ABSTRACT
Efficient assembly of multiple DNA fragments is a pivotal technology for synthetic biology. A scarless and sequence-independent DNA assembly method (DATEL) using thermal exonucleases has been developed recently. Here, we present a simplified DATEL (sDATEL) for efficient assembly of unphosphorylated DNA fragments with low cost. The sDATEL method is only dependent on Taq DNA polymerase and Taq DNA ligase. After optimizing the committed parameters of the reaction system such as pH and the concentration of Mg2+ and NAD+, the assembly efficiency was increased by 32-fold. To further improve the assembly capacity, the number of thermal cycles was optimized, resulting in successful assembly 4 unphosphorylated DNA fragments with an accuracy of 75%. sDATEL could be a desirable method for routine manual and automated assembly.
KEYWORDS: DNA assembly, PCR, Simplified DATEL, Synthetic biology, Unphosphorylated DNA fragments
Introduction
Efficient assembly of multiple small DNA fragments into large constructs is a pivotal technology in biotechnology and synthetic biology.1,2 Many DNA recombinant techniques that depend on traditional restriction endonucleases and DNA ligase have been developed and widely used. However, due to the tedious procedures and low capacity, the development of easy-to-use, sequence-independent assembly methods with high efficiency and accuracy is always needed to meet the increasingly high demand. In this regard, many novel scarless DNA assembly methods have been presented, for instance, the restriction enzyme dependent methods (LBS,3 GoldenGate,4 GoldenBraid,5 MoClo6 and MASTER7); the end-homology dependent methods (SLIC,8 NE-LIC,9 CPEC,10 Gibson assembly11 and In-Fusion12); the uracil excision dependent methods13-15 and in vivo DNA assembly methods with Saccharomyces cerevisia16-19 and Bacillus subtilis.20
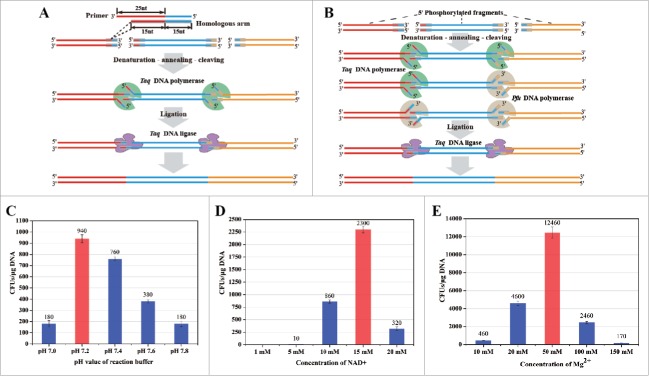
Recently, we reported a scarless and sequence-independent DNA assembly method (DATEL) by recruiting thermal exonucleases (Taq and Pfu DNA polymerase) and Taq DNA ligase.21 Under the optimized conditions, DATEL allows rapid assembly of 2–10 DNA fragments with high accuracy. As shown in Fig. 1B, all the displaced single stranded overhangs were completely digested by Taq (5′–3′ exonuclease activity) and Pfu (3′–5′ exonuclease activity) DNA polymerases and the nicks were joined by the Taq DNA ligase. However, all the DNA fragments required phosphorylation for achieving high assembly efficiency. To simply the workflow and decrease the cost, a sDATEL technique for efficient assembly of unphosphorylated DNA fragments was developed by using Taq DNA polymerase and Taq DNA ligase.
Figure 1.
Principle and optimization of sDATEL assembly method. (A) Schematic diagram of sDATEL. The length of overhang was set as 30 nt while the primers for amplifying DNA fragments were designed with 40 nt. After denaturation and annealing, the displaced overhangs at the fork structures were cleaved by Taq DNA polymerase and the nicks were joined by Taq DNA ligase; (B) Schematic diagram of DATEL;21 (C), (D) and (E) were the effects on the assembly efficiency that caused by pH, NAD+ and Mg2+, respectively.
