Abstract
Several families of plant viruses evolved cap-independent translation enhancers (3′CITE) in the 3′ untranslated regions of their genomic (g)RNAs to compete with ongoing cap-dependent translation of cellular mRNAs. Umbravirus Pea enation mosaic virus (PEMV)2 is the only example where three 3′CITEs enhance translation: the eIF4E-binding Panicum mosaic virus-like translational enhancer (PTE) and ribosome-binding 3′ T-shaped structure (TSS) have been found in viruses of different genera, while the ribosome-binding kl-TSS that provides a long-distance interaction with the 5′ end is unique. We report that the PTE is the key translation promoting element, but inhibits translation in cis and in trans in the absence of the kl-TSS by sequestering initiation factor eIF4G. PEMV2 strongly outcompeted a cellular mRNA mimic for translation, indicating that the combination of kl-TSS and PTE is highly efficient. Transferring the 3′–5′ interaction from the kl-TSS to the PTE (to fulfill its functionality as found in other viruses) supported translationin vitro, but gRNA did not accumulate to detectable levels in protoplasts in the absence of the kl-TSS. It was shown that the PTE in conjunction with the kl-TSS did not markedly affect the translation initiation rate but rather increased the number of gRNAs available for translation. A model is proposed to explain how 3′CITE-based regulation of ribosome recruitment enhances virus fitness.
INTRODUCTION
Protein synthesis is generally divided into four phases: initiation, elongation, termination and recycling, with translation efficiency mainly depending on initiation, a rate-limiting step. The conventional paradigm of translation initiation in eukaryotes involves binding of the 5′-m7GpppN cap to initiation factor 4E (eIF4E), part of the eIF4F protein complex, which along with several other ligands play a central role in the further accommodation of 40S ribosomal subunits at the 5′ ends of mRNAs. The resulting 43S pre-initiation complex scans through the 5′ untranslated region (UTR) seeking an initiation codon in a proper context. If successful, the 40S subunit joins with the 60S subunit forming the complete 80S ribosome, which starts elongation of the polypeptide chain (1). Whereas this model has been the major accepted mechanism of translation initiation in eukaryotes for the past 30 years, recent transcriptome-wide studies reveal a far more complex picture (2). Highly cap/eIF4E-dependent templates were found to be a relatively small percentage of cellular mRNAs, while making up the major portion of mRNAs within polysomes of proliferating cells. The majority of the transcriptome comprised mRNAs with weak, if any, requirement for cap/eIF4E but with a strong dependence on eIF4G, which is another subunit of eIF4F. It was suggested that mRNAs of this type may contain elements with a high affinity to eIF4G (3) and these cap-independent translation enhancers (CITEs) can provide initiation levels sufficient to compete with downregulated capped mRNAs, especially in non-dividing cells. The main difference between well-studied internal ribosome entry sites (IRES) of viruses (4,5) and CITEs is that, the latter are not capable of promoting internal initiation and yet can still direct translation that is 5′ end dependent (6–8).
A common feature of all viruses is the employment of cellular ribosomes for synthesis of viral proteins. To ensure rapid synthesis of proteins required for replication, RNA viruses employ one or more of four general strategies to gain preferential access to the translation machinery: (i) inactivation of specific host proteins required for different stages of cap-dependent translation (9); (ii) reducing the abundance of cellular mRNAs by stimulating their decay (10,11); (iii) outcompeting on-going host translation, presumably by robust acquisition of translation factors by cis-acting RNA elements in the viral genome (5); and (iv) generating non-coding (nc)RNAs that bind and inhibit critical translation factors such as eIF4G (12–14). Whereas some animal RNA viruses can apparently compete advantageously with cellular mRNAs (15), they generally employ the first two strategies, which can severely impact the condition of the cell. In contrast, plant RNA viruses, many of which lack a 5′ cap and/or poly(A) tail, require living infected cells to spread to adjacent cells via interconnecting plasmodesmata. Thus, they generally employ the third and fourth strategies of containing cis-acting elements, and generating trans-acting ncRNAs containing these elements, which confer a translational advantage to the viral genome over cellular mRNAs. Such elements have mainly been studied in viruses from the Tombusviridae and Luteoviridae, where they occupy a location in the genomic (g)RNA that is either within the 3′UTR or in nearby coding sequence and the term ‘3′CITE’ was coined in this connection.
3′CITEs are categorized according to their well-defined secondary structures and include the translation enhancer domain (TED), barley yellow dwarf virus (BYDV)-like element (BTE), panicum mosaic virus-like translational enhancer (PTE), T-shaped structure (TSS), I-shaped structure (ISS) and Y-shaped structure (YSS) (5). Functional characterization of these 3′CITEs has mainly been limited to delineating their interaction with translation components and defining how bound components are delivered to the 5′ end. Translation enhancement by most 3′CITEs requires binding to translation initiation factors eIF4G (BTE) (16); eIF4E (PTE and TED), which can subsequently interact with eIF4G (17,18); or eIF4F (ISS and YSS) (19–21), which in plants is composed of eIF4E and eIF4G (22), making these elements similar to conjectural 5′CITEs of animal cellular mRNAs and the internal cap-assisted translation element of histone H4 (23). Furthermore, nearly all plant virus 3′CITEs participate in a long-distance RNA–RNA interaction with the apical loop of a hairpin in the gRNA 5–′ region (either UTR or coding). This long-distance bridge is thought to mimic circularization of capped cellular mRNAs produced by interaction of eIF4F with poly(A)-binding protein (PABP) bound to the polyadenylated 3′ end (24), which enhances multiple steps in translation (25). An exception is the TSS located near the 3′ end of carmovirus Turnip crinkle virus (TCV), which directly binds 80S and 60S ribosomal subunits (26–28), and contains no discernable RNA–RNA bridge connecting the 3′CITE to the 5′ end. The lack of a long-distance interaction between TCV UTRs led to a ribosome bridging model, which posits that 3′TSS-bound 60S subunits join with 40S ribosomal subunits bound to a pyrimidine-rich sequence in the 5′UTR to facilitate translation (29).
