Abstract
Viruses that infect bacteria (phages) can influence bacterial community dynamics, bacterial genome evolution and ecosystem biogeochemistry. These influences differ depending on whether phages establish lytic, chronic or lysogenic infections. Although the first two produce virion progeny, with lytic infections resulting in cell destruction, phages undergoing lysogenic infections replicate with cells without producing virions. The impacts of lysogeny are numerous and well-studied at the cellular level, but ecosystem-level consequences remain underexplored compared to those of lytic infections. Here, we review lysogeny from molecular mechanisms to ecological patterns to emerging approaches of investigation. Our goal is to highlight both its diversity and importance in complex communities. Altogether, using a combined viral ecology toolkit that is applied across broad model systems and environments will help us understand more of the diverse lifestyles and ecological impacts of lysogens in nature.
Why study lysogeny?
Bacteria significantly alter the biosphere, affecting global biogeochemical cycles and the biology of other organisms biology (Alivisatos et al., 2015), but do so under the constraints of bacteriophage (phage) infection. Such phage infections range from productive to lysogenic (see Concept Box and Figure 1a) dependent on, for example, phage genetics, host genetics, phage concentration, host physiology and environmental conditions. Temperate phages in particular can replicate either lysogenically as prophages or, instead, produce virions. Although prophages in lysogenic cycles largely have been viewed as dormant entities, both prophages and their subsequent productive cycles can affect individual cells as well as entire communities. The phenomenon of lysogeny is well known, early reviews of lysogeny date to over half a century ago (Lwoff, 1953), though mostly temperate phages infecting E. coli have been characterized in depth including λ, Mu, P1 or N15. In nature, phages have been detected wherever their host microbes exist (Weinbauer, 2004), with reviews focusing on total viral communities from soil, aquatic and host-associated systems (Chibani-Chennoufi et al., 2004; Suttle, 2005; Sawstrom et al., 2008; Srinivasiah et al., 2008; Williamson, 2011; Marcó et al., 2012; Reyes et al., 2012; Abeles and Pride, 2014; Sime-Ngando, 2014; Virgin, 2014; Ogilvie and Jones, 2015; Zablocki et al., 2016). Assessing predominant infection dynamics and associated driving factors, however, can be challenging due to difficulties in determining lysogeny abundance, diversity and ecology (Paul and Weinbauer, 2010).
Concept Box.
Bacteriophage (phage): Virus that infects a bacterial host.
Lytic cycle/infection: Virus reproduction that destroys its host cell to release virion progeny.
Lysogenic cycle/infection: Non-bactericidal phage infection with phage genome replication but no virion production.
Induction: Virus infection changes from a lysogenic cycle to a productive cycle.
Chronic cycle/infection: Generally non-bactericidal phage infection, where virions are produced and continuously released.
Productive cycle/infection: Virus reproduction with production of virion particles.
Virulent phage: Phage which displays only lytic cycles (no chronic or lysogenic cycles).
Temperate phage: Phage which can undergo either virion-productive or lysogenic cycles.
Prophage: Phage genome that replicates with its host cell while not generating virion progeny.
Cryptic prophage: Prophage that has mutationally lost its ability to enter a virion-productive cycle.
Lysogen: Bacterial cell that harbors at least one prophage.
Polylysogen: Bacterial cell that harbors more than one prophage.
Transduction: Virion-mediated transfer of bacterial DNA to new bacteria either with associated temperate phage genome (specialized transduction) or not in association with phage genome (generalized transduction).
Virome: Metagenomic sequences of viral communities.
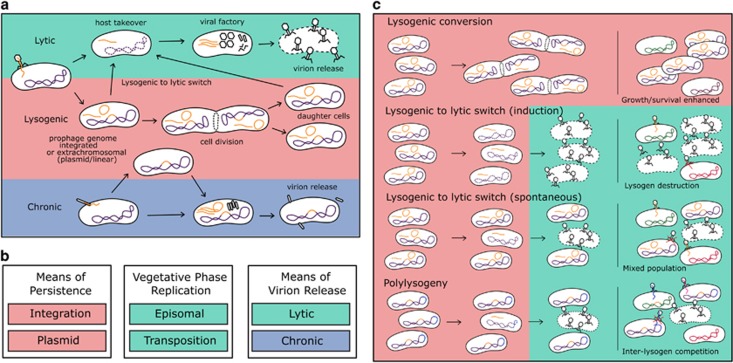
Figure 1.
Modes of temperate phage infection, from cell to community. (a) A temperate phage can infect a cell through either virion-productive or lysogenic cycles, where it either hijacks its host’s metabolism to produce new virion progeny or instead replicates its genome alongside the host without producing new virions, respectively. The production of virion particles can occur either following phage adsorption (productive cycle) or instead following a switch from a lysogenic cycle to a productive cycle (lytic or instead chronic infection cycles depending on the temperate phage). Although these are generalized dynamics of infection, details can vary with specific phage-host types, ranging from efficient to inefficient infections, where the dynamics and outcome of the infection may vary. (b) Summary of phage infection strategies by stage. Persistence describes the prophage stage during the lysogenic cycle, replication describes the phage-genome state during productive cycles and release refers not just to the means by which progeny phage virions transition from the intracellular to extracellular state but also the impact of productive cycles on the phage-infected bacterium (i.e., lytic but not chronic results in host-bacterium physiological death). (c) Implications of modes of temperate phage infection on bacterial communities. Lysogenic conversion constitutes the phenotypic effects of prophage carriage to its host cell. The lysogenic to lytic switch changes community structure by creating a mixed cell community where some lysogens are removed via lysis and the released virions can infect surrounding cells. Polylysogeny occurs when a bacterium possesses multiple prophages.
Here, we complement efforts to specifically review lysogeny which have largely focused on prophage genomics and impacts of lysogeny on either microbial cells (Casjens, 2003; Brussow et al., 2004; Canchaya et al., 2004; Abedon and LeJeune, 2005; Fortier and Sekulovic, 2013; Hargreaves et al., 2014; Feiner et al., 2015; Menouni et al., 2015; Davies et al., 2016) or particular environments (for example, aquatic (Paul, 2008)). We aim to establish an integrative view across model systems and environments, whereas also highlighting emerging tools for the study of lysogeny in nature.
