Abstract
The epidermis is maintained by multiple stem cell populations whose progeny differentiate along diverse, and spatially distinct, lineages. Here we show that the transcription factor Gata6 controls the identity of the previously uncharacterized sebaceous duct (SD) lineage and identify the Gata6 down-stream transcription factor network that specifies a lineage switch between sebocytes and SD cells. During wound healing differentiated Gata6+ cells migrate from the SD into the interfollicular epidermis and dedifferentiate, acquiring the ability to undergo long-term self-renewal and differentiate into a much wider range of epidermal lineages than in undamaged tissue. Our data not only demonstrate that the structural and functional complexity of the junctional zone is regulated by Gata6, but also reveal that dedifferentiation is a previously unrecognized property of post-mitotic, terminally differentiated cells that have lost contact with the basement membrane. This resolves the long-standing debate about the contribution of terminally differentiated cells to epidermal wound repair.
Introduction
It is now recognised that the regenerative potential of a tissue relies on cellular plasticity, which involves the loss of homeostatic restrictions and the acquisition of new features by both adult stem cells and committed cells1–7. In the simple epithelia of intestine and lung, dedifferentiation following injury has recently been reported. In the gut, villin-negative cells revert to a stem cell state2, while in the lung committed airway epithelial cells revert to stem cells that are able to persist in the tissue following repair1.
In the epidermis there are several stem cell populations that maintain distinct compartments during homeostasis, but are able to contribute to additional lineages during wound repair. For example, in the junctional zone (JZ), the intersection between the hair follicle (HF), sebaceous gland (SG) and interfollicular epidermis (IFE), the resident Lrig1+ stem cells normally maintain the sebaceous gland and the infundibulum, but they can also form the IFE, SG and HF following injury or transplantation3, 8. Nevertheless, epidermal dedifferentiation has not been observed, potentially because is a rarer or lineage-specific event compared to other epithelia or because of the architectural complexity of the tissue. In the epidermis, in contrast to simple epithelia, stem cells detach from the basement membrane during terminal differentiation9–11. In the lower growing HF, committed stem cell progeny are not able to revert into stem cells upon injury, even when the stem cell reservoir is depleted12.
Given the architectural complexity of the JZ we set out to identify transcriptional regulators of lineage differentiation within this region and subsequently to investigate the fate of the differentiated cells during wound healing. In so doing, we have revealed a role for Gata6 in regulating the sebaceous duct lineage and shown that Gata6 lineage cells are able to undergo dedifferentiation into stem cells following injury.
Results
Gata6 is a marker of the sebaceous duct lineage
We have previously described a transgenic mouse line, K14ΔNLef1, in which inhibition of epidermal Wnt signaling results in conversion of hair follicles into sebocytes and multilayered epidermal cysts13,14 (Supplementary Fig.1a). One of the earliest changes to occur in K14ΔNLef1 mice is an expansion of the JZ (Fig. 1a). We compared previously published signature genes for Lrig1+ cells in wild type mice3 with genes that were upregulated in K14ΔNLef1 epidermal cells compared to littermate controls (Fig. 1b and Supplementary Fig. 1b). Transcription factor enrichment analysis of the promoters of genes that were upregulated in both datasets identified the GATA motif as being the most significantly enriched (Fig. 1b).
Figure 1. Identification of Gata6 as a marker of the sebaceous duct lineage.
a. Wild type (WT) and K14ΔNLef1 hair follicles from back skin labelled with anti- Lrig1 antibody. Asterisk indicates nonspecific staining of hair shaft. b, DNA motif enrichment analysis on the promoters of up-regulated genes in K14ΔNLef1 vs WT epidermal cells and in WT Lrig1+ cells. TF = Transcription Factor. c, WT and K14ΔNLef1 back skin sections stained for Krt14 and Gata6. d, Gata6 expression in the JZ/SD. Gata6-tdTomato reporter epidermal tail whole mount showing tdTomato and lipid labeling. e, Back skin section of WT hair follicle stained with antibodies to Gata6 and Itga6 (bottom panel). f, Gene Ontology enrichment analysis of Gata6 direct target genes is dependent on the distance between the transcription start sites (TSS) and Gata6 ChIP-Seq peaks. Kbp=kilobase pair. g-i, Overexpression of Gata6 decreases proliferation rate and promotes cell migration. g, h % confluence (g) and % scratch wound re-epithelialisation (h) of mouse primary keratinocytes infected with empty vector or Gata6 lentivirus. Data are means of n=4 (g) or n=3 (h) independent replicates ± s.e.m. Representative images from n=2 independent experiments of scratch wound assay are shown (i). Scale bars: 50 μm
[AU: in fig.2 is not clear whether the panels below (e) and (f) belong to (e), (f) or represent an independent panel altogether. Please clarify restructuring or adding labels to the figure. Error bars for some of the panels are missing too (e.g. magnification in Fig.2f, etc). Please also specify how many experiments the images are representative of.]
Since Gata factors are known to regulate differentiation15, 16 and cell migration17, we hypothesized that Gata factors might regulate exit from the JZ stem cell compartment. Although Gata6 expression has been recently reported in the IFE and lower HF18, we found that Gata6 expression was confined to the upper SG and JZ in adult epidermis (Fig. 1c) consistent with single cell transcriptomic data19. In K14ΔNLef1 epidermis Gata6 was highly upregulated and was expressed in the expanded JZ and in developing cysts (Fig. 1c and Supplementary Fig. 1c). No other Gata family members, such as Gata4 or Gata316 were detected.