Very little is understood about how 3′CITEs enhance translation and confer a selective advantage to the viral gRNA although it is generally assumed to involve enhancing ribosome recycling and/or the translation initiation rate. The association of most 3′CITEs with eIF4F by binding to either eIF4E or eIF4G, and participating in a long-distance interaction suggests that most 3′CITEs will share a common process for enhancing translation (21,30). The best studied 3′CITE, the BYDV BTE, attracts 40S subunits in the presence of eIF4F, helicase factors eIF4A and eIF4B, and adenosine triphosphate, followed by delivery of 40S to the 5′ end through its long-distance interaction. Subsequent scanning of the 40S subunit from the 5′ end has been postulated for 3′CITEs based on insertion of AUGs upstream of the initiation site causing translational repression (31,32). Since the BTE also functions when translocated to the 5′UTR (7), one explanation for positioning CITEs at the 3′ end is for enhancing translation of both the gRNA and any sgRNAs via a single cis-element. However, how this mode of non-canonical translation initiation affects the initiation rate and polysome profile in comparison with canonical translation of capped mRNAs and transcripts containing 5′CITEs was unknown.
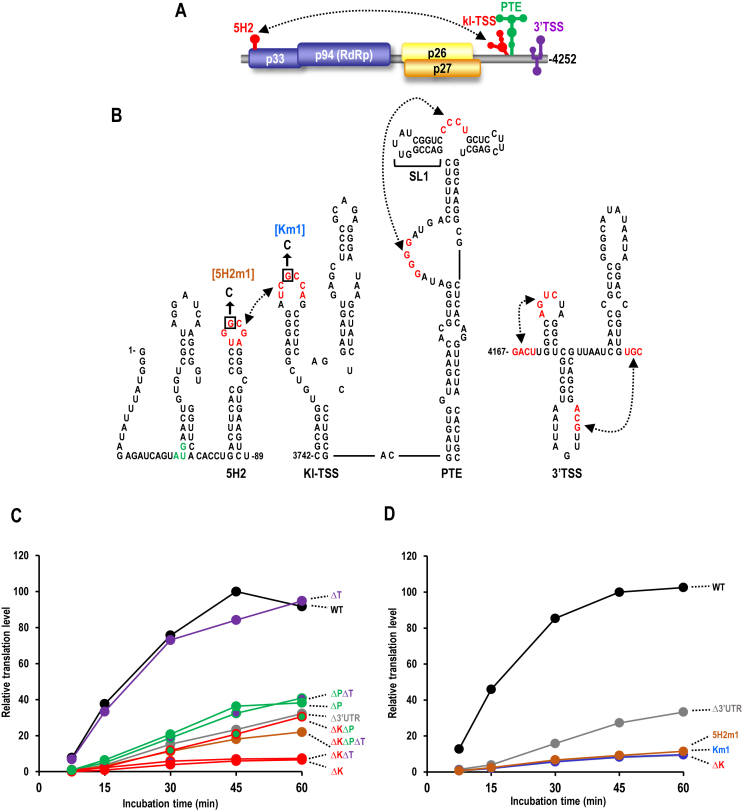
Plant viruses whose translation depends on 3′CITEs usually contain a single identified CITE. The two exceptions are carmovirus Melon necrotic spot virus N strain, which contains two 3′CITEs—an ISS element and an unclassified element that was likely acquired through recombination with a luteovirus (33), and RNA2 of Pea enation mosaic virus (PEMV), which contains three 3′CITEs (26,27,34). PEMV is composed of two taxonomically distinct, positive-strand RNAs: enamovirus PEMV1 and umbravirus PEMV2. PEMV1 provides the coat protein for packaging both RNAs whereas PEMV2 supplies movement proteins for both viruses (35). PEMV2, which has been used extensively as a model to study non-canonical translation (26,27,34,36), has a single gRNA of 4252 nt that lacks a 5′ cap and 3′ poly(A) tail and encodes four proteins: p33 and its −1 frameshifting extension product p94 (the viral RNA-dependent RNA polymerase [RdRp]) are translated from the gRNA; and p26 and p27, which are associated with stability and/or movement of the gRNA and are expressed from overlapping open reading frames (ORFs) on a subgenomic (sg)RNA (37) (Figure 1A). The three PEMV2 3′CITEs in the unusually long (703 nt) 3′UTR are shown in Figure 1B. The 3′TSS, which binds to ribosomes and is structurally similar to the TCV TSS, is located near the 3′ end and has organizational counterparts in several other umbraviruses (our unpublished data), as well as in several carmoviruses (27,38). The PTE, located in the central portion of the 3′UTR, has also been found in viruses in several genera within the Tombusviridae (17,18). PTEs are composed of two hairpins separated by a C-rich loop and a supporting stem with a large G-rich bulge. The pseudoknot that forms between the G-rich bulge and the C-rich loop is critical for binding to translation initiation factor eIF4E (18). Strikingly, unlike PTEs found in other viruses (39), the 5′ side hairpin of the PEMV2 PTE (SL1) does not engage in a long-distance RNA–RNA interaction with the 5′ end. Instead, the PTE relies on the unique adjacent upstream kl-TSS for delivery of bound eIF4E and factors that interact with eIF4E (e.g. eIF4G) to the 5′ end (26). The kl-TSS, which can also directly bind to 80S ribosomes and both ribosomal subunits, has a two hairpin, three-way branched structure and a short supporting stem, similar to the upper portion of the PTE. The loop of the 5′ side kl-TSS hairpin engages in a kissing-loop interaction with the apical loop of 5′ coding-region hairpin 5H2 (26) and this long-distance interaction is not affected when ribosomes are bound to the kl-TSS (34) (Figure 1B). Although it was shown that all three 3′CITEs are important for translation of various luciferase reporter constructs in vivo, it remains unresolved: (i) whether all or a subset of these 3′CITEs are important for translation of the full-length gRNA and sgRNA; (ii) why the PEMV2 PTE evolved to eliminate its long-distance interaction; and (iii) the mechanisms by which these 3′CITEs enhance translation.