Lysogeny mechanistic diversity
Most molecular knowledge of lysogeny has been derived from a handful of E. coli phages, such as λ and Mu, which integrate into the bacterial chromosome via site-specific recombination (Casjens and Hendrix, 2015) or random transposition (Harshey, 2014), respectively. In contrast, other phages are maintained extrachromosomally with either circular (for example, P1, (Lobocka et al., 2004) or linear (for example, N15, (Ravin, 2015)) genomes. Some temperate phages, such as satellite phage P4, require other temperate phages, such as P2, to complete infection cycles (Christie and Calendar, 2016). Other temperate phages, for example,Vibrio cholera phage CTXphi, chronically infect their host during productive cycles and integrate during lysogenic cycles (McLeod et al., 2005). Although infection details differ, lysogeny generally proceeds through three steps: (i) establishment, (ii) maintenance and also, potentially, (iii) induction of productive cycles (Figure 1). Focusing on λ as a reference, we highlight how these mechanistic factors both serve as a knowledge baseline for further study and can contribute to lysogen ecology.
Establishment
Given evasion of host resistance mechanisms (Samson et al., 2013), temperate phages ‘decide’ whether to produce virions (productive cycle) or instead establish lysogenic cycles as prophages. In λ, the ‘decision’ to enter lysogeny is driven by genetic compatibility (for example, host attB integration sites), host physiological state (for example, nutrient depletion increases lysogeny) and phage density (for example, higher MOIs increase lysogeny) (Casjens and Hendrix, 2015). Integration is driven via recombinases acting on phage (attP) and bacterial (attB) attachment site sequences that determine specificity (Fogg et al., 2014), or prophages may integrate randomly (for example, Mu) or not at all (for example, P1).
Maintenance
Once established, integrated prophages replicate as part of the bacterial chromosome, whereas extrachromosomal prophages require genes for plasmid inheritance (for example, ParA/ParB in P1 or SopA/SopB in N15) and persistence (for example, toxin–antitoxin proteins that kill plasmid-lacking cells) (Casjens and Hendrix, 2015; Ravin, 2015). Either state has the potential to impact on host gene regulation and resulting biology, including growth rate, development and phenotype (Feiner et al., 2015). In λ, maintenance is tightly regulated by its repressor, CI, via a complex genetic feedback circuit (Bednarz et al., 2014). During this stage, prophages not only can alter cellular processes, but are subject to evolutionary change, with selection presumably balancing phage versus cell needs (Bobay et al., 2013).
Induction
Activation of the lytic-lysogenic switch occurs either spontaneously at low frequency (10−8–10−5 per cell for λ (Czyz et al., 2001)) or as a result of external stressors such as those triggering the cell’s DNA damage response (the SOS response) (Casjens and Hendrix, 2015), leading to inactivation of CI. Stressors include changes in nutrients, pH or temperature, and exposure to antibiotics, hydrogen peroxide, foreign DNA (Banks et al., 2003; Mell and Redfield, 2014; Casjens and Hendrix, 2015) or DNA damaging agents (Cochran et al., 1998). Alternatively, prophages can influence the induction of other phages. For example, satellite temperate phage P4 is induced via Cox, an anti-repressor encoded by temperate phage P2 (Christie and Dokland, 2012), whereas Enterococcus prophages pp1, pp3 and pp5 inhibit the induction of co-infecting prophages pp4 and pp6 (Matos et al., 2013). This view of intracellular phage-mediated competition highlights the complexity of interactions among phages of likely ecological relevance.
Once induced, prophages replicate either episomally (for example, λ, P1, N15) or by transposition (for example, Mu). Later, virion particles assemble and are packaged with phage DNA via endonucleolytic enzymes that either cut DNA at specific sites (for example, cos phages) or non-specifically after filling up the capsid (for example, headful packaging by pac phages) (Rao and Feiss, 2015). Specialized transduction (by cos temperate phages) and generalized transduction (by pac phages generally) can differentially impact bacterial genome evolution (Rao and Feiss, 2015).
Such models of temperate phage infection (Figure 1) offer a comparative baseline for discovering variations in lysogeny in nature. For example, as observed in Staphylococcus aureus, temperate phage can integrate into one host genome but exist extrachromosomally in others (Utter et al., 2014), or as found in Salmonella, be asymmetrically inherited by only one daughter cell (Cenens et al., 2013a). As conditions in nature are highly variable, it is also critical to distinguish lysogeny from delayed or inefficient lytic infections (Dang et al., 2015), as well as determine how natural infections can differ from those in the laboratory (Chibani-Chennoufi et al., 2004). Establishment of new phage-host model systems also will be instrumental towards furthering our understanding of lysogen ecology.
Benefits and consequences of lysogeny
Temperate phages alter the biology of their hosts and, in turn, influence the surrounding community of host and non-host cells (Figure 1c). We briefly summarize four such effects, which have been previously reviewed. First, integrating prophages can engineer a host’s genome (Menouni et al., 2015) and help to regulate gene expression and function such that integration and excision alters cellular processes (Feiner et al., 2015). Second, prophages can change cell physiology by introducing novel functions or altering pre-existing ones, such as virulence factor production, metabolism, cell development and immunity to phages (Hargreaves et al., 2014). Third, through transduction, temperate phages can facilitate the transfer of bacterial DNA that potentially confer new phenotypes such as antibiotic resistance (Davies et al., 2016). Fourth, induced and released temperate phages can modify bacterial communities by lysing competitor bacterial strains (Duerkop et al., 2012), lysogenizing other cells (Gama et al., 2013), and liberating intracellular contents for neighboring cells to use as nutrients (Nanda et al., 2015).
