Abstract
Cardiac stem cells (CSCs) are the most promising and effective candidates for the therapy of cardiac regenerative diseases; however, they have marked limitations. For instance, the implantation of CSCs is hampered by factors such as their sustainability and long-term durability. Gene modification appears to be the most effective method of optimizing CSCs and gene therapy trials have demonstrated that efficient gene transfer is key to achieving therapeutic efficacy. However, the transduction ability of adenovirus (Ad) is limited. Previous studies have reported that low expression of coxsackie and adenovirus receptor (CAR) in target cells decreases the transduction efficiency. A promising method for improving Ad-mediated gene transfer is to increase CAR expression in target cells. The present study investigated the effect of the Raf-mitogen-associated protein kinase (MAPK) kinase (MEK)-extracellular signal-associated protein kinase (ERK) signaling pathway on the expression of CAR on CSCs, as this pathway decreases cell-cell adhesion via cell surface molecules. The results demonstrated that interference with the Raf-MEK-ERK signaling pathway by knockdown of ERK1/2 upregulated the expression of CAR. The entry of the Ad into the cells was increased following inhibition of ERK1/2. Moreover, following knockdown of CAR, the entry of Ad into cells was decreased. However, knockdown of c-Jun N-terminal kinase and p38 as other components of the MAPK pathway did not affect CAR expression. Therefore, CAR expression in CSCs may be mediated via the Raf-MEK-ERK signaling pathway. Upregulation of CAR by knockdown of ERK1/2 may significantly improve Ad-mediated genetic modification of CSCs in the treatment of cardiovascular diseases.
Keywords: coxsackie and adenovirus receptor, gene therapy, extracellular signal-regulated kinase 1/2, cardiac stem cells
Introduction
The efficacy of cardiac stem cell (CSC) transplantation for cardiomyoplasty is limited due to poor survival of the implanted cells in ischemic hearts (1,2). Furthermore, strategies aiming to improve the efficiency of CSC transplantation have shown little success.
Genetically modified or re-constructed adenoviral vectors have been widely used to optimize CSCs for cellular cardiomyoplasty in the treatment of cardiovascular disease (3–7). McCormick (8) reported that improved adenoviruses effectively deliver therapeutic genes or drugs into target cells. Recombinant adenoviral vectors have been constructed, which have shown promising effects in clinical and pre-clinical studies (9). Furthermore, the efficiency of gene therapy was found to mainly be based on the binding ability of the internalized adenovirus and its target.
The coxsackie and adenovirus receptor (CAR) is a transmembrane protein with a molecular weight of 46-kDa. CAR interacts with tight junction protein complexes in normal human cells (10). It has also been shown to enhance the viral attachment capability by interacting with the adenovirus fiber-knob protein in cells (11). The loss or decrease in expression of CAR may significantly decrease viral infectivity, however, this decrease in viral infectivity may be rescued by ectopic expression of CAR protein (12). It has therefore been suggested that the amount and location of CAR expression may determine viral infectivity. Internalization of adenoviruses in cells is primarily mediated by binding of the virus penton base to αvβ3- and αvβ5-integrins (13). Studies have also indicated that decreased expression of CAR is observed in highly malignant tumors (14–16). Molecular cues and signaling pathways modulating CAR expression in CSCs have remained elusive. While certain studies have investigated the application of CAR in adenoviral vector-mediated expression for the regeneration of transplanted CSCs, the potential molecular mechanisms of the regulation of CAR expression remain to be fully elucidated. The expression of tight junction proteins is mediated by triggering the oncogenic signaling pathway, for instance, by triggering the Ras-activated extracellular signal-regulated kinase (ERK) signaling pathway in cells (16,17). The Raf-mitogen-activated protein kinase (MAPK) kinase (MEK) -ERK signaling pathway may serve a central role in regulating CAR and viral uptake, by which adenoviral vector therapy may be optimized (18,19). The present study demonstrated that vector-mediated inhibition of the Raf-MEK-ERK pathway enhanced CAR expression in mouse CSCs, therefore providing potential targets for improving adenovirus-based treatments. The results indicated that knockdown of ERK1/2 expression led to an upregulation of CAR on the CSC surface, which may be associated with increased adenoviral uptake into cells.