Figure 1.
Contributions of 3′CITEs for translation of PEMV2 gRNA in vitro. (A) Genome organization of PEMV2. Dashed line denotes the kissing-loop interaction between the kl-TSS and 5′ proximal hairpin 5H2. The approximate locations of the three 3′CITEs are shown. (B) Secondary structures of the 3′CITEs and the 5′ terminal 89 nt. Dashed lines denote tertiary interactions including the long-distance interaction (left) and pseudoknots (middle and right) between residues shown in red. Mutations introduced into the apical loops involved in the kissing-loop interaction are shown. (C) In vitro translation of wild-type (WT) and mutant gRNAs in wheat germ extracts (WGE). ΔK, ΔP, and ΔT are deletions of the kl-TSS, PTE and TSS, respectively. (D) In vitro translation of WT and mutant gRNAs that are missing the long-distant interaction. Assays in (C) and (D) were repeated once, with similar results.
We now report that the PTE is the key 3′CITE for translation of PEMV2 gRNA, but requires the long-distance interaction of the kl-TSS for function in wheat germ extracts (WGE) and protoplasts. In WGE, the 3′TSS did not contribute to translation of the gRNA. When the kl-TSS long-distance interaction was abolished or not present, the PTE inhibited translation of the gRNA by sequestering eIF4G. Transferring the long-distance interaction from the kl-TSS to SL1 of the PTE to mimic PTE in other viruses generated a functional PTE translational enhancer in vitro, but complete elimination of the kl-TSS significantly reduced accumulation of the gRNA in protoplasts. Pronounced trans-inhibitory effects of the PTE and finding that PEMV2 strongly outcompeted a cellular mRNA mimic for translation suggests an additional PTE function: competitive inhibition of host mRNA translation by depletion of eIF4G. We also found that the main effect of the cooperative PTE and kl-TSS action was not the previously assumed increase in the translation initiation rate, but rather there was an increase of the number of translated gRNA molecules. We propose a model of how the cyclicity of CITE-driven ribosome recruitment ensures a steady initiation rate optimal for virus fitness.
MATERIALS AND METHODS
Construction of virus mutants and RNA transcription
pUC19-PEMV2, which contains full-length wild-type (WT) PEMV2 gRNA downstream of a T7 promoter, has been described previously (26). PEMV2 mutations were generated using DNA oligonucleotides and one-step site-directed mutagenesis (40). Reporter construct 5′89+3U, in which the firefly luciferase gene is flanked by PEMV2 5′ 89 nt and 3′UTR at its 5′ and 3′ ends, respectively, and its mutant 5′89+3Ukm2, containing mutations in the kissing loop of the kl-TSS, have been previously reported (26). All mutations were verified by sequencing. Plasmids were linearized and used as templates for in vitro transcription using T7 RNA polymerase that generated uncapped RNA transcripts. RNA transcripts were quality assessed by subjecting to electrophoresis through 1% agarose gels in 0.5× Tris-borate-EDTA (TBE) and quantified using nanodrop (Thermo Sci).
RNA labeling
RNA transcripts (40 μg) were incubated in 40 μl of 62.5 mM NaOAc pH5.5, 62.5 μM NaIO4 for 90 min at 4°C. Excess NaIO4 was neutralized with 2 μl of 2.5 mM NaSO3. After 15 min at 4°C, RNA was ethanol precipitated and dissolved in 40 μl of labeling solution (62.5 mM NaOAc pH 5.5, 0.1 mM Fluorescein-5-thiosemicarbazide). After 4 h at 4°C, excess dye was removed by spin filtration using a MicroSpin G-25 column (GE Healthcare). Labeled RNA was ethanol precipitated, dissolved in 20 μl of RNAse-free water and quantified.
In vitro translation assays
Uncapped viral gRNAs synthesized in vitro (0.5 pmol) were subjected to translation in a 10 μl WGE reaction (Promega) in the presence of 35S-methionine and 100 mM potassium acetate according to the manufacturer’s instructions. Translation mixes were incubated at 25°C for 45 min unless noted. Translation products were separated on 10% sodium dodecylsulphate-polyacrylamide electrophoresis gels, which were subsequently dried and exposed to a phosphorimaging screen. Phosphorimaging screens were scanned using a FLA-5100 fluorescent image analyzer (Fujifilm). The radioactive band corresponding to p33 was quantified using Multi Gauge 3.0 software (Fujifilm). For trans-inhibition assays, 10- or 20-fold molar excess of WT or mutant PTE [positions 3815–3912] competitor RNAs or 2.5- to 80-fold excess of Cricket paralysis virus intergenic IRES (CrPV IGR-IRES; positions 6026–6230] were added to WGE just prior to incubation. For trans-action of translation initiation factors, Arabidopsis translation initiation factors eIF4E, eIF4G and eIF4F expressed in Escherichia coli and purified as described previously (41), were individually added to WGE at a final concentration of 200 nM just prior to incubation. Translation initiation factors were the kind gift of Karen Browning.