Prophages can be ‘domesticated’ by losing genes, including those necessary for virion production (Wang et al., 2010; Bobay et al., 2014). Resulting prophage-derived genomic elements can be selectively maintained by cells should they still confer some advantage. Such cryptic prophages also can directly impact the wider ecosystem. R-type bacteriocins, which resemble phage tail-like particles, can, for example, kill neighboring competitor bacteria (Gebhart et al., 2012). Phage tail-like metamorphosis-associated contractile structures in the biofilm formed by Pseudoaltermonas luteoviolacae, by contrast, can trigger the settlement of eukaryotic tube worm larvae to surfaces (Shikuma et al., 2014), though exactly how P. luteoviolacae might benefit from this process is unclear. Prophage decay also can result in repetitive sequences that facilitate chromosomal insertions, creating niche-defining genomic islands. In Cyanobacteria, these have been hypothesized to influence expression of adjacent nitrogen stress response genes (Sullivan et al., 2009). Decayed prophages can still be inducible as well as capable of lysing their host cell, as illustrated by R-type bacteriocins (Gebhart et al., 2012), and their genes may re-enter the virus gene pool via recombination during co-infection with other phages (De Paepe et al., 2014a).
Genetic, ecological and functional insights into lysogeny
Viral ecology has been extensively studied and reviewed, providing insights into lysogeny and its influencing factors that we synthesize here (Figure 2). Genetically, temperate phages have been identified in ~40–50% of microbial genomes (Canchaya et al., 2003; Casjens, 2003; Fouts, 2006; Paul, 2008; Touchon et al., 2016), and more recently from 21/30 phyla for which complete isolate genomes were available (Touchon et al., 2016). Ecologically, lysogens are widespread (Chibani-Chennoufi et al., 2004), occurring in terrestrial (Williamson et al., 2007; Srinivasiah et al., 2008), aquatic (Wommack and Colwell, 2000; Weinbauer, 2004; Suttle, 2005; Paul, 2008; Sawstrom et al., 2008; Paul and Weinbauer, 2010; Sime-Ngando, 2014) and host-associated ecosystems (Reyes et al., 2012; Abeles and Pride, 2014; De Paepe et al., 2014b; Virgin, 2014; Edlund et al., 2015; Ogilvie and Jones, 2015), though frequencies vary.
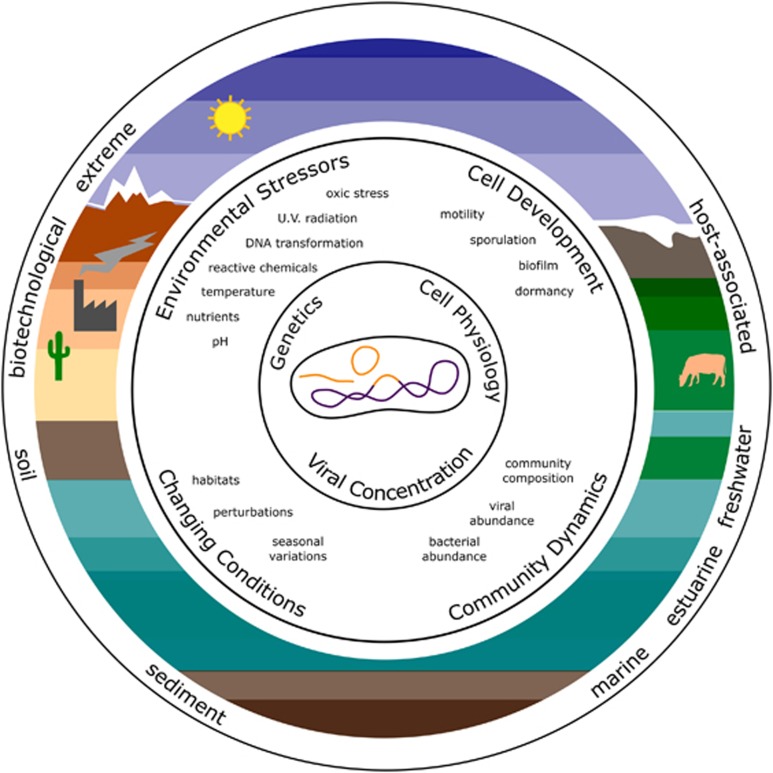
Figure 2.
Ecology of lysogeny. The establishment and maintenance of lysogeny in the cell depends on the genotype of phage and host, the physiological status of the cell, and phage concentration. In nature, temperate phage-host cell interactions are influenced by multiple factors, ranging from biological (e.g., cell development and community dynamics) to environmental (e.g., factors that cause stress and environmental fluctuations). Altogether, the interplay of all of these factors may help to explain the incidence of lysogeny across different systems, including aquatic, terrestrial and microbiome-associated (e.g., as associated with humans, animals and so on).
Functionally, a paradigm of lysogeny is that it enhances phage and host survival, particularly under adverse conditions, with most data deriving from aquatic and soil environments (Paul, 2008; Sawstrom et al., 2008; Srinivasiah et al., 2008; Williamson, 2011; Sime-Ngando, 2014). Specifically, lysogen frequency generally is inversely correlated with primary microbial productivity levels during specific seasons or environments (Sawstrom et al., 2008). Modeling (Stewart and Levin, 1984) suggests that the lysogenic state should be favored under conditions that cause reduced host cell number and activity (for example, low productivity, low nutrients or reduced host fitness) or when viral particle decay rates are high (for example, from heat or UV exposure), as postulated in soil and aquatic environments (Sime-Ngando, 2014).
Virus-to-microbe-ratios (VMR) have been associated with lysogeny such that lower VMRs (due to, for example, high rates of virion decay and/or low virion production) may be indicative of conditions that could favor lysogeny (Williamson, 2011). VMRs vary considerably, from 1.4 to 160 in marine waters (Wigington et al., 2016) and between ~0.001 and >1000 in different soils (Williamson, 2011). Generally where lower VMRs occur, for example, human gut lumen (0.019 to 0.209; (Kim et al., 2011)) and some soils (for example, ⩾12 in wetland soils; (Williamson et al., 2007)), lysogeny is hypothesized to be common. Conversely, where higher VMRs occur, for example, marine oligotrophic waters and cold desert soils, lysogeny is considered to be less prevalent (Paul, 2008; Zablocki et al., 2016). Recent research builds on this, finding that low VMRs occurred where microbial cell numbers are most abundant (Knowles et al., 2016). How specific virus-host abundances fit into either the previous view of where low host cell numbers would increase lysogeny over lytic infections, to the alternative that high host cell numbers promotes lysogeny, invites renewed effort to develop experimental hypothesis-testing approaches, as exemplified by the few but important mesocosm experiments available (Srinivasiah et al., 2008; Pradeep Ram and Sime-Ngando, 2010).
