Abstract
Kalirin7 (Kal7), a postsynaptic Rho GDP/GTP exchange factor (RhoGEF), plays a crucial role in long term potentiation and in the effects of cocaine on behavior and spine morphology. The KALRN gene has been linked to schizophrenia and other disorders of synaptic function. Mass spectrometry was used to quantify phosphorylation at 26 sites in Kal7 from individual adult rat nucleus accumbens and prefrontal cortex before and after exposure to acute or chronic cocaine. Region- and isoform-specific phosphorylation was observed along with region-specific effects of cocaine on Kal7 phosphorylation. Evaluation of the functional significance of multi-site phosphorylation in a complex protein like Kalirin is difficult. With the identification of five tyrosine phosphorylation (pY) sites, a panel of 71 SH2 domains was screened, identifying subsets that interacted with multiple pY sites in Kal7. In addition to this type of reversible interaction, endoproteolytic cleavage by calpain plays an essential role in long-term potentiation. Calpain cleaved Kal7 at two sites, separating the N-terminal domain, which affects spine length, and the PDZ binding motif from the GEF domain. Mutations preventing phosphorylation did not affect calpain sensitivity or GEF activity; phosphomimetic mutations at specific sites altered protein stability, increased calpain sensitivity and reduced GEF activity.
Keywords: RhoGEF, mass spectrometry, nucleus accumbens, prefrontal cortex, cocaine, tyrosine phosphorylation
Introduction
Genetic studies implicate the KALRN gene in schizophrenia, stroke, autism, substance abuse and intellectual disability1-9. KALRN is highly conserved from C. elegans and D. melanogaster to humans10. Alternative splicing results in three major protein isoforms (Fig.1A); expression of Kal9 and Kal12 predominates during early development and Kal7 is the most abundant isoform in the adult central nervous system11. While attention has focused on its GEF domains and their ability to activate Rac1 and RhoA, other domains clearly play essential roles. ΔKal7, a minor splice variant lacking the Sec14 domain and first four spectrin repeats (Fig.1A), is unable to support normal spine formation; synaptic cluster density, area and localization all require Kal712.
Figure 1. Kalirin isoforms and sample preparation.
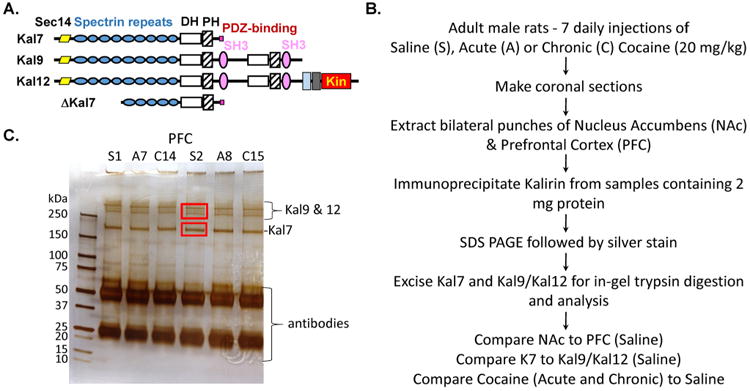
A. Kal7 consists of a Sec14 domain, 9 spectrin repeats (SR1 to SR9), a DH domain, a PH domain, a short, unstructured linker region and a PDZ binding motif42. Biophysical studies predict a compact structure for Kal710, 22. The structures of the other major isoforms are indicated; Kin, kinase. B. Adult male rats (n = 6-7/group) were subjected to saline, acute or chronic cocaine treatment. Animals were sacrificed 30 min after the final injection. Immunoprecipitates prepared from individual PFC and NAc samples (S1, A7, C14, etc.) were separated by SDS PAGE. C. Protein bands were visualized by silver staining; Kal7, Kal9 and Kal12 are indicated. Gel fragments containing Kal7 and a pool of Kal9 and Kal12 were excised and processed.
Behavioral studies of mice lacking Kal7 (Kal7KO) revealed its essential role in fear-based learning and in the response to chronic cocaine13-16. KalSRKO (full Kalirin knockout) mice show similar behavioral and drug-dependent deficits in addition to endocrine, cardiac and skeletal deficiencies17-19. Electrophysiological studies demonstrated an essential role for Kal7 in forms of long-term potentiation and long-term depression that require glutamate receptors containing a GluN2B subunit (Grin2b)20. The Kal7 PH1 domain binds directly to GluN2B and Kal7 has been localized with GluN receptors at both synaptic and perisynaptic sites12. Studied intensively as models of learning and memory, the changes in synaptic function that accompany long-term potentiation and long-term depression require rapid, reversible changes in ion fluxes, protein localization and protein phosphorylation, as well as more lasting alterations in transcription and translation. In addition, limited proteolytic cleavage of synaptic proteins such as spectrin, MARCKS, and drebrin by calpains, neutral calcium-activated cysteine proteases, is essential for long-term potentiation involving GluN2B-containing receptors21.
While inhibition of its GEF activity interferes with Kalirin-dependent synaptic plasticity14, Kal7 also exerts GEF-independent effects. Its Sec14 domain binds phosphoinositides and alters spine length and spine-head size10, 12,22. Its spectrin repeat regions have many binding partners and affect spine density, spine length and synaptic size4, 12 The PDZ-binding motif, which is unique to Kal7, interacts with PDZ domains in PSD95 and afadin, key components of the PSD23, 24 . Consistent with this, a cell permeant peptide encompassing this PDZ binding motif blocks the phosphorylation of Kal7 that occurs in response GluN receptor activation25 and blocks the ability of serotonin to modulate spine morphology23.
Genetic screens identified dTrio, the Drosophila homolog of Kalirin, and placed it in the same pathway as Abl, a non-receptor tyrosine kinase26, 27. Following up on this observation, we demonstrated that phosphorylation by Abl1 of a fragment of Kalirin containing spectrin repeats 4 through 6 increased its sensitivity to calpain cleavage12. Selective pharmacological inhibition of Abl in primary neuronal cultures reduced spine density in WT neurons, but not in KalSRKO neurons, linking the role of Abl to Kalirin in controlling dendritic spines12. Biochemical studies demonstrated a more compact structure for Kal7 than for AKal710, and neither Kal7 nor ΔKal7 was as active as its isolated GEF1 domain. We wanted to test the hypothesis that Kal7 phosphorylation altered its sensitivity to calpain cleavage, perhaps separating its active domains or activating its GEF1 domain.
The rapid phosphorylation and dephosphorylation of many PSD proteins is critical for spine formation and function28-35. After identifying 22 phosphorylation sites in mouse brain Kal736 and demonstrating that Kalirin is a substrate for multiple kinases36, 37, we suggested that Kal7 served as a postsynaptic signaling hub. The goal of this study was to provide a comprehensive analysis of the phosphorylation sites utilized in tissues where Kalirin plays a significant role, notably nucleus accumbens and prefrontal cortex, which are crucial to the biological responses to cocaine38-41. Questions to be addressed included whether phosphorylation differed in different brain regions, whether phosphorylation patterns were unique to different isoforms of Kalirin, and whether the extent of phosphorylation was modulated by cocaine use. The answers to all three questions were affirmative. The abundance of phosphotyrosine residues identified in Kal7 suggested a role for interactions mediated by SH2 domains and a high throughput screen identified multiple interactors. By mutating single phosphorylation sites, we demonstrated that the presence of a single negative charge could alter GEF activity and the ability of calpain to cleave Kal7 into smaller, biologically active fragments24, 25, 43.
Results and Discussion
Kalirin phosphorylation is brain region specific
Because Kalirin plays an essential role in the behavioral response of rodents to cocaine and cocaine-stimulated alterations in dendritic spine morphology, we focused our analyses on the nucleus accumbens (NAc) and prefrontal cortex (PFC), two very different regions known to play an essential role in the addictive response39, 44. Animals received seven daily injections of saline (Saline, control), six daily injections of saline followed by a single injection of cocaine (Acute) or seven daily injections of cocaine (Chronic) (Fig. 1B). Tissue from individual animals was homogenized in buffer containing multiple phosphatase inhibitors. Kal7, Kal9 and Kal12 were isolated by immunoprecipitation with antibody to their common N-terminal Sec14 domain; after SDS-PAGE and silver staining, a gel fragment containing Kal7 and a gel fragment containing both Kal9 and Kal12 were excised (Fig. 1C). Gel fragments were processed individually for analysis by mass spectrometry.
Using information-dependent SWATH acquisition, we identified a total of 26 phosphorylated residues in Kal7 isolated from control PFC or NAc. For most of the phosphopeptides, manual validation analyses of the b- and y-ions from the MS/MS fragmentation spectra allowed identification of the phosphorylation site36 (summarized in Fig. 2). The pattern for five of the peptides was more complex, and could not be fully resolved with the b- and y-ions obtained. Ambiguous sites are denoted with an ampersand (&). Composite Kal7 data for individual control animals are shown in Fig. 2. The extent of phosphorylation at each site was determined by averaging the values determined for the m, m+1 and m+2 isotopic mass spectral peaks from each animal. Data for all of the animals in a given group were in excellent agreement, with an average standard deviation within each treatment group or tissue of 7.6%.
Figure 2. Identification of 26 phosphorylation sites in Kal7.
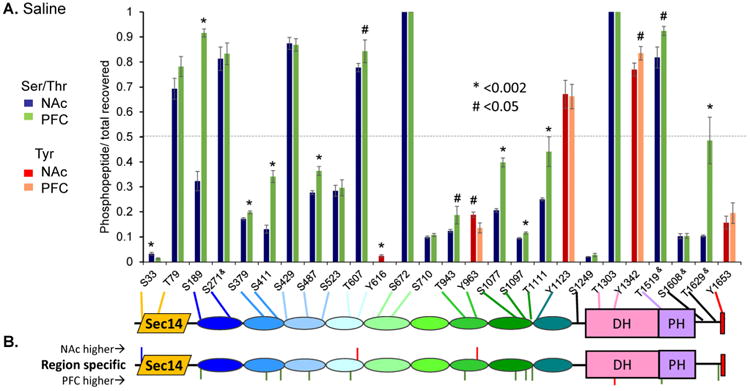
A. Bar graph showing degree of phosphorylation of each site relative to the total amount of peptide recovered. Blue and green bars show phosphorylation of Ser/Thr residues in NAc and PFC, respectively; red and pink bars indicate the same for Tyr residues. Residues are indicated on the X axis, where ampersands (&) indicate indefinite assignment. The grey dashed line indicates 50% phosphorylation. Sites that differ significantly in extent of phosphorylation in NAc vs. PFC are indicated by a # (p <0.05) or * (p <0.002) above the bar. A schematic of Kal7 indicating the location of each phosphorylation site is shown below the bar graph. Sites are numbered using AF230644_1 for rat cKal7. B. Schematic identifies residues where phosphorylation was significantly different between NAc and PFC in saline injected control animals.
As observed in mouse brain36, phosphorylation sites were identified throughout Kal7 (Fig. 2A). About two-thirds of the sites were in the spectrin repeat region, which is known to interact with multiple proteins, including peptidylglycine α-amidating monooxygenase, Huntingtin-associated protein, inducible nitric oxide synthase, Arf6 and deleted in schizophrenia-14, 42, 45. Based on the Russell/Linding definition of disorder46, the vast majority of the phosphorylation sites were within a structural domain. Except for SR6, phosphorylation sites were found in each structural domain. Half of the 18 phosphorylation sites in the spectrin repeat region were in loops connecting the helices forming each spectrin repeat10. Six other phosphorylation sites fell into disordered regions predicted at the N- and C-termini of Kal7 and in the linker between SR9 and the catalytic DH domain. Interestingly, tyrosine phosphorylation was far more common in Kal7 (5/26 sites, or 19%) than is typical of the average phosphoprotein (<2% phosphotyrosine)47-50. Nine of the 26 sites reported here were previously identified in rat aKal7 expressed transiently in HEK cells or exposed in vitro to purified kinases36. Although phosphorylated in vitro by purified kinases, phosphorylated S83 and T1590 have not yet been identified in Kal7 isolated from rat brain. Phosphorylated T1590 was identified in transfected HEK cells and both sites are included in Supplemental Table S1. Given that Kal7 accounts for only about 0.01% of total brain protein20, 36, enrichment by immunoprecipitation was essential to the success of this study; even large, global phosphoproteomic studies have identified only a few phosphorylation sites in Kalirin48, 49, 51.
