Abstract
Adult and fetal hematopoietic stem cells (HSCs) display a glycolytic phenotype, which is required for maintenance of stemness; however, whether mitochondrial respiration is required to maintain HSC function is not known. Here we report that loss of the mitochondrial complex III subunit Rieske iron sulfur protein (RISP) in fetal mouse HSCs allows them to proliferate but impairs their differentiation, resulting in anemia and prenatal death. RISP null fetal HSCs displayed impaired respiration resulting in a decreased NAD+/NADH ratio. RISP null fetal HSCs and progenitors exhibited an increase in both DNA and histone methylation associated with increases in 2-hydroxyglutarate (2-HG), a metabolite known to inhibit DNA and histone demethylases. RISP inactivation in adult HSCs also impaired respiration resulting in loss of quiescence concomitant with severe pancytopenia and lethality. Thus, respiration is dispensable for adult or fetal HSC proliferation, but essential for fetal HSC differentiation and maintenance of adult HSC quiescence.
Introduction
Hematopoietic stem cells (HSCs) require tight regulation of cell cycle, apoptosis and oxidative stress to sustain a balance between HSC quiescence, self-renewal and differentiation. Deregulation of this balance can result in the development of blood disorders such as anemia and leukemia. Emerging evidence suggests that the metabolic state of HSCs regulates hematopoiesis, although the mechanisms that link metabolism to HSC function remain poorly understood1, 2. Many of the metabolic studies on hematopoiesis have focused on the role of glycolysis and its major regulator HIF-1 in maintaining functional stem and progenitor cell populations3,4, however, it is not known whether mitochondrial respiration is essential for HSCs function.
HSCs have low mitochondrial mass compared to more differentiated progenitors 5, 6, reside in hypoxic niches in the bone marrow, 5, 7, 8 and are highly sensitive to oxidative stress 9-14. The mitochondrial respiratory chain is one of the main generators of reactive oxygen species (ROS), and accumulation of ROS-induced mitochondrial DNA (mtDNA) mutations in HSCs impairs appropriate multi-lineage differentiation6, 15. Furthermore, increasing mitochondrial metabolism in HSCs triggers aberrant entry into the cell cycle and ultimately stem cell exhaustion16. Aberrant increase in mitochondrial function can also result in loss of adult HSC quiescence11, 16. Deletion of the kinase LKB1 and the PTEN-like mitochondrial phosphatase (PTPMT1) in HSCs results in defective hematopoiesis with accompanying decreases of mitochondrial metabolism17-20. Loss of AMPK in HSCs causes a similar decrease in mitochondrial metabolism as LKB1 and PTPMT1 inactivation, yet does not incur any hematopoiesis defects20. LKB1 and PTPMT1 likely affect multiple processes within HSCs, and it is not clear whether primary mitochondrial defects underlie the phenotypes observed in these knockouts. In the present study, we directly tested whether mitochondrial respiration is necessary for proliferation and maintenance of HSC and progenitor function by deleting Uqcrfs1 in fetal and adult HSCs. Uqcrfs1 encodes for the Rieske iron-sulfur protein (RISP), an essential subunit of the mitochondrial complex III21.
Results
RISP deficiency in fetal HSCs impairs respiration and causes severe anemia
We first investigated whether mitochondrial metabolism in HSCs was necessary for fetal development.
Uqcrfs1 floxed mice were crossed to Vav-iCre mice (RISP KO), which constitutively express the codon-improved Cre (iCre) driven by Vav regulatory elements, resulting in hematopoietic stem cell specific gene deletion in mid-gestation. RISP KO mice displayed normal Mendelian ratios at embryonic day 15.5 (E15.5) (Fig. 1a). RISP expression was efficiently reduced in the lineage negative (Lin-) cells isolated from RISP KO fetal livers at day E15.5 (Fig. 1b). RISP KO mice displayed paler embryos at E18.5 indicative of anemia and died prior to completing gestation (Fig. 1c). The fetal liver (FL) is the main hematopoietic organ between E12.5 and E16.5, a period characterized by the expansion of the hematopoietic pool and massive production of erythrocytes22. RISP KO FLs were smaller and displayed a decrease in cellularity associated with an increase in cell death (Fig. 1c-e). Peripheral blood analysis at E18.5 confirmed the anemic phenotype, with severe reductions in red and white blood cells (RBC, WBC), hemoglobin (HB), hematocrit (HCT) and platelet (PLT) numbers in the RISP KO fetuses (Fig. 1f). We obtained similar results from Vav-iCre+ Tfamfl/fl (TFAM KO) mice (Supplementary Fig. 1). The deletion of Tfam (mitochondrial transcription factor A) results in loss of mtDNA-encoded proteins essential for mitochondrial electron transport chain function23.
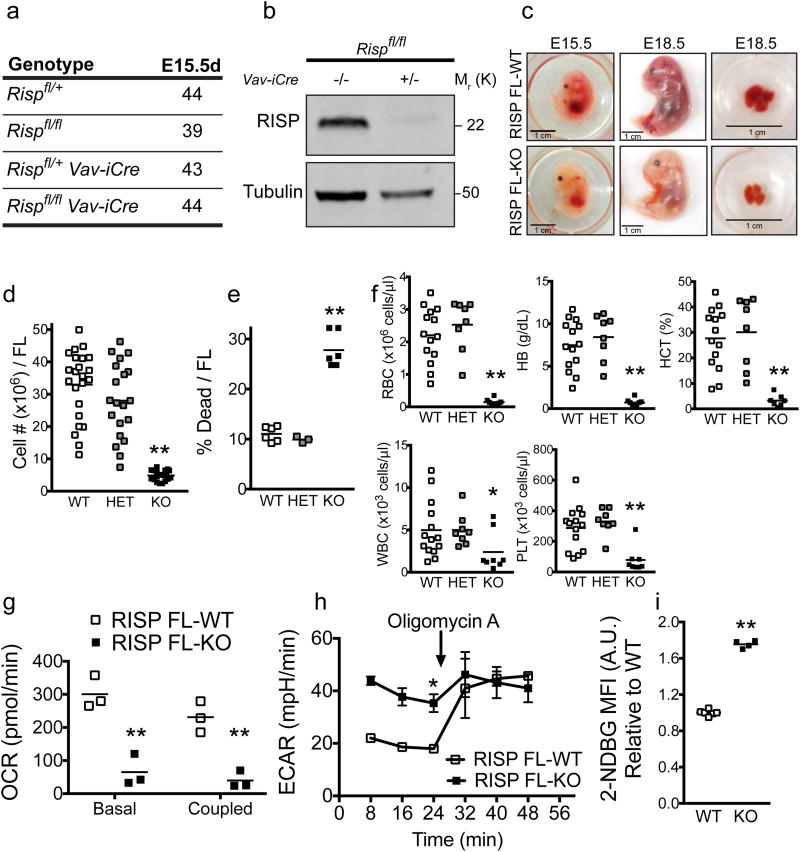
Figure 1. Loss of RISP in hematopoietic cells impairs mitochondrial oxidative metabolism and results in severe fetal anemia.
(a) Genotypes from crosses between Rispfl/fl and Rispfl/fl Vav-iCre mice.
(b) Protein expression of RISP in lineage negative (Lin- ) fetal liver cells analyzed by Western blot. The experiment was performed three times. The figure displayed is one representative experiment. Image of the whole blot is displayed in Supplementary Fig. 8.
(c) Representative pictures at E15.5 and E18.5 days of Rispfl/fl and Rispfl/fl Vav-iCre fetuses, and fetal livers at E18.5 days.
(d) Total cell number per fetal liver (FL) at E15.5 days. (WT n=23; HET n=20; KO n=20).
(e) Percentage of cell death per FL at E15.5 days. (WT n=6; HET n=3; KO n=6).
(f) Peripheral blood cell parameters from E18.5 days embryos, including RBC (red blood cells), HB (hemoglobin), HCT (hematocrit), WBC (white blood cells) and PLT (platelets) (WT n=14; HET n=8; KO n=8).
(g) Oxygen consumption rate (OCR) of Lin- FL cells in basal and coupled conditions. (WT n=3; KO n=3).
(h) Extracellular acidification rate (ECAR) of Lin- FL cells in basal conditions and after exposure to 1 μM oligomycin A. Figure is representative of three independent experiments. (WT n=3; KO n=3).
(i) 2-NBDG uptake in Lin- FL cells. Results are expressed as relative to WT (WT n=6; KO n=4), Data in Figure 1 are mean ± SEM. *p ≤ 0.05; **p ≤ 0.01. All p values calculated using Student's t-test.
We measured biochemical parameters in Lin- FL cells. Biochemical analysis requires ample cell numbers, so we focused on this population to measure respiration. Lin- cells isolated from RISP KO mice displayed a decrease in OCR (Fig. 1g) with an increase in extracellular acidification rate (ECAR), a measurement of glycolytic flux (Fig. 1h). Oligomycin, an inhibitor of mitochondrial ATP synthase, increased ECAR in RISP WT Lin- cells, indicating that these cells can increase glycolysis in response to inhibition of mitochondrial ATP generation (Fig. 1h). In contrast, RISP KO Lin- cells were maximally glycolytic at baseline and did not further increase ECAR upon addition of oligomycin (Fig. 1h). RISP KO Lin- cells also displayed an increase in glucose uptake consistent with the increase in glycolytic flux (Fig. 1i).
