PI(4)P directly interacts with the cytosolic domain of yeast Golgi vacuolar H+-ATPase (V-ATPase) a-isoform, Stv1, and the human Golgi a-subunit isoform. Lys-84 of Stv1 is essential for PI(4)P interaction, and localization of Stv1-containing V-ATPases in vivo requires the PI(4)P interaction. We propose that phosphatidylinositol binding exerts organelle-specific control over V-ATPases.
Abstract
Luminal pH and phosphoinositide content are fundamental features of organelle identity. Vacuolar H+-ATPases (V-ATPases) drive organelle acidification in all eukaryotes, and membrane-bound a-subunit isoforms of the V-ATPase are implicated in organelle-specific targeting and regulation. Earlier work demonstrated that the endolysosomal lipid PI(3,5)P2 activates V-ATPases containing the vacuolar a-subunit isoform in Saccharomyces cerevisiae. Here we demonstrate that PI(4)P, the predominant Golgi phosphatidylinositol (PI) species, directly interacts with the cytosolic amino terminal (NT) domain of the yeast Golgi V-ATPase a-isoform Stv1. Lysine-84 of Stv1NT is essential for interaction with PI(4)P in vitro and in vivo, and interaction with PI(4)P is required for efficient localization of Stv1-containing V-ATPases. The cytosolic NT domain of the human V-ATPase a2 isoform specifically interacts with PI(4)P in vitro, consistent with its Golgi localization and function. We propose that NT domains of Vo a-subunit isoforms interact specifically with PI lipids in their organelles of residence. These interactions can transmit organelle-specific targeting or regulation information to V-ATPases.
INTRODUCTION
Eukaryotic cells have distinct membrane-bound organelles that perform specific functions essential for cellular homeostasis and fitness of the organism. Organelles of the secretory and endocytic pathways direct protein and membrane trafficking, degrade macromolecular waste, secrete enzymes and hormones, recycle receptors, and support neuronal communication. These compartmentalized functions require organelles to maintain distinct chemical identities, and luminal pH is an important component of organelle identity. In contrast to the neutral to slightly alkaline cytosolic pH, several of the organelles of the secretory and biosynthetic pathway are acidic, including the yeast vacuole (pH 5.7–6.2) and mammalian lysosomes (pH 4.7–4.9), Golgi apparatus (pH 6.4–6.8), and endosomes (pH 5.5–6.5) (Casey et al., 2010; Brett et al., 2011; Diakov et al., 2013) The vacuolar H+-ATPase (V-ATPase) is a highly conserved ATP-driven H+-pump that is primarily responsible for organelle acidification (Kane, 2006). The V-ATPase is composed of 14 different subunits (Kane, 2006) arranged into a cytosolic V1 sector and a membrane-bound Vo sector. ATP hydrolysis in the V1 sector is coupled to proton translocation through the Vo sector (Kane, 2006).
Higher organisms, including mammals, require V-ATPase activity to survive (Sun-Wada et al., 2000; Nelson, 2003) However, there is relatively little information about how V-ATPases control organelle-specific acidification. In higher organisms, tissue- and organelle-enriched isoforms of several subunits target specific V-ATPase subpopulations and help mediate organelle-specific properties. In particular, mammals have four isoforms of the Vo a-subunit (a1–a4), distributed in a tissue- and organelle-enriched manner (Forgac, 2007). V-ATPase localization has been strongly linked to Vo a-subunit content (Toyomura et al., 2000). Furthermore, several V-ATPase–linked diseases are directly correlated with mutations in the Vo a-subunit isoforms. The congenital disease autosomal recessive cutis laxa-type II, caused by glycosylation defects in the Golgi, is linked to mutations in the human a2 isoform (Kornak et al., 2008). Osteopetrosis and distal renal tubule acidosis have been directly correlated to mutations in plasma membrane–localized a3 and a4 isoforms, respectively (Frattini et al., 2000; Smith et al., 2000).
In the budding yeast Saccharomyces cerevisiae, the V-ATPase is not essential for growth, but loss of V-ATPase function is conditionally lethal. V-ATPase–deficient yeast mutants (vma mutants) can grow in acidic media (pH 5) but fail to grow in alkaline (pH 7.5) media or media containing elevated Ca2+ (Nelson and Nelson, 1990; Ohya et al., 1991). Only the Vo a-subunit is present as two organelle-specific isoforms in yeast, suggesting that this subunit must account for differences in targeting, regulation, and catalytic properties of V-ATPases containing the different a-subunit isoforms (Forgac, 2007). The Vph1 isoform targets V-ATPases to vacuoles, and Stv1 targets V-ATPases to the Golgi apparatus and endosomes (Manolson et al., 1994; Kawasaki-Nishi et al., 2001a). Deletion of both STV1 and VPH1 is required for cells to exhibit the full Vma− phenotype, characterized by failure to grow at high pH and calcium concentrations. vph1∆ mutants survive but grow slowly under these conditions, and fail to grow on a medium containing Zn2+, suggesting Zn2+ sensitivity is a vacuole-specific phenotype (Finnigan et al., 2011). stv1∆ mutants have no obvious growth defect, possibly because Vph1 inhabits the same compartments en route to the vacuole and thus can compensate for loss of STV1 (Graham et al., 1998; Finnigan et al., 2012). Functional effects of STV1 mutations have been analyzed in vph1∆ mutant background (Finnigan et al., 2012).
In addition to subunit isoforms, other organelle-specific factors help govern V-ATPase properties (Qi and Forgac, 2007), but neither the identity of these factors nor the nature of their interactions with V-ATPases were well understood. Phosphatidylinositol (PI) lipids exhibit significant organelle-specific enrichment in all eukaryotic cells, as well as identifying microdomains within organelle membranes (Strahl and Thorner, 2007; Idevall-Hagren and De Camilli, 2015). PI lipid species are generated by reversible phosphorylation of the inositol head group mediated by specific kinases and phosphatases (Balla, 2013). In addition to regulating soluble, cytosolic enzymes, PI lipids can regulate the activity of membrane-bound ion channels and transporters in space and in time (Hilgemann, 2004; Zhang et al., 2012; Hille et al., 2015). PI(3,5)P2 is a low-level, signaling lipid present predominantly at the vacuoles in yeast and late endosomes and lysosomes in mammals (Ho et al., 2012). It is synthesized from PI(3)P by the PI(3)P-dependent-5-kinase, Fab1/PIKfyve, acting in complex with the 5-phosphatase Fig 4 and the scaffolding protein Vac14 (Dove et al., 2002; Duex et al., 2006; Zolov et al., 2012). PI(3,5)P2 activates Vph1-containing V-ATPases in yeast by promoting tight assembly of the V1 and Vo sectors of the enzyme (Li et al., 2014). Recognition of PI(3,5)P2 by the Vo domain of V-ATPase was attributed to the cytosolic N-terminal domain of Vph1 (Vph1NT), and Vph1NT could be recruited to membranes by high PI(3,5)P2 levels in the absence of other V-ATPase subunits (Li et al., 2014). V-ATPase has also been linked to vacuolation defects in mammalian cells arising from reduction in PI(3,5)P2 content (Compton et al., 2016).
FIGURE 4:
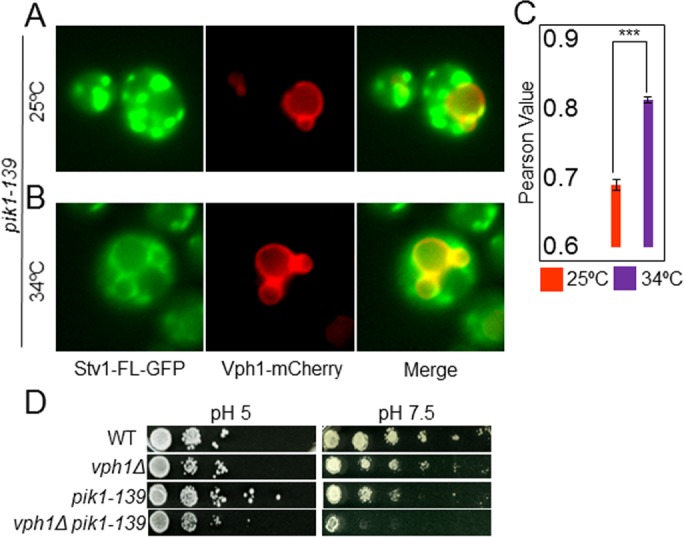
Down-regulation of intracellular PI(4)P level induces mislocalization and impaired function of V-ATPases containing Stv1. (A, B) Localization of Stv1-GFP and Vph1-mCherry in pik1-39 mutant. (A) Localization at the permissive temperature of 25°C (representative of n = 511 yeast cells) or (B) after a 2-h incubation at the restrictive temperature of 34°C (representative of n = 1203 yeast cells). Vacuoles are marked by Vph1-mCherry. (C) Mean ± SEM of Pearson correlation coefficient for colocalization of GFP and mCherry from three independent experiments is represented in a histogram (p < 0.00005). (D) Growth assay of congenic WT, vph1Δ, pik1-139, and vph1Δ pik1-139 cells in media buffered to pH 5.0 (left) and pH 7.5 (right). Cells from each strain were diluted to the same density, then 10-fold serial dilutions (left to right) were made and pinned to YEPD plates buffered to the indicated pH. The growth assay shown is representative of two independent experiments.
In contrast to PI(3,5)P2, PI(4)P is highly enriched in the Golgi network and plasma membrane of yeast and mammals (D’Angelo et al., 2008). We hypothesized that PI lipids enriched in specific organelle membranes provide localized regulation to V-ATPases by binding to organelle-specific Vo a-subunit isoforms. In this work, we find that Stv1NT interacts specifically with PI(4)P in vitro and demonstrate that the Golgi pool of PI(4)P helps to localize V-ATPases containing full-length Stv1, as well as supporting their function. Additionally, we show that the N-terminal domain of the human Golgi/endosomal Vo a-subunit isoform, a2 (ATP6V0A2), preferentially interacts with PI(4)P, suggesting that PI interactions may be a conserved feature of V-ATPases.
RESULTS
Cytosolic NT of Stv1 localizes constitutively to puncta, independent of the Vo membrane sector and the vacuolar lipid PI(3,5)P2
The soluble cytosolic NT domain of the vacuolar isoform of Vo a-subunit (Vph1NT) was reversibly recruited to membranes when levels of the late endosome/vacuole-specific lipid PI(3,5)P2 were elevated (Li et al., 2014). To assess the localization of the cytosolic NT domain of the Golgi/endosome Vo a-isoform, Stv1 (Stv1NT), we replaced the membrane domain of Stv1 with mCherry to create Stv1NT-mCherry. Interestingly, Stv1NT-mCherry localized constitutively to puncta that appeared to be nonvacuolar (Figure 1A). Although Stv1NT-mCherry cannot assemble into intact Vo sectors because it lacks its membrane domain, V-ATPases with intact Vo sectors containing Vph1 are still present in the Stv1NT strain. We therefore introduced Stv1NT-mCherry into vph1Δ cells to remove both intact Vo a-subunit isoforms and prevent any assembly of Vo sectors (Manolson et al., 1992; Leng et al., 1998). Stv1NT-mCherry still recruited to puncta in the absence of Vo, suggesting that the membrane-bound Vo sectors are not required to recruit Stv1NT from the cytosol to puncta (Figure 1B). vac14Δ cells synthesize only 2% of the wild-type levels of PI(3,5)P2 (Duex et al., 2006), and this mutant does not recruit Vph1NT–green fluorescent protein (Vph1NT-GFP) to membranes, even under osmotic shock (Li et al., 2014). Stv1NT-mCherry, however, did not require Vac14 to form puncta (Figure 1C). PI(3,5)P2 is therefore not required for Stv1NT localization in these foci.