Materials and methods
Bacterial strains and culture conditions
E. coli strain JM109 (ATCC No. 53323) was used as a host for all the transformations. Luria-Bertani (LB) solid medium (2% agar) and liquid medium (tryptone 10 g/L, yeast extract 5 g/L and NaCl 10 g/L) with Ampicillin (50 μg/mL) or with Ampicillin (50 μg/mL) and Kanamycin (50 μg/mL) in 37°C were used for cell growth and transformation screening.
Chemicals and kits
SanPrep Column Plasmid Mini-Preps kit, Ampicillin sodium and kanamycin sulfate were purchased from Sangon Biotech, Shanghai, China. Gel Purification kit, DpnI, 2 × PrimeSTAR HS DNA polymerase and 2 × Taq DNA polymerase were purchased from Takara, Shanghai, China. Taq DNA ligase and Taq DNA polymerase were purchased from New England Biolabs, USA. Other reagents were purchased from Sinopharm Group Co., Ltd.
Preparation of DNA fragments
All the primers (Sangon Biotech, Shanghai, China) used for amplifying DNA fragments are listed in Table 1. For assembly of 2 fragments, primers pUC19-F and pUC19-R, gfp-P30F and gfp-P30R were used for amplification of the pUC19 backbone and gfp, respectively. For assembly of 3 fragments, primers GK/pUC19-F and GK/pUC19-R, GK /gfp-F and GK /gfp-R, GK /kan-F and GK /kan-R were used for amplification of the pUC19 backbone, gfp and kan, respectively. For assembly of 4 fragments, primers EBI/pUC19-F and EBI/pUC19-R, EBI/crtE-F and EBI/crtE-R, EBI/crtB-F and EBI/crtB-R, EBI/crtI-F and EBI/crtI-R were used for amplification of the pUC19 backbone, crtE, crtB and crtI, respectively. Specifically, the overlapping fragments in the primers were designed with a consistent Toa (50°C). Polymerase chain reaction (PCR) was performed in 50-μL final volume with 25 μL 2 × PrimeSTAR HS DNA polymerase, 1 μL DNA template, 10 pmol of each appropriate primer individually. After amplification with thermal cycles (95°C for 3 min; 30 cycles of 95°C for 30 s, 55°C for 30 s and 72°C for 1 min/kb, 72°C for 5 min), the DNA templates in PCR products were digested by DpnI (Fermentas FastDigest). Then, the PCR products were individually purified for next assembly.
Table 1.
Primers used in this study.
| Primers | Sequence (5′–3′) |
|---|---|
| puc19-F | TTCTTCTCCCTTACCCATGGCGTAATCATGGTCATAGCTGTTTCCT |
| puc19-R | TGGATGAACTATACAAATAACTGGCCGTCGTTTTACAACGTCG |
| gfp-P30F | CATGATTACGCCATGGGTAAGGGAGAAGAA |
| gfp-P30R | CGACGGCCAGTTATTTGTATAGTTCATCCA |
| GK/pUC19-F | TTCTTCTCCCTTACCCATGGCGTAATCATGGTCATAGCTGTTTCCT |
| GK/pUC19-R | CTCGATGAGTTCTTCTAACTGGCCGTCGTTTTACAACGTCG |
| GK /gfp-F | CATGATTACGCCATGGGTAAGGGAGAAGAA |
| GK /gfp-R | TTGAATATGGCTCATTTATTTGTATAGTTCATCCA |
| GK /kan-F | TGGATGAACTATACAAATAAATGAGCCATATTCAACGGGAAAC |
| GK /kan-R | TGTAAAACGACGGCCAGTTAGAAGAACTCATCGAGCATC |
| EBI/pUC19-F | GAGGATCTGGCTTGACTCGAATTCACTGGCCGTCGTTTTA |
| EBI/pUC19-R | ACAGACCGTCATCATAGTCGACCTGCAGGCATGCAAGCTT |
| EBI/crtE-F | GCCTGCAGGTCGACTATGATGACGGTCTGTGCAGAACAAC |
| EBI/crtE-R | ATCCCACCTCCATTTAACTGACGGCAGCGAGTTTTTTC |
| EBI/crtB-F | TGCCGTCAGTTAAATGGAGGTGGGATCGAAAAGCTTTG |
| EBI/crtB-R | TTCATTTTTTTTCCTCCTTTAAACGGGGCGCTGCCAGAGATCA |
| EBI/crtI-F | CCCGTTTAAAGGAGGAAAAAAAATGAATAGAACTACAGTAATTGGCG |
| EBI/crtI-R | GCCAGTGAATTCGAGTCAAGCCAGATCCTCCAGCATCAAT |
Workflow of sDATEL
The sDATEL reaction was performed 15-μL system containing 100 ng of each DNA fragment, 1.5 μL modified reaction buffer, 1 U Taq DNA polymerase, 40 U Taq DNA ligase. The PCR reaction was performed with the following conditions: 94°C for 2 min; 3 cycles of 94°C for 30 s, 50°C for 1 min, 68°C for 30 min and 50°C for 5 min; 10 min at 66°C.
