014) [31] | Selected patients from a cohort | 28 | 49 (18–91) | 23 (82.1%) | ICU admission | GCS ≥14 (14) GCS ≤13 (14) AIS 0–2 (10) AIS 3–5 (18) | |
| Yurgel et al. (2007) [32] | Selected patients from a cohort | 41 | 34 (18–64) | 41 (100%) | Study entry, 24 h later | GCS survivors: 6.5 GCS nonsurvivors: 5 APACHE II survivors: 12.5 APACHE II nonsurvivors: 18.3 | 2 |
| Filho et al. (2014) [33] | Prospective cohort and healthy controls | 188 | 34.8 (13.9) | 165 (88%) | ICU admission | GCS survivors: 6.3 GCS nonsurvivors: 5.2 | 4 |
| Wang et al. (2014) [16] | Prospective cohort | 88 | 36 (20–53.75) | 55 (62.5%) | ICU admission, 4 days, 7 days | GCS: 15 [13-15] ISS: 16 [11-20] | 4 |
Values are given as mean ± SD or median (interquartile range) unless indicated otherwise
AIS abbreviated injury scale, APACHE Acute Physiology and Chronic Health Evaluation, GCS Glasgow Coma Scale, ICU intensive care unit, ISS Injury Severity Score, NA not available, SOFA Sepsis-related Organ Failure Assessment, SIRS systemic inflammatory response syndrome
aEvery study was assessed for cfDNA sampling and analysis: 1 point was given if circulating DNA was analyzed in plasma; 1 point was given if blood was collected in either an EDTA tube or cell-free DNA tube; 1 point was given if blood was processed before 4 h; 1 point was given if blood was centrifuged one or more times; 1 point was given if blood was frozen at –80 °C or –20 °C depending on whether cfDNA analysis was based on specific sequence or cfDNA quantification, respectively
The patient population differed to some extent between the studies. Ten studies included all trauma patients in the ICU [14, 15, 29, 34–39], while five studies required patients to be diagnosed with TBI [16, 30–33]. One study only included male patients [32]. Treatment strategies during ICU admission were not described, which mean that different treatments might have been used. There is a risk of bacterial DNA contamination in DNA assessment methods, but only one study reported on bacterial DNA analyses [38].
Risk of bias within studies
Risk of bias assessments are presented in Additional file 3. According to the adapted version of the SIGN checklist (Additional file 4), one study was rated as a high-quality study [16] while 13 studies were rated as being of acceptable quality and one was rated as of unacceptable quality. Eleven of 15 studies had control groups [14, 16, 30–33, 35–39]; 12 studies failed to deliver adjusted confidence intervals [14, 15, 29–31, 34, 36–39], and a total of five studies were deemed to have a high risk of selection bias as a consequence of a retrospective design [14], failure to provide exclusion criteria [39], and nonconsecutive patient inclusion [29, 32]. A possibility of detection bias was assessed in 11 of 15 groups as a consequence of these not providing information regarding the blinding of essential risk factors such as severity of trauma or clinical outcome [14, 15, 30–32, 34–39].
Two studies were considered as high-quality studies based on DNA processing [15, 39] while nine were deemed of acceptable quality [14, 16, 29, 33, 35–38] and three presented with an unacceptable quality of DNA processing [30, 32, 34]. One study was not assessed as no information regarding blood sampling and processing was available [31]. Twelve of 15 studies used plasma [14–16, 29, 32–39]. All studies centrifuged blood samples prior to freezing. None of the TBI studies stored the samples at –80 °C while 9 of 10 nonspecific trauma studies did [15, 29, 34–39].
Results of individual studies
Severity of trauma and post-traumatic complications
The main outcomes are presented in Table 2. In a study of 56 patients, no correlation was found between severity of trauma scored with ISS and levels of cfDNA [37]; however, four other studies found a significant correlation between cfDNA and ISS score. Patients diagnosed with acute lung injury (ALI) or acute respiratory distress syndrome (ARDS) had significantly higher cfDNA compared to patients without these diagnoses [14]. This corresponds to a study which proved that repeatedly measured levels of cfDNA 1 h, 2 h, and 3 h after ICU admission in 25 patients at all time points were significantly correlated with a higher degree of injury [29]. ISS scores were also correlated to cfDNA in a prospective study from 2008 on 37 patients [15] which investigated cfDNA originating from neutrophils using a fluorescent method, which was able to identify three different types of cfDNA kinetics. Type 1 kinetics were characterized by an initial value of cfDNA below 800 ng/mL followed by a rapid decrease. Type 2 kinetics were characterized by initial values above 800 ng/mL followed by a rapid decrease. Type 3 kinetics of cfDNA was characterized by high initial values above 800 ng/mL and a rapid decrease on days 1–5 followed by a significant increase of cfDNA on days 7–10 compared to type 1 and 2 kinetics. Type 3 kinetics were also associated with a higher ISS score, but not significantly. A significant association between type 3 kinetics and sepsis was found.