Materials and methods
Cell lines and CSC isolation and culture
In the present study, human endometrial cancer cells from the ishikawa (ISK) cell line were donated by the Second Affiliated Hospital of Harbin Medical University (Harbin, China). CSCs were isolated from the extracted hearts of Balb/c mice (18–25 g) using a method previously published by Beltrami et al (20), Liu et al (21,22) and Wang et al (23). All Balb/c mice were purchased from the Department of Laboratory Animal Science of the Second Affiliated Hospital of Harbin Medical University (Harbin, China). In brief, heparin (5,000 IU/kg) was intraperitoneally injected into the mice 20 min prior to the experiments. All mice were subsequently sacrificed by cervical dislocation. The heart was excised and the aorta was rapidly cannulated. Subsequently, the cannulated hearts were mounted and observed using a Langendorff perfusion system (Hugo Sachs Elektronik-Harvard Apparatus GmbH, March, Germany) with constant flow. The perfusion pressure of the hearts was observed and monitored. In order to remove the blood from the mouse hearts, they were treated and perfused with Ca2+-free Tyrode's solution (BSS-350; Boston BioProducts, Ashland, MA, USA) for 5–10 min. Subsequently, the hearts were digested with collagenase (0.5 mg/ml; Sigma-Aldrich; Merck Millipore, Darmstadt, Germany) and trypsin (0.5 mg/ml; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) for 30 min at 37°C. The heart tissue was subsequently extracted and cleaved, and the cell suspension was filtered with a prepared Steriflip instrument (SCNY00100-1EA; EMD Millipore, Billerica, MA, USA). The obtained suspension was treated with fluorescein isothiocyanate (FITC)-labeled rat anti-mouse CD117/c-kit polyclonal antibody (1:100, catalog no. 553354, BD Biosciences, Franklin Lakes, NJ, USA) and the suspension was separated using MACS anti-FITC microbeads (Miltenyi Biotech, Bergisch Gladbach, Germany). Small round cells, containing most of the c-kit+ sub-population, were harvested. The small round cells were cultured in HyClone Dulbecco's Modified Eagle's medium (DMEM)/F12 (Thermo Fisher Scientific, Inc.) supplemented with 10 ng/ml bovine fibroblast growth factor (PeproTech, Rocky Hill, NJ, USA), 10% fetal bovine serum (FBS, Hyclone; GE Healthcare Life Sciences, Logan, UT, USA), epidermal growth factor (PeproTech), 10 ng/ml insulin growth factor (PeproTech) and 10 ng/ml leukemia inhibitory factor (Sigma-Aldrich; Merck Millipore) for 3–5 days at 37°C. The cells were subjected to subsequent examinations or experiments whenever they were recovered. The experimental animal and cell experimental processes and protocols of the present study were approved by the Ethics Committee of Harbin Medical University (Harbin, China).
Small interfering RNA (siRNA) silencing
Gene silencing by siRNA involves a small double-stranded RNA that degrades target mRNA. The siRNA duplex was synthesized at Shanghai GenePharma Co., Ltd (Shanghai, China). To detect the regulatory roles of MAPKs in mediating CAR expression, additional CSCs were transfected with 100 pmol siRNA against; c-Jun N-terminal kinase (JNK), forward, 5′-UCAAGGAAUAGUGUGUGCAGCUUAU-3′, and reverse 5′-CAGCCCAGTAATATAGTAGTA-3′; p38 forward, 5′-GGACCUCCUUAUAGACGAAUU-3′ and reverse, 5′-GGACCUCCUUAUAGACGAAUU-3′; ERK1/2 forward, 5′-GACCGGAUGUUAACCUUUAUU-3′ and reverse, 5′-GACCGGAUGUUAACCUUUAUU-3′; CAR forward, 5′-GACGCAUCUAUAAAUGUGAUU-3′ and reverse, 5′-GACGCAUCUAUAAAUGUGAUU-3′, using Lipofectamine® RNAiMAX (Invitrogen; Thermo Fisher Scientific, Inc.) for 48 h. In the control group, cells were treated with either a transfection reagent (vehicle) or a non-targeting siRNA (siRNA-NC), forward, 5′-UUCUCCGAACGUGUCACG-3′ and reverse, 5′-ACGUGACACGUUCGGAGAATT-3′ (Shanghai GenePharma Co., Ltd.).
In vitro adenoviral vector transduction
Recombinant adenovirus-green fluorescence protein (Ad-GFP) was purchased from Shanghai GenePharma Co., Ltd. Ad transductions were performed in six-well plates, with cells seeded at 2–5×105/well 48 h previously. Virus particles (109/well) were diluted in serum-free DMEM/F12 (1 ml/well; Thermo Fisher Scientific, Inc.) and then incubated at 37°C with the cells for 90 min. Subsequently, the viral suspension was replaced with complete culture medium (containing FBS). Following 72 h transduction, cells were washed with phosphate-buffered saline (PBS) and subjected to flow cytometric and fluorescence microscopic analysis.