FIGURE 1:

Localization of Stv1NT-mCherry to puncta. Stv1NT-mCherry (in which the transmembrane C-terminal domain of Stv1 is replaced by mCherry) was expressed from the genomic STV1 locus in (A) S. cerevisiae wild-type strain BY4741; (B) BY4741 vph1∆ cells, which lack the second Vo a-subunit isoform; and (C) BY4741 vac14∆ cells, which lack PI(3,5)P2. For each set of panels, differential interference contrast images are shown on the left and mCherry fluorescence is shown on the right.
Stv1NT specifically interacts with the Golgi lipid PI(4)P in vitro
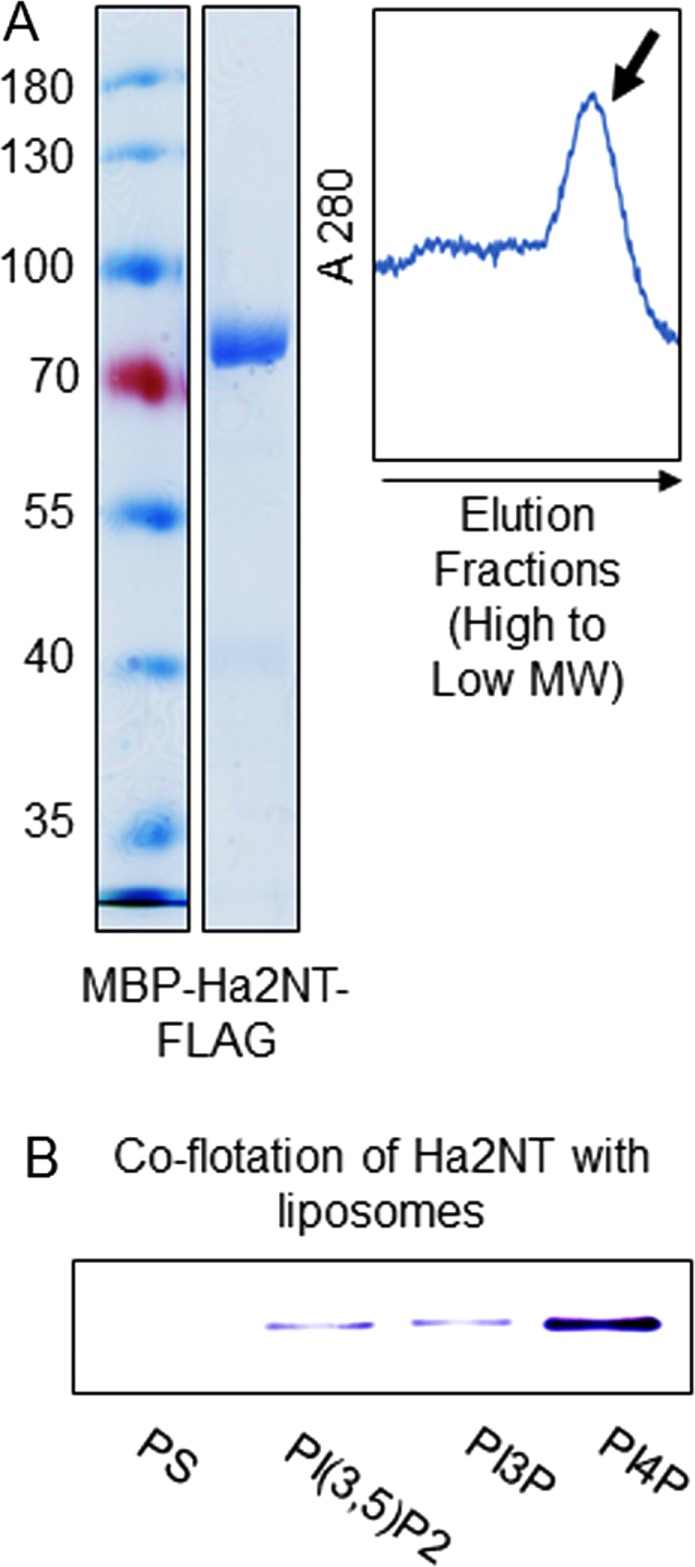
A soluble form of Vph1NT (aa 1–372) was previously expressed and purified from Escherichia coli as a mix of a monomer–dimer that could be separated by size-exclusion chromatography (Oot and Wilkens, 2012; Couoh-Cardel et al., 2015). We designed a similar construct of Stv1NT(aa 1–418) by alignment with Vph1NT and expressed it in E. coli with an N-terminal maltose-binding protein (MBP) fusion and a C-terminal FLAG tag. We obtained highly pure MBP-Stv1NT-FLAG after double-affinity chromatography, followed by-size exclusion chromatography (Figure 2A). This Stv1NT construct also eluted as monomer and higher oligomer fractions (Figure 2A, right) from the gel-filtration column. We used the monomer fraction of purified MBP-Stv1NT-FLAG to test for lipid interactions on a PIP blot in vitro. Interestingly, MBP-Stv1NT-FLAG interacted strongly and directly only with PI(4)P (Figure 2B; a control MBP blot showed no interactions). PI(4)P is the predominant PI lipid species in the Golgi (Gillooly et al., 2000; Levine and Munro, 2002). This result strongly suggested that Stv1, the Golgi isoform of Vo a-subunit, selectively interacts with the Golgi-specific phosphoinositide, PI(4)P.
FIGURE 2:
Stv1NT specifically interacts with the Golgi lipid PI(4)P in vitro. (A) Left, Coomassie-stained SDS–PAGE showing purified MBP-Stv1NT- FLAG and molecular mass markers. Right, the elution profile of MBP-Stv1NT-FLAG from a Sephadex 200 gel-filtration column. The sharp lowest molecular weight fraction, indicated by M, corresponds to the monomer and was used for the lipid–protein binding assays. (B) PIP strips probed with MBP-Stv1NT-FLAG and MBP. Left, MBP-Stv1NT-FLAG interacts specifically with PI(4)P. Right, MBP alone does not interact with any of the phospholipid head groups. PIP blots shown are representative of at least three different blots probed with protein from at least two independent protein purifications. (C) Schematic diagram describing the liposome flotation assay. (D) Anti-FLAG immunoblot of MBP-Stv1NT-FLAG in fractions obtained from flotation with liposomes containing 5% of the indicated PI lipid or PS as a control. Equal amounts of MBP-Stv1NT-FLAG were mixed with liposomes before flotation. After flotation, protein was precipitated from each fraction and resuspended, and equal volumes were separated by SDS–PAGE and transferred to nitrocellulose. The numbers indicate the fractions from top to bottom of the flotation tube (indicated as 1–6 in C). The third fraction (arrow) represents the position of liposomes after flotation.
Next we tested for Stv1NT-PI lipid interactions in liposomes of defined PI lipid content (Figure 2C). In a liposome flotation assay, liposomes float from high to low density in a sucrose density gradient upon centrifugation because of their relatively low density. Protein, on the other hand, can float only when bound to the liposomes (Busse et al., 2013). We made liposomes as described in Materials and Methods, combined liposomes of varied PI lipid content with equal amounts of protein in the 35% sucrose layer, and assessed coflotation of protein with the liposomes to lower-density fractions after centrifugation. Liposomes were found at the top of the 25% sucrose layer, in fraction 3, after centrifugation. Most of the Stv1NT cofloated with PI(4)P-containing liposomes. Significantly, Stv1NT does not float with control liposomes (containing no PI, but with an equivalent percentage of phosphatidylserine [PS] compensating for the charge; Figure 2D). Little interaction was found with liposomes containing PI(3)P (Figure 2D). Some protein entered the 25% sucrose layer with the PI(3,5)P2 liposomes (fraction 4), but did not end up with the liposomes, suggesting weak binding. In combination, these results indicate that Stv1NT directly and specifically interacts with the Golgi-enriched PI lipid PI(4)P and has, at best, a very weak interaction with the endosomal lipid PI(3)P or vacuolar lipid PI(3,5)P2.
Localization of Stv1-NT is regulated by the Golgi PI4-kinase Pik1 and the PI(4)P-phosphatase Sac1
S. cerevisiae contains three nonredundant kinases that synthesize PI(4)P from phosphatidylinositol, namely, Pik1, Stt4, and Lsb6, all of which have known homologues in mammals (D’Angelo et al., 2008). PIK1 and STT4 are essential for cell survival and encode organelle-specific kinases (Hama et al., 1999; Audhya et al., 2000; Audhya and Emr, 2002). Pik1 maintains PI(4)P levels at the Golgi (Walch-Solimena and Novick, 1999). We hypothesized that recruitment of cytosolic Stv1NT-mCherry to puncta might depend on the Pik1-dependent, Golgi-localized pool of PI(4)P. Cells containing a temperature-sensitive allele of PIK1, pik1-139, fail to grow at 34°C or higher (Sciorra et al., 2005). We used this strain to address whether Stv1NT localization to foci is Pik1 dependent. We found that pik1-139 yeast cells recruited Stv1NT to foci at permissive (25°C) and semipermissive (30°C) temperatures (Figure 3, A and C), but that Stv1NT-mCherry remained cytosolic when the cells were exposed to a restrictive temperature (34°C) (Figure 3, B and C), suggesting that recruitment of Stv1NT from cytosol to membranes requires Golgi PI(4)P.
FIGURE 3:

Localization of Stv1-NT is regulated by the Golgi PI4-kinase Pik1 and the PI(4)P-phosphatase Sac1. (A) Stv1NT-mCherry localization in pik1-139 cells at permissive temperature (25°C). (B) Stv1NT-mCherry localization in pik1-139 cells following by a shift to restrictive temperature (34°C) for 2 h. (C) Percentage of cells exhibiting punctate localization of Stv1NT-mCherry at permissive temperature (n = 284) and restrictive temperature (n = 277) (p < 0.001). (D, E) Localization of Stv1NT-mCherry in wild-type cells (D) and sac1Δ cells (E). (F) Box-and-whisker plot showing normalized median intensity of mCherry signal at puncta in WT (n = 68) cells and sac1Δ (n = 52) cells (p < 0.00005). (G) Percentage of cells with larger puncta in wild-type and sac1∆ cells was determined as described in Materials and Methods (p < 0.05).
