Abstract
This study reports a microfluidic device for generating 2-dimensional concentration gradients of biomolecules along the width and length of a chamber and conventional 1-dimensional gradients along the width of its lateral parallel channels. The gradient profile can be precisely controlled by the applied flow rate. The proposed design is simple and straightforward, has a small footprint size compared to previously reported devices such as tree-shape designs, and for the first time, provides capability of generating desired 2D and 1D gradients, simultaneously. The finite element simulation analysis proves the feasibility of the microfluidic device, and the fluorescently labelled IgG antibody is used to demonstrate generated chemical gradients. This simple microfluidic device can be implemented for a wide range of high-throughput concentration gradient applications such as chemotaxis, drug screening, and organs-on-chips.
I. INTRODUCTION
Generating precise chemical concentration gradients is greatly momentous in several research areas such as cell migration and chemotaxis,1 cancer metastasis,2,3 stem cell,4 drug screening,5 and microfabrication.6 From a biological perspective, it is quite challenging to create and control a consistent chemical gradient in vitro which can exactly mimic in vivo conditions—an enclosed environment introducing minute volumes of biomolecules which can adhere to creeping laminar flow while resisting diffusion forces. However, lab-on-a-chip (LOC) technology represents unique solutions to create stable and controlled gradients of different biomolecules by means of microfluidic principles and has been utilized for several applications such as drug screening and studying cell interactions.7–12
The free-diffusion of soluble molecules from the “source” to “sink” is one way to generate concentration gradients in microfluidic devices.13–16 In this system, concentration gradients evolve as the molecules diffuse along the length of a microfluidic channel or a reservoir connecting source to sink and reach a steady state. These gradient generators are simple and can be used in high throughput applications requiring a low volume of reagents.17 Nevertheless, gradients usually require a long time to be created inside the microfluidic channel and can be maintained for a limited time since the size of the source and sink is very small in microfluidic systems.11 External pumping components may be required to address this problem, which brings about the complexity of the system.
Another gradient generation method is based on the diffusion of species across the interface of two adjacent laminar flows inside a microfluidic channel. Many of the flow-based microfluidic gradient generators have been relied on the tree-shape design introduced by Jeon et al. in 2000.18–21 The main limitation of the tree-shape design is its considerably large footprint size which minimizes the space available for the targeted test. Moreover, long diffusion time is required to generate the desired gradient in channels, which limits either the flow rate or the channel length.22–28
Although the designs that produce one-dimensional (1D) gradients, a change in the concentration perpendicular to the fluid flow input, have been well established, there are very few studies on microfluidic devices generating two-dimensional (2D) gradients. For instance, Jang et al.29 and Cooksey et al.30 utilized micro-wells and multifaceted tree-shape design, respectively, to achieve microfluidic-based 2D gradient generators. These devices are not only demanding to fabricate but also involve a multilayer control for fluid handling which is complex to operate. Microfluidic devices with 2D chemical gradients could be powerful tools in a wide range of applications such as antibiotic susceptibility testing, characterization of cellular responses, and chemotaxis when high-throughput experiments with a low volume of reagents are required.
Here, we report a simple microfluidic design for generating both 2D and 1D concentration gradients of biomolecules based on the hydrodynamic resistances induced by channel geometries. The proposed design generates two-dimensional gradients of biomolecules across both the width and length of a main chamber which can be well controlled by the applied flow rate. The main chamber seeds five parallel channels with different hydrodynamic resistances. These parallel channels can also produce one-dimensional gradients. Table SI (supplementary material) highlights the novelty and advantages of our microfluidic design in comparison to those already reported in the literature.