RISP deficient fetal HSCs are able to proliferate but have impaired differentiation
Next, we analyzed HSC and multipotent progenitor population (MPP) numbers in RISP KO mice (Supplementary Fig. 2). The absolute numbers of phenotypic fetal HSCs were maintained at both E15.5 and E17.5 (Fig. 2a). By contrast, RISP KO FLs displayed a decrease in MPPs suggesting a defect in HSC differentiation (Fig. 2b). RISP null fetal HSCs displayed an increase in glucose uptake (Fig. 2c). Unlike adult stem cells, which are mainly quiescent, fetal HSCs undergo rapid cycling. To determine whether loss of RISP affects proliferation, intraperitoneal injection of BrdU was administered to pregnant mothers carrying E15.5 embryos 6 hours before analysis. RISP KO fetal HSCs displayed an increased percentage of BrdU positive cells, indicative of an increase in cell proliferation (Fig. 2d). Also, annexin V staining showed a slight increase in apoptosis (Fig. 2e). RISP deficiency resulted in diminished mROS, measured by MitoSOX oxidation, without changes in total ROS, as assessed by DCFH oxidation (Fig. 2f). The loss of respiration did not induce significant changes in mitochondrial membrane potential or mitochondrial mass (Fig. 2g). Mitochondrial membrane potential can be maintained in respiratory deficient cells by reversing the F1FoATPase to proton pump by utilizing glycolytic ATP24. Although RISP KO fetal HSCs are able to proliferate, they might not be able to retain their proper stem cell functions. Indeed, we observed a decrease in a panel of genes implicated in maintenance of fetal HSC function in RISP KO fetal HSCs (Fig. 2h).
Figure 2. RISP is essential for fetal HSC differentiation.
(a) Total number of HSCs per FL at E15.5d and E17.5d in RISP FL-WT and FL-KO mice (E15.5 WT n=19 and KO n=6; E17.5 WT n=8 and KO n=7).
(b) Total number of MPPs per FL at E15.5d and E17.5d in RISP FL-WT and FL-KO mice (E15.5 WT n=19 and KO n=7; E17.5 WT n=6 and KO n=7).
(c) 2-NDBG MFI of FL HSCs at E15.5d in RISP FL-WT and FL-KO mice. Results are expressed as relative to WT (WT n=6; KO n=7).
(d) Percentage of BrdU-positive FL HSCs from E15.5d in RISP FL-WT and FL-KO mice (WT n=6; KO n=5).
(e) Percentage of annexin V positive FL HSCs from E15.5d in RISP FL-WT and FL-KO mice (WT n=6; KO n=9).
(f) MitoSox and DCFH MFI of FL HSCs from E15.5d in RISP FL-WT and FL-KO mice. Results are expressed as relative to WT (MitoSOX WT n=4; KO n=6) (DCFH WT n=8; KO n=6).
(g) TMRE and MTG MFI of FL HSCs from E15.5d in RISP FL-WT and FL-KO mice. TMRE values were CCCP corrected. Results are expressed as relative to WT (TMRE WT n=4; KO n=6) (MTG WT n=11; KO n=12).
(h) Quantification of genes implicated in fetal HSC maintenance from fetal HSC cells isolated from RISP FL-WT and FL-KO mice. Results are expressed as relative fold change of KO compared to WT for sorted HSCs. (WT n=4; KO n=4 or n=3 for HoxA9).
Data in Figure 2 are mean ± SEM. *p ≤ 0.05; **p ≤ 0.01. All p values calculated using Student's t-test.
RISP deficiency in fetal HSCs causes depletion of myeloid progenitors
As severe anemia was the main phenotype of the RISP KO fetuses, we decided to investigate the underlying mechanisms by examining various myeloid progenitor (MP) populations. The total number of myeloid progenitors in the RISP KO FL was drastically reduced (Fig. 3a), while the frequency of myeloid progenitors was maintained (Fig. 3b). Common myeloid progenitors (CMPs) were increased in frequency (Fig. 3b). Interestingly, we observed a decrease in the frequency of MEPs with a concomitant increase in GMPs (Fig. 3b). The differences in MEPs versus GMPs were reflected in the expression profile of several lineage-specific transcription factors (Fig. 3c). GATA1 upregulation with a concomitant decrease in GATA2 is required for MEPs to differentiate into megakaryocytes and erythrocytes. PU.1 is essential for GMPs to differentiate into granulocytes and monocyte/macrophages, while C/EBPα is required for granulocyte differentiation25. GATA-1, but not PU.1, mRNA, was diminished in RISP KO myeloid progenitors, while CEBPα and GATA2 mRNA was upregulated in sorted fetal liver MPs (Fig. 3c).
Figure 3. Ablation of RISP depletes fetal myeloid progenitors.
(a and b) Frequency and total cell number of MP (myeloid progenitors), CMP (common myeloid progenitor), GMP (granulocyte macrophage progenitor) and MEP (megakaryocyte-erythrocyte progenitor) in fetal livers from RISP FL-WT and FL-KO mice. (WT n=5; KO n=5).
(c) Quantitative RT-PCR analysis of pro-myeloid cell lineage commitment genes in MP cells isolated from RISP FL-WT and FL-KO mice. Results are expressed as relative fold change of KO compared to WT. (WT n=7; KO n=7).
(d) Cell frequency and cell number of the erythroid population maturation in the fetal defined by staining against surface markers CD71 and Ter119 in RISP FL-WT and FL-KO mice (WT n=13; KO n=11).
(e) Cell frequency and cell number of the CD41/CD42 populations from FL cells at E15.5d in RISP FL-WT and FL-KO mice. (WT n=8; KO n=6).
(f) Cell frequency and cell number of the Mac-1/Gr-1 populations from FL cells at E15.5d in RISP FL-WT and FL-KO mice. (WT n=8; KO n=6).
(g) Quantification of CFU-GM, BFU-E and CFU-GEMM colonies from Methocult cultures of 105 FL RISP WT and KO cells cells under normoxic (21% O2) and hypoxic (1.5% O2) conditions. (WT n=12; KO n=6).
(h) Quantification of CFU-Mk colonies from MegaCult-collagen gels of 105 FL RISP WT and KO cells under normoxic (21% O2) and hypoxic (1.5% O2) conditions. (WT n=8; KO n=5).
Data in Figure 3 are mean ± SEM. *p ≤ 0.05; **p ≤ 0.01. All p values calculated using Student's t-test.
Next, we examined the maturation of different myeloid lineages in the RISP KO fetal livers. Erythrocyte maturation was impaired in the RISP KO shown by an increase in the frequency of proerythroblasts (R1) and a concomitant reduction in the more mature progenitors (Fig. 3d). Absolute numbers of erythroid cells at every stage of maturation were diminished in the RISP KO, consistent with the severe anemia phenotype (Fig. 3d). In contrast to the differentiation defects observed in erythroid lineages, the frequencies of immature (CD41+/CD42-), and mature (CD41+/CD42+) and megakaryocytes, monocytes (Mac1+ Gr1-) were increased due to loss of RISP, while granulocytes (Mac1+ Gr1+) were unchanged (Fig. 3e-f). Again, the absolute numbers of megakaryocytes, granulocytes, and monocytes were diminished due to the low numbers of total progenitors (Fig. 3e-f). To test whether loss of RISP impairs progenitor function, we performed in vitro colony forming unit (CFU) assays. Methylcellulose cultures were scored for burst-forming unit-erythroid (BFU-E), granulocyte–erythrocyte–megakaryocyte–macrophage (CFU-GEMM), and granulocyte–macrophage (CFU-GM) colonies. Cells lacking RISP gave rise to significantly fewer BFU-E and CFU-GEMM colonies compared to CFU-GM colonies (Fig. 3g). Hypoxia, which is known to stimulate erythropoiesis, increased BFU-E colonies in RISP WT but not in RISP KO (Fig. 3g). Megakaryocytic colony (CFU-Mks) formation was also impaired (Fig. 3h).
To test whether mROS levels were altered in MPs following loss of RISP, we assessed mitochondrial ROS by measuring MitoSOX fluorescence. We did not detect any difference in MitoSOX fluorescence between RISP KO and RISP WT MPs (Supplementary Fig. 3a). Furthermore, we also did not observe any changes in mitochondrial membrane potential between RISP KO and RISP WT MPs (Supplementary Fig. 3b). To directly test whether changes in ROS affect erythrocyte differentiation, we treated pregnant mice with either pan-antioxidant NAC (to decrease ROS) or the glutathione depleting agent BSO (to increase ROS). The concentration of NAC utilized has previously rescued ROS-induced phenotypes observed due to the deletion of FOXOs9. Neither NAC nor BSO could rescue differentiation of RISP KO erythrocytes in vivo or in vitro (Supplementary Fig. 3c-f). BSO was able to increase the levels of the ROS-sensitive p16 mRNA in Lin- progenitors, indicating that BSO effectively generated ROS (Supplementary Fig. 3g). Higher concentrations of BSO did not rescue the erythrocyte maturation (Supplementary Fig.3h). Thus, the impairment of erythropoiesis due to loss of RISP is likely independent of ROS.