Kal7 expression is largely limited to neurons, with Kal7 concentrated at the PSD11, 12. Excitatory glutamatergic neurons predominate in the PFC while inhibitory GABAergic neurons predominate in the NAc39, 44, 52. We hypothesized that this difference in cell type would influence the phosphorylation state of Kal7 (Fig. 2B). Three sites were more heavily phosphorylated in Kal7 isolated from NAc than from PFC; two of these sites were tyrosine residues (Y616 and Y963). While not extensively phosphorylated in NAc, phosphorylation of Tyr616 was not detectable in PFC. Twelve sites were more heavily phosphorylated in Kal7 isolated from PFC than from NAc; only one of the sites was a tyrosine residue (Y1342).
The GPS 3.0 program (http://gps.biocuckoo.org/)53 was unique in its ability to accurately predict the Kal7 phosphorylation sites identified. In addition, previous in vitro phosphorylation studies using protein kinase C, CaMKII, casein kinase 2, Abl1, TrkB and Fyn support the GPS 3.0 kinase predictions (Table 1)36. Recent reports suggest that CaMKII phosphorylation of T95 controls Kal7 function25, 37, 43. However, GPS3.0 identifies T79, not T95, as a CaMKII site and our in vitro studies demonstrate that CaMKII phosphorylates Kal7 at T79, not at T9536. Importantly, pT95-containing peptides were not detected in any of the 80 samples analyzed; peptides containing non-phosphorylated T95 were always abundantly recovered36.
Previous studies implicated specific kinases in the regulation of Kal7. Serum-inducible, glucocorticoid-inducible kinase 1 (SGK1) mediates neuropathic pain in the spinal cord, and does so by increasing Kalirin phosphorylation35. SGK1 is predicted to phosphorylate a single site in SR3, S523 (Table S1). Similarly, Kal7 was identified as a MAPKAPK5 interactor and substrate (S487)54. MAPKAPK5 is predicted to phosphorylate two sites in Kal7, S487 and S1249 (Table S1). S487 is located in an SH3 binding motif that can form an intramolecular interaction with the first SH3 domain of the longer Kalirin isoforms, inhibiting GEF activity55. Although T1590 can be phosphorylated by Cdk5 in vitro and phosphorylation of T1590 affects function in PC12 cells56, phosphorylation at this site has not been detected in rat or mouse brain36.
Kalirin phosphorylation is isoform-specific
Kal9 and Kal12, the major isoforms expressed during development, are less prevalent than Kal7 in the adult brain. Six phosphorylation sites (5 Ser/Thr and 1 Tyr) were identified in the region common to Kal9 and Kal12 but absent from Kal7 (Fig. 3A). The region unique to Kal12 contained 2 more Ser/Thr phosphorylation sites (Fig. 3A), and more sites may be identified when greater amounts of Kal9/Kal12 are available for analysis.
Figure 3. Identification of 30 phosphorylation sites in Kal9/12 from rat PFC.

A. Schematic showing phosphorylation sites identified in Kal9/Kal12 from Saline-injected control rat PFC. Residue numbers are shown only for pY sites. Sites shared by Kal9/Kal12 are bracketed, as are sites unique to Kal12. B. Isoform-specific phosphorylation sites are depicted. Phosphorylation sites identified in Kal7 but absent from Kal9/Kal12 are indicated above the schematic. Phosphorylation sites identified in Kal9/Kal12 but absent from Kal7 are shown below the schematic.
While Kal7 is largely particulate, much of the Kal9/Kal12 is soluble11, 22. Since a difference in subcellular localization could result in exposure to different protein kinases, we asked whether the extent of phosphorylation at sites common to Kal7, Kal9 and Kal12 differed. Twenty-two of the 30 phosphorylation sites identified in Kal9/12 isolated from control PFC fell in the region shared with Kal7 (Fig. 3A). Although present in the shared spectrin repeat region, phosphorylation of three Ser residues (S283, S924, S1044) was observed only in Kal9/Kal12 (Fig. 3B). Six phosphorylation sites identified in Kal7 (1 Tyr and 5 Ser/Thr) were not phosphorylated in Kal9/12 (Fig. 3B).
The Sec14 domain, which plays an essential role in regulating spine length12, contains only one phosphorylation site (T79). Strikingly, although heavily phosphorylated in Kal7, T79 was not detectably phosphorylated in Kal9/12. Selective usage of the Kal-C promoter, which places an amphipathic helix at the N-terminus of Kal7 but not Kal9/Kal12, contributes to their differing subcellular localizations and could alter their exposure to kinases and phosphatases11, 12, 22. Since the ability of the isolated Sec14 domain to interact with phosphoinositides is increased in a T79E mutant, phosphorylation at this site may be of functional significance12.
Phosphotyrosine sites in Kalirin exhibit unique SH2 domain binding profiles
Coupled with the established role for Kalirin in downstream signaling from multiple receptor tyrosine kinases57, 58, the marked preponderance of phosphotyrosine residues in Kalirin suggested that an exploration of its interactions with phosphotyrosine binding domains would be informative. To systemically identify these interactions, we utilized a reverse-phase Src homology 2 (SH2) screening platform59, 60. Peptides corresponding to each of the four internal phosphotyrosine sites in rat brain Kal7 were synthesized with a biotinylated N-terminus and an amidated C-terminus (Fig. 4A). The effect of phosphorylating Tyr1653, which is located within the PDZ-binding motif, was not tested since it would not be expected to interact with SH2 domains61-63. Based on their expression in the nervous system and their stability, we selected 71 of the 120 SH2 domains in the human and mouse genomes for analysis; one phosphotyrosine binding (PTB) domain and two mutant substrate-trapping protein tyrosine phosphatase (PTP) catalytic domains were also analyzed (Fig. 4A) (Supplemental Table S2). Each was expressed as a glutathione S-transferase (GST) fusion protein and applied to a membrane onto which the peptides had been spotted (Fig. 4B). The immobilized peptides were visualized using streptavidin and a phosphotyrosine antibody; bound probe was visualized using antibody to GST (Fig. 4C, D).
Figure 4. Screening pY peptides with multiple GST-SH2 constructs.
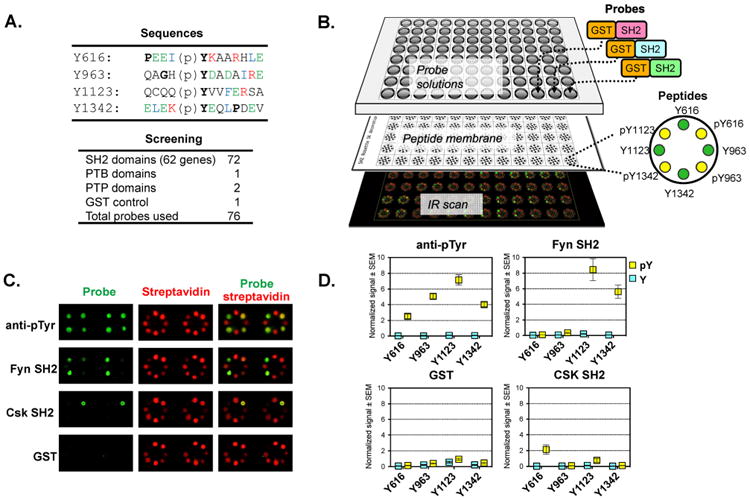
A. Seventy-four phosphotyrosine interacting domains (SH2, PTP, and PTB) and a GST control were tested. The probe list is provided in Supplemental Table S2. The sequences of the four internal Kal7 pY peptides tested are shown. B. Overview of SH2 screen. Four pairs of phosphorylated and non-phosphorylated peptides were immobilized in a rosette pattern; after incubation with the GST-tagged probes, signal was detected with infrared (IR) dye labeled secondary antibodies. C. Representative IR scan data are shown; green signal indicates probe binding and red signal monitors peptide loading using streptavidin. The pY antibody control confirms the phosphorylation state of the biotinylated peptide. The GST control assesses background. In this panel, Fyn-SH2 bound strongly to pY1123 and pY1342, while Csk-SH2 bound only to pY616. D. Quantified results for pY, Fyn-SH2, Csk-SH2, and GST probes. Signal for each peptide was normalized to the streptavidin signal for that peptide; values plotted are the average of quadruplicates with SEM. As seen in the streptavidin signal, the pY1123 peptide bound relatively poorly to the membrane; the ratiometric quantification corrects for this.
The SH2-peptide interactions were phosphorylation dependent, with each phosphopeptide exhibiting a unique profile; data from quadruplicate assays are shown as a heatmap in Fig. 5A and in Supplemental Table S3. Each probe, including non-binders, was validated previously60. Many probes bound to more than one Kalirin phosphotyrosine peptide; the pY1123 peptide exhibited phosphorylation dependent binding to almost half of the probes. In contrast, the Src inhibitor Csk SH2 probe bound only to the pY616 peptide. The Rin2 probe showed a preference for pY963 and pY1342 peptides, exhibiting little interaction with the pY616 peptide. SH2 domains that recognize multiple pY sites in Kal7 may have an advantage, given the brief dwell times (∼100 msec) of SH2 domains on pY sites and the short lifetimes (∼15 sec) of many pY residues64.
Figure 5. Comparison of GST-SH2 binding data for different pY sites.
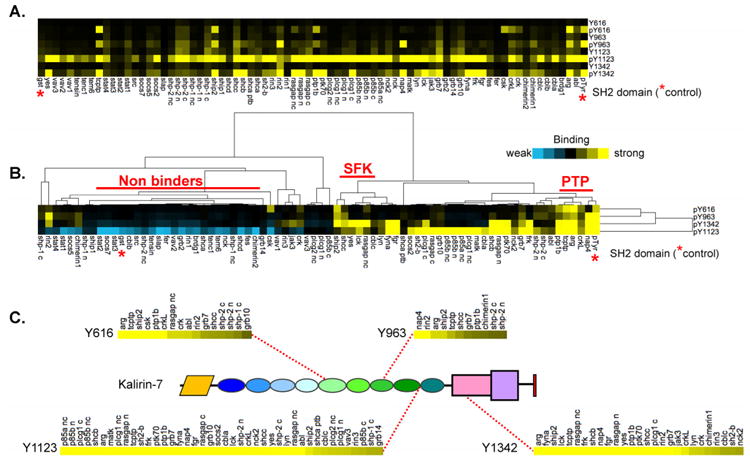
A. Normalized SH2 binding to the four internal Kal7 pY sites are presented as a heatmap (positive binding in yellow); probes appear in reverse alphabetical order, left to right. Overall the pY peptides showed enhanced signal compared to their non-phosphorylated Y peptide counterparts, indicating phosphorylation-dependent binding of the probes. Notably, pY1123 was recognized by more probes than the other peptides. Red asterisks indicate the pTyr and GST controls. B. Hierarchical clustering was used to identify probes exhibiting similar binding patterns; two subclusters were identified (SFK: Src family kinases; PTP: protein tyrosine phosphatases). C. Interactors for each pY peptide were ordered by signal intensity (strong interactors on the left) and the position of each pY in Kal7 is shown. The cut-off for positive binders was defined as >3× the GST background.