We next tested whether recruitment of Stv1NT from cytosol to puncta is increased with elevated intracellular PI(4)P levels. Sac1 is a nonessential PI(4)P phosphatase that converts PI(4)P to PI in both yeast and mammals (Guo et al., 1999). Deletion of SAC1 compromises PI(4)P turnover, resulting in higher intracellular levels of PI(4)P (Roy and Levine, 2004). When Stv1NT-mCherry was expressed in cells lacking Sac1, Stv1NT recruitment to foci increased (Figure 3, D–F). A significant increase in the size of Stv1NT-mCherry foci in sac1Δ cells relative to wild type was also observed (Figure 3G). This suggests that an increase in the levels of PI(4)P can draw more Stv1NT from cytosol to membranes.
Down-regulation of intracellular PI(4)P level mislocalizes and impairs the function of V-ATPases containing full-length Stv1
We hypothesized that PI(4)P might be required to localize V-ATPases assembled with full-length Stv1 to the Golgi. We tagged full-length Stv1 with GFP (Stv1FL-GFP) in the pik1-139 strain. Stv1FL-GFP maintained its Golgi/endosomal localization at a permissive temperature (Figure 4A). However, a portion of the Stv1FL-GFP escaped to the vacuolar membrane, where it colocalized with mCherry-tagged Vph1, after a 2-h exposure to the restrictive temperature (34°C; Figure 4, B and C). This indicates that PI(4)P is required for full retention of Stv1-containing V-ATPases in the Golgi. Next we examined whether the activity of V-ATPases containing Stv1FL was compromised when intracellular PI(4)P levels were reduced. Loss of Stv1 function is only discernible in a vph1Δ background (Manolson et al., 1994), so we constructed a double-mutant strain, vph1Δpik1-139. When grown at semipermissive temperature (30°C), the vph1Δpik1-139 double mutant phenocopied vma or vph1Δstv1Δ mutants by growing much better at pH 5 than at pH 7.5. This indicates that V-ATPases containing the Stv1 isoform may be functionally impaired when intracellular levels of PI(4)P are reduced (Figure 4D).
Stv1-NT requires Lys-84 (K84) residue to bind to PI(4)P
Neither the primary sequence nor structures obtained by homology modeling of Stv1NT indicate any canonical PI binding domain. However, homology models of Stv1NT based on the cryo–electron microscopy (cryo-EM) structure of the assembled yeast V-ATPase containing Vph1 (Zhao et al., 2015) and a crystal structure of the related Meiothermus ruber aNT (Srinivasan et al., 2011) allowed us to model Stv1NT and position it relative to the membrane (Figure 5A). Membrane proteins often bind PI species without canonical PI binding domains, with the help of positively charged residues, sometimes facilitated by nearby aromatic residues (Lemmon, 2008; Baskaran et al., 2012; Lenoir et al., 2015). We sought basic amino acids in the Stv1NT sequence that were 1) predicted to be exposed, 2) conserved among closely related fungal species but not conserved in Vph1NT, and 3) near aromatic amino acid residues. One site in Stv1NT meeting these criteria is shown on a model of the V1-Vo interface in Figure 5A. Mutagenesis of Lys-84 to alanine (Stv1NT(K84A)) yielded an MBP-Stv1NT(K84A)-FLAG fusion protein that behaved similarly to the wild-type protein during purification (Figure 5B). Stv1NT(K84A) did not cofloat with PI(4)P liposomes like wild-type Stv1NT and instead stayed predominantly at the bottom fraction of the sucrose density gradient (Figure 5C). This suggested that there is a significant decrease in binding of the K84A mutant protein to PI(4)P-containing liposomes. We next tested whether the K84A mutation altered recruitment of Stv1NT-mCherry to the puncta in vivo (Figure 5, D and E). Interestingly, Stv1NT(K84A)-mCherry exhibited diffuse cytosolic staining and almost completely lost its ability to recruit to puncta (Figure 5E). The localization of the mutant Stv1NT is similar to wild-type Stv1NT in pik1-139 cells at restrictive temperature (Figure 3, B and C). In combination, these results indicate that Lys-84 is required for Stv1NT binding to PI(4)P both in vitro and in vivo.
FIGURE 5:

Stv1-NT requires Lys-84 (K84) to bind to PI(4)P. (A) Model of the yeast V1–Vo interface based on PDB 3J9T, with Stv1NT substituted for Vph1NT. (Note that Vph1CT was not modeled in 3J9T.) V1 subunits are shown in gray; Vo subunits are shown in cyan. The Stv1NT homology model shown in red was based on Vph1NT in PDB 3J9T. Stv1(K84) is shown as a space-filling model in bright blue, and W83 and Y85 side chains are also colored blue. (B) Coomassie-stained SDS–PAGE of expressed and purified MBP-Stv1NT(K84A)-FLAG (top). Elution pattern of MBP-Stv1NT(K84A)-FLAG from a Sephadex S200 gel-filtration column (bottom). The black arrow indicates elution of high to low molecular weight protein. The monomer fraction used for liposome flotation assay is indicated by an orange arrow. (C) Western blot of fraction 3 (liposome-containing fraction; Figure 2) and fraction 6 (bottom fraction; Figure 2) from flotation of wild-type MBP-Stv1NT-FLAG and MBP-Stv1NT(K84A)-FLAG with PI(4)P-containing liposomes in a liposome flotation assay. (D, E) Localization of Stv1NT-mCherry in cells expressing wild-type Stv1NT-mCherry (D) or Stv1NT(K84A)-mCherry (E). (F) Histogram showing the percentage of cells localizing Stv1NT-mCherry to puncta in cells expressing wild-type Stv1NT-mCherry (orange) or Stv1NT(K84A)-mCherry (purple). Mean (%) ± range from two experiments is presented (p < 0.05). In these two experiments, 77 and 29 cells with wild-type Stv1NT were counted and 188 and 69 mutant cells with mutant Stv1NT were counted.
The K84A mutation causes partial mislocalization of Stv1FL-GFP
We next tested whether the K84A mutation in Stv1 compromises localization and function of V-ATPases containing full-length Stv1 in vivo. We introduced the K84A mutation into full-length Stv1-GFP to make Stv1FL(K84A). It was previously reported that the W83KY sequence in Stv1 is required for its localization to the Golgi (Finnigan et al., 2012). We found that Stv1FL(K84A)-GFP localized to puncta but showed some mislocalization to the vacuole, as judged from partial colocalization with vacuolar FM4-64 (Figure 6, A–C). The proportion of cells with a vacuolar GFP signal increased from 18% to 70% in the presence of K84A mutation. Zn2+ sensitivity is specific to loss of V-ATPase activity at the vacuole (Finnigan et al., 2012). However, Zn2+ sensitivity can be suppressed by overexpression of wild-type STV1, which results in localization of excess Stv1 to the vacuole (Figure 6D), or by stv1 mutations that interfere with Golgi retention or retrieval (Finnigan et al., 2012). When Stv1FL(K84A) was introduced in a vph1∆ mutant, it restored growth of the vph1∆ mutant at high Zn2+ concentrations, without STV1 overexpression (Figure 6D). This result suggested that V-ATPases containing Stv1(K84A) not only are mislocalized to the vacuole but also are functional there. However, a double stv1(K84A)vph1Δ mutant grows more weakly than a vph1Δ single mutant at pH 7.5 (Figure 6D). Growth at alkaline pH requires V-ATPase activity in both the Golgi and the vacuole (Manolson et al., 1994; Finnigan et al., 2011). The results suggest that the K84A mutation compromises Stv1FL function at the Golgi. Thus the K84A mutation in STV1 not only allows a partial escape of Stv1-containing V-ATPases to the vacuole but also impairs or depletes V-ATPase activity at the Golgi, enhancing the growth defect of vph1Δ cells at pH 7.5. These results indicate that Stv1(K84)-dependent binding to PI(4)P is required for efficient retention and function of Stv1-containing V-ATPases at the Golgi.
FIGURE 6:
K84A mutation in Stv1FL causes partial mislocalization of Stv1FL-GFP and impairs function of V-ATPases bearing Stv1. (A) Localization of wild-type Stv1FL-GFP in yeast cells stained with vacuolar dye FM 4-64 (representative of n = 410). (B) Localization of Stv1FL(K84A)-GFP in yeast cells stained with FM 4-64 (n = 204). (C) Histogram representing mean percentage ± SEM of cells with Stv1FL-GFP (WT or K84A) localized to the vacuole (p < 0.0005). (D) Serial-dilution growth assay of congenic WT, vma2∆ (representing a total loss of V-ATPase function), vph1Δ, vph1∆ stv1(K84A), and vph1Δ 2µ-STV1 (overexpressing Stv1) cells on YEPD, pH 5 (left), YEPD + 4 mM Zn2+ (middle), and YEPD, pH 7.5 (right). The growth assay shown is representative of two independent experiments.
The NT domain of human Golgi/endosomal Vo a-subunit isoform a2 interacts with PI(4)P in vitro
We next asked whether PI(4)P recognition is a general property of Golgi-localized Vo a-subunit isoforms. The human a2 isoform is ubiquitously expressed across tissues and is present in Golgi/endosomal compartments of mammalian cells (Hurtado-Lorenzo et al., 2006; Fischer et al., 2012). Loss-of-function mutations in the a2 isoform result in Golgi-specific glycosylation-related disorders in humans, giving rise to rare diseases including autosomal recessive cutis laxa type 2A (Kornak et al., 2008). We tagged, expressed, and purified the human MBP-a2NT-FLAG (Hu a2NT) from E. coli (Figure 7A), as described for Stv1NT. The MBP-a2NT-FLAG construct purified predominantly as a monomer (Figure 7A). We divided the expressed protein equally among liposomes containing different PI species or control PS liposomes, and centrifuged the mixture to float the liposomes as shown in Figure 2. Immunoblots of the floated liposome fraction indicate that Hu a2NT cofloated with liposomes containing PI(4)P in a highly specific manner (Figure 7B). Little if any Hu a2NT floated with liposomes containing PI(3)P or PI(3,5)P2. This indicates that the Golgi/endosomal human isoform of Vo a-subunit, a2, can directly interact with the Golgi-enriched PI lipid PI(4)P, similar to the yeast Golgi isoform Stv1.
FIGURE 7:
The N-terminal domain of human Golgi/endosomal Vo a-subunit isoform, Hu a2, interacts with PI(4)P in vitro. (A) Expression and purification of human a2NT (Hu a2NT) as MBP-Hu a2NT-FLAG from E. coli. Coomassie-stained SDS–PAGE with molecular mass markers (left). S200 elution pattern of MBP-Hu a2NT-FLAG (right). Arrow indicates monomer fraction used for liposome flotation assay. (B) Immunoblot of MBP-Hu a2NT-FLAG protein from the liposome-containing fraction obtained after flotation of equal amounts of protein with liposomes with the same lipid content, except for 5% of PS, PI(3)P, PI(3,5)P2, or PI(4)P, as indicated. Protein was precipitated from each fraction, separated by SDS–PAGE, and detected with anti-FLAG antibody as described in Figure 2.