Loss of RISP in fetal HSCs impairs their repopulating capacity
We evaluated the long-term repopulating capacity of RISP WT or RISP KO FL cells in vivo (Supplementary Fig. 4). RISP deficient fetal HSCs did not conduct long-term multi-lineage reconstitution of irradiated mice possibly due to engraftment failure (Supplementary Fig. 4a-c). To circumvent the loss of engraftment by RISP KO FL cells, we crossed Mx1-cre mice to Uqcrfs1 floxed mice and transplanted donor CD45.2 Rispfl/fl or Rispflllf; Mx1-Cre fetal liver cells along with competitor CD45.1 wild-type bone marrow cells, into CD45.1 recipient mice. After 8 weeks of engraftment, Poly (I:C) was administered to induce Cre expression and loss of RISP, which resulted in a sharp decline in CD45.2 cells in the peripheral blood of Rispfl/fl; Mx1-Cre mice (Figure 4a). Specifically, we observed a drastic reduction in CD45.2 B cells, T cells, and myeloid cells in the peripheral blood upon loss of RISP (Fig. 4b). Moreover, there was a substantial loss of RISP deficient CD45.2 HSCs and progenitor populations in the bone marrow 16 weeks after Cre induction with Poly (I:C) (Fig. 4c). These results were confirmed with a second independent competitive transplantation assay (Supplementary Fig. 4d-e). Thus, there is an intrinsic requirement for RISP in fetal HSCs to maintain hematopoiesis.
Figure 4. RISP deficiency impairs fetal HSC competitive repopulation capacity.
(a) The total contribution of CD45.2 fetal liver (FL) donor cells to the peripheral blood of irradiated CD45.1 recipient mice in a competitive transplantation assay following engraftment and subsequent Poly (I:C) treatment. Data represent the percentage of CD45.2 donor cells relative to the initial chimerism.
(b) The contribution of CD45.2 FL donor cells to specific lineages in the peripheral blood of irradiated CD45.1 recipient mice in a competitive transplantation assay following engraftment and subsequent Poly (I:C) treatment. B-cell lineage (B220+), T-cell lineage (CD3+) and myeloid lineage (Mac-1+, Gr-1+). Data represent the percentage of CD45.2 donor cells relative to the initial chimerism.
(c) The contribution of CD45.2 FL donor cells to specific lineages (LSK, ST-HSC, LT-HSC, MPP, MP, CMP, GMP, and MEP) in the bone marrow of irradiated CD45.1 recipient mice in a competitive transplantation assay after engraftment and subsequent Poly (I:C) treatment. Bone marrow was assayed four months post-Poly (I:C) treatment.
Data represent the percentage of CD45.2 donor cells relative to the initial chimerism. Data represent the average contribution for Rispfl/fl (RISP FL-WT; n=9 mice) and Rispfl/fl Mx1-Cre (RISP FL-KO; n=4 mice) in Figure 4a-c. Data in Figure 4a-c are mean ± SEM. *p ≤ 0.05; **p ≤ 0.01. All p values calculated using Student's t-test.
Loss of RISP in fetal HSCs causes gene expression and epigenetic changes
To investigate the cause underlying the inability of fetal HSCs to generate progenitors, we conducted RNA-seq analysis. We observed widespread gene deregulation due to loss of RISP in fetal HSCs. Specifically, 588 and 438 genes were significantly up- and down-regulated in RISP deficient HSCs (fold change ≥ 1.5, p-value ≤ 0.01; Fig. 5a). Pathway analysis revealed a significant increase in the expression of genes associated with unfolded protein response and MYC pathways, and decrease of genes associated with the immune response, ROS, and heme metabolism (Fig. 5b and 5c). Recently, we demonstrated that impairment of respiration due to mtDNA loss causes hypo-acetylation of certain histone marks in cancer cells24. Furthermore, fatty acid oxidation by mitochondria can also generate the acetyl-CoA necessary for histone acetylation26. We therefore conducted an unbiased mass spectrometry-based analysis of all potential histone modifications. We observed histone hypo-acetylation and hypermethylation of several histone lysine modifications, as well as increased global DNA methylation as assessed by elevated 5-methylcytosine levels in RISP KO fetal Lin- cells (Fig. 5d-f) and Supplementary Table 1). The key histone methylation marks hyper-elevated were confirmed in a second independent experiment (Supplementary Table 2). Collectively, these results indicate that mitochondrial complex III is essential to maintain proper gene expression and epigenetic homeostasis in fetal HSCs and progenitors.
Figure 5. Loss of RISP in fetal HSCs leads to deregulation of gene expression, DNA and histone hypermethylation, and histone hypoacetylation.
(a-c) Heat map, top enriched gene ontology terms and gene signatures from RNA sequence analysis reveals deregulation of gene expression in RISP null fetal HSCs. RNA sequence analysis was done on fetal HSCs isolated from 3 different RISP FL-WT and FL-KO mice.
(d) Percentage of 5-methylcytosine of total cytosine in Lin- fetal liver cells from RISP FL-WT and FL-KO mice. n=4 ± SEM *p ≤ 0.05. p-value calculated using Student's t-test.
(e, f) Mass spectrometry-based epigenetic analysis of various histone lysine methylation and acetylation modifications. Data represents the analysis of pooled lin- FL cells from 4 WT and 7 RISP KO animals. Supplementary Table 2 contains the data from a second independent mass spectrometry-based epigenetic analysis using pooled lin- FL cells from 3 WT and 3 RISP KO animals.
Loss of RISP leads to increased levels of 2-hydroxyglutrate, concomitant with a decreased NAD+/NADH ratio
The increase in DNA and histone methylation upon loss of RISP (Fig. 5) led us to examine the levels of key metabolites in RISP WT versus KO cells. The alpha-ketoglutarate (α-KG)-dependent dioxygenases, including JmJC domain-containing histone lysine demethylases (KDMs) and the TET (ten-eleven translocation) family of 5-methlycytosine (5mC) hydroxylases, which are involved in oxidizing 5-methylcytosine into 5-hydroxymethylcytosine, utilize oxygen, Fe(II), and α-KG to catalyze the hydroxylation of substrates, resulting in the decarboxylation of α-KG to succinate and release of carbon dioxide27. The TCA cycle metabolites succinate and fumarate, as well as the oncometabolite 2-hydroxyglutrate (2HG), are antagonists of α-KG, competitively inhibiting the activity of α-KG-dependent dioxygenases, including KDMs and TETs28, 29. Cancer cells harboring mutations in IDH1 and IDH2 have increased levels of D(R)-2-HG, concomitant with hypermethylation of histones and DNA27. Cancer cells deficient in fumarate hydratase display elevated levels of fumarate associated with increased histone and DNA methylation30, 31. Normal cells produce L(S)-2-HG from promiscuous substrate usage of α-KG by lactate dehydrogenase or malate dehydrogenase under conditions with a decreased NAD+/NADH ratio (e.g. oxygen limitation32, 33). L(S)-2-HG is a more potent inhibitor of α-KG-dependent dioxygenases than D(R)-2-HG or succinate34. We examined the NAD+/NADH ratio, as well as the levels of α-KG, succinate, fumarate and 2-HG in WT and RISP null fetal liver Lin- cells. As expected, the loss of complex III function due to RISP deficiency results in a decrease in the NAD+/NADH ratio (Fig. 6a). 2HG, α-KG, succinate, and fumarate were elevated in RISP null Lin- cells compared to WT cells (Fig. 6b). Furthermore, the 2-HG/α-KG ratio was significantly increased, while the succinate/α-KG and fumarate/αKG ratios were elevated (Fig. 6c-e). Since L(S)-2-HG is a known inhibitor of histone demethylases and TETs, these results suggest that accumulation of L(S)-2-HG, fumarate, and succinate in RISP null cells results in the hypermethylation of histone and DNA observed in Fig. 5. Glycolytic (Fig. 6f), pentose phosphate (Fig. 6g), and TCA cycle intermediates (Fig. 6h), as well as the overall amino acid and nucleotide pools, were maintained in RISP null Lin-cells (Supplementary Fig. 5) indicating that the proliferating RISP null cells are able to maintain many of the biosynthetic intermediates to support growth. Aspartate, a metabolite previously necessary for in vitro cellular proliferation and dependent on high NAD+/NADH ratio35, 36 was significantly diminished (Figure 6h).
Figure 6. Risp deficiency decreases NAD+/NADH ratio and increases 2-HG.
(a) The ratio of NAD+/NADH in RISP WT and KO Lin- FL cells. (WT n=12 mice; KO n=7 mice).
(b) Concentrations of a-ketoglutarate (αKG), succinate, fumarate, and 2-hydroxyglutrate (2-HG) in RISP WT and KO Lin- FL cells. The cellular concentration of 2-HG in RISP KO Lin- FL cells ranged from 1.75mM to 2.61mM with an average concentration of 2.21mM.
(c-e) The ratio of 2-HG, succinate, and fumarate to aKG in RISP WT and KO Lin- FL cells.
(f-g) Relative abundance of glycolytic and TCA cycle metabolites extracted from RISP WT and KO Lin- FL cells. Peak areas of each metabolite were normalized to cell number. Metabolites in Figure 6b-h were measured from WT n=7 mice; KO n=5 mice. Raw metabolite data is available as Supplementary Table 3. Data in Figure 6 are mean ± SEM. *p ≤ 0.05; **p ≤ 0.01. All p values calculated using Student's t-test.
