Abstract
Objective
The dismal prognosis of pancreatic cancer has been linked to poor tumor differentiation. However, molecular basis of pancreatic cancer differentiation and potential therapeutic value of the underlying molecules remain unknown. We investigated the mechanistic underexpression of Krüppel-like factor 4 (KLF4) in pancreatic cancer and defined a novel epigenetic pathway of its activation for pancreatic cancer differentiation and treatment.
Design
Expressions of KLF4 and DNMT1 in pancreatic cancer tissues were determined by immunohistochemistry and the genetic and epigenetic alterations of KLF4 in and KLF4’s impact on differentiation of pancreatic cancer were examined using molecular biology techniques. The function of dietary 3,3’-diindolylmethane (DIM) on miR-152/DNMT1/KLF4 signaling in pancreatic cancer was evaluated using both cell culture and animal model.
Results
Overexpression of DNMT1 and promoter hypermethylation contributed to decreased KLF4 expression in and associated with poor differentiation of pancreatic cancer. Manipulation of KLF4 expression significantly affected differentiation marker expressions in pancreatic cancer cells. DIM treatment significantly induced miR-152 expression, which blocked DNMT1 protein expression and its binding to KLF4 promoter region, and consequently, reduced promoter DNA methylation and activated KLF4 expression in pancreatic cancer cells. Additionally, DIM treatment caused significant inhibition of cell growth in vitro and tumorigenesis in animal model of pancreatic cancer.
Conclusions
This is the first demonstration that dysregulated KLF4 expression associates with poor differentiation of pancreatic cancer. Epigenetic activation of miR-152/DNMT1/KLF4 signaling pathway by dietary DIM causes differentiation and significant growth inhibition of pancreatic cancer cells, highlighting its translational implications for pancreatic and other cancers.
Keywords: microRNA-152; DNMT1; KLF4; 3,3’-diindolylmethane; dedifferentiation; pancreatic cancer
Introduction
Pancreatic cancer, mainly pancreatic adenocarcinoma (PDAC), is currently the third leading cause of cancer-related death in the United States, with a 5-year survival rate of less than 8% (1). The dismal prognosis of pancreatic cancer has been linked to poor tumor differentiation (2, 3), a widely accepted index for prediction of therapeutic response and prognosis of many cancers including pancreatic cancer. Despite significant clinical implications, however, the molecules and signaling pathways that regulate the differentiation of pancreatic cancer remain poorly understood.
Cancer is a disease that results from the successive accumulation of genetic and epigenetic alterations, while epigenetic changes frequently precede and can induce genetic mutations that cause cancer (4, 5). DNA hypermethylation is one of the best-understood epigenetic mechanisms that contribute to cancer development and progression (6, 7), which is attributed to either excessive DNA methylation or deficient demethylation; the former is regulated by a family of DNA methyltransferase enzymes (DNMT1, DNMT3A, and DNMT3B) that mediate the transfer of methyl groups from S-adenosylmethionine to the 5 position of cytosine (5-methylcytosine, 5mC) bases in the dinucleotide sequence CpG. Hypermethylation of tumor suppressor genes has been observed in all kinds of cancers and is generally assumed to be functionally equivalent to genetic loss-of-function mutations (8). The potential reversion of epigenetically silenced tumor suppressors has emerged as a promising strategy for cancer prevention and treatment (9, 10). However, drugs that can effectively reactivate silent tumor suppressors by targeting aberrant DNA methylation with high specificity and low toxicity have yet to be identified. Other epigenetic mechanisms involved in cancer include histone modifications and altered expression of microRNAs (4, 5), a group of small non-coding, single-strand RNAs of 19–25 nucleotides in length that have profound implications in cancer development, progression, diagnosis and, in particular, treatment (11, 12). Thus, identification of novel and nontoxic compounds that can modulate microRNA expressions for cancer target therapy has drawn much attention (12).
KLF4 is a zinc-finger transcription factor. Physiologically, KLF4 is expressed primarily in terminally differentiated epithelial cells in organs such as the skin, gastrointestinal tract and pancreatic duct , whereas disruption of klf4 in mice selectively perturbs late-stage differentiation of skin structures, colonic goblet cells, and causes precancerous changes in the adult stomach because of altered gastric epithelial cell proliferation and differentiation (13). Clinically, reduced or loss of KLF4 expression has been found in various tumors and associates with poor tumor differentiation (14–17), and genetic and epigenetic alterations have been attributed to loss of KLF4 expression in these cancer cells (13, 18, 19). We and others have shown previously that KLF4 has tumor suppressive function in pancreatic cancer (20–22), although elevated KLF4 expression was found in premalignant lesion of pancreas (23, 24). However, the characteristic alterations of KLF4 and associated mechanisms in pancreatic cancer have not been well understood. In addition, whether KLF4 is involved in the regulation of pancreatic cancer differentiation and whether KLF4 could be a potential target for pancreatic cancer treatment await for further investigation.
It has long been known that certain foods contain bioactive molecules that can modulate epigenome and have chemopreventive and/or anti-tumor activities. Mounting evidence showed that 3,3’-diindolylmethane (DIM), the major bioactive metabolite of nutritional component indole-3-carbinol found relatively high in cruciferous vegetables, is a promising cancer preventative and anti-tumor agent in various cancers including breast, prostate, and cervical cancers (25, 26), and several clinical trials have been approved in healthy subjects or patients with premalignant or malignant lesion (see clinical trials: NCT01391689, NCT01022333, NCT02197000, NCT01726127, NCT00392652, NCT00784394) (27, 28). Available preliminary data indicate that supplementation with I3C or the related dimer 3,3’-diindolylmethane (DIM) may have beneficial effects in treating conditions related to human papilloma virus infection, such as cervical intraepithelial neoplasia and recurrent respiratory papillomatosis (25, 29, 30). However, whether DIM has antitumor effect in pancreatic cancer and if so, the exact molecular mechanisms behind remain poorly understood.
In present study, we demonstrated for the first time that reduced or loss of KLF4 expression associated with increased DNMT1 expression and poor differentiation of pancreatic cancer; dietary DIM significantly induced KLF4 expression in and differentiation of pancreatic cancer cells, in which a novel epigenetic signaling pathway of miR-152/DNMT1/KLF4 was discovered. This pathway may have translational implications for pancreatic and other cancers.
Materials and Methods
Detailed information about cell proliferation and Spheroid colony formation assay, Western and southern blot analysis, Methylation specific and real-time PCR, immunocytochemistry, and Chromatin immunoprecipitation assay are described in the Supplementary Materials and Methods.
Cell lines and culture conditions
Human pancreatic adenocarcinoma cell lines CaPan-1, CaPan-2, AsPC-1, BxPC-3, Hs766T, MiaPaCa-2, PANC-1, PANC-28, PANC-48, PaTu8902, FG, Colo357 and L3.7, and gastric cancer cell lines GT5 and N87 were purchased from the American Type Culture Collection (Manassas, VA) or obtained as described previously (21). The HCT-116 parental colorectal cells and the cells with DNA methyltransferase 1 genetic disruption (HCT-116-DT1-KO) were gifts from Dr. Bert Vogelstein (The Johns Hopkins University School of Medicine, Baltimore, MD). All cancer cell lines were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% fetal bovine serum (FBS) or otherwise described.
miRNA array hybridization and analysis
RNA was extracted using Trizol reagent. Total RNA (5 µg) was reverse transcribed using biotin end-labelled random random octamer oligonucleotide primers. Hybridization of biotin-labelled complementary DNA was performed using a custom miRNA microarray chip (ncRNA Program at Center for Targeted Therapy, M.D. Anderson Cancer Center, USA). A beta-uniform mixture (BUM) model was used for data analysis and unpaired t-test was used to identify differentially expressed genes when the false discovery rate (FDR) was set at 20% (31).
Luciferase reporter construction and activity assay
A 325-bp fragment of DNMT1 3’-UTR was amplified and subcloned into Spe I/Hind III sites of pMir-Report Luciferase vector (Ambion, Austin, TX). The constructed vector was co-transfected with miR-152 mature minics, anti-miR-152 (Ambion, Austin, TX), or non-target control (NC) oligos. In some experiments, transfected cells were also treated with 15 µMol DIM for 36h. Luciferase activity was measured at 48 hr after transfection using a Dual-Glo Luciferase Assay kit (Promega, Madison, WI) and expressed as the relative ratio of firefly luciferase activity to Renilla luciferase activity (32).
Human tissue samples and immunohistochemical analysis
Expression of KLF4 and DNMT1 was analyzed using human pancreatic cancer and normal tissue arrays (US Biomax, Inc., Rockville, MD). The use of the tissue samples was approved by the Institutional Review Board of The University of Texas MD Anderson Cancer Center. Standard immunohistochemical procedures were carried out using anti-KLF4 (Abgent, San Diego, CA) or anti-DNMT1 antibody (Cell Signaling Technology), and the staining results were scored by 2 investigators who were blinded to the clinical data, as described previously (32).