DISCUSSION
A V-ATPase isoform-specific phosphoinositide code hypothesis
We have shown that the yeast Golgi–resident a-subunit isoform, Stv1, and the human Golgi–associated isoform a2, directly interact with PI(4)P, the PI species enriched in the Golgi (Levine and Munro, 2002). Supporting the physiological significance of this interaction, mutations disrupting the yeast Stv1-PI(4)P interaction or reducing PI(4)P levels compromise localization of Stv1-containing V-ATPases. Previous work has shown that V-ATPases containing the vacuolar isoform of the yeast Vo a-subunit, Vph1, are activated by the vacuolar PI lipid PI(3,5)P2, and Vph1NT can be recruited to membranes in a PI(3,5)P2-dependent manner (Li et al., 2014). In combination, these data strongly suggest that a PI recognition code resides within the cytosolic N-terminal domain of Vo a-subunits that helps them to recognize PI lipids in specific membranes. We hypothesize that this is a direct and physiologically significant interaction that can affect the localization, activity, and/or regulation of V-ATPases in a compartment-specific manner.
Interactions of PI lipids with the cytosolic N-terminal domain of the a-subunit must exert their regulatory effects in the context of assembled V-ATPase and/or the Vo subcomplex, both of which are physiologically relevant complexes (Kane, 1995). V-ATPases are obligate proton pumps that couple ATP hydrolysis in the V1 domain to proton transport through the Vo domain, and the integral membrane CT domain of the Vo a subunit forms part of the proton pore (Forgac, 2007; Zhao et al., 2015). The aNT domain bridges the V1 and Vo complexes, serving a critical role as a stator subunit for rotational catalysis and as an important target for multiple modes of regulation (Kawasaki-Nishi et al., 2001a; Lu et al., 2004; Oot and Wilkens, 2012; Hosokawa et al., 2013). The mammalian a-subunit isoforms have previously been shown to encode localization information (Oka et al., 2001; Toyomura et al., 2003). Experiments with chimeras of the two yeast a-subunit isoforms indicated that the aNT domain contains information for both subcellular localization and regulation of V-ATPase activity by reversible disassembly (Kawasaki-Nishi et al., 2001a). Interactions between a-subunit isoforms and PI lipids may impact both the regulatory and localization aspects of aNT function and help to account for functional differences between a-subunit isoforms.
Recent structures for fully assembled yeast V-ATPase and Vo complexes suggest potential mechanisms for V-ATPase regulation by PI binding (Zhao et al., 2015; Mazhab-Jafari et al., 2016). Interestingly, the Stv1 K84 residue lies in a sequence that was previously implicated in Golgi localization of Stv1-containing V-ATPase complexes (Finnigan et al., 2012). Finnigan and colleagues determined that mutations in the W83KY sequence resulted in escape of Stv1-containing V-ATPases from the Golgi to the vacuole; such an escape could arise from a failure in either Golgi retention or retrieval from endosomal compartments (Bryant and Stevens, 1997; Finnigan et al., 2012). Our results with the Stv1(K84A) mutation also indicate that this mutation results in a partial escape to the vacuole in vivo, consistent with previous results (Finnigan et al., 2012), and furthermore demonstrate that K84 is essential for PI(4)P binding in vitro. Depletion of Golgi PI(4)P in the pik1-139 mutant has similar effects on Stv1 localization, indicating that the PI(4)P binding capacity of Stv1 is important for localization of Stv1-containing V-ATPases at the Golgi. The PI(4)P binding site in Stv1 does not resemble canonical PI(4)P sites, but many membrane proteins have noncanonical PI lipid–binding sites (Hille et al., 2015). PI binding sites frequently consist of basic amino acids capable of interacting with PI head groups, often surrounded by aromatic and hydrophobic amino acids that insert into the membrane (Lemmon, 2008; Baskaran et al., 2012; Lenoir et al., 2015). The Stv1 W83 and Y85 residues were essential for Stv1 localization in experiments by Finnigan et al. (2012), and these residues could reinforce PI(4)P binding initiated by Stv1 K84. Additionally, the ability of Stv1NT to specifically recognize the Golgi pool of PI(4)P over the plasma membrane pool indicates that Stv1NT must be recognizing membrane features beyond the PI head group. Other PI(4)P-binding proteins have been shown to distinguish between PI(4)P pools via secondary binding sites (Roy and Levine, 2004; Luo et al., 2015). Neither our experiments nor those of Finnigan and colleagues, directly distinguish failure of retention from diminished retrieval of Stv1-containing V-ATPases in the mutants. By quantitating the levels of Stv1NT bound to PI(4)P-containing liposomes, versus the “free” Stv1NT remaining at the bottom on the gradient in Figure 2D, we can estimate a Kd in the micromolar range for Stv1NT binding. Current models suggest that, in full-length Stv1, the K84 residue is likely to be in proximity to the membrane (Figure 5A), which would increase the local concentration and the apparent affinity. Unless this region of Stv1NT is somehow masked in the cell, we would therefore predict that full-length Stv1 binds to Golgi PI(4)P tightly and that PI(4)P thus could promote retention in the Golgi or in PI(4)P-containing microdomains of the Golgi (Lenoir et al., 2015). A Stv1-PI(4)P interaction would likely be less important for retrieval from the lower PI(4)P environment of the endosome. However, it is possible that the region around K84 could participate in retrieval from low-PI(4)P compartments, such as endosomes, through other mechanisms.
It has been hypothesized that PI binding to membrane channels and transporters could control their activity in space and/or in time (Hille et al., 2015). PI control of V-ATPases appears to invoke both spatial and temporal regulation. PI(4)P-dependent retention of Stv1-containing V-ATPases in the Golgi is a “spatial” regulation that helps ensure distribution of this population of V-ATPases to the Golgi and optimizes their activity there. In contrast, PI(3,5)P2 regulation of Vph1-containing V-ATPases appears to represent a reversible, time-dependent activation. Consistent with temporal regulation of Vph1-containing V-ATPases, but not Stv1-containing V-ATPases, Stv1NT constitutively binds to PI(4)P-containing membranes when expressed in yeast (Figure 1A), while Vph1NT is reversibly recruited to membranes in parallel with increases in the signaling lipid PI(3,5)P2 (Li et al., 2014). Vacuolar localization of Vo sectors containing full-length Vph1 is retained in mutants lacking PI(3,5)P2 (Li et al., 2014). However, V-ATPase activity and V1-Vo assembly increase under conditions in which PI(3,5)P2 levels reversibly rise, such as salt stress (Bonangelino et al., 2002; Duex et al., 2006; Li et al., 2014). We hypothesize that PI(3,5)P2 exerts a temporal regulation of V-ATPases at the vacuole, in which challenges such as salt stress transiently activate the V-ATPase through PI(3,5)P2 interaction (Bonangelino et al., 2002; Duex et al., 2006; Li et al., 2014). The current yeast V-ATPase structures suggest that there are major conformational differences between Vph1NT in assembled V-ATPases and in free Vo sectors (Zhao et al., 2015; Mazhab-Jafari et al., 2016), as well as movements in Vph1NT during catalytic rotation of the active enzyme (Zhao et al., 2015). By analogy to the stabilization of specific conformations of membrane channels by binding to PI(4,5)P2 (Hansen et al., 2011), it is possible that binding to PI(3,5)P2 transiently traps Vph1NT in a conformation that promotes binding to the V1 sector. We did not directly test the effects of loss of PI(4)P binding on the activity or assembly of Stv1-containing ATPases, but growth assays (Figures 4D and 6D) indicate that their activity is decreased. Significantly, Stv1-containing V-ATPases are much less susceptible to reversible disassembly than Vph1-containing complexes (Kawasaki-Nishi et al., 2001b), so PI lipid effects on V1-Vo interactions may be less pronounced. Nevertheless, it is still possible that V-ATPase activity is affected through other mechanisms, or that escape of complexes from the Golgi in the mutants leaves too few V-ATPases to support proper Golgi acidification. Further experiments are needed to assess these possibilities and to define the full range of compartment-specific PI effects on V-ATPase activity.
Implications of PI lipid interactions for control of organelle acidification
There is substantial evidence indicating that PI lipids can regulate organelle acidification in higher eukaryotes; direct isoform-specific control of V-ATPase activity could support this regulation. Phagosomes in macrophages require a “second wave” of PI(4)P to mature, and this secondary increase in PI(4)P levels is accompanied by phagosome acidification (Levin et al., 2017). PI(4)P-deficient phagosomes fail to acidify through an unknown mechanism (Levin et al., 2017). Although loss of the human a2 isoform has been associated with Golgi-related phenotypes (Kornak et al., 2008), this isoform can also inhabit early endosomes (Hurtado-Lorenzo et al., 2006), suggesting that the a2 isoform could be susceptible to PI(4)P levels in maturing phagosomes. V-ATPase activity has also been linked to vacuolization of podocytes arising from altered PI(3,5)P2 metabolism (Schulze et al., 2017), but the a-subunit isoform present in the vacuoles is unknown. Overall, binding to specific PI lipids on different compartment membranes could impart distinct properties on mammalian V-ATPases containing different a-subunit isoforms. For example, in human cells, the a1 isoform has been found in the late endosomes and lysosomes (Zoncu et al., 2011) and the a2 isoform in the Golgi and early endosomes (Hurtado-Lorenzo et al., 2006; Kornak et al., 2008). This distribution of different isoforms is likely to influence the final pH of these organelles, with lysosomes and late endosomes achieving a lower luminal pH than Golgi and early endosomes, and specific PI binding could reinforce isoform distribution. Conversely, dependence on a PI lipid present in a later compartment of the biosynthetic or endocytic pathway would limit activities of V-ATPases as they transit through earlier steps of the pathway. In general, PI and a-subunit isoform interactions have the potential to bestow distinct activities and regulatory properties upon V-ATPases in different organelles.
Pan V-ATPase inhibitors tend to be cytotoxic, which has limited their usage therapeutically (Huss and Wieczorek, 2009; Toro et al., 2012). Understanding the working principles of subunit isoforms of V-ATPase, including their PI dependence, could pave the way for therapeutic control of V-ATPase subpopulations, which is a goal in treatment of osteoporosis, cancer metastasis, and certain neurodegenerative diseases (Toro et al., 2012; Smith et al., 2016; Bagh et al., 2017). Among the steps needed to achieve this goal are a more complete understanding of the PI specificities of different a-subunit isoforms and the effects of PI binding on localization and activity of V-ATPases containing these isoforms. Our in vitro experiments with the expressed human a2NT represent a step in this direction. Defining binding sites in other a-subunits, as we have for Stv1NT, will facilitate assessment of functional roles of PI binding in vivo. Mechanistic insights from this type of experiment can be expanded by structural studies of conformational changes generated by PI binding. Taken together, these experiments will permit us to read and manipulate the PI codes residing within the V-ATPase.