In adult epidermis the stem cell populations in the JZ are characterized by expression of Lgr620 and Lrig1, both of which produce daughter cells that differentiate into SG. A subpopulation of Gata6+ cells co-expressed these markers (Supplementary Fig. 1d, e). Genetic lineage tracing experiments using Lgr6-EGFP-ires-CreERT221 crossed with Rosa26-fl/STOP/fl-tdTomato reporter mice confirmed the progeny of Lgr6+ stem cells included Gata6+ cells (Supplementary Fig. 1d). Gata6 was not expressed by CD34 expressing stem cells in the hair follicle bulge, which lies below the JZ (Supplementary Fig. 1e, i), and there was no co-expression of Gata6 with Tcf3/4 transcription factors required for bulge homeostasis22. However there was some co-expression of Gata6 with Sox9, a crucial transcription factor for maintenance of the sebaceous gland and hair follicle23 (Supplementary Fig. 1e)
To study Gata6+ cells we used a Gata6 reporter mouse in which the endogenous Gata6 promoter drives expression of tdTomato24 (Supplementary Fig. 1f-i). By characterizing the location of tdTomato+ cells in Z stacks of tail epidermal whole mounts, we found that Gata6 was not only expressed in the JZ but also in the SG duct25 that connects the upper SG to the JZ and acts as a conduit for release of sebum onto the skin surface (Fig. 1d and Supplementary Movie 1). We also observed that Gata6 was strongly upregulated in cells that had detached from the basement membrane (Fig. 1e).
Gata6 regulates sebaceous duct differentiation
To identify Gata6 target genes we performed ChIP-Seq with two different Gata6 antibodies. We found 12317 genomic targets with the expected Gata6 DNA motif (Supplementary Fig. 1j-m). We discovered that Gata6 regulates the promoters of genes involved in cellular processes such as “cell cycle” or “RNA splicing” and binds the distant regions of genes involved in processes such as “tube development” and “cell motility” (Fig. 1f). Consistent with the Gene ontology (GO), we found that Gata6 negatively regulates proliferation of cultured mouse epidermal cells and promotes migration in a scratch wound assay (Fig 1g-i).
We then compared the gene expression profiles of four subpopulations of epidermal cells from wild type mice in the resting (telogen) phase of the hair cycle: Gata6+Itga6+, Gata6+Itga6-, CD34+Itga6+ cells and all remaining Itga6+ cells (Fig. 2a, b; Supplementary Fig. 2a). GO analysis of differentially expressed gene clusters indicated Gata6+Itga6- cells were non-dividing cells (Fig. 1c) that expressed several terminal differentiation markers, including Involucrin and Blimp126 (expressed in the IFE, JZ and sebaceous duct), Plet1, Krt79 and Atp6v1c2 (expressed in SD but not IFE) (Fig. 2d-f, Supplementary Fig. 2b-j). The vast majority of Gata6+ cells had a suprabasal location. Those cells that were present in the basal layer showed little or no Ki67 labelling, and expressed terminal differentiation markers such as Fabp5 (Fig. 2f). Whereas the majority of Gata6+ cells in the SD and JZ expressed Blimp1, Blimp1+ cells in the differentiating layers of the IFE were Gata6-(Supplementary Fig. 2b, c). These results are summarized schematically in Fig. 2h.
Figure 2. Architectural characterization of the junctional zone.
a, Flow sorting of basal JZ/SD cells (green), suprabasal duct cells (violet), bulge cells (yellow) and all remaining basal cells (grey) (see schematic in b). b, c, Transcriptome analysis of the 4 sorted populations from (a) (n=3 mice) and Gene Ontology analysis of the 3 gene clusters highlighted in the heat map (c). d-f, Gata6 positive cells express the late differentiation markers Blimp1 (d) and Krt79 (e) and the early differentiation marker Fabp541 (f, g). Wild type skin sections were stained with the antibodies shown. Boxed areas are shown at higher magnification. Single fluorescent channel images are also shown (g). Red arrows mark Fabp5-positive basal cells; yellow arrows Fabp5-negative basal cells in the JZ/SD and IFE (g). Basal Gata6+ cells in the JZ/SD coexpress Fabp5 (red arrow and framed magnification view of the Fabp5 single fluorescent channel) while some Gata6- cells do not (yellow arrow and framed magnification view of the Fabp5 single fluorescent channel). Representative images of n=3 independent experiments. h, Wild type back skin sections in telogen (left panel) and anagen (central panel) stained for Gata6 and Ki67. Right hand panels show single fluorescent channel images at higher magnification (from left: DAPI, Gata6, Ki67) of the white inserts. Representative images of n=3 independent experiments i, Schematic of Gata6 expressing cells in the JZ/SD area. Dashed lines demarcate epidermal-dermal boundaries. Scale bars: 50 μm (d,h), 25 μm (e-g).
Our analysis predicted that Gata6 regulates differentiation of JZ cells into the SD lineage. To test this we performed gain and loss of function experiments. In Gata6-tdTomato reporter mice insertion of tdTomato leads to loss of one Gata6 allele. By crossing the reporter mice with mice that are heterozygous for epidermal-specific loss of Gata6 (K5cre x Gata6 Flox27 - Supplementary Fig. 3a) it was possible to analyse the Gata6+ cell population in the absence of Gata6 expression. By flow cytometry we showed that in the absence of Gata6 the number of bulge cells (CD34+Itga6+) was unaffected. In contrast, the numbers of dtTomato+Gata6+Itga6+ JZ cells and tdTomato+Gata6+Itga6- SD cells were reduced by 50% (Fig. 3a, Supplementary Fig. 3b). These results were confirmed by immunofluorescence and RT-qPCR (Supplementary Fig. 3c-e). Consistent with our in vitro data, Gata6 genetic ablation in K14ΔNLef1 mice led to increased expression of several direct target genes involved in mitosis (Supplementary Fig. 3f-h).
