Abstract
The aurora kinases (AURK) comprise an evolutionarily conserved family of serine/threonine kinases involved in mitosis and meiosis. While most mitotic cells express two AURK isoforms (AURKA and AURKB), mammalian germ cells also express a third, AURKC. Although much is known about the functions of the kinases in mitosis, less is known about how the three isoforms function to coordinate meiosis. This review is aimed at describing what is known about the three isoforms in female meiosis, the similarities and differences between kinase functions, and speculates as to why mammalian germ cells require expression of three AURKs instead of two.
Keywords: Meiosis;;;, Aurora kinase, oocyte, chromosomal passenger complex
Meiosis, a specialized cellular division
Meiosis differs fundamentally from mitosis because through two subsequent divisions, and no intervening round of DNA replication, haploid gametes form that are genetically and biochemically distinct from the starting diploid precursor cell. Homologous recombination (see Glossary) and reduction of the chromosome content in half generates genetically distinct cells. Coupled to meiosis is the formation of gametes (gametogenesis) a developmental program that varies between species and sex that makes the gamete biochemically distinct from the starting cell. The first meiotic division (meiosis I (MI)) is unique because homologous chromosomes segregate while sister chromatids remain associated with one another. The subsequent meiotic division (meiosis II (MII)) resembles that of mitosis where sister chromatids segregate. In mammals, meiotic maturation is the process that couples completion of meiosis I with the acquisition of developmental competence to support fertilization (Fig. 1). In females there are several hallmark maturation events that are linked to the cell cycle. First, meiosis is not continuous, it is initiated during fetal development where DNA replication and homologous recombination occur, followed by a prolonged arrest at the dictyate stage of prophase I. During the arrest, oocyte growth occurs. This arrest lasts until the organism is reproductively mature, after which a subset of the oocytes will resume meiosis in response to species-specific regulatory cues. The second hallmark is that once oocytes reach full size, they cease transcription and remain silent throughout completion of meiosis, relying solely on mRNA transcribed during oocyte growth. Therefore, oocytes rely on recruitment of stored maternal transcripts to assist in completing meiosis and subsequent developmental changes that occur in pre-implantation embryos prior to zygotic genome activation [1]. Finally, vertebrate oocytes arrest at metaphase of MII, and will not complete MII until fertilized by sperm.
Figure 1. Stages of meiotic maturation in mouse oocytes.
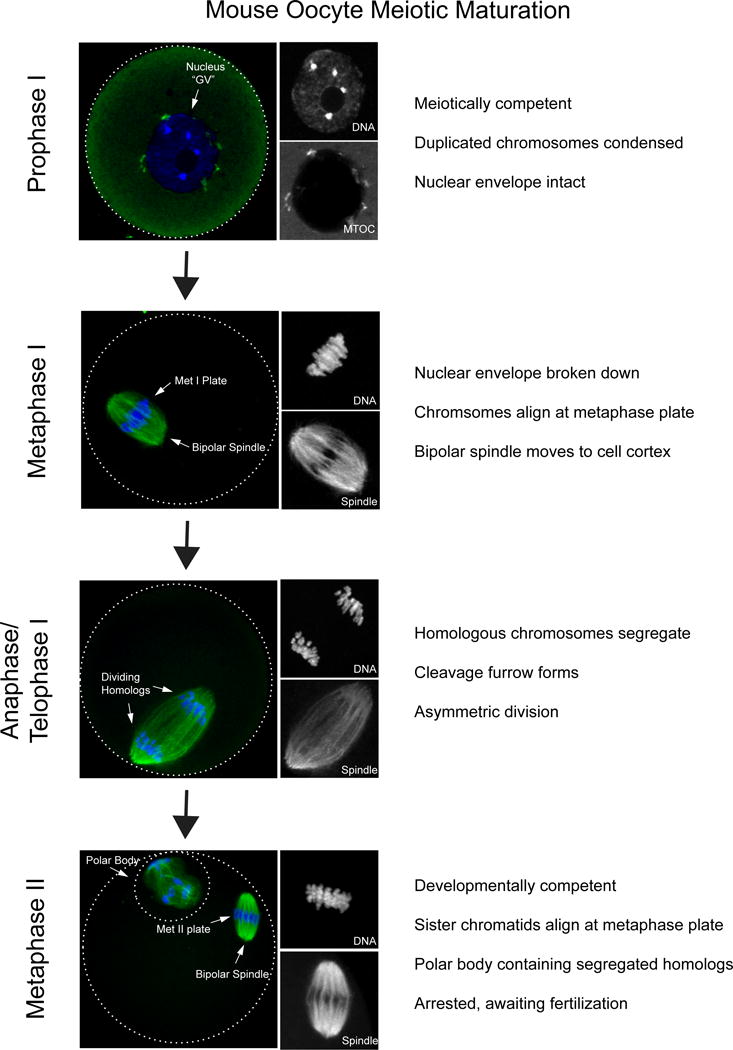
Prophase I-arrested oocytes from CF1 mice were matured in vitro to the various stages in meiosis; prophase I (0h), Metaphase I (7h), Anaphase (10h), Metaphase II (16h) prior to fixation and immunocytochemistry to detect spindle (α-tubulin; green in merge) and DNA (DAPI, blue in merge). Optical zoom images of DNA and spindle are shown on the right in grey. In prophase I the nucleus, classically referred to as the germinal vesicle (GV), remains intact. The nucleolus is visible at the center of the nucleus. Microtubule organizing centers (MTOCs; green) are attached to the nuclear membrane. Once meiosis resumes chromosomes align along the metaphase plate, organized by a bipolar spindle that moves to the cell cortex. In anaphase/telophase I homologous chromosomes are segregated. In metaphase II a polar body is visible, containing half of the chromosome compliment. Within the main body of the egg sister chromatids align along a metaphase plate and the cell arrests until fertilized by sperm.
It is critical that the meiotic divisions occur accurately, as errors in chromosome segregation cause aneuploidy. Aneuploidies are frequently incompatible with life and are the leading genetic cause of infertility and failure of in vitro fertilization in humans [2]. Although aneuploidy is rare in most organisms, humans are particularly prone to chromosomal abnormalities, with 10-30% of fertilized eggs being aneuploid, accounting for nearly one third of miscarriages [2].
Aurora Kinases
The precise regulation of meiosis is a choreographed dance involving numerous proteins that ensure formation of a healthy gamete. The aurora protein kinases (Aurks) are a conserved family of protein kinases, key in coordinating this dance in mitosis and meiosis [3–5]. The kinases act as molecular switches, regulating multiple processes in cell division including but not limited to; spindle organization, chromosome alignment, the spindle assembly checkpoint, cytokinesis, and the abscission checkpoint (Table 1) [3].
Table 1. Summary of aurora kinase localization and function in mitosis and meiosis.
INCENP, inner centromere protein; ICA, inner chromatid axis; K-MT, kinetochore microtubule; MTOC, microtubule organizing center; SAC, Spindle assembly checkpoint; TPX2, targeting protein for Xklp2.
| Mitosis | Meiosis | |||||
|---|---|---|---|---|---|---|
| Aurora Kinase | Binding Partner(s) | Localization | Function(s) | Binding Partner(s) | Localization | Established Function(s) |
| - AURKA |
- TPX2 Bora |
- Spindle Poles |
- Spindle organization |
- TPX2 Bora |
- Spindle Poles |
- Spindle organization K-MT correction |
| AURKB | INCENP | Centromeres | Chromosome alignment K-MT correction Cohesion SAC Cytokinesis Abcission Checkpoint |
? | Microtubules | Chromsome alignment |
| AURKC | N/A | N/A | N/A | INCENP | Spindle Poles Centromeres/ICA | MTOC clustering Chromsome alignment Chromsome condensation K-MT correction |
The mammalian genome encodes three AURK isoforms (AURKA, AURKB, and AURKC) (Fig. 2, Key Figure). AURKA is expressed in mitotic and meiotic cells and localizes to spindle poles to regulate spindle mechanics [6, 7]. AURKB is also expressed in mitosis and meiosis with dynamic protein localization: first, localizing to chromosomes at metaphase where it regulates chromosome alignment and kinetochore-microtubule (K-MT) attachments and then in anaphase localizing to the spindle midzone to assist in cytokinesis [3, 8–10]. AURKC expression is primarily restricted to germ cells and it has higher sequence similarity to AURKB than to AURKA [11–13]. AURKC localization is a hybrid of AURKA and AURKB in mitosis, because it localizes to spindle poles and chromosomes in metaphase, and the spindle midzone in anaphase [14–16]. Found only in mammals, AURKC may have arisen from a genome duplication event of an ancestral AURKB/C gene found in cold-blooded vertebrates [5]. The conservation of a third AURK in mammalian meiosis has been a mystery in gamete biology for decades. Why do gametes require the presence of an additional AURK compared to their mitotic counterparts?
Figure 2. Aurora kinase localization in mitosis and meiosis.