RISP deficiency in adult HSCs impairs respiration and hematopoiesis
Adult bone marrow HSCs are more quiescent than fetal liver HSCs, which undergo continuous proliferation to generate blood cells in utero. We examined whether respiration was essential for adult HSCs by administering Poly (I:C) to Mx1-cre mice that were crossed to Uqcrfs1 floxed mice. This generated RISP deficient adult Lin- cells that exhibit impaired respiration enhanced glycolysis and a decrease in the NAD+/NADH ratio (Fig. 7a-c). Poly (I:C) induction resulted in pancytopenia and loss of bone marrow (BM) cellularity within 2 weeks, with subsequent lethality of animals within 3 weeks (Fig. 7d-f). To assess the repopulating capacity of adult RISP null HSCs, we performed a competitive reconstitution assay in which Rispfl/fl or Rispfl/fl; Mx1-Cre bone marrow cells (CD45.2) competed against an equal number of bone marrow cells from a congenic (CD45.1) mouse to reconstitute the hematopoietic compartment of an irradiated recipient mouse (CD45.1). Poly (I:C) was administered after 8 weeks of engraftment, which induced a rapid decline in Cre+ CD45.2 cells in the peripheral blood of mice, including T cells, B cells, and myeloid cells (Fig. 7g and Supplementary Fig. 6a). At 16 weeks post Poly (I:C), there was a substantial loss of RISP-deficient CD45.2 HSCs and progenitor populations in the bone marrow (Supplementary Fig. 6b). These results were confirmed with a second independent competitive transplantation assay Supplementary Fig. 6c-d). Thus, RISP is necessary to maintain adult HSC function in a cell-autonomous manner.
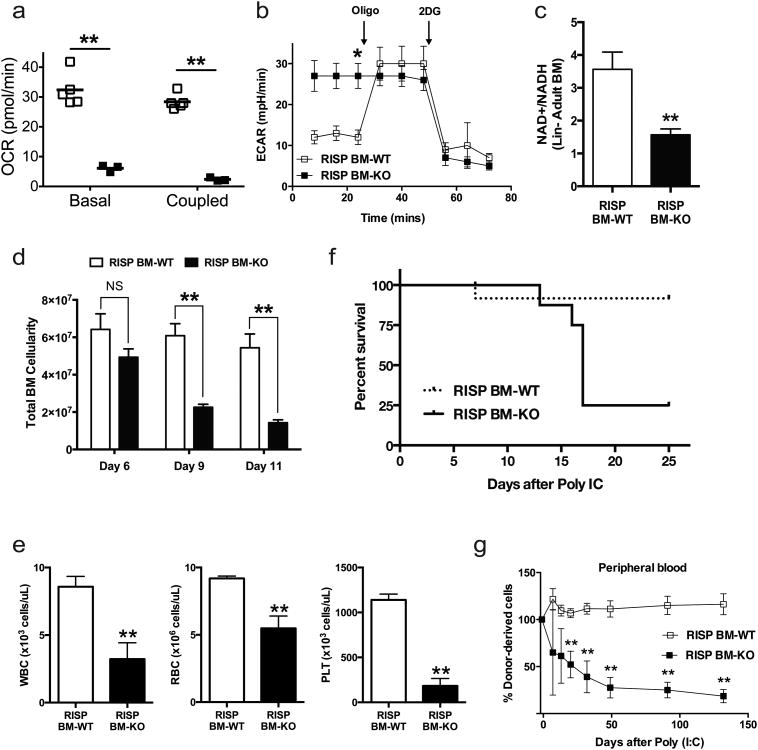
Figure 7. Loss of RISP in adult hematopoietic stem cells impairs mitochondrial oxidative metabolism and leads to a severe pancytopenia phenotype.
(a) Basal and coupled oxygen consumption rate (OCR) of Lin- BM cells from adult Rispfl/fl (RISP BM-WT) or Rispfl/fl Mx1-Cre (RISP BM-KO) mice 6 days after PolyI:C treatment. Data represent average of RISP BM-WT (n=5) or RISP BM-KO (n=3) ± SEM.
(b) Extracellular acidification rate (ECAR) of Lin- BM cells from RISP BM-WT and RISP BM-KO mice 6 days after Poly (I:C) treatment under basal conditions, treatment with 1uM oligomycin A (Oligo) or 20mM 2-deoxy-glucose (2DG). Data represents the average of RISP BM-WT (n=5) or RISP BM-KO (n=3) ± SEM.
(c) The ratio of NAD+/NADH in LSK cells sorted from BM of adult RISP BM-WT and RISP BM-KO mice 6 days after Poly (I:C) treatment. Data represent average of RISP BM-WT (n=6) or RISP BM-KO (n=6) ± SEM.
(d) Total cellularity of BM taken from two femurs and two tibias for each mouse, either 6, 9 or 11 days after Poly (I:C) treatment. Data represents average of RISP BM-WT (day 6: n=9, day 9: n=6, day 11: n=8 mice) or RISP BM-KO ((day 6: n=10, day 9: n=14, day 11: n=11mice) ± SEM.
(e) Peripheral blood cell counts in adult RISP BM-WT or RISP BM-KO mice 13 days after Poly (I:C) treatment. WBC (white blood cells), RBC (red blood cells), PLT (platelets). Data represent average of RISP BM-WT (n=10 mice) or RISP BM-KO (n=8 mice) ± SEM.
(f) Survival curve of adult RISP BM-WT or RISP BM-KO mice treated with Poly (I:C). Data represent RISP BM-WT (n=10 mice) or RISP BM-KO (n=9 mice) ± SEM. Surviving RISP BM-KO mice treated with Poly (I:C) failed to delete RISP.
(g) Total contribution of CD45.2 bone marrow (BM) donor cells to the peripheral blood of irradiated CD45.1 recipient mice in a competitive transplantation assay following engraftment and subsequent Poly (I:C) treatment. Data represent the percentage of CD45.2 donor cells relative to the initial chimerism. Data represent the average contribution for Rispfl/fl (RISP BM-WT; n=4 mice) and Rispfl/fl Mx1-Cre (RISP BM-KO; n=6 mice).
Data in Figure 7 are mean ± SEM. *p ≤ 0.05; **p ≤ 0.01. All p values calculated using Student's t-test.
RISP maintains adult HSC quiescence
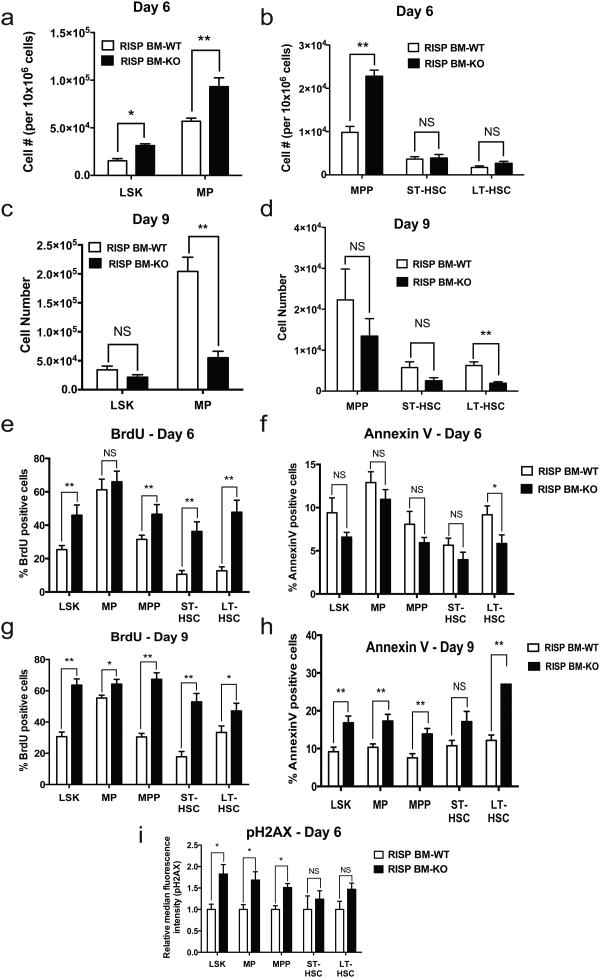
To examine the acute impact of RISP loss on HSCs and progenitor populations, we analyzed the bone marrow at 6 and 9 days after administering Poly (I:C) (Supplementary Fig. 7) LSK and myeloid progenitor populations were increased at day 6, but decreased by day 9 (Fig. 8 a-d). Loss of HSC quiescence and aberrant entry into cell cycle can lead to HSC exhaustion, hence we examined BrdU incorporation into HSCs at day 6 (Fig. 8e). Indeed RISP deficient adult HSCs and progenitors displayed increased BrdU incorporation, indicating entry into cell cycle. Importantly, there was no significant decrease in viability at day 6 (Fig. 8f). By day 9 however, despite the increase in BrdU labeling, there was significant HSC and progenitor cell death (Fig. 8g-h). The increase in proliferation of LSK at day 6 was accompanied by elevated phospho-H2AX staining indicative of cellular stress (Fig. 8i). Taken together these results indicate that RISP is essential for maintaining adult HSC quiescence.
Figure 8. RISP deficiency in adult HSCs leads to impaired differentiation, loss of quiescence, increased apoptosis and increased DNA damage.
(a and b) Cell number (per 10×106 cells) of indicated populations in adult BM of RISP BM-WT or RISP BM-KO mice 6 days after Poly (I:C) treatment. Data represent average of RISP BM-WT (n=7 mice) or RISP BM-KO (n=8 mice) ± SEM.
(c and d) Cell number of indicated populations in adult BM of RISP BM-WT or RISP BM-KO mice 9 days after Poly (I:C) treatment. Data represent average of RISP BM-WT (n=10 mice) or RISP BM-KO (n=14 mice) ± SEM.
(e and f) Percent of BrdU (e) or AnnexinV (f) positive cells in indicated populations in adult RISP BM-WT or RISP BM-KO mice 6 days after Poly (I:C) treatment. Data represents the average of RISP BM-WT (n=10 mice) or RISP BM-KO (n=9 mice) ± SEM.
(g and h) Percent of BrdU (e) or AnnexinV (f) positive cells in indicated populations in adult RISP BM-WT or RISP BM-KO mice 9 days after Poly (I:C) treatment. Data represents the average of RISP BM-WT (n=8 mice) or RISP BM-KO (n=9 mice) ± SEM.