Identification of positive colonies
After transformation, all the colonies on LB plates were counted for calculation of Colony forming units (CFUs) per microgram DNA. After colony PCR, all the positive colonies were picked and cultured in 15 mL of LB medium. Then, the plasmids were extracted by SanPrep Column Plasmid Mini-Preps Kit for DNA sequencing (Sangon Biotech, Shanghai, China).
Results and discussion
Operating principle of sDATEL
To simplify the operation process and decrease the cost, the sDATEL assembly system was established by only using Taq DNA polymerase (5′–3′exonuclease activity) and Taq DNA ligase. Specifically, the oligonucleotides for amplifying DNA fragments were designed with a length of 40 nt while the overhangs for annealing and assembly were set as 30 nt (Fig. 1A). After amplification and purification, the DNA fragments without phosphorylation were directly mixed in the reaction system. After denaturation and annealing, the overlaps of adjacent DNA fragments hybridized while the displaced single stranded overhangs with 5′–3′ directions were completely digested by Taq DNA polymerase. Then, all the nicks between 5′ phosphate group and 3′ hydroxyl group were joined by the thermostable Taq DNA ligase. Compared with the 3-enzyme system, this 2-enzyme system should be much easier for operation and optimization. Although only (1/2)n hybridization chance would be completed by Taq DNA polymerase when assembling n DNA fragments, all the digested fragments with unpaired overhangs should be assembled with more compatible reaction conditions.
Optimization of reaction buffer to improve assembly efficiency
Initially, the previously used DATEL system buffer (10 × Taq buffer, 200 mM Tris-HCl, 250 mM Potassium acetate, 100 mM Magnesium acetate, 10 mM NAD+, 1% Triton X-100, pH 7.6) were evaluated for assembling unphosphorylated DNA fragments with Taq DNA polymerase and Taq DNA ligase, however, only 2 fragments were assembled with a low assembly efficiency. Consequently, the reaction system was further optimized for assembling the gfp gene (717 bp) and the pUC19 backbone (2686 bp).
In consideration of the different optimal pH values for Taq DNA polymerase (pH 8.3) and Taq DNA ligase (pH 7.6), pH gradient ranging from 7.0 to 8.0 was set to examine and compare for DNA assembly. As shown in Fig. 1C, the assembly efficiency was significantly enhanced (from 380 to 940 CFUs/μg DNA) when pH value was maintained at 7.2. The results suggest that the optimal pHs for the exonuclease activity and polymerase activity of Taq DNA polymerase were quite different. At the same time, the effects of different concentrations of NAD+ (from 1 to 20 mM) on the assembly efficiency was investigated since NAD+ is a cofactor of Taq DNA ligase.22-24 Obviously, NAD+ played a critical role on assembly efficiency and the highest value (2300 CFUs/μg DNA) was achieved with 15 mM NAD+ (Fig. 1D). In addition, the concentration of Mg2+ (a cofactor of Taq DNA polymerase) was further investigated and optimized to reconcile the activities of Taq DNA polymerase (exonuclease activity) and Taq DNA ligase. As shown in Fig. 1E, when the concentration of Mg2+ was increased to 50 mM, the assembly efficiency was increased to 12460 CFUs/μg DNA which was 32-times of the initial value (380 CFUs/μg DNA). More excitingly, the assembly accuracy (colonies correct %) was 100% after confirmation with colony PCR (Fig. 2B) and DNA sequencing analysis (data not shown). As a result, the reaction buffer was optimized as follow: 200 mM Tris-HCl, 250 mM Potassium acetate, 50 mM Magnesium acetate, 15 mM NAD+, 1% Triton X-100, pH 7.2 (10 × buffer).