Table 2.
CfDNA’s ability to predict outcomes
| Article/outcomes | Survival | Complications | Severity score | Cut-off |
|---|---|---|---|---|
| Nonspecific trauma | ||||
| Ren et al. [37] | NA | Not able to predict secondary infection | No significant correlation between cfDNA and ISS | 700 copies/L plasma sensitivity: 41.1% specificity: 96.7% PPV: 95.85% NPV: 46.8% |
| Margraf et al. [15] | Lower levels of cfDNA had a NPV of 100% for MOF and death | Higher levels of cfDNA on admission and on days 7–10 was significantly associated with sepsis | Higher levels of cfDNA correlated with higher ISS scores. | NA |
| Lo et al. [14] | All patients undergoing death had significantly higher cfDNA than survivors | ALI and ARDS patients had significantly higher cfDNA than patients without this diagnosis | Higher levels of cfDNA correlated with higher ISS scores. | 232.719 KE/L had a sensitivity and specificity for death of 78% and 82%, respectively |
| Wijeratne et al. [39] | 2.3-fold higher cfDNA in nonsurvivors than in survivors (no p value) | Ventilated patients had significantly higher cfDNA values than nonventilated patients | Higher levels of cfDNA correlated with higher SOFA scores but not with APACHE II | 6.109 GE/mL sensitivity and specificity at 85% and 80%, respectively, for death |
| Lam et al. [29] | Study 1: NA Study 2: NA | Study 1: statistical difference between patients with OF and nonOF patients Study 2: on days 2, 3, 4, and 5 patients with MODS had significantly higher cfDNA that nonMODS patients | Study 1: At 1 h, 2 h, 3 h cfDNA significantly higher in patients with severe injury compared to less severely injured patients. Study 2: NA | NA |
| McIlroy et al. [43] | Not powered | No correlation between levels of mtDNA or nDNA in relation to SIRS or MOF | Not powered | Not powered |
| Yamanouchi et al. [34] | Significantly higher mtDNA in nonsurvivors compared to survivors (2 patients died) | No correlation with SOFA or APACHE II scores | High mtDNA significantly correlated with high ISS score (p < 0.05) | NA |
| Lam et al. [36] | Significantly higher mtDNA in nonsurvivors compared to survivors (2 patients died) | NA | Patients with severe injury (ISS >16) had significantly higher mtDNA than patients with minor/moderate injury (ISS < 16). Similar results for nDNA | NA |
| Gu et al. [35] | NA | mtDNA on ICU admission able to predict SIRS (p < 0.001) | mtDNA significantly correlated to high APACHE II score(p = 0.034) and ISS score (p < 0.001) | Cut-off value of mtDNA 1.3185 μg/ml with a sensitivity of 67% and specificity of 75% |
| Traumatic brain injury | ||||
| Macher et al. [30] | Significantly higher decrease of cfDNA from 0 to 24 h in survivors than in nonsurvivors | Patients with high GCS score (11–15) had significantly lower serum DNA levels at admission and 24 h than severe TBI patients | Patients with high APACHE II (<15) score and ISS had significantly higher cfDNA that patients with lower APACHE II score and ISS at 24 h | A cut-off ratio of 1.95 had a sensitivity and specificity of 70% and 66%, respectively |
| Shaked et al. [31] | Significantly higher levels of cfDNA in nonsurvivors than in survivors | Significantly higher cfDNA in patients with GOS score ≤4 vs patients with GOS score 5 | A cut-off ratio of 700 ng/ml had a sensitivity and specificity of 82% and 59% to predict GOS <5 | |
| Yurgel et al. [32] | Significantly higher levels of cfDNA at 24 h in nonsurvivors than in survivors | No difference in cfDNA between isolated TBI and TBI with extracranial injuries | 77,883,5 KE/L at 24 h with a sensitivity and specificity of 67% and 76% for mortality | |
| Filho et al. [33] | Significantly higher levels of cfDNA in nonsurvivors than in survivors | High cfDNA levels on admission siginificantly associated with death | Significantly higher cfDNA in patients with lower GCS score compared with patients with higher GCS score | 171.381 KE/L plasma sensitivity of 43%, specificity of 90% |