Where to explore lysogeny next
Next-generation sequencing is beginning to map viral diversity across less-explored ecosystems, using genomes derived from viral particles (Reyes et al., 2012) or total microbial communities (Paez-Espino et al., 2016). The abundance, diversity and activity of lysogeny in many systems, however, remains underexplored.
In applied settings, understanding lytic-lysogenic dynamics may aid in improving biotechnological processes, such as in agricultural or food production, but such knowledge is in its infancy. For example, prophages are diverse among lactic acid bacteria responsible for fermentation processes such as wine (for example, Oenococcus) or dairy (for example, Lactococcus and Lactobacillus) (Marcó et al., 2012; Kot et al., 2014). These offer models of microbial succession during fermentations that are both ecologically and economically interesting given that prophages can be both beneficial (improving host fitness) and detrimental (lysing cells upon induction).
In host-associated systems, lysogeny studies have largely focused on microbial virulence (Brussow et al., 2004; Abedon and LeJeune, 2005; Fortier and Sekulovic, 2013; Hargreaves et al., 2014; Feiner et al., 2015; Davies et al., 2016). In humans, temperate phages appear common (Abeles and Pride, 2014; De Paepe et al., 2014b; Edlund et al., 2015; Ogilvie and Jones, 2015). In addition, infection dynamics of the human gut virome (Waller et al., 2014) show low host cell abundances when temperate phages are in lytic cycles, presumably lysing those hosts. Another study found increased expression of phage genes involved in lytic cycles during periodontal disease (Santiago-Rodriguez et al., 2015), suggesting a connection between phages and disease, though it was not confirmed that these genes were associated with temperate phages. Given the importance of microbiota in health and disease (Alivisatos et al., 2015), and the added consequences of the use of antibiotics, which are stressors that impact on lysogen induction (Allen et al., 2011), host-associated systems are ideal for exploring the implications of the lytic-lysogenic switch. Further, investigation of specific temperate phage–host interaction dynamics can reveal key players driving ecosystem change (Allen et al., 2011; Waller et al., 2014).
Across host genomes, patterns are emerging in both the types of bacteria that are commonly lysogenized and prophage diversity. For example, the detection of λ-like, P1-like or Mu-like phages varies across microbial genomes, with Mu-like and P1-like being least reported (Casjens, 2003; Fouts, 2006; Bobay et al., 2013; Roux et al., 2015b). In addition, prophages appear to predominate across some microbial phyla (for example, 74% of Firmicutes, 41% of Actinobacteria and 22% of Cyanobacteria), but not others (for example, 0% of Chlamydiae) (Touchon et al., 2016). Correlations between prophage frequency and bacterial growth rates, genome size and pathogenicity have been found (Touchon et al., 2016) and could be driven by a microbe’s ecology. For example, prophages have been associated only with invasive Staphylococcus aureus strains (Goerke et al., 2009), and are absent from the intracellular pathogen, Chlamydiae (Touchon et al., 2016).
Emerging approaches to study lysogeny in nature
A combination of approaches for investigating lysogeny in bacterial isolates and complex communities (Figure 3) can be used to assess its abundance, diversity and activity. Experimental methods for detecting lysogeny have provided foundational mechanistic and ecological knowledge, but are recognized to have methodological issues. First, they often require exposure to stressors or lysogen dilution (Paul and Weinbauer, 2010), which can have variable induction efficiencies (Braid et al., 2004; Hanh et al., 2015; Niu et al., 2015). Second, quantifying induction is problematic as virion enumeration is challenging on field samples (Forterre et al., 2013) and highly sensitive to burst size estimates (Parada et al., 2006). Here, we focus on how sequencing-based methods can complement more traditional experimental approaches and help formulate hypotheses to experimentally test.
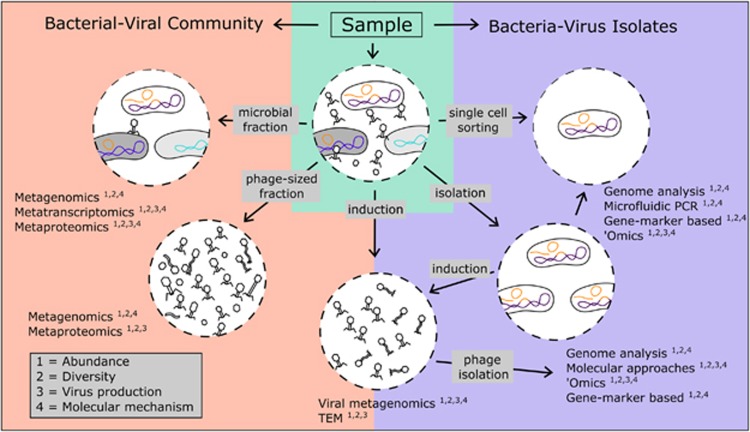
Figure 3.
Approaches and methods pipeline for characterizing lysogeny. Multiple approaches to investigating lysogeny can be applied to a bacterial community either prior to induction, following induction or to isolate samples. Community samples may be divided into bacterial and viral fractions, where the DNA can be sequenced (metagenomics) and prophages analyzed bioinformatically. In addition, sequencing RNA (metatranscriptomics) and protein (metaproteomics) may provide information on abundance and activity. Treatment of samples with inducing agents (e.g., mitomycin C, UV radiation) can measure the proportion of lysogens that are sensitive to such treatment by detecting the switch from lysogenic to productive (generally lytic) cycles. Information of phage-cell interactions can be obtained by culturing and isolating lysogens from environmental samples, which can then be treated for induction and/or analyzed in detail genetically, molecularly and structurally (microscopy). Techniques such as ‘omics (transcriptomics, proteomics, metabolomics) allow characterization of molecular changes during infection. When paired with single-cell resolution, infection dynamics can be followed to determine the prophage type (e.g., integrated versus extrachromosomal) and the fraction of infected and lysed cells. A combination of these approaches can inform the distribution, abundance and types of temperate phages, lysogens, and uninfected hosts, as well as increase our mechanistic understanding of the establishment, maintenance and dynamics of temperate phage infections.