(i) Median fluorescence intensity of pH2AX in indicated cell population in RISP BM-KO relative to RISP BM-WT mice. Data represents average RISP BM-WT (n=8 mice) or RISP BM-KO (n=8 mice) ± SEM.
Data in Figure 8 are mean ± SEM. *p<0.05; **p<0.01. All p values calculated using Student's t-test.
Discussion
Our present study identifies that completely disrupting respiration, which enhances glycolysis, diminishes fetal and adult HSC function in a cell-autonomous manner, impairing hematopoiesis, and leading to severe pancytopenia and subsequent lethality. Mitochondria, in addition to generating ATP, are biosynthetic and signaling organelles37. The TCA cycle generates intermediates that are essential for lipid, amino acid, and nucleotide synthesis, as well as for histone acetylation24. Fetal HSCs undergo continuous cycling and expand rapidly during development. Based on the critical role of mitochondria in various biosynthetic processes, the loss of respiration due to RISP depletion was predicted to decrease HSC proliferation. Surprisingly, we observed that RISP null fetal HSCs are able to proliferate and maintain mitochondrial membrane potential; however, they lose their stem cell properties, as indicated by a decrease in stem cell genes and a loss of multilineage repopulation potential. HSCs give rise to multipotent progenitors that are not able to self-renew but are able to undergo a complete hematopoietic lineage differentiation. The RISP null fetal HSCs were not able to generate sufficient numbers of multipotent progenitors indicating a defect in HSC differentiation. We and other investigators have noted that mitochondrial metabolism is essential for cellular differentiation, and our current work is consistent with these previous findings38-41.
A potential explanation for impaired differentiation of RISP null HSCs is the hypo-acetylation due to the diminished levels of citrate generated in the mitochondria as well as the increased 2-HG levels due to a decreased NAD+/NADH ratio, which cause the hypermethylation of DNA and histones. Interestingly, L(S)-2-HG is oxidized in the mitochondria and converted back to α-KG by L(S)-2-HG dehydrogenases (L2HGDH), a FAD-containing enzyme that is dependent on active complex III32. Thus, loss of complex III activity both decreases NAD+/NADH and impairs L2HGDH activity to favor 2-HG production. Our data are consistent with the previous finding that cancer cells harboring mutations in the cytochrome b subunit of complex III have elevated 2-HG levels42. L-2HG is known to drive epigenetics changes in renal cell carcinoma43. It is important to note that succinate and fumarate were elevated in our RISP null cells thus could also participate in inhibiting histone demethylases and TETs. Taken together, these data suggest that respiration controls HSC function through metabolites that regulate epigenetics, consistent with the idea that mitochondria can serve as signaling organelles.
While fetal HSCs cycle rapidly, adult HSCs are largely quiescent. Respiratory impairment in the adult HSCs caused loss of quiescence and entry into the cell cycle resulting in a depleted progenitor cell pool and lethality. Thus respiration is not necessary for cell proliferation in either fetal or adult HSCs. This is in contrast to the observation that cancer cells with respiratory deficiency due to the loss of mtDNA, display diminished tumorigenesis in vivo44 . Additionally, the anti-diabetic drug, metformin, exerts anti-tumorigenic activity by inhibiting mitochondrial complex I, highlighting the essential requirement of the functional respiratory chain for tumor growth45. It is not fully understood why a functional respiratory chain is necessary for tumor cell proliferation in vivo. One model based on in vitro studies indicates that impairment of respiration decreases the NAD+/NADH ratio to limit intracellular aspartate levels required for cell proliferation 35, 36. Moreover, a critical step in pyrimidine synthesis is the dihydroorotate dehydrogenase (DHOD) enzyme, which requires active complex III activity46. Thus, complex III deficient proliferating cancer cells require exogenous pyruvate to maintain NAD+/NADH ratio for aspartate synthesis and exogenous uridine to support pyrimidine synthesis44. Our current findings suggest that complex III deficient mouse HSCs, despite decreased levels of NAD+/NADH, are able to proliferate, suggesting that they exhibit remarkable metabolic plasticity that allows them to acquire the necessary metabolites for cell proliferation in the absence of a functional respiratory chain. It is important to note that respiratory deficient cells are able to conduct reductive TCA cycle metabolism rather than the canonical oxidative TCA metabolism to maintain metabolites levels47. We speculate that HSCs are not impaired in their ability to proliferate in the absence of respiration, in part because they are able to conduct reductive TCA cycle metabolism to provide adequate building blocks for proliferation. Alternatively, they may acquire metabolites from the HSC environmental niche to support proliferation.
In conclusion, we have identified three important functions of respiration in maintenance of hematopoiesis homeostasis. First, respiration is dispensable for HSC proliferation. Second, respiration is required to maintain fetal HSC differentiation into progenitors and adult HSC quiescence. Third, respiration maintains the epigenetic landscape by likely repressing 2-HG, fumarate and succinate levels in HSCs. These findings indicate that mitochondrial metabolism plays a causal role in hematopoiesis by influencing fate decisions of HSCs possibly through 2-HG rather than simply fulfilling the biosynthetic and bioenergetics requirements of HSCs. It will be interesting to learn whether our findings extend to other stem cell models.
Methods
Mice
C57BL/6 Uqcrfs1 (RISPfl/fl)48 and C57BL/6 Tfamfl/fl,48 mice were described previously. Vav-iCre (Stock #008610) and Mx-1 Cre (Stock #003556) mice were obtained from the Jackson laboratory. Recipients in transplantation assays were adult C57BL/6 CD45.1 mice (Jackson Laboratory, stock#002014). L-buthionine-sulfoximine (Sigma) at 2 or 20 mM and N-acetylcysteine (Sigma) at 1 mg/mL were administered in drinking water to 8.5 d.p.c (days post coitum) pregnant females until the day fetuses were extracted. Poly I:C (GE) was injected intraperitoneally every other day for five days at a dose of 12mg/kg. All animal procedures were approved by Institutional Animal Care and Use Committee (IACUC) at Northwestern University.
Flow cytometry
Fetal livers from 15.5 days embryos were minced with scissors in PBS containing 0.1% BSA. To obtain single cells suspension the tissue was triturated through a 23.5G needle and filtered through a nylon strainer (70 or 100 μm). HSC and MPP studies were performed by staining FL cells with a lineage antibody cocktail biotin-conjugated (including CD45R/B220, CD5, Gr-1, Ter119, CD8, CD19, 7-4, Stem Cell Technologies) and posterior staining with streptavidin-eFluor 450 (eBioscience), CD48 (HM48-1, BD Biosciences), CD150 (TC15-12F12.2, BioLegend), Sca-1 (D7, eBioscience) and Mac-1 (M1/70, BD Biosciences). Staining with fixable viability dye eFluor®506 (eBioscience) was previously done to distinguish live from dead cells. Staining with MitoSox or DCFH was performed at 5 μM, MitoTracker Green (MTG) at 20 nM and tetramethylrhodamine ethyl ester perchlorate (TMRE) at 5 nM (Sigma), all for 30 minutes at 37°C after surface antigen staining (all from Molecular Probes). In some studies, BrdU at 100 mg/Kg was injected intraperitoneally into pregnant females 6 hours before extraction of the fetuses. FITC-BrdU staining was performed following manufacturing instructions (BD Biosciences). Annexin V analyses were performed with a FITC conjugated antibody (BD Biosciences) by staining the cells in annexin V buffer (BD Biosciences) at RT after antigen surface staining. After that, surface antigen staining was performed as described and cells were fixed for 30 minutes with fixation buffer (BD Biosciences). Then, cells were incubated with pyronin Y (Sigma) at 1 μg/ml for 30 minutes at 4°C. Lineage negative cells were purified following manufacturer's instruction. Lin- or HSCs were incubated with 2-NDBG (Invitrogen) at 100 μM for 1 hour at 37°C in DMEM containing SCF, IL-3, and IL-6 at 10 ng/mL, EPO at 3U/mL and uridine at 100 μg/ml. In the case of the HSCs, cell death and surface antigen stainings were performed after 2-NDBG incubation.
To study lineage maturation FL cells were stained with conjugated antibodies against specific surface markers including Ter119 (BD Biosciences), CD71 (R17217, BioLegend), Mac-1 (M1/70, BD Biosciences), Gr-1 (RB6-8C5, BD Biosciences), CD41 (MWreg30, BD Biosciences) and CD42 (Xia.G5, Emfret).
Myeloid progenitors were analyzed by staining with a lineage antibody cocktail biotin-conjugated as done for HSCs, and posterior staining with streptavidin (eBiosciences), Sca-1 (D7, eBioscience), c-kit (2B8, eBiosciences), CD34 (RAM34, eBiosciences) and FgIIIcR (2.4G2, BD Biosciences). MitoSOX and TMRE incubations were performed as described for HSCs.