Figure 3. Gata6 controls the identity of the sebaceous duct (SD) lineage and distinguishes the sebocyte and SD lineages.
a, Flow cytometry analysis (dot plots and quantification) of basal JZ cells (Itga6+Gata6+), differentiated SD cells (Itga6-Gata6+) and bulge cells (Itga6+Cd34+), shows loss of the Gata6 expressing duct lineage upon genetic ablation of Gata6. Data are means ± s.d. from n=3 mice. *P<0.05, by unpaired Student’s t-test. b, Heat map representing hierarchical clustering of differentially expressed genes in microdissected interfollicular epidermis (IFE), hair follicle (HF) and sebaceous gland (SG) (see schematic) (n=3 biological replicates). c, d mRNA expression of markers specific for different cell compartments in Gata6 overexpressing primary keratinocytes compared with empty vector control cells collected at the indicated number of days following calcium-induced differentiation (c) or hours following suspension-induced differentiation (d). Heat map (log2 fold change = FC) (c) and RT-qPCR (d) are shown. Data are means ± s.d. from n=3 independent biological replicates. e, Cytoscape visualization of the Gata6-centric transcription factor network built by merging the features identified by different genomic approaches (see Methods). Magnification of selected elements from SG and JZ/SD sub-networks is shown. f, Plots of ChIP-Seq reads aligned to the Ar and Blimp1 loci identifying them as Gata6 direct targets. g, Blimp1 is induced while Ar is repressed by Gata6 overexpression. RT-qPCR of mRNAs from Gata6 overexpressing or control (empty vector) primary mouse keratinocytes collected at the indicated number of days after switching to high calcium medium. Data are means ± s.d. from n=3 independent biological samples.
To examine whether Gata6 transcriptional activity regulates epidermal lineage identity and differentiation in a gain of function experiment, we compared the relative abundance of lineage markers (including direct target genes) in control versus Gata6 overexpressing primary mouse keratinocytes during suspension or calcium-induced differentiation in culture. Read out genes were obtained from the expression profile of Gata6 expressing SD cells (Fig. 2b) and from an additional transcriptome analysis performed to discriminate the gene signatures in IFE, SG and HF microdissected from tail epidermis (Fig. 3b and Supplementary Fig. 3i-k). RT-qPCR experiments showed that SD markers were induced by Gata6 transcriptional activity, while IFE markers were not (Fig. 3c, d).
To achieve a more complete understanding of how the different epidermal compartments are maintained, we built a comprehensive transcription factor network (Fig. 3e) merging data from our genomics approaches (see Methods). During differentiation Gata6 induced all SG duct genes tested and, in contrast, down regulated several genes characteristic of differentiated sebocytes (Fig. 3c). This led us to focus on the SG and JZ/sebaceous ducts as distinct TF sub-networks, with the Androgen receptor (Ar) featuring in the SG network and Blimp-1 in the JZ/sebaceous ducts network (Fig. 3e). Both genes are known to play a role in the SG. Endogenous Gata6 and Blimp-1 co-localized in terminally differentiated SG duct cells (Fig. 2d) while Ar localized in lower SG28.
Chip-Seq revealed Blimp1 and Ar as Gata6 direct targets (Fig. 3f). However, overexpression of Gata6 led to induction of Blimp-1 while repressing Ar expression (Fig. 3g). The analysis of the epidermal transcription factor network together with our in vivo observations (Supplementary Fig. 3l-p) led us to conclude that Gata6 transcriptional activity defines the SD and is functionally distinct from the lower SG.
Fate of the Gata6 lineage during homeostasis and wound healing
To examine the fate of Gata6+ progeny, we created a mouse model in which EGFPCreERT2 is inserted in the Gata6 endogenous locus. By crossing Gata6EGFPCreERT2 mice with Rosa26-fl/STOP/fl-tdTomato mice we could genetically label (GL) Gata6 expressing cells by application of 4-hydroxy-Tamoxifen (4OHT) and follow the fate of their progeny. In undamaged skin the progeny of Gata6+ cells were restricted to the JZ and SG duct (Supplementary Fig. 4a). In agreement with our functional data (Fig. 3a, c), this indicates that Gata6 expressing cells represent a JZ cell population that is restricted to the SD lineage. In contrast, the progeny of Lrig1+ stem cells are found in the entire SG during epidermal homeostasis and extend into the lower anagen HF3.
It has previously been shown that Lrig1+ stem cells can contribute to the IFE following wounding3. In order to examine the role of lineage-restricted, differentiated Gata6+ cells in wound healing, we performed lineage-tracing experiments in parallel with Gata6EGFPCreERT2 and Lrig1EGFPCreERT23 mice crossed to the Rosa26-fl/STOP/fl-tdTomato strain. 9-week-old mice were treated once with 4OHT to induce tdTomato genetic labeling and two days later a full thickness circular skin wound was created (day 0).