| Wang et al. [16] | nDNA had significantly higher levels on days 1, 4, and 7 in patients with poor outcome compared to patients with good outcome | High nDNA significantly correlated with low GCS | High nDNA significantly correlated with high ISS score | 72.95 ng/ml sensitivity of 87.5%, specificity of 86.2% |
AIS abbreviated injury scale, ALI acute lung injury, APACHE Acute Physiology and Chronic Health Evaluation, ARDS acute respiratory distress syndrome, cfDNA cell-free DNA, GCS Glasgow Coma Scale, GE genome equivalent, GOS Glasgow outcome scale, ICU intensive care unit, ISS Injury Severity Score, KE kilo equivalent, MODS multiple organ dysfunction syndrome, MOF multi-organ failure, mtDNA circulating mitochondrial DNA, NA not available, nDNA circulating nuclear DNA, NPV negative predictive value, OF organ failure, PPV positive predictive value, SOFA Sepsis-related Organ Failure Assessment, SIRS systemic inflammatory response syndrome, TBI traumatic brain injury
SOFA and APACHE II scores were investigated in a study on 96 patients which found that higher levels of cfDNA significantly correlated with higher SOFA scores but not with APACHE II score [39]. A study measuring mtDNA in trauma patients found a significant correlation between mtDNA levels in the plasma and APACHE II and ISS scores [35]. Similar results were found in two other studies [34, 36], but one study did not find an association between mtDNA levels and APACHE II score [34].
In a study on patients with severe TBI, high APACHE II and ISS scores were significantly correlated with higher levels of cfDNA 24 h after ICU admission. This study also showed that patients with high GCS score (11–15) had significantly lower cfDNA at both ICU admission and after 24 h [30], which was confirmed in a similar study [33]. However, a significant correlation between patients with isolated TBI and patients with extracranial injuries was not found for cfDNA levels in 41 male patients with TBI [32].
Regarding nDNA, similar results have been presented where patients with high nDNA had lower GCS scores and significantly higher ISS scores [16]. However, one study, reporting on both nDNA and mtDNA, found no correlation between levels of nDNA and mtDNA in relation to SIRS or multi-organ failure (MOF) [38].
Survival
Survival data in relation to cfDNA was presented in ten studies. One study reported not being powered to present survival data [38], while three references did not present survival data analyzed in relation to cfDNA levels [29, 35, 37]. In the earliest study measuring cfDNA in a retrospective population of trauma patients, it was found that all patients who died in the ICU had significantly higher cfDNA than survivors [14]. Likewise, it has been reported that nonsurvivors had a 2.3-fold higher cfDNA level than survivors [39]. In a prospective cohort of 37 patients, lower cfDNA levels at ICU admission had a negative predictive value (NPV) of 100% in relation to death [15]. In another study, survivors presented significantly lower cfDNA levels on ICU admission and 24 h after, but not 48 h after, ICU admission [30]. Other authors only found this significant correlation at 24 h after ICU admission [31–33]. Two of the studies measuring mtDNA presented survival data, both showing nonsurviving patients with significantly higher mtDNA than survivors [34, 36]. Finally, in a prospective study of 88 patients where nDNA was measured at ICU admission and on days 4 and 7, patients with a fatal outcome had significantly higher nDNA at all time points [16].
Discussion
This systematic review found strong evidence in favor of using cfDNA as a prognostic tool in relation to mortality in trauma patients admitted to the ICU. We found that cfDNA can be used as a prognostic tool for the outcome of post-traumatic complications and the severity of trauma to a moderate extent.