Animal model of tumor growth
PANC-1 and PANC-28 tumor cells in exponential growth phase were prepared, and then 1.5×106 cells were injected into the subcutis of 7~8-week-old female athymic BALB/c nude mice (NCI, Fredrick), respectively. When tumors became palpable, the mice were fed with AIN-93G control diet or the diet containing 2000 ppm of Bio-DIM (from BioResponse, Boulder, CO; the food was produced by Research Diets, Inc) for 6 weeks according to a previous report (33). For tumorigenic assay using orthotopic mouse model, PANC-28 cells (1.5×106 cells/50 µL in growth factor reduced matrigel (Corning Life Sciences, Tewksbury, MA) were injected into the pancreases of nude mice, the mice were fed with control or Bio-DIM containing diet for 5 weeks beginning on Days 4 after tumor cell injection. Then, the animals were killed and the primary tumor tissues were harvested, weighed, and processed for further analysis. All animal experiments were reviewed and approved by the University of Texas MD Anderson Cancer Center Institutional Animal Care and Use Committee.
Statistical analysis
The significance of the in vitro data and in vivo data was determined by Student t test (2-tailed), Mann–Whitney test (2-tailed), one-way ANOVA or Fisher’s exact test. P < 0.05 was considered significant.
Results
Reduced or loss of KLF4 expression predicts poor differentiation of pancreatic cancer
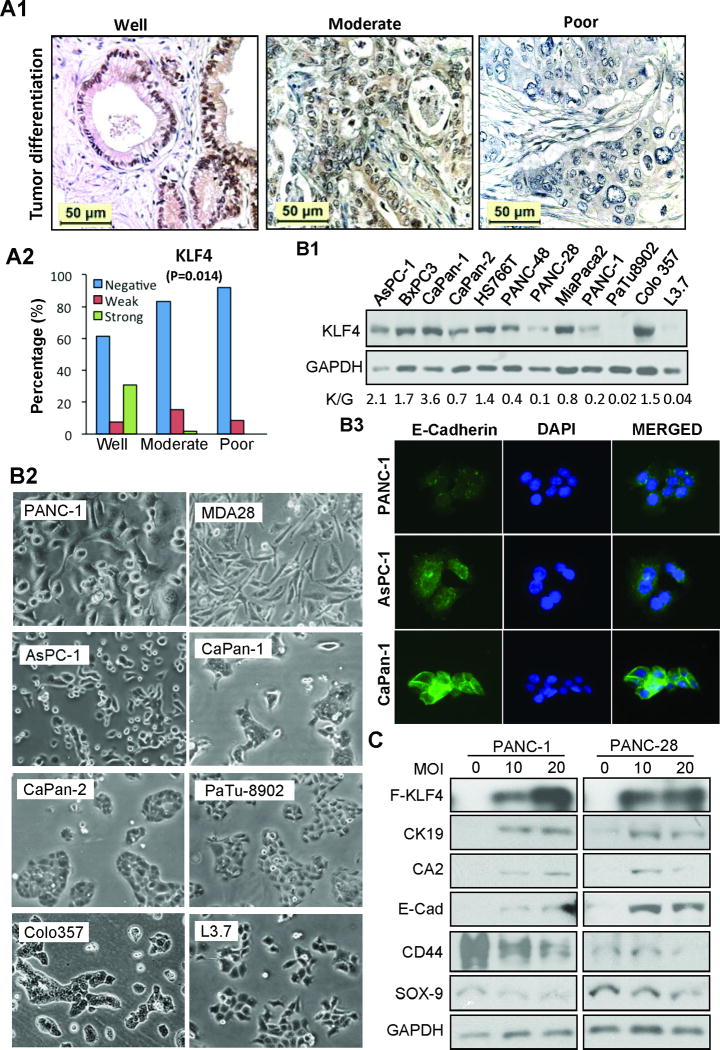
Previously, our and other studies have shown that the level of KLF4 expression is significantly reduced in pancreatic cancer tissues when compared with that in normal tissues (20–22). The clinicopathologic impact of altered KLF4 expression on pancreatic cancer was further analyzed using a pancreatic cancer tissue microarray (TMA). Strikingly, reduced or loss of KLF4 expression predicted poor tumor differentiation (P=.014) (Figure 1A &Supplementary Table 1), which is consistent with our previous study (34). Similarly, poorly differentiated pancreatic cancer cell lines (i.e., PANC-1, PANC-28, L3.7) expressed lower levels of KLF4 protein than well (CaPan-1, COLO357) or moderately differentiated (CaPan-2, MiaPaCa-2) cells (Figure 1B: panels B1 & B2) (35), which was consistent with the results of immunofluorescent (IF) staining showing that cells with relative higher KLF4 expression had higher E-cadherin staining (Figure 1B: panel B3). Significantly, transduction of KLF4 gene dose-dependently induced the expression of pancreatic ductal epithelial markers while reduced progenitor cell marker expressions in poorly differentiated PANC-1 and PANC-28 cells (Figure 1C). These results demonstrated that KLF4 regulated pancreatic cancer cell differentiation.
Figure 1.
Relationship between KLF4 expression and pancreatic cancer differentiation. (A) IHC analysis of KLF4 expression in pancreatic cancer tissues. Representative images of KLF4 expression in different tumor grades (panel A1) and the overall KLF4 expression associated with tumor differentiation (panel A2, Fisher’s exact Test, P=0.014). (B) Western blot analysis of KLF4 expression in pancreatic cancer cell lines, with relative KLF4 level expressed as K/G (KLF4/GAPDH) shown in below (panel B1); morphological images of related cell lines (panel B2); immunofluorescent (IF) staining of E-Cadherin expression in related cell lines (B3). (C) Western blot analysis of Flag-KLF4 and related protein expressions in pancreatic cancer cells at 48 hr after transduction of Ad-KLF4 viral vectors. Ad-GFP viral vectors were used as a control or to make the total multiplicity of infection (MOI) equal in each group.
Genetic and epigenetic alterations of KLF4 in pancreatic cancer
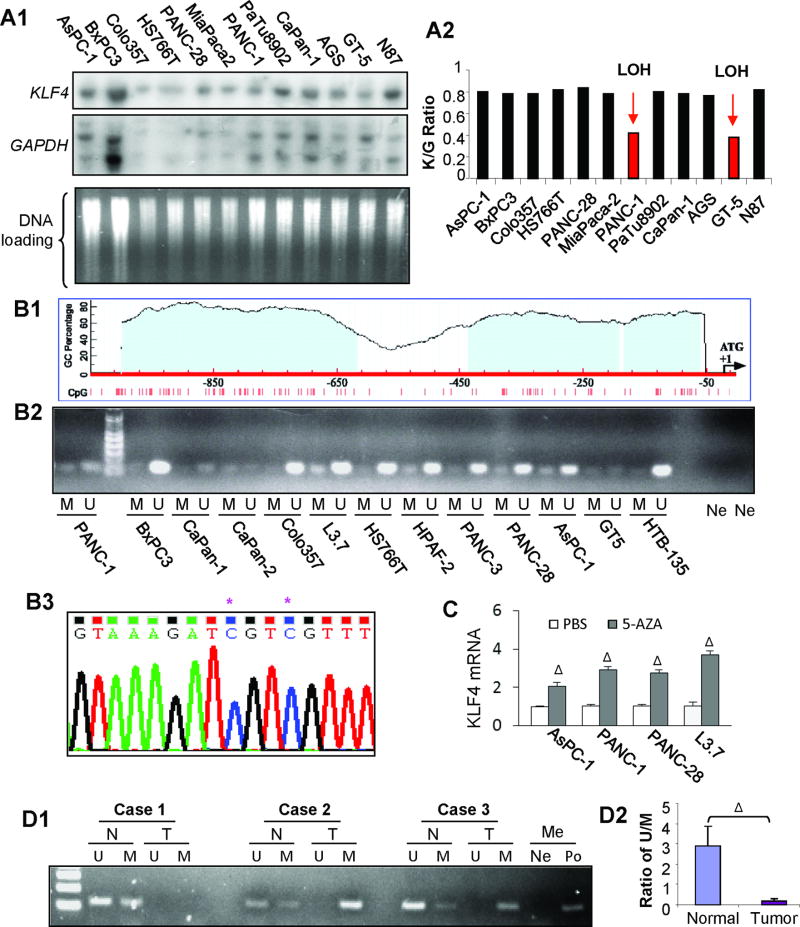
To understand the mechanisms of KLF4 underexpression in pancreatic cancer, we first investigated KLF4 gene status in pancreatic cancer cell lines by Southern blot analysis. We identified an LOH in PANC-1 cells (Figure 2A), which was consistent with the result of Western blot analysis, showing that PANC-1 cells had a relative low level of KLF4 expression (Figure 1B: panel B1). Although a previous study of in situ hybridization using microsatellite marker (D9S105) claims a high frequency (46.9%) of LOH at the loci close to KLF4 gene in PDAC (22), a recent TCGA data indicates a much lower frequency (less than 15%) of loss KLF4 gene in PDAC (Supplementary Figure 1A), suggesting that there are other mechanisms responsible for reduced or absence of KLF4 protein expression in PDAC (20, 21). Additionally, we also identified a KLF4 point mutation in BxPC3 pancreatic cancer cell line, but this mutation did not significantly affect the subcellular localization and function of KLF4 protein (Supplementary Figure 1B).