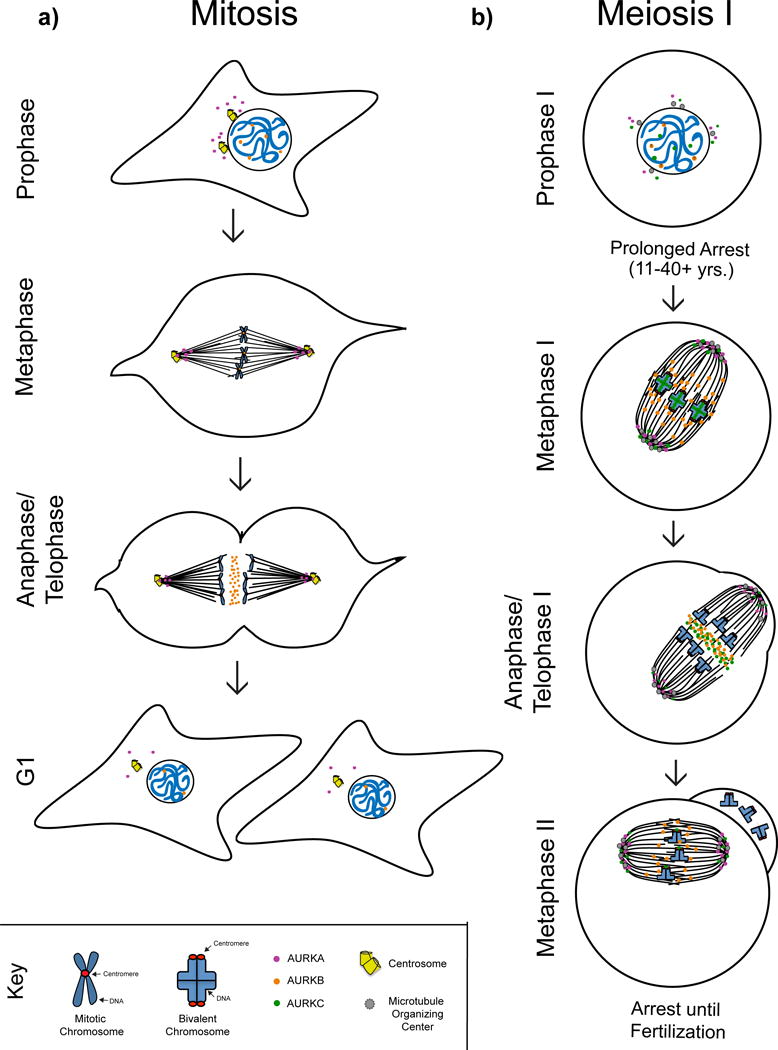
Schematic representation of AURKA, AURKB, and AURKC localization in mitosis and meiosis in mammalian cells. a) In mitotic prophase AURKA (purple circles) is concentrated around duplicated centrosomes while AURKB (orange circles) is nuclear. In metaphase AURKA is found at spindle poles while AURKB is located on centromeres. In anaphase AURKA remains at spindle poles whereas AURKB concentrates at the spindle midzone. Daughter cells enter G1 stage where the expression of both kinases is substantially reduced. b) In prophase of meiosis I AURKA clusters around microtubule organizing centers in the cytoplasm while the location of AURKB remains unknown. AURKC (green circles) can be found both in the nucleus and cytoplasm. In metaphase I AURKA localizes to spindle poles while AURKB is on spindle microtubules and potentially kinetochores. AURKC is found both at spindle poles and at the interchromatid axis of MI bivalents. In anaphase I AURKA and AURKC localize to spindle poles while AURKB and a subset of AURKC concentrate at the spindle midzone. Mammalian oocytes arrest at metaphase of meiosis II until fertilization with AURKA and AURKC at spindle poles, AURKB on the spindle microtubules and AURKC concentrated at the centromere.
The unique MI division and prolonged periods of cell-cycle arrest, make it enticing to imagine that a third AURK is required to mediate specific roles unique to oocyte meiotic maturation. Understanding these functions, and how AURKC interacts with the other two homologs, will be critical for understanding the complex inner workings of meiosis and hopefully will shed light on why aneuploidies are so common in humans. The purpose of this review is to highlight the current understanding of the functions of the AURKs in female meiosis, to speculate why meiotic cells use three AURKs, and to identify significant remaining questions in the field. To remain concise this review will not elaborate on the details of key regulatory pathways including spindle morphogenesis, the spindle assemble checkpoint, cytokinesis, and the abscission checkpoint. Many other reviews do a thorough job covering these topics in mitosis in detail, and instead, here we will focus on the known requirements for the Aurora kinase family members as they relate to these pathways[10, 17–28].
Regulation of localization and activity
AURKA
In metaphase I (Met I) and II (Met II), AURKA localizes to spindle poles where its activity is important for regulating spindle organization (Table 1) (Fig. 2, Key Figure) [3, 29, 30]. Work in Xenopus egg extracts showed AURKA localization and activation requires the microtubule-associated protein TPX2 [31–33]. Knockout of Tpx2 in mice using a conditional gene trap strategy is lethal, causing arrest at the morula stage of pre-implantation embryonic development [34]. Inspection of these arrested embryos revealed loss of AURKA at spindles and defective microtubule nucleation. In addition to TPX2, Bora, another AURKA binding partner, is critical for the localization and activation of AURKA at spindle poles in mouse oocytes [35]. In wild-type oocytes Bora co-localizes with AURKA at spindle poles and depletion of Bora using siRNA disrupts AURKA localization and results in abnormal spindle morphologies and misaligned chromosomes at Met I. These data suggest that Bora also regulates AURKA, however, whether Bora acts upstream of the kinase or the mechanism by which it is involved in microtubule organization in oocytes remains to be determined.
AURKB
In mitosis, AURKB is an essential regulator of cell division, with known functions including; regulation of chromosome alignment, the spindle assembly checkpoint (SAC), erroneous K-MT attachment detection, chromatid condensation, cohesion, the abscission checkpoint, and cytokinesis (Table 1) [17, 19, 36–40]. While the requirement for AURKB in mitosis is fairly well defined, the role of AURKB in meiosis is less understood. One reason for this lack of understanding is the conflicting evidence about the presence of AURKB protein in mouse oocytes in the literature. The AURKB antibodies that detect AURKB in somatic cells may not be sensitive enough to routinely detect AURKB in mammalian gametes making examination of the endogenous protein localization difficult [14, 41]. However, some groups have successfully detected AURKB by immunocytochemistry and western blotting [14, 42]. These results are consistent with the presence of Aurkb transcripts in mouse oocytes [43] and of the protein in human oocytes and embryos [13, 44–46]. Unlike mitosis where AURKB localizes to centromeres, endogenous AURKB protein localizes to the spindle during Met I and Met II in mouse oocytes, and may be absent from the chromosomes (Fig. 2, Key Figure) [14]. However, while not detectable on chromosomes, AURKB may still regulate chromosome alignment. Treatment of oocytes with a low concentration of the AURKB/C small molecule inhibitor ZM447439, results in chromosome misalignment in MI [44]. Importantly, the dose of ZM447439 utilized (1.5 μM) was at the threshold of having a phenotypic effect. This dose was therefore selected for a rescue experiment aimed to determine which AURK was required for chromosome alignment. Only overexpression of AURKB, but not AURKA or AURKC, rescues this alignment defect. These results further provide evidence for the endogenous expression of AURKB and suggest the kinase regulates chromosome alignment [44]. However, the precise AURKB function(s) in meiosis is unknown and is an active area of exploration.
AURKC
AURKC is the most elusive of the AURKs, with expression primarily restricted to germ cells in mammals, a few somatic tissue types, and many human cancer lines [47–52]. AURKC is most similar in sequence to AURKB, differing most significantly in the N-terminus, with AURKC lacking destruction motifs found in AURKA and AURKB.
Similar to AURKB in mitosis, AURKC acts as catalytic subunit of the chromosomal passenger complex (CPC) in meiosis [53]. The CPC is a multi-protein complex comprised of INCENP, Survivin, Borealin, and in some cases TIP60. AURKB/C activity is dependent upon binding INCENP, the scaffolding unit of the complex, which stimulates auto-phosphorylation and activation of the kinase [54]. Survivin and Borealin are critical for determining localization of the complex to the chromosomes via recognition of histone phospho-marks [55–58].
Phosphorylation of centromeric Histone H3 at threonine 3 (H3pT3) by Haspin kinase is recognized by Survivin, thereby driving the CPC to centromeres and the interchromatid axes (ICA) in metaphase of meiosis I [16, 59–66]. Concomitantly, phosphorylation of Histone 2A at threonine 120 (H2ApT120) by BUB1 kinase also positively regulate chromosomal CPC localization [66–68]. The role of TIP60 in meiosis remains unknown.