Figure 2.
Performance of sDATEL for assembling multiple fragments. (A) Schematic diagram of the assembly of 2, 3 and 4 DNA fragments; (B), (C) and (D) were the gel electrophoresis analysis results for assembly of 2, 3 and 4 DNA fragments, respectively, and (E), (F) and (G) were the corresponding plates.
Optimization of thermal cycles for improving assembly capacity
After construction and optimization of the assembly reaction system, the capacity and accuracy of sDATEL for assembling multiple fragments were evaluated. As shown in Fig. 2A, the reporter genes (the gfp gene of 717 bp and the kanamycin encoding gene kan of 819 bp), the lycopene biosynthesis pathway genes (crtE, crtI and crtB which were 912 bp, 891 bp and 1479 bp, respectively) and the linear cloning vector pUC19 (2686 bp) were assembled as designed 3 and 4 fragments, respectively. When assembling 3 fragments, more than 830 CFUs/μg ligated DNA were achieved while the accuracy was 82 % (Fig. 2C). In contrast, sDATEL failed to assemble 4 fragments with 3 thermal cycles (Fig. 3). Inspired by the commonly used GoldenGate method,4 the thermal cycles were further investigated since the mixed DNA fragments were not fully digested by Taq DNA polymerase or the digested fragments were not completely ligated by Taq DNA ligase. As expected, when the cycle number was increased to 10, the assembly efficiency was about 350 CFUs/μg ligated DNA with an accuracy value of 36.8%. Interestingly, when further increased the cycle number to 15, the assembly efficiency dropped to 190 CFUs/μg ligated DNA while a higher accuracy value (75%) was achieved (Fig. 2DG, Fig. 3). The results indicated that the number of thermal cycles as a variate should be examined and optimized in each experiment. In addition, the assembly capacity was not enhanced when further increasing thermal cycles (overnight). Thus, to improve the assembly capacity of sDATEL, discovery and application of novel thermal 5′-3′ exonucleases should be an alternative desirable approach in future.
Figure 3.

Assembly capacity and accuracy of sDATEL method.
Conclusion
In the present study, we developed a sDATEL method for efficient assembly of unphosphorylated DNA fragments. Specifically, 2–4 DNA fragments could be efficiently assembled with high accuracy values (from 75% to 100%). Because of its simple workflow and low cost (Table 2), the constructed sDATEL could be competent for routine DNA assembly with manual or automated operations.
Table 2.
Comparison of sDATEL with the commercial Gibson method.
| Methods | Assembly capacity | Enzymes | Cost of each reaction | Assembly accuracy |
|---|---|---|---|---|
| Gibson11 | 2–6 | T5 exonuclease (NEB) | $18.5 | 35% ∼100% |
| Phusion polymerase (NEB) | ||||
| Taq DNA ligase (NEB) | ||||
| DATEL21 | 2–10 | Pfu DNA polymerase (Biosci) | $2.8 | 74% ∼100% |
| Taq DNA polymerase (NEB) | ||||
| Taq DNA ligase (NEB) | ||||
| T4 PNK (Takara) | ||||
| sDATEL | 2–4 | Taq DNA polymerase (NEB) | $1.7 | 75% ∼100% |
| Taq DNA ligase (NEB) |
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This work was financially supported by the National Natural Science Foundation of China (31670092), the Major State Basic Research Development Program of China (973 Program, 2014CB745103), the Natural Science Foundation of Jiangsu Province (BK20141107), a grant from the Key Technologies R&D Program of Jiangsu Province, China (BE2014607) and Program for Changjiang Scholars and Innovative Research Team in University (No. IRT_15R26).