II. MATERIALS AND METHODS
The microfluidic device was designed using AutoCAD® software (Autodesk Inc., CA, USA) and printed on a chrome mask. Standard photolithography was then applied to produce an SU-8 based mold on a silicon wafer using the printed mask. Next, soft lithography was applied to create the microfluidic channels on the polydimethylsiloxane (PDMS, Essex Chemical, Boston, MA) substrate. The PDMS layer was peeled off from the mold, and the inlet and outlet ports were punched. The protocol for covalently functionalizing the surface with generated concentration gradients and the assembly of the microfluidic device has been introduced elsewhere.31,32 Briefly, both PDMS and glass substrates were oxygen plasma treated, and then, (3-Aminopropyl) triethoxysilane (APTES, Sigma-Aldrich) was micro-contact printed onto the glass substrates using a flat PDMS stamp. Instantly, the functionalized glass substrate was kept in contact with the PDMS layer to create irreversible bonds. The platform was then dried at 60 °C, and N-hydroxysuccinimide (NHS, Sigma-Aldrich) together with 1-ethyl-3–(3-dimethylaminopropyl) carbodiimide (EDC, Sigma-Aldrich) and phosphate buffered saline (PBS, Sigma-Aldrich) were used to activate the printed amine groups so as to covalently attach biomolecules onto the surface. To demonstrate generated gradients, fluorescein isothiocyanate (FITC) conjugated-IgG antibody and Cy3 conjugated IgG (Abcam, Cambridge, MA) were flown into the inlets by a multi-stage syringe pump. Furthermore, an inverted fluorescence microscope (ZEISS-Axio Observer Inverted Microscope) was used to image the generated gradients.
COMSOL software (COMSOL Inc., Burlington, MA) was used to perform the finite element simulation analysis. The 3D model of the microfluidic device was designed to process the full scale simulation of the experimental setup. The governing equations used for the computational fluid dynamic (CFD) analysis were Navier-Stokes and continuity equations assuming incompressible steady state conditions
| (1) |
| (2) |
where u is the velocity field, p is the pressure, η is the dynamic viscosity, and ρ is the liquid density. The fluid inside the channels was assumed to be a Newtonian fluid and to have the physical properties of water.
Species diffusion in the solution was modelled using multi-physics capabilities of COMSOL. The velocity components from the CFD results were simultaneously implemented for concentration distribution using the convection and diffusion module. Species diffusion in flow conditions was solved by applying Eq. (3)
| (3) |
where c represents each species concentration, t is the time, and D is the diffusion coefficient.
III. RESULTS AND DISCUSSION
Figure 1(a) shows the microfluidic design. It begins with two opposite inlet channels which then merge into a single channel perpendicular to the inlet axis. This channel joins the main chamber (1600 μm width × 800 μm length) which feeds into five parallel channels with different maximum widths as follows: channel 1st = 50 μm, channel 2nd = 100 μm, channel 3rd = 200 μm, channel 4th = 300 μm, and channel 5th = 500 μm. The entrance of all these channels is equal to 50 μm, and the height of all channels and chamber is 60 μm.
FIG. 1.
(a) Schematic representation of the microfluidic design. (b) Simulation results for velocity streamlines throughout the chamber by applying the flow rate of 1 μl min−1. (c) Velocity field at the entrance of the five parallel channels. (d) Simulation results for the concentration gradient inside the main chamber and side channels.
The size of the device has been optimized to be a minute footprint size of 4 mm × 8 mm, allowing us to fabricate a high throughput device by adding replicas of the same design in parallel for high throughput experiments (Fig. S1, supplementary material).
By imposing certain hydrodynamic resistances via 5 parallel channels, the microfluidic device is able to provide 2D gradients in the main chamber and 1D gradients inside channels 4 and 5 (Fig. 3). From the electronic-hydrodynamic analogy, the hydrodynamic resistance (Rh) of a straight microchannel in a laminar incompressible steady state flow with a rectangular cross section can be defined as follows:33,34
| (4) |
where L, w, and h are the length, width, and height of the channel, respectively, and μ is the dynamic viscosity. According to Eq. (4), changing the width of the microchannel results in different hydrodynamic resistances. Consequently, in order to achieve the desired hydrodynamic resistance in our microfluidic design, the geometry of the channels was adjusted based on Eq. (4).
FIG. 3.