CPC localization is dynamic, localizing to centromeres and the ICA of chromosomes in Met I and the spindle midzone in anaphase (Table 1) (Fig. 2, Key Figure) [14]. However, whether both AURKB and AURKC act as CPC members in meiosis is unknown. Additionally, like AURKA, AURKC localizes to the spindle poles throughout meiosis in mouse oocytes, detectable by AURKC specific antibodies and by exogenous expression of fluorescently labeled AURKC-Gfp [15]. Most of our understanding about the function of AURKB/C in meiosis stems from studies that utilize small molecule inhibitors (AZD1152 and ZM447439). However, due to their high sequence similarity, these inhibitors are not selective for AURKB over AURKC at doses required to elicit strong phenotypes [44, 45, 69–71]. In addition, siRNA approaches are reportedly inefficient and non-specific [69, 70], and expression of a dominant-negative Aurkc allele disrupts both AURKC and AURKB function [14, 72]. These molecular constraints have made deciphering whether there are non-overlapping AURKB and AURKC functions in meiosis a challenge. In an attempt to tackle this problem, researchers expressed a dominant-negative allele of Aurkc that mutates the kinase ATP pocket (Aurkc-L93A), that does not affect AURKB activity [14]. The selectivity of the mutant was confirmed by a genetic experiment. Using oocytes from either Aurkc−/− or Aurkb−/− mice, INCENP phosphorylation (pINCENP) levels were monitored. This was chosen as a marker of AURK activity because it is an AURKB/C substrate and binding partner. After exogenous expression of Aurkc-L93A in oocytes from Aurkb−/− mice that only express AURKC, pINCENP levels were significantly reduced compared to wild-type oocytes, whereas Aurkc−/− oocytes that only express AURKB expressing the mutant had wild-type levels of pINCENP. These results were interpreted as AURKC-L93A only inhibits AURKC, and cannot inhibit AURKB. Therefore, when expressed in wild-type oocytes, the resulting phenotypes revealed that AURKC is the dominant CPC kinase, consistent with other suggestions in the literature that AURKC has replaced AURKB-CPC chromosome function in meiosis [14, 43, 69]. Loss of AURKC activity resulted in misaligned chromosomes likely due to improper K-MT attachments, a reduction in the ability to successfully complete MI, and aneuploid MII eggs. While AURKB and AURKC sequence similarities are well documented, we would be remiss to not acknowledge that sequence similarities between AURKA and AURKB also exist [73–75]. In mitotic cells, a single amino acid substitution in AURKA (in human Gly198 to Asn) enables the kinase to interact with INCENP instead of TPX2 in vivo and to phosphorylate AURKB substrates. Expression of the Aurka mutant in Aurkb-depleted cells rescued the chromosome alignment defects and promoted mitotic progression [73]. The authors went on to show that binding partner specificity is dictated by length and hydrophilicity of the side chain in the binding domain [73]. Although, the same substitution in AURKB (in human Asn 142 to Gly) did allow localization to spindle poles, some protein remained CPC-bound at centromeres. Functional complementation for loss of AURKA was not fully evaluated for this AURKB mutant because of the residual centromere population. This residue resides in kinase subdomain IV, where AURKC is more homologous to AURKB at this site (Asn 108 in human). However, other residues in this subdomain are homologous to AURKA (ie. His 111 in human AURKC and His 201 in human AURKA). Therefore, it is tempting to speculate that AURKC localizes to chromosomes and poles because of this hybrid binding domain sequence.
In female mice, germline deletion of Aurkc causes subfertility, and oocytes have similar, albeit less severe, phenotypes as those expressing the selective dominant-negative Aurkc allele. These phenotypes include a reduction in oocytes capable of progressing to Met II [13]. Using a live-cell imaging assay to monitor protein destruction, AURKB-mCherry turns over 50% more than AURKC-Gfp during meiotic maturation, suggesting that AURKC is more stable than AURKB [13]. The authors concluded that this increased stability might compensate for the inherent instability of AURKB in a prolonged cell division that does not have active transcription. In addition to being more stable, Aurkc is a maternally recruited message, with significant translation occurring during meiotic maturation. This increase contrasts that of AURKB protein, which declines throughout meiosis. Aurkc recruitment may provide a boost of AURK activity to support meiotic progression when AURKB protein is limited [76]. However, while loss of AURKC leads to MI arrest, about one-half of AURKC knockout oocytes successfully complete meiosis. Upon further examination of these AURKC knockout oocytes, AURKB localized to the centromeres/kinetochore and ICA and INCENP was phosphorylated, demonstrating the ability for AURKB to compensate for the loss of AURKC in oocytes [13].
Separating AURKB and AURKC function
AURKB’s ability to compensate for the loss of AURKC implies these kinases have some functional equivalence. However, this observation raises an important question; if the two kinases have non-overlapping functions within the oocyte, how are these roles mediated if the kinases are so similar? One way non-overlapping functions could be carried out is through two CPC complexes that differ in the catalytic component. To test this hypothesis recent experiments used co-immunoprecipitation, RNAi, and sucrose sedimentation analyses of a prostate cancer cell line (PC-3) that expresses both AURKB and AURKC. The authors found that while both kinases bind INCENP in vivo, AURKB and AURKC cannot bind within the same complex [77]. The next question to answer will be how two kinases, so similar in functional ability, could have separate roles and potentially localization when in CPC complexes.
A hint at how the two kinases perform separate functions could come from the discovery of two CPC subpopulations in mitotic cells. Dimerization of Borealin plays a critical role in localization of a proportion of the CPC to kinetochores [78]. This localization is independent from an inner centromeric CPC pool regulated by Survivin recognition of H3pT3 [78]. Importantly, these two spatially distinct CPC pools are responsible for regulating different functions within the cell. Specifically inner-centromere-localized CPC was important for destabilizing improper K-MT attachments and activation of the SAC, while the function of the kinetochore population is still under investigation.
Differentially regulated CPC populations are also present in meiosis. In mouse oocytes, inhibition of Haspin results in a loss of AURKC-CPC from the centromeres and ICA but not from the kinetochores [16]. Loss of centromere/ICA-localized CPC resulted in an increase of stable improper K-MT attachments, suggesting this population is important for K-MT error correction [16]. Experiments aimed at perturbing kinetochore-localized CPC will be critical to determining the specific roles of this subpopulation of the CPC and whether it contains AURKB or AURKC. We therefore hypothesize similar spatial distributions on mouse oocyte bivalents could support this division of labor (Fig. 3).
Figure 3. Hypothesized mechanism of AURKB-CPC and AURKC-CPC spatial distribution to explain separation of function in germ cells.
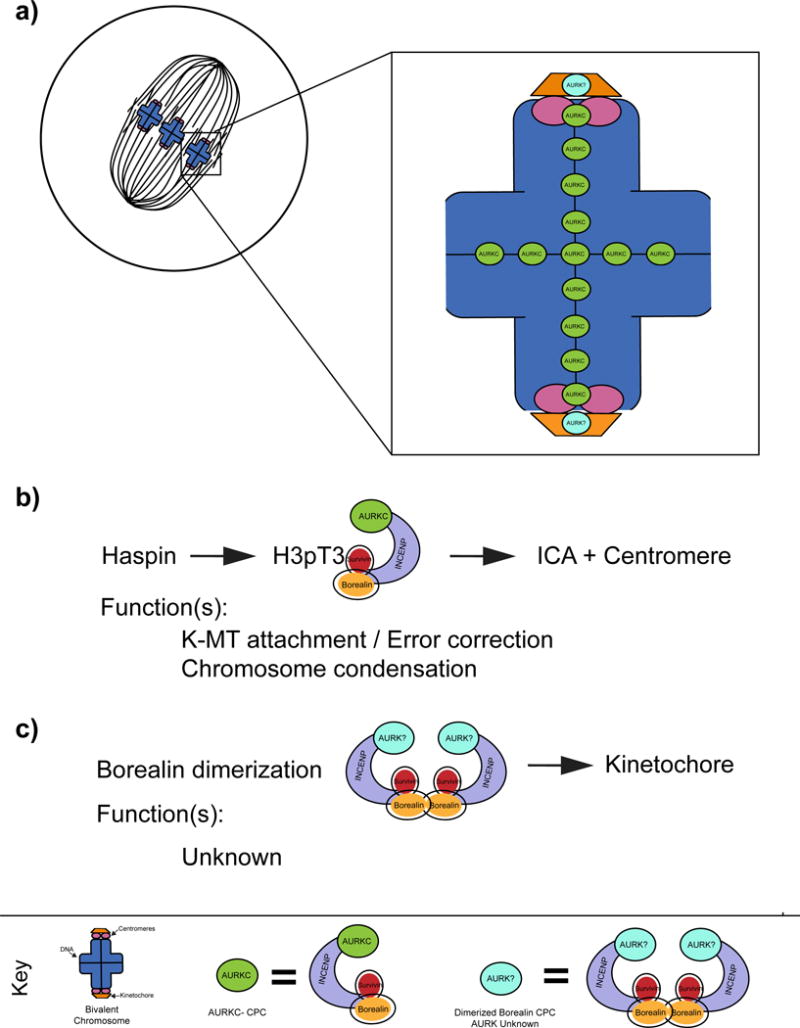
a) Schematic representation of hypothesized CPC sub-populations in mouse oocytes on MI bivalents. b) Survivin recognition of phosphorylated histone 3 at threonine 3 (H3pT3) drives AURKC-CPC to the interchromatid axis (ICA) and centromeres. This CPC subpopulation is responsible for error correction in meiosis I via destabilization of erroneous kinetochore-microtubule (K-MT) attachments and regulating chromosome condensation. c) Hypothesized Borealin dimerization drives AURKB/C-CPC localization to kinetochores. This CPC sub-populations functions remain unknown and may require AURKB and/or AURKC.