Immunofluorescence
The cells were incubated with 4% (v/v) formaldehyde for 15 min at room temperature. Subsequently, the cells were washed three times with PBS for 5 min each and blocked with 10% bovine serum albumin (HyClone; GE Healthcare Life Sciences). The cells were then incubated with rabbit anti-CAR antibody (1:100 dilution; cat. no. H-300; Santa Cruz Biotechnology, Inc., Dallas, TX, USA) for 1 h at 37°C. The cells were washed again three times with PBS (5 min each) and treated with Alexa Fluor 555-labeled goat anti-rabbit antibody (1:1,000 dilution; cat. no., A-21428, Invitrogen; Thermo Fisher Scientific, Inc.) at 37°C in the dark for 1 h. Subsequent to washing with PBS, the nuclei were counter-stained with the 4′, 6-diamidino-2-phenylindole (Sigma-Aldrich; Merck Millipore). Finally, the cells were observed and examined under a fluorescence microscope (DMI4000B; Leica Microsystems, Wetzlar, Germany).
Western blot analysis
The cells were lysed following hypertrophy induction using buffer radioimmunopreciptation assay buffer (Abcam, Cambridge, MA, USA) by adding a protein inhibitor cocktail (Roche Diagnostics, Indianapolis, IN, USA) to recover the extract. To separate proteins, samples were denatured at 97.5°C for >2 min. A total of 20 µg of protein was resolved by SDS-PAGE at 12.5% and transferred onto polyvinylidene difluoride membranes (EMD Millipore), which were then blocked with 5% non-fat milk in Tris-buffered saline containing Tween 20 (TBST) for 1 h at room temperature. Mouse anti-phosphorylated (p)-JNK (1:1,000; CST#9255), anti-p-p38 (1:1,000; CST#9216), anti-p-ERK1/2 (1:1,000; CST#9106), anti-total (t)-JNK (1:1,000; CST#3708), anti-t-p38 (1:1,000; CST#9217) and anti-t-ERK1/2 (1:1,000; CST#4696) were all purchased from Cell Signaling Technology, Inc. (Danvers, MA, USA). Rabbit anti-β-actin (1:200; PR-0255) was purchased from Zhongshan Goldenbridge Biotechnology Co. Ltd, Beijing, China and mouse anti-CAR monoclonal antibody (1:200; sc-373791) was purchased from Santa Cruz Biotechnology, Inc. The membranes were incubated with the above antibodies overnight at 4°C. Subsequently, the membranes were incubated with horseradish peroxidase (HRP)-conjugated Affinipure goat anti-rabbit immunoglobulin (Ig) G (H+L) polyclonal antibody (1:2,000; ab6721, Abcam) and HRP-conjugated rabbit anti-mouse IgG (H+L)-labeled polyclonal antibody (HRP; 1:2,000; ab6728, Abcam) at 37°C 1 h and washed with TBST solution. Bands were then visualized by enhanced chemiluminescence (ECL) detection using BeyoECL Plus (Beyotime Institute of Biotechnology, Inc., Haimen, China). Images of the gels were captured on X-ray film, following which the bands were stripped with 5 ml stripping buffer (CoWin Biotech, Beijing, China) at room temperature for 15 min and hybridized with β-actin (1:200; TA890010, OriGene Technologies, Inc., Beijing, China) at room temperature for 15 min for normalization. Protein bands were quantified by densitometry using the Tanon Gel Imaging System (Shanghai Tanon Co. Ltd., Shanghai, China).