Figure 2.
Genetic and epigenetic alterations of KLF4 in pancreatic cancer. (A) Southern blot analysis of KLF4 gene status in pancreatic cancer cell lines and previously examined gastric cancer cell lines were included for controls. Representative blot images with related probes (panel A1) and the Ratio of KLF4/GAPDH signal intensity for normalization and comparison (panel A2). (B) Diagram of CpG sites and CpG islands in the promoter region of KLF4 gene (panel B1). Methylation-specific PCR analysis of DNA methylation using primers specific for the unmethylated (U) or methylated (M) KLF4 promoter region using the genomic DNA extracted from pancreatic and gastric cancer cell lines (panel B2). Note: Ne, H20 was used as a negative control for the primers of U and M, respectively. Bisulfite–DNA sequence analysis confirmed the existence of KLF4 promoter methylation in L3.7 cells (panel B3). Note: * indicates methylated C in CpG sites. (C) Poorly differentiated pancreatic cancer cells were cultured in the presence of AZA (1 µmol/L) or not (PBS) for 3 days. Total RNA was extracted and used for RT-qPCR analysis of KLF4 mRNA expression. (D) Methylation-specific PCR analysis of genomic DNA extracted from laser-capture microdissection samples of paired pancreatic normal (N) and tumor (T) tissues (panel D1), with an average ratio of U/M shown in Panel D2Note: H20 (Ne) and methylation-positive DNA sample (Po) were used with methylation specific primers, respectively; ΔP < .01 vs control. (See also supplementary Figures 1&2).
Given that the promoter region of KLF4 contains typical CpG islands (Figure 2B: panel B1) (32), methylation-specific PCR method was used to determine the methylation status of KLF4 promoter region in pancreatic cancer cell lines, and found that pancreatic cancer cell lines exhibited differential hypermethylation, especially in L3.7, PANC-1, PANC-28 cells (Figure 2B : panel B2), and furthermore, KLF4 promoter DNA methylation was confirmed by bisulfate DNA sequence analysis with an example showing in Figure 2B (panel B3; L3.7 cells). Additionally, treatment of pancreatic cancer cells with 5-Aza-2′-Deoxycytidine (5-AZA), an inhibitor of DNA methyltransferase, significantly induced KLF4 expression at both mRNA and protein levels (Figure 2C & Supplementary 1C). Consistently, significant KLF4 promoter methylation was detected in human pancreatic cancer tissues when compared with that in matched adjacent normal tissue specimens (Figure 2D & Supplementary Figure 2). These results suggest that promoter hypermethylation contributes to reduced or loss of KLF4 expression in a subset of pancreatic cancer.
DNMT1 expression inversely correlates with KLF4 expression in pancreatic cancer
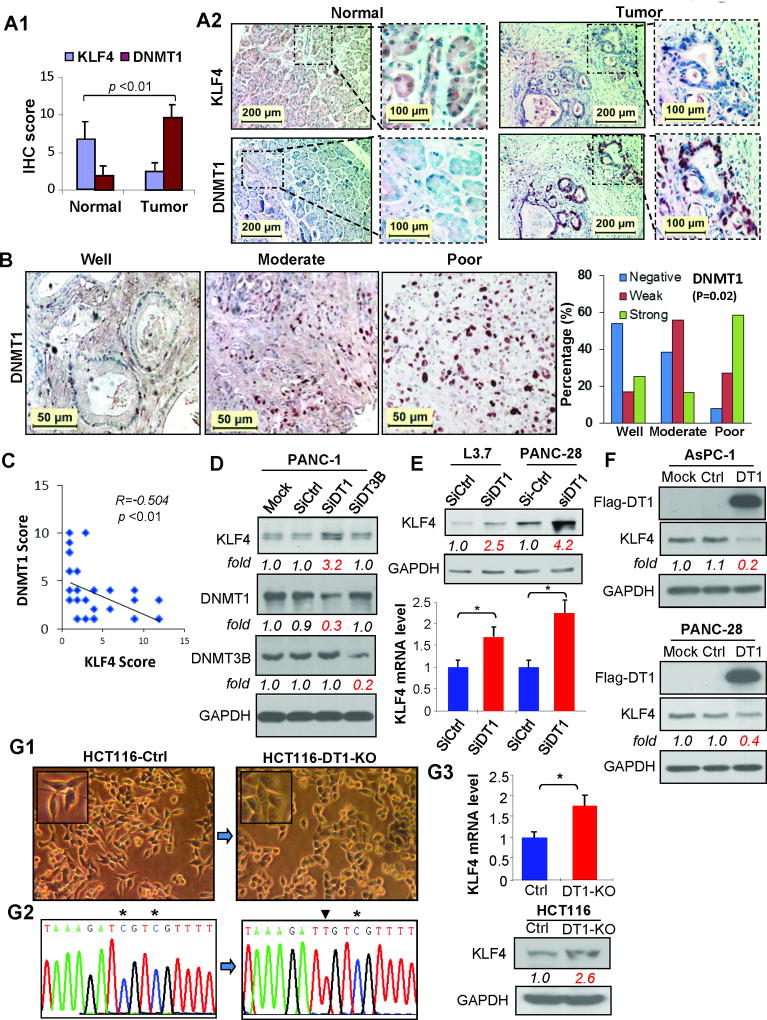
To explore possible mechanisms underlying KLF4 hypermethylation in pancreatic cancer, we first examined the expressions of DNMT1 and KLF4 by IHC analysis of consecutive sections of TMAs containing 6 paired pancreatic normal and cancer tissues. We observed that pancreatic normal tissues, in general, exhibited moderate KLF4 but negative or weak DNMT1 staining, whereas pancreatic cancer tissues exhibited negative or weak KLF4 but moderate or strong positive DNMT1 staining (Figure 3 A; Supplementary Figure 3). In sharp contrast to KLF4 expression (Figure 1A), increased DNMT1 expression correlated with poor differentiation of pancreatic cancer (Figure 3B), which was consistent with previous studies (36, 37). An inverse correlation between KLF4 and DNMT1 expressions in pancreatic cancer tissues was statistically significant (Figure 3C; P<0.01, r=−0.504). To provide causal evidence of the impact of DNA methyltransferase on KLF4 expression regulation, both DNMT1 and DNMT3B siRNAs were used to transfect PANC-1 cells, respectively, and found that knockdown of DNMT1 expression increased KLF4 protein expression, while knockdown of DNMT3B expression had no significant effect (Figure 3D). Consistently, transduction of DNMT1 siRNA into L3.7 and PANC-28 cells resulted in upregulation of KLF4 expression at both protein and mRNA levels (Figure 3E). In contrast, forced expression of DNMT1 led to reduced KLF4 protein expression in AsPC-1 and PANC-28 cells (Figure 3F). Additionally, increased KLF4 mRNA and protein expressions, reduced KLF4 promoter methylation and less spindle-like cellular morphology were observed in HCT116 colon cancer cells with DNMT1 gene knockout when compared with that in HCT116 control cells (DNMT1 gene intact) (Figure 7G). These results suggest a close relationship between DNMT1 overexpression and KLF4 underexpression in and its important role in poor differentiation of pancreatic cancer.
Figure 3.
Comparison of DNMT1 and KLF4 expressions in pancreatic cancer tissues determined by IHC analysis. (A) Average scores of KLF4 and DNMT1 immunostaining in tissue arrays containing paired pancreatic normal and cancer tissue sections (ΔP<.01 vs normal) (panel A1), and representative imaging of IHC staining (panel A2). (B) Representative images of DNMT1 expression in different tumor grades (left 3 panels) and the overall DNMT1 expression associated with tumor differentiation (right panel, Fisher’s exact Test, P=0.019). (C) Correlation analysis of overall KLF4 and DNMT1 expressions in pancreatic cancer tissue array. (D) Western blot analysis of KLF4 protein expression in PANC-1 cells at 48 hr post-transfection of DNMT1 or DNMT3 siRNA. (E) Western blot (uper panel) or RT-qPCR (lower panel) analysis of KLF4 expression at 48 hr post-transfection of DNMT1 siRNA in the cell lines indicated. (F) Western blot analysis of KLF4 protein expression at 48 hr post-transfection of DNMT1 expressing vector in the cell lines indicated. (G) Morphorlogical images of HCT116 Ctr or with DNMT1 gene knockout (HCT116-DT1-KO) cells (G1); Bisulfite–DNA sequence analysis confirmed reduced KLF4 promoter DNA methylation in HCT116-DT1-KO cells when compared to HCT116-Ctr cells (G2), which is consistant with the results of RT-qPCR or Western blot analysis of KLF4 expression in HCT116-Ctr and HCT116-DT1-KO cells (G3). (See also supplementary Figure 3).