Translation competition assays
The two templates competing for translation with PEMV2 were: (i) GloLuc, containing the 5′ leader sequence of human β-globin, the firefly luciferase ORF, the pGL3 vector-derived 3′UTR and a 50 nt poly(A) tail; and (ii) TZ10ΩLuc, containing the firefly luciferase ORF downstream of the TMV Ω sequence and followed by the 3′UTR of TMV (42). Following in vitro translation using T7 RNA polymerase, GloLuc transcripts were capped at their 5′ ends using the Vaccinia Capping System (NEB) according to the manufacturer’s instructions. Each of these reporter mRNAs (0.5 pmol) were translated alone or with 0.5 pmol of PEMV2, as described above.
Virus accumulation in protoplasts
Preparation and transfection of Arabidopsis protoplasts were as described previously (43). Briefly, protoplasts were prepared from seed-started callus that was grown on solid MS media with appropriate hormones. Protoplasts (5 × 106) were transfected with 20 μg of in vitro synthesized viral gRNA using a polyethylene glycol-mediated transformation method. Total RNA was extracted from protoplasts at 24 h post-transfection and quantified using a nanodrop (Thermo Sci). Three micrograms of total RNA was separated on 1.5% denaturing agarose gels and transferred to positively charged nylon membranes (GE), as described previously (44). The membrane was subsequently probed using three 32P-labeled DNA oligonucleotides complementary to positions 2731–2771, 2969–3004 and 3229–3270 of PEMV2 gRNA. After hybridization, the membrane was exposed overnight to a phosphorimaging screen.
Ribosome toeprinting
Toeprinting assays were performed as described (32). Briefly, 0.5 pmol of viral gRNA transcripts was incubated in a 10 μl of WGE reaction in the presence of 5 mM cycloheximide (CHX) for 30 min at 25°C. A total of 3 μl of the WGE reaction was added to annealing buffer (1×: 10 mM Dithiothrietol, 0.5 mM dNTP, 5 mM CHX and 10 U RNaseOUT). After 2 min incubation at 55°C, 1 pmol of 32P-labeled primer (complementary to position 98–114 of PEMV2 gRNA) was added and incubation continued at 37°C for 5 min. The primer was then extended using Superscript III (Thermo Sci) for 30 min at 50°C. cDNA synthesized was subjected to electrophoresis through a 10% polyacrylamide 8 M urea gel, followed by exposure overnight to a phosphorimaging screen.
SHAPE structural probing
Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE) RNA structure probing was performed as previously described (45). Briefly, viral gRNA transcripts were added to folding buffer (80 mM Tris–HCl pH 8.0, 160 mM NH4Cl and 11 mM MgOAc), denatured at 65°C for 5 min and then incubated at 37°C for 20 min. The RNA was then chemically modified by incubation with either 17.5 mM 1-methyl-7-nitroisatoic anhydride (dissolved in dimethyl sulfoxide [DMSO]) or DMSO as a control at 37°C for 40 min. Subsequently, the modified RNA was ethanol precipitated, dissolved in nuclease-free water and subjected to electrophoresis through an 8% polyacrylamide 8 M urea gel. The gel was dried and exposed to a phosphorimaging screen. Intensity of each band was quantified using the semiautomated footprinting analysis software (46,47).
Statistical analysis
Data from three independent experiments were statistically analyzed using ANOVA or t-test, as indicated (Graphpad prism 7.0).
In situ monitoring of protein synthesis and calculation of the timing of translation
Translation reaction components described above were mixed on ice, adjusted to 80% of the final volume and incubated for 2 min at 25°C. Pre-heated 5-fold concentrated luciferase mRNA (2 μl) was diluted with 8 μl of the prepared reaction mixture and immediately put into the temperature-controlled cell of a Chemilum-12 multichannel luminometer. The intensity of light emission generated through luciferase activity was measured continuously by collecting the streaming data on the computer. The first 15 min of the measured kinetic data were fitted by the numerical solutions of a parameterized differential equation system describing normal Gauss distribution that represents the approximation model for the second time-derivative of the kinetic curve. The position of the Gaussian peak indicates the average ‘full-translation’ time needed to synthesize a full-length luciferase molecule (42). All calculations were made using IgorPro 6.3 data analysis software (WaveMetrics, Inc.).
Sedimentation analysis of polyribosomes
WGE translation reaction mixtures with labeled reporter RNAs (20 μl) were collected after 25 min of translation, chilled on ice, supplemented with CHX up to 0.01 mg/ml and layered atop a linear 15–45% sucrose gradient in 12 ml Ultra-Clear Beckman tubes, containing 25 mM Tris–HCl pH 7.6, 5 mM MgCl2, 100 mM KCl, 0.1 mM ethylenediaminetetraacetic acid (EDTA) and 0.01 mg/ml CHX. Samples were subjected to centrifugation for 2 h 45 min in a SW-41 rotor in an Optima L-90K (Beckman-Coulter) ultracentrifuge at 37 000 RPM at 4°C. Gradients were fractionated from the bottom of the tubes at 0.5 ml/min with continuous measurement of the optical density at 254 nm with a UVCord 2238 (Pharmacia) and fluorescence (λex = 492 nm and λem = 518 nm) with an RF-5031PC fluorometer (Shimadzu) equipped with a 120 μl liquid chromotography flow cell.