Our SH2 profiling of Kal7 peptides identified nearly a hundred previously undocumented interactions. The binding patterns were analyzed by hierarchical clustering, separating binders from non-binders and grouping SH2 domains that exhibited similar interaction patterns (Fig. 5B). Two Tyr residues located in or near the DH/PH domain (Y1123, Y1342) were favored by a broad range of SH2 domains, perhaps due to the presence of hydrophobic residues at the Y+3 position, a general consensus for SH2 ligands65. A small cluster favoring the pY1342 peptide was dominated by Src family kinases (SFK: Yes, Lck, Lyn, Fyn, Fgr), consistent with the presence of Glu at Y+266. The two Tyr residues in the middle of the spectrin repeat region (Y616, Y963) were bound by a smaller number of SH2 domains. Y963 (QAGHpYDADAIRE) is a partial match for the Nck SH2 domain binding consensus (pY-D/y-d/e/l/y-V)66, but Nck binding was not detected, possibly due to the absence of Val at the Y+3 position. Both of the protein tyrosine phosphatase (PTP) domains tested exhibited binding profiles resembling that of the non-receptor tyrosine kinase Abl2/Arg. The unique sets of SH2 domains targeting each phosphotyrosine site are identified in Fig. 5C, emphasizing the strength of the SH2 domain interactions with pY1123 in the 9th spectrin repeat and with pY1342 in the DH domain.
TrkB-mediated phosphorylation of Kal7 facilitates SH2 domain interactions
We next assessed phosphorylation-dependent SH2 domain binding to intact Kal7. We expressed and purified Kal7 using Baculovirus and Hi5 cells (Fig. 6A). Taking into account the strength of the pY1123 interaction with multiple SH2 domains, the prediction that TrkB can phosphorylate Kalirin at Y1123 (Table S1) and the essential role of Kalirin in BDNF-triggered neurite outgrowth and branching58, we incubated purified Kal7 with or without recombinant TrkB kinase and ATP. Before incubation, recombinant Kal7 (191 kDa) was not detected by immunoblotting with antibodies to phosphotyrosine (Fig. 6B) or phosphothreonine (not shown). After incubation with TrkB kinase (66 kDa), both Kal7 and TrkB were detected by phosphotyrosine antibodies; mass spectrometric analysis identified Y1123 as the major site phosphorylated by TrkB (not shown). Phosphopeptide analysis following in vitro phosphorylation of SR7:9 by TrkB also identified Y1123 as the preferred site; less extensive phosphorylation of five additional Tyr residues was also detected (not shown).
Figure 6. Far-Western analysis of selected interactors.
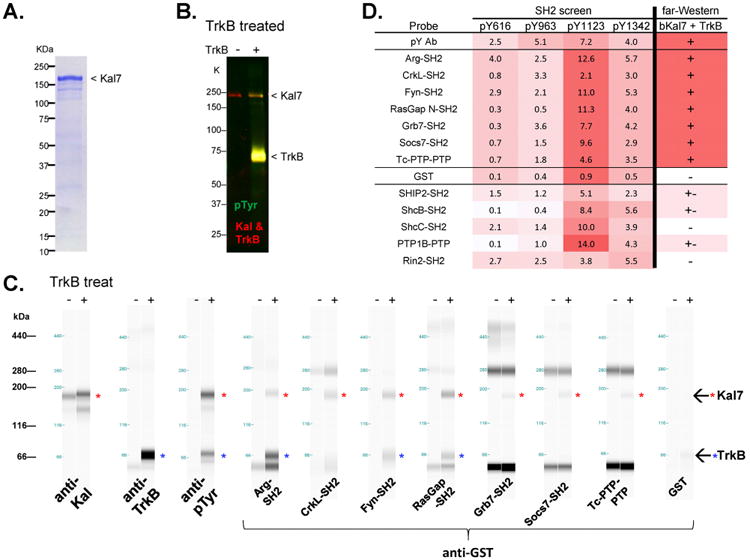
A. Coomassie gel of purified bKal7. B. Purified bKal7 was tyrosine phosphorylated using TrkB. Presence of purified bKal7 protein (∼190 KDa), TrkB protein (∼70 KDa), and their tyrosine phosphorylation state were assessed by Western blot analysis. C. Validation of SH2 screen. Capillary far-Western assay was performed to confirm direct interactions between the probe and purified, phosphorylated bKal7. Red asterisks in blot view indicate the presence of phospho-Kal7 (∼190 KDa) with TrkB treatment; blue asterisks indicate TrkB (∼70 KDa). The higher molecular weight bands detected by several probes are non-specific. D. Summary of SH2 screen and capillary far-Western results for 12 SH2/PTP domains and control probes. Positive interactions are indicated, with red color proportional to strength.
The ability of TrkB-phosphorylated Kal7 to interact with 12 of the probes that bound to the pY1123 peptide and to additional Kalirin pY peptides was assessed62, 63. Traditional far-Western blotting experiments were not successful, likely due to insensitivity. Hence, we employed the capillary Western ProteinSimple system for a sensitive alternative far-Western method, to assess SH2 probe binding to intact Kal7. Kal7 phosphorylated by TrkB interacted with the SH2 domains of Abl2 (Arg), CrkL, Fyn, Grb7, Socs7, RasGap (Rasa1) and Ptpn2 (Tcptp) (Fig. 6C, D). A weak interaction was observed between TrkB-phosphorylated Kal7 and Ship2-SH2, ShcB-SH2 and Ptp1b PTP. No interaction was detected between phosphorylated Kal7 and Rin2-SH2 or ShcC-SH2. The far Western results with intact Kal7 indicate that many of the interactions detected in the SH2 screen could occur in vivo.
Based on their established roles in TrkB signaling, several SH2-domain interactors (Fyn, CrkL) could contribute to signaling disruptions downstream of activated TrkB in neurons lacking Kalirin. The SH2 domain of Fyn can interact with autophosphorylated TrkB and the ability of BDNF to stimulate TrkB movement into lipid rafts is dependent on Fyn67. In addition, synaptic NR2B-containing NMDARs, which interact with Kalirin20, are stabilized after phosphorylation by Fyn68. The ability of Kalirin pY1123 to interact with CrkL may also contribute to altered TrkB signaling kinetics and signal integration. Phosphorylation of a single Tyr in ARMS following TrkB activation switches its interaction from the CrkL SH3 domain to the CrkL SH2 domain; now available, its SH3 domain can bind targets involved in sustained MAPK signaling69.
Drosophila Trio (equally related to KALRN and TRIO) plays an essential role in Abl-mediated axonal outgrowth26, leading to the prediction that Abl and/or Abl2/Arg play similar roles in vertebrates. The Abl tyrosine kinases play an essential role in Rac activation at adherens junctions and inhibitors disrupt neurite branching after integrin-mediated adhesion to laminin70. With its well established roles in integrin signaling, activation of cortactin and N-WASp and binding to F-actin, an interaction of pY1123 in Kalirin with Abl2/Arg could integrate TrkB and integrin receptor signaling.
Phosphorylation may target Kal7 for calpain-mediated cleavage
Calpains, like Kal7, play an essential role in synaptic plasticity21. Calpains catalyze limited cleavage of their target proteins, which include cytoskeletal proteins known to localize to spines and proteins involved in dendritic protein synthesis12, 21, 71, 72. Several observations suggest that phosphorylation-regulated limited cleavage of Kalirin by calpain could play an essential role in controlling Kalirin function. First, fragments of Kal7 exert GEF-independent effects on spine formation and morphology12. Kal7 binds directly to the GluN2B subunit of GluN receptors, placing it where it could be cleaved by μ-calpain activated by Ca2+ entering via this ion channel or by m-calpain activated by BDNF stimulation of Erk21. Kal7 levels in the hippocampus decrease in response to transient forebrain ischemia/reperfusion, a condition known to activate calpain7. Finally, μ-calpain cleavage of a fragment of Kalirin containing only spectrin repeats 4 through 6 (SR4:6) is increased following phosphorylation of Y591 by Abl112
To evaluate its sensitivity to μ-calpain, Kal7 was expressed transiently in HEK293 cells. Kal7-containing lysates were incubated with increasing amounts of activated μ-calpain and then subjected to Western blot analysis (Fig. 7A). In the absence of exogenous calpain, Kal7 (191K) was remarkably stable; with increasing amounts of activated μ-calpain, products of 147K, 109K and 72K were detected by polyclonal antibodies to SR4:7 (Fig. 7A). A Sec14 domain antibody also identified the 147K and 72K fragments (Fig. 7B). A C-terminal antibody identified the 109K fragment and small amounts of a 45K fragment (Fig. 7B). As observed for many other substrates, μ-calpain cleaved Kal7 at a limited number of sites.
Figure 7. Calpain cleavage of Kal7.

A. Lysates (30 μl; ∼60 μg protein) of HEK cells expressing Kal7 were analyzed directly (Input, In) or digested with the indicated dose of μ-calpain (0 to 120 ng) at 37C for 1 h before denaturation with SDS and Western blot analysis using an affinity-purified antibody that recognizes SR4:7; markers are shown to the left and observed masses to the right. B. In additional experiments, Input and μ-calpain treated samples prepared with the highest amount of μ-calpain were analyzed using antibodies to the C-terminus (product masses shown in red) and Sec14 domain (product masses shown in blue) of Kal7. C. Input and μ-calpain-digested samples were analyzed using the SR4:7 antibody and the C-term antibody; after normalizing the C-term/SR4:7 ratio for each Input sample to 1.0, the ratio calculated for the corresponding calpain-digested sample was significantly less than 1.0 (p < 0.001; t-test), suggesting that intact Kal7 and a C-terminally truncated μ-calpain product were not being resolved by this gel system. D. Vector encoding the MycHis-tagged GEF1-end protein shown in the schematic, which extends from the GEF1 domain to the C-terminus56 was transiently expressed and analyzed as described for Panels A and B; the red arrow indicates the location of a high probability calpain site. Proteins were visualized with Myc antibody (left) or C-term antibody (right), demonstrating calpain-catalyzed removal of the C-terminal epitope from the 41K product. E. For each μ-calpain digest, the total amount of signal obtained with the SR4:7 or C-term antibody was determined; the % of that signal accounted for by each major product was determined (n = 4; error bars are std dev). F. μ-Calpain cleavage sites in rat Kal7 were predicted using GPS-CCD74 the 5 top sites, with scores > 1.0 are shown in thick red lines; sites with scores of 0.8 to 1.0 are shown in black. Cleavage at the most C-terminal calpain site generates a product just 3 amino acid longer than the unique region of Kal7. G. HEK293 cells transiently transfected with vector encoding aKal7 were treated with the indicated dose of staurosporine for 4 h before extraction for μ-calpain digestion. Staurosporine treatment increased the amount of intact Kal7 and reduced product formation.
Digests and input samples were routinely analyzed using the SR4:7 antibody and the C-terminal antibody. When normalized to the appropriate input, it became clear that C-terminal antigenicity was lost from the 191 K band following calpain digestion (Fig. 7C). With the ratio of the C-terminal to SR4:7 antibody signals for the 191 K band set to 1.0, the ratio for the calpain treated sample was less than 1.0 (Fig.7C), indicating that μ-calpain cleaved at a site near the C-terminus of Kal7. Since attempts to resolve full-length Kal7 (191K) from a form lacking the C-terminal epitope (188K) were not successful, we expressed a much smaller, epitope-tagged, N-terminally truncated protein lacking the Sec14 domain and all 9 spectrin repeats. Following calpain cleavage, Myc antibody detected the intact protein along with a single smaller product (Fig. 7D). Visualization of the same samples with the C-terminal antibody revealed only the upper band. Thus we concluded that cleavage by μ-calpain removed the unique PDZ binding motif from the C-terminus of this truncated variant of Kal7 (red arrow in Fig.7D).