Differences in catalytic activity could provide additional functional diversity. Previously, the only direct interaction with AURKs and the CPC subunits was detected with INCENP, however, immunoprecipitation of endogenous AURKC from PC-3 cells revealed the presence of an AURKC-Survivin dimer that did not contain INCENP [77]. Interestingly, this interaction positively regulates AURKC, but not AURKB, activity. In vitro kinase assays found that AURKC, and not AURKB, phosphorylates Survivin at serine 20, promoting auto-activation of the kinase. This new substrate was identified as playing an important role in regulating correction of K-MT attachments and cytokinesis [11]. These data, taken together with findings of differentially localized CPC subpopulations, could provide the answer to how AURKB and AURKC maintain non-overlapping functions in oocytes. It is tempting to speculate that AURKC-CPC, the predominant AURK localized to the centromeres and ICA, is responsible for regulating K-MT attachments while AURKB could comprise the kinetochore CPC population that monitors the SAC (Fig. 3).
AURK Functions
Regulation of spindle dynamics: AURKA and AURKC
Unlike mitotic cells that build a spindle using centriole-containing centrosomes, oocytes of many organisms, including mammals, lack centrioles[79]. Mouse oocytes instead rely on many microtubule-organizing centers (MTOCs) that nucleate microtubules and cluster together to form two spindle poles (Fig. 2, Key Figure). AURKA co-localizes with MTOCs throughout meiosis and is required to recruit γ-tubulin, a key MTOC component [80]. Knockdown of Aurka in mouse oocytes results in the loss of γ-tubulin and pericentrin localization leading to MTOC scattering [45].
In mitotic prophase, newly nucleated microtubules are stabilized via phosphorylation of the centrosome-associated protein TACC3 that is mediated by AURKA [81]. Stable microtubules are then assembled into a bipolar structure via the molecular motor protein Kinesin 5 (Eg5), another AURKA substrate [6, 82–86]. Pharmacological inhibition of AURKA in bovine oocytes, and the subsequent loss of TACC3 phosphorylation, leads to abnormal meiotic spindles and misaligned chromosomes [81]. Similarly, knockdown of Aurka (air-1) in Caenorhabditis elegans embryos results in a failure to assemble microtubules into a bipolar spindle. This spindle-building function is dependent on the activity of the kinase because expression of a catalytically inactive form of AURKA (air-1K73R, T201A) failed to rescue this phenotype. While cells fail to form a bipolar spindle when AURKA is depleted, overexpression of the protein leads to MTOC multiplication and abnormally long, distorted microtubules, suggesting proper regulation of AURKA expression is critical to spindle formation in meiosis [30].
A new function for AURKC in bipolar spindle assembly was recently described in mouse oocytes. By changing fixation conditions to preserve MTOC structure, active AURKC was discovered to also co-localize to MTOCs during meiotic maturation (Fig. 2, Key Figure) [15]. Similar to localization at the ICA, this localization required Haspin kinase activity. MTOC-localized AURKC is required for MTOC clustering because perturbation of its localization by inhibition of Haspin, or by expression of the selective dominant-negative Aurkc allele resulted in the inability to cluster MTOCs [15]. This phenotype could be rescued only through the overexpression of AURKC, and not AURKB or AURKA, highlighting a unique function for AURKC at MTOCs [15]. Interestingly, this AURKC function appears to not require INCENP because depletion of INCENP from oocytes does not result in MTOC clustering defects. The mechanism by which AURKC regulates MTOC clustering is not yet understood, however this function could be relevant to understanding how some cancers, particularly those that express AURKC, cluster supernumerary centrosomes into a pseudo bipolar spindle to drive cell proliferation.
Chromosome condensation and cohesion: AURKB and AURKC
Chromosome compaction is essential for chromosome segregation and requires a multi-protein complex called condensin, that uses SMC1 and SMC2 as hinges to structurally organize the chromosomes into tight bundles [87]. In C. elegans, AURKB (AIR-2) is required for the localization of condensin in pro-metaphase along the short arm of bivalents [88]. In oocytes, loss of AURKC-CPC from the ICA delocalizes condensin, indicating this function may be AURKC-specific in gametes [16]. However, there is still much work needed to fully understand the role of the AURKs in regulating chromosome condensation.
One key difference between MI and mitosis is the separation of homologous chromosomes, instead of sister chromatids. For oocytes, this segregation pattern requires that sister chromatids remain tightly associated in MI, behaving as a single unit when binding spindle microtubules. Cohesins hold sister chromatids together, localizing along the bivalents and at centromeres during Met I, promoting their tight association. For homologs to separate in anaphase I, separase must cleave the cohesin localized between bivalents but centromeric cohesin must remain intact. To protect centromeric cohesin, Shugoshin-2 (SGO2) localizes to centromeres and prevents phosphorylation of the meiotic cohesin subunit REC8 [89–92]. In mitotic cells, phosphorylation of SGO2 by AURKB is essential for cohesion protection [93, 94]. In mouse oocytes loss of active AURKC-CPC does not perturb SGO2 localization or cause premature separation of sister chromatids suggesting that AURKB-CPC is responsible for this function [14]. Further work to decipher if protection of centromeric cohesion during MI is regulated by AURKB and not AURKC-CPC will be imperative to understand how MI is regulated.
Kinetochore-microtubule attachments and error correction: AURKB and AURKC, and surprisingly AURKA
Bi-orientation of homologous chromosomes at the metaphase plate is critical for accurate chromosome segregation. Chromosome alignment is mediated by the attachment of kinetochores to spindle fibers. In mammalian oocytes, K-MT attachments begin to form in pro-metaphase I as spindle fibers nucleate from MTOCs and search for kinetochores [95, 96]. These fibers capture the kinetochores, orienting chromosomes towards the center spindle. Poleward ejection forces emanating from opposite spindle poles stabilize the chromosomes along the metaphase plate.
Improper attachments can lead to inaccurate chromosome segregation in anaphase, and ultimately aneuploidy. To ensure the timely and accurate segregation of chromosomes, the cell identifies the presence of unattached and improperly attached kinetochores. The latter is likely the most challenging because oocytes with one improper attachment can evade detection [96–99]. K-MT attachments of bi-oriented chromosomes are stabilized while incorrectly attached chromatids are released, allowing correction. Almost all kinetochores will undergo this process multiple times before finally stably attaching to a spindle pole [100]. The Aurora kinases are critical regulators of this process, however, the manner in which they regulate attachment status is not fully understood. Two models have been proposed for the mechanism in which the AURKs mediate K-MT attachment regulation. The first model relies on proximity of centromeric-localized AURK-CPC to its kinetochore-bound substrates. In mitosis, as sister kinetochores bind microtubules from opposing spindle poles, tension is generated, actively stretching the kinetochore away from the centromere. This tension generates an AURK-CPC “activity gradient” as the kinase is positioned farther from its substrates. Chromosomes that are bi-oriented contain the most tension, physically limiting AURKB-CPC from phosphorylating kinetochore-bound substrates. Improperly attached chromatids lack tension, allowing AURKB-CPC to phosphorylate its substrates and ultimately destabilizing the attachment. This model is problematic however, as low tension does not immediately result in microtubule release in meiosis [96]. Work in mouse oocytes showed that cells containing many unaligned bivalents fail to undergo anaphase, while those containing a small number of mis-attached bivalents were not [96].
The inability to respond to chromosomes that are attached but not bi-oriented (low tension) suggests that kinetochore proximity and tension alone are insufficient to explain the complex error correction mechanism employed by meiotic cells. The second model addresses this problem, suggesting that proximity to the spindle poles, rather than tension drives K-MT release. However, uncoupling tension from chromosome position to test this hypothesis is challenging. To solve this issue, researchers crossed mouse strains that exhibit homologous chromosomes with differing centromere strengths, generating a model system where chromosomes are not aligned at the metaphase plate but remain under tension [101]. In these oocytes, kinetochores closest to spindle poles more often were unattached, while those closest to the metaphase plate almost always contained stable attachments. Interestingly, the pole-ward destabilization of K-MTs was dependent upon AURKA [101]. Because AURKA sequence in the catalytic domain is similar to its homologs, it is perhaps not surprising that it can target presumably the same substrates to destabilize K-MTs. However, a caveat to these findings is the discovery of AURKC at the spindle poles (Fig. 2, Key Figure) [15]. Whether both AURKA and AURKC regulate the pole-ward destabilization of K-MTs or this role is dependent solely on one AURK remains unknown.
The Spindle Assembly Checkpoint: AURKB and AURKC
The CPC also regulates the SAC, a signaling cascade triggered by unattached kinetochores that delays anaphase onset until all chromosomes are aligned at the metaphase plate [17]. In mitosis, AURKB-CPC plays a direct role in the SAC by recruiting proteins important for its function [102, 103]. In meiosis, the mechanism by which the CPC regulates the SAC is not as clear. In particular, one question is whether the CPC has a direct role in recruiting SAC components to kinetochores or if it acts indirectly through tension-dependent error-correction that would transiently generate unattached kinetochores.
To distinguish a requirement for AURKC and/or AURKB in the SAC in oocytes, researchers expressed a dominant-negative allele of Aurkc in mouse oocytes [14]. These oocytes arrested at Met I when exposed to nocodazole, providing evidence for a responsive SAC signal, suggesting that the SAC may be regulated by AURKB in meiosis. Further work is required to uncover whether this role is specific to AURKB or AURKC in wild-type oocytes. One possible model would be that AURKC participates indirectly by creating unattached kinetochores by sensing and destabilizing improper K-MTs while AURKB is responsible for recruiting SAC proteins to kinetochores.