RESULTS
3′CITE involvement in translation of PEMV2 gRNA in vitro
The three PEMV2 3′CITEs were previously identified using luciferase reporter constructs (17,26,27). To better understand the importance and functional association of the 3′CITEs in facilitating translation of the natural template, full-length PEMV2 gRNA lacking either the 3′UTR, or one or more of the 3′CITEs were assayed for synthesis of p33 in WGE (Figure 1C and D). Time-course assays showed that mutant gRNA missing all three CITEs (ΔKΔPΔT) reduced translation by 82% at 45 min, similar to gRNA lacking the entire 3′UTR (Δ3′UTR) (Figure 1D). Although deletion of the 3′TSS alone (ΔT) had no discernable impact on p33 levels, the 3′TSS does contribute to translation of the PEMV2 sgRNA (Gao, F., and Simon, A.E. (2017) Differential use of 3’ CITEs by the subgenomic RNA of Pea enation mosaic virus 2. Virology, in press). Deletion of the PTE alone (ΔP) or ΔPΔT reduced synthesis of p33 to a similar extent as ΔKΔPΔT or Δ3′UTR, suggesting that the PTE is the key 3′CITE in this assay. However, deletion of the kl-TSS alone (ΔK) or ΔKΔT reduced translation 3-fold more than ΔP, ΔKΔPΔT or Δ3′UTR. Mutations that disrupted the long-distance interaction either in the kl-TSS (Km1) or in 5′ hairpin 5H2 (5H2m1) reduced translation to a similar extent as ΔK, indicating that the kissing-loop interaction is critical for translation enhancement in the gRNA (Figure 1D). Deletion of the kl-TSS and the PTE (ΔKΔP) eliminated the negative effect of deleting only the kl-TSS, with translation now similar to ΔP, ΔKΔPΔT or Δ3′UTR. These results suggest that in WGE: (i) there are no additional translation elements independent of these 3′CITEs; (ii) the 3′TSS does not contribute to gRNA translation; (iii) the kl-TSS does not enhance translation in the absence of the PTE; (iv) the PTE inhibits translation in the absence of the kl-TSS; and (v) the kl-TSS and PTE function in tandem to enhance translation.
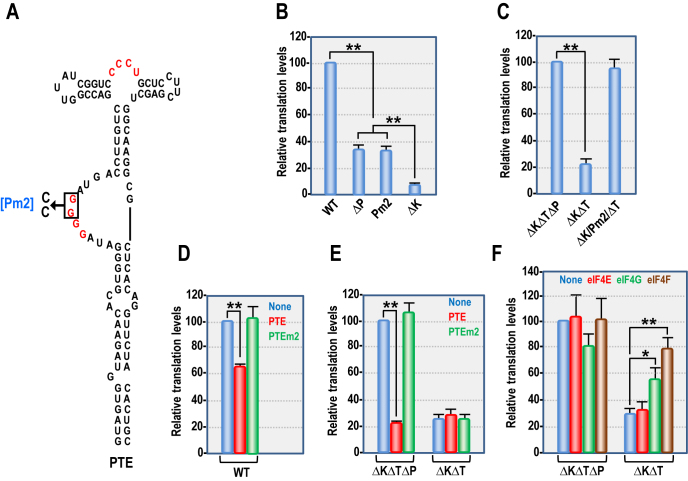
These findings suggest that the kl-TSS ribosome-binding ability and participation in the long-distance interaction (that circularizes the template) do not contribute positively to translation in vitro in the absence of the PTE. However, an alternative explanation is that deletion of the PTE affects the structure of the kl-TSS, which abrogates its ability to connect with the 5′ end. To investigate this possibility, gRNA containing only two mutations in the G-rich bulge of the PTE (Pm2) that individually significantly reduced eIF4E binding (17) was substituted for gRNA containing a full deletion of the PTE (ΔP) in a single time point WGE translation assay (Figure 2A). Pm2 reduced p33 synthesis to 34% of WT, similar to ΔP levels of translation (35%) (Figure 2B) and ΔK reduced p33 synthesis to 9% of WT. These results suggest that the kl-TSS-mediated long-distance interaction and ribosome-binding abilities do not enhance translation of PEMV2 gRNA in WGE independent of the PTE.
Figure 2.
PTE can inhibit translation by sequestering eIF4G. (A) Pm2 mutation that disrupts eIF4E binding (17) is shown. Residues in red participate in the internal pseudoknot. (B) In vitro translation of WT and mutant gRNAs in WGE. (C) In vitro translation of ΔKΔPΔT, ΔKΔT and ΔKΔT/Pm2 gRNAs. (D) Trans-inhibition of WT gRNA translation in WGE. Fragments (10-fold molar ratio) containing either the WT PTE or PTE Pm2 were added to reaction mixtures containing WT gRNA. None, no added fragment. (E) Trans-inhibition of mutant gRNA translation in WGE. (F) Purified Arabidopsis eIF4E, eIF4G or eIF4F (200 nM) were added to WGE translation mixes containing ΔKΔPΔT or ΔKΔT gRNAs. None, no added translation factors. Data are from three independent experiments. Unpaired t-test was used to analyze the statistical significance for B and C, and one-way ANOVA was used to analyze the significance of D–F. One or two asterisks indicate statistical difference with P < 0.05 or 0.01, respectively.
The PTE inhibits translation by sequestering eIF4G
Although eIF4E is not a rate-limiting translation initiation factor in WGE, eIF4G is present at low levels and virtually all is tightly complexed with eIF4E (48) (K. Browning, personal communication). One possibility for why the PTE inhibited translation in the absence of the kl-TSS long-distance connection with the 5′ end was its ability to sequester eIF4G through interaction with PTE-bound eIF4E, leaving insufficient eIF4G for 3′CITE-independent translation. To determine the validity of this hypothesis, translation of ΔKΔPΔT was compared with translation of ΔKΔT with Pm2 (ΔK/Pm2/ΔT). As before, ΔKΔT reduced translation of p33 by 76% compared with ΔKΔPΔT. In contrast, translation of ΔK/Pm2/ΔT was not significantly reduced compared with ΔKΔPΔT (Figure 2C). This suggests that ability to bind eIF4E is needed for inhibition of translation by the PTE in cis. When fragments containing the PTE were added in trans (10-fold molar excess) to WT or mutant gRNAs, translation of WT gRNA was reduced by 34%, indicating that there was a limited supply of either eIF4E or (more likely) eIF4G (Figure 2D). ΔKΔPΔT translation was reduced by 77%, but translation of ΔKΔT was not significantly impacted since the PTE was also present in cis to bind to eIF4E (Figure 2E). In contrast, Pm2 fragments added in trans had no significant effect on translation of either WT gRNA, ΔKΔPΔT or ΔKΔT.