The major Kal7 cleavage products detected by the SR4:7 and C-terminal antibodies were quantified with respect to the undigested input analyzed on the same gel (Fig. 7E). A 109K product was detected by both antibodies. The 72K product detected by the SR4:7 antibody was also detected by the Sec14 antibody. Cleavage within SR5, as observed previously for SR4:612, would generate both products. Calpain specificity is determined by the 11 residues surrounding the cleavage site, but is also sensitive to tertiary structure73, 74. Potential calpain cleavage sites in Kal7 were identified using the GPS-CCD1 program74 with the top 5 sites indicated by red arrows and 14 additional sites by black lines (Fig. 7F) (Suppl. Table S4). The sites are clustered, with five in SR5. Cleavage at the most C-terminal of the predicted calpain cleavage sites would remove the 20 amino acid sequence unique to Kal7, reducing its mass by only 2.5 kDa, an amount too small to resolve by SDS-PAGE (191K vs. 188K). Cleavage at this site is consistent with the products identified in Fig. 7D and would have multiple consequences: released from Kal7, PDZ domain interactors like PSD-95 would be more mobile; the subcellular localization of the truncated Kal7 protein, which is expected to be fully active, would be altered; if released, the C-terminal peptide might disrupt other PDZ domain interactions, as demonstrated using the cell-permeable peptide matching the COOH-terminus of Kal775. Although their substrate specificities are generally similar, μ-and m-calpain play distinct roles in the nervous system and it will be important to determine whether both recognize the two major cleavage sites in Kal7 equally well.
Kal7 expressed in HEK293 cells is extensively phosphorated35. We first asked whether treatment of transfected HEK cells with staurosporine76 which inhibits multiple protein kinases, could alter the sensitivity of Kal7 to cleavage by calpain (Fig. 7G). Cleavage was evaluated using the C-terminal and SR4:7 antibodies; the cleavage products detected by both antibodies were quantified and the prevalence of each product was calculated. Three experiments were conducted, treating duplicate wells of HEK cells transiently expressing Kal7 with 50 nM to 10 μM staurosporine for 30 min to 14 h. Exposure to 1 or 10 μM staurosporine for 4 h diminished calpain cleavage of Kal7 to the same extent (Fig. 7G); more Kal7 remained intact and the amount of 109K protein decreased. A similar effect was seen when cells were treated with 1 μM staurosporine for 14 h (not shown). Taken together, these experiments led us to examine the effect of mutating known phosphorylation sites on the ability of calpain to cleave Kal7.
Phosphomimetic mutations alter sensitivity of Kal7 to calpain
To identify individual sites that might affect the sensitivity of Kal7 to calpain, we introduced mutations that prevent phosphorylation (Tyr→Phe; Thr/Ser→Ala) or introduce a negative charge (Tyr→Glu; Thr/Ser→Glu/Asp). Although far from a perfect mimic of phosphorylation, examining these mutants let us focus on Kal7 modified in a predictable manner. Initial expression levels, recovery of signal after cleavage by μ-calpain and cleavage fragments recognized by the SR4:7 and C-terminal antibodies were monitored. A screen of nine sites (Fig. 8A) revealed that mutations at six sites were without effect (black lettering) and identified Y591, Y1123 and T1519& as sites of interest (red lettering). For the SR1 and Y591 mutants, transfection with equal amounts of vector encoding the phosphomimetic and non-phosphorylatable mutant yielded equivalent levels of 191K protein recognized by the SR4:7 antibody (Fig. 8B). In contrast, for the Y1123 and T1519& mutants, levels of 191K phosphomimetic mutant were significantly lower than levels of 191K non-phosphorylatable mutant, suggesting that introduction of a negative charge resulted in instability.
Figure 8. Effect of single site mutations on calpain cleavage of Kal7.
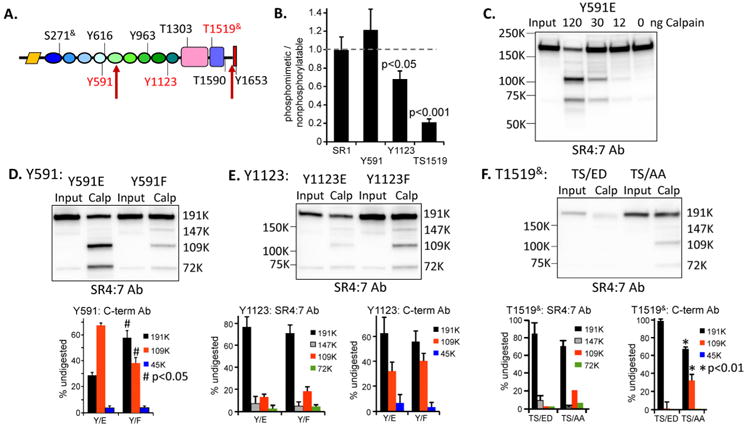
A. Diagram indicating the sites targeted for mutagenesis and the major μ-calpain cleavage sites; mutation of the sites indicated in red affected stability and/or cleavage. B. Ratio of 191K band intensity (SR4:7 Ab) for phosphomimetic vs. nonphosphorylatable Kal7 mutants expressed in HEK cells (n = 3 to 7; One-Way ANOVA by ranks). C. Lysates prepared from HEK cells expressing Kal7/Y591E were digested using different amounts of μ calpain, as in Fig. 7A. D. Lysates containing Kal7/Y591E or Kal7/Y591F were digested with μ-calpain under identical conditions, revealing more extensive cleavage of the Y591E mutant. Quantitative analysis of μ-calpain products of Kal7/Y591E and Kal7/Y591F detected using the C-term antibody; the digestion patterns differed significantly (One-Way ANOVA on Ranks; p<0.05 for 191K, 109K and 45K). E. Analysis of Kal7/Y1123E and Kal7/Y1123F mutants. Although consistently expressed less well than Kal7/Y1123F, Kal7/Y1123E yielded similar digestion products. F. Analysis of Kal7 with T1519S1520 mutated to ED or to AA. Kal7/TS/ED was poorly expressed compared to Kal7/TS/AA, and the stable products of μ-calpain digestion were barely detected in digests of Kal7/TS/ED (p<0.01, both antibodies, One-Way ANOVA by ranks).
As observed for Kal7, the Y591E mutant was stable in the absence of exogenous calpain and the μ calpain concentration curve resembled that observed for Kal7 (Fig. 8C, 7A). Although antigenicity with the SR4:7 antibody was retained during cleavage, replacing Y591 in SR4 with an acidic residue produced a protein that was more susceptible to cleavage by μ-calpain within SR5, producing more of the 109K fragment (diagram in Fig.7F); the behavior of the Y591F mutant was indistinguishable from Kal7 (Fig. 8D).
Cells expressing Kal7 with a glutamic acid residue replacing Y1123 (Y1123E) consistently contained less exogenous protein than cells expressing the Y1123F mutant, suggesting increased degradation of the Y1123E protein (Fig. 8E). Assessment of the products of μ-calpain cleavage revealed accumulation of the 109K product containing the C-terminal PDZ binding motif and the 72K product containing the N-terminus; while less protein was present, any changes in structure were not recognized by μ-calpain (Fig.8E).
Although replacing the Thr1519-Ser1520 sequence in the PH domain of Kal7 with Ala-Ala was without effect on stability or cleavage pattern, placing adjacent negatively charged residues at these position (Glu-Asp) yielded what appeared to be a very unstable protein that was not recognized in the normal manner by μ-calpain and instead simply disappeared upon calpain digestion (Fig.8F).
The ability of Kalirin to activate Rac1 is remarkably insensitive to phosphorylation site mutation
Many RhoGEFs are activated by phosphorylation within the catalytic domain or at an adjacent site, relieving inhibition77-80. Since the GEF domain of Kalirin is extensively phosphorylated (T1303, Y1342, T1519&), we first examined the consequences of preventing phosphorylation at these four sites. A cell-based Rac activation assay was used to obtain quantitative data. Simultaneously mutating all four phosphorylation sites in the GEF1 domain to non-phosphorylatable residues (GEF1-quad) had no effect on the ability of the isolated GEF domain to activate Rac1 (excitation at 420 nm, FRET peak at 525 nm; Fig. 9A). Replacing Y1653 with F or E was also without effect on GEF activity in constructs extending from the first GEF domain of Kalirin to the C-terminus of Kal7 (Fig. 9B; GEF1-end); the protein:FRET dose-response was quantified using the FRET peak at 525 nm vs. protein expression level determined by Western analysis (inset). Similarly, preventing phosphorylation at these sites had no effect on the ability of full-length Kal7 to activate Rac1 (Fig. 9C). We obtained similar results in assays using the Rac1 biosensor in AtT-20 mouse pituitary tumor cells (not shown). Since these sites are heavily phosphorylated in HEK cells36 our data indicate that phosphorylation of these sites does not regulate GEF activity in this system.
Figure 9. Effect of phosphorylation on GEF activity.

A. Cell-based DoraRac biosensor FRET assay. GEF1: WT vs. non-phosphorylatable mutant (quad: T1303A, Y1342F, TS/AA). B. GEF1→ end. WT vs. Y1653E and Y1653F, plotted using densitized protein values from Western analyses; inactive Kal-GEF/ND/AA gave no activity above Biosensor alone. C. Kal7 mutants: Y1653F and Quad (non-phosphorylatable) mutant, plotted using densitized protein values from Western analyses. D-F. Biosensor assays demonstrated that Kal7/Y591F and /Y591E were equally active in 3 assays. GEF assays demonstrated that Kal7 and Kal7/Y1123F were equally active in 5 assays in duplicate (p=0.85), while Kal7/Y1123E was significantly less active in 4 assays in duplicate (p <0.001). Kal7/TS/AA was fully active in 3 assays, but the TS/ED version of Kal7 was 3-20 times less active in 3 assays.
We next assessed the GEF activity of Kal7 mutated at the three sites shown in Fig. 8D, E, F to affect stability and calpain sensitivity (Fig. 9D, E, F). The GEF activity of Kal7/Y591E and Kal7/Y591F did not differ from that of Kal7 (Fig. 9D). Although more sensitive than Kal7 to calpain cleavage in SR5, Kal7/Y591 was not further degraded by calpain and the spectrin repeats present in the major 109K product would be expected to exert an inhibitory effect on the GEF domain. While replacement of Y1123 with Phe had no effect on GEF activity, replacement with Glu reduced GEF activity to half; since the Rac activation data are plotted against the amount of Kal7 protein in the lysate, the lowered stability of Kal7/Y1123E does not explain its decreased Rac activation activity (Fig. 9E). Consistent with its stability and sensitivity to calpain digestion, Kal7 with non-phosphorylatable residues at TS1519/20 was as active as Kal7. Even after adjusting for its limited stability (Fig. 8F), Kal7 with adjacent acidic residues at the TS1519/20 position was much less active than Kal7 (Fig. 9F). The inability of calpain to produce its characteristic set of cleavage products from the TS/ED mutant is consistent with an altered overall conformation and diminished catalytic activity.
Although Fyn phosphorylation of Y1653, the penultimate residue of Kal7, was reported to have a major stimulatory effect on Rac1 activation by Kal724 the Rac1 biosensor assay showed no effect of preventing or mimicking phosphorylation at this site (Fig. 9B, C), suggesting that the effect did not result from direct effects of phosphorylation on Rac1 GEF activity. In a separate series of experiments, the ability of PSD-95 in brain extracts to bind to synthetic peptides encompassing the COOH-terminus of Kal7 was also unaffected by the phosphorylation state of Y1653 (not shown).
While the isolated GEF1 domain of Kalirin is substantially more active than Kal7, the structural basis for this inhibition is not yet clear; simply appending spectrin repeats 5 through 9 to the GEF1 domain is sufficient to mimic the inhibition observed in Kal7. Like several other GEFs for RhoA, the activity of the GEF2 domain of Kalirin is inhibited by an intramolecular interaction between its DH and PH domains. After G protein coupled receptor-mediated activation, Gαq interacts with a highly conserved C-terminal extension of the PH domain, relieving the intramolecular inhibition81.