Cytokinesis
While the role of the AURKs in cytokinesis in meiosis remains an active area of exploration, details from overexpression studies provide hints that AURKB and AURKC may be involved in these processes in germ cells. Overexpression of Aurkc mRNA in mouse oocytes results in cytokinesis failure and ultimately the formation of polyploid gametes [69]. By contrast, oocytes injected with Aurkb mRNA caused arrest in Met I and prolonged APC/C activation, suggesting that the kinases are important for regulating these processes but may operate in different or even opposing ways [69].
Human Variants
Work in mouse oocytes and other meiotic systems reveal essential functions for the AURKs. Ultimately we, and others, aim to understand the reproductive implications of humans harboring variants in the Aurk genes. Similar to mouse, human oocytes and pre-implantation embryos express both AURKB and AURKC [46]. However, whether loss or gain-of-function mutations alter fertility in women is currently unknown. It is well-established that loss-of-function alleles of AURKC cause male sterility [104–109] and there is some evidence that variants in AURKB may also reduce male fertility [110]. Given the redundancies and compensatory abilities of the AURKs in female mouse meiosis, it is possible that human oocytes can tolerate AURK variants and still make viable eggs. Until we start to make connections to female reproductive fitness and the genome, these questions remain unanswered.
Concluding remarks
The AURKs are critical regulators of cell division, however, little is known about the mechanisms by which these kinases function in meiosis. Recent work has begun to chip away at the specific roles each of the kinases play, however, as more is uncovered, additional questions arise. If AURKB and AURKC do have non-overlapping functions in meiosis, how are they differentially regulated (See Outstanding Questions)? One potential source of differential regulation could be proximity to substrates. Based on data from mitotic cells that express both AURKB and AURKC, they exist in separate complexes [77]. Additionally, the kinases differ in catalytic activity levels, with AURKC, and not AURKB, capable of binding Survivin, increasing its auto-phosphorylation [77]. While AURKB and AURKC are similar in sequence and structure, catalytic activity, stability, and binding partner affinity differences are likely key to their separate functions. It is tempting to hypothesize that one requirement for two independent CPC pools could act to restrict the amount of Survivin available to bind AURKC, thus limiting its activity, as overexpression of AURKC results in atypical mitotic progression in cancer cells [111].
The high sequence similarity among the AURK family members has made discerning the individual kinase functions in meiosis challenging. Technical limitations in specifically targeting AURKB or AURKC using small molecule inhibitors and compensatory abilities in single knockout animals add to this challenge. Novel techniques will be required to discern the individual functions of these kinases in meiosis including methods to identify the endogenous localization of the kinases and targeted inhibition. Pinpointing the mechanism that restricts the isoforms to their respective signaling networks will be critical for unraveling their complex functional regulation.
Outstanding Questions.
Can AURKC compensate for loss of AURKB in mammalian oocytes?
Do AURKB-CPC and AURKC-CPC exist in distinct chromosome localized pools and do they target different substrates in meiosis?
What are the substrates of these kinases in meiotic cells?
What regulates the spatial temporal differences between the AURKs?
Are all three auroras interchangeable in their functional abilities?
What regulates AURK binding partner specificity in meiosis?
Is pole-directed chromosome alignment mediated by AURKA or AURKC?
Is the requirement for the AURKs and compensatory abilities sexually dimorphic?
Trends Box.
AURKC is the primary CPC catalytic subunit, required for meiotic cellular events similar to those of AURKB in mitotic cells.
A subpopulation of AURKC localizes to spindle poles and regulates microtubule-organizing center clustering.
Aurkc is a maternal transcript recruited for translation during meiotic maturation.
AURKC protein is more stable than AURKB protein during meiotic maturation.
Haspin phosphorylation of histone H3 at T3 is required for the localization of centromeric, and not kinetochore AURKC-CPC.
AURKB is expressed in mouse oocytes and localizes to spindle microtubules.
AURKB can compensate for loss of AURKC in mouse oocytes.
AURKA, the polar AURK, can destabilize improper kinetochore-microtubule attachments.
Acknowledgments
The authors apologize for not being able to cite all the primary work due to space limitations. The authors would like to thank Drs. Cecilia Blengini and Suzanne Quartuccio for editorial comments. Work documented in this review was supported by grants from the NIH (F31HD089591 to A.L.N.) and (R01GM112801-02 to K.S.).
Glossary
- Aneuploidy
Presence of an abnormal number of chromosomes in a cell
- Aurora protein kinases (AURKs)
Refers to any of the aurora kinase family members. Family of evolutionarily conserved serine/threonine kinases found in mitosis and meiosis
- AURKA
Aurora kinase A. Also known as the polar kinase, involved in bipolar spindle formation and chromosome alignment in mitosis and meiosis
- AURKB
Aurora kinase B. Chromosome localized AURK expressed in mitosis and meiosis, involved in multiple processes in cell division including chromosome alignment, the spindle assembly checkpoint, and cytokinesis
- AURKC
Aurora Kinase C. Chromosome and spindle pole localized AURK expressed in mammalian meiosis. Involved in multiple processes in meiotic cell division including chromosome alignment and meiotic progression
- Aurkc-L93A
Mutation of the gatekeeper leucine residue in the ATP-binding pocket of AURKC in mouse oocytes that inactivates the kinase and does not disrupt AURKB activity
- AZD1152 (Barasertib)
Highly selective AURKB/C inhibitor. IC50 of 0.37nM in a cell-free assay, ~3700 fold more selective for AURKB over AURKA
- BORA
AURKA binding protein. Involved in microtubule nucleation and building of the mitotic and meiotic spindles
- Borealin
Cell division cycle-associated protein 8. Docking subunit of the chromosomal passenger complex. Dimerization of Borealin subunits drives inner centromeric chromosomal passenger complex localization in mitosis
- BUB1
Serine/threonine protein kinase expressed in mitosis and meiosis. Important for localization of chromosomal passenger complex via phosphorylation of Histone 2A at threonine 120 and spindle assembly checkpoint proteins to chromosomes
- Chromosomal passenger complex (CPC)
Protein complex comprised of INCENP, Survivin, Borealin, and a single aurora kinase. Involved in many processes in mitotic and meiotic divisions
- Condensin
Large protein complex comprised of two ATPases of the SMC family, a kleisin, and one or two additional subunits. Structurally organizes chromosomes into tight bundles
- Haspin
Germ-cell specific gene 2 (GSG2). Serine/threonine protein kinase expressed in mitosis and meiosis. Important for localization of the chromosomal passenger complex to chromosomes via phosphorylation of Histone 3 at threonine 3
- Homologous recombination
Genetic exchange and subsequent recombination of DNA between DNA molecules in meiosis
- INCENP
Inner Centromere Protein. Core structural component of the chromosomal passenger complex. Binds to AURK, Survivin, and Borealin
- Interchromatid axis (ICA)
Axis between homologous chromosomes in metaphase of meiosis I
- Kinetochore
A multi-protein complex assembled at chromosome centromeres, attaches to spindle fibers to mediate chromosome alignment
- Kinetochore-microtubule (K-MT)
Spindle microtubules that make direct interaction and attachment to the kinetochore complex at centromeres
- Kinesin 5 (Eg5)
Molecular motor protein that slides antiparallel microtubules during spindle assembly
- MAD2
Mitotic Arrest Deficient 2. Spindle checkpoint protein
- Maternally recruited message
A mechanism to increase protein levels in oocytes while transcription is not occurring. Stored mRNAs are translated during meiotic maturation. This recruitment of messages for translation is regulated via cytoplasmic polyadenylation element and DazL binding sequences present in 3s UTRs
- Meiosis I (MI)
Segregation of homologous chromosomes; also called the reductional segregation.
- Meiosis II (MII)
Segregation of sister chromatids; also called the equational segregation.