Bone marrow was recovered by sequential smashing of the recovered bones using a mortar and pestle in 5ml of HBSS + 2%FBS. The mixture was filtered through a 70μM filter and washed 3 times with an additional 5ml of HBSS + 2%FBS. Viability staining was performed following red blood cell lysis (eBioscience) using Live/Dead Fixable Blue Dead Cell Stain (ThermoFisher Scientific). Surface staining was performed in PBS + 2%FBS. For lineage identification cells were stained with Anti-TER119 (Clone: TER-119), Anti-NK1.1 (Clone: PK136), Anti-CD45R (Clone: RA3-6B2), Anti-CD4 (Clone: GK1.5), Anti-CD8α (Clone: 53-6.7), Anti-CD11b (Clone: M1/70), and Anti-Ly6G & Ly6C (RB6-8C5). LSK and MP were identified using Anti-Sca-1 (Clone: D7) and Anti-CD117 (Clone 2B8). MPPs and HSCs were further classified using anti-CD150 (Clone: Q38-480) and anti-CD48 (Clone: HM48-1). Measurement of apoptotic cells was performed by staining cells with Annexin V, Alexa Fluor® 350 conjugate (ThermoFisher) for 30 minutes following surface staining. Measurement of BrdU incorporation in adult mice in vivo was analyzed using the BrdU Staining Kit for Flow Cytometry PerCP-eFluor® 710 (eBioscience), and following the manufacturer's instructions. Mice were injected intraperitoneally with 170mg/kg of BrdU (Sigma) dissolved in PBS 24hrs prior to isolation of bone marrow. Following BrdU injections, mice were kept on drinking water containing 1mg/ml of BrdU until isolation.
For the bone marrow transplantation assays, repopulation contribution was studied by staining with CD45.1 (A20, BD Biosciences), CD45.2 (104, BD Biosciences), Gr1 (RB6-8C5, BD Biosciences), Mac1 (M1/70, BD Biosciences), CD3 (17A2, BD Biosciences), B220 (RA3-6B2, BD Biosciences), lineage-streptavidin, Sca-1, c-kit and CD34. All flow cytometry analyses were performed in a BDLSRII or BD Fortessa analyzers and data were analyzed with the FlowJo software. Sorting of myeloid progenitors cells and HSCs was performed in a MoFlo FACS (Beckman-Coulter).
Protein extraction and western blot
Lineage negative cells were purified following manufacturer's instruction (Stem Cell Technologies). Then, cells were lysed in cell lysis buffer (Cell Signaling), resolved on 4 to 20% polyacrylamide gels (BioRad) and transferred to nitrocellulose membranes. Membranes were blocked using 5% Milk for 1 hour and then incubated in the primary antibody overnight. Antibodies used were anti-RISP (Mitosciences; Catalog Number: ab14746. Used at a 1:500 dilution), anti-TFAM (a gift from Gerald Shadel, Yale University. Used at a 1:500 dilution), anti-β-actin (Sigma; Catalog Number: A2066. Used at 1:10,000 dilution) and anti-α-Tubulin (Sigma; Catalog Number: T9026. Used at a 1:2000 dilution). Membranes were washed 4 times with TBST and then incubated with secondary antibody. Secondary antibodies used were Anti-mouse IgG, HRP-Linked (Cell Signaling; Catalog Number: 7076S. 1:10,000 Dilution, used for anti-TFAM), Anti-rabbit IgG, HRP-Linked (Cell Signaling; Catalog Number: 7074S. 1:10,000 Dilution, used for anti-β-actin), and Goat Anti-Mouse IRDye 680RD (Li-cor; Catalog Number: C50721. 1:5,000 dilution. Used for Anti-RISP and Anti-Tubulin). TFAM and actin blots were then treated wth ECL (Pierce) and developed using film. The RISP and tubulin blots were directly imaged using the Odyssey Fc Analyzer (Licor).
Blood count
Peripheral blood from 18.5 days embryos or 8 to 12-week old adult mice was collected in EDTA-treated tubes (SARSDTED) and cell blood parameters were analyzed in a HEMAVET cell blood counter machine.
Colony formation unit assays
Colony formation units for CFU-GM, BFU-E and CFU-GEMM were performed in Methocult media and CFU-Mk in MegaCult-collagen gels following manufacturing instructions (Stem Cell Technologies). 105 FL cells were plated per well and colonies were counted 8 to 10 days after. Normoxic condition (21% O2), hypoxic conditions (1.5% O2), or treatments with NAC at 500 μM or BSO at 500 μM were performed.
RNA-seq analysis
Total RNA was isolated using RNeasy Plus Micro Kit (Qiagen) following manufacturer's protocol. RNA-seq library was prepared using the SMARTer® Stranded Total RNA-Seq Kit – Pico Input Mammalian (Takara-Clontech). Libraries were pooled and sequenced on an Illumina Nextseq500 platform using the 75bp high output sequencing kit. The sequencing reads were aligned to mouse (mm9) reference genome by TopHat v2.0.1350 with the parameters: --solexaquals --no-novel-juncs. The raw read counts were calculated using HTSeq51. Differential gene expression analyses were performed by Cufflinks v2.2.152 using fold change ≥ 1.5 and p-value ≤ 0.01 as the cutoff. For specific gene expression, 1 μg was reverse transcribed with RETROscript first strand synthesis kit (Ambion) and qRT-PCR was performed on a Bio-Rad iCycler iQ with iQSYBR Green Supermix (Bio-Rad). The primers used were p16Ink4a forward: GTACCCCGATTCAGGTGATG and reverse: GGAGAAGGTAGTGGGGTCCT; Gata-2 forward: TGCTGTGGCTGGCTGAATCC and reverse: TCGTCTCCCTGGGCTCACTC; Gata-1 forward: CCTTGACCTTGTGGCAGAG and reverse: AGGTAGAGGCAGGAGAATGG; PU.1 forward: AGGGCAACCGCAAGAAGATGAC and reverse: GCACCTCGCCGCTGAACTG; CEBPα forward: CCGCTGTTGCTGAAGGAACTTG and reverse: CACCGCTGGGACACAGAGAC; Risp forward: CTGCTTCTGTCCGTTTTTCC and reverse: GCATAAGCAACACCCACAGT; Actin forward: ATTGTTACCAACTGGGACATG and reverse: CGAAGTCTAGAGCAACATAGCACA; HRPT forward: CCTCATGGACTGATTATGGACA and reverse: ATGTAATCCAGCAGGTCAGCAA; HoxA9 forward: CGCCGACGCTGCTGATGAG and reverse: CACCGCCGCCTTGGACTG; MOZ forward: GTGTCAGTTTGGGGCATCTC and reverse: CTTAAGCCAGTCTAGGGGGT; Cited2 forward: CGCATCATCACCAGCAGCAG and reverse: CGCTCGTGGCATTCATGTTG; Runx1 forward: CTCACTAGGCAGTGGGTTCTTT and reverse: GACCATCAGAAATCTCCATCTCAT; Slc forward: CTCACTAGGCAGTGGGTTCTTT and reverse: GACCATCAGAAATCTCCATCTCAT; Pbx forward: AGCTAACTCGCCCTCTACTC and reverse: TTGACATATCACTGATGCCCTG.
Oxygen consumption and extracellular acidification rate measurements
OCR and ECAR were measured in a XF24 or XF96 extracellular flux analyzers (Seahorse Bioscience). Basal mitochondrial respiration was measured by subtracting the OCR values after treatment with 1 μM antimycin A and 1 μM rotenone (sigma). Coupled respiration was determined by treatment with 1 μM oligomycin A (Sigma), by the subtraction of oligomycin A values from basal respiration. Experiments were performed in Seahorse media (DMEM containing 0.5% dialyzed serum, 10 mM glucose and 2 mM glutamine, lacking bicarbonate and HEPES according to Seahorse Bioscience's instructions. The media was supplemented with SCF, IL-3 and IL-6 at 10 ng/mL, EPO at 3U/mL and uridine at 100 μg/ml). Glycolytic flux was determined by ECAR sensitivity to 2-deoxyglucose (2-DG, 40mM).
Metabolite measurements and analysis
Frozen cell pellets were thawed on wet ice and extracted with 80/20 MeOH/H2O (v/v) at a ratio of 225 μL extraction solution to 1X106 cells. Extracted samples were centrifuged at 14,000 RPMs for 15 min at 4 ° C. A volume of supernatant equivalent to 450X103 cells per well (100 μL) was transferred to v-bottom 96-well plates and evaporated under reduced pressure. Prior to injection, dried extracts were reconstituted in LC-MS grade water (30 μL). The extracted samples were analyzed with LC-MS metabolomics. Metabolites were analyzed for relative abundance by high-resolution accurate mass detection (HRAM). HRAM data was acquired using a QExactive™ Orbitrap mass spectrometer (Thermo Fisher Scientific) operated in negative ion mode coupled to an Accela pump. LCMS in negative ion mode was achieved by usage of a Phenomenex Synergi Hydro-RP 100Å (2.5 μm, 2.0 mm x 100 mm) column and by implementation of a gradient elution program as described53. Ionization source working parameters were optimized. The heater temperature was set to 400 °C, ion spray voltage was set to 2.75 kV. A 22 min full-scan method was used to acquire data. An m/z scan range from 80 to 1200 was chosen and the resolution was set at 70,000. The automatic gain control (AGC) target was set at 1X106 and the maximum injection time was 500 ms. Instrument control and acquisition were carried out by Xcalibur 2.2 software (Thermo Fisher Scientific) and Tracefinder™ 2.1 software, respectively (Thermo Fisher Scientific). The elution gradient was carried out with a binary solvent system consisting of 3% methanol in water, 10 mM tributylamine, 15mM acetic acid, brought to pH of 5.0 +/- 0.05 using acetic acid (solvent A) and 100% methanol (solvent B) at a constant flow rate of 200 μL min−1. The linear gradient employed was as follows: 0–2.5 min 100% A, 2.5–5 min increase from 0 to 20% B, 5–7.5 min maintain 80% A and 20% B, 7.5–13 min increase from 20 to 55% B, 13–15.5 min increase from 55 to 95% B, 15.5–18.5 min maintain 5% A and 95% B, 18.5–19 min decrease from 95 – 0% B, followed by 6 min of re-equilibration at 100% A. The column temperature was maintained at 25 °C and sample volumes of 10 μL were injected. For cellular concentrations of 2-HG, lineage negative cells were isolated as described above, and cell diameter was measured using Cellometer Vision CBA Image Cytometer (Nexcelom). The average cell diameter was measured as 9.2μM and the cells were assumed to be spherical and contain 70% water.