MATERIALS AND METHODS
Media and cell growth
Rich medium for yeast culture contained 1% yeast extract/2% peptone/2% dextrose (YEPD). YEPD was buffered to pH 5.0 (YEPD, pH 5) or pH 7.5 (YEPD, pH 7.5) using 50 mM potassium phosphate and 50 mM potassium succinate. YEPD + Zn2+ was prepared by adding 4 mM zinc chloride to unbuffered YEPD. Auxotrophic selections were performed on fully supplemented minimal medium with 2% dextrose (SD), from which individual nutrients were removed. For antibiotic selections in yeast, YEPD contained 200 µg/ml G418 (Life Technologies Genticin, Thermo Fisher Scientific) or 100 µg/ml nourseothricin (Jena Bioscience) or 200 µg/ml hygromycin B (Invitrogen).
For dilution growth assays, the indicated wild-type or mutant yeast cells were grown to log phase under suitable conditions. Cell suspensions were adjusted to a single density, and 10-fold serial dilutions were made and then pinned onto the desired plates. The Vma− phenotype was assessed by comparing growth on YEPD, pH 5 and YEPD, pH 7.5 plates (Nelson and Nelson, 1990). YEPD + Zn2+ plates were used to examine a lack or rescue of Vph1 (Finnigan et al., 2012).
Liquid medium for E. coli culture consisted of 2.5% Luria–Bertani broth (LB), Miller powder (Fisher BioReagents), adjusted to pH 7.0. For expression of proteins from E. coli 0.2% dextrose was added to LB. To maintain plasmids carrying an ampicillin- or chloramphenicol-resistance genes, we added 125 µg/ml ampicillin (Sigma-Aldrich) or 34 µg/ml chloramphenicol (Sigma-Aldrich), respectively.
Yeast strains
Strains containing the vph1Δ::kanMX, sac1Δ::kanMX and vac14Δ::kanMX mutations in the BY4714 background (Table 1) were obtained from the MATa haploid yeast deletion library (Winzeler et al., 1999) purchased from Research Genetics/Open Biosystems. pik1-139::kanMX strain was obtained from a library of temperature-sensitive mutants (Li et al., 2011).
TABLE 1:
Yeast strains used in this study.
| Strain | Description | Reference |
|---|---|---|
| BY4742 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 | Brachmann et al., 1998 |
| BY4741 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | |
| vph1Δ::kanMX | BY4741-vph1Δ::KanR | Winzeler et al., 1999 |
| vac14Δ::kanMX | BY4741-vac14Δ::KanR | |
| sac1Δ::kanMX | BY4741-sac1Δ::KanR | |
| vma2Δ::kanMX | BY4741-vma2Δ::KanR | |
| pik1-139 | BY4741-pik1-139::KanR | Sciorra et al., 2005; Li et al., 2011 |
| Stv1NT-mCherry | BY4742-STV1(1-1356 ORF)::mCherry::HygR | This study |
| vph1Δ Stv1NT-mCherry | BY4741-vph1Δ::KanR STV1(1-1356 ORF)::mCherry::HygR | This study |
| vac14Δ Stv1NT-mCherry | BY4741-vac14Δ::KanR STV1(1-1356 ORF)::mCherry::HygR | This study |
| sac1Δ Stv1NT-mCherry | BY4741-sac1Δ::KanR STV1(1-1356 ORF)::mCherry::HygR | This study |
| pik1-139 Stv1NT-mCherry | BY4741-pik1-139::KanR STV1(1-1356 ORF)::mCherry::HygR | This study |
| pik1-139 Stv1FL-GFP Vph1-mCherry | BY4741-pik1-139::KanR STV1::GFP::HIS3MX6 VPH1::mCherry::HygR | This study |
| vph1Δpik1-139 | BY4741-pik1-139::KanR vph1Δ::NatR | This study |
| Stv1NT(K84A)-mCherry | BY4742-STV1(1-1356 ORF; K84A)::mCherry::HygR | This study |
| EY0986 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 (S288C) | Huh et al., 2003 |
| Stv1FL-GFP | EY0986-STV1::GFP::HIS3MX6 | |
| Stv1FL(K84A)-GFP | EY0986-STV1(K84A)::GFP::HIS3MX6 | This study |
| STV1(K84A) | BY4741-STV1(K84A) | This study |
| vph1ΔSTV1(K84A) | BY4741-STV1(K84A) vph1Δ::KanR | This study |
Amino acids 453–890 of Stv1 were replaced by mCherry to yield genomic expression of Stv1NT(1-452)-mCherry fusion from the STV1 promoter in yeast. A linear DNA fragment was PCR amplified from plasmid pBS35 (containing mCherry-hygromycin) (Hailey et al., 2002) using the oligonucleotides Stv1-50F2 and Stv1-50R1 (Table 2), which had added flanking sequences of 50 base pairs homologous to sequences at the end of the Stv1NT and 3′ end of STV1 (Longtine et al., 1998). vph1Δ Stv1NT-mCherry, vac14Δ Stv1NT-mCherry, sac1Δ Stv1NT-mCherry, and pik1-139 Stv1NT-mCherry strains were generated similarly (Table 1). Full-length Vph1 was tagged with mCherry in the same manner using oligonucleotides Vph1FL-F2 and Vph1FL-R1 (Table 2). BY4742 Stv1FL-GFP (Stv1-GFP::HIS3) was purchased from Invitrogen (now Thermo Scientific). A linear DNA fragment was amplified from the genomic DNA of Stv1FL-GFP yeast strain (Table 1) using oligonucleotides STV1 2391ORF and STV1+500 (Table 2). The product was transformed into pik1-139 Vph1-mCherry cells to obtain pik1-139 Stv1FL-GFP Vph1-mCherry (Table 1).
TABLE 2:
Oligonucleotides used in this study.
| Primer name | Sequence (5′→3′) |
|---|---|
| Stv1-50F2 | GGCCTTCCAATCGATTGTGGATGCATACGGTATCGCAACATATAAAGAAATCAATCGGATCCCCGGGTTAATTAA |
| Stv1-50R1 | GGACATTGCCTCCACCCAATGCAAACGTAGCGCATGCAACATTGCGGAAGGAATTCGAGCTCGTTTAAAC |
| Vph1FL-F2 | GAGTATAAAGACATGGAAGTCGCTGTTGCTAGTGCAAGCTCTTCCGCTTCAAGCCGGATCCCCGGGTTAATTAA |
| Vph1FL-R1 | CTCACTATATAGTAGCATCATTTATTATTTAATGAAGTACTTAAATGTTTCGCGAATTCGAGCTCGTTTAAAC |
| STV1-URA-UP | CCAGTTGAGGCGTTTCGATGAAGTGGAAAGGATGGTAGGCTTCTTGAATGAGGTACTGAGAGTGCACCACGCT |
| STV1-URA-DOWN | GGATCAATTCTGTGGAAGGCACCCAACCTTCGGCTATTAGACCCTGTGATCAGTTTTTTAGTTTTGCTGGCC |
| STV1 2391ORF | GGCACTATCGTTGGCGCATG |
| STV1+500 | TACAGCAGAGATTTATGGTATGCC |
| Vph1-1 | GAGGTATTTAGAAGTGAAGAGAC |
| Vph1-C4 | AACGTTTCATGAGATAAGTTTGGC |
| Stv1NT-FLAG-1 | GCGGATCCATGAATCAAGAAGAGGCTATATTC |
| Stv1NT-FLAG-2 | GCGTGCAGTTACTTGTCATCGTCATCCTTATAGTCTGTTGCGATACGCTATGCATC |
| Stv1-418a | GTCATCGTCATCCTTATAGTCTAGGATCACATTAAAGACGGT |
| Stv1-418b | ACCGTCTTTAATGTGATCCTAGACTATAAGGATGACGATGAC |
| a2_1-364(KpnI)F | TTAGCCGGTACCAATGGGGTCCCTGTTCCG |
| a2_1-364(HindIII)R | TTACCAAAGCTTCTATTCTTTTGTGGGGATTATATTCATGAATGAGGGGATTG |
| Hua2-FLAG-For | AAGGACGACGATGACAAGTAGAAGCTTGGCACTGGCCGTCGT |
| Hua2-FLAG-Rev | GTCATCGTCGTCCTTGTAATCTTCTTTTGTGGGGATTATATTCATG |
The K84A mutation was introduced into cells expressing Stv1NT-mCherry, Stv1FL-GFP, and vph1Δ::kanMX strains (Table 1) using positive and negative selection for URA3. Specifically, a URA3 fragment with ends homologous to Stv1NT was obtained by PCR amplification from a pRS316 template (Kitazono, 2011) using oligonucleotides STV1-URA-UP and STV1-URA-DOWN (Table 2). This fragment was used to transform BY4742 Stv1NT-mCherry cells, and transformants were selected on SD lacking uracil, resulting in a strain in which nucleotides 221–1130 of STV1 were replaced with the URA3 gene. A fragment containing Stv1(K84A) was then PCR amplified from an expression plasmid carrying the K84A mutation in Stv1-NT using the oligonucleotides Stv1-mut-UP and Stv1-mut-DOWN (Table 2). Transformants capable of growth in the presence of 5-fluoro-orotic acid were selected, analyzed for replacement of the URA3 gene, and sequenced to confirm introduction of the mutation (Storici et al., 2001).
To obtain the vph1Δpik1-139 double mutant (Table 1), the vph1Δ::NatR allele was PCR amplified from the corresponding vph1∆ strain using oligonucleotides Vph1-1 and Vph1-C4 (Table 2) and transformed into pik1-139 cells. A 2-micron high copy number plasmid (YEp352 vector) expressing hemagglutinin-tagged Stv1 (2µSTV1) (Kawasaki-Nishi et al., 2001b) was used to overexpress Stv1 in vph1Δ strain.
Cloning, expression, and purification of Stv1NT and Hu a2NT from E. coli
A sequence corresponding to aa 1–448 of Stv1NT was PCR amplified from yeast genomic DNA with oligonucleotides Stv1NT-FLAG-1 and Stv1NT-FLAG-2 (Table 2) and cloned into a pMal-C2E-Enterokinase vector using the restriction enzymes BamHI and PstI. The MBP-Stv1NT-FLAG fusion construct used for expression contained aa 1–418 and was created by the deletion of 30 amino acids from the aforementioned construct using the oligonucleotides Stv1-418a and Stv1-418b (Table 2). Stv1NT (1-418) was then cloned into the BamHI and PstI sites of the pMal-C2E-Precission protease (pMal-PPase) vector, which places a Precission protease site between MBP and Stv1NT.
The coding sequence for human a2 isoform (ATP6V0A2) was obtained in pLDNT7_nFLAG vector (Center for Personalized Diagnostics). The NT of human a2 (Hu a2NT) cloned into the pMal:PPase vector was a kind gift from the laboratory of Stephan Wilkens (SUNY Upstate Medical University). Hu a2NT was amplified using the oligonucleotides a2_1-364(KpnI)F and a2_1-364(HindIII)R (Table 2), and introduced into the KpnI and HindIII sites of pMal:PPase vector by restriction digest and ligation. A FLAG tag was added at the C-terminus of Hu a2NT using the oligonucleotides Hua2-FLAG-For and Hua2-FLAG-Rev (Table 2) for expression of the MBP-Hu a2NT-FLAG fusion protein.