- Metaphase I (Met I)
Metaphase of Meiosis I. Alignment of bivalents along the metaphase plate in meiosis I
- Metaphase II (Met II)
Metaphase of Meiosis II. Alignment of univalents along the metaphase plate in meiosis II
- Microtubule-organizing center (MTOC)
Acentriolar centrosomes that nucleates microtubules to generate a meiotic spindle in oocytes
- Nuclear envelope breakdown (NEBD)
The breakdown of the nuclear lamina at the resumption of meiosis I
- REC8
Meiosis specific kleisin component of the cohesin complex. Assists in binding sister chromatids together
- Separase
Cysteine protease that hydrolyses cohesin, driving its removal from chromosome arms and separation of sister chromatids
- Shugoshin-2 (SGO2)
Localizes to centromeres and blocks cohesion removal in meiosis I
- SMC1/SMC2
ATPase hinge subunits of the condensing complex
- Spindle assembly checkpoint (SAC)
Signaling checkpoint that regulates the metaphase-anaphase transition. When the SAC signal is active, the cell remains in metaphase until all chromosomes have correct attachments to the spindle apparatus
- Subfertility
Reduced fertility compared to normal. In mouse, this refers to smaller than normal litter sizes or fewer litters over a reproductive life-span
- Survivin
Baculoviral inhibitor of apoptosis repeat-containing 5, BIRC5. Docking subunit of the chromosomal passenger complex. Recognizes phosphorylation of histone H3 at threonine 3. Binds AURKC and INCENP
- TACC3
Transforming Acidic Coiled-Coil Containing Protein 3. Motor spindle protein involved in spindle stability in mitosis and meiosis
- TIP60
Histone acetyl transferase. Sometimes a subunit of the chromosomal passenger complex
- TPX2
Microtubule associate protein that binds AURKA
- ZM447439
Highly selective AURKB/C inhibitor with IC50 of 130nM. Selectivity for AURKA at 110nM. Demonstrated to disrupt AURKB/C localization/function, and not AURKA [112]
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Zuccotti M, et al. Fully-mature antral mouse oocytes are transcriptionally silent but their heterochromatin maintains a transcriptional permissive histone acetylation profile. J Assist Reprod Genet. 2011;28(12):1193–6. doi: 10.1007/s10815-011-9562-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hassold T, Hunt P. To err (meiotically) is human: the genesis of human aneuploidy. Nat Rev Genet. 2001;2(4):280–91. doi: 10.1038/35066065. [DOI] [PubMed] [Google Scholar]
- 3.Carmena M, Earnshaw WC. The cellular geography of aurora kinases. Nat Rev Mol Cell Biol. 2003;4(11):842–54. doi: 10.1038/nrm1245. [DOI] [PubMed] [Google Scholar]
- 4.Nigg EA. Mitotic kinases as regulators of cell division and its checkpoints. Nat Rev Mol Cell Biol. 2001;2(1):21–32. doi: 10.1038/35048096. [DOI] [PubMed] [Google Scholar]
- 5.Brown JR, et al. Evolutionary relationships of Aurora kinases: implications for model organism studies and the development of anti-cancer drugs. BMC Evol Biol. 2004;4:39. doi: 10.1186/1471-2148-4-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barr AR, Gergely F. Aurora-A: the maker and breaker of spindle poles. J Cell Sci. 2007;120(Pt 17):2987–96. doi: 10.1242/jcs.013136. [DOI] [PubMed] [Google Scholar]
- 7.Giet R, Petretti C, Prigent C. Aurora kinases, aneuploidy and cancer, a coincidence or a real link? Trends Cell Biol. 2005;15(5):241–50. doi: 10.1016/j.tcb.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 8.Green RA, Paluch E, Oegema K. Cytokinesis in animal cells. Annu Rev Cell Dev Biol. 2012;28:29–58. doi: 10.1146/annurev-cellbio-101011-155718. [DOI] [PubMed] [Google Scholar]
- 9.Glotzer M. The 3Ms of central spindle assembly: microtubules, motors and MAPs. Nat Rev Mol Cell Biol. 2009;10(1):9–20. doi: 10.1038/nrm2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mierzwa B, Gerlich DW. Cytokinetic abscission: molecular mechanisms and temporal control. Dev Cell. 2014;31(5):525–38. doi: 10.1016/j.devcel.2014.11.006. [DOI] [PubMed] [Google Scholar]
- 11.Sasai K, et al. Aurora-C kinase is a novel chromosomal passenger protein that can complement Aurora-B kinase function in mitotic cells. Cell Motil Cytoskeleton. 2004;59(4):249–63. doi: 10.1002/cm.20039. [DOI] [PubMed] [Google Scholar]
- 12.Slattery SD, et al. Aurora-C kinase supports mitotic progression in the absence of Aurora-B. Cell Cycle. 2014;8(18):2986–2997. [PubMed] [Google Scholar]
- 13.Schindler K, et al. Maternally recruited Aurora C kinase is more stable than Aurora B to support mouse oocyte maturation and early development. Proc Natl Acad Sci U S A. 2012;109(33):E2215–22. doi: 10.1073/pnas.1120517109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Balboula AZ, Schindler K. Selective disruption of aurora C kinase reveals distinct functions from aurora B kinase during meiosis in mouse oocytes. PLoS Genet. 2014;10(2):e1004194. doi: 10.1371/journal.pgen.1004194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Balboula AZ, et al. Haspin kinase regulates microtubule-organizing center clustering and stability through Aurora kinase C in mouse oocytes. J Cell Sci. 2016;129(19):3648–3660. doi: 10.1242/jcs.189340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nguyen AL, et al. Phosphorylation of threonine 3 on histone H3 by haspin kinase is required for meiosis I in mouse oocytes. J Cell Sci. 2014;127(Pt 23):5066–78. doi: 10.1242/jcs.158840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carmena M. Abscission checkpoint control: stuck in the middle with Aurora B. Open Biol. 2012;2(7):120095. doi: 10.1098/rsob.120095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nahse V, et al. The Abscission Checkpoint: Making It to the Final Cut. Trends Cell Biol. 2017;27(1):1–11. doi: 10.1016/j.tcb.2016.10.001. [DOI] [PubMed] [Google Scholar]
- 19.Steigemann P, et al. Aurora B-mediated abscission checkpoint protects against tetraploidization. Cell. 2009;136(3):473–84. doi: 10.1016/j.cell.2008.12.020. [DOI] [PubMed] [Google Scholar]
- 20.van der Waal MS, et al. Cell division control by the Chromosomal Passenger Complex. Exp Cell Res. 2012;318(12):1407–20. doi: 10.1016/j.yexcr.2012.03.015. [DOI] [PubMed] [Google Scholar]
- 21.Agromayor M, Martin-Serrano J. Knowing when to cut and run: mechanisms that control cytokinetic abscission. Trends Cell Biol. 2013;23(9):433–41. doi: 10.1016/j.tcb.2013.04.006. [DOI] [PubMed] [Google Scholar]
- 22.Kitagawa M, Lee SH. The chromosomal passenger complex (CPC) as a key orchestrator of orderly mitotic exit and cytokinesis. Front Cell Dev Biol. 2015;3:14. doi: 10.3389/fcell.2015.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.D’Avino PP, Capalbo L. New Auroras on the Roles of the Chromosomal Passenger Complex in Cytokinesis: Implications for Cancer Therapies. Front Oncol. 2015;5:221. doi: 10.3389/fonc.2015.00221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krenn V, Musacchio A. The Aurora B Kinase in Chromosome Bi-Orientation and Spindle Checkpoint Signaling. Front Oncol. 2015;5:225. doi: 10.3389/fonc.2015.00225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carmena M, et al. The chromosomal passenger complex (CPC): from easy rider to the godfather of mitosis. Nat Rev Mol Cell Biol. 2012;13(12):789–803. doi: 10.1038/nrm3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Asteriti IA, De Mattia F, Guarguaglini G. Cross-Talk between AURKA and Plk1 in Mitotic Entry and Spindle Assembly. Front Oncol. 2015;5:283. doi: 10.3389/fonc.2015.00283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang G, Jiang Q, Zhang C. The role of mitotic kinases in coupling the centrosome cycle with the assembly of the mitotic spindle. J Cell Sci. 2014;127(Pt 19):4111–22. doi: 10.1242/jcs.151753. [DOI] [PubMed] [Google Scholar]
- 28.Hochegger H, Hegarat N, Pereira-Leal JB. Aurora at the pole and equator: overlapping functions of Aurora kinases in the mitotic spindle. Open Biol. 2013;3(3):120185. doi: 10.1098/rsob.120185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yao LJ, et al. Aurora-A is a critical regulator of microtubule assembly and nuclear activity in mouse oocytes, fertilized eggs, and early embryos. Biol Reprod. 2004;70(5):1392–9. doi: 10.1095/biolreprod.103.025155. [DOI] [PubMed] [Google Scholar]
- 30.Saskova A, et al. Aurora kinase A controls meiosis I progression in mouse oocytes. Cell Cycle. 2008;7(15):2368–76. doi: 10.4161/cc.6361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eyers PA, et al. A novel mechanism for activation of the protein kinase Aurora A. Curr Biol. 2003;13(8):691–7. doi: 10.1016/s0960-9822(03)00166-0. [DOI] [PubMed] [Google Scholar]
- 32.Eyers PA, Maller JL. Regulation of Xenopus Aurora A activation by TPX2. J Biol Chem. 2004;279(10):9008–15. doi: 10.1074/jbc.M312424200. [DOI] [PubMed] [Google Scholar]
- 33.Tsai MY, Zheng Y. Aurora A kinase-coated beads function as microtubule-organizing centers and enhance RanGTP-induced spindle assembly. Curr Biol. 2005;15(23):2156–63. doi: 10.1016/j.cub.2005.10.054. [DOI] [PubMed] [Google Scholar]