With regard to linking cfDNA levels to clinical outcomes such as mortality and complications, we found that all of the included studies that presented survival data concluded that higher levels of cfDNA were significantly correlated to a higher mortality rate. It seems that cfDNA levels measured even at the earliest stages of ICU admission were able to predict a higher mortality rate [14, 31, 36, 39]. This finding was confirmed in 108 patients with sepsis [7]. Likewise, increasing levels of nDNA and mtDNA were significantly associated with increasing mortality [30, 31]. With regard to associating cfDNA levels with development of complications, the results were not conclusive. One study found that patients with high initial cfDNA and elevated levels of cfDNA on days 7 to 10 after ICU admission were significantly associated with sepsis [15]. Similar results were found in a study on a mixed population of ICU patients where cfDNA was measured on ICU admission, which found that cfDNA was higher in patients developing sepsis (p = 0.03) and in nonsurviving patients (p > 0.001) [40]. However, another prospective study on 160 consecutive ICU patients could not confirm these results [8], which may be due to the cfDNA processing. Interestingly, regarding the prognostic capabilities of cfDNA, it was found that cfDNA levels were superior to classification systems such as multiple organ dysfunction syndrome (MODS) regarding complications or APACHE II scores regarding mortality [41].
This systematic review has limitations as well as strengths. A strength of this review is that all of the included studies calculated severity score analysis of the trauma patients, and that the cfDNA levels were correlated to these scores. In general, high severity scores correlated to higher levels of cfDNA. A limitation is that even though we searched widely in four databases, the result of our search strategy may have been subject to publications bias, which we were not able to adjust for. Another limitation was that the patient populations differed to some extent between the included studies, which rendered it impossible to perform meta-analysis on the outcomes. No information regarding treatment strategies or regimes in the respective hospitals were available, which could lead to differences in outcome based on different treatments. Heterogeneity in cfDNA sampling and processing were found, which made direct comparison of the results difficult. Only one study reported on bacterial DNA analysis.
An interesting aspect of mtDNA is the hypothesis of mtDNA acting as a DAMP. In the light of the study by Xiao and collegues [42], where circulating leukocyte transcriptome after severe trauma and burn injury was measured, it could be argued that mtDNA is a cornerstone of the immune reaction. They found that following severe trauma and burn injury a global reprioritization occurs in >80% of the cellular functions and pathways, a phenomenon they call a “genomic storm”. The formation of neutrophil extracellular traps by mtDNA has also been proposed as another factor in immune reaction following trauma [43]. Another study where nDNA and mtDNA was associated with immune function was the study by Timmermanns et al. [44], who found that nDNA but not mtDNA correlated negatively with human leukocyte antigen (HLA-DR) expression, suggesting that nDNA could possibly function as a DAMP.
With regard to the future of cfDNA research in the trauma field it is of utmost importance that the correct methods of cfDNA sampling and processing are used so that cfDNA is correctly analyzed. It could be argued that cfDNA should be measured pre- and postsurgery as some of the same mechanics comes into action during surgery as in trauma. Ren et al. found that two patients who repeatedly underwent surgery had increased cfDNA levels after each surgery [37]. This aspect of cfDNA could be very interesting if the same prediction and prognostic pattern could be observed in patients undergoing surgery. cfDNA could also be viewed as a tool in identifying trauma patients who need immediate treatment.
Conclusion
This systematic review provides an overview of the possible benefits of measuring cfDNA in relation to mortality, severity of trauma, and post-traumatic complications in trauma patients in the ICU. Moreover, it illustrates important aspects of DNA sampling and processing that should be considered when implementing cfDNA as measurement of injury severity. Finally, it encourages further research in this area with the aforementioned guidelines for DNA processing. The value of cfDNA, nDNA, and mtDNA should be investigated in patients undergoing major elective surgery.
Acknowledgements
Not applicable.
Funding
No funding was received for this study.
Availability of data and materials
Not applicable.