Quantitative reverse transcription quantitative polymerase chain reaction (RT-qPCR)
Total RNA was isolated from the cells subjected to the different experimental conditions with TRIzol (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's instructions. Following pretreatment with RNase-free DNase I, 2-µg miRNA was subjected to the protocols of the miScript Reverse Transcription kit and the miScript SYBR Green PCR kit (Qiagen Benelux B.V, Venlo, the Netherlands). RNA was converted into cDNA by reverse transcription with the Transcriptor First Strand cDNA Synthesis kit (Invitrogen; Thermo Fisher Scientific, Inc.) qPCR was performed using a FastStart Universal SYBR Green Master Mix (ROX) (Roche Applied Science, Mannheim, Germany) in a 10 µl reaction volume on an ABI 7900 Genome Analyzer System (5 µl SYBR green Mix, 200 µM forward and reverse primers, 1 µl cDNA template) The primer sequences are presented in Table I. The PCR reaction conditions were initial denaturation at 94°C for 5 min; degeneration at 94°C for 15 sec, annealing at 55°C for 30 sec and 72°C for 30 sec, for a total of 35 cycles; followed by extension at 72°C for 5 min. Gene amplification was confirmed after calculating the melting temperatures (Tm) for the products from the melting peak curve (2dF/dT vs. temperature). All amplicons were collected and 10 ml of each reaction was confirmed by 1.5% agarose gel electrophoresis and sequencing. A cross-point vs. logarithmic concentration standard curve was generated using serial dilutions of one of the cDNA samples or known concentrations of plasmid DNA. Negative controls were included, using cDNAs synthesized in the same manner as described above, but without reverse transcriptase. Each cDNA sample was run in triplicate. Relative mRNA expression levels were calculated using the −2ΔΔCq method (24) based on the threshold cycle (Cq) values and were normalized to the internal control of GAPDH. The primers (GenePharma Co., Ltd.) utilized in the present study are listed in Table I.
Table I.
Primers for polymerase chain reaction.
| Gene | Primer sequences |
|---|---|
| CAR | F: 5′-GCATCACTACACCCGAACAGA-3′; |
| R: 5′-CAAGAACGGTCAGCAGGAAT-3′; | |
| GAPDH | F: 5′-GGCACAGTCAAGGCTGAGAATG-3′; |
| R: 5′-ATGGTGGTGAAGACGCCAGTA-3′; |
CAR, coxsackie and adenovirus receptor; F, forward; R, reverse.
Flow cytometric analysis
The Ad suspension was transfected for 24 h and cells were subsequently cultured for 48 h and harvested. Then cells were re-suspended at a density of 1×106 cells in 500 µl binding buffer. A total of 10,000 events were collected and the proportion of GFP cells was analyzed with a FACSCalibur flow cytometer (BD Biosciences).
Statistical analysis
Statistical analysis was performed using SPSS 18.0 software (SPSS Inc., Chicago, IL, USA). Values are expressed as the mean ± standard error. Comparisons between the parameters were analyzed using Student's t-test. P<0.05 was considered to indicate a statistically significant difference.
Results
Characterization of CAR expression on mCSCs
In a preliminary study by our group, immunofluorescent microscopic analysis only demonstrated small amounts of CAR expression at the surface of SW480 cells. In the present study, CAR protein was also located in the cell cytoplasm, while small amounts were located at the cell surface, which may have posed a disadvantage regarding Ad uptake in mCSC and ISK cells (Fig. 1).
Figure 1.

Characteristics of CAR expression and inhibition of mitogen-activated protein kinase on murine CSCs. Immunofluorescence analysis of CAR expression on CSCs and ISK cells. The nuclei were counter-stained with 4′,6-diamidino-2-phenylindole (original magnification, ×400). ISK, ishikawa cells; CSC, cardiac stem cells; CAR, coxsackie and adenovirus receptor.
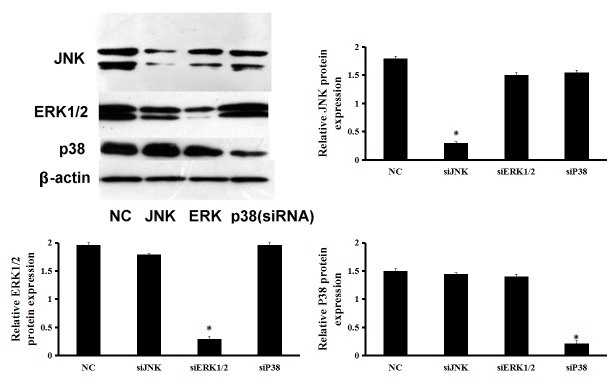
MAPK pathway silencing in mCSCs
In order to further explore the influence of the MAPK pathway on CAR expression in mCSCs, ERK1/2, JNK and P38 were individually silenced with siRNA (Fig. 2). Following 48 h transfection, a knockdown efficiency of >80% of the targeted proteins was observed in the cells transfected with specific siRNAs against ERK1/2, JNK and p38 (Fig. 2), which was also sustained for up to 72 h (data not shown).
Figure 2.
Cellular protein was collected after individual transfection with non-targeting, JNK, ERK1/2 or p38 siRNAs for 48 h, followed by detection of ERK1/2, JNK and p38 protein expression by western blot analysis. *P<0.05 vs. NC. ISK, ishikawa cells; CSC, cardiac stem cell; JNK, c-Jun N-terminal kinase; ERK, extracellular signal-regulated kinase; siRNA, small interfering RNA; NC, negative control.