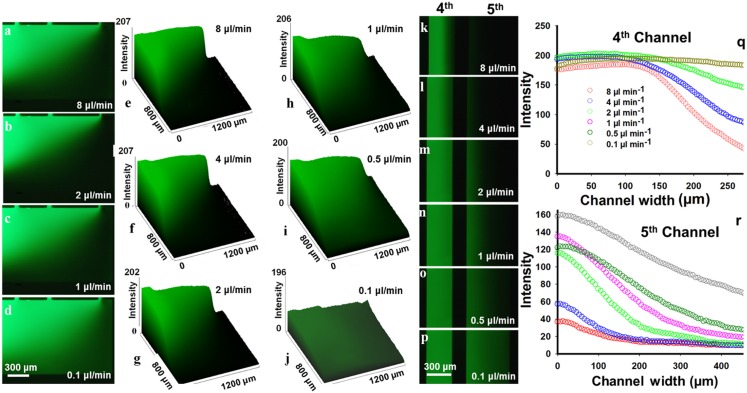
Fluorescence microscopy results using FITC conjugated IgG at different inlet flow rates. (a)–(d) Generated 2D gradient patterns inside the chamber. (e)–(j) 3D intensity plots quantifying the fluorescence intensity of the 2D concentration gradients in the main chamber. (k)–(p) Generated 1D gradient in channels 4 and 5. (q) and (r) Intensity diagram of channels 4 and 5 as a function of the channel width showing the 1D gradient inside the channels.
A corresponding computational fluid dynamics analysis and simulation of the concentration distribution were performed using COMSOL software. In the simulation process, to generate the 2D gradient, the biomolecule solution and buffer merge into the single channel until they enter the gradient chamber with applying desired flow rates. The biomolecule solution enters the chamber from the side that is furthest to the channel with the lowest resistance.
Figure 1(b) depicts the simulation results for streamlines inside the chamber applying a flow rate of 1 μl/min. It can be seen that the density of streamlines increases at the entrance of the chamber and at the entrance of the 5 parallel channels. Moreover, there is a gradual increase in the streamline density moving from the 1st channel to the 5th channel. The velocity field at the entrance of the five parallel channels is plotted in Fig. 1(c). Each of the parallel channels aspirates the fluid with different flow rates, which brings about the biomolecule distribution inside the chamber and inside the side channels.
Figure 1(d) demonstrates the simulation results for concentration distribution in the microfluidic device. As the width of the channel decreases, the channel hydrodynamic resistance increases. Thus, there is a higher concentration of biomolecules at the entrance of the first channel and the concentration is reduced along the length and width of the chamber due to lower hydrodynamic resistance and more fluid velocity. Moreover, Fig. 1(d) shows 1D gradients in the parallel channels perpendicular to the inlet flow. Furthermore, changing the arrangement and configuration of hydrodynamic resistances in these parallel channels leads to different gradient patterns shown in Figs. S2–S5 (supplementary material).
In addition to the 2D concentration gradient, there is a velocity gradient inside the main chamber as shown in Figs. 2(a) and 2(b) together with their arc-length directions. The directions of X and Y components of the velocity field are based on the coordinate systems shown in Fig. 1(a). According to Fig. 2(a), at the chamber's entrance (shown by a yellow line), there is a high velocity peak, which is due to the inlet flow rate. By getting far from the entrance and approaching the 5 parallel channels, the first sharp velocity peak decreases and other peaks gradually appear along the arc-length direction. Different fluid velocities inside the 5 parallel channels induced by their different hydrodynamic resistances are the main reason for the generation of these new velocity peaks. At the end side of the chamber close to the entrances of the 5 parallel channels, the velocity peaks related to each channel could be readily distinguished. Regarding the velocity pattern along the width directions [Fig. 2(b)], the velocity curve becomes smooth by getting far from the chamber's entrance since there are no channels at the width side of the chamber. Figures 2(c) and 2(d) illustrate the simulation outcome of the concentration along the length and width of the chamber when two different bimolecular solutions (indicated as green and red lines) enter through two opposite inlets of the microfluidic device. These patterns confirm the appropriate 2D concentration gradient of two different biomolecules throughout the chamber.
FIG. 2.
Simulation results of velocity and concentration gradients. (a) Y-component of velocity fields along represented arc-length directions with different colors (right image) inside the main chamber. (b) X-component of velocity fields along represented arc-length directions (right image). (c) Concentration gradients of two separate biomolecular solutions along the length of the chamber with represented arc-length directions (right image). (d) Concentration gradients along the width of the chamber with represented arc-length directions (right image).