Kal7 phosphorylation varies in a tissue-specific manner with cocaine exposure
To assess physiologically relevant changes in Kal7 phosphorylation, we evaluated the effects of experimenter administered acute and chronic cocaine vs. saline on Kal7 immunoprecipitated from NAc and PFC. The ability of repeated cocaine exposure to increase spine density in NAc neurons requires the presence of Kal715. Consistent with this, spine formation in cultures of rat medium spiny neurons is increased in response to expression of exogenous Kal782. Perhaps more strikingly, Kal7KO mice self-administer cocaine at a much higher rate than WT mice, and exhibit significantly elevated locomotor responses to injected cocaine13, 15. We identified different sites in Kal7 from each brain region at which the extent of phosphorylation was cocaine responsive (Fig. 10). All four cocaine-responsive sites in NAc Kal7 were in the C-terminal half of the protein and two were Tyr sites. Phosphorylation at Y963, Y1342 and T1629& was decreased by chronic cocaine treatment, while Kal7 phosphorylation was increased by acute cocaine at a single site, S1608& (Fig. 10B); PI3K-like protein kinases that share specificity for SQ sites (ATM, ATR, and DNA-PK)83 are candidate kinases for this site (Table S1). Phosphorylation of Y1342 was reduced by chronic cocaine treatment in both brain regions, but the GEF activity of Kal7/Quad mutant (which includes Y1342F) did not differ from that of Kal7 (Fig. 9C). By contrast, in PFC, four of the five sites affected by cocaine were near the N-terminus of Kal7. Phosphorylation of Kal7 was increased only at S33 and was decreased at S379, S411, S429 and Y1342 following acute or chronic cocaine treatment. Cell-type specific features of signaling pathway activation or kinase and phosphatase expression may contribute to the differences observed.
Figure 10. Cocaine-mediated changes in Kal7 phosphorylation in NAc and PFC.
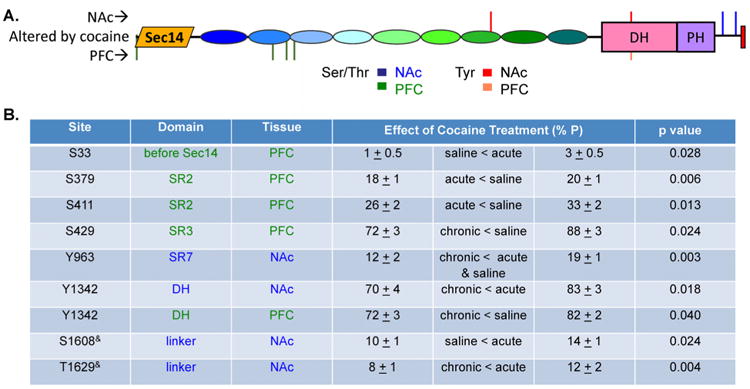
A. Diagram showing location of cocaine regulated phosphorylation sites in Kal7 isolated from NAc (above schematic) and PFC (below schematic). B. Table summarizing significant changes in degree of phosphorylation at cocaine-regulated sites in NAc and PFC Kal7.
Conclusions
Like many PSD proteins, Kal7 is phosphorylated at multiple sites. Tissue-specific alterations in its phosphorylation and in the effects of acute and chronic cocaine on phosphorylation at several sites were observed, consistent with the essential role played by Kal7 in the morphological and behavioral effects of chronic cocaine. Understanding the combinatorial effects of multisite phosphorylation is an essential part of understanding how a complex protein like Kal7 integrates inputs from receptor and non-receptor tyrosine kinases, GPCRs and ligand gated ion channels. Consistent with genetic studies in Drosophila, which linked ABL family non-receptor tyrosine kinases to Rho GEFs of the KALRN/TRIO family, four internal pY sites were shown to interact with overlapping sets of SH2 domain proteins. Phosphorylation of Kal7 by TrkB kinase facilitated its interaction with the SH2 domain of ABL2/ARG, consistent with a major role for Kalrn in TrkB-mediated control of neuronal morphology. In addition to rapidly reversible interactions, long-term potentiation requires changes that are not readily reversible. Calpain-mediated cleavage of several cytoskeletal and PSD proteins plays an essential role, along with changes in gene transcription and translation. Calpain cleavage of Kal7 released an N-terminal region known to increase spine length and the C-terminal PDZ binding motif unique to Kal7. Placement of an acidic charge at a single site in its spectrin repeat region increased the sensitivity of Kal7 to calpain cleavage. Although mutation of phosphorylation sites in the Kalirin GEF1 domain had no effect on the GEF activity, further studies will be required to determine if simultaneous mutations of other phosphorylation sites identified in this study have any downstream signaling and physiological effects in neurons.
Experimental Procedures
Animal Treatment
All of the procedures used were approved by our Institutional Animal Care and Use Committee and are consistent with ARRIVE guidelines (http://www.elsevier.com/__data/promis_misc/622936arrive_guidelines.pdf). Adult male Sprague Dawley rats purchased from Charles River and allowed to acclimate for one week were split into three groups of six or seven. Animals were weighed (average 260 ± 5 gm) and each animal received an intraperitoneal injection every day for the next seven days; immediately after each injection, animals were placed singly into clean cages without food, water or bedding (with orange scent), where they remained for 45 min before being returned to their home cage; this experimental paradigm provides unique context pairing with cocaine, important for locomotor sensitization and dendritic spine growth. Animals in the Saline group received 1.0 ml saline each day. Cocaine was dissolved in phosphate buffered saline (4 mg/ml). Animals in the Acute Cocaine group received six daily injections of saline; on Day 7 they received a single 20 mg/kg injection of cocaine and were sacrificed 30 min later. Animals in the Chronic Cocaine group received seven daily injections of 20 mg/kg cocaine and were sacrificed 30 min after the final injection.
Tissue (PFC and NAc) was harvested from coronal sections using a tissue punch, weighed, sonicated in 10 volumes of SDS-P lysis buffer (50 mM Tris, 1% SDS, 130 mM NaCl, 10 mM Na pyrophosphate, 50 mM NaF, 5 mM EDTA, pH 7.6) containing 1 mM PMSF, 1 mM orthovanadate and one PhosSTOP tablet (Roche)/10ml and heated at 95C for 5 min (Fig.1B). Samples were then centrifuged at 14,000 × g for 15 min; supernatants were transferred to new tubes, aliquots were taken for protein determination and samples were stored at -80C. Protein concentration was determined using the bicinchoninic acid assay (Pierce) with bovine serum albumin as a standard and ranged from 7 to 10 mg/ml.
Immunoprecipitation
Immunoprecipitation was carried out with slight modifications of the protocol described previously36 using affinity-purified antibody to the Sec14 domain of Kalirin22 For each sample, 0.8 volumes of 10% NP-40 (Pierce, SurfActs) treated with orthovanadate and PhosSTOP was added to an aliquot containing 2 mg protein; samples were allowed to rotate end over end in the cold room for 20 min and then centrifuged at 4C for 15 min at 14,000 × g to remove any insoluble material. Supernatants were then further diluted with 3.5 volumes TM-P (20 mM Na TES, 10 mM mannitol, 5 mM EDTA, 50 mM NaF, 10 mM Na pyrophosphate, pH 7.4) containing 1 mM orthovanadate, 1 tablet PhosSTOP/10 ml and Calbiochem Phosphatase Inhibitor Cocktail I. Each sample then rotated end over end overnight at 4C with ∼3 μg affinity-purified CT302 antibody (Sec14 specificity) and 10 μ Protein A beads that had been washed with TM-P-IPT buffer (TM-P buffer containing 1% TX-100). Beads were pelleted and washed twice with TM-P-IPT buffer and once with TM-P buffer. Bound protein was eluted by boiling the beads in Laemmli sample buffer treated with orthovanadate and Calbiochem Phosphatase Inhibitor Cocktail I. Samples fractionated on 10-lane BioRad 4-15% gels were visualized using the Pierce SilverSNAP kit; each band of Kal7 and Kal9/12 was excised and frozen until processing for mass spectroscopic analysis (Fig.1C). Simultaneous analysis of 50, 100 and 200 ng bovine serum albumin confirmed the recovery of about 200 ng (1 pmol) Kal7 in immunoprecipitates from 2 mg total protein (0.01% total protein); over 90% of the Kal7 in lysates was generally recovered in the immunoprecipitates.
Identification and quantitative analysis of phosphorylation sites
Immunopurified Kal7 samples were cleaved using in-gel tryptic digestion (V51111; Promega, Madison WI) and subjected to information-dependent acquisition and SWATH acquisition, as described36, 84-86. Skyline (v2.4)87 was employed for precursor ion quantification using SWATH data for the various phosphorylation sites. The approach to estimating the extent of phosphorylation at each site was the method described by Baucum et al.88 bearing in mind the cautions about absolute values due to ionization efficiencies. Depending on the peptide, various modifications were identified through the spectral library created in Skyline, most notably cysteine carboxamido-methylation, methionine oxidation and variable tryptic cleavage (2 miscleavages). Modifications were verified by manual inspection of the raw MS/MS fragmentation mass spectra for every assigned precursor mass containing the phosphorylation site. The areas under the isotopic envelope for the M, M+1 and M+2 isotopic peaks were calculated for each sample for all variants of each phosphorylated peptide (with all Cys, Met and tryptic variations included), and the ratio (phosphorylated/[phosphorylated + nonphosphorylated]) was calculated for the M, M+1 and M+2 ions for each sample. The data are expressed as the ratio of phosphorylated/total recovered peptide and statistical analyses were performed using a one-way ANOVA (SigmaPlot), as in88. Sites are numbered using AF230644_1 for rat cKal7. The data set is deposited at https://yped.med.yale.edu:8443/repository/ViewSeriesMenu.do?series_id=5016&series_name=Kal7+Phosphorylation+is+tissue-specific+and+responsive+to+cocaine.
Expression of Kalirin phosphorylation site mutants
Starting with the pEAK.HisMycKal7(a) expression vector12, the Stratagene QuikChange protocol (La Jolla, CA) was used to replace individual residues identified as phosphorylation sites with either a potentially phosphomimetic, acidic residue (E for T or Y; D for S) or a non-phosphorylatable residue (F for Y; A for S or T). Residues studied included: SS271,276 in spectrin repeat 1 (SR1); Y591 in SR4; Y616 in SR4; Y963 in SR7 (a known Abl1 site)12; Y1123 in SR9 (a predicted site for TrkB); T1303, Y1342 and TS1519,1520 in the GEF1 domain; and Y1653 (a known Fyn site)36 in the PDZ-binding motif. Several vectors were constructed with 3 and 4 phosphorylation sites mutated. All constructs were verified by DNA sequencing. Protein mass and stability were verified by Western blot analysis of transiently transfected HEK293 (pEAK Rapid) cells.
Use of Rac1 biosensor to quantify GEF activity
Dora-Rac1, a FRET biosensor for Rac1, was used to evaluate Rac1 activation in cells expressing each Kal7 mutant58, 89. Briefly, vector encoding the Dora-Rac1 biosensor and various amounts of vector encoding Myc-tagged Kalirin or KalGEF1 were transfected into HEK293 cells. Thirty hours after transfection, FRET ratios were measured at room temperature for each well, exciting at 420 nm and collecting spectra from 440 to 600 nm. Cells were harvested and protein expression levels were determined by Western blot analysis using antibodies against Myc or Kalirin. The FRET ratio (525/475) for each mutant was plotted against the observed protein expression level.
SH2 screening
Twelve amino acid peptides corresponding to the 4 internal sites of tyrosine phosphorylation (phosphorylated and non-phosphorylated) were synthesized with a biotinylated NH2-terminus and an amidated C-terminus (Biomatik, Wilmington DE): PEEI(p)Y616KAARHLEa; QAGH(p)Y963DADAIREa; QCQQ(p)Y1123VVFERSAa; ELEK(p)Y1342EQLPDEVa. SH2 domain screening was performed as described previously59, 60. Briefly, dissolved peptides were diluted with 2x spotting solution (100 mM Tris–HCl, pH6.8, 30% glycerol, 2% SDS) to approximately 10 μg/μL and spotted in duplicate in a rosette pattern on gelatin-coated nitrocellulose membranes. Membranes were blocked with 5% milk in TBST (25 mM Tris–HCl, pH 8.0, 150 mM NaCl, and 0.05 % (v/v) Tween-20) and incubated with 200 nM GST fusion proteins (71 SH2, 2 PTP, 1 PTB and GST control) for 2 h in a 96-well chamber plate, and subsequently with Dylight 800-conjugated anti-GST antibody (Rockland Immunochemicals) and IRDye 680RD-conjugated streptavidin (LI-COR Biotechnology). Membranes were washed with TBST and signal was quantified using an Odyssey IR scanner with ImageStudio software (LI-COR Biotechnology). The intensity of the biotin signal was used for normalization of SH2 binding values from the quadruplicate spots. The list of GST fusion probes used is provided in Supplemental Table 2. To classify SH2 domains by clustering analysis, the normalized values were imported to Cluster 3.0, median centered for each peptide, and processed with the average linkage algorithm. Dendrograms were constructed using TreeView.