- 34.Aguirre-Portoles C, et al. Tpx2 controls spindle integrity, genome stability, and tumor development. Cancer Res. 2012;72(6):1518–28. doi: 10.1158/0008-5472.CAN-11-1971. [DOI] [PubMed] [Google Scholar]
- 35.Zhai R, et al. Bora regulates meiotic spindle assembly and cell cycle during mouse oocyte meiosis. Mol Reprod Dev. 2013;80(6):474–87. doi: 10.1002/mrd.22185. [DOI] [PubMed] [Google Scholar]
- 36.Kelly AE, Funabiki H. Correcting aberrant kinetochore microtubule attachments: an Aurora B-centric view. Curr Opin Cell Biol. 2009;21(1):51–8. doi: 10.1016/j.ceb.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guse A, Mishima M, Glotzer M. Phosphorylation of ZEN-4/MKLP1 by aurora B regulates completion of cytokinesis. Curr Biol. 2005;15(8):778–86. doi: 10.1016/j.cub.2005.03.041. [DOI] [PubMed] [Google Scholar]
- 38.Nezi L, Musacchio A. Sister chromatid tension and the spindle assembly checkpoint. Curr Opin Cell Biol. 2009;21(6):785–95. doi: 10.1016/j.ceb.2009.09.007. [DOI] [PubMed] [Google Scholar]
- 39.Johansen KM, Johansen J. Regulation of chromatin structure by histone H3S10 phosphorylation. Chromosome Res. 2006;14(4):393–404. doi: 10.1007/s10577-006-1063-4. [DOI] [PubMed] [Google Scholar]
- 40.Santaguida S, et al. Evidence that Aurora B is implicated in spindle checkpoint signalling independently of error correction. EMBO J. 2011;30(8):1508–19. doi: 10.1038/emboj.2011.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang K-t, et al. Aurora-C Kinase Deficiency Causes Cytokinesis Failure in Meiosis I and Production of Large Polyploid Oocytes in Mice. 2010;21:2371–2383. doi: 10.1091/mbc.E10-02-0170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vogt E, Kipp A, Eichenlaub-Ritter U. Aurora kinase B, epigenetic state of centromeric heterochromatin and chiasma resolution in oocytes. Reprod Biomed Online. 2009;19(3):352–68. doi: 10.1016/s1472-6483(10)60169-1. [DOI] [PubMed] [Google Scholar]
- 43.Yang KT, et al. Aurora-C kinase deficiency causes cytokinesis failure in meiosis I and production of large polyploid oocytes in mice. Mol Biol Cell. 2010;21(14):2371–83. doi: 10.1091/mbc.E10-02-0170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shuda K, et al. Aurora kinase B modulates chromosome alignment in mouse oocytes. Mol Reprod Dev. 2009;76(11):1094–105. doi: 10.1002/mrd.21075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Swain JE, et al. Regulation of spindle and chromatin dynamics during early and late stages of oocyte maturation by aurora kinases. Mol Hum Reprod. 2008;14(5):291–9. doi: 10.1093/molehr/gan015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Avo Santos M, et al. A role for Aurora C in the chromosomal passenger complex during human preimplantation embryo development. Hum Reprod. 2011;26(7):1868–81. doi: 10.1093/humrep/der111. [DOI] [PubMed] [Google Scholar]
- 47.Tseng TC, et al. Protein kinase profile of sperm and eggs: cloning and characterization of two novel testis-specific protein kinases (AIE1, AIE2) related to yeast and fly chromosome segregation regulators. DNA Cell Biol. 1998;17(10):823–33. doi: 10.1089/dna.1998.17.823. [DOI] [PubMed] [Google Scholar]
- 48.Gopalan G, Chan CS, Donovan PJ. A novel mammalian, mitotic spindle-associated kinase is related to yeast and fly chromosome segregation regulators. J Cell Biol. 1997;138(3):643–56. doi: 10.1083/jcb.138.3.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yanai A, et al. ayk1, a novel mammalian gene related to Drosophila aurora centrosome separation kinase, is specifically expressed during meiosis. Oncogene. 1997;14(24):2943–50. doi: 10.1038/sj.onc.1201144. [DOI] [PubMed] [Google Scholar]
- 50.Yan X, et al. Aurora C is directly associated with Survivin and required for cytokinesis. Genes Cells. 2005;10(6):617–26. doi: 10.1111/j.1365-2443.2005.00863.x. [DOI] [PubMed] [Google Scholar]
- 51.Yan X, et al. Cloning and characterization of a novel human Aurora C splicing variant. Biochem Biophys Res Commun. 2005;328(1):353–61. doi: 10.1016/j.bbrc.2004.12.168. [DOI] [PubMed] [Google Scholar]
- 52.Price DM, et al. Nocturnal activation of aurora C in rat pineal gland: its role in the norepinephrine-induced phosphorylation of histone H3 and gene expression. Endocrinology. 2009;150(5):2334–41. doi: 10.1210/en.2008-1507. [DOI] [PubMed] [Google Scholar]
- 53.Li X, et al. Direct association with inner centromere protein (INCENP) activates the novel chromosomal passenger protein, Aurora-C. J Biol Chem. 2004;279(45):47201–11. doi: 10.1074/jbc.M403029200. [DOI] [PubMed] [Google Scholar]
- 54.Goldenson B, et al. Aurora kinase A is required for hematopoiesis but is dispensable for murine megakaryocyte endomitosis and differentiation. Blood. 2015;125(13):2141–50. doi: 10.1182/blood-2014-12-615401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vader G, Maia AF, Lens SM. The chromosomal passenger complex and the spindle assembly checkpoint: kinetochore-microtubule error correction and beyond. Cell Div. 2008;3:10. doi: 10.1186/1747-1028-3-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jeyaprakash AA, et al. Structural basis for the recognition of phosphorylated histone h3 by the survivin subunit of the chromosomal passenger complex. Structure. 2011;19(11):1625–34. doi: 10.1016/j.str.2011.09.002. [DOI] [PubMed] [Google Scholar]
- 57.Wu Y, et al. A new small-molecule Aurora B inhibitor shows antitumor activity in human cancer cell lines. Mol Biol Rep. 2015;42(2):517–24. doi: 10.1007/s11033-014-3795-0. [DOI] [PubMed] [Google Scholar]
- 58.Gassmann R, et al. Borealin: a novel chromosomal passenger required for stability of the bipolar mitotic spindle. J Cell Biol. 2004;166(2):179–91. doi: 10.1083/jcb.200404001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dai J, Higgins JM. Haspin: a mitotic histone kinase required for metaphase chromosome alignment. Cell Cycle. 2005;4(5):665–8. doi: 10.4161/cc.4.5.1683. [DOI] [PubMed] [Google Scholar]
- 60.Dai J, Kateneva AV, Higgins JM. Studies of haspin-depleted cells reveal that spindle-pole integrity in mitosis requires chromosome cohesion. J Cell Sci. 2009;122(Pt 22):4168–76. doi: 10.1242/jcs.054122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang F, et al. Histone H3 Thr-3 phosphorylation by Haspin positions Aurora B at centromeres in mitosis. Science. 2010;330(6001):231–5. doi: 10.1126/science.1189435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dai J, Sullivan BA, Higgins JM. Regulation of mitotic chromosome cohesion by Haspin and Aurora B. Dev Cell. 2006;11(5):741–50. doi: 10.1016/j.devcel.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 63.Higgins JM. Haspin: a newly discovered regulator of mitotic chromosome behavior. Chromosoma. 2010;119(2):137–47. doi: 10.1007/s00412-009-0250-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Higgins JM. Structure, function and evolution of haspin and haspin-related proteins, a distinctive group of eukaryotic protein kinases. Cell Mol Life Sci. 2003;60(3):446–62. doi: 10.1007/s000180300038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dai J, et al. The kinase haspin is required for mitotic histone H3 Thr 3 phosphorylation and normal metaphase chromosome alignment. Genes Dev. 2005;19(4):472–88. doi: 10.1101/gad.1267105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang F, et al. A positive feedback loop involving Haspin and Aurora B promotes CPC accumulation at centromeres in mitosis. Curr Biol. 2011;21(12):1061–9. doi: 10.1016/j.cub.2011.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tsukahara T, Tanno Y, Watanabe Y. Phosphorylation of the CPC by Cdk1 promotes chromosome bi-orientation. Nature. 2010;467(7316):719–23. doi: 10.1038/nature09390. [DOI] [PubMed] [Google Scholar]
- 68.Kawashima SA, et al. Phosphorylation of H2A by Bub1 Prevents Chromosomal Instability Through Localizing Shugoshin. Science. 2009;327(5962):172–177. doi: 10.1126/science.1180189. [DOI] [PubMed] [Google Scholar]
- 69.Sharif B, et al. The chromosome passenger complex is required for fidelity of chromosome transmission and cytokinesis in meiosis of mouse oocytes. J Cell Sci. 2010;123(Pt 24):4292–300. doi: 10.1242/jcs.067447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen HL, et al. Overexpression of an Aurora-C kinase-deficient mutant disrupts the Aurora-B/INCENP complex and induces polyploidy. J Biomed Sci. 2005;12(2):297–310. doi: 10.1007/s11373-005-0980-0. [DOI] [PubMed] [Google Scholar]
- 71.Lane SI, et al. The Aurora kinase inhibitor ZM447439 accelerates first meiosis in mouse oocytes by overriding the spindle assembly checkpoint. Reproduction. 2010;140(4):521–30. doi: 10.1530/REP-10-0223. [DOI] [PubMed] [Google Scholar]
- 72.Chen HL, et al. Overexpression of an Aurora-C kinase-deficient mutant disrupts the Aurora-B/INCENP complex and induces polyploidy. J Biomed Sci. 2005;12(2):297–310. doi: 10.1007/s11373-005-0980-0. [DOI] [PubMed] [Google Scholar]