Authors’ contributions
MG and IG formed the conceptual framework of this article. MG conducted the literature search, and drafted the manuscript. JB helped to form the conceptual framework of this article and to conduct the literature search and revised the manuscript for important intellectual content. All authors read and approved the final version of the manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Abbreviations
- AIS
Abbreviated injury scale
- ALI
Acute lung injury
- APACHE
Acute Physiology and Chronic Health Evaluation
- ARDS
Acute respiratory distress syndrome
- cfDNA
Cell-free DNA
- DAMP
Damage-associated molecular pattern
- GCS
Glasgow Coma Scale
- ICU
Intensive care unit
- ISS
Injury Severity Score
- MOF
Multi-organ failure
- mtDNA
Circulating mitochondrial DNA
- nDNA
Circulating nuclear DNA
- NPV
Negative predictive value
- SIRS
Systemic inflammatory response syndrome
- SOFA
Sepsis-related Organ Failure Assessment
- TBI
Traumatic brain injury
Additional files
cfDNA measurement. CfDNA assessment checkpoints. (DOCX 14 kb)
CfDNA measurement results. Results of cfDNA assessment checkpoints for each study. One point was given for if circulating DNA was analyzed in plasma. One point was given if blood was collected in either an EDTA tube or cell-free DNA tube. One point was given if blood was processed before 4 h. One point was given if blood was centrifuged one or more times. One point was given if blood was frozen at –80 °C or –20 °C depending on whether cfDNA analysis was based on specific sequence or cfDNA quantification, respectively. GAPDH glyceraldehyde 3-phosphate dehydrogenase, NADH nicotinamide adenine dinucleotide. (DOCX 25 kb)
Risk of bias assessment. Results of risk of bias assessment for each study. (DOCX 18 kb)
Simplified SIGN methodology checklist of COHORT studies. Risk of bias assessment checklist. (DOCX 17 kb)
Contributor Information
Mikail Gögenur, Email: mgog@regionsjaelland.dk.
Jakob Burcharth, Email: jakobburcharth@gmail.com.
Ismail Gögenur, Email: igo@regionsjaelland.dk.
References
- 1.Kim K, Shin DG, Park MK, Baik SH, Kim TH, Kim S, Lee S. Circulating cell-free DNA as a promising biomarker in patients with gastric cancer: diagnostic validity and significant reduction of cfDNA after surgical resection. Ann Surg Treat Res. 2014;86:136–42. doi: 10.4174/astr.2014.86.3.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Szpechcinski A, Chorostowska-Wynimko J, Struniawski R, Kupis W, Rudzinski P, Langfort R, Puscinska E, Bielen P, Sliwinski P, Orlowski T. Cell-free DNA levels in plasma of patients with non-small-cell lung cancer and inflammatory lung disease. Br J Cancer. 2015;113:476–83. doi: 10.1038/bjc.2015.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reinert T, Scholer LV, Thomsen R, Tobiasen H, Vang S, Nordentoft I, Lamy P, Kannerup AS, Mortensen FV, Stribolt K, Hamilton-Dutoit S, Nielsen HJ, Laurberg S, Pallisgaard N, Pedersen JS, Orntoft TF, Andersen CL. Analysis of circulating tumour DNA to monitor disease burden following colorectal cancer surgery. Gut. 2016;65:625–34. doi: 10.1136/gutjnl-2014-308859. [DOI] [PubMed] [Google Scholar]
- 4.Norton M, Jacobsson B, Swamy G, Laurent L, Ranzini A, Brar H, Tomlinson M, Pereira L, Spitz J, Hollemon D, Cuckle H, Musci T, Wapner R. Cell-free DNA analysis for noninvasive examination of trisomy. N Engl J Med. 2015;372:1589–97. doi: 10.1056/NEJMoa1407349. [DOI] [PubMed] [Google Scholar]