Far-Western analysis of GST-SH2 binding to TrkB phosphorylated bKal7
Purified SR7:910 (0.5 μM) was phosphorylated by TrkB (50 nM), using the protocol specified by the manufacturer (SignalChem, Richmond, Canada). The reaction was terminated by adding 5X SDS sample loading buffer (10% SDS, 25% β-mercaptoethanol, 50% glycerol, 0.125 M Tris-HCl, pH6.8) and heating at 95C for 5 min. After SilverSNAP staining, a gel slice containing SR7:9 was excised and processed for mass spec analysis; pY1123 was the most abundant site identified, along with Y963, Y1000, Y1017, Y1165 and Y1211. His6-bKal711, 22 expressed in Hi5 cells was purified using Talon Superflow Metal Affinity Resin (#635506; Clontech Laboratories) equilibrated with 50 mM Na phosphate, 300 mM NaCl, 0.05% TX-100, pH 8.0, eluted with a gradient to 1.0 M imidazole in the same buffer titrated to pH 7.0 and dialyzed into 20 mM Na TES, 0.3 M NaCl, pH 8.0. Purified bKal7 was phosphorylated as described above but using 100 nM TrkB and 200 nM bKal7 with 4 mM ATP; TrkB was not added to the buffer control. Phosphorylated bKal7 was prepared for mass spec analysis as described for SR7:9; pY1123 was the major site identified. Based on the rosette screen and the biologically significant role played by Kalirin in TrkB signaling58, a subset of 12 GST-SH2 and -PTB domains were tested for their ability to bind to bKal7 phosphorylated in vitro by recombinant TrkB kinase. For Western blotting, TrkB-treated and untreated bKal7 proteins were separated on 4-12 % NuPAGE gels (Invitrogen), transferred to nitrocellulose, and probed with antibodies to phosphotyrosine (Cell Signaling), Kalirin22 and phosphorylated TrkB /pY816 (Ab75173, Abcam). Capillary far-Western analyses were performed using the WES Capillary Western instrument (ProteinSimple) following the manufacturer's instructions and the previously established far-Western blotting protocol60, 62, 90. The protein samples were diluted to 8 ng/μL and loaded into a 25-lane capillary cartridge, immobilized, and subsequently incubated with GST-SH2 probes and HRP-conjugated anti-GST antibody followed by chemiluminescent detection. Increased band peaks of tyrosine phosphorylated bKal7 were confirmed by Graph View (Compass 2.7.1).
Calpain sensitivity
HEK293 cells plated in 12-well dishes and transiently transfected (1 μg vector) 24 h earlier using TransIT-2020 (Mirus Bio, LLC) were harvested by scraping into the spent medium; cell pellets subjected to three freeze/thaw cycles in 200 μl phosphate buffered saline containing 1% TX-100 were clarified by centrifugation at 17,000 × g for 20 min. After addition of CaCl2 to 0.5 mM, μ-calpain (200 ng) was added to a 50 μl aliquot of the lysate and samples were incubated for 1 h at 37C. Proteolysis was stopped by the addition of Laemmli sample buffer and heating for 5 min at 95C. Aliquots (10 μl) of the original lysate and the calpain digested sample were fractionated on 4 to 15% gradient gels (BioRad); separate gels were used to analyze recovery of SR4:7 and C-terminal antigenicity. Non-saturated images were quantified using SynGene software. Predicted calpain cleavage sites are listed in Supplemental Table 4.
Statistics
Statistical tests (t-tests; RM-ANOVA) were performed using OpenOffice 4.0, Sigma Plot 11.0 or GraphPad Prism 7.0.
Supplementary Material
Supplemental Table S1. Kinase Prediction for Identified Kal7 Phosphorylation Sites. Predictions from GPS 3.0 web server phosphorylation site prediction program (http://gps.biocuckoo.org/) are shown, using only mammalian enzymes and human gene names for clarity. Enzymes in red have been shown to phosphorylate the sites indicated12, 36 (this work). Pale green shading denotes Kal7-only; tan shading denotes Kal9/12-only; blue shading denotes Kal12-only.
Supplemental Table S2. SH2 and other domain probe list. From left to right columns, Probe name, Official gene symbol, NCBI Gene site ID, amino acid position of the domain insert, and GST fusion structure are provided. The two phosphotyrosine phosphatase constructs are substrate-trapping mutants91.
Supplemental Table S3. SH2 screen data. Normalized binding signal values with SEM are given for all peptides and domain probes.
Supplemental Table S4. Calpain site predictions for Kal7. Predictions utilized the GPS-CCD program of Liu Z et al74.
Acknowledgments
We thank Darlene D'Amato, Yanping Wang, Edward Voss, and Jean Kanyo for invaluable technical support.
This work was supported by NIH grants DK032948 (BAE, REM), DA23082 (REM, BAE), CA1154966 (KM), Quest for CURES (Leukemia & Lymphoma Society; KM), 2 P30 DA018343 (TTL), GM117061 (YIW), CA133346 (AJK), NS089662 (AJK) and F31 MH105043 (ADL) and a grant from the Stanley Center for Psychiatric Research (AJK, BAE, REM).
Abbreviations
- Kal7
Kalirin7
- Kal9/12
Kalirin9 plus Kalirin12
- NAc
nucleus accumbens
- PFC
prefrontal cortex
- PTP
Protein Tyrosine Phosphatase
- PTB
Protein Tyrosine Binding
- SH2
Src Homology
Footnotes
Conflict of interest: None to declare.
Author contributions: MBM and YY performed experiments and wrote the manuscript
KM planned and performed SH2 profiling experiments and wrote the manuscript
DDK planned and performed experiments
TA and TTL performed the mass spectrometry
YIW created and validated key reagents
ADL and AJK expressed Kalirin using baculovirus vectors
BAE and REM planned and performed experiments and wrote the manuscript
References
- 1.Russell TA, Blizinsky KD, Cobia DJ, Cahill ME, Xie Z, Sweet RA, Duan J, Gejman PV, Wang L, Csernansky JG, Penzes P. A sequence variant in human KALRN impairs protein function and coincides with reduced cortical thickness. Nat Commun. 2014;5:4858. doi: 10.1038/ncomms5858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kushima I, Nakamura Y, Aleksic B, Ikeda M, Ito Y, Ozaki N. Resequencing and association analysis of the KALRN and EPHB1 genes and their contribution to schizophrenia susceptibility. Schizo Bull. 2012;38:552–560. doi: 10.1093/schbul/sbq118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deo AJ, Cahill ME, Li S, Goldszer I, Lewis DA, Penzes P, Sweet DA. Increased expression of Kalirin-9 in the auditory cortex of schizophrenia subjects: Its role in dendritic pathology. Neurobiol Disease. 2012;45:796–803. doi: 10.1016/j.nbd.2011.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hayashi-Takagi A, Takaki M, Graziane N, Seshadri S, Murdoch H, Dunlop AJ, Makino Y, Seshadri AJ, Ishizuka K, Srivastava DP, Xie Z, Baraban JM, Houslay MD, Tomoda T, Brandon NJ, Kamiya A, Yan Z, Penzes P, Sawa A. Disrupted-in-Schizophrenia 1 (DISC1) regulates spines of the glutamate synapse via Rac1. Nature Neurosci. 2010;13:327–332. doi: 10.1038/nn.2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hill JJ, Hashimoto T, Lewis DA. Molecular mechanisms contributing to dendritic spine alterations in the prefrontal cortex of subjects with schizophrenia. Mol Psychiatry. 2006;11:557–566. doi: 10.1038/sj.mp.4001792. [DOI] [PubMed] [Google Scholar]
- 6.Krug T, Manso H, Gouveia L, Sobral J, Xavier JM, Albergaria I, Gaspar G, Correia M, Viana-Baptista M, Simões RM, Pinto AN, Taipa R, Ferreira C, Fontes JR, Silva MR, Gabriel JP, Matos I, Lopes G, Ferro JM, Vicente AM, Oliveira SA. Kalirin: a novel genetic risk factor for ischemic stroke. Hum Genet. 2010;127:513–523. doi: 10.1007/s00439-010-0790-y. [DOI] [PubMed] [Google Scholar]
- 7.Beresewicz M, Kowalczyk JE, Zablocka B. Kalirin-7, a protein enriched in postsynaptic density, is involved in ischemic signal transduction. Neurochem Res. 2008;33:1789–1794. doi: 10.1007/s11064-008-9631-y. [DOI] [PubMed] [Google Scholar]
- 8.Makrythanasis P, Guipponi M, Santoni FA, Zaki M, Issa MY, Ansar M, Hamamy H, Antonarakis SE. Exome sequencing discloses KALRN homozygous variant as likely cause of intellectual disability and short stature in a consanguineous pedigree. Hum Genomics. 2016;10:26. doi: 10.1186/s40246-40016-40082-40242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peña-Oliver Y, Carvalho FM, Sanchez-Roige S, Quinlan EB, Jia T, Walker-Tilley T, Rulten SL, Pearl FM, Banaschewski T, Barker GJ, Bokde AL, Büchel C, Conrod PJ, Flor H, Gallinat J, Garavan H, Heinz A, Gowland P, Paillere Martinot ML, Paus T, Rietschel M, Robbins TW, Smolka MN, Schumann G, Stephens DN Consortium I. Mouse and Human Genetic Analyses Associate Kalirin with Ventral Striatal Activation during Impulsivity and with Alcohol Misuse. Front Genetics. 2016;7:52. doi: 10.3389/fgene.2017.00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vishwanatha KS, Wang Y, Keutmann HT, Mains RE, Eipper BA. Structural organization of the nine spectrin repeats of kalirin. Biochemistry. 2012;51:5663–5673. doi: 10.1021/bi300583s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miller MB, Yan Y, Wu Y, Hao B, Mains RE, Eipper BA. Alternate promoter usage generates two subpopulations of the neuronal RhoGEF Kalirin-7. J Neurochem. 2016;140:889–902. doi: 10.1111/jnc.13749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ma XM, Miller MB, Vishwanatha KS, Gross MJ, Wang Y, Abbott T, Lam TT, Mains RE, Eipper BA. Nonenzymatic domains of Kalirin7 contribute to spine morphogenesis through interactions with phosphoinositides and Abl. Mol Biol Cell. 2014;25:1458–1471. doi: 10.1091/mbc.E13-04-0215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mazzone CM, Larese TP, Kiraly DD, Eipper BA, Mains RE. Analysis of Kalirin-7 knockout mice reveals different effects in female mice. Mol Pharmacol. 2012;82:1241–1249. doi: 10.1124/mol.112.080838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lemtiri-Chlieh F, Zhao L, Kiraly DD, Eipper BA, Mains RE, Levine ES. Kalirin-7 is necessary for normal NMDA receptor-dependent synaptic plasticity. BMC Neurosci. 2011;12:126. doi: 10.1186/1471-2202-1112-1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kiraly DD, Ma XM, Mazzone CM, Xin X, Mains RE. Behavioral and Morphological Responses to Cocaine Require Kalirin7. Biol Psychiatry. 2010;68:249–255. doi: 10.1016/j.biopsych.2010.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ma XM, Kiraly DD, Gaier ED, Wang Y, Kim EJ, Levine ES, Eipper BA, Mains RE. Kalirin-7 Is Required for Synaptic Structure and Function. J Neurosci. 2008;28:12368–12382. doi: 10.1523/JNEUROSCI.4269-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mandela P, Yan Y, Larese T, Eipper BA, Mains RE. Elimination of Kalrn Expression in POMC Cells Reduces Anxiety-Like Behavior and Contextual Fear Learning. Horm Behav. 2014;66:430–438. doi: 10.1016/j.yhbeh.2014.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang S, Elensite PP, Wayakanon K, Mandela P, Eipper BA, Mains RE, Allen MR, Bruzzaniti A. The Rho-GEF Kalirin regulates bone mass and the function of osteoblasts and osteoclasts. Bone. 2013;60:235–245. doi: 10.1016/j.bone.2013.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu JH, Fanaroff AC, Sharma KC, Smith LS, Brian L, Eipper BA, Mains RE, Freedman NJ, Zhang L. Kalirin promotes neointimal hyperplasia by activating Rac in smooth muscle cells. Arterioscler Thromb Vasc Biol. 2013;33:702–708. doi: 10.1161/ATVBAHA.112.300234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kiraly DD, Lemtiri-Chlieh F, Levine ES, Mains RE, Eipper BA. Kalirin Binds the NR2B Subunit of the NMDA Receptor, Altering Its Synaptic Localization and Function. J Neurosci. 2011;31:12554–12565. doi: 10.1523/JNEUROSCI.3143-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Briz V, Baudry M. Calpains: Master Regulators of Synaptic Plasticity. Neuroscientist. 2016;2016 doi: 10.1177/1073858416649178. [DOI] [PubMed] [Google Scholar]