Sequencing-enabled approaches for identifying and quantifying temperate phages:
Prophage sequences can be identified from whole or draft microbial isolate genomes (Lima-Mendez et al., 2008; Zhou et al., 2011; Akhter et al., 2012; Roux et al., 2015a); theoretically from single-cell genomes (Roux et al., 2014; Labonte et al., 2015b); or from microbial metagenomes (Waller et al., 2014; Roux et al., 2015a). From viromes, temperate phages can be identified by marker genes (for example, integrases or ParA/B genes (Emerson et al., 2012)); phylogenic analysis of conserved genes (for example, DNA polymerase gene (Schmidt et al., 2014)); similarity to isolate phages or by cross-comparing viral and bacterial sequence datasets (Waller et al., 2014); and by predicting phage/host lifestyle through in silico genome analysis (for example, with PHACTS (McNair et al., 2012)). Such sequence-based approaches can even help identify hosts (Waller et al., 2014; Hannigan et al., 2015; Labonte et al., 2015a, b; Edwards et al., 2016) and the relative proportions of temperate phages undergoing lysogenic or lytic infections (Waller et al., 2014). Prophage functionality, however, from in silico prediction requires experimental validation. In addition, activity can be inferred from presence in metatranscriptomes (Dupont et al., 2015; Engelhardt et al., 2015; Santiago-Rodriguez et al., 2015) and metaproteomes (Ogilvie et al., 2013) or by coupling viromics to induction experiments (McDaniel et al., 2008). Although confirming activity depends on experimental induction, this latter approach revealed seasonal patterns in lysogen frequency, inversely correlated to bacterial productivity in Antarctic Ocean waters (Brum et al., 2015).
Improving sequence-based and experimental characterization of lysogeny:
Sequence-based approaches can be improved with better technology to obtain (Brown et al., 2014), assemble (Bankevich et al., 2012) and identify temperate phages either by circumventing reference database limitations (for example, via k-mer analysis (Hurwitz et al., 2014)) or expanding known prophage sequence diversity (Roux et al., 2015b; Paez-Espino et al., 2016).
Experimentally, there is critical need for developing both additional experimental approaches that can help test in silico-derived hypotheses, and new model systems that can capture the diversity of lysogenic infections in nature. Here, methods for gene marker-based approaches are emerging for single-cell resolution including microfluidic digital PCR (Tadmor et al., 2011), fluorescently labeled probes (Allers et al., 2013), fluorescently labeled phages (Zeng et al., 2010)), and fluorescent reporters of prophage gene expression and genome inheritance (Cenens et al., 2013b). These can help discriminate between lysogeny and poorly characterized lysogenic (Abedon, 2009) or inefficiently lytic (Dang et al., 2015) infections. Although such methods could be improved, as discussed in (Dang and Sullivan, 2014), they nevertheless still should be helpful for characterizing lysogenic infections.
Conclusions
Temperate phages can switch between infection modes that have different but significant affects on microbial communities. Lytic cycles both kill and solubilize host bacteria, whereas lysogenic cycles impact host cells more subtly, both physiologically and genetically. Though lysogeny has been long recognized, fundamental questions nonetheless remain: (i) How do lysogen abundances change across space, time and taxa? (ii) What are the consequences and impacts on prophage-host interaction dynamics and ecosystem function? (iii) What drives these patterns? (iv) From the phage perspective, what types of prophages predominate, and why? (v) From the host perspective, why are some bacteria more prone to lysogeny than others? Numerous emerging approaches offer opportunities to develop model systems, as well as study the diverse ecological roles that lysogens have in natural microbial communities. Given these new approaches, we posit that as lytic phages have dominated the viral ecology literature to date, temperate phages should soon share the spotlight.
Acknowledgments
Anca Segall, Bob Blasdel, Claudia Igler, Bentley Fane, Martha Clokie and members of the OSU VirusLab, especially Simon Roux, Joanne Emerson and Sheri Floge, are thanked for shaping our thinking on temperate phage biology, although the latter are also thanked for comments on the manuscript. Support for this synthesis effort was provided by a Gordon and Betty Moore Foundation Investigator Award (#3790) and National Science Foundation award (OCE# 1536989) to MBS.
Footnotes
The authors declare no conflict of interest.
References
- Abedon S, LeJeune J. (2005). Why bacteriophage encode exotoxins and other virulence factors. Evol Bioinform Online 1: 97. [PMC free article] [PubMed] [Google Scholar]
- Abedon ST. (2009). Disambiguating bacteriophage pseudolysogeny: an historical analysis of pseudolysogeny, and the phage carrier state. In: Adams HT (ed). Contemporary Trends in Bacteriophage Research. Nova Science Publishers: New York, NY, USA. [Google Scholar]
- Abeles SR, Pride DT. (2014). Molecular bases and role of viruses in the human microbiome. J Mol Biol 426: 3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhter S, Aziz RK, Edwards RA. (2012). PhiSpy: a novel algorithm for finding prophages in bacterial genomes that combines similarity- and composition-based strategies. Nucleic Acids Res 40: e126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alivisatos AP, Blaser MJ, Brodie EL, Chun M, Dangl JL, Donohue TJ et al. (2015). MICROBIOME. A unified initiative to harness Earth's microbiomes. Science 350: 507. [DOI] [PubMed] [Google Scholar]