Knockdown of ERK1/2 increases expression of CAR protein and its abundance at the surface of mCSCs
Following the silencing of ERK1/2 with siRNA, mCSCs exhibited significantly increased CAR protein expression, while there was no such effect in siRNA-NC (Fig. 3A). Immunofluorescence microscopy was also performed to investigate whether inhibition of Raf-MEK-ERK signaling affected not only CAR expression but also its sub-cellular localization. In control cells, CAR was only weakly expressed in the cytoplasm and CAR expression at the cell surface was low. However, after ERK1/2 silencing with siRNA-ERK1/2, this expression pattern clearly changed, with CAR being mainly located at the cell surface at sites of cell-cell contact (Fig. 3B). Furthermore, results from RT-qPCR indicated that in mCSCs, CAR mRNA levels were significantly increased following the silencing of ERK1/2 with siRNA (Fig. 3C). These results showed that the changes of CAR levels are specifically associated with signaling through the ERK1/2 pathway.
Figure 3.
Effects of mitogen-activated protein kinase silencing on CAR expression in mCSCs. mCSCs were individually treated with siRNA-ERK1/2, -JNK, -P38 or -NC (control) for 48 h. (A) Western blot analysis of CAR protein expression. Representative immunoblots and densitometrically quantified CAR protein levels. (B) Effect of ERK1/2 silencing on the subcellular CAR protein distribution observed by immunofluorescence microscopy. DNA was visualized by 4′,6-diamidino-2-phenylindole staining. (C) CAR mRNA expression determined by reverse-transcription quantitative polymerase chain reaction with normalization to GAPDH. The bar graphs represent the mean values of triplicate measurements ± standard error of the mean. *P<0.05 vs. NC. mCSC, murine cardiac stem cell; JNK, c-Jun N-terminal kinase; ERK, extracellular signal-regulated kinase; siRNA, small interfering RNA; CAR, coxsackie and adenovirus receptor; NC, negative control.
Furthermore, the present study investigated the effects of JNK and p38 silencing on CAR expression. As well as ERK1/2, the JNK and p38 pathways are the other two major MAPK signal transduction pathways. The results indicated that transfection of JNK- and P38-siRNA had no effect on CAR protein expression (Fig. 3A). In order to evaluate whether the CAR protein changes were caused by changes in CAR gene transcription, RT-qPCR was performed. In mCSCs, CAR mRNA levels were unchanged after silencing of JNK or p38 with siRNA (Fig. 3C). The above results suggested that the ERK1/2 but not the JNK and p38 signaling transduction pathways affect CAR expression. However, these effects require investigation in other cell types.
ERK1/2 silencing increases Ad uptake in mCSCs, which is inhibited by CAR siRNA
As CAR acts as the primary cellular binding site for the Ad (19), the present study hypothesized that increased CAR expression at the cardiac stem cell surface may lead to enhanced viral uptake. In order to verify this hypothesis, CSCs were first infected with a non-replicating Ad-GFP following treatment with ERK1/2-, JNK- or P38-siRNA. The FACS assay showed significantly increased infectivity of CSCs treated with the ERK1/2-siRNA compared with control cells. However, treatment with JNK- and P38-siRNA did not affect the infectivity of CSCs (Fig. 4A).
Figure 4.

Changes in viral uptake following ERK1/2 silencing. The infectivity of a GFP-expressing, non-replicating adenovirus at a multiplicity of infection of 10 was evaluated after 48 h treatment with siRNA-ERK1/2 or siRNA-NC (control group) (A) using fluorescence-assisted cell sorting analysis. Values are expressed as the mean of triplicate measurements ± standard error. *P<0.05 vs. NC. (B) The infectivity was also assessed using fluorescence microscopy (scale bar, 200 µm). Under the same conditions, the left panel shows transmitted light images and the right panels show green fluorescence. JNK, c-Jun N-terminal kinase; ERK, extracellular signal-regulated kinase; siRNA, small interfering RNA; NC, negative control; GFP, green fluorescence protein. CAR, coxsackie and adenovirus receptor; mCSC, murine cardiac stem cell.
To evaluate whether ERK1/2-siRNA increases the Ad transgene expression in mCSCs, fluorescence microscopy was used. The results showed that in the ERK1/2-siRNA transfected mCSCs, GFP expression was markedly increased (Fig. 4B).