- 22.Miller MB, Vishwanatha KS, Mains RE, Eipper BA. An N-terminal Amphipathic Helix Binds Phosphoinositides and Enhances Kalirin Sec14 Domain-mediated Membrane Interactions. J Biol Chem. 2015;290:13541–13555. doi: 10.1074/jbc.M115.636746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones KA, Srivastava DP, Allen JA, Strachan RT, Roth BL, Penzes P. Rapid modulation of spine morphology by the 5-HT2A serotonin receptor through kalirin-7 signaling. Proc Natl Acad Sci USA. 2009;106:19575–19580. doi: 10.1073/pnas.0905884106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cahill ME, Jones KA, Rafalovich I, Xie Z, Barros CS, Müller U, Penzes P. Control of interneuron dendritic growth through NRG1/erbB4-mediated kalirin-7 disinhibition. Mol Psych. 2012;17:99–107. doi: 10.1038/mp.2011.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xie Z, Srivastava DP, Photowala H, Kai L, Cahill ME, Woolfrey KM, Shum CY, Surmeier DJ, Penzes P. Kalirin-7 controls activity-dependent structural and functional plasticity of dendritic spines. Neuron. 2007;56:640–656. doi: 10.1016/j.neuron.2007.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liebl EC, Forsthoefel DJ, Franco LS, Sample SH, Hess JE, Cowger JA, Chandler MP, Shupert AM, Seeger MA. Dosage-sensitive, reciprocal genetic interactions between the Abl tyrosine kinase and the putative GEF trio reveal trio's role in axon pathfinding. Neuron. 2000;26:107–118. doi: 10.1016/s0896-6273(00)81142-3. [DOI] [PubMed] [Google Scholar]
- 27.Bateman J, Shu H, Van Vactor D. The Guanine Nucleotide Exchange Factor Trio Mediates Axonal Development in the Drosophila Embryo. Neuron. 2000;26:93–106. doi: 10.1016/s0896-6273(00)81141-1. [DOI] [PubMed] [Google Scholar]
- 28.Vickers CA, Stephens B, Bowen J, Arbuthnotta GW, Grant SG, Ingham CA. Neurone specific regulation of dendritic spines in vivo by post synaptic density 95 protein (PSD-95) Brain Res. 2006;1090:89–98. doi: 10.1016/j.brainres.2006.03.075. [DOI] [PubMed] [Google Scholar]
- 29.Elias GM, Funke L, Stein V, Grant SG, Bredt DS, Nicoll RA. Synapse-specific and developmentally regulated targeting of AMPA receptors by a family of MAGUK scaffolding proteins. Neuron. 2006;52:307–320. doi: 10.1016/j.neuron.2006.09.012. [DOI] [PubMed] [Google Scholar]
- 30.Collins MO, Husi H, Yu L, Brandon JM, Anderson CN, Blackstock WP, Choudhary JS, Grant SG. Molecular characterization and comparison of the components and multiprotein complexes in the postsynatpic proteome. J Neurochem. 2006;96(1):16–23. doi: 10.1111/j.1471-4159.2005.03507.x. [DOI] [PubMed] [Google Scholar]
- 31.Pocklington AJ, Cumiskey M, Armstrong JD, Grant SG. The proteomes of neurotransmitter receptor complexes form modular networks with distributed functionality underlying plasticity and behavior. Mol Syst Biol. 2006;2:1–14. doi: 10.1038/msb4100041. 2006.0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grant SG. The synapse proteome and phosphoproteome: a new paradigm for synapse biology. Biochem Soc Trans. 2006;34:59–63. doi: 10.1042/BST0340059. [DOI] [PubMed] [Google Scholar]
- 33.Carlin RK, Grab DJ, Cohen RS, Siekevitz P. Isolation and characterization of postsynaptic densities from various brain regions: enrichment of different types of postsynaptic densities. J Cell Biol. 1980;86:831–845. doi: 10.1083/jcb.86.3.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen Q, Zhu X, Zhang Y, Wetsel WC, Lee TH, Zhang X. Integrin-linked kinase is involved in cocaine sensitization by regulating PSD-95 and Synapsin 1 expression and GluR1 Ser845 phosphorylation. J Mol Neurosci. 2010;40:284–294. doi: 10.1007/s12031-009-9218-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peng HY, Chen GD, Lai CY, Hsieh MC, Lin TB. Spinal serum-inducible and glucocorticoid-inducible kinase 1 mediates neuropathic pain via kalirin and downstream PSD-95- dependent NR2B phosphorylation in rats. J Neurosci. 2013;33:5227–5240. doi: 10.1523/JNEUROSCI.4452-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kiraly DD, Stone KL, Colangelo CM, Abbott T, Wang Y, Mains RE, Eipper BA. Identification of kalirin-7 as a potential post-synaptic density signaling hub. J Proteom Res. 2011;10:2828–2841. doi: 10.1021/pr200088w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang X, Cahill ME, Werner CT, Christoffel DJ, Golden SA, Xie Z, Loweth JA, Marinelli M, Russo SJ, Penzes P, Wolf ME. Kalirin-7 mediates cocaine-induced AMPA receptor and spine plasticity, enabling incentive sensitization. J Neurosci. 2013;33:11012–11022. doi: 10.1523/JNEUROSCI.1097-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hodes GE, Pfau ML, Purushothaman I, Ahn HF, Golden SA, Christoffel DJ, Magida J, Brancato A, Takahashi A, Flanigan ME, Ménard C, Aleyasin H, Koo JW, Lorsch ZS, Feng J, Heshmati M, Wang M, Turecki G, Neve R, Zhang B, Shen L, Nestler EJ, Russo SJ. Sex Differences in Nucleus Accumbens Transcriptome Profiles Associated with Susceptibility versus Resilience to Subchronic Variable Stress. J Neurosci. 2015;35:16362–16376. doi: 10.1523/JNEUROSCI.1392-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nestler EJ. Cellular basis of memory for addiction. Dialogues Clin Neurosci. 2013;15:431–443. doi: 10.31887/DCNS.2013.15.4/enestler. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gipson CD, Kupchik YM, Kalivas PW. Rapid, transient synaptic plasticity in addiction. Neuropharmacology. 2014;76 pt B:276–286. doi: 10.1016/j.neuropharm.2013.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li X, Wolf ME. Multiple faces of BDNF in cocaine addiction. Behav Brain Res. 2015;279:240–254. doi: 10.1016/j.bbr.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miller MB, Yan Y, Eipper BA, Mains RE. Neuronal Rho GEFs in Synaptic Physiology and Behavior. Neuroscientist. 2013;19:255–273. doi: 10.1177/1073858413475486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Herring BE, Nicoll RA. Kalirin and Trio proteins serve critical roles in excitatory synaptic transmission and LTP. Proc Natl Acad Sci USA. 2016;113:2264–2269. doi: 10.1073/pnas.1600179113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Russo SJ, Dietz DM, Dumitriu D, Morrison JH, Malenka RC, Nestler EJ. The addicted synapse: mechanisms of synaptic and structural plasticity in nucleus accumbens. Trends Neurosci. 2010;33:267–276. doi: 10.1016/j.tins.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Koo TH, Eipper BA, Donaldson JG. Arf6 recruits the Rac GEF Kalirin to the plasma membrane facilitating Rac activation. BMC CellBiol. 2007;8:29. doi: 10.1186/1471-2121-1188-1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Linding R, Jensen LJ, Diella F, Bork P, Gibson TJ, Russell RB. Protein disorder prediction: implications for structural proteomics. Structure. 2003;11:1453–1459. doi: 10.1016/j.str.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 47.Gnad F, Ren S, Cox J, Olson JV, Macek B, Oroshi M, Mann M. PHOSIDA (phosphorylation site database): management, structural and evolutionary investigation, and prediction of phosphosites. Genome Biol. 2007;8:R250. doi: 10.1186/gb-2007-1188-1111-r1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liao L, Sando RC, Farnum JB, Vanderklish PW, Maximov A, Yates JR. 15N- labeled brain enables quantification of proteome and phosphoproteome in cultured primary neurons. J Proteom Res. 2012;11:1341–1353. doi: 10.1021/pr200987h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu P, Pisitkun T, Wang G, Wang R, Katagiri Y, Gucek M, Knepper MA, Geller HM. Global analysis of neuronal phosphoproteome regulation by chondroitin sulfate proteoglycans. PLoS One. 2013;8:e59285. doi: 10.1371/journal.pone.0059285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huttlin EL, Jedrychowski MP, Elias JE, Goswami T, Rad R, Beausoleil SA, LVillen J, Haas W, Sowa ME, Gygi SP. A tissue-specific atlas of mouse protein phosphorylation and expression. Cell. 2010;143:1174–1189. doi: 10.1016/j.cell.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang F, Blanchard AP, Elisma F, Granger M, Xu H, Bennett SA, Figeys D, Zou H. Phosphoproteome analysis of an early onset mouse model (TgCRND8) of Alzheimer's disease reveals temporal changes in neuronal and glia signaling pathways. Proteomics. 2013;13:1292–1305. doi: 10.1002/pmic.201200415. [DOI] [PubMed] [Google Scholar]
- 52.Wolf ME, Tseng KY. Calcium-permeable AMPA receptors in the VTA and nucleus accumbens after cocaine exposure: when, how, and why? Front Mol Neurosci. 2012;5:72. doi: 10.3389/fnmol.2012.00072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xue Y, Liu Z, Cao J, Ma Q, Gao X, Wang Q, Jin C, Zhou Y, Wen L, Ren J. GPS 2.1: enhanced prediction of kinase-specific phosphorylation sites with an algorithm of motif length selection. Protein Eng Des Sel. 2011;24:255–260. doi: 10.1093/protein/gzq094. [DOI] [PubMed] [Google Scholar]
- 54.Brand F, Schumacher S, Kant S, Menon MB, Simon R, Turgeon B, Britsch S, Meloche S, Gaestel M, Kotlyarov A. The Extracellular Signal-Regulated Kinase 3 (Mitogen- Activated Protein Kinase 6 [MAPK6])-MAPK-Activated Protein Kinase 5 Signaling Complex Regulates Septin Function and Dendrite Morphology. Mol Cell Biol. 2012;32:2467–2478. doi: 10.1128/MCB.06633-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schiller MR, Chakrabarti K, King GF, Schiller NI, Eipper BA, Maciejewski MW. Regulation of RhoGEF activity by intramolecular and intermolecular SH3 interactions. J Biol Chem. 2006;281:18774–18786. doi: 10.1074/jbc.M512482200. [DOI] [PubMed] [Google Scholar]