- 5.De Vlaminck I, Valantine HA, Snyder TM, Strehl C, Cohen G, Luikart H, Neff NF, Okamoto J, Bernstein D, Weisshaar D, Quake SR, Khush KK. Circulating cell-free DNA enables noninvasive diagnosis of heart transplant rejection. Sci Transl Med. 2014;6:241ra277. doi: 10.1126/scitranslmed.3007803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shoham Y, Krieger Y, Perry ZH, Shaked G, Bogdanov-Berezovsky A, Silberstein E, Sagi A, Douvdevani A. Admission cell free DNA as a prognostic factor in burns: quantification by use of a direct rapid fluorometric technique. Biomed Res Int. 2014;2014:306580. doi: 10.1155/2014/306580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Avriel A, Paryente Wiessman M, Almog Y, Perl Y, Novack V, Galante O, Klein M, Pencina MJ, Douvdevani A. Admission cell free DNA levels predict 28-day mortality in patients with severe sepsis in intensive care. PLoS One. 2014;9:e100514. doi: 10.1371/journal.pone.0100514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garnacho-Montero J, Huici-Moreno MJ, Gutierrez-Pizarraya A, Lopez I, Marquez-Vacaro JA, Macher H, Guerrero JM, Puppo-Moreno A. Prognostic and diagnostic value of eosinopenia, C-reactive protein, procalcitonin, and circulating cell-free DNA in critically ill patients admitted with suspicion of sepsis. Crit Care. 2014;18:R116. doi: 10.1186/cc13908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Spindler KL, Pallisgaard N, Appelt AL, Andersen RF, Schou JV, Nielsen D, Pfeiffer P, Yilmaz M, Johansen JS, Hoegdall EV, Jakobsen A, Jensen BV. Clinical utility of KRAS status in circulating plasma DNA compared to archival tumour tissue from patients with metastatic colorectal cancer treated with anti-epidermal growth factor receptor therapy. Eur J Cancer. 2015;51:2678–85. doi: 10.1016/j.ejca.2015.06.118. [DOI] [PubMed] [Google Scholar]
- 10.Spindler KL, Appelt AL, Pallisgaard N, Andersen RF, Brandslund I, Jakobsen A. Cell-free DNA in healthy individuals, noncancerous disease and strong prognostic value in colorectal cancer. Int J Cancer. 2014;135:2984–91. doi: 10.1002/ijc.28946. [DOI] [PubMed] [Google Scholar]
- 11.Spindler KL, Pallisgaard N, Andersen RF, Brandslund I, Jakobsen A. Circulating free DNA as biomarker and source for mutation detection in metastatic colorectal cancer. PLoS One. 2015;10:e0108247. doi: 10.1371/journal.pone.0108247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fournie G, Courtin J, Laval F, Chalé J, Pourrat J, Pujazon M, Laugue D, Carles P. Plasma DNA as a marker of cancerous cell death. Investigations in patients suffering from lung cancer and in nude mice bearing human tumours. Cancer Lett. 1995;91:221–7. doi: 10.1016/0304-3835(95)03742-F. [DOI] [PubMed] [Google Scholar]
- 13.Fournie G, Martres F, Pourrat J, Alary C, Rumeau M. Plasma DNA as cell death marker in elderly patients. Gerontology. 1993;39:215–21. doi: 10.1159/000213536. [DOI] [PubMed] [Google Scholar]
- 14.Lo YMD, Rainer TH, Chan LYS, Hjelm NM, Cocks RA. Plasma DNA as a prognostic marker in trauma patients. Clin Chem. 2000;46:319–23. [PubMed] [Google Scholar]
- 15.Margraf S, Lögters T, Reipen J, Altrichter J, Scholz M, Windolf J. Neutrophil-derived circulating free DNA (CF-DNA/NETS): a potential prognostic marker for posttraumatic development of inflammatory second hit and sepsis. Shock. 2008;30:352–8. doi: 10.1097/SHK.0b013e31816a6bb1. [DOI] [PubMed] [Google Scholar]
- 16.Wang H-C, Lin Y-J, Tsai N-W, Su BY-J, Kung C-T, Chen W-F, Kwan A-L, Lu C-H. Serial plasma deoxyribonucleic acid levels as predictors of outcome in acute traumatic brain injury. J Neurotrauma. 2014;31:1039–45. doi: 10.1089/neu.2013.3070. [DOI] [PubMed] [Google Scholar]
- 17.Diehl F, Schmidt K, Choti M, Romans K, Goodman S, Li M, Thornton K, Agrawal N, Sokoll L, Szabo S, Kinzler K, Vogelstein B, Diaz LJ. Circulating mutant DNA to assess tumor dynamics. Nat Med. 2008;14:985–90. doi: 10.1038/nm.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rubartelli A, Lotze M. Inside, outside, upside down: damage-associated molecular-pattern molecules (DAMPs) and redox. Trends Immunol. 2007;28:429–36. doi: 10.1016/j.it.2007.08.004. [DOI] [PubMed] [Google Scholar]