Within 3 days of wounding Gata6 and Lrig1 tdTomato labelled cells exited the JZ and began colonizing the IFE in the direction of the wound (Fig. 4a, b). The progeny of Lrig1+ cells exited the JZ by upward migration as a continuous tongue of basal and suprabasal cells (Fig. 4a, b, d). In contrast, Gata6 labelled cells only exited into the suprabasal epidermal layers and did so as individual cells interspersed with unlabelled cells (Fig 4a, b, c). Unexpectedly, in intact hair follicles in the proximity of the wound site Gata6 labelled cells were no longer confined to the JZ and SD, but extended into the base of the SG (in 63% of pilosebaceous units) and, rarely, the lower HF (Fig. 4e-g and 5a). It was previously observed that epidermal cells are recruited to move out of the JZ by vitamin A treatment in the absence of wounding8. Since Gata6 is induced by vitamin A (Supplementary Fig. 4b-i), we speculate that Gata6+ progeny of Lrig1+ stem cells are responsible for this effect and suggest that the vitamin A signaling might be involved in the migration of Gata6+ cells into intact HF and SG adjacent to wounds.
Figure 4. Fate of Lrig1 and Gata6 lineages during wound healing.
a, Sections of skin close to a wound (left panels - tail) or at the wound edges (right panels - back) stained with antibodies to Gata6, Lrig1 and tdTomato showing the fate of Gata6 (top panels) and Lrig1 (bottom panels) genetically labeled (GL) cells in the early stage of re-epithelialization 3 days after wounding. Representative images of n=3 independent experiments. b, Schematic and quantification of cells exiting the JZ of the HF and migrating into the IFE wound site. The presence of labelled basal and suprabasal cells in ten IFE positions (from “a” to “l”) is shown. Cell position “a” is assigned to the first IFE nucleus out of the HF as indicated by a straight yellow line and yellow arrow in the left panels in (a). Data are means ± s.e.m. from n=3 (Lrig1, HF=24) or n=4 (Gata6, HF=30) lineage-traced mice. c, d, Whole mounts of tail epidermis showing the contribution of Gata6 (c) and Lrig1 (d) tdTomato GL cells to wound healing. Right panels are higher magnification views. Representative images of n=3 independent experiments. e, f, Invasion of Gata6 GL cells into uninjured epithelial HF niches (white arrows) close to the wound site. Left hand panels show SG cross sections while right hand panels show tail epidermal whole mounts. Gata6 tdTomato GL cells in late stages of re-epithelialization in IFE wound proximity (f) or in distant areas (e). Asterisk indicates Gata6 GL cells involved in IFE re-epithelialization. Yellow brackets indicate the bulge areas. Representative images of n=3 independent experiments. g, Percentage of tdTomato+ lower SG close to or far from the wound, determined from histological sections of back skin. Data are means ± s.d. from 3 mice (48 SG). Dashed lines indicate epidermal-dermal boundary (a) or SG (e, f). Scale bars: 50 μm.
Figure 5. Dedifferentiation of Gata6 lineage SD cells and acquisition of stem cell properties.
a, Tail skin sections 28 days after wounding stained with antibodies to Gata6 or Lrig1 showing the fate of Gata6 (left panel) and Lrig1 (right panel) tdTomato labeled cells. Both cell populations colonize the IFE wound bed. Gata6 labeled cells close to the wound also invade uninjured niches in HF and lower SG (white arrows), while further from the wound (yellow arrows) they remain in the JZ and SD. b, c, Sections of tail skin stained with antibodies to Itga6 and tdTomato. Two days after tamoxifen treatment (day 0 of wounding) of Gata6EGFPCreERT2 Rosa26-fl/STOP/fl-tdTomato tail skin, differentiated suprabasal JZ/SG duct cells are labeled (top left panel in b). By day 6 differentiated JZ/SD cells have left the HF and are migrating suprabasally (b); by day 12 they are present in the basal layer of the wound bed (b). In contrast, Lrig1 genetically labeled (GL) cells exit the JZ as basal and suprabasal cells (c). Boxed regions (b, c) are also shown at higher magnification. d, Quantification of basal tdTomato labeled cells derived from the Gata6 or Lrig1 lineage at the indicated time points in tail skin after wounding. Box-and-whisker plots: mid-line, median; box, 25th to 75th percentiles; and whiskers, minimum and maximum. Data are from n=3 to n=4 mice per time point (average of 10 wound bed sections per time point). e, Sections of re-epithelized tail IFE showing tdTomato Gata6 GL cells stained with antibodies to Ki67. f, g, Wound bed tail skin sections showing columns of IFE cells derived from dedifferentiation of Gata6 GL cells (tdTomato) stained with Krt14 (f) and Ki67(g). Ki67 quantification at the indicated time points is shown. Data are means ± s.e.m. from n=6 to n=17 epidermal proliferative unit (EPU) per time point. **P<0.005, by unpaired Student’s t-test. Dashed lines indicate dermal-epidermal junction. ND = not detected. Scale bars: 50 μm (a-c); 25 μm (e–g).
Dedifferentiation of Gata6 lineage cells
By treating tail epidermis with a low concentration of Tamoxifen we could selectively mark non-dividing (EdU-), differentiated Gata6 expressing cells (Supplementary Fig. 5a-c and Fig. 5b). After these labelled cells exited the JZ, they initially remained suprabasal while migrating into the wound bed (Fig. 5b, d). However, by day 12 labelled progeny of Gata6+ cells were detected in the re-epithelialised basal layer (Fig. 5b, d, e and Supplementary Fig. 5d, e). Progeny of Lrig1 expressing cells were found in the basal and suprabasal layers at all the time points examined, including 124 days after wounding (Fig. 5c, d and Supplementary Fig. 5e). Not only did Gata6 labelled cells enter the basal layer of the healing wound, they also began, like Lrig1 GL cells, to proliferate (Fig. 5e, Supplementary Fig. 5f). Furthermore, Gata6 progeny began to express Krt14 (Fig. 5f) suggesting an unprecedented ability of terminally differentiated epidermal cells to de-differentiate and acquire stem cell properties following wounding. Even 124 days post wounding columns of tdTomato labelled Gata6 progeny extending from the basal to the outermost epidermal layers were readily detectable (Fig. 5d, g). Ki67 labelling of Gata6 progeny was higher at 124 days than at earlier time points (Fig. 5g).