- 56.Xin X, Wang Y, Ma XM, Rompolas P, Keutmann HT, Mains RE, Eipper BA. Regulation of kalirin by cdk5. J Cell Sci. 2008;121:2601–2611. doi: 10.1242/jcs.016089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chakrabarti K, Lin R, Schiller NI, Wang Y, Koubi D, Fan YX, Rudbin BB, Johnson GR, Schiller MR. Critical role for Kalirin in nerve growth factor signaling through TrkA. Mol Cell Biol. 2005;25:5106–5118. doi: 10.1128/MCB.25.12.5106-5118.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yan Y, Eipper BA, Mains RE. Kalirin is required for BDNF-TrkB stimulated neurite outgrowth and branching. Neuropharmacology. 2016;107:227–238. doi: 10.1016/j.neuropharm.2016.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ng KY, Machida K. Rosette assay: highly customizable dot-blot for SH2 domain screening. Mtds Mol Biol. 2017;1555:437–451. doi: 10.1007/978-1-4939-6762-9_26. [DOI] [PubMed] [Google Scholar]
- 60.Machida K, Thompson CM, Dierck K, Jablonowski K, Kärkkäinen S, Liu B, Zhang H, Nash PD, Newman DK, Nollau P, Pawson T, Renkema GH, Saksela K, Schiller MR, Shin DG, Mayer BJ. High throughput phosphotyrosine profiling using SH2 domains. Mol Cell. 2007;26:899–915. doi: 10.1016/j.molcel.2007.05.031. [DOI] [PubMed] [Google Scholar]
- 61.Liu BA, Jablonowski K, Raina M, Arcé M, Pawson T, Nash PD. The Human and Mouse Complement of SH2 Domain Proteins—Establishing the Boundaries of Phosphotyrosine Signaling. Mol Cell. 2006;22:851–868. doi: 10.1016/j.molcel.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 62.Jadwin JS, Mayer BJ, Machida K. Detection and Quantification of Protein–Protein Interactions by Far-Western Blotting. Mtds Mol Biol. 2015;1312:379–398. doi: 10.1007/978-1-4939-2694-7_38. [DOI] [PubMed] [Google Scholar]
- 63.Machida K, Mayer BJ. Detection of Protein–Protein Interactions by Far-Western Blotting. Mtds Mol Biol. 2009;536:313–329. doi: 10.1007/978-1-59745-542-8_34. [DOI] [PubMed] [Google Scholar]
- 64.Mayer BJ. Perspective: Dynamics of receptor tyrosine kinase signaling complexes. FEBS Lett. 2012;586:2575–2579. doi: 10.1016/j.febslet.2012.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Huang H, Li L, Wu C, Schibli D, Gao Y, Li SS. Defining the specificity space of the human SCR homology 2 domain. Mol Cell Proteomics. 2008;7:768–784. doi: 10.1074/mcp.M700312-MCP200. [DOI] [PubMed] [Google Scholar]
- 66.Jones N, Blasutig IM, Eremina V, Ruston JM, Bladt F, Li H, Huang H, Larose L, Li SS, Takano T, Quaggin SE, Pawson T. Nck adaptor proteins link nephrin to the actin cytoskeleton of kidney podocytes. Nature. 2006;440:818–823. doi: 10.1038/nature04662. [DOI] [PubMed] [Google Scholar]
- 67.Pereira DB, Chao MV. The tyrosine kinase Fyn determines the localization of TrkB receptors in lipid rafts. J Neurosci. 2007;27:4859–4869. doi: 10.1523/JNEUROSCI.4587-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Trepanier CH, Jackson MF, MacDonald JF. Regulation of NMDA receptors by the tyrosine kinase Fyn. FEBS J. 2012;279:12–19. doi: 10.1111/j.1742-4658.2011.08391.x. [DOI] [PubMed] [Google Scholar]
- 69.Arévalo JC, Pereira DB, Yano H, Teng KK, Chao MV. Identification of a switch in neurotrophin signaling by selective tyrosine phosphorylation. J Biol Chem. 2006;281:1001–1007. doi: 10.1074/jbc.M504163200. [DOI] [PubMed] [Google Scholar]
- 70.Simpson MA, Bradley WD, Harburger D, Parsons M, Calderwood DA, Koleske AJ. Direct interactions with the integrin β1 cytoplasmic tail activate the Abl2/Arg kinase. J Biol Chem. 2015;290:8360–8372. doi: 10.1074/jbc.M115.638874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wu HY, Lynch DR. Calpain and Synaptic Function. Mol Neurobiol. 2006;33:215–236. doi: 10.1385/MN:33:3:215. [DOI] [PubMed] [Google Scholar]
- 72.Khoutorsky A, Spira ME. Activity-dependent calpain activation plays a critical role in synaptic facilitation and post-tetanic potentiation. Learn Mem. 2009;16:129–141. doi: 10.1101/lm.1275709. [DOI] [PubMed] [Google Scholar]
- 73.Tompa P, Buzder-Lantos P, Tantos A, Farkas A, Szilágyi A, Bánóczi Z, Hudecz F, Friedrich P. On the sequential determinants of calpain cleavage. J Biol Chem. 2004;279:20775–20785. doi: 10.1074/jbc.M313873200. [DOI] [PubMed] [Google Scholar]
- 74.Liu Z, Cao J, Gao X, Ma Q, Ren J, Xue Y. GPS-CCD: a novel computational program for the prediction of calpain cleavage sites. PLoS ONE. 2011;6:e19001. doi: 10.1371/journal.pone.0019001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ma XM, Wang Y, Ferraro F, Mains RE, Eipper BA. Kalirin-7 is an essential component of both shaft and spine excitatory synapses in hippocampal interneurons. J Neurosci. 2008;28:711–724. doi: 10.1523/JNEUROSCI.5283-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Okubo-Suzuki R, Okada D, Sekiguchi M, Inokuchi K. Synaptopodin maintains the neural activity-dependent enlargement of dendritic spines in hippocampal neurons. Mol Cell Neurosci. 2008;38:266–276. doi: 10.1016/j.mcn.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 77.Sato K, Sugiyama T, Nagase T, Kitade Y, Ueda H. Threonine 680 phosphorylation of FLJ00018/PLEKHG2, a Rho family-specific guanine nucleotide exchange factor, by epidermal growth factor receptor signaling regulates cell morphology of Neuro-2a cells. J Biol Chem. 2014;289:10045–10056. doi: 10.1074/jbc.M113.521880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gupta M, Qi X, Thakur V, Manor D. Tyrosine phosphorylation of Dbl regulates GTPase signaling. J Biol Chem. 2014;289:17195–17202. doi: 10.1074/jbc.M114.573782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Llavero F, Urzelai B, Osinalde N, Gálvez P, Lacerda HM, Parada LA, Zugaza JL. Guanine nucleotide exchange factor aPIX leads to activation of the Rac 1 GTPase/glycogen phosphorylase pathway in interleukin (IL)-2-stimulated T cells. J Biol Chem. 2015;290:9171–9182. doi: 10.1074/jbc.M114.608414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang J, Ren J, Wu B, Feng S, Cai G, Tuluc F, Peränen J, Guo W. Activation of Rab8 guanine nucleotide exchange factor Rabin8 by ERK1/2 in response to EGF signaling. Proc Natl Acad Sci USA. 2015;112:148–153. doi: 10.1073/pnas.1412089112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lutz S, Shankaranarayanan A, Coco C, Ridilla M, Wieland T, Tesmer JJG. Structure of G-alpha-q-p63RhoGEF-RhoA Complex Reveals a Pathway for the Activation of RhoA by GPCRs. Science. 2007;318:1923–1927. doi: 10.1126/science.1147554. [DOI] [PubMed] [Google Scholar]
- 82.Ma XM, Huang JP, Xin X, Wang Y, Mains RE, Eipper BA. A Role for Kalirin in the Response of Rat Medium Spiny Neurons to Cocaine. Mol Pharmacol. 2012;82:738–745. doi: 10.1124/mol.112.080044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Siddoway B, Hou H, Yang H, Petralia R, Xia H. Synaptic activity bidirectionally regulates a novel sequence-specific S-Q phosphoproteome in neurons. J Neurochem. 2014;128:841–851. doi: 10.1111/jnc.12487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wu JX, Song X, Pascovici D, Zaw T, Care N, Krisp C, Molloy MP. SWATH Mass Spectrometry Performance Using Extended Peptide MS/MS Assay Libraries. Mol Cell Proteomics. 2016;15:2501–2514. doi: 10.1074/mcp.M115.055558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schubert OT, Gillet LC, Collins BC, Navarro P, Rosenberger G, Wolski WE, Lam H, Amodei D, Mallick P, MacLean B, Aebersold R. Building high-quality assay libraries for targeted analysis of SWATH MS data. Nat Protoc. 2015;10:426–441. doi: 10.1038/nprot.2015.015. [DOI] [PubMed] [Google Scholar]
- 86.Kelstrup CD, Jersie-Christensen RR, Batth TS, Arrey TN, Kuehn A, Kellmann M, Olsen JV. Rapid and Deep Proteomes by Faster Sequencing on a Benchtop Quadrupole Ultra-High-Field Orbitrap Mass Spectrometer. J Proteome Res. 2014;13:6187–6195. doi: 10.1021/pr500985w. [DOI] [PubMed] [Google Scholar]
- 87.MacLean B, Tomazela DM, Shulman N, Chambers M, Finney GL, Frewen B, Kern R, Tabb DL, Liebler DC, MacCoss MJ. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26:966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Baucum AJ, Shonesy BC, Rose KL, Colbran RJ. Quantitative proteomics analysis of CaMKII phosphorylation and the CaMKII interactome in the mouse forebrain. ACS Chem Neurosci. 2015;6:615–631. doi: 10.1021/cn500337u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Das S, Yin T, Yang Q, Zhang J, Wu YI, Yu J. Single-molecule tracking of small GTPase Rac1 uncovers spatial regulation of membrane translocation and mechanism for polarized signaling. Proc Natl Acad Sci USA. 2015;112:E6067–E6076. doi: 10.1073/pnas.1409667112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Jadwin JA, Oh D, Curran TG, Ogiue-Ikeda M, Jia L, White FM, Machida K, Yu J, Mayer BJ. Time-resolved multimodal analysis of Src Homology 2 (SH2) domain binding in signaling by receptor tyrosine kinases. eLife. 2016;5:e11835. doi: 10.7554/eLife.11835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Brady-Kalnay SM, Rimm DL, Tonks NK. Receptor Protein Tyrosine Phosphatase PTPμ Associates with Cadherins and Catenins In Vivo. J Cell Biol. 1995;130:977–986. doi: 10.1083/jcb.130.4.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table S1. Kinase Prediction for Identified Kal7 Phosphorylation Sites. Predictions from GPS 3.0 web server phosphorylation site prediction program (http://gps.biocuckoo.org/) are shown, using only mammalian enzymes and human gene names for clarity. Enzymes in red have been shown to phosphorylate the sites indicated12, 36 (this work). Pale green shading denotes Kal7-only; tan shading denotes Kal9/12-only; blue shading denotes Kal12-only.
Supplemental Table S2. SH2 and other domain probe list. From left to right columns, Probe name, Official gene symbol, NCBI Gene site ID, amino acid position of the domain insert, and GST fusion structure are provided. The two phosphotyrosine phosphatase constructs are substrate-trapping mutants91.
Supplemental Table S3. SH2 screen data. Normalized binding signal values with SEM are given for all peptides and domain probes.
Supplemental Table S4. Calpain site predictions for Kal7. Predictions utilized the GPS-CCD program of Liu Z et al74.