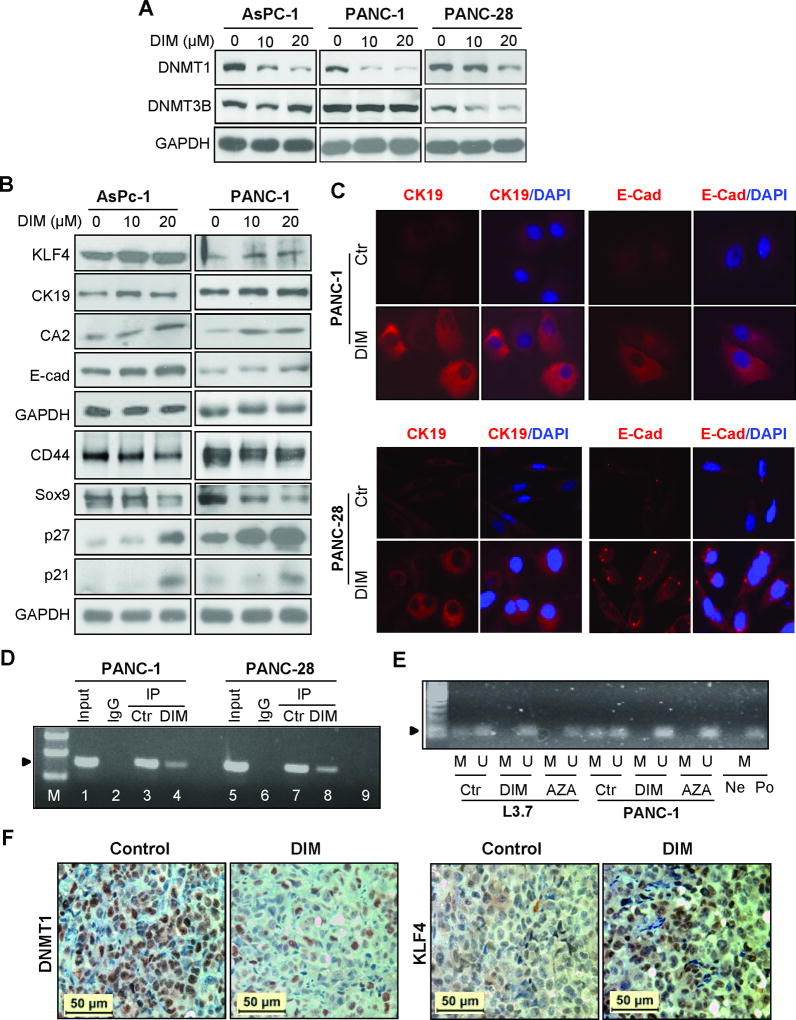
Induction of growth inhibition and differentiation of pancreatic cancer cells by dietary DIM treatment
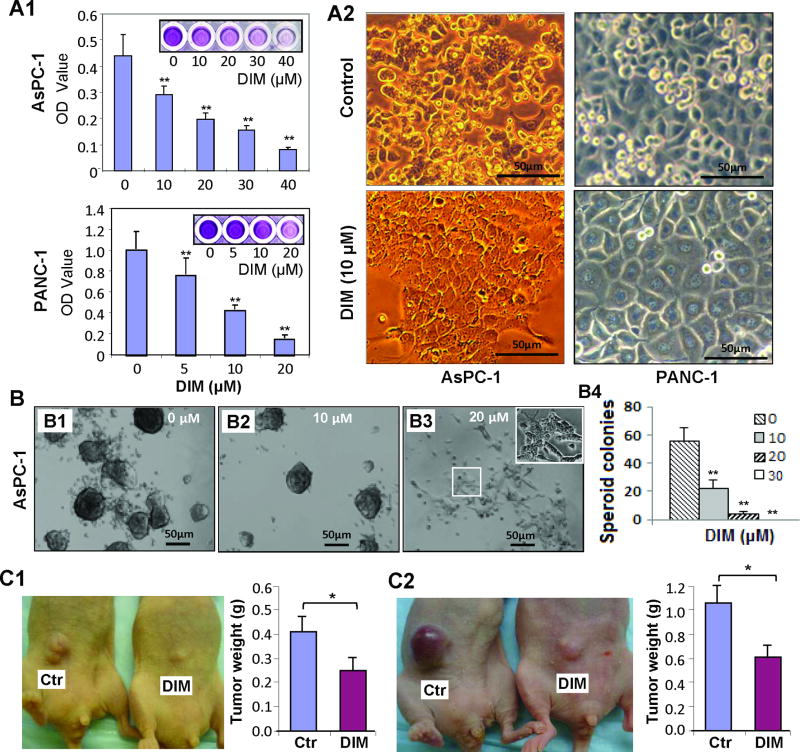
To explore whether DIM has novel function of modulating epigenome and inducing cell differentiation in pancreatic cancer, we first examined the effect of DIM on cell growth (since induction of cell differentiation requires cell growth inhibition or cell cycle arrest). As shown in Figure 4A, DIM treatment dose-dependently inhibited the growth and cell cycle progression in AsPC-1 and PANC-1 cells in vitro (Figure 4A & Supplementary 4), which was associated with significant cell morphological changes as indicated by flat and monolayer cell growth (Figure 4A, panel A2), suggesting that DIM can induce pancreatic cancer cell differentiation. Consistently, we found that DIM treatment significantly inhibited spheroid colony formation of stem-like AsPC-1 and L3.7 cells in a dose-dependent manner (Figure 4B & Supplementary Figure 5). When the dose of DIM increased to 20 µM, the spheroid colony formation was drastically reduced with few live cells growing in a flat and monolayer pattern (Figure 4B: panel B3, insert), suggesting that DIM can also inhibit the growth and induce the differentiation of spheroid-forming pancreatic cancer cells. Next, we asked whether DIM has antitumor effect in animal model. Consistent with in vitro data, DIM treatment significantly inhibited tumor growth in nude mice bearing PANC-1 and PANC-28 xenograft tumors (Figure 4C & Supplementary Figure 6), clearly indicating the potential clinical application of DIM for pancreatic cancer treatment.
Figure 4.
Induction of growth inhibition and differentiation by DIM in pancreatic cancer cells. (A) Dose-dependent inhibition of cell proliferation by MTT assay in AsPC-1 and PANC-1 cells after DIM treament (panel A1), and concomitant with significant cellular morphological change (at 60 hr after DIM treatment) (panel A2). (B) Secondary-spheroid colony formation of spheroid forming cells isolated from typical spheres of AsPC-1 cells and treated with different doses of DIM (panels B1-3) for 2 weeks, and the statistical results are shown (panel B4) (C) Inhibitory effect of DIM treatment on tumorigenesis of PANC-1 (panel C1) and PANC-28 (Panel C2) cells in xenograft mouse model. *P < .05; **P < .01 versus control. Scale bar: 50µm. (See also supplementary Figures 4, 5, and 6).
Modulation of cellular differentiation related gene expressions in pancreatic cancer cells by DIM treatment
To explore the potential mechanism of DIM treatment on pancreatic cancer cell differentiation and growth inhibition, we asked whether DIM treatment affects DNA methyltransferase expression given the profound effect of DIM on epigenetic modification, particularly DNA methylation (38). We first treated poorly differentiated AsPC-1, PANC-1, and PANC-28 cells with DIM, and found that DIM treatment dose-dependently inhibited DNMT1 but had no significant effect on DNMT3B expression (Figure 5A). Next, we examined the effect of DIM on cell differentiation marker expression. As shown in Figure 5B, DIM treatment dose-dependently increased KLF4, CK-19, CA2, E-Cad, P27, and P21, but decreased pancreatic progenitor or cancer stem cell markers CD44 and SOX-9 expressions. Immunofluorescent staining also supported this observation (Figure 5C & Supplementary 6). Although P27 and P21 are two key cell cycle negative regulators essential for cell growth inhibition, they are also required for cell differentiation (39, 40). Since KLF4 plays an important role in the regulation of pancreatic ductal epithelial differentiation and those cell differentiation markers have been approved to be downstream target genes of KLF4 (41, 42), we assumed that DIM’s effect on inhibition of DNMT1 expression may be involved in upregulation of KLF4 expression after DIM treatment, and thus, we performed Chromatin Immunoprecipitation (CHIP) assay. We found that DIM treatment significantly reduced the binding of DNMT1 to KLF4 promoter region rich in CpG islands (Figure 5D); and concomitantly, significantly inhibited KLF4 promoter hypermethylation (Figure 5E). Similarly, tumor bearing mice treated with DIM resulted in significant reduction of DNMT1 but induction of KLF4 expression in tumor tissues as determined by IHC analysis (Figure 5F & Figure 4C). These results discover a novel antitumor function of DIM in pancreatic cancer by inducing cancer cell differentiation (43).
Figure 5.