To further test the hypothesis that the Ad uptake is improved by upregulation of CAR, mCSCs were transfected with CAR siRNA. Western blot analysis revealed that after 48 h transfection, siRNA-CAR had a >80% knockdown efficiency (Fig. 5A), which was sustained for up to 72 h (data not shown). Following treatment with CAR siRNA, the infectivity of CSCs by Ad-GFP was decreased and remained at low levels when ERK1/2 siRNA was co-transfected (Fig. 5B). These results indicated that decreases in CAR expression at the cardiac stem cell surface inhibited viral uptake and that the presence of CAR is required for Ad uptake, which may be increased via inhibition of the ERK1/2 pathway, rather than being affected by the other two branches of the MAPK pathway.
Figure 5.

Changes in viral uptake and CAR expression after silencing of ERK1/2. mCSCs were treated with siRNA-CAR alone or siRNA-CAR with siRNA-ERK1/2 for 48 h. siRNA-NC-treated mCSCs were used as a control group. (A) CAR protein expression was detected by immunoblot analysis and densitometrically quantified. The bar graphs represent the mean values of triplicate measurements ± standard error of the mean. *P<0.05 vs. NC. (B) The infectivity of a GFP-expressing, non-replicating adenovirus at a multiplicity of infection of 10 was evaluated by fluorescence microscopy (scale bar, 100 µm). Under the same conditions, the left panel shows transmitted light images and the right panels show fluorescence. ERK, extracellular signal-regulated kinase; siRNA, small interfering RNA; NC, negative control; GFP, green fluorescence protein; CAR, coxsackie and adenovirus receptor; mCSC, murine cardiac stem cell.
Discussion
The present study aimed to explore the effects of ERK1/2 signaling on CAR expression in mCSCs as a potential target for Ad-based therapies. The main findings obtained were the following: i) CAR protein is located in the cell cytoplasm, while a small portion of the protein is located at the surface of mCSCs and ISK cells; ii) knockdown of ERK1/2 increases the expression of CAR protein in mCSCs but not JNK and P38; iii) silencing of ERK1/2 leads to upregulation of CAR protein at the cell surface in mCSCs, while silencing of JNK and P38 does not; and iv) ERK1/2 siRNA increases Ad uptake in mCSCs, which is abrogated by co-transfection with CAR siRNA. Overall, the results showed that in mCSCs, inhibition of ERK1/2 phosphorylation with siRNA led to elevated CAR expression, which increased Ad uptake. However, other branches of the MAPK pathway (JNK and P38) had no effect on CAR expression and Ad uptake in mCSCs.
In a number of cases, the low expression of CAR in target cells limits the efficacy of gene therapy approaches. A previous study demonstrated that the CAR receptor acts as the key factor for sufficient viral uptake (25). In normal cells, CAR is an adhesion molecule that interacts with the adherens and tight junction proteins (10). Loss of CAR on the cell surface significantly decreases viral infectivity, which may be reversed by the ectopic CAR expression. The present study focused on the regulatory mechanisms of CAR expression at the molecular level in order to explore how it modulates viral infectivity.
Firstly, CAR expression in mCSCs and the ISK cell line was detected by immunofluorescence. The findings were consistent with those of a previous study, showing that CAR protein is located in the cell cytoplasm while a small amount is present at the cell surface, which poses a disadvantage regarding Ad uptake (18).
The Raf-MEK-ERK signaling pathway downregulates cell-cell adhesion complexes. For instance, tight- and adherens junctions (17) and CAR act as cell adhesion molecules, which also participate in the formation of the apical junction complex and the tight junction (10,26–28). In the present study, it was therefore hypothesized that the Raf-MEK-ERK signaling pathway contributes to the downregulation of CAR in cardiac stem cells. To test this hypothesis, CSCs were treated with siRNA-ERK1/2, -JNK and -p38, and the effects on CAR expression and Ad uptake were assessed.
Treatment of CSCs with siRNA-ERK1/2 led to an increase in CAR protein levels at the cell membrane, with marked upregulation observed at sites of cell-cell contact. Relocation of CAR at the plasma membrane following the inhibition of ERK was sufficient to enhance the uptake of Ad. The aforementioned effects appear to depend on CAR specifically, as silencing of CAR significantly reduced Ad uptake, which could not be rescued by co-transfection with ERK1/2-siRNA. Furthermore, the effects of the other branches of the MAPK pathway were examined; however, neither the expression of CAR nor Ad uptake were affected by the silencing of JNK or P38. These results suggest that inhibition of Raf-MEK-ERK signaling may affect not only the expression of CAR, but also its sub-cellular localization on the membrane to improve the uptake of Ad by mCSCs.