To explore whether dedifferentiation was a unique property of Gata6+ cells, we performed lineage tracing experiments to examine the fate of cells expressing Blimp1, which is a marker of terminally differentiated cells within the IFE, SG and HF26,29 (Fig. 2b, e; Supplementary Fig. 2a-c, g; Supplementary Fig. 3l; Supplementary Fig. 6a-d). By 6 days after wounding tdTomato labelled cells were found in a basal position at the wound edge (Supplementary Fig. 6e). Columns of cells derived from Blimp1+ cells were detected in the wound bed at day 12 (Supplementary Fig. 6f, g) and day 120 (Supplementary Fig. 6h). Not withstanding the limitations of using a non-inducible Cre line and our inability to distinguish between Blimp1 GL cells originating in the IFE, HF and SG, we speculate that dedifferentiation may be a general property of terminally differentiated epidermal cells following wounding.
Plasticity of Gata6 lineage cells
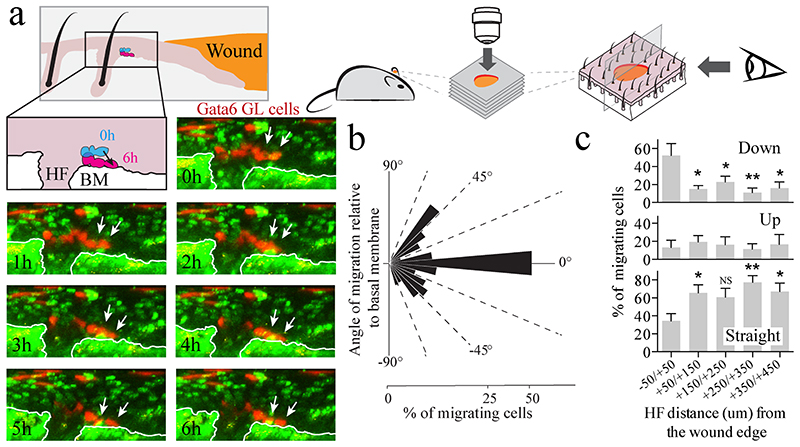
As an independent test of the plasticity of differentiated Gata6 GL cells, we examined their contribution to epidermis reconstituted from disaggregated cells in a chamber graft assay30 (Fig. 6). Flow sorted differentiated (Itga6 low) Gata6 GL cells were found in the IFE, SG and HF of 4/6 grafts examined, where they contributed to both the differentiated compartments and to the Krt14+ compartment attached to the basement membrane (Fig. 6a-f, Supplementary Fig. 7a, b). Thus Gata6 GL cells were able to dedifferentiate in the skin reconstitution assay as well as following wounding. To monitor the earliest events in dedifferentiation we followed the fate of individual Gata6 GL cells by in vivo live imaging for 8 to 16 hours, 5 days after wounding ear skin (Fig. 7a and Supplementary Fig. 8). We observed Gata6 labelled cells exiting a total of 49 hair follicles from 6 individual wounds. We measured the angle at which individual labelled cells moved relative to the position of the basement membrane (Fig. 8b). Most cells remained in a suprabasal position and moved parallel to the basement membrane (Fig. 8b, ‘straight’ in Fig. 8c). However a small proportion of labeled cells were displaced upwards (Fig. 8b, ‘up’ in Fig. 8c) or moved downwards towards the basal layer (Fig. 8b, ‘down’ in Fig. 8c). In 6 out of 25 hair follicles in which downward migration occurred we could observe the entire process of suprabasal to basal transition. The proportion of cells exhibiting downward migration was significantly higher at the wound edge than distal areas (Fig. 8c). This suggests that dedifferentiation is more likely to happen in close proximity to the wound.
Figure 6. Contribution of Gata6+ cells to reconstituted skin.
a, Epidermal cells isolated from the back skin of Gata6-tdTomato reporter/GFP-expressing adult mice in telogen were sorted into differentiated Gata6+ cells (Itga6 low/tdTomato positive; blue gate) and Itga6 high (red gate) cells. These two populations were compared in a skin reconstitution assay. b, Chamber in place one 1 week after implantation. c, d, Grafts containing differentiated Gata6+ cells and epidermal stem cell enriched Itga6 high cells showed similar numbers of reconstituted hair follicles after 5 weeks. Macroscopic views of representative grafts (left panels) and quantification (right panel) are shown (N=3 Itga6 high; N=6 differentiated Gata6 cells). Data are means ± s.d e, f, Dedifferentiation of Gata6+ cells in grafts. Graft skin sections were stained for Gfp (green) and Krt14 (red). Higher magnification views and single fluorescent channel images of the white inserts are shown. Yellow arrows indicate Gfp positive basal cells derived from Gata6+ or Itga6 high sorted cells in the interfollicular epidermis. Dashed lines demarcate epidermal-dermal boundaries. Representative images from n=3 to n=6 chamber graft assays (Itga6 high and Itga6 low/tdTomato positive cells respectively). Scale bars: 25 μm.