Effects of DIM on DNMT1, KLF4, and cell differentiation marker expressions in pancreatic cancer cells. (A) Western blot analysis of DNMT1 and DNM3B protein expression at 48 hr after different doses of DIM treatment in pancreatic cancer cells. GAPDH expression served as the loading control. (B) Western blot analysis of KLF4 and related marker expressions at 48 hr after different doses of DIM treatment. (C) IF staining of CK-19 and E-Cadherin expressions at 48 hr after DIM treatment. (D) ChIP analysis of DNMT1 binding to the promoter region of KLF4 in pancreatic cancer cells treated with control (DMSO) or DIM (20 µM) for 2 days. (E) Methylation-specific PCR analysis of DNA derived from pancreatic cancer cells treated with DMSO control, 15 µM DIM, or 2.0 µM AZA for 5 days. Note: H20 (Ne) and methylation-positive DNA sample (Po) were added, respectively, into PCR reactions containing methylation-specific primers. (F) IHC analysis of DNMT1 and KLF4 expression in xenograft tumor tissue samples of PANC-1 cells derived from mice fed with DIM or control food, respectively. (See also supplementary Figure 7).
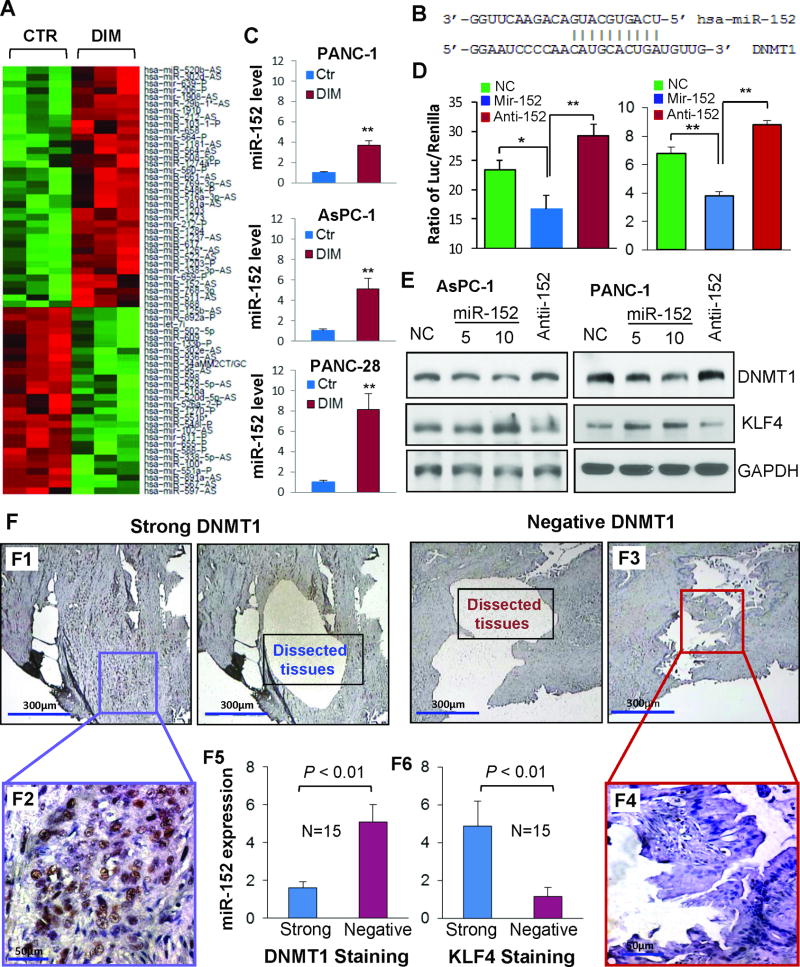
Upregulation of miR-152 expression correlates with reduced DNMT1 but increased KLF4 expression
Given that microRNA plays an important role in regulation of gene expression in physiological and pathological conditions(44–46) and in order to further understand epigenetic mechanism behind DIM’s regulatory effect on DNMT1 expression, we performed microRNA microarray analysis and identified 64 differentially expressed microRNAs (Figure 6A), of which 35 were unregulated and 29 were down-regulated (Supplementary table 3 & 4). Based on bioinformatic target prediction (Figure 6B), miR-152, a previously identified tumor suppressor microRNA (47, 48), was selected for further functional validation. Consistent with microarray data, RT-qPCR analysis conformed the significant upregulation of miR-152 in the RNA samples used for microarray analysis or in newly extracted RNA samples from DIM treated cells in repeat experiments (Figure 6C). To validate the regulatory function of miR-152 on DNMT1 expression, DNMT1 3’UTR luciferase reporter construct containing the miR-152 target site was cotransfected with mature miR-152 mimics or anti-miR-152 oligos into PANC-1 and AsPC-1 cells, respectively, the results showed that miR-152 mimics significantly downregulated while anti-miR-152 upregulated DNMT1 3’UTR luciferase reporter activity (Figure 6D). Additionally, DIM treatment significantly attenuated anti-miR-152’s action on DNMT1 3’UTR luciferase reporter activity (Supplementary Figure 8). Causally, transfection of miR-152 mature mimics resulted in decrease of DNMT1 but increase of KLF4 protein expression, whereas transfection of anti-miR-152 did the opposite (Figure 6E). We also observed that forced expression of miR-152 significantly regulated the expressions of E-Cadherin, CK-19, and Vimentin expression in PANC-28 cells (Supplementary Figure 7B), which were identified as DNMT1 and/or KLF4 downstream target genes and are consistent with the results showing in Figure 5A–B & Figure 6E. Clinically, pancreatic cancer tissues with positive DNMT1 expression had lower miR-152 and KLF4 expressions than the tissues with negative DNMT1 expression (Figure 6F). All these data suggest that upregulation of miR-152 contributes to the decreased DNMT1 and increased KLF4 expression induced by DIM in pancreatic cancer cells.
Figure 6.
Regulation of miR-152 and its downstream gene expressions by DIM in pancreatic cancer cells. (A) Microarray analysis identification of differentially expressed microRNAs (heatmap) in PANC-1 cells treated with DIM (15 µM) or control (DMSO) for 48 hr. (B) microRNA target prediction indicates that miR-152 seed sequence can bind to 3’-UTR region of DNMT1 mRNA. (C) RT-qPCR validation of miR-152 expression in the RNA samples used for microarray analysis (upper panel) or newly extracted RNA samples from cells treated with DIM (15 µM) or control (DMSO) for 48 hr (lower two panels). (D) Effect of cotransfection of pMIR-DNMT1-3’UTR reporter with non-targeting control (NC), miR-152 mimic or inhibitor oligos on luciferase activity in AsPC-1 (left panel) and PANC-1 (right panel) cells. (E) Western blot analysis of DNMT1 and KLF4 expressions at 48 hr after transfection of NC, miR-152 mimic or inhibitor oligos in AsPC-1 and PANC-1 cells. (F) Representative images of human pancreatic cancer tissues with strong (panels F1 & F2: higer magnification) or negative (panels F3 & F4: higer magnification) DNMT1 staining were microdissected and used for RT-qPCR analysis of miR-152 expression with quantitative results showing in panel F5 Similarly, microdissection samples with strong or negative KLF4 staining were used for RT-qPCR analysis of miR-152 expression, with quantitative results showing in panel F6. *P < .05; **P < .01 versus control. (See also supplementary Figure 8).
Discussion
In present study, we demonstrated for the first time that overexpression of DNMT1 contributed to promoter DNA hypermethylation and reduced or loss of KLF4 expression, which associated with poor differentiation of pancreatic cancer; and dietary DIM significantly induced KLF4 expression in and differentiation of pancreatic cancer cells, in which a novel epigenetic signaling axis of miR-152/DNMT1/KLF4 was discovered. Thus, our study not only provides new insight into the alterations of KLF4 in pancreatic cancer but also discoveres a novel mechanistic action of DIM’s antitumor activity, which may have translational implications for pancreatic and other cancers.
Pancreatic cancer is generally thought to arise from pancreatic acinar epithelial cells, although ductal and other differentiated or progenitor/stem cells have also been suggested as potential cells of origin , KLF4 has been demonstrated to play a critical role in induction of pancreatic ductal epithelial differentiation, and a context-dependent function has been recently identified in pancreatic cancer (23, 41, 49),. However, the molecular mechanisms underlying KLF4 loss of expression or inactivation in developed PDAC remain largely unknown. In the present study, we found that loss of KLF4 expression correlated with poor tumor differentiation, and forced expression of KLF4 not only significantly increased the expression of the pancreatic ductal epithelial markers such as CK19, CA2, and E-cadherin, but also decreased the expression of the pancreatic progenitor or cancer stem cell markers such as SOX-9 and CD44, which substantiates the critical role of KLF4 in regulating the differentiation of pancreatic cancer cells and further support the notion of tumor suppressive function of KLF4 in pancreatic cancer (20, 50, 51). In addition, our data clearly showed that KLF4 gene promoter methylation contributed to reduced or loss of KLF4 expression in pancreatic cancer, which is consistent with previous findings in gastric, colon and hepatocellular cancers, and lymphoma or medulloblastoma (16, 18, 32, 52–54). The epigenetic mechanism of KLF4 inactivation prompted us to explore the possibility of targeted activation of KLF4 expression for pancreatic cancer treatment. Interestingly, in the present study, we found that dietary DIM effectively activated KLF4 expression, and this activation associated with significant induction of differentiation and growth inhibition of pancreatic cancer cells. This novel finding not only provides new insight into the mechanisms of DIM’s cancer prevention and therapeutic effects but also suggest that targeted activation of KLF4 might be a promising differentiation therapy for pancreatic and other cancers, and such attempt has been proposed in clinical trials for solid tumors and acute myeloid leukemia (AML) (NCT01281592).