- 19.Zhang Q, Raoof M, Chen Y, Sumi Y, Sursal T, Junger W, Brohi K, Itagaki K, Hauser CJ. Circulating mitochondrial DAMPs cause inflammatory responses to injury. Nature. 2010;464:104–7. doi: 10.1038/nature08780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moher D, Liberati A, Tatzlaff J, et al. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 2009;6: doi:10.1371/journal.pmed.1000097. [DOI] [PMC free article] [PubMed]
- 21.El Messaoudi S, Rolet F, Mouliere F, Thierry AR. Circulating cell free DNA: preanalytical considerations. Clin Chim Acta. 2013;424:222–30. doi: 10.1016/j.cca.2013.05.022. [DOI] [PubMed] [Google Scholar]
- 22.Gosselt AN, Slooter AJ, Boere PR, Zaal IJ. Risk factors for delirium after on-pump cardiac surgery: a systematic review. Crit Care. 2015;19:346. doi: 10.1186/s13054-015-1060-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rainer TH, Chan AK, Lee LL, Yim VW, Lam NY, Yeung SW, Graham CA, Lo DY. Use of plasma DNA to predict mortality and need for intensive care in patients with abdominal pain. Clin Chim Acta. 2008;398:113–7. doi: 10.1016/j.cca.2008.08.022. [DOI] [PubMed] [Google Scholar]
- 24.Rainer TH, Lam NY, Man CY, Chiu RW, Woo KS, Lo YM. Plasma beta-globin DNA as a prognostic marker in chest pain patients. Clin Chim Acta. 2006;368:110–3. doi: 10.1016/j.cca.2005.12.021. [DOI] [PubMed] [Google Scholar]
- 25.Potenza L, Calcabrini C, Bellis RD, Mancini U, Polidori E, Zeppa S, Alloni R, Cucchiarini L, Dacha M. Effect of surgical stress on nuclear and mitochondrial DNA from healthy sections of colon and rectum of patients with colorectal cancer. J Biosci. 2011;36:243–51. doi: 10.1007/s12038-011-9064-7. [DOI] [PubMed] [Google Scholar]
- 26.Levy M, Benesova L, Lipska L, Belsanova B, Minarikova P, Veprekova G, Zavoral M, Minarik M. Utility of cell-free tumour DNA for post-surgical follow-up of colorectal cancer patients. Anticancer Res. 2012;32:1621–6. [PubMed] [Google Scholar]
- 27.Nakahira K, Kyung SY, Rogers AJ, Gazourian L, Youn S, Massaro AF, Quintana C, Osorio JC, Wang Z, Zhao Y, Lawler LA, Christie JD, Meyer NJ, Mc Causland FR, Waikar SS, Waxman AB, Chung RT, Bueno R, Rosas IO, Fredenburgh LE, Baron RM, Christiani DC, Hunninghake GM, Choi AM. Circulating mitochondrial DNA in patients in the ICU as a marker of mortality: derivation and validation. PLoS Med. 2013;10:e1001577. doi: 10.1371/journal.pmed.1001577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saukkonen K, Lakkisto P, Varpula M, Varpula T, Voipio-Pulkki LM, Pettila V, Pulkki K. Association of cell-free plasma DNA with hospital mortality and organ dysfunction in intensive care unit patients. Intensive Care Med. 2007;33:1624–7. doi: 10.1007/s00134-007-0686-z. [DOI] [PubMed] [Google Scholar]
- 29.Lam NYL, Rainer TH, Chan LYS, Joynt GM, Lo YMD. Time course of early and late changes in plasma DNA in trauma patients. Clin Chem. 2003;49:1286–91. doi: 10.1373/49.8.1286. [DOI] [PubMed] [Google Scholar]
- 30.Macher H, Egea-Guerrero JJ, Revuelto-Rey J, Gordillo-Escobar E, Enamorado-Enamorado J, Boza A, Rodriguez A, Molinero P, Guerrero JM, Dominguez-Roldán JM, Murillo-Cabezas F, Rubio A. Role of early cell-free DNA levels decrease as a predictive marker of fatal outcome after severe traumatic brain injury. Clin Chim Acta. 2012;414:12–7. doi: 10.1016/j.cca.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 31.Shaked G, Douvdevani A, Yair S, Zlotnik A, Czeiger D. The role of cell-free DNA measured by a fluorescent test in the management of isolated traumatic head injuries. Scand J Trauma Resusc Emerg Med. 2014;22: doi:10.1186/1757-7241-1122-1121. [DOI] [PMC free article] [PubMed]