References
- [1].Ellis T, Adie T, Baldwin GS. DNA assembly for synthetic biology: from parts to pathways and beyond. Integr Biol-UK 2011; 3:109-18; PMID:21246151; https://doi.org/ 10.1039/c0ib00070a [DOI] [PubMed] [Google Scholar]
- [2].Kang Z, Zhang J, Jin P, Yang S. Directed evolution combined with synthetic biology strategies expedite semi-rational engineering of genes and genomes. Bioengineered 2015; 6:136-40; PMID:25621864; https://doi.org/ 10.4161/bioe.34347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Kodumal SJ, Patel KG, Reid R, Menzella HG, Welch M, Santi DV. Total synthesis of long DNA sequences: synthesis of a contiguous 32-kb polyketide synthase gene cluster. Proc Natl Acad Sci U S A 2004; 101:15573-8; PMID:15496466; https://doi.org/ 10.1073/pnas.0406911101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Engler C, Gruetzner R, Kandzia R, Marillonnet S. Golden gate shuffling: a one-pot DNA shuffling method based on type IIs restriction enzymes. PLoS One 2009; 4:e5553; PMID:19436741; https://doi.org/ 10.1371/journal.pone.0005553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Sarrion-Perdigones A, Falconi EE, Zandalinas SI, Juarez P, Fernandez-del-Carmen A, Granell A, Orzaez D. GoldenBraid: an iterative cloning system for standardized assembly of reusable genetic modules. PLoS One 2011; 6:e21622; PMID:21750718; https://doi.org/ 10.1371/journal.pone.0021622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Weber E, Engler C, Gruetzner R, Werner S, Marillonnet S. A modular cloning system for standardized assembly of multigene constructs. PLoS One 2011; 6:e16765; PMID:21364738; https://doi.org/ 10.1371/journal.pone.0016765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Chen WH, Qin ZJ, Wang J, Zhao GP. The MASTER (methylation-assisted tailorable ends rational) ligation method for seamless DNA assembly. Nucleic Acids Res 2013; 41; PMID:23444142; https://doi.org/ 10.1093/nar/gkt122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Li MZ, Elledge SJ. Harnessing homologous recombination in vitro to generate recombinant DNA via SLIC. Nat Methods 2007; 4:251-6; PMID:17293868; https://doi.org/ 10.1038/nmeth1010 [DOI] [PubMed] [Google Scholar]
- [9].Wang RY, Shi ZY, Guo YY, Chen JC, Chen GQ. DNA fragments assembly based on nicking enzyme system. PLoS One 2013; 8:e57943; PMID:23483947; https://doi.org/ 10.1371/journal.pone.0057943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Quan JY, Tian JD. Circular polymerase extension cloning for high-throughput cloning of complex and combinatorial DNA libraries. Nat Protoc 2011; 6:242-51; PMID:21293463; https://doi.org/ 10.1038/nprot.2010.181 [DOI] [PubMed] [Google Scholar]
- [11].Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA, Smith HO. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 2009; 6:343-U41; PMID:19363495; https://doi.org/ 10.1038/nmeth.1318 [DOI] [PubMed] [Google Scholar]
- [12].Sleight SC, Bartley BA, Lieviant JA, Sauro HM. In-Fusion BioBrick assembly and re-engineering. Nucleic Acids Res 2010; 38:2624-36; PMID:20385581; https://doi.org/ 10.1093/nar/gkq179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Nour-Eldin HH, Hansen BG, Norholm MHH, Jensen JK, Halkier BA. Advancing uracil-excision based cloning towards an ideal technique for cloning PCR fragments. Nucleic Acids Res 2006; 34:e122; PMID:17000637; https://doi.org/ 10.1093/nar/gkl635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Cavaleiro AM, Kim SH, Seppala S, Nielsen MT, Norholm MHH. Accurate DNA assembly and genome engineering with optimized uracil excision cloning. ACS Synth Biol 2015; 4:1042-6; PMID:26263045; https://doi.org/ 10.1021/acssynbio.5b00113 [DOI] [PubMed] [Google Scholar]