The CFD analysis was quite analogous to the experimental results demonstrated in Fig. 3. This experiment was performed via employing a FITC conjugated-IgG antibody and PBS solution flown into the microfluidic device through the two opposite inlets. Figures 3(a)–(3d) show the generated 2D gradients of the FITC conjugated IgG antibody inside the chamber at various inlet flow rates. The 3D fluorescence intensities of the generated chemical gradients are plotted in Figs. 3(e)–3(j) for the applied flow rates using Image J software. It is obvious that the inlet flow rate has a substantial effect on the generated gradient's pattern. At high flow rates of 8 and 2 μl min−1, there was a steep slope of reduction in fluorescence intensity along the chamber's diagonal; however, at lower flow rates, the fluorescence intensity decreased more gently. Moreover, there was no significant gradient at a very low flow rate of 0.1 μl min−1. Produced convection in the Y-direction, because of the varying hydrodynamic resistances in parallel channels, is the main factor in the formation of gradients, and the generated gradients could easily be covalently attached to the aminosilanized surface using EDC-NHS chemistry.32
The 1D gradients generated in the channels 4 and 5 are shown in Figs. 3(k)–3(p) at different inlet flow rates, and the related intensity diagrams are plotted in Figs. 3(q) and 3(r). The 1D gradient perpendicular to the parallel channel direction is due to the effect of the generated 2D-gradient in the main chamber. At a flow rate of 8 μl min−1, very high fluid velocity of channel 5 caused a very low fluorescence concentration at the very left side of the channel. The generated 1D gradient is more distinct in channel 4 with higher hydrodynamic resistance compared to channel 5. At lower inlet flow rates, the laminar flow rates inside the channel 5 were in an appropriate range to create a 1D gradient. As a proof of concept, we immobilized Escherichia coli (ATCC8739) bacteria inside the microfluidic device and demonstrated successful gradient generation using FITC labelled IgG solution (Fig. S6, supplementary material). This provides a powerful tool for performing rapid and high-throughput antibiotic susceptibility tests.
To further demonstrate the design principle, we examined flowing two different fluorescently labelled IgG antibodies from two different inlets. Figure 4 illustrates these results where FITC conjugated IgG and Cy3 conjugated IgG were flown into the opposite inlets and proceeded to the chamber through the entrance narrow channel as two coflowing streams—the Cy3 conjugated antibody stream on the right and the FITC conjugated antibody on the left side of the entrance channel. In this system, two conjugated antibodies entered the main chamber at the center of the chamber length. The low intensity of red fluorescence determines the minor generated 2D gradients of the Cy3 conjugated IgG, while the FITC conjugated IgG represents well-established 2D gradients, confirming the effect of implemented different hydrodynamic resistances in the design where a smooth 2D gradient is generated towards the channels with lower hydrodynamic resistance. At a flow rate of 8 μl min−1, as the Cy3 conjugated antibody could rapidly reach the low hydrodynamic resistant channels 4 and 5, there is not enough time for antibodies to diffuse, thereby forming a steep concentration gradient pattern. Decreasing the flow rate, however, brought about more gradual 2D concentration gradients throughout the main chamber.
FIG. 4.
Generated 2D gradients of FITC conjugated IgG and Cy3 conjugated IgG inside the chamber together with their 3D intensity plots at different inlet flow rates of (a) 0.1 μl min−1 (b) 1 μl min−1 (c) 2 μl min−1 and (d) 8 μl min−1, obtained using Image J software.
IV. CONCLUSIONS
In conclusion, a simple and straightforward microfluidic design was introduced to generate 2D and 1D gradients of biomolecules. Having remarkably small footprint size, the device is capable of generating two-dimensional concentration gradients throughout its chamber and one-dimensional gradients inside its parallel channels. The simulation results demonstrated the creation of 2D velocity field and particle concentration gradients inside the chamber owing to various hydrodynamic resistances of the 5 parallel lateral channels. Fluorescence microscopy experiments successfully illustrated different 2D and 1D gradient patterns dependent on the inlet flow rates. Due to its small footprint, several designs can be integrated into a single chip (Fig. S1, supplementary material) for a wide range of high-throughput concentration gradient applications such as antibiotic susceptibility tests, chemotaxis, drug screening, and organs-on-chips where 2D or 1D chemical gradients of biomolecules are needed.