- 32.Yurgel VC, Ikuta N, Rocha ABD, Lunge VR, Schneider RF, Fonseca ASK, Grivicich I, Zanoni C, Regner A. Role of plasma DNA as a predictive marker of fatal outcome following severe head injury in males. J Neurotrauma. 2007;24:1172–81. doi: 10.1089/neu.2006.0160. [DOI] [PubMed] [Google Scholar]
- 33.Filho EMR, Simon D, Ikuta N, Klovan C, Dannebrock FA, Oliveira CO, Regner A. Elevated cell-free plasma DNA level as an independent predictor of mortality in patients with severe traumatic brain injury. J Neurotrauma. 2014;31:1639–46. doi: 10.1089/neu.2013.3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamanouchi S, Kudo D, Yamada M, Miyagawa N, Furukawa H, Kushimoto S. Plasma mitochondrial DNA levels in patients with trauma and severe sepsis: time course and the association with clinical status. J Crit Care. 2013;28:1027–31. doi: 10.1016/j.jcrc.2013.05.006. [DOI] [PubMed] [Google Scholar]
- 35.Gu X, Yao Y, Wu G, Lv T, Luo L, Song Y. The plasma mitochondrial DNA is an independent predictor for post-traumatic systemic inflammatory response syndrome. Plos One. 2013;8:e72834. doi: 10.1371/journal.pone.0072834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lam NY, Rainer TH, Chiu RW, Joynt GM, Lo YM. Plasma mitochondrial DNA concentrations after trauma. Clin Chem. 2004;50:213–6. doi: 10.1373/clinchem.2003.025783. [DOI] [PubMed] [Google Scholar]
- 37.Ren B, Liu F, Xu F, He J, Zhu H, Zou G. Is plasma cell-free DNA really a useful marker for diagnosis and treatment of trauma patients? Clin Chim Acta. 2013;424:109–13. doi: 10.1016/j.cca.2013.05.015. [DOI] [PubMed] [Google Scholar]
- 38.McIlroy DJ, Bigland M, White AE, Hardy BM, Lott N, Smith DW, Balogh ZJ. Cell necrosis-independent sustained mitochondrial and nuclear DNA release following trauma surgery. J Trauma Acute Care Surg. 2015;78:282–8. doi: 10.1097/TA.0000000000000519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wijeratne S, Butt A, Burns S, Sherwood K, Boyd O, Swaminathan R. Cell-free plasma DNA as a prognostic marker in intensive treatment unit patients. Ann N Y Acad Sci. 2004;1022:232–8. doi: 10.1196/annals.1318.036. [DOI] [PubMed] [Google Scholar]
- 40.Rhodes A, Wort SJ, Thomas H, Collinson P, Bennett ED. Plasma DNA concentration as a predictor of mortality and sepsis in critically ill patients. Crit Care. 2006;10:R60. doi: 10.1186/cc4894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dwidevi DJ, Toltl LJ, Swystun LL, et al. Prognostic utility and characterization of cell-free DNA in patients with severe sepsis. Crit Care. 2012;16:R151. doi: 10.1186/cc11466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xiao W, Mindrinos MN, Seok J, Cuschieri J, Cuenca AG, Gao H, Hayden DL, Hennessy L, Moore EE, Minei JP, Bankey PE, Johnson JL, Sperry J, Nathens AB, Billiar TR, West MA, Brownstein BH, Mason PH, Baker HV, Finnerty CC, Jeschke MG, Lopez MC, Klein MB, Gamelli RL, Gibran NS, Arnoldo B, Xu W, Zhang Y, Calvano SE, McDonald-Smith GP, Schoenfeld DA, Storey JD, Cobb JP, Warren HS, Moldawer LL, Herndon DN, Lowry SF, Maier RV, Davis RW, Tompkins RG, Inflammation, Host Response to Injury Large-Scale Collaborative Research P A genomic storm in critically injured humans. J Exp Med. 2011;208:2581–90. doi: 10.1084/jem.20111354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McIlroy DJ, Jarnicki AG, Au GG, Lott N, Smith DW, Hansbro PM, Balogh ZJ. Mitochondrial DNA neutrophil extracellular traps are formed after trauma and subsequent surgery. J Crit Care. 2014;29:1133. doi: 10.1016/j.jcrc.2014.07.013. [DOI] [PubMed] [Google Scholar]
- 44.Timmermanns K, Kox M, Vaneker M, Berg M, John A, Laarhoven A, Hoeven H, Scheffer GJ, Pickkers P. Plasma levels of danger-associated molecular patterns are associated with immune suppression in trauma patients. Intensive Care Med. 2016;42:551–61. doi: 10.1007/s00134-015-4205-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.