- [15].Nielsen MT, Madsen KM, Seppala S, Christensen U, Riisberg L, Harrison SJ, Møller BL, Nørholm MH. Assembly of highly standardized gene fragments for high-level production of porphyrins in E. coli. ACS Synth Biol 2015; 4:274-82; PMID:24905856; https://doi.org/ 10.1021/sb500055u [DOI] [PubMed] [Google Scholar]
- [16].Shao ZY, Zhao H, Zhao HM. DNA assembler, an in vivo genetic method for rapid construction of biochemical pathways. Nucleic Acids Res 2009; 37:e16; PMID:19074487; https://doi.org/ 10.1093/nar/gkn991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Gibson DG, Benders GA, Axelrod KC, Zaveri J, Algire MA, Moodie M, Montague MG, Venter JC, Smith HO, Hutchison CA 3rd. One-step assembly in yeast of 25 overlapping DNA fragments to form a complete synthetic Mycoplasma genitalium genome. Proc Natl Acad Sci U S A 2008; 105:20404-9; PMID:19073939; https://doi.org/ 10.1073/pnas.0811011106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Lin QH, Jia B, Mitchell LA, Luo JC, Yang K, Zeller KI, Zhang W, Xu Z, Stracquadanio G, Bader JS, et al.. RADOM, an efficient in vivo method for assembling designed DNA fragments up to 10 kb long in Saccharomyces cerevisiae. ACS Synth Biol 2015; 4:213-20; PMID:24895839; https://doi.org/ 10.1021/sb500241e [DOI] [PubMed] [Google Scholar]
- [19].Zhou JT, Wu RH, Xue XL, Qin ZJ. CasHRA (Cas9-facilitated homologous recombination assembly) method of constructing megabase-sized DNA. Nucleic Acids Res 2016; 44:e124; PMID:27220470; https://doi.org/ 10.1093/nar/gkw475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Itaya M, Fujita K, Kuroki A, Tsuge K. Bottom-up genome assembly using the Bacillus subtilis genome vector. Nat Methods 2008; 5:41-3; PMID:18066072; https://doi.org/ 10.1038/nmeth1143 [DOI] [PubMed] [Google Scholar]
- [21].Jin P, Ding W, Du G, Chen J, Kang Z. DATEL: a scarless and sequence-independent DNA assembly method using thermostable exonucleases and ligase. ACS Synth Biol 2016; 5:1028-32; PMID:27230689; https://doi.org/ 10.1021/acssynbio.6b00078 [DOI] [PubMed] [Google Scholar]
- [22].Barany F. Genetic disease detection and DNA amplification using cloned thermostable ligase. Proc Natl Acad Sci U S A 1991; 88:189-93; PMID:1986365; https://doi.org/ 10.1073/pnas.88.1.189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Housby JN, Thorbjarnardóttir SH, Jónsson ZO, Southern EM. Optimised ligation of oligonucleotides by thermal ligases: comparison of Thermus scotoductus and Rhodothermus marinus DNA ligases to other thermophilic ligases. Nucleic Acids Res 2000; 28:e10; PMID:10637340; https://doi.org/ 10.1093/nar/28.3.e10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Takahashi M, Yamaguchi E, Uchida T. Thermophilic DNA ligase. Purification and properties of the enzyme from Thermus thermophilus HB8. J Bio Chem 1984; 259:10041-7; PMID:6469954; https://doi.org/ 10.1093/oxfordjournals.jbchem.a130975 [DOI] [PubMed] [Google Scholar]