Figure 7. Live imaging of suprabasal Gata6 GL cells migrating to the basal layer during wound healing.
a, Time lapse live imaging strategy (top) for visualizing suprabasal Gata6 GL cells migrating towards the wound edge in ear skin. Bottom: a series of Z stacks was taken each hour and the orthogonal images were reconstructed to follow the migration angle with respect to the basal membrane (collagen). Downward migration of Gata6 GL cells (white arrows) is shown: orthogonal images at the indicated time points from the beginning of image acquisition showing GL cells (red) and nuclei, collagen (green). White lines indicate the basal membrane. Transition from suprabasal to basal layer is summarised schematically. Scale bars: 25 μm. b, Quantification of the angle of migration (bins of 10 degrees) with respect to the basement membrane of all the migratory events of the GL Gata6 cells per HF detected in n=6 mice (49 HF, 670 cells) during time lapse imaging for 12.5±3.8 hours. c, Percentage of labelled cells at the indicated distances from the wound edge showing downward (-90/-10 degrees), straight (-10/+10 degrees) or upward (+10/+90 degrees) migration. From n=7 to n=12 hair follicles per bin. Data are means ± s.e.m. *P<0.05; **P<0.005, by unpaired Student’s t-test.
Figure 8. Wound induced plasticity of Gata6 lineage linked to loss of JZ identity and acquisition of multi-lineage differentiation potential.
a, Sections of tail skin at the indicated time points after wounding showing tdTomato labelled cells, derived from differentiated Gata6 SD cells, stained with antibodies to the SD markers Krt79 and Plet1. White arrows indicate rare Krt79+ cells at the wound edge. Representative images of n=3 independent experiments. b, Wound bed section of tdTomato labeled columns of IFE cells derived from differentiated SD cells, stained with antibodies to scale (Krt31) and interscale (Filaggrin - Flg) markers. The percentage of IFE cell columns (EPU) labelled with antibodies to Krt31 and Flg is shown for Gata6 GL and Lrig1 GL cells (bottom panel). N=5 mice (39-41 EPU). Data are means ± s.e.m. *P<0.05, by unpaired Student’s t-test. c, Sections of tail skin showing Gata6 GL cells invading uninjured niches. Co-expression of tdTomato and the lower HF markers Lef1 and Krt31 or lipid marker of differentiated sebocytes (white arrow) is shown. Representative images of n=3 independent experiments. Dashed or solid lines indicate dermal-epidermal boundary Scale bars: 25 μm.
Since epidermal cells increase in size during terminal differentiation31, 32 we examined whether dedifferentiation correlated with a reduction in cell size. As shown in Supplementary Fig. 8e, f, Gata6 GL cells that had newly attached to the basement membrane were indistinguishable in size from unlabelled basal cells and were smaller that suprabasal Gata6 GL cells. In addition, basal layer Gata6 GL cells at the wound margins exhibited a characteristic elongation in the direction of the wound, consistent with ongoing cell migration (Supplementary Fig. 16f).
Gata6 labelled cells maintained Gata6 expression in the infundibulum but as soon as they entered the IFE on wounding they no longer expressed Gata6 (Fig. 4a) or the SD-specific markers Krt79 and Plet1 (Fig. 8a), with the exception of a few Krt79+ cells at the wound edge (Fig, 8a). Gata6 and SD lineage markers were not expressed in the proliferative columns founded by Gata6 labelled cells in the reconstituted IFE of wounds. In healed tail wounds Gata6 labelled cells gave rise to both the scale (Krt31+) and interscale (Flg+) lineages of the IFE33. In contrast, Lrig1 labelled progeny primarily gave rise to the interscale tail lineage (Fig. 8b). Not only did Gata6 labelled cells give rise to both IFE lineages, they also - in those HF in which Gata6 progeny entered the lower HF and SG – expressed Lef1 and Krt31 in the hair bulb and produced lipid, a characteristic of differentiated sebocytes (Fig. 8c).
Discussion
It was previously believed that in multilayered epithelia such as the epidermis terminal differentiation is irreversible9, 12. Combining genomics, transcriptomics, imaging techniques in live mice, skin reconstitution assays and histological analysis of lineage tracing experiments, our study has not only identified Gata6 as a key regulator of the SD lineage, but has also revealed that differentiated Gata6+ cells can dedifferentiate in response to wounding, exhibiting sustained self-renewal, indicative of re-entry into a stem cell state. Although such plasticity of differentiated cells in the epidermis has not been reported10, 11, it is consistent with the observation that Blimp1+ sebocytes are post-mitotic in vivo yet capable of undergoing self-renewal in culture26.
Upon dedifferentiation Gata6 GL cells had similar self-renewal ability to Lrig1 GL epidermal stem cells but superior multi-lineage differentiation potential34. We speculate that the explanation lies in the Gata6 target genes we identified by ChIP-Seq. As shown for Gata435, Gata6 may act as a pioneer transcription factor, opening heterochromatin to favor the access of other transcription factors. We discovered that Gata6 regulates genes involved in cell motility and, consistent with this, that Gata6 promotes migration of epidermal cells in culture. The results are consistent with a model whereby under homeostatic conditions Gata6 not only promotes SD differentiation but also maintains the migratory potential of the differentiated duct cells, providing a reservoir of cells that can be mobilized for wound repair. It is interesting that in Gata6+ JZ cells the acquisition of plasticity on injury is linked to loss of the cells’ original identity and acquisition of the identity of each new epidermal niche in which they are located4.