Mass spectrometry to identify histone modifications
Nuclei were isolated using gentle detergent treatment (0.3% NP-40 in NIB-250 buffer) of cells and centrifugation at 0.6g. Detergent was removed by 2x washing of obtained pellets with NIB-250, no NP-40 buffer. Histones from isolated nuclei were acid-extracted and derivatized with propionic anhydride both prior to and following trypsin digestion as previously described (Garcia, B. A. et al. Nat. Protocols, 2007). Propionylated histone peptides were resuspended in 50μL water with 0.1% TFA and 3μL were injected in 3 technical replicates on nanoLC/triple quadrupole MS which consisted of a Dionex UltiMate 3000 coupled to a ThermoFisher Scientific TSQ Quantiva triple quadrupole mass spectrometer. Buffer A was 100% LC-MS grade water with 0.1% formic acid and buffer B was 100% ACN. The propionylated peptides were loaded onto an in-house packed C18 trapping column (4 cm × 150 μm; Magic AQ C18, 3μm, 200 Å -Michrom) for 10 min at a flow rate of 2.5 μL/min in 0.1% TFA loading buffer. The peptides were separated by a gradient from 1 to 35% buffer B from 5 to 45 min. The analytical column was 10 cm × 75 μm PicoChip (1PCH7515-105H253-NV New Objective) consisted of the same C18 material as the trapping column. The triple quadrupole settings were as follows: collision gas pressure of 1.5 m Torr; Q1 peak width of 0.7 (FWHM); cycle time of 3 s; skimmer offset of 10 V; electrospray voltage of 2.5kV. SRM mass spectrometer transitions were developed as described previously54, 55. Data were analyzed using Skyline software (v3.5; MacCoss Lab, University of Washington) with Savitzky–Golay smoothing of peaks56. Automatic peak assignment and retention times were checked manually. Final results are shown as a relative percentage of given modification in the total pool of given peptide which was quantified. The mass spectrometry experiment measurement of all the epigenetic marks requires an abundance of cells. Thus, we pooled lineage negative fetal liver cells from WT and KO samples in order to achieve an adequate cell number for analysis.
Cytosine Methylation Analysis
DNA methylation was measured using the MethylFlash Methylated DNA 5-mC Quantification Kit-Colorimetric (Epigentek) following the manufacturers instructions. In brief, DNA was isolated from purified lin- cells using the QIAamp DNA mini kit (Qiagen). DNA concentrations were measured using the Qubit® dsDNA HS Assay Kit (ThermoFisher). 50ng of DNA was loaded per well of the MethylFlash kit. The absolute quantity of methyl-cytosine in samples was obtained through the generation of a standard curve of known concentrations of methyl-cytosine (0, .5, 1, 2, 5 and 10ng). Color change was measured using the SpectraMax M Series Multi-Mode Microplate Reader (Molecular Probes).
Competitive repopulation assays
Competitive transplantation assays using Rispfl/fl Vav-iCre were performed by coinjection of 2×105 FL cells from E15.5d Rispfl/fl Vav-iCre or RISPfl/fl CD45.2 donors with 5×105 adult bone marrow cells from CD45.1 mice into lethally irradiated CD45.1 recipients. Four, eight and twelve weeks after transplantation peripheral blood was extracted and overall, B, T and myeloid lineage reconstitution was analyzed. Additionally, competitive transplantation assays using RISPfl/fl Mx-1 Cre animals were performed as follows. 1×105 FL cells from E15.5d RISPfl/fl Mx-1 Cre or RISPfl/fl CD45.2 donors and 9×105 adult bone marrow cells from CD45.1 mice or 5×105 adult bone marrow cells from RISPfl/fl Mx-1 Cre or RISPfl/fl CD45.2 donors and 5×105 adult bone marrow cells from CD45.1 mice were injected retro-orbitally into lethally irradiated CD45.1 recipients. 8 weeks post-transplant mice were treated with 10mg/kg of poly I:C (GE) every other day for five days. Levels of reconstitution in the peripheral blood were measured for 4 months using Anti-CD45.1 (Clone: A20), Anti-CD45.2 (Clone: 104), Anti-CD11b (Clone: M1/70), Anti-Ly6G (RB6-8C5), Anti-CD45R (Clone: RA3-6B2), and Anti-CD3 (Clone: 17A2). 4 months following poly I:C administration, the relative contributions to the HSC pool were analyzed using the same staining protocol described above.
Cell Lines
No cell lines were used in this study. No cell lines used in this study were found in the database of commonly misidentified cell lines that is maintained by ICLAC and NCBI Biosample. The cell lines were not authenticated. The cell lines were not tested for mycoplasma contamination.
Statistics and reproducibility
P-values were calculated by using an unpaired Student's t-test using Graphpad Prism 7 (GraphpadSoftware). Data are present as mean ± s.e.m unless specifically stated in the figure legend. Numbers of biological or technical replicates are stated in the figure legends. Randomization was not performed in animal experiments. The investigators were not blinded to allocation during experiments and outcome assessment. All images are representative of at least 3 three independent experiments or mice of the same genotype. Western blots are representative of at least 3 independent experiments, and whole gel images of published blots are displayed in supplementary figure 8. No statistical method was used to predetermine sample size and experiments were not randomized.
Data availability
RNA-seq data seen in figure 5 have been deposited Gene Expression Omnibus (GEO) under accession code GSE95341. The mass spectrometry proteomics data in Figure 5 and Supplementary Table 1 and 2 have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD006054. Raw metabolomics data supporting Figure 6 have been included in Supplementary Table 3. All other data from manuscript are available from the corresponding author upon reasonable request.
Supplementary Material
Acknowledgments
This work was supported by NIH (R35CA197532) to NSC, NIH (T32 GM008061) to LPD, NIH (T32 T32HL076139) to SEW. J.X. is supported by the NIH/NIDDK grants K01DK093543 and R01DK111430 and the Cancer Prevention and Research Institute of Texas (CPRIT) New Investigator award (RR140025). We thank Robert H. Lurie Cancer Center Flow Cytometry facility supported by NCI CCSG P30 CA060553 for their invaluable assistance. Proteomics services were performed by the Northwestern Proteomics Core Facility, generously supported by NCI CCSG P30 CA060553 awarded to the Robert H Lurie Comprehensive Cancer Center and the National Resource for Translational and Developmental Proteomics supported by P41 GM108569.
Footnotes
Author Contributions: E.A, S.E.W. and L.P.D carried out most of the experiments in the paper. B.T and S.M. provided technical expertise and carried out some of the initial experiments. -X.L, Y.Z, and Z.S did RNA sequence analysis. M.S and K.M conducted and analyzed metabolomics. P.T.S generated the RISP KO mice. E.A, S.E.W, J.X., L.P.D, J.D.C and N.S.C provided intellectual input and wrote the paper.