Acknowledgements
The present study was funded by the Key Laboratory of Myocardial Ischemia Mechanism and Treatment (grant no. KF201304), the Heilongjiang Postdoctoral Foundation Assistance (grant no. LBH-Z14218) and the China Postdoctoral Science Foundation Grant (no. 2014M56137). The authors would like to thank Dr Zhaohua Zhong (Department of Microbiology, Harbin Medical University, Harbin, China) for their help.
References
- 1.Bergmann O, Bhardwaj RD, Bernard S, Zdunek S, Barnabé-Heider F, Walsh S, Zupicich J, Alkass K, Buchholz BA, Druid H. Evidence for cardiomyocyte renewal in humans. Science. 2009;324:98–102. doi: 10.1126/science.1164680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hsieh PC, Segers VF, Davis ME, MacGillivray C, Gannon J, Molkentin JD, Robbins J, Lee RT. Evidence from a genetic fate-mapping study that stem cells refresh adult mammalian cardiomyocytes after injury. Nat Med. 2007;13:970–974. doi: 10.1038/nm1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ylä-Herttuala S, Martin JF. Cardiovascular gene therapy. Lancet. 2000;355:213–222. doi: 10.1016/S0140-6736(99)04180-X. [DOI] [PubMed] [Google Scholar]
- 4.Schneider MD, French BA. The advent of adenovirus. Gene therapy for cardiovascular disease. Circulation. 1993;88:1937–1942. doi: 10.1161/01.CIR.88.4.1937. [DOI] [PubMed] [Google Scholar]
- 5.Nabel EG. Gene therapy for cardiovascular disease. Circulation. 1995;91:541–548. doi: 10.1161/01.CIR.91.2.541. [DOI] [PubMed] [Google Scholar]
- 6.Kalka C, Baumgartner I. Gene and stem cell therapy in peripheral arterial occlusive disease. Vasc Med. 2008;13:157–172. doi: 10.1177/1358863x08088616. [DOI] [PubMed] [Google Scholar]
- 7.Ly H, Kawase Y, Yoneyama R, Hajjar RJ. Gene therapy in the treatment of heart failure. Physiology (Bethesda) 2007;22:81–96. doi: 10.1152/physiol.00037.2006. [DOI] [PubMed] [Google Scholar]
- 8.McCormick F. Cancer gene therapy: Fringe or cutting edge? Nat Rev Cancer. 2001;1:130–141. doi: 10.1038/35101008. [DOI] [PubMed] [Google Scholar]
- 9.Steorge J. Gene therapy progress and prospects: Adenoviral vectors. Gene Ther. 2003;10:1135–1141. doi: 10.1038/sj.gt.3302071. [DOI] [PubMed] [Google Scholar]
- 10.Cohen CJ, Gaetz J, Ohman T, Bergelson JM. Multiple regions within the coxsackievirus and adenovirus receptor cytoplasmic domain are required for basolateral sorting. J Biol Chem. 2001;276:25392–25398. doi: 10.1074/jbc.M009531200. [DOI] [PubMed] [Google Scholar]
- 11.Krasnykh V, Belousova N, Korokhov N, Mikheeva G, Curiel DT. Genetic targeting of an adenovirus vector via replacement of the fiber protein with the phage T4 fibritin. J Viro. 2001;75:4176–4183. doi: 10.1128/JVI.75.9.4176-4183.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Okegawa T, Pong RC, Li Y, Bergelson JM, Sagalowsky AI, Hsieh JT. The mechanism of the growth-inhibitory effect of coxsackie and adenovirus receptor (CAR) on human bladder cancer: A functional analysis of car protein structure. Cancer Res. 2001;61:6592–6600. [PubMed] [Google Scholar]
- 13.Wickham TJ, Mathias P, Cheresh DA, Nemerow GR. Integrins alpha v beta 3 and alpha v beta 5 promote adenovirus internalization but not virus attachment. Cell. 1993;73:309–319. doi: 10.1016/0092-8674(93)90231-E. [DOI] [PubMed] [Google Scholar]
- 14.Li Y, Pong RC, Bergelson JM, Hall MC, Sagalowsky AI, Tseng CP, Wang Z, Hsieh JT. Loss of adenoviral receptor expression in human bladder cancer cells: A potential impact on the efficacy of gene therapy. Cancer Res. 1999;59:325–330. [PubMed] [Google Scholar]