Both experimental and animal studies have shown that DIM has preventive and/or therapeutic effects in various cancers, which has led to clinical trials of DIM in healthy volunteers or for the treatment of cervical diseases, laryngeal papilloma, and prostate cancers (55, 56). However, how DIM triggers these biological effects and the underlying signaling cascades involved have remained unclear. In the present study, by using siRNA, adenoviral vectors, and functional analyses, we identified KLF4 as a critical mediator of DIM’s antitumor activity in pancreatic cancer, which may help to explain the dependency of 5,5′-dibromo-bis(3′-indolyl)methane, an analogue of DIM, on KLF4 for induction of p21CIP1 expression in colon cancer cells (57). Additionally, DIM’s novel functions of inducing pancreatic cancer cell differentiation, suppressing the expression of CD44, a pancreatic cancer stem cell (CSC) marker and a downstream target gene of KLF4 and then inhibiting the spheroid colony formation of pancreatic cancer cells are of clinical significance. These findings are also in line with a recent study showing that pancreatic CSCs expressed higher DNMT1 levels than non-CSC, and thus, had a higher level of DNA methylation; and pharmacologic or genetic targeting of DNMT1 in CSCs reduced their self-renewal and in vivo tumorigenic potential, defining DNMT1 as a candidate CSC therapeutic target (58). Given that pancreatic CSCs are mainly responsible for therapeutic resistance and relapse, our findings may help the development of novel anti-CSC strategies to improve the poor outcome of PDAC patients.
Furthermore, our findings add to the accumulating evidence demonstrating the important role of DNMT1 in pancreatic cancer development and progression. DNMT1 is required to maintain CpG methylation and aberrant silencing of many tumor-suppressor genes including the p16Ink4A gene, which is essential for cancer cell proliferation and survival (59), while inaction of p16 is observed in more than 95% of human pancreatic cancer cells (60, 61). Reduced DNMT1 levels resulted in a decrease in both early- and late-stage lesions and reduced tumorigenesis of pancreatic cancer in an animal model (62), whereas elevated DNMT1 expression was observed in nearly 80% of human pancreatic cancer tissues and cell lines (37, 63). In present study, we found that elevated DNMT1 expression associated with poor differentiation of and contributed to the promoter methylation and reduced expression of KLF4 in pancreatic cancer. Although the molecular mechanisms underlying DNMT1 overexpression in various cancers still remain unclear, a negative feedback regulatory loop between miR-148a/152 and DNMT1 was identified in breast cancer (64), and reduced miR-152 expression may contribute to DNMT1 overexpression in various cancers including pancreatic cancer (47, 48, 65), and our data support this notion. Therapeutically, several DNMT inhibitors are currently being evaluated in preclinical and clinical studies, which include various analogues of adenosine, cytidine or deoxycytidine (66). However, such drugs have had limited clinical success, perhaps because of their cytotoxicity associated with their incorporation into DNA and their instability in vivo (66). Development of related, stable, non-nucleoside compounds that can facilitate activation of tumor-suppressive genes in cancer cells by directly inhibiting DNMTs without being incorporated into DNA would be ideal for chemoprevention and chemotherapy. Our novel finding that dietary DIM, a low toxic, non-nucleoside and naturally occurred compound that can potently suppress DNMT1 expression, should have immediate applications in cancer prevention and therapy. This study is also the first demonstration that dietary DIM can significantly induce miR-152 expression, which provides a novel mechanism through which dietary DIM modulates epigenome. Nevertheless, there are still many questions remaining unanswered, such as how DIM activates miR-152 expression and whether there are other mechanisms that are involved in DIM’s regulation of DNMT1 and KLF4 expressions.
In summary, this study provides critical insight into dysregulated KLF4 expression in and its impact on the differentiation of pancreatic cancer. Epigenetic activation of miR-152/DNMT1/KLF4 signaling pathway by dietary DIM causes differentiation and significant growth inhibition of pancreatic cancer cells, highlighting its translational implications for pancreatic and other cancers.
Supplementary Material
Translational significance.
We have used human pancreatic adenocarcinoma (PDAC) specimens and molecular biology and animal models to evaluate the activation and function of miR-152/DNMT1/KLF4 pathway in human PDAC differentiation. In primary human PDAC, increased expression of DNMT1 correlated with decreased expression of KLF4, while increased DNMT1 expression inversely correlated with decreased expression of miR-152. Our mechanistic findings indicate that DNMT1 is a direct transcriptional target of miR-152 and that frequently downregulated miR-152 expression leads to DNMT1 overexpression. Moreover, DNMT1 overexpression causes hypermethylation of KLF4 promoter and suppresses the expression of KLF4 and promotes PDAC cell dedifferentiation, while treatment of dietary 3,3’-diindolylmethane (DIM) reverses this process in vitro and animal models, suggesting a novel molecular basis for the critical role of miR-152/DNMT1/KLF4 pathway in PDAC progression and its potential as a target to design effective therapy. Therefore, our findings may have a significant effect on clinical management of patients with PDAC.
Acknowledgments
The authors thank Dawn Chalaire for editorial comments, and thank Trupti Mehta for technical support with LCM.
Financial Support: The work was supported in part by grants R01CA172233, 1R01CA195651 and 1R01CA198090; and grants R03-CA124523 from the National Cancer Institute, NIH and grant #10A073 from the American Institute for Cancer Research, Lockton Fund for Pancreatic Cancer Research and the University Cancer Foundation via the Institutional Research Grant program at the University of Texas MD Anderson Cancer Center.
Glossary
Abbreviations used in this paper
- DIM
3,3’-diindolylmethane
- DMEM
Dulbecco’s modified Eagle’s medium
- DMSO
dimethyl sulfoxide
- DNMT1
DNA (cytosine-5)-methyltransferase 1
- DNMT3B
DNA (cytosine-5-)-methyltransferase 3 beta
- FBS
fetal bovine serum
- GAPDH
glyceraldehyde 3-phosphate dehydrogenase
- HPRT1
hypoxanthine-guanine phosphoribosyltransferase 1
- KLF4
Krüppel-like factor 4
- miR-152
microRNA-152, MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- LCM
Laser capture microdissection
- PCR
polymerase chain reaction
- SiRNA
small interfering RNA;
Footnotes
Authors’ Contributors
Conception and design: DW, VKX.
Development of methodology: DW, VKX, ZL, YY
Acquisition of data: VKX, DW, ZL, YY
Analysis and interpretation of data: DW, VKX, ZL,YY, XZ, JZ, ZJu, JW, HW.
Writing, review and/or revision of the manuscript: DW, VKX.
Administrative, technical, or material support: DW, KX, XZ, ZJ
Study supervision: DW
Obtaining funding: DW, KX.