Rosetta (DE3) competent cells (Novagen), (genotype: F− ompT hsdSB(rB− mB−) gal dcm (DE3) pRARE (CamR)) were used to express both Stv1NT and Hu a2NT. Amylose resin was purchased from New England BioLabs (NEB). The E. coli cells were freshly transformed and grown to an A600 of 0.6 in rich broth containing ampicillin and chloramphenicol. Expression was induced by addition of 0.3 mM isopropyl β-d-1-thiogalactopyranoside followed by a 16-h incubation at 19°C. Cells were pelleted and frozen in 25 ml of amylose column buffer (ACB; 20 mM Tris-HCl, pH 7.4, 0.2 M NaCl, and 1 mM EDTA) per liter of bacterial culture. Pellets were thawed at 4°C; 1 mM lysozyme, 2 mM MgCl2, 20 mM DNase I, and 10 mM dithiothreitol (DTT) were added; and the mixture was rocked for 30 min at 4°C. Just before cell lysis, 1 mM phenylmethylsulfonyl fluoride (PMSF) was added, followed by sonication on ice. The lysate was centrifuged at 17,640 × g for 30 min at 4°C, and the collected supernatant was diluted by adding 3 ml of ACB containing 10 mM DTT per 1 ml of the supernatant. The first round of purification was done using a 5 ml amylose column and included a wash with 10 column volumes of ACB and elution with 10 mM maltose, 5 mM DTT in ACB. Fractions containing protein (A280 > 0.5) was pooled together and dialyzed overnight at 4°C against FLAG buffer (50 mM Tris-HCl, pH 7.4, and 150 mM NaCl). A second round of affinity purification was performed using anti-FLAG M2 Affinity Gel (Sigma). Peak fractions were pooled, and 10 mM DTT was added immediately. After being concentrated to at least 1 mg/ml, the protein solution was run on a Sephadex 200 (S200) gel-filtration fast protein liquid chromatography (FPLC) column on an AKTA FPLC system. The peak corresponding to the monomeric molecular mass was collected and used for additional experiments.
PIP blot
PIP strip membranes (Life Technologies) were blocked in blocking buffer (3% fat-free bovine serum albumin [Sigma] in TBS [20 mM Tris-HCl, pH 7.5, 500 mM NaCl]) for 1 h at room temperature with mild agitation. Blocked PIP strips were then incubated in 0.4 µM MBP-Stv1NT-FLAG in blocking buffer for 1 h. Bound protein was detected by incubation with mouse monoclonal anti-FLAG M2 antibody (Sigma) followed by horseradish peroxide–conjugated anti-mouse antibody (Bio-Rad). All incubations were for 1 h at room temperature and were followed by three TBS washes for 10 min. Finally, the strip was treated with a chemiluminescent substrate (Thermo Scientific) for 5 min and developed by exposure to film for 5–20 s. The control MBP strip was probed with mouse monoclonal anti-MBP antibody (NEB).
Liposome preparation and flotation assay
Lipids were obtained in solution or as lyophilized powder from Avanti Polar Lipids. Powdered lipids were dissolved in CH3OH:CHCl3:H2O at a 9:20:1 ratio. Liposomes were composed of the stipulated concentrations of the following (x:y) phospholipids, where x indicates the length of carbon chain and y indicates the number of unsaturated (C=C) linkages. Liposomes contained 55% (mol %) 16:0 phosphatidylcholine (PC), 25% 16:0 PS, 18% 16:0 phosphatidylethnolamine (PE), and 5% 18:1 phosphatidylinositol phosphates (PI(4)P, PI(3)P, or PI(3,5)P2) and 2% 16:0 NBD-PE (for visualization of liposomes). Control liposomes had no PIPs but had an additional 5% of 16:0 PS. The mixed lipids were dried using a centrivap lyophilizer with a vacuum pump at 35°C for 30–40 min. The dried lipid film was rehydrated at ice-cold temperature using liposome-making buffer (25 mM NaCl, pH 7.4, 50 mM Tris-HCl), then transferred to 55°C for 20 min, which was followed by five freeze (dry ice)-thaw (42°C water bath) cycles. Uniform-sized liposomes were prepared by extruding at least 20 times through a 100-nm filter in a mini-extruder (Avanti Polar Lipids).
Protein was mixed with 100 µl of control (0% PIP) or experimental (5% PIP) liposomes in 100 mM NaCl (200 mM for MBP-Hu a2NT-FLAG), 25 mM Tris-HCl (pH 7.4) and incubated at 30°C for 5 min. Sucrose was added to a final concentration of 35% (wt/vol) and a final volume of 300 µl. Final concentration of protein was 0.3 µM, and final total lipid concentration was 0.33 mM. This was topped with 300 µl steps of 25% and 0% sucrose containing no protein. The sucrose density gradient was centrifuged for 90 min at 55,000 rpm at 20°C in a TLS-55 swinging bucket rotor using a Beckman Coulter mini-ultracentrifuge.
Six equivolume fractions were collected from top to bottom of the centrifuge tubes (corresponding to fractions 1–6 in Figure 2, C and D) using a Hamilton syringe. The colored liposomes floated to the junction of 0% and 25% sucrose and were collected in the third fraction. Protein from the collected fractions was precipitated using 10% trichloroacetic acid. The pellet was washed with cold acetone and resuspended in 50 µl of cracking buffer (50 mM Tris-HCl, pH 6.8, 8M urea, 5% SDS, and 1 mM EDTA). Equal volumes of each fraction were separated by SDS–PAGE. Protein was transferred overnight to a nitrocellulose membrane and immunoblotted using a mouse monoclonal anti-FLAG antibody. Liposome flotations shown in Figures 2, 5, and 7 are representative of at least three independent flotations conducted with protein from at least two separate protein purifications.
Fluorescence microscopy and image processing
Cells were grown overnight at 30°C in fully supplemented minimal medium to log phase. Cells expressing Stv1NT-mCherry were quick fixed using 4% formaldehyde for 1 h at 30°C (25°C for temperature-sensitive strains) before imaging. Images for Stv1-FL-GFP were acquired from live cells. Images were taken using a Zeiss Axio fluorescence microscope attached to a Hamamatsu EMCCD camera. For each different tagged protein, images were obtained at the same exposure time and processed in parallel using ImageJ (National Institutes of Health) and Adobe Photoshop Elements 14. All image quantification was done using ImageJ.
pik1-139 cells expressing Stv1NT-mCherry or Stv1FL-GFP were grown overnight at 25°C in fully supplemented minimal medium and shifted to 25°C (permissive) or 34°C (restrictive) at log phase. Pictures of live or quick-fixed cells were acquired and processed as described earlier. Additionally, cells were categorized and quantified for whether they formed puncta and were represented as percentage of cells forming puncta. Similar quantification was performed to compare cells expressing WT and K84A Stv1NT-mCherry. sac1Δ cells expressing Stv1NT-mCherry were quantified for localized intensity of mCherry in the puncta by normalized maximum fluorescence. A standard 0.5 µm × 0.5 µm punctum in a cell expressing wild-type Sac1 was used to compare puncta size in both sac1Δ and wild-type-sac1 cells. Cells containing one or more visibly bigger puncta were considered as a unit and represented as percentage of cells containing a bigger puncta.
Vph1-mCherry (Finnigan et al., 2011) was used to identify vacuoles in the pik1-139 cells. A Pearson correlation coefficient was calculated for mislocalization of Stv1FL-GFP to the vacuoles stained by Vph1-mCherry in pik1-139 cells. In Stv1(K84A)FL-GFP or WT-Stv1-GFP cells, vacuoles were stained with an 8 µM pulse of FM 4-64 for 30 min (Jin et al., 2008), followed by a wash with YEPD, pH 5 and a 90-min chase, all at 30°C. Cells were spun down and resuspended in fully supplemented minimal medium before being imaged. The extent of vacuolar mislocalization was determined by counting the number of cells with Stv1FL-GFP only in puncta versus the number of cells containing both puncta and vacuolar GFP staining overlapping with FM4-64.
Protein structure prediction
The homology model of Stv1NT (1-448) was obtained by submission of the sequence to the Phyre 2.0 suite of software (Kelley et al., 2015). A 100% confidence model covering 73% of the submitted sequence was obtained based on Vph1NT in the state 1 rotational structure of the yeast V-ATPase (PDB 3j9t) obtained by cryo-EM (Zhao et al., 2015). (Vph1NT had been modeled into the cryo-EM structure based on the crystal structure of the cytosolic N-terminal domain of a homologous subunit in the M. ruber V-ATPase [PDB 3rrk; Srinivasan et al., 2011].) The Stv1NT model was substituted for Vph1NT in the corresponding V-ATPase structure using UCSF Chimera (Pettersen et al., 2004).
Statistical analysis
Data presented as mean ± SEM were from independent averages of biological replicates. Error bars represent SEM of averages from individual experiments. The normalized fluorescence intensity of Stv1NT-mCherry at a puncta for WT and sac1Δ cells has been represented as a box-and-whisker plot. The middle line represents a median value, the box limits denote first and third quartiles, and the ends of the whiskers represent maximum and minimum values. These data are drawn from a combined data set of two biological replicates. An unpaired t test was performed to examine whether two mean values were significantly different by using a criterion of p < 0.05. p values are included in the figures: *, p < 0.05; **, p < 0.005; ***, p < 0.0005.
Acknowledgments
This work was supported by National Institutes of Health R01 GM50322 to P.M.K. We thank Maureen Tarsio, Thomas Benz, and Cameron Smith for excellent technical assistance and Stephan Wilkens and the Wilkens lab for sharing the human a2 expression plasmid, for use of the FPLC, and for helpful discussions.
Abbreviations used:
- ACB
amylose column buffer
- DTT
dithiothreitol
- EM
electron microscopy
- FPLC
fast protein liquid chromatography
- GFP
green fluorescent protein
- MBP
maltose-binding protein
- PC
phosphatidylcholine
- PE
phosphatidylethnolamine
- PI
phosphatidylinositol
- PS
phosphatidylserine
- SD
supplemented minimal medium with 2% dextrose
- V-ATPase
vacuolar H+-ATPase
- YEPD
1% yeast extract/2% peptone/2% dextrose.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E17-05-0316) on July 18, 2017.