There are two main theories of how re-epithelialization of skin wounds occurs: the leap-frog model, in which suprabasal cells migrate on top of basal cells and dedifferentiate to become the new leading cells at the wound edge36, 37, and the sliding model in which layered epidermal cells move towards the wound margin in a cohesive manner38–40. Our results reconcile these hypotheses, showing the coexistence of both repair mechanisms, with Lrig1+ stem cells entering the wound as a cohesive population, and Gata6+ cells migrating individually within the suprabasal cells until they make contact with the interface with the dermis and revert to a stem cell state.
Online Content
Methods, along with any additional Supplementary Data display items and Source Data, are available in the online version of the paper; references unique to these sections appear only in the online paper.
Supplementary Material
Acknowledgements
We thanks Hiro Fujiwara, Sam Woodhouse, Beate M Lichtenberger, Ajay Mishra, Valentina Proserpio, Matteo Cereda and all the members of the Watt laboratory for helpful discussions and for excellent suggestions; Xiangang Zou (CR-UK Cambridge Research Institute, Cambridge, UK) and the Mary Lyon Centre at MRC Harwell MRC for assistance in generating the Gata6 KI mouse model; Denny Cottle and Charlotte Collins for access to animal samples; the core facilities of CRI and KCL and the NIHR GSTT/KCL Biomedical Research Centre for superb technical assistance; the Paterson Institute Microarray Facility for performing arrays; Dr. Hiroshi Hamada for kindly sharing the Gata6 reporter mouse; Barry Rosen and Fabio Amaral, (Wellcome Trust Sanger Institute, Cambridge, UK) for the eGFPCreERT2 cassette. This work was funded by the UK Medical Research Council, the Wellcome Trust and the European Union FP7 programme. Toru Hiratsuka is the recipient of a EU Marie Curie Fellowship; Emanuel Rognoni is the recipient of an EMBO long term fellowship.
References
- 1.Tata PR, et al. Dedifferentiation of committed epithelial cells into stem cells in vivo. Nature. 2013;503:218–223. doi: 10.1038/nature12777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van Es JH, et al. Dll1+ secretory progenitor cells revert to stem cells upon crypt damage. Nature cell biology. 2012;14:1099–1104. doi: 10.1038/ncb2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Page ME, Lombard P, Ng F, Gottgens B, Jensen KB. The epidermis comprises autonomous compartments maintained by distinct stem cell populations. Cell stem cell. 2013;13:471–482. doi: 10.1016/j.stem.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Donati G, Watt FM. Stem cell heterogeneity and plasticity in epithelia. Cell stem cell. 2015;16:465–476. doi: 10.1016/j.stem.2015.04.014. [DOI] [PubMed] [Google Scholar]
- 5.Tetteh PW, Farin HF, Clevers H. Plasticity within stem cell hierarchies in mammalian epithelia. Trends in cell biology. 2015;25:100–108. doi: 10.1016/j.tcb.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 6.Blanpain C, Fuchs E. Stem cell plasticity. Plasticity of epithelial stem cells in tissue regeneration. Science. 2014;344:1242281. doi: 10.1126/science.1242281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ito M, et al. Stem cells in the hair follicle bulge contribute to wound repair but not to homeostasis of the epidermis. Nature medicine. 2005;11:1351–1354. doi: 10.1038/nm1328. [DOI] [PubMed] [Google Scholar]
- 8.Jensen KB, et al. Lrig1 expression defines a distinct multipotent stem cell population in mammalian epidermis. Cell stem cell. 2009;4:427–439. doi: 10.1016/j.stem.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Watt FM. Mammalian skin cell biology: at the interface between laboratory and clinic. Science. 2014;346:937–940. doi: 10.1126/science.1253734. [DOI] [PubMed] [Google Scholar]
- 10.Park S, et al. Tissue-scale coordination of cellular behaviour promotes epidermal wound repair in live mice. Nature cell biology. 2017;19:155–163. doi: 10.1038/ncb3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aragona M, et al. Defining stem cell dynamics and migration during wound healing in mouse skin epidermis. Nature communications. 2017;8:14684. doi: 10.1038/ncomms14684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hsu YC, Pasolli HA, Fuchs E. Dynamics between stem cells, niche, and progeny in the hair follicle. Cell. 2011;144:92–105. doi: 10.1016/j.cell.2010.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Niemann C, Owens DM, Hulsken J, Birchmeier W, Watt FM. Expression of DeltaNLef1 in mouse epidermis results in differentiation of hair follicles into squamous epidermal cysts and formation of skin tumours. Development. 2002;129:95–109. doi: 10.1242/dev.129.1.95. [DOI] [PubMed] [Google Scholar]
- 14.Donati G, et al. Epidermal Wnt/beta-catenin signaling regulates adipocyte differentiation via secretion of adipogenic factors. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:E1501–1509. doi: 10.1073/pnas.1312880111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheung WK, et al. Control of alveolar differentiation by the lineage transcription factors GATA6 and HOPX inhibits lung adenocarcinoma metastasis. Cancer cell. 2013;23:725–738. doi: 10.1016/j.ccr.2013.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaufman CK, et al. GATA-3: an unexpected regulator of cell lineage determination in skin. Genes & development. 2003;17:2108–2122. doi: 10.1101/gad.1115203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Campbell K, Whissell G, Franch-Marro X, Batlle E, Casanova J. Specific GATA factors act as conserved inducers of an endodermal-EMT. Developmental cell. 2011;21:1051–1061. doi: 10.1016/j.devcel.2011.10.005. [DOI] [PubMed] [Google Scholar]