Disclosure of Potential Conflicts of Interest: The authors disclose no potential conflicts of interest.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66(1):7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Wasif N, Ko CY, Farrell J, Wainberg Z, Hines OJ, Reber H, et al. Impact of tumor grade on prognosis in pancreatic cancer: should we include grade in AJCC staging? Ann Surg Oncol. 2010;17(9):2312–20. doi: 10.1245/s10434-010-1071-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sheng W, Dong M, Zhou J, Li X, Dong Q. Down regulation of CAII is associated with tumor differentiation and poor prognosis in patients with pancreatic cancer. J Surg Oncol. 2013;107(5):536–43. doi: 10.1002/jso.23282. [DOI] [PubMed] [Google Scholar]
- 4.Brower V. Epigenetics: Unravelling the cancer code. Nature. 2011;471(7339):S12–3. doi: 10.1038/471S12a. [DOI] [PubMed] [Google Scholar]
- 5.You JS, Jones PA. Cancer genetics and epigenetics: two sides of the same coin? Cancer Cell. 2012;22(1):9–20. doi: 10.1016/j.ccr.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pellacani D, Kestoras D, Droop AP, Frame FM, Berry PA, Lawrence MG, et al. DNA hypermethylation in prostate cancer is a consequence of aberrant epithelial differentiation and hyperproliferation. Cell Death Differ. 2014;21(5):761–73. doi: 10.1038/cdd.2013.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baylin SB, Ohm JE. Epigenetic gene silencing in cancer-a mechanism for early oncogenic pathway addiction? Nat Rev Cancer. 2006;6(2):107–16. doi: 10.1038/nrc1799. [DOI] [PubMed] [Google Scholar]
- 8.Baylin SB, Herman JG. DNA hypermethylation in tumorigenesis: epigenetics joins genetics. Trends Genet. 2000;16(4):168–74. doi: 10.1016/s0168-9525(99)01971-x. [DOI] [PubMed] [Google Scholar]
- 9.Tenen DG. Disruption of differentiation in human cancer: AML shows the way. Nat Rev Cancer. 2003;3(2):89–101. doi: 10.1038/nrc989. [DOI] [PubMed] [Google Scholar]
- 10.Lomberk GA, Iovanna J, Urrutia R. The promise of epigenomic therapeutics in pancreatic cancer. Epigenomics. 2016;8(6):831–42. doi: 10.2217/epi-2015-0016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6(11):857–66. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 12.Costa PM, Pedroso de Lima MC. MicroRNAs as Molecular Targets for Cancer Therapy: On the Modulation of MicroRNA Expression. Pharmaceuticals (Basel, Switzerland) 2013;6(10):1195–220. doi: 10.3390/ph6101195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Katz JP, Perreault N, Goldstein BG, Actman L, McNally SR, Silberg DG, et al. Loss of Klf4 in mice causes altered proliferation and differentiation and precancerous changes in the adult stomach. Gastroenterology. 2005;128(4):935–45. doi: 10.1053/j.gastro.2005.02.022. [DOI] [PubMed] [Google Scholar]
- 14.Guo X, Tang Y. KLF4 translation level is associated with differentiation stage of different pediatric leukemias in both cell lines and primary samples. Clin Exp Med. 2013;13(2):99–107. doi: 10.1007/s10238-012-0187-4. [DOI] [PubMed] [Google Scholar]
- 15.Hu R, Zuo Y, Zuo L, Liu C, Zhang S, Wu Q, et al. KLF4 Expression Correlates with the Degree of Differentiation in Colorectal Cancer. Gut Liver. 2011;5(2):154–9. doi: 10.5009/gnl.2011.5.2.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tetreault MP, Yang Y, Katz JP. Kruppel-like factors in cancer. Nat Rev Cancer. 2013;13(10):701–13. doi: 10.1038/nrc3582. [DOI] [PubMed] [Google Scholar]
- 17.Shum CK, Lau ST, Tsoi LL, Chan LK, Yam JW, Ohira M, et al. Kruppel-like factor 4 (KLF4) suppresses neuroblastoma cell growth and determines non-tumorigenic lineage differentiation. Oncogene. 2013;32(35):4086–99. doi: 10.1038/onc.2012.437. [DOI] [PubMed] [Google Scholar]
- 18.Zhao W, Hisamuddin IM, Nandan MO, Babbin BA, Lamb NE, Yang VW. Identification of Kruppel-like factor 4 as a potential tumor suppressor gene in colorectal cancer. Oncogene. 2004;23(2):395–402. doi: 10.1038/sj.onc.1207067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang W, Chen X, Kato Y, Evans PM, Yuan S, Yang J, et al. Novel cross talk of Kruppel-like factor 4 and beta-catenin regulates normal intestinal homeostasis and tumor repression. Mol Cell Biol. 2006;26(6):2055–64. doi: 10.1128/MCB.26.6.2055-2064.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zammarchi F, Morelli M, Menicagli M, Di Cristofano C, Zavaglia K, Paolucci A, et al. KLF4 is a novel candidate tumor suppressor gene in pancreatic ductal carcinoma. Am J Pathol. 2011;178(1):361–72. doi: 10.1016/j.ajpath.2010.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wei D, Wang L, Kanai M, Jia Z, Le X, Li Q, et al. KLF4alpha up-regulation promotes cell cycle progression and reduces survival time of patients with pancreatic cancer. Gastroenterology. 2010;139(6):2135–45. doi: 10.1053/j.gastro.2010.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Funel N, Morelli M, Giovannetti E, Del Chiaro M, Pollina LE, Mosca F, et al. Loss of heterozygosity status of D9S105 marker is associated with downregulation of Kruppel-like factor 4 expression in pancreatic ductal adenocarcinoma and pancreatic intraepithelial lesions. Pancreatology. 2011;11(1):30–42. doi: 10.1159/000322990. [DOI] [PubMed] [Google Scholar]
- 23.Wei D, Wang L, Yan Y, Jia Z, Gagea M, Li Z, et al. KLF4 Is Essential for Induction of Cellular Identity Change and Acinar-to-Ductal Reprogramming during Early Pancreatic Carcinogenesis. Cancer cell. 2016;29(3):324–38. doi: 10.1016/j.ccell.2016.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prasad NB, Biankin AV, Fukushima N, Maitra A, Dhara S, Elkahloun AG, et al. Gene expression profiles in pancreatic intraepithelial neoplasia reflect the effects of Hedgehog signaling on pancreatic ductal epithelial cells. Cancer research. 2005;65(5):1619–26. doi: 10.1158/0008-5472.CAN-04-1413. [DOI] [PubMed] [Google Scholar]
- 25.Higdon JV, Delage B, Williams DE, Dashwood RH. Cruciferous vegetables and human cancer risk: epidemiologic evidence and mechanistic basis. Pharmacological research. 2007;55(3):224–36. doi: 10.1016/j.phrs.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thomson CA, Ho E, Strom MB. Chemopreventive properties of 3,3’-diindolylmethane in breast cancer: evidence from experimental and human studies. Nutrition reviews. 2016;74(7):432–43. doi: 10.1093/nutrit/nuw010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fujioka N, Ainslie-Waldman CE, Upadhyaya P, Carmella SG, Fritz VA, Rohwer C, et al. Urinary 3,3’-diindolylmethane: a biomarker of glucobrassicin exposure and indole-3-carbinol uptake in humans. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2014;23(2):282–7. doi: 10.1158/1055-9965.EPI-13-0645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reed GA, Peterson KS, Smith HJ, Gray JC, Sullivan DK, Mayo MS, et al. A phase I study of indole-3-carbinol in women: tolerability and effects. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2005;14(8):1953–60. doi: 10.1158/1055-9965.EPI-05-0121. [DOI] [PubMed] [Google Scholar]
- 29.Gee JR, Saltzstein DR, Messing E, Kim K, Kolesar J, Huang W, et al. Phase Ib placebo-controlled, tissue biomarker trial of diindolylmethane (BR-DIMNG) in patients with prostate cancer who are undergoing prostatectomy. European journal of cancer prevention : the official journal of the European Cancer Prevention Organisation (ECP) 2016;25(4):312–20. doi: 10.1097/CEJ.0000000000000189. [DOI] [PubMed] [Google Scholar]
- 30.Paltsev M, Kiselev V, Drukh V, Muyzhnek E, Kuznetsov I, Andrianova E, et al. First results of the double-blind randomized placebo-controlled multicenter clinical trial of DIM-based therapy designed as personalized approach to reverse prostatic intraepithelial neoplasia (PIN) The EPMA journal. 2016;7:5. doi: 10.1186/s13167-016-0057-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pounds S, Morris SW. Estimating the occurrence of false positives and false negatives in microarray studies by approximating and partitioning the empirical distribution of p-values. Bioinformatics (Oxford, England) 2003;19(10):1236–42. doi: 10.1093/bioinformatics/btg148. [DOI] [PubMed] [Google Scholar]
- 32.Wei D, Gong W, Kanai M, Schlunk C, Wang L, Yao JC, et al. Drastic down-regulation of Kruppel-like factor 4 expression is critical in human gastric cancer development and progression. Cancer research. 2005;65(7):2746–54. doi: 10.1158/0008-5472.CAN-04-3619. [DOI] [PubMed] [Google Scholar]