References
- 1.Chandel NS, Jasper H, Ho TT, Passegue E. Metabolic regulation of stem cell function in tissue homeostasis and organismal ageing. Nature cell biology. 2016;18:823–832. doi: 10.1038/ncb3385. [DOI] [PubMed] [Google Scholar]
- 2.Ito K, Suda T. Metabolic requirements for the maintenance of self-renewing stem cells. Nature reviews Molecular cell biology. 2014;15:243–256. doi: 10.1038/nrm3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Takubo K, et al. Regulation of Glycolysis by Pdk Functions as a Metabolic Checkpoint for Cell Cycle Quiescence in Hematopoietic Stem Cells. Cell stem cell. 2013;12:49–61. doi: 10.1016/j.stem.2012.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Adelman DM, Maltepe E, Simon MC. Multilineage embryonic hematopoiesis requires hypoxic ARNT activity. Genes & development. 1999;13:2478–2483. doi: 10.1101/gad.13.19.2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Simsek T, et al. The Distinct Metabolic Profile of Hematopoietic Stem Cells Reflects Their Location in a Hypoxic Niche. Cell stem cell. 2010;7:380–390. doi: 10.1016/j.stem.2010.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Norddahl Gudmundur L, et al. Accumulating Mitochondrial DNA Mutations Drive Premature Hematopoietic Aging Phenotypes Distinct from Physiological Stem Cell Aging. Cell stem cell. 2011;8:499–510. doi: 10.1016/j.stem.2011.03.009. [DOI] [PubMed] [Google Scholar]
- 7.Parmar K, Mauch P, Vergilio JA, Sackstein R, Down JD. Distribution of hematopoietic stem cells in the bone marrow according to regional hypoxia. Proceedings of the National Academy of Sciences. 2007;104:5431–5436. doi: 10.1073/pnas.0701152104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Spencer JA, et al. Direct measurement of local oxygen concentration in the bone marrow of live animals. Nature. 2014;508:269–273. doi: 10.1038/nature13034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tothova Z, et al. FoxOs Are Critical Mediators of Hematopoietic Stem Cell Resistance to Physiologic Oxidative Stress. Cell. 2007;128:325–339. doi: 10.1016/j.cell.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 10.Ito K, et al. Reactive oxygen species act through p38 MAPK to limit the lifespan of hematopoietic stem cells. Nat Med. 2006;12:446–451. doi: 10.1038/nm1388. [DOI] [PubMed] [Google Scholar]
- 11.Chen C, et al. TSC–mTOR maintains quiescence and function of hematopoietic stem cells by repressing mitochondrial biogenesis and reactive oxygen species. The Journal of experimental medicine. 2008;205:2397–2408. doi: 10.1084/jem.20081297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jung H, et al. TXNIP Maintains the Hematopoietic Cell Pool by Switching the Function of p53 under Oxidative Stress. Cell metabolism. 2013;18:75–85. doi: 10.1016/j.cmet.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 13.Ito K, et al. Regulation of oxidative stress by ATM is required for self-renewal of haematopoietic stem cells. Nature. 2004;431:997–1002. doi: 10.1038/nature02989. [DOI] [PubMed] [Google Scholar]
- 14.Maryanovich M, et al. The ATM-BID pathway regulates quiescence and survival of haematopoietic stem cells. Nature cell biology. 2012;14:535–541. doi: 10.1038/ncb2468. [DOI] [PubMed] [Google Scholar]
- 15.Ahlqvist KJ, et al. Somatic progenitor cell vulnerability to mitochondrial DNA mutagenesis underlies progeroid phenotypes in Polg mutator mice. Cell metabolism. 2012;15:100–109. doi: 10.1016/j.cmet.2011.11.012. [DOI] [PubMed] [Google Scholar]
- 16.Maryanovich M, et al. An MTCH2 pathway repressing mitochondria metabolism regulates haematopoietic stem cell fate. Nature communications. 2015;6:7901. doi: 10.1038/ncomms8901. [DOI] [PubMed] [Google Scholar]
- 17.Yu WM, et al. Metabolic Regulation by the Mitochondrial Phosphatase PTPMT1 Is Required for Hematopoietic Stem Cell Differentiation. Cell stem cell. 2013;12:62–74. doi: 10.1016/j.stem.2012.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gan B, et al. Lkb1 regulates quiescence and metabolic homeostasis of haematopoietic stem cells. Nature. 2010;468:701–704. doi: 10.1038/nature09595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gurumurthy S, et al. The Lkb1 metabolic sensor maintains haematopoietic stem cell survival. Nature. 2010;468:659–663. doi: 10.1038/nature09572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakada D, Saunders TL, Morrison SJ. Lkb1 regulates cell cycle and energy metabolism in haematopoietic stem cells. Nature. 2010;468:653–658. doi: 10.1038/nature09571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sena Laura A, et al. Mitochondria Are Required for Antigen-Specific T Cell Activation through Reactive Oxygen Species Signaling. Immunity. 2013;38:225–236. doi: 10.1016/j.immuni.2012.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ema H, Nakauchi H. Expansion of hematopoietic stem cells in the developing liver of a mouse embryo. Blood. 2000;95:2284–2288. [PubMed] [Google Scholar]
- 23.Larsson NG, et al. Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nat Genet. 1998;18:231–236. doi: 10.1038/ng0398-231. [DOI] [PubMed] [Google Scholar]
- 24.Martinez-Reyes I, et al. TCA Cycle and Mitochondrial Membrane Potential Are Necessary for Diverse Biological Functions. Molecular cell. 2015 doi: 10.1016/j.molcel.2015.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Orkin SH, Zon LI. Hematopoiesis: an evolving paradigm for stem cell biology. Cell. 2008;132:631–644. doi: 10.1016/j.cell.2008.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McDonnell E, et al. Lipids Reprogram Metabolism to Become a Major Carbon Source for Histone Acetylation. Cell reports. 2016;17:1463–1472. doi: 10.1016/j.celrep.2016.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Losman JA, Kaelin WG., Jr What a difference a hydroxyl makes: mutant IDH, (R)-2-hydroxyglutarate, and cancer. Genes & development. 2013;27:836–852. doi: 10.1101/gad.217406.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu W, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases. Cancer cell. 2011;19:17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xiao M, et al. Inhibition of alpha-KG-dependent histone and DNA demethylases by fumarate and succinate that are accumulated in mutations of FH and SDH tumor suppressors. Genes & development. 2012;26:1326–1338. doi: 10.1101/gad.191056.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sullivan Lucas B, et al. The Proto-oncometabolite Fumarate Binds Glutathione to Amplify ROS-Dependent Signaling. Molecular cell. doi: 10.1016/j.molcel.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sciacovelli M, et al. Fumarate is an epigenetic modifier that elicits epithelial-to-mesenchymal transition. Nature. 2016;537:544–547. doi: 10.1038/nature19353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Engqvist MK, Esser C, Maier A, Lercher MJ, Maurino VG. Mitochondrial 2-hydroxyglutarate metabolism. Mitochondrion. 2014;19(Pt B):275–281. doi: 10.1016/j.mito.2014.02.009. [DOI] [PubMed] [Google Scholar]
- 33.Oldham WM, Clish CB, Yang Y, Loscalzo J. Hypoxia-Mediated Increases in L-2-hydroxyglutarate Coordinate the Metabolic Response to Reductive Stress. Cell metabolism. 2015;22:291–303. doi: 10.1016/j.cmet.2015.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chowdhury R, et al. The oncometabolite 2-hydroxyglutarate inhibits histone lysine demethylases. EMBO reports. 2011;12:463–469. doi: 10.1038/embor.2011.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Birsoy K, et al. An Essential Role of the Mitochondrial Electron Transport Chain in Cell Proliferation Is to Enable Aspartate Synthesis. Cell. 2015;162:540–551. doi: 10.1016/j.cell.2015.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sullivan LB, et al. Supporting Aspartate Biosynthesis Is an Essential Function of Respiration in Proliferating Cells. Cell. 2015;162:552–563. doi: 10.1016/j.cell.2015.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chandel NS. Evolution of Mitochondria as Signaling Organelles. Cell metabolism. 2015;22:204–206. doi: 10.1016/j.cmet.2015.05.013. [DOI] [PubMed] [Google Scholar]
- 38.Tormos Kathryn V, et al. Mitochondrial Complex III ROS Regulate Adipocyte Differentiation. Cell metabolism. 2011;14:537–544. doi: 10.1016/j.cmet.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hamanaka RB, et al. Mitochondrial Reactive Oxygen Species Promote Epidermal Differentiation and Hair Follicle Development. Sci Signal. 2013;6:ra8. doi: 10.1126/scisignal.2003638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Owusu-Ansah E, Banerjee U. Reactive oxygen species prime Drosophila haematopoietic progenitors for differentiation. Nature. 2009;461:537–541. doi: 10.1038/nature08313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang J, et al. UCP2 regulates energy metabolism and differentiation potential of human pluripotent stem cells. The EMBO journal. 2011;30:4860–4873. doi: 10.1038/emboj.2011.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mullen AR, et al. Oxidation of alpha-ketoglutarate is required for reductive carboxylation in cancer cells with mitochondrial defects. Cell Rep. 2014;7:1679–1690. doi: 10.1016/j.celrep.2014.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shim EH, et al. L-2-Hydroxyglutarate: an epigenetic modifier and putative oncometabolite in renal cancer. Cancer discovery. 2014;4:1290–1298. doi: 10.1158/2159-8290.CD-13-0696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weinberg F, et al. Mitochondrial metabolism and ROS generation are essential for Kras-mediated tumorigenicity. Proceedings of the National Academy of Sciences. 2010;107:8788–8793. doi: 10.1073/pnas.1003428107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wheaton WW, et al. Metformin inhibits mitochondrial complex I of cancer cells to reduce tumorigenesis. eLife. 2014;3:e02242. doi: 10.7554/eLife.02242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Evans DR, Guy HI. Mammalian pyrimidine biosynthesis: fresh insights into an ancient pathway. The Journal of biological chemistry. 2004;279:33035–33038. doi: 10.1074/jbc.R400007200. [DOI] [PubMed] [Google Scholar]
- 47.Mullen AR, et al. Reductive carboxylation supports growth in tumour cells with defective mitochondria. Nature. 2012;481:385–388. doi: 10.1038/nature10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sena Laura A, et al. Mitochondria Are Required for Antigen-Specific T Cell Activation through Reactive Oxygen Species Signaling. Immunity. 2013;38:225–236. doi: 10.1016/j.immuni.2012.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hamanaka RB, et al. Mitochondrial Reactive Oxygen Species Promote Epidermal Differentiation and Hair Follicle Development. Sci Signal. 2013;6:ra8. doi: 10.1126/scisignal.2003638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature biotechnology. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lu W, et al. Metabolomic analysis via reversed-phase ion-pairing liquid chromatography coupled to a stand alone orbitrap mass spectrometer. Analytical chemistry. 2010;82:3212–3221. doi: 10.1021/ac902837x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zheng Y, Tipton JD, Thomas PM, Kelleher NL, Sweet SM. Site-specific human histone H3 methylation stability: fast K4me3 turnover. Proteomics. 2014;14:2190–2199. doi: 10.1002/pmic.201400060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zheng Y, Thomas PM, Kelleher NL. Measurement of acetylation turnover at distinct lysines in human histones identifies long-lived acetylation sites. Nature communications. 2013;4:2203. doi: 10.1038/ncomms3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.MacLean B, et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26:966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data seen in figure 5 have been deposited Gene Expression Omnibus (GEO) under accession code GSE95341. The mass spectrometry proteomics data in Figure 5 and Supplementary Table 1 and 2 have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD006054. Raw metabolomics data supporting Figure 6 have been included in Supplementary Table 3. All other data from manuscript are available from the corresponding author upon reasonable request.