To determine if sequestration of eIF4E and/or eIF4G was responsible for translation inhibition by the PTE in cis, synthesis of p33 was assayed using templates ΔKΔPΔT and ΔKΔT supplemented with 200 nM of either Arabidopsis thaliana eIF4E, eIF4G or eIF4F (Figure 2F). None of the added eIFs had a significant impact on translation of ΔKΔPΔT, suggesting that, in the absence of the PTE, none of these eIFs were limiting in WGE. Addition of eIF4E had no significant effect on translation of ΔKΔT, but addition of eIF4G or eIF4F improved translation of ΔKΔT by 27 and 49%, respectively. These results suggest the WT PTE in trans or in cis (when the long-distance interaction is absent) inhibits translation due to sequestration of eIF4G through interaction with bound eIF4E.
PEMV2 outcompetes a cellular mRNA mimic in translation.
To test how efficiently PEMV2 with its 3′ kl-TSS/PTE combination competes for translation with other templates, competition assays were conducted with two well-characterized luciferase reporter constructs that translate efficiently in WGE but have different translation requirements (42,49). GloLuc contains the human β-globin 5′UTR and a 3′ 50 nt poly(A) tail, and transcripts were capped following synthesis thus mimicking a cellular mRNA. TZ10ΩLuc contains the TMV omega (Ω) translation enhancer sequence in its 5′UTR, which can direct cap-independent translation in an eIF4F-dependent manner (50) and the 3′UTR of tobacco mosaic virus (TMV) (42,50), which functionally replaces a poly(A) tail by binding to PABP (51,52).
The two reporter templates were translated in WGE either alone or in combination with PEMV2 at a 1:1 molar ratio. When PEMV2 was co-translated with capped GloLuc, GloLuc translation was reduced by 89%, whereas levels of p33 were reduced by only 26%. This indicates that one or more translation factor(s) required by both templates was limiting, and that under these conditions, PEMV2 is more efficiently translated than this capped/polyadenylated template (Figure 3, left panel). In contrast, co-translation of PEMV2 with TZ10ΩLuc reduced p33 and luciferase to a similar extent (34 and 46%, respectively) (Figure 3, right panel), suggesting that the 3′ PTE and 5′ Ω translation enhancers have similar efficiencies of translation. These data suggest that the kl-TSS/PTE of PEMV2 provides a competitive advantage to the viral template over canonical capped mRNAs, probably due to a higher affinity of the PTE for eIF4F.
Figure 3.
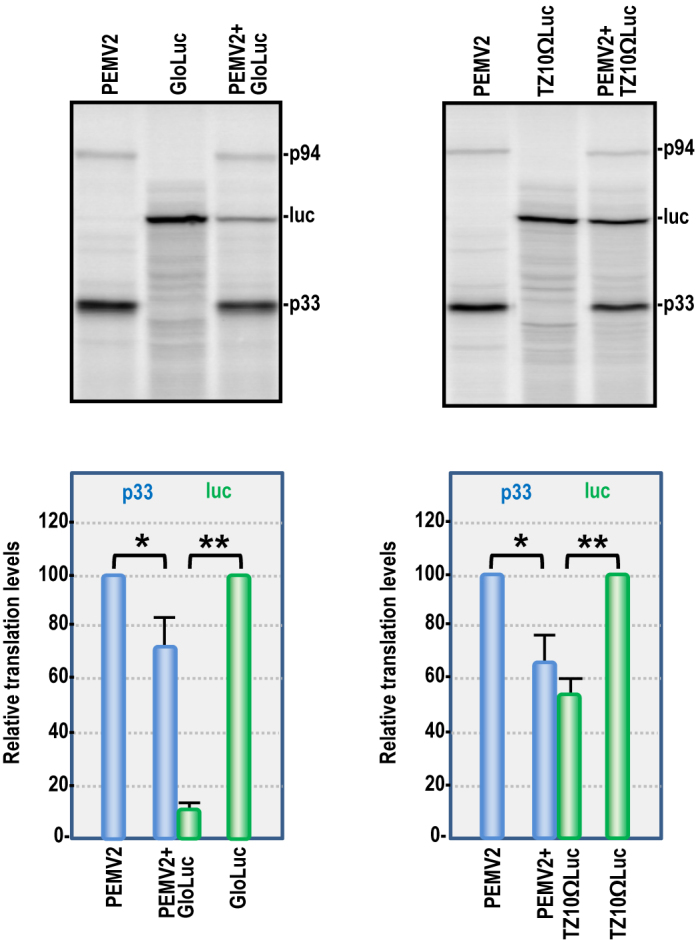
Competition for translation between PEMV2 and capped or uncapped reporter transcripts in WGE. PEMV2 (0.5 pmol) was translated either alone or with (0.5 pmol) of capped GloLuc (left) or uncapped TZ10ΩLuc (right). Data are from three independent experiments. Unpaired t-test was used for the statistical analysis. One or two asterisks indicate statistical difference with P < 0.05 or 0.01, respectively.