- 18.Wang AB, Zhang YV, Tumbar T. Gata6 promotes hair follicle progenitor cell renewal by genome maintenance during proliferation. The EMBO journal. 2016 doi: 10.15252/embj.201694572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Joost S, et al. Single-Cell Transcriptomics Reveals that Differentiation and Spatial Signatures Shape Epidermal and Hair Follicle Heterogeneity. Cell systems. 2016;3:221–237.:e229. doi: 10.1016/j.cels.2016.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fullgrabe A, et al. Dynamics of Lgr6(+) Progenitor Cells in the Hair Follicle, Sebaceous Gland, and Interfollicular Epidermis. Stem cell reports. 2015;5:843–855. doi: 10.1016/j.stemcr.2015.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Snippert HJ, et al. Lgr6 marks stem cells in the hair follicle that generate all cell lineages of the skin. Science. 2010;327:1385–1389. doi: 10.1126/science.1184733. [DOI] [PubMed] [Google Scholar]
- 22.Nguyen H, Rendl M, Fuchs E. Tcf3 governs stem cell features and represses cell fate determination in skin. Cell. 2006;127:171–183. doi: 10.1016/j.cell.2006.07.036. [DOI] [PubMed] [Google Scholar]
- 23.Nowak JA, Polak L, Pasolli HA, Fuchs E. Hair follicle stem cells are specified and function in early skin morphogenesis. Cell stem cell. 2008;3:33–43. doi: 10.1016/j.stem.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takaoka K, Yamamoto M, Hamada H. Origin and role of distal visceral endoderm, a group of cells that determines anterior-posterior polarity of the mouse embryo. Nature cell biology. 2011;13:743–752. doi: 10.1038/ncb2251. [DOI] [PubMed] [Google Scholar]
- 25.Veniaminova NA, et al. Keratin 79 identifies a novel population of migratory epithelial cells that initiates hair canal morphogenesis and regeneration. Development. 2013;140:4870–4880. doi: 10.1242/dev.101725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kretzschmar K, et al. BLIMP1 is required for postnatal epidermal homeostasis but does not define a sebaceous gland progenitor under steady-state conditions. Stem cell reports. 2014;3:620–633. doi: 10.1016/j.stemcr.2014.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sodhi CP, Li J, Duncan SA. Generation of mice harbouring a conditional loss-of-function allele of Gata6. BMC developmental biology. 2006;6:19. doi: 10.1186/1471-213X-6-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cottle DL, et al. c-MYC-induced sebaceous gland differentiation is controlled by an androgen receptor/p53 axis. Cell reports. 2013;3:427–441. doi: 10.1016/j.celrep.2013.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Magnusdottir E, et al. Epidermal terminal differentiation depends on B lymphocyte-induced maturation protein-1. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:14988–14993. doi: 10.1073/pnas.0707323104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Driskell RR, et al. Distinct fibroblast lineages determine dermal architecture in skin development and repair. Nature. 2013;504:277–281. doi: 10.1038/nature12783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Watt FM, Green H. Involucrin synthesis is correlated with cell size in human epidermal cultures. The Journal of cell biology. 1981;90:738–742. doi: 10.1083/jcb.90.3.738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jones PH, Watt FM. Separation of human epidermal stem cells from transit amplifying cells on the basis of differences in integrin function and expression. Cell. 1993;73:713–724. doi: 10.1016/0092-8674(93)90251-k. [DOI] [PubMed] [Google Scholar]
- 33.Gomez C, et al. The interfollicular epidermis of adult mouse tail comprises two distinct cell lineages that are differentially regulated by Wnt, Edaradd, and Lrig1. Stem cell reports. 2013;1:19–27. doi: 10.1016/j.stemcr.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kretzschmar K, Watt FM. Markers of epidermal stem cell subpopulations in adult mammalian skin. Cold Spring Harbor perspectives in medicine. 2014;4 doi: 10.1101/cshperspect.a013631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zaret KS, Carroll JS. Pioneer transcription factors: establishing competence for gene expression. Genes & development. 2011;25:2227–2241. doi: 10.1101/gad.176826.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krawczyk WS. A pattern of epidermal cell migration during wound healing. The Journal of cell biology. 1971;49:247–263. doi: 10.1083/jcb.49.2.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paladini RD, Takahashi K, Bravo NS, Coulombe PA. Onset of re-epithelialization after skin injury correlates with a reorganization of keratin filaments in wound edge keratinocytes: defining a potential role for keratin 16. The Journal of cell biology. 1996;132:381–397. doi: 10.1083/jcb.132.3.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Radice GP. The spreading of epithelial cells during wound closure in Xenopus larvae. Developmental biology. 1980;76:26–46. doi: 10.1016/0012-1606(80)90360-7. [DOI] [PubMed] [Google Scholar]
- 39.Woodley DT. Reepithelialization. In: Clark RAF, editor. The Molecular and Cellular Biology of Wound Repair. Second edition. Plenum Press; New York: 1996. pp. 339–354. [Google Scholar]
- 40.Safferling K, et al. Wound healing revised: a novel reepithelialization mechanism revealed by in vitro and in silico models. The Journal of cell biology. 2013;203:691–709. doi: 10.1083/jcb.201212020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Collins CA, Watt FM. Dynamic regulation of retinoic acid-binding proteins in developing, adult and neoplastic skin reveals roles for beta-catenin and Notch signalling. Developmental biology. 2008;324:55–67. doi: 10.1016/j.ydbio.2008.08.034. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.