- 15.Okegawa T, Li Y, Pong RC, Bergelson JM, Zhou J, Hsieh JT. The dual impact of coxsackie and adenovirus receptor expression on human prostate cancer gene therapy. Cancer Res. 2000;60:5031–5036. [PubMed] [Google Scholar]
- 16.Rauen KA, Sudilovsky D, Le JL, Chew KL, Hann B, Weinberg V, Schmitt LD, McCormick F. Expression of the coxsackie adenovirus receptor in normal prostate and in primary and metastatic prostate carcinoma: Potential relevance to gene therapy. Cancer Res. 2002;62:3812–3818. [PubMed] [Google Scholar]
- 17.Chen Yh, Lu Q, Schneeberger EE, Goodenough DA. Restoration of tight junction structure and barrier function by down-regulation of the mitogen-activated protein kinase pathway in ras-transformed Madin-Darby canine kidney cells. Mol Biol Cell. 2000;11:849–862. doi: 10.1091/mbc.11.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hemminki A, Kanerva A, Liu B, Wang M, Alvarez RD, Siegal GP, Curiel DT. Modulation of coxsackie-adenovirus receptor expression for increased adenoviral transgene expression. Cancer Res. 2003;63:847–853. [PubMed] [Google Scholar]
- 19.Farmer C, Morton PE, Snippe M, Santis G, Parsons M. Coxsackie adenovirus receptor (CAR) regulates integrin function through activation of p44/42 MAPK. Exp Cell Res. 2009;315:2637–2647. doi: 10.1016/j.yexcr.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 20.Beltrami AP, Barlucchi L, Torella D, Baker M, Limana F, Chimenti S, Kasahara H, Rota M, Musso E, Urbanek K. Adult cardiac stem cells are multipotent and support myocardial regeneration. Cell. 2003;114:763–776. doi: 10.1016/S0092-8674(03)00687-1. [DOI] [PubMed] [Google Scholar]
- 21.Liu J, Wang Y, Du W, Liu W, Liu F, Zhang L, Zhang M, Hou M, Liu K, Zhang S, Yu B. Wnt1 inhibits hydrogen peroxide-induced apoptosis in mouse cardiac stem cells. PloS One. 2013;8:e58883. doi: 10.1371/journal.pone.0058883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu J, Wang Y, Du W, Yu B. Sca-1-positive cardiac stem cell migration in a cardiac infarction model. Inflammation. 2013;36:738–749. doi: 10.1007/s10753-013-9600-8. [DOI] [PubMed] [Google Scholar]
- 23.Wang Y, Liu J, Cui J, Sun M, Du W, Chen T, Ming X, Zhang L, Tian J, Li J, et al. MiR218 modulates wnt signaling in mouse cardiac stem cells by promoting Proliferation and Inhibiting Differentiation through a Positive Feedback Loop. Sci Rep. 2016;6:20968. doi: 10.1038/srep20968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 25.Bergelson JM, Cunningham JA, Droguett G, Kurt-Jones EA, Krithivas A, Hong JS, Horwitz MS, Crowell RL, Finberg RW. Isolation of a common receptor for Coxsackie B viruses and adenoviruses 2 and 5. Science. 1997;275:1320–1323. doi: 10.1126/science.275.5304.1320. [DOI] [PubMed] [Google Scholar]
- 26.van Raaij MJ, Chouin E, van der Zandt H, Bergelson JM, Cusack S. Dimeric structure of the coxsackievirus and adenovirus receptor D1 domain at 1.7 Å resolution. Structure. 2000;8:1147–1155. doi: 10.1016/S0969-2126(00)00528-1. [DOI] [PubMed] [Google Scholar]
- 27.Honda T, Saitoh H, Masuko M, Katagiri-Abe T, Tominaga K, Kozakai I, Kobayashi K, Kumanishi T, Watanabe YG, Odani S, Kuwano R. The coxsackievirus-adenovirus receptor protein as a cell adhesion molecule in the developing mouse brain. Brain Res Mol Brain Res. 2000;77:19–28. doi: 10.1016/S0169-328X(00)00036-X. [DOI] [PubMed] [Google Scholar]
- 28.Walters RW, Freimuth P, Moninger TO, Ganske I, Zabner J, Welsh MJ. Adenovirus fiber disrupts CAR-mediated intercellular adhesion allowing virus escape. Cell. 2002;110:789–799. doi: 10.1016/S0092-8674(02)00912-1. [DOI] [PubMed] [Google Scholar]