Transferring the long-distance interaction from the kl-TSS to the PTE
The PEMV2 PTE is unique compared with other Tombusvirid PTEs in that it is not directly involved in a long-distance interaction with the 5′ end (5,39). This suggests either that the PEMV2 PTE may be functionally incapable of participating in the kissing-loop interaction or that the ability may have been lost after the virus acquired the kl-TSS. We were interested in addressing whether the PEMV2 PTE is capable of a functional long-distance interaction, and if so, whether the remaining ribosome-binding activity of the kl-TSS is important for translation in vitro or in vivo. The PTE sequence that normally participates in the long-distance interaction is the apical loop of its 5′ SL1 hairpin (Figure 4A). The SL1 loop sequence in the six Tombusvirid PTEs that are known or predicted to participate in a long-distance interaction is ‘CUGCCA’ or the complement ‘UGGCA’, and three additional Tombusvirids have the shortened ‘GCCA’ or the complement ‘UGGC’ (5). The PEMV2 kl-TSS loop sequence contains the similar ‘UCGCCA’, and thus replacing the PEMV2 SL1 loop with the kl-TSS loop should generate a PTE with an interacting sequence similar to that found in other known PTEs.
Figure 4.
Transferring the long-distance interaction from the kl-TSS to the PTE. (A) Diagram of the alterations made to 5H2, kl-TSS and PTE. PTE with the kl-TSS loop sequence is denoted as PTE*. WT interacting sequences are denoted as red circles and presence of a mutation in the sequence is denoted by a blue outline. The PTE mutation (Pm2) that disrupts eIF4E binding is denoted by a gray circle with a blue outline. (B) Diagram of WT PEMV2 with interaction between the kl-TSS and 5H2 shown. (C) Diagrams of gRNA mutants generated. (D) In vitro translation of mutant gRNAs depicted in (C). Relative levels of p33 are quantified. (E) Relative virus accumulation in protoplasts. Total RNA was extracted at 24 h post-inoculation and analyzed by northern blotting. Data are from three independent experiments. For (D) and (E), unpaired t-test was used to analyze statistical significance. One or two asterisks indicate statistical difference with P < 0.05 or 0.01, respectively.
The PTE in ΔK or Km1 gRNA backgrounds was altered to contain the loop sequence of the kl-TSS 5′ hairpin (PTE*; Figure 4A), generating gRNAs ΔK/PTE* and Km1/PTE* (Figure 4C). In vitro translation assays indicated no significant difference between translation of WT gRNA, ΔK/PTE* or Km1/PTE* (Figure 4D), indicating that PTE* was fully capable of engaging in the long-distance interaction with 5′ end hairpin 5H2. In addition, the lack of significant difference in translation efficiency between ΔK/PTE* and Km1/PTE* indicates that the ribosome-binding activity of the kl-TSS is not contributing to translation in WGE. When the PTE* SL1 interacting sequence contained a single base mutation that eliminates the long-distance interaction (PTE*m1; Figure 4A) in the ΔK or Km1 gRNA backgrounds (ΔK/PTE*m1 and Km1/PTE*m1), translation was reduced to ΔK or Km1 levels (Figure 4D). When PTE* was altered to include mutations disrupting eIF4E binding (PTE*m2), translation in both the ΔK (ΔK/PTE*m2) or Km1 (Km1/PTE*m2) backgrounds was reduced by ∼4-fold.
As described above, both ΔK/PTE* and Km1/PTE* gRNAs were efficiently translated in WGE. To determine if ΔK/PTE* and Km1/PTE* were viable viruses in plant cells, both mutant gRNAs were inoculated independently into Arabidopsis protoplasts (Figure 4E). Twenty-four hours post-inoculation, ΔK/PTE* was undetectable by northern blots, whereas Km1/PTE* accumulated to 27% of WT PEMV2 levels (Figure 4E). Since disruption of the long-distance interaction in WT gRNA reduces virus accumulation to background levels (26), this result suggests that PTE* is capable of participating in the long-distance interaction in vivo, and that the kl-TSS (in the absence of its long-distance interacting capacity) contributes to virus viability.
To test whether the ribosome-binding activity of the kl-TSS and 3′TSS were not needed for translation in WGE because the extracts contained an abundance of ribosomes (39), in vitro translation reactions were performed in WGE that were partially depleted of ribosomes by addition of the CrPV IGR-IRES. CrPV IGR-IRES, which binds to 40S subunits with high affinity and subsequently can recruit the 60S subunit (53,54), is capable of directing translation in WGE (55). When the molar ratio of the CrPV IGR-IRES to WT PEMV2 gRNA in WGE was 5-fold or less, no inhibition of translation was observed (Supplementary Figure S1). However, when the molar ratio was increased to 20-fold, translation of WT PEMV2 gRNA was reduced by nearly 80% (Supplementary Figure S1). Using this level of added CrPV IGR-IRES, translation of PEMV2 gRNA in WGE remained unaffected by deletion of the 3′TSS (compare WT and ΔT, Supplementary Figure S1B) or deletion of the kl-TSS when the long-distance interaction was transferred to the PTE (Km1/PTE* versus ΔK/PTE*). These results indicate that ribosome-binding activity of the kl-TSS and 3′TSS are not important in WGE even in the presence of a limiting amount of ribosomes.