- 73.Fu J, et al. A single amino acid change converts Aurora-A into Aurora-B-like kinase in terms of partner specificity and cellular function. Proc Natl Acad Sci U S A. 2009;106(17):6939–44. doi: 10.1073/pnas.0900833106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hans F, et al. Molecular distinctions between Aurora A and B: a single residue change transforms Aurora A into correctly localized and functional Aurora B. Mol Biol Cell. 2009;20(15):3491–502. doi: 10.1091/mbc.E09-05-0370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Eyers PA, Churchill ME, Maller JL. The Aurora A and Aurora B protein kinases: a single amino acid difference controls intrinsic activity and activation by TPX2. Cell Cycle. 2005;4(6):784–9. doi: 10.4161/cc.4.6.1693. [DOI] [PubMed] [Google Scholar]
- 76.Schindler K, et al. Maternally recruited Aurora C kinase is more stable than Aurora B to support mouse oocyte maturation and early development. Proceedings of the National Academy of Sciences. 2012;109(33):E2215–E2222. doi: 10.1073/pnas.1120517109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sasai K, et al. Aurora-C Interactions with Survivin and INCENP Reveal Shared and Distinct Features Compared with Aurora-B Chromosome Passenger Protein Complex. PLoS One. 2016;11(6):e0157305. doi: 10.1371/journal.pone.0157305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bekier ME, et al. Borealin dimerization mediates optimal CPC checkpoint function by enhancing localization to centromeres and kinetochores. Nat Commun. 2015;6:6775. doi: 10.1038/ncomms7775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Breuer M, et al. HURP permits MTOC sorting for robust meiotic spindle bipolarity, similar to extra centrosome clustering in cancer cells. J Cell Biol. 2010;191(7):1251–60. doi: 10.1083/jcb.201005065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Solc P, et al. Aurora kinase A drives MTOC biogenesis but does not trigger resumption of meiosis in mouse oocytes matured in vivo. Biol Reprod. 2012;87(4):85. doi: 10.1095/biolreprod.112.101014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mahdipour M, et al. TACC3 Is Important for Correct Progression of Meiosis in Bovine Oocytes. PLoS One. 2015;10(7):e0132591. doi: 10.1371/journal.pone.0132591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Castro A, et al. APC/Fizzy-Related targets Aurora-A kinase for proteolysis. EMBO Rep. 2002;3(5):457–62. doi: 10.1093/embo-reports/kvf095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Crane R, et al. Aurora A, meiosis and mitosis. Biol Cell. 2004;96(3):215–29. doi: 10.1016/j.biolcel.2003.09.008. [DOI] [PubMed] [Google Scholar]
- 84.Giet R, Prigent C. Aurora/Ipl1p-related kinases, a new oncogenic family of mitotic serine-threonine kinases. J Cell Sci. 1999;112(Pt 21):3591–601. doi: 10.1242/jcs.112.21.3591. [DOI] [PubMed] [Google Scholar]
- 85.Sumiyoshi E, et al. Caenorhabditis elegans Aurora A kinase is required for the formation of spindle microtubules in female meiosis. Mol Biol Cell. 2015;26(23):4187–96. doi: 10.1091/mbc.E15-05-0258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Schuh M, Ellenberg J. Self-organization of MTOCs replaces centrosome function during acentrosomal spindle assembly in live mouse oocytes. Cell. 2007;130(3):484–98. doi: 10.1016/j.cell.2007.06.025. [DOI] [PubMed] [Google Scholar]
- 87.Nasmyth K, Haering CH. The structure and function of SMC and kleisin complexes. Annu Rev Biochem. 2005;74:595–648. doi: 10.1146/annurev.biochem.74.082803.133219. [DOI] [PubMed] [Google Scholar]
- 88.Collette KS, et al. Different roles for Aurora B in condensin targeting during mitosis and meiosis. J Cell Sci. 2011;124(Pt 21):3684–94. doi: 10.1242/jcs.088336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Brar GA, et al. Rec8 phosphorylation and recombination promote the stepwise loss of cohesins in meiosis. Nature. 2006;441(7092):532–6. doi: 10.1038/nature04794. [DOI] [PubMed] [Google Scholar]
- 90.Kitajima TS, et al. Shugoshin collaborates with protein phosphatase 2A to protect cohesin. Nature. 2006;441(7089):46–52. doi: 10.1038/nature04663. [DOI] [PubMed] [Google Scholar]
- 91.Riedel CG, et al. Protein phosphatase 2A protects centromeric sister chromatid cohesion during meiosis I. Nature. 2006;441(7089):53–61. doi: 10.1038/nature04664. [DOI] [PubMed] [Google Scholar]
- 92.Clift D, Marston AL. The role of shugoshin in meiotic chromosome segregation. Cytogenet Genome Res. 2011;133(2–4):234–42. doi: 10.1159/000323793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tanno Y, et al. Phosphorylation of mammalian Sgo2 by Aurora B recruits PP2A and MCAK to centromeres. Genes Dev. 2010;24(19):2169–79. doi: 10.1101/gad.1945310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Meppelink A, et al. Shugoshin-1 balances Aurora B kinase activity via PP2A to promote chromosome bi-orientation. Cell Rep. 2015;11(4):508–15. doi: 10.1016/j.celrep.2015.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hauf S, Watanabe Y. Kinetochore orientation in mitosis and meiosis. Cell. 2004;119(3):317–27. doi: 10.1016/j.cell.2004.10.014. [DOI] [PubMed] [Google Scholar]
- 96.Lane SI, Yun Y, Jones KT. Timing of anaphase-promoting complex activation in mouse oocytes is predicted by microtubule-kinetochore attachment but not by bivalent alignment or tension. Development. 2012;139(11):1947–55. doi: 10.1242/dev.077040. [DOI] [PubMed] [Google Scholar]
- 97.Kolano A, et al. Error-prone mammalian female meiosis from silencing the spindle assembly checkpoint without normal interkinetochore tension. Proc Natl Acad Sci U S A. 2012;109(27):E1858–67. doi: 10.1073/pnas.1204686109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sebestova J, et al. Lack of response to unaligned chromosomes in mammalian female gametes. Cell Cycle. 2012;11(16):3011–8. doi: 10.4161/cc.21398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gui L, Homer H. Spindle assembly checkpoint signalling is uncoupled from chromosomal position in mouse oocytes. Development. 2012;139(11):1941–6. doi: 10.1242/dev.078352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kitajima TS, Ohsugi M, Ellenberg J. Complete kinetochore tracking reveals error-prone homologous chromosome biorientation in mammalian oocytes. Cell. 2011;146(4):568–81. doi: 10.1016/j.cell.2011.07.031. [DOI] [PubMed] [Google Scholar]
- 101.Chmatal L, et al. Spatial Regulation of Kinetochore Microtubule Attachments by Destabilization at Spindle Poles in Meiosis I. Curr Biol. 2015;25(14):1835–41. doi: 10.1016/j.cub.2015.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.DeAntoni A, Sala V, Musacchio A. Explaining the oligomerization properties of the spindle assembly checkpoint protein Mad2. Philos Trans R Soc Lond B Biol Sci. 2005;360(1455):637–47. doi: 10.1098/rstb.2004.1618. discussion 447–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lens SM, et al. Survivin is required for a sustained spindle checkpoint arrest in response to lack of tension. EMBO J. 2003;22(12):2934–47. doi: 10.1093/emboj/cdg307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Dieterich K, et al. Homozygous mutation of AURKC yields large-headed polyploid spermatozoa and causes male infertility. Nat Genet. 2007;39(5):661–5. doi: 10.1038/ng2027. [DOI] [PubMed] [Google Scholar]
- 105.Kimmins S, et al. Differential functions of the Aurora-B and Aurora-C kinases in mammalian spermatogenesis. Mol Endocrinol. 2007;21(3):726–39. doi: 10.1210/me.2006-0332. [DOI] [PubMed] [Google Scholar]
- 106.Miyamoto T, et al. Male infertility and its genetic causes. J Obstet Gynaecol Res. 2015;41(10):1501–5. doi: 10.1111/jog.12765. [DOI] [PubMed] [Google Scholar]
- 107.Ray PF. Deciphering the genetics of male infertility: progress and challenges. J Urol. 2011;186(4):1183–4. doi: 10.1016/j.juro.2011.07.047. [DOI] [PubMed] [Google Scholar]
- 108.Ounis L, et al. Mutations of the aurora kinase C gene causing macrozoospermia are the most frequent genetic cause of male infertility in Algerian men. Asian J Androl. 2015;17(1):68–73. doi: 10.4103/1008-682X.136441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Coutton C, et al. [Genetics of male infertility: the new players] Med Sci (Paris) 2012;28(5):497–502. doi: 10.1051/medsci/2012285014. [DOI] [PubMed] [Google Scholar]
- 110.Lopez-Carrasco A, et al. Mutation screening of AURKB and SYCP3 in patients with reproductive problems. Mol Hum Reprod. 2013;19(2):102–8. doi: 10.1093/molehr/gas047. [DOI] [PubMed] [Google Scholar]
- 111.Khan J, et al. Overexpression of active Aurora-C kinase results in cell transformation and tumour formation. PLoS One. 2011;6(10):e26512. doi: 10.1371/journal.pone.0026512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ditchfield C, et al. Aurora B couples chromosome alignment with anaphase by targeting BubR1, Mad2, and Cenp-E to kinetochores. J Cell Biol. 2003;161 doi: 10.1083/jcb.200208091. [DOI] [PMC free article] [PubMed] [Google Scholar]