REFERENCES
- Audhya A, Emr SD. Stt4 PI 4-kinase localizes to the plasma membrane and functions in the Pkc1-mediated MAP kinase cascade. Dev Cell. 2002;2:593–605. doi: 10.1016/s1534-5807(02)00168-5. [DOI] [PubMed] [Google Scholar]
- Audhya A, Foti M, Emr SD. Distinct roles for the yeast phosphatidylinositol 4-kinases, Stt4p and Pik1p, in secretion, cell growth, and organelle membrane dynamics. Mol Biol Cell. 2000;11:2673–2689. doi: 10.1091/mbc.11.8.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagh MB, Peng S, Chandra G, Zhang Z, Singh SP, Pattabiraman N, Liu A, Mukherjee AB. Misrouting of v-ATPase subunit V0a1 dysregulates lysosomal acidification in a neurodegenerative lysosomal storage disease model. Nat Commun. 2017;8:14612. doi: 10.1038/ncomms14612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balla T. Phosphoinositides: tiny lipids with giant impact on cell regulation. Physiol Rev. 2013;93:1019–1137. doi: 10.1152/physrev.00028.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskaran S, Ragusa MJ, Boura E, Hurley JH. Two-site recognition of phosphatidylinositol 3-phosphate by PROPPINs in autophagy. Mol Cell. 2012;47:339–348. doi: 10.1016/j.molcel.2012.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonangelino CJ, Nau JJ, Duex JE, Brinkman M, Wurmser AE, Gary JD, Emr SD, Weisman LS. Osmotic stress-induced increase of phosphatidylinositol 3,5-bisphosphate requires Vac14p, an activator of the lipid kinase Fab1p. J Cell Biol. 2002;156:1015–1028. doi: 10.1083/jcb.200201002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast (Chichester, England) 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Brett CL, Kallay L, Hua Z, Green R, Chyou A, Zhang Y, Graham TR, Donowitz M, Rao R. Genome-wide analysis reveals the vacuolar pH-stat of Saccharomyces cerevisiae. PLoS One. 2011;6:e17619. doi: 10.1371/journal.pone.0017619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant NJ, Stevens TH. Two separate signals act independently to localize a yeast late Golgi membrane protein through a combination of retrieval and retention. J Cell Biol. 1997;136:287–297. doi: 10.1083/jcb.136.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busse RA, Scacioc A, Hernandez JM, Krick R, Stephan M, Janshoff A, Thumm M, Kuhnel K. Qualitative and quantitative characterization of protein-phosphoinositide interactions with liposome-based methods. Autophagy. 2013;9:770–777. doi: 10.4161/auto.23978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey JR, Grinstein S, Orlowski J. Sensors and regulators of intracellular pH. Nat Rev Mol Cell Biol. 2010;11:50–61. doi: 10.1038/nrm2820. [DOI] [PubMed] [Google Scholar]
- Compton LM, Ikonomov OC, Sbrissa D, Garg P, Shisheva A. Active vacuolar H+ ATPase and functional cycle of Rab5 are required for the vacuolation defect triggered by PtdIns(3,5)P2 loss under PIKfyve or Vps34 deficiency. Am J Physiol Cell Physiol. 2016;311:C366–C377. doi: 10.1152/ajpcell.00104.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couoh-Cardel S, Milgrom E, Wilkens S. Affinity purification and structural features of the yeast vacuolar ATPase Vo membrane sector. J Biol Chem. 2015;290:27959–27971. doi: 10.1074/jbc.M115.662494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Angelo G, Vicinanza M, Di Campli A, De Matteis MA. The multiple roles of PtdIns(4)P—not just the precursor of PtdIns(4,5)P2. J Cell Sci. 2008;121:1955–1963. doi: 10.1242/jcs.023630. [DOI] [PubMed] [Google Scholar]
- Diakov TT, Tarsio M, Kane PM. Measurement of vacuolar and cytosolic pH in vivo in yeast cell suspensions. J Vis Exp. 2013;2013:50261. doi: 10.3791/50261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dove SK, McEwen RK, Mayes A, Hughes DC, Beggs JD, Michell RH. Vac14 controls PtdIns(3,5)P(2) synthesis and Fab1-dependent protein trafficking to the multivesicular body. Curr Biol. 2002;12:885–893. doi: 10.1016/s0960-9822(02)00891-6. [DOI] [PubMed] [Google Scholar]
- Duex JE, Nau JJ, Kauffman EJ, Weisman LS. Phosphoinositide 5-phosphatase Fig 4p is required for both acute rise and subsequent fall in stress-induced phosphatidylinositol 3,5-bisphosphate levels. Eukaryot Cell. 2006;5:723–731. doi: 10.1128/EC.5.4.723-731.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnigan GC, Cronan GE, Park HJ, Srinivasan S, Quiocho FA, Stevens TH. Sorting of the yeast vacuolar-type, proton-translocating ATPase enzyme complex (V-ATPase): identification of a necessary and sufficient Golgi/endosomal retention signal in Stv1p. J Biol Chem. 2012;287:19487–19500. doi: 10.1074/jbc.M112.343814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnigan GC, Hanson-Smith V, Houser BD, Park HJ, Stevens TH. The reconstructed ancestral subunit a functions as both V-ATPase isoforms Vph1p and Stv1p in Saccharomyces cerevisiae. Mol Biol Cell. 2011;22:3176–3191. doi: 10.1091/mbc.E11-03-0244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer B, Dimopoulou A, Egerer J, Gardeitchik T, Kidd A, Jost D, Kayserili H, Alanay Y, Tantcheva-Poor I, Mangold E, et al. Further characterization of ATP6V0A2-related autosomal recessive cutis laxa. Hum Genet. 2012;131:1761–1773. doi: 10.1007/s00439-012-1197-8. [DOI] [PubMed] [Google Scholar]
- Forgac M. Vacuolar ATPases: rotary proton pumps in physiology and pathophysiology. Nat Rev Mol Cell Biol. 2007;8:917–929. doi: 10.1038/nrm2272. [DOI] [PubMed] [Google Scholar]
- Frattini A, Orchard PJ, Sobacchi C, Giliani S, Abinun M, Mattsson JP, Keeling DJ, Andersson AK, Wallbrandt P, Zecca L, et al. Defects in TCIRG1 subunit of the vacuolar proton pump are responsible for a subset of human autosomal recessive osteopetrosis. Nat Genet. 2000;25:343–346. doi: 10.1038/77131. [DOI] [PubMed] [Google Scholar]
- Gillooly DJ, Morrow IC, Lindsay M, Gould R, Bryant NJ, Gaullier JM, Parton RG, Stenmark H. Localization of phosphatidylinositol 3-phosphate in yeast and mammalian cells. EMBO J. 2000;19:4577–4588. doi: 10.1093/emboj/19.17.4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham LA, Hill KJ, Stevens TH. Assembly of the yeast vacuolar H+-ATPase occurs in the endoplasmic reticulum and requires a Vma12p/Vma22p assembly complex. J Cell Biol. 1998;142:39–49. doi: 10.1083/jcb.142.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S, Stolz LE, Lemrow SM, York JD. SAC1-like domains of yeast SAC1, INP52, and INP53 and of human synaptojanin encode polyphosphoinositide phosphatases. J Biol Chem. 1999;274:12990–12995. doi: 10.1074/jbc.274.19.12990. [DOI] [PubMed] [Google Scholar]
- Hailey DW, Davis TN, Muller EG. Fluorescence resonance energy transfer using color variants of green fluorescent protein. Methods Enzymol. 2002;351:34–49. doi: 10.1016/s0076-6879(02)51840-1. [DOI] [PubMed] [Google Scholar]
- Hama H, Schnieders EA, Thorner J, Takemoto JY, DeWald DB. Direct involvement of phosphatidylinositol 4-phosphate in secretion in the yeast Saccharomyces cerevisiae. J Biol Chem. 1999;274:34294–34300. doi: 10.1074/jbc.274.48.34294. [DOI] [PubMed] [Google Scholar]
- Hansen SB, Tao X, MacKinnon R. Structural basis of PIP2 activation of the classical inward rectifier K+ channel Kir2.2. Nature. 2011;477:495–498. doi: 10.1038/nature10370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilgemann DW. Biochemistry. Oily barbarians breach ion channel gates. Science. 2004;304:223–224. doi: 10.1126/science.1097439. [DOI] [PubMed] [Google Scholar]
- Hille B, Dickson EJ, Kruse M, Vivas O, Suh BC. Phosphoinositides regulate ion channels. Biochim Biophys Acta. 2015;1851:844–856. doi: 10.1016/j.bbalip.2014.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho CY, Alghamdi TA, Botelho RJ. Phosphatidylinositol-3,5-bisphosphate: no longer the poor PIP2. Traffic. 2012;13:1–8. doi: 10.1111/j.1600-0854.2011.01246.x. [DOI] [PubMed] [Google Scholar]
- Hosokawa H, Dip PV, Merkulova M, Bakulina A, Zhuang Z, Khatri A, Jian X, Keating SM, Bueler SA, Rubinstein JL, et al. The N termini of a-subunit isoforms are involved in signaling between vacuolar H+-ATPase (V-ATPase) and cytohesin-2. J Biol Chem. 2013;288:5896–5913. doi: 10.1074/jbc.M112.409169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O’Shea EK. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- Hurtado-Lorenzo A, Skinner M, El Annan J, Futai M, Sun-Wada GH, Bourgoin S, Casanova J, Wildeman A, Bechoua S, Ausiello DA, et al. V-ATPase interacts with ARNO and Arf6 in early endosomes and regulates the protein degradative pathway. Nat Cell Biol. 2006;8:124–136. doi: 10.1038/ncb1348. [DOI] [PubMed] [Google Scholar]
- Huss M, Wieczorek H. Inhibitors of V-ATPases: old and new players. J Exp Biol. 2009;212:341–346. doi: 10.1242/jeb.024067. [DOI] [PubMed] [Google Scholar]
- Idevall-Hagren O, De Camilli P. Detection and manipulation of phosphoinositides. Biochim Biophys Acta. 2015;1851:736–745. doi: 10.1016/j.bbalip.2014.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin N, Chow CY, Liu L, Zolov SN, Bronson R, Davisson M, Petersen JL, Zhang Y, Park S, Duex JE, et al. VAC14 nucleates a protein complex essential for the acute interconversion of PI3P and PI(3,5)P(2) in yeast and mouse. EMBO J. 2008;27:3221–3234. doi: 10.1038/emboj.2008.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane PM. Disassembly and reassembly of the yeast vacuolar H+-ATPase in vivo. J Biol Chem. 1995;270:17025–17032. [PubMed] [Google Scholar]
- Kane PM. The where, when, and how of organelle acidification by the yeast vacuolar H+-ATPase. Microbiol Mol Biol Rev. 2006;70:177–191. doi: 10.1128/MMBR.70.1.177-191.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki-Nishi S, Bowers K, Nishi T, Forgac M, Stevens TH. The amino-terminal domain of the vacuolar proton-translocating ATPase a subunit controls targeting and in vivo dissociation, and the carboxyl-terminal domain affects coupling of proton transport and ATP hydrolysis. J Biol Chem. 2001a;276:47411–47420. doi: 10.1074/jbc.M108310200. [DOI] [PubMed] [Google Scholar]