- Allen HK, Looft T, Bayles DO, Humphrey S, Levine UY, Alt D et al. (2011). Antibiotics in feed induce prophages in swine fecal microbiomes. MBio 2: e00260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allers E, Moraru C, Duhaime MB, Beneze E, Solonenko N, Barrero-Canosa J et al. (2013). Single-cell and population level viral infection dynamics revealed by phageFISH, a method to visualize intracellular and free viruses. Environ Microbiol 15: 2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS et al. (2012). SPAdes: A new genome assembly algorithm and Its applications to single-cell sequencing. J Comput Biol 19: 455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banks DJ, Lei BF, Musser JM. (2003). Prophage induction and expression of prophage-encoded virulence factors in group A Streptococcus serotype M3 strain MGAS315. Infect Immun 71: 7079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bednarz M, Halliday JA, Herman C, Golding I. (2014). Revisiting bistability in the lysis/lysogeny circuit of bacteriophage lambda. PLoS One 9: e100876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobay L-M, Rocha EPC, Touchon M. (2013). The adaptation of temperate bacteriophages to their host genomes. Mol Biol Evol 30: 737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobay L-M, Touchon M, Rocha EPC. (2014). Pervasive domestication of defective prophages by bacteria. Proc Natl Acad Sci USA 111: 12127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braid MD, Silhavy JL, Kitts CL, Cano RJ, Howe MM. (2004). Complete genomic sequence of bacteriophage B3, a Mu-like phage of Pseudomonas aeruginosa. J Bacteriol 186: 6560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown SD, Nagaraju S, Utturkar S, De Tissera S, Segovia S, Mitchell W et al. (2014). Comparison of single-molecule sequencing and hybrid approaches for finishing the genome of Clostridium autoethanogenum and analysis of CRISPR systems in industrial relevant Clostridia. Biotechnol Biofuels 7: 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brum JR, Hurwitz BL, Schofield O, Ducklow HW, Sullivan MB. (2015). Seasonal time bombs: dominant temperate viruses affect Southern Ocean microbial dynamics. ISME J 10: 437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brussow H, Canchaya C, Hardt WD. (2004). Phages and the evolution of bacterial pathogens: From genomic rearrangements to lysogenic conversion. Microbiol Mol Biol Rev 68: 560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canchaya C, Fournous G, Brüssow H. (2004). The impact of prophages on bacterial chromosomes. Mol Microbiol 53: 9. [DOI] [PubMed] [Google Scholar]
- Canchaya C, Proux C, Fournous G, Bruttin A, Brussow H. (2003). Prophage genomics. Microbiol Mol Biol Rev 67: 238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casjens S. (2003). Prophages and bacterial genomics: what have we learned so far? Mol Microbiol 49: 277. [DOI] [PubMed] [Google Scholar]
- Casjens SR, Hendrix RW. (2015). Bacteriophage lambda: early pioneer and still relevant. Virology 479: 310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cenens W, Makumi A, Mebrhatu MT, Lavigne R, Aertsen A. (2013. a). Phage–host interactions during pseudolysogeny. Bacteriophage 3: e25029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cenens W, Mebrhatu MT, Makumi A, Ceyssens P-J, Lavigne R, Van Houdt R et al. (2013. b). Expression of a novel P22 ORFan gene reveals the phage carrier state in Salmonella typhimurium. PLoS Genet 9: e1003269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chibani-Chennoufi S, Bruttin A, Dillmann ML, Brussow H. (2004). Phage–host interaction: an ecological perspective. J Bacteriol 186: 3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie GE, Calendar R. (2016). Bacteriophage P2. Bacteriophage 6: e1145782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie GE, Dokland T. (2012). Pirates of the Caudovirales. Virology 434: 210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochran PK, Kellogg CA, Paul JH. (1998). Prophage induction of indigenous marine lysogenic bacteria by environmental pollutants. Mar Ecol Prog Ser 164: 125. [Google Scholar]
- Czyz A, Los M, Wrobel B, Wegrzyn G. (2001). Inhibition of spontaneous induction of lambdoid prophages in Escherichia coli cultures: simple procedures with possible biotechnological applications. BMC Biotechnol 1: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang VT, Howard-Varona C, Schwenck S, Sullivan MB. (2015). Variably lytic infection dynamics of large Bacteroidetes podovirus phi38:1 against two Cellulophaga baltica host strains. Environ Microbiol 17: 4659. [DOI] [PubMed] [Google Scholar]
- Dang VT, Sullivan MB. (2014). Emerging methods to study bacteriophage infection at the single-cell level. Front Microbiol 5: 724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies EV, Winstanley C, Fothergill JL, James CE. (2016). The role of temperate bacteriophages in bacterial infection. FEMS Microbiol Lett 363: fnw015. [DOI] [PubMed] [Google Scholar]
- De Paepe M, Hutinet G, Son O, Amarir-Bouhram J, Schbath S, Petit M-A. (2014. a). Temperate phages acquire DNA from defective prophages by relaxed homologous recombination: the role of Rad52-Like recombinases. PLoS Genet 10: e1004181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Paepe M, Leclerc M, Tinsley CR, Petit M-A. (2014. b). Bacteriophages: an underestimated role in human and animal health? Front Cell Infect Microbiol 4: 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duerkop BA, Clements CV, Rollins D, Rodrigues JLM, Hooper LV. (2012). A composite bacteriophage alters colonization by an intestinal commensal bacterium. Proc Natl Acad Sci USA 109: 17621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont CL, McCrow JP, Valas R, Moustafa A, Walworth N, Goodenough U et al. (2015). Genomes and gene expression across light and productivity gradients in eastern subtropical Pacific microbial communities. ISME J 9: 1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edlund A, Santiago-Rodriguez TM, Boehm TK, Pride DT. (2015). Bacteriophage and their potential roles in the human oral cavity. J Oral Microbiol 7: 27423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards RA, McNair K, Faust K, Raes J, Dutilh BE. (2016). Computational approaches to predict bacteriophage–host relationships. FEMS Microbiol Rev 40: 258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson JB, Thomas BC, Andrade K, Allen EE, Heidelberg KB, Banfield JF. (2012). Dynamic viral populations in hypersaline systems as revealed by metagenomic assembly. Appl Environ Microbiol 78: 6309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelhardt T, Orsi WD, Jorgensen BB. (2015). Viral activities and life cycles in deep subseafloor sediments. Environ Microbiol Rep 7: 868. [DOI] [PubMed] [Google Scholar]
- Feiner R, Argov T, Rabinovich L, Sigal N, Borovok I, Herskovits AA. (2015). A new perspective on lysogeny: prophages as active regulatory switches of bacteria. Nat Rev Microbiol 13: 641. [DOI] [PubMed] [Google Scholar]