- 33.Sepkovic DW, Stein J, Carlisle AD, Ksieski HB, Auborn K, Bradlow HL. Diindolylmethane inhibits cervical dysplasia, alters estrogen metabolism, and enhances immune response in the K14-HPV16 transgenic mouse model. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2009;18(11):2957–64. doi: 10.1158/1055-9965.EPI-09-0698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shi M, Cui J, Du J, Wei D, Jia Z, Zhang J, et al. A novel KLF4/LDHA signaling pathway regulates aerobic glycolysis in and progression of pancreatic cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2014;20(16):4370–80. doi: 10.1158/1078-0432.CCR-14-0186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Loukopoulos P, Kanetaka K, Takamura M, Shibata T, Sakamoto M, Hirohashi S. Orthotopic transplantation models of pancreatic adenocarcinoma derived from cell lines and primary tumors and displaying varying metastatic activity. Pancreas. 2004;29(3):193–203. doi: 10.1097/00006676-200410000-00004. [DOI] [PubMed] [Google Scholar]
- 36.Peng DF, Kanai Y, Sawada M, Ushijima S, Hiraoka N, Kosuge T, et al. Increased DNA methyltransferase 1 (DNMT1) protein expression in precancerous conditions and ductal carcinomas of the pancreas. Cancer Sci. 2005;96(7):403–8. doi: 10.1111/j.1349-7006.2005.00071.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peng DF, Kanai Y, Sawada M, Ushijima S, Hiraoka N, Kitazawa S, et al. DNA methylation of multiple tumor-related genes in association with overexpression of DNA methyltransferase 1 (DNMT1) during multistage carcinogenesis of the pancreas. Carcinogenesis. 2006;27(6):1160–8. doi: 10.1093/carcin/bgi361. [DOI] [PubMed] [Google Scholar]
- 38.Wu TY, Khor TO, Su ZY, Saw CL, Shu L, Cheung KL, et al. Epigenetic modifications of Nrf2 by 3,3’-diindolylmethane in vitro in TRAMP C1 cell line and in vivo TRAMP prostate tumors. The AAPS journal. 2013;15(3):864–74. doi: 10.1208/s12248-013-9493-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hauser PJ, Agrawal D, Flanagan M, Pledger WJ. The role of p27kip1 in the in vitro differentiation of murine keratinocytes. Cell growth & differentiation : the molecular biology journal of the American Association for Cancer Research. 1997;8(2):203–11. [PubMed] [Google Scholar]
- 40.Zezula J, Casaccia-Bonnefil P, Ezhevsky SA, Osterhout DJ, Levine JM, Dowdy SF, et al. p21cip1 is required for the differentiation of oligodendrocytes independently of cell cycle withdrawal. EMBO reports. 2001;2(1):27–34. doi: 10.1093/embo-reports/kve008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brembeck FH, Rustgi AK. The tissue-dependent keratin 19 gene transcription is regulated by GKLF/KLF4 and Sp1. J Biol Chem. 2000;275(36):28230–9. doi: 10.1074/jbc.M004013200. [DOI] [PubMed] [Google Scholar]
- 42.Yori JL, Johnson E, Zhou G, Jain MK, Keri RA. Kruppel-like factor 4 inhibits epithelial-to-mesenchymal transition through regulation of E-cadherin gene expression. J Biol Chem. 2010;285(22):16854–63. doi: 10.1074/jbc.M110.114546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li C, Heidt DG, Dalerba P, Burant CF, Zhang L, Adsay V, et al. Identification of pancreatic cancer stem cells. Cancer research. 2007;67(3):1030–7. doi: 10.1158/0008-5472.CAN-06-2030. [DOI] [PubMed] [Google Scholar]
- 44.Das S, Foley N, Bryan K, Watters KM, Bray I, Murphy DM, et al. MicroRNA mediates DNA demethylation events triggered by retinoic acid during neuroblastoma cell differentiation. Cancer research. 2010;70(20):7874–81. doi: 10.1158/0008-5472.CAN-10-1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12(12):861–74. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 46.Lujambio A, Lowe SW. The microcosmos of cancer. Nature. 2012;482(7385):347–55. doi: 10.1038/nature10888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tsuruta T, Kozaki K, Uesugi A, Furuta M, Hirasawa A, Imoto I, et al. miR-152 is a tumor suppressor microRNA that is silenced by DNA hypermethylation in endometrial cancer. Cancer research. 2011;71(20):6450–62. doi: 10.1158/0008-5472.CAN-11-0364. [DOI] [PubMed] [Google Scholar]
- 48.Huang J, Wang Y, Guo Y, Sun S. Down-regulated microRNA-152 induces aberrant DNA methylation in hepatitis B virus-related hepatocellular carcinoma by targeting DNA methyltransferase 1. Hepatology. 2010;52(1):60–70. doi: 10.1002/hep.23660. [DOI] [PubMed] [Google Scholar]
- 49.Brembeck FH, Moffett J, Wang TC, Rustgi AK. The keratin 19 promoter is potent for cell-specific targeting of genes in transgenic mice. Gastroenterology. 2001;120(7):1720–8. doi: 10.1053/gast.2001.24846. [DOI] [PubMed] [Google Scholar]
- 50.Wei D, Kanai M, Jia Z, Le X, Xie K. Kruppel-like factor 4 induces p27Kip1 expression in and suppresses the growth and metastasis of human pancreatic cancer cells. Cancer research. 2008;68(12):4631–9. doi: 10.1158/0008-5472.CAN-07-5953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yan Y, Li Z, Kong X, Jia Z, Zuo X, Gagea M, et al. KLF4-Mediated Suppression of CD44 Signaling Negatively Impacts Pancreatic Cancer Stemness and Metastasis. Cancer research. 2016;76(8):2419–31. doi: 10.1158/0008-5472.CAN-15-1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li Q, Gao Y, Jia Z, Mishra L, Guo K, Li Z, et al. Dysregulated Kruppel-like factor 4 and vitamin D receptor signaling contribute to progression of hepatocellular carcinoma. Gastroenterology. 2012;143(3):799–810. doi: 10.1053/j.gastro.2012.05.043. e1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guan H, Xie L, Leithauser F, Flossbach L, Moller P, Wirth T, et al. KLF4 is a tumor suppressor in B-cell non-Hodgkin lymphoma and in classic Hodgkin lymphoma. Blood. 2010;116(9):1469–78. doi: 10.1182/blood-2009-12-256446. [DOI] [PubMed] [Google Scholar]
- 54.Faber K, Bullinger L, Ragu C, Garding A, Mertens D, Miller C, et al. CDX2-driven leukemogenesis involves KLF4 repression and deregulated PPARgamma signaling. J Clin Invest. 2013;123(1):299–314. doi: 10.1172/JCI64745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Del Priore G, Gudipudi DK, Montemarano N, Restivo AM, Malanowska-Stega J, Arslan AA. Oral diindolylmethane (DIM): pilot evaluation of a nonsurgical treatment for cervical dysplasia. Gynecol Oncol. 2010;116(3):464–7. doi: 10.1016/j.ygyno.2009.10.060. [DOI] [PubMed] [Google Scholar]
- 56.Heath EI, Heilbrun LK, Li J, Vaishampayan U, Harper F, Pemberton P, et al. A phase I dose-escalation study of oral BR-DIM (BioResponse 3,3’- Diindolylmethane) in castrate-resistant, non-metastatic prostate cancer. Am J Transl Res. 2010;2(4):402–11. [PMC free article] [PubMed] [Google Scholar]
- 57.Cho SD, Chintharlapalli S, Abdelrahim M, Papineni S, Liu S, Guo J, et al. 5,5’-Dibromo-bis(3’-indolyl)methane induces Kruppel-like factor 4 and p21 in colon cancer cells. Mol Cancer Ther. 2008;7(7):2109–20. doi: 10.1158/1535-7163.MCT-07-2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zagorac S, Alcala S, Fernandez Bayon G, Bou Kheir T, Schoenhals M, Gonzalez-Neira A, et al. DNMT1 Inhibition Reprograms Pancreatic Cancer Stem Cells via Upregulation of the miR-17-92 Cluster. Cancer research. 2016;76(15):4546–58. doi: 10.1158/0008-5472.CAN-15-3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Robert MF, Morin S, Beaulieu N, Gauthier F, Chute IC, Barsalou A, et al. DNMT1 is required to maintain CpG methylation and aberrant gene silencing in human cancer cells. Nat Genet. 2003;33(1):61–5. doi: 10.1038/ng1068. [DOI] [PubMed] [Google Scholar]
- 60.Ueki T, Toyota M, Sohn T, Yeo CJ, Issa JP, Hruban RH, et al. Hypermethylation of multiple genes in pancreatic adenocarcinoma. Cancer research. 2000;60(7):1835–9. [PubMed] [Google Scholar]
- 61.Schutte M, Hruban RH, Geradts J, Maynard R, Hilgers W, Rabindran SK, et al. Abrogation of the Rb/p16 tumor-suppressive pathway in virtually all pancreatic carcinomas. Cancer research. 1997;57(15):3126–30. [PubMed] [Google Scholar]
- 62.Oghamian S, Sodir NM, Bashir MU, Shen H, Cullins AE, Carroll CA, et al. Reduction of pancreatic acinar cell tumor multiplicity in Dnmt1 hypomorphic mice. Carcinogenesis. 2011;32(6):829–35. doi: 10.1093/carcin/bgr039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li A, Omura N, Hong SM, Goggins M. Pancreatic cancer DNMT1 expression and sensitivity to DNMT1 inhibitors. Cancer Biol Ther. 2010;9(4) doi: 10.4161/cbt.9.4.10750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xu Q, Jiang Y, Yin Y, Li Q, He J, Jing Y, et al. A regulatory circuit of miR-148a/152 and DNMT1 in modulating cell transformation and tumor angiogenesis through IGF-IR and IRS1. J Mol Cell Biol. 2013;5(1):3–13. doi: 10.1093/jmcb/mjs049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Azizi M, Teimoori-Toolabi L, Arzanani MK, Azadmanesh K, Fard-Esfahani P, Zeinali S. MicroRNA-148b and microRNA-152 reactivate tumor suppressor genes through suppression of DNA methyltransferase-1 gene in pancreatic cancer cell lines. Cancer Biol Ther. 2014;15(4):419–27. doi: 10.4161/cbt.27630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Qiu W, Su GH. Refining methylation-targeting therapies for pancreatic cancer-focusing on DNMT1. Cancer Biol Ther. 2010;9(4) doi: 10.4161/cbt.9.4.11167. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.