V. SUPPLEMENTARY MATERIAL
See supplementary material for more information on the proposed microfluidic design and other microfluidic gradient generators already published in the literature.
ACKNOWLEDGMENTS
The financial support for this work was provided by the Natural Science and Engineering Research Council of Canada (NSERC) Discovery Grant and start-up funds from McMaster Faculty of Engineering.
References
- 1. Jeon N. L., Baskaran H., Dertinger S. K. W., Whitesides G. M., Van De Water L., and Toner M., Nat. Biotechnol. 20, 826 (2002). 10.1038/nbt712 [DOI] [PubMed] [Google Scholar]
- 2. Wang S.-J., Saadi W., Lin F., Nguyen C. M.-C., and Li Jeon N., Exp. Cell Res. 300, 180 (2004). 10.1016/j.yexcr.2004.06.030 [DOI] [PubMed] [Google Scholar]
- 3. Saadi W., Wang S.-J., Lin F., and Jeon N. L., Biomed. Microdevices 8, 109 (2006). 10.1007/s10544-006-7706-6 [DOI] [PubMed] [Google Scholar]
- 4. Chung B. G., Flanagan L. A., Rhee S. W., Schwartz P. H., Lee A. P., Monuki E. S., and Jeon N. L., Lab Chip 5, 401 (2005). 10.1039/b417651k [DOI] [PubMed] [Google Scholar]
- 5. Hong B., Xue P., Wu Y., Bao J., Chuah Y. J., and Kang Y., Biomed. Microdevices 18, 21 (2016). 10.1007/s10544-016-0054-2 [DOI] [PubMed] [Google Scholar]
- 6. Kenis P. J. A., Ismagilov R. F., Takayama S., Whitesides G. M., Li S., and White H. S., Acc. Chem. Res. 33, 841 (2000). 10.1021/ar000062u [DOI] [PubMed] [Google Scholar]
- 7. Barata D., Van Blitterswijk C., and Habibovic P., Acta Biomater. 34, 1 (2016). 10.1016/j.actbio.2015.09.009 [DOI] [PubMed] [Google Scholar]
- 8. Du G., Fang Q., and den Toonder J. M. J., Anal. Chim. Acta 903, 36 (2016). 10.1016/j.aca.2015.11.023 [DOI] [PubMed] [Google Scholar]
- 9. Halldorsson S., Lucumi E., Gómez-Sjöberg R., and Fleming R. M. T., Biosens. Bioelectron. 63, 218 (2015). 10.1016/j.bios.2014.07.029 [DOI] [PubMed] [Google Scholar]
- 10. Du Y., Shim J., Vidula M., Hancock M. J., Lo E., Chung B. G., Borenstein J. T., Khabiry M., Cropek D. M., and Khademhosseini A., Lab Chip 9, 761 (2009). 10.1039/B815990D [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kim S., Kim H. J., and Jeon N. L., Integr. Biol. 2, 584 (2010). 10.1039/c0ib00055h [DOI] [PubMed] [Google Scholar]
- 12. Toh A. G. G., Wang Z. P., Yang C., and Nguyen N. T., Microfluid. Nanofluid. 16, 1 (2014). 10.1007/s10404-013-1236-3 [DOI] [Google Scholar]
- 13. Abhyankar V. V., Lokuta M. A., Huttenlocher A., and Beebe D. J., Lab Chip 6, 389 (2006). 10.1039/b514133h [DOI] [PubMed] [Google Scholar]
- 14. Wu H., Huang B., and Zare R. N., J. Am. Chem. Soc. 128, 4194 (2006). 10.1021/ja058530o [DOI] [PubMed] [Google Scholar]
- 15. Saka Y., Macpherson M., and Giuraniuc C. V., Physica A 470, 132 (2017). 10.1016/j.physa.2016.11.134 [DOI] [Google Scholar]
- 16. Luo X., Vo T., Jambi F., Pham P., and Choy J. S., Lab Chip 16, 3815 (2016). 10.1039/C6LC00742B [DOI] [PubMed] [Google Scholar]