- Kawasaki-Nishi S, Nishi T, Forgac M. Yeast V-ATPase complexes containing different isoforms of the 100-kDa a-subunit differ in coupling efficiency and in vivo dissociation. J Biol Chem. 2001b;276:17941–17948. doi: 10.1074/jbc.M010790200. [DOI] [PubMed] [Google Scholar]
- Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. The Phyre2 Web portal for protein modeling, prediction and analysis. Nat Protoc. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitazono AA. Optimized protocols and plasmids for in vivo cloning in yeast. Gene. 2011;484:86–89. doi: 10.1016/j.gene.2011.06.006. [DOI] [PubMed] [Google Scholar]
- Kornak U, Reynders E, Dimopoulou A, van Reeuwijk J, Fischer B, Rajab A, Budde B, Nurnberg P, Foulquier F, Lefeber D, et al. Impaired glycosylation and cutis laxa caused by mutations in the vesicular H+-ATPase subunit ATP6V0A2. Nat Genet. 2008;40:32–34. doi: 10.1038/ng.2007.45. [DOI] [PubMed] [Google Scholar]
- Lemmon MA. Membrane recognition by phospholipid-binding domains. Nat Rev Mol Cell Biol. 2008;9:99–111. doi: 10.1038/nrm2328. [DOI] [PubMed] [Google Scholar]
- Leng XH, Manolson MF, Forgac M. Function of the COOH-terminal domain of Vph1p in activity and assembly of the yeast V-ATPase. J Biol Chem. 1998;273:6717–6723. doi: 10.1074/jbc.273.12.6717. [DOI] [PubMed] [Google Scholar]
- Lenoir M, Grzybek M, Majkowski M, Rajesh S, Kaur J, Whittaker SB, Coskun U, Overduin M. Structural basis of dynamic membrane recognition by trans-Golgi network specific FAPP proteins. J Mol Biol. 2015;427:966–981. doi: 10.1016/j.jmb.2014.12.023. [DOI] [PubMed] [Google Scholar]
- Levin R, Hammond GR, Balla T, De Camilli P, Fairn GD, Grinstein S. Multiphasic dynamics of phosphatidylinositol 4-phosphate during phagocytosis. Mol Biol Cell. 2017;28:128–140. doi: 10.1091/mbc.E16-06-0451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine TP, Munro S. Targeting of Golgi-specific pleckstrin homology domains involves both PtdIns 4-kinase-dependent and -independent components. Curr Biol. 2002;12:695–704. doi: 10.1016/s0960-9822(02)00779-0. [DOI] [PubMed] [Google Scholar]
- Li SC, Diakov TT, Xu T, Tarsio M, Zhu W, Couoh-Cardel S, Weisman LS, Kane PM. The signaling lipid PI(3,5)P(2) stabilizes V(1)-V(o) sector interactions and activates the V-ATPase. Mol Biol Cell. 2014;25:1251–1262. doi: 10.1091/mbc.E13-10-0563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Vizeacoumar FJ, Bahr S, Li J, Warringer J, Vizeacoumar FS, Min R, Vandersluis B, Bellay J, Devit M, et al. Systematic exploration of essential yeast gene function with temperature-sensitive mutants. Nat Biotechnol. 2011;29:361–367. doi: 10.1038/nbt.1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Lu M, Sautin YY, Holliday LS, Gluck SL. The glycolytic enzyme aldolase mediates assembly, expression, and activity of vacuolar H+-ATPase. J Biol Chem. 2004;279:8732–8739. doi: 10.1074/jbc.M303871200. [DOI] [PubMed] [Google Scholar]
- Luo X, Wasilko DJ, Liu Y, Sun J, Wu X, Luo ZQ, Mao Y. Structure of the Legionella virulence factor, SidC reveals a unique PI(4)P-specific binding domain essential for its targeting to the bacterial phagosome. PLoS Pathog. 2015;11:e1004965. doi: 10.1371/journal.ppat.1004965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolson MF, Proteau D, Preston RA, Stenbit A, Roberts BT, Hoyt MA, Preuss D, Mulholland J, Botstein D, Jones EW. The VPH1 gene encodes a 95-kDa integral membrane polypeptide required for in vivo assembly and activity of the yeast vacuolar H+-ATPase. J Biol Chem. 1992;267:14294–14303. [PubMed] [Google Scholar]
- Manolson MF, Wu B, Proteau D, Taillon BE, Roberts BT, Hoyt MA, Jones EW. STV1 gene encodes functional homologue of 95-kDa yeast vacuolar H+-ATPase subunit Vph1p. J Biol Chem. 1994;269:14064–14074. [PubMed] [Google Scholar]
- Mazhab-Jafari MT, Rohou A, Schmidt C, Bueler SA, Benlekbir S, Robinson CV, Rubinstein JL. Atomic model for the membrane-embedded VO motor of a eukaryotic V-ATPase. Nature. 2016;539:118–122. doi: 10.1038/nature19828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson H, Nelson N. Disruption of genes encoding subunits of yeast vacuolar H+-ATPase causes conditional lethality. Proc Natl Acad Sci USA. 1990;87:3503–3507. doi: 10.1073/pnas.87.9.3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson N. A journey from mammals to yeast with vacuolar H+-ATPase (V-ATPase) J Bioenerg Biomembr. 2003;35:281–289. doi: 10.1023/a:1025768529677. [DOI] [PubMed] [Google Scholar]
- Ohya Y, Umemoto N, Tanida I, Ohta A, Iida H, Anraku Y. Calcium-sensitive cls mutants of Saccharomyces cerevisiae showing a Pet− phenotype are ascribable to defects of vacuolar membrane H+-ATPase activity. J Biol Chem. 1991;266:13971–13977. [PubMed] [Google Scholar]
- Oka T, Murata Y, Namba M, Yoshimizu T, Toyomura T, Yamamoto A, Sun-Wada GH, Hamasaki N, Wada Y, Futai M. a4, a unique kidney-specific isoform of mouse vacuolar H+-ATPase subunit a. J Biol Chem. 2001;276:40050–40054. doi: 10.1074/jbc.M106488200. [DOI] [PubMed] [Google Scholar]
- Oot RA, Wilkens S. Subunit interactions at the V1-Vo interface in yeast vacuolar ATPase. J Biol Chem. 2012;287:13396–13406. doi: 10.1074/jbc.M112.343962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Qi J, Forgac M. Cellular environment is important in controlling V-ATPase dissociation and its dependence on activity. J Biol Chem. 2007;282:24743–24751. doi: 10.1074/jbc.M700663200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A, Levine TP. Multiple pools of phosphatidylinositol 4-phosphate detected using the pleckstrin homology domain of Osh2p. J Biol Chem. 2004;279:44683–44689. doi: 10.1074/jbc.M401583200. [DOI] [PubMed] [Google Scholar]
- Schulze U, Vollenbroker B, Kuhnl A, Granado D, Bayraktar S, Rescher U, Pavenstadt H, Weide T. Cellular vacuolization caused by overexpression of the PIKfyve-binding deficient Vac14L156R is rescued by starvation and inhibition of vacuolar-ATPase. Biochim Biophys Acta. 2017;1864:749–759. doi: 10.1016/j.bbamcr.2017.02.012. [DOI] [PubMed] [Google Scholar]
- Sciorra VA, Audhya A, Parsons AB, Segev N, Boone C, Emr SD. Synthetic genetic array analysis of the PtdIns 4-kinase Pik1p identifies components in a Golgi-specific Ypt31/rab-GTPase signaling pathway. Mol Biol Cell. 2005;16:776–793. doi: 10.1091/mbc.E04-08-0700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AN, Skaug J, Choate KA, Nayir A, Bakkaloglu A, Ozen S, Hulton SA, Sanjad SA, Al-Sabban EA, Lifton RP, et al. Mutations in ATP6N1B, encoding a new kidney vacuolar proton pump 116-kD subunit, cause recessive distal renal tubular acidosis with preserved hearing. Nat Genet. 2000;26:71–75. doi: 10.1038/79208. [DOI] [PubMed] [Google Scholar]
- Smith GA, Howell GJ, Phillips C, Muench SP, Ponnambalam S, Harrison MA. Extracellular and luminal pH regulation by vacuolar H+-ATPase isoform expression and targeting to the plasma membrane and endosomes. J Biol Chem. 2016;291:8500–8515. doi: 10.1074/jbc.M116.723395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan S, Vyas NK, Baker ML, Quiocho FA. Crystal structure of the cytoplasmic N-terminal domain of subunit I, a homolog of subunit a, of V-ATPase. J Mol Biol. 2011;412:14–21. doi: 10.1016/j.jmb.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storici F, Lewis LK, Resnick MA. In vivo site-directed mutagenesis using oligonucleotides. Nat Biotechnol. 2001;19:773–776. doi: 10.1038/90837. [DOI] [PubMed] [Google Scholar]
- Strahl T, Thorner J. Synthesis and function of membrane phosphoinositides in budding yeast, Saccharomyces cerevisiae. Biochim Biophys Acta. 2007;1771:353–404. doi: 10.1016/j.bbalip.2007.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun-Wada G, Murata Y, Yamamoto A, Kanazawa H, Wada Y, Futai M. Acidic endomembrane organelles are required for mouse postimplantation development. Dev Biol. 2000;228:315–325. doi: 10.1006/dbio.2000.9963. [DOI] [PubMed] [Google Scholar]
- Toro EJ, Ostrov DA, Wronski TJ, Holliday LS. Rational identification of enoxacin as a novel V-ATPase-directed osteoclast inhibitor. Curr Protein Pept Sci. 2012;13:180–191. doi: 10.2174/138920312800493151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyomura T, Murata Y, Yamamoto A, Oka T, Sun-Wada GH, Wada Y, Futai M. From lysosomes to the plasma membrane: localization of vacuolar-type H+-ATPase with the a3 isoform during osteoclast differentiation. J Biol Chem. 2003;278:22023–22030. doi: 10.1074/jbc.M302436200. [DOI] [PubMed] [Google Scholar]
- Toyomura T, Oka T, Yamaguchi C, Wada Y, Futai M. Three subunit a isoforms of mouse vacuolar H+-ATPase. Preferential expression of the a3 isoform during osteoclast differentiation. J Biol Chem. 2000;275:8760–8765. doi: 10.1074/jbc.275.12.8760. [DOI] [PubMed] [Google Scholar]
- Walch-Solimena C, Novick P. The yeast phosphatidylinositol-4-OH kinase pik1 regulates secretion at the Golgi. Nat Cell Biol. 1999;1:523–525. doi: 10.1038/70319. [DOI] [PubMed] [Google Scholar]
- Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke JD, Bussey H, et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- Zhang X, Li X, Xu H. Phosphoinositide isoforms determine compartment-specific ion channel activity. Proc Natl Acad Sci USA. 2012;109:11384–11389. doi: 10.1073/pnas.1202194109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Benlekbir S, Rubinstein JL. Electron cryomicroscopy observation of rotational states in a eukaryotic V-ATPase. Nature. 2015;521:241–245. doi: 10.1038/nature14365. [DOI] [PubMed] [Google Scholar]
- Zolov SN, Bridges D, Zhang Y, Lee WW, Riehle E, Verma R, Lenk GM, Converso-Baran K, Weide T, Albin RL, et al. In vivo, Pikfyve generates PI(3,5)P2, which serves as both a signaling lipid and the major precursor for PI5P. Proc Natl Acad Sci USA. 2012;109:17472–17477. doi: 10.1073/pnas.1203106109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoncu R, Bar-Peled L, Efeyan A, Wang S, Sancak Y, Sabatini DM. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H+-ATPase. Science. 2011;334:678–683. doi: 10.1126/science.1207056. [DOI] [PMC free article] [PubMed] [Google Scholar]