- Fogg PCM, Colloms S, Rosser S, Stark M, Smith MCM. (2014). New applications for phage integrases. J Mol Biol 426: 2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forterre P, Soler N, Krupovic M, Marguet E, Ackermann H-W. (2013). Fake virus particles generated by fluorescence microscopy. Trends Microbiol 21: 1. [DOI] [PubMed] [Google Scholar]
- Fortier L-C, Sekulovic O. (2013). Importance of prophages to evolution and virulence of bacterial pathogens. Virulence 4: 354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouts DE. (2006). Phage_Finder: automated identification and classification of prophage regions in complete bacterial genome sequences. Nucleic Acids Res 34: 5839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gama JA, Reis AM, Domingues I, Mendes-Soares H, Matos AM, Dionisio F. (2013). Temperate bacterial viruses as double-edged swords in bacterial warfare. PLoS One 8: e59043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebhart D, Williams SR, Bishop-Lilly KA, Govoni GR, Willner KM, Butani A et al. (2012). Novel high-molecular-weight, R-type bacteriocins of Clostridium difficile. J Bacteriol 194: 6240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goerke C, Pantucek R, Holtfreter S, Schulte B, Zink M, Grumann D et al. (2009). Diversity of prophages in dominant Staphylococcus aureus clonal lineages. J Bact 191: 3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanh N-K, Bettarel Y, Bouvier T, Bouvier C, Hai D-N, Lam N-N et al. (2015). Coral mucus is a hot spot for viral infections. Appl Environ Microbiol 81: 5773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannigan GD, Meisel JS, Tyldsley AS, Zheng Q, Hodkinson BP, SanMiguel AJ et al. (2015). The human skin double-stranded DNA virome: topographical and temporal diversity, genetic enrichment, and dynamic associations with the host microbiome. MBio 6: e01578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargreaves KR, Kropinski AM, Clokie MRC. (2014). Bacteriophage behavioural ecology: How phages alter their bacterial host’s habits. Bacteriophage 4: e29866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harshey RM. (2014). Transposable phage mu. Microbiology spectrum 2: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurwitz BL, Westveld AH, Brum JR, Sullivan MB. (2014). Modeling ecological drivers in marine viral communities using comparative metagenomics and network analyses. Proc Natl Acad Sci USA 111: 10714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M-S, Park E-J, Roh SW, Bae J-W. (2011). Diversity and abundance of single-stranded DNA viruses in human feces. Appl Environ Microbiol 77: 8062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles B, Silveira CB, Bailey BA, Barott K, Cantu VA, Cobián-Güemes AG et al. (2016). Lytic to temperate switching of viral communities. Nature 531: 466. [DOI] [PubMed] [Google Scholar]
- Kot W, Neve H, Heller KJ, Vogensen FK. (2014). Bacteriophages of LeuconostocOenococcus, and Weissella. Front Microbiol 5: 186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labonte JM, Field EK, Lau M, Chivian D, Van Heerden E, Wommace KE et al. (2015. a). Single cell genomics indicates horizontal gene transfer and viral infections in a deep subsurface Firmicutes population. Front Microbiol 6: 349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labonte JM, Swan BK, Poulos B, Luo H, Koren S, Hallam SJ et al. (2015. b). Single-cell genomics-based analysis of virus-host interactions in marine surface bacterioplankton. ISME J 9: 2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima-Mendez G, Van Helden J, Toussaint A, Leplae R. (2008). Prophinder: a computational tool for prophage prediction in prokaryotic genomes. Bioinformatics 24: 863. [DOI] [PubMed] [Google Scholar]
- Lobocka MB, Rose DJ, Plunkett G, Rusin M, Samojedny A, Lehnherr H et al. (2004). Genome of bacteriophage P1. J Bacteriol 186: 7032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lwoff A. (1953). Lysogeny. Bacteriol Rev 17: 269–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcó M, Moineau S, Quiberoni A. (2012). Bacteriophages and diary fermentations. Bacteriophage 2: 149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matos RC, Lapaque N, Rigottier-Gois L, Debarbieux L, Meylheuc T, Gonzalez-Zorn B et al. (2013). Enterococcus faecalisprophage dynamics and contributions to pathogenic traits. PLoS Genet 9: e1003539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDaniel L, Breitbart M, Mobberley J, Long A, Haynes M, Rohwer F et al. (2008). Metagenomic analysis of lysogeny in Tampa Bay: implications for prophage gene expression. PLoS One 3: e3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLeod SM, Kimsey HH, Davis BM, Waldor MK. (2005). CTX phi and Vibrio cholerae: exploring a newly recognized type of phage–host cell relationship. Mol Microbiol 57: 347. [DOI] [PubMed] [Google Scholar]
- McNair K, Bailey BA, Edwards RA. (2012). PHACTS, a computational approach to classifying the lifestyle of phages. Bioinformatics 28: 614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mell JC, Redfield RJ. (2014). Natural competence and the evolution of DNA uptake specificity. J Bacteriol 196: 1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menouni R, Hutinet G, Petit M-A, Ansaldi M. (2015). Bacterial genome remodeling through bacteriophage recombination. FEMS Microbiol Lett 362: 1. [DOI] [PubMed] [Google Scholar]
- Nanda AM, Thormann K, Frunzke J. (2015). Impact of spontaneous prophage induction on the fitness of bacterial populations and host–microbe interactions. J Bacteriol 197: 410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu YD, Cook SR, Wang JY, Klima CL, Hsu YH, Kropinski AM et al. (2015). Comparative analysis of multiple inducible phages from Mannheimia haemolytica. BMC Microbiol 15: 175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogilvie LA, Bowler LD, Caplin J, Dedi C, Diston D, Cheek E et al. (2013). Genome signature-based dissection of human gut metagenomes to extract subliminal viral sequences. Nat Commun 4: 2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogilvie LA, Jones BV. (2015). The human gut virome: a multifaceted majority. Front Microbiol 6: 918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paez-Espino D, Eloe-Fadrosh EA, Pavlopoulos GA, Thomas AD, Huntemann M, Mikhailova N et al. (2016). Uncovering Earth’s virome. Nature 536: 425. [DOI] [PubMed] [Google Scholar]
- Parada V, Herndl GJ, Weinbauer MG. (2006). Viral burst size of heterotrophic prokaryotes in aquatic systems. J Mar Biol Assoc UK 86: 613. [Google Scholar]