- 17. Atencia J., Morrow J., and Locascio L. E., Lab Chip 9, 2707 (2009). 10.1039/B902113B [DOI] [PubMed] [Google Scholar]
- 18. Jeon N. L., Dertinger S. K. W., Chiu D. T., Choi I. S., Stroock A. D., and Whitesides G., Langmuir 16, 8311 (2000). 10.1021/la000600b [DOI] [Google Scholar]
- 19. Chen X., Wu D., Mei X., Zhou Z., Wang L., Zhao Y., Zheng G., and Sun D., in 2016 IEEE 10th International Conference on Nano/Molecular Medicine and Engineering (2016), pp. 104–108. [Google Scholar]
- 20. Ko K.-Y. and Kim I.-H., Biotechnol. Bioprocess Eng. 21, 453 (2016). 10.1007/s12257-015-0796-3 [DOI] [Google Scholar]
- 21. Wang H., Chen C., Xiang Z., Wang M., and Lee C., Lab Chip 15, 1445 (2015). 10.1039/C4LC01451K [DOI] [PubMed] [Google Scholar]
- 22. Shi X., Ostrovidov S., Shu Y., Liang X., Nakajima K., Wu H., and Khademhosseini A., Langmuir 30, 832 (2014). 10.1021/la4041216 [DOI] [PubMed] [Google Scholar]
- 23. Ruan J., Wang L., Xu M., Cui D., Zhou X., and Liu D., Mater. Sci. Eng. C 29, 674 (2009). 10.1016/j.msec.2008.12.009 [DOI] [Google Scholar]
- 24. Hung P. J., Lee P. J., Sabounchi P., Lin R., and Lee L. P., Biotechnol. Bioeng. 89, 1 (2005). 10.1002/bit.20289 [DOI] [PubMed] [Google Scholar]
- 25. Park S., Kim D., Mitchell R. J., and Kim T., Lab Chip 11, 2916 (2011). 10.1039/c1lc20230h [DOI] [PubMed] [Google Scholar]
- 26. Yoo H. J., Park J., and Yoon T. H., Cytometry, Part A 83, 356 (2013). 10.1002/cyto.a.22261 [DOI] [PubMed] [Google Scholar]
- 27. Campbell K. and Groisman A., Lab Chip 7, 264 (2007). 10.1039/B610011B [DOI] [PubMed] [Google Scholar]
- 28. Dertinger S. K. W., Jiang X., Li Z., Murthy V. N., and Whitesides G. M., Proc. Natl. Acad. Sci. U.S.A. 99, 12542 (2002). 10.1073/pnas.192457199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jang Y.-H., Hancock M. J., Kim S. B., Selimović S., Sim W. Y., Bae H., and Khademhosseini A., Lab Chip 11, 3277 (2011). 10.1039/c1lc20449a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cooksey G. A., Sip C. G., and Folch A., Lab Chip 9, 417 (2009). 10.1039/B806803H [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Didar T. F. and Tabrizian M., Lab Chip 12, 4363 (2012). 10.1039/c2lc40233e [DOI] [PubMed] [Google Scholar]
- 32. Didar T. F., Foudeh A. M., and Tabrizian M., Anal. Chem. 84, 1012 (2012). 10.1021/ac2025877 [DOI] [PubMed] [Google Scholar]
- 33. Oh K. W., Lee K., Ahn B., and Furlani E. P., Lab Chip 12, 515 (2012). 10.1039/C2LC20799K [DOI] [PubMed] [Google Scholar]
- 34. Beebe D. J., Mensing G. A., and Walker G. M., Annu. Rev. Biomed. Eng. 4, 261 (2002). 10.1146/annurev.bioeng.4.112601.125916 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
See supplementary material for more information on the proposed microfluidic design and other microfluidic gradient generators already published in the literature.