- Paul JH. (2008). Prophages in marine bacteria: dangerous molecular time bombs or the key to survival in the seas? ISME J 2: 579. [DOI] [PubMed] [Google Scholar]
- Paul JH, Weinbauer MG. (2010). Detection of lysogeny in marine environments. In: Wilhelm SW et al. (eds). Manual of Aquatic Viral Ecology. ASLO: Waco, TX, USA. [Google Scholar]
- Pradeep Ram AS, Sime-Ngando T. (2010). Resources drive trade-off between viral lifestyles in the plankton: evidence from freshwater microbial microcosms. Environ Microbiol 12: 467. [DOI] [PubMed] [Google Scholar]
- Rao VB, Feiss M. (2015). Mechanisms of DNA packaging by large double-stranded DNA viruses. Annu Rev Virol 2: 351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravin NV. (2015). Replication and maintenance of linear phage-plasmid N15. Microbiol Spectr 3: 0032. [DOI] [PubMed] [Google Scholar]
- Reyes A, Semenkovich NP, Whiteson K, Rohwer F, Gordon JI. (2012). Going viral: next-generation sequencing applied to phage populations in the human gut. Nat Rev Microbiol 10: 607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux S, Enault F, Hurwitz B, Sullivan M. (2015. a). VirSorter: mining viral signal from microbial genomic data. PeerJ 3: e985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux S, Hallam SJ, Woyke T, Sullivan MB. (2015. b). Viral dark matter and virus–host interactions resolved from publicly available microbial genomes. eLife 4: e08490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux S, Hawley AK, Torres Beltran M, Scofield M, Schwientek P, Stepanauskas R et al. (2014). Ecology and evolution of viruses infecting uncultivated SUP05 bacteria as revealed by single-cell- and meta-genomics. Elife 3: e03125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson JE, Magadan AH, Sabri M, Moineau S. (2013). Revenge of the phages: defeating bacterial defences. Nat Rev Microbiol 11: 675. [DOI] [PubMed] [Google Scholar]
- Santiago-Rodriguez TM, Naidu M, Abeles SR, Boehm TK, Ly M, Pride DT. (2015). Transcriptome analysis of bacteriophage communities in periodontal health and disease. BMC Genomics 16: 549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawstrom C, Lisle J, Anesio AM, Priscu JC, Laybourn-Parry J. (2008). Bacteriophage in polar inland waters. Extremophiles 12: 167. [DOI] [PubMed] [Google Scholar]
- Schmidt HF, Sakowski EG, Williamson SJ, Polson SW, Wommack KE. (2014). Shotgun metagenomics indicates novel family A DNA polymerases predominate within marine virioplankton. ISME J 8: 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikuma NJ, Pilhofer M, Weiss GL, Hadfield MG, Jensen GJ, Newman DK. (2014). Marine tubeworm metamorphosis induced by arrays of bacterial phage tail-like structures. Science 343: 529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sime-Ngando T. (2014). Environmental bacteriophages: viruses of microbes in aquatic ecosystems. Front Microbiol 5: 355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasiah S, Bhavsar J, Thapar K, Liles M, Schoenfeld T, Wommack KE. (2008). Phages across the biosphere: contrasts of viruses in soil and aquatic environments. Res Microbiol 159: 349. [DOI] [PubMed] [Google Scholar]
- Stewart FM, Levin BR. (1984). The population biology of bacterial viruses: why be temperate. Theor Popul Biol 26: 93. [DOI] [PubMed] [Google Scholar]
- Sullivan MB, Krastins B, Hughes JL, Kelly L, Chase M, Sarracino D et al. (2009). The genome and structural proteome of an ocean siphovirus: a new window into the cyanobacterial 'mobilome’. Environ Microbiol 11: 2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suttle CA. (2005). Viruses in the sea. Nature 437: 356. [DOI] [PubMed] [Google Scholar]
- Tadmor AD, Ottesen EA, Leadbetter JR, Phillips R. (2011). Probing individual environmental bacteria for viruses by using microfluidic digital PCR. Science 333: 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touchon M, Bernheim A, Rocha EPC. (2016). Genetic and life-history traits associated with the distribution of prophages in bacteria. ISME J 10: 2744–2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utter B, Deutsch DR, Schuch R, Winer BY, Verratti K, Bishop-Lilly K et al. (2014). Beyond the chromosome: the prevalence of unique extra-chromosomal bacteriophages with integrated virulence genes in pathogenic Staphylococcus aureus. PLoS One 9: e100502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virgin HW. (2014). The virome in mammalian physiology and disease. Cell 157: 142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waller AS, Yamada T, Kristensen DM, Kultima JR, Sunagawa S, Koonin EV et al. (2014). Classification and quantification of bacteriophage taxa in human gut metagenomes. ISME J 8: 1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Kim Y, Ma Q, Hong SH, Pokusaeva K, Sturino JM et al. (2010). Cryptic prophages help bacteria cope with adverse environments. Nat Commun 1: 147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinbauer MG. (2004). Ecology of prokaryotic viruses. FEMS Microbiol Rev 28: 127. [DOI] [PubMed] [Google Scholar]
- Wigington CH, Sonderegger D, Brussaard CPD, Buchan A, Finke JF, Fuhrman JA et al. (2016). Re-examination of the relationship between marine virus and microbial cell abundances. Nature Microbiology 1: 15024. [DOI] [PubMed] [Google Scholar]
- Williamson KE. (2011). Soil phage ecology: abundance, distribution, and interactions with bacterial hosts. In: Witzany G (ed). Biocommunication in Soil Microorganisms. Springer: Berlin, Germany. [Google Scholar]
- Williamson KE, Radosevich M, Smith DW, Wommack KE. (2007). Incidence of lysogeny within temperate and extreme soil environments. Environ Microbiol 9: 2563. [DOI] [PubMed] [Google Scholar]
- Wommack KE, Colwell RR. (2000). Virioplankton: viruses in aquatic ecosystems. Microbiol Mol Biol Rev 64: 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zablocki O, Adriaenssens EM, Cowan D. (2016). Diversity and ecology of viruses in hyperarid desert soils. Appl Environ Microbiol 82: 770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L, Skinner SO, Zong C, Sippy J, Feiss M, Golding I. (2010). Decision making at a subcellular level determines the outcome of bacteriophage infection. Cell 141: 682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Liang Y, Lynch KH, Dennis JJ, Wishart DS. (2011). PHAST: a fast phage search tool. Nucleic Acids Res 39: W347. [DOI] [PMC free article] [PubMed] [Google Scholar]