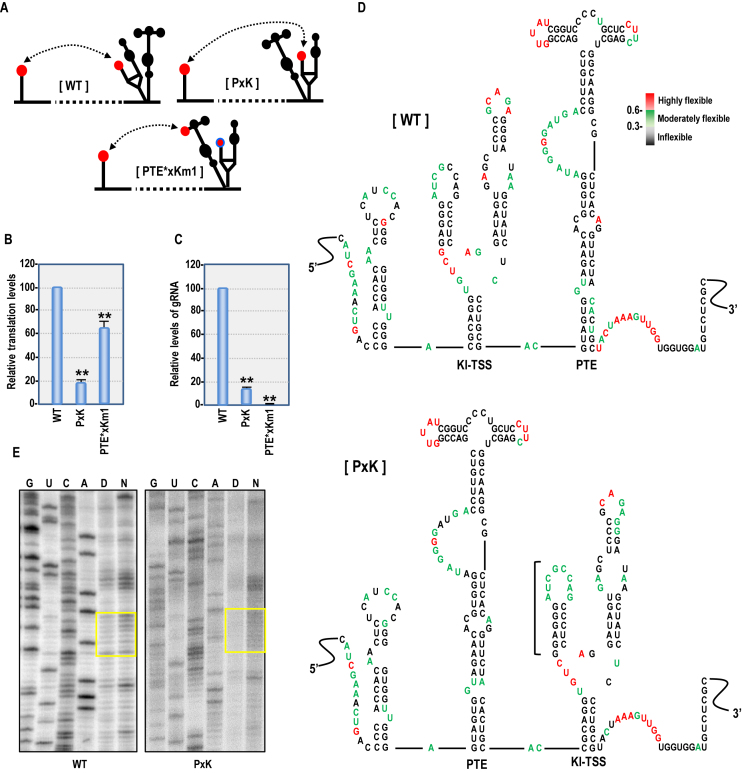
Importance of the kl-TSS and PTE spatial locations
To determine if relative location affects translation enhancement by the kl-TSS and PTE, their positions were reversed, generating mutant PxK (Figure 5A). PxK, which retained the kl-TSS long-distance interacting sequence, showed a 5-fold reduction in p33 synthesis in WGE compared with WT (Figure 5B), and a 7.8-fold reduction in levels of gRNA accumulating in protoplasts (Figure 4C). When PxK was altered to transfer the longdistance interaction from the kl-TSS to the now upstream PTE (PTE*xKm1), translation improved 3.6-fold in WGE (Figure 5B), but the mutant gRNA was not detectable in protoplasts (Figure 5C). Since reversal of the kl-TSS and PTE may have affected the structure of the two elements, SHAPE (selective 2′-hydroxyl acylation analyzed by primer extension) structure probing was used to investigate differences in nucleotide flexibility in the full-length WT and PxK gRNAs. The reactivity of each nucleotide to NMIA, which correlates with flexibility of the nucleotide, was quantified and colored in red (medium to high reactivity), green (low to medium reactivity), or black (little to no reactivity) (Figure 5D). Exchanging the positions of the PTE and kl-TSS caused mainly minor differences in flexibility in residues within the elements and surrounding sequences and did not alter their secondary structures (Figure 5D). However, the flexibilities of kl-TSS loop residues that participate in the long-distance interaction were significantly changed (Figure 5D and E). In WT gRNA, four of the eight kl-TSS residues in the loop that engages in the long-distance interaction exhibited low to moderate flexibility whereas in PxK, all eight residues showed low to moderately flexibility. This suggests that placing the kl-TSS downstream of the PTE reduced the ability of the loop to participate in the long-distance interaction with 5H2, and that specific spatial requirements may exist for 3′CITEs that connect with the 5′ end.
Figure 5.
Effect of exchanging positions of the kl-TSS and PTE. (A) Diagrams of WT and mutant gRNAs with exchanged positions of the kl-TSS and PTE. WT interacting sequences are denoted as red circles and mutant sequences have a blue outline. (B) In vitro translation of gRNAs in WGE. (C) Relative virus accumulation in protoplasts (see legend to Figure 4). (D) Secondary structures in the 3′UTR of WT and PxK gRNAs. Nucleotide flexibilities were determined using SHAPE structural probing and quantified using semiautomated footprinting analysis software. Nucleotides with reactivity of ≥0.6 are in red (medium to high flexibility), those with values ranging from 0.3 to 0.6 are green (low to medium flexibility) and those with values <0.3 are black (inflexible). (E) SHAPE phophorimages showing flexibility alterations in the terminal loop of the 5′ hairpin in the kl-TSS (boxed). G, U, C, A nucleotide ladder lanes; D, DMSO-treated control; N, NIMA-treated.
The kl-TSS and PTE enhance ribosome association with the initiation codon
To determine if the 3′CITEs affect translation by increasing ribosome association with the initiation site, toeprinting assays were conducted in the presence of the protein synthesis inhibitor CHX. Ribosomes stalled at the p33 initiation codon were monitored for their obstruction of reverse transcriptase-mediated primer extension, leading to characteristic cDNA toeprint band(s) 16–18 nt downstream of the initiation codon at the leading edge of the ribosome. WT gRNA produced a toeprint in the expected location that was absent when the initiation codon was altered from AUG to ACG (Figure 6). A second toeprint band was also found 7–8 nt downstream of the AUG, which may reflect improper fixing of the gRNA in the 40S binding cleft, allowing for further extension by the reverse transcriptase (56,57). Δ3′UTR and ΔKΔPΔT reduced ribosome toeprints to 27 and 18% of WT, respectively, similar to their reduction in translation efficiency (Figure 1C). ΔT, which did not affect p33 synthesis in WGE, had no significant effect on ribosome toeprinting. ΔP reduced toeprinting to 40% of WT levels whereas ΔK reduced toeprinting to 10% of WT. Overall, the toeprinting data were consistent with the in vitro translation results, strongly suggesting that translation enhancement by the kl-TSS and PTE is associated with translation initiation and ribosome occupancy at the p33 initiation codon.
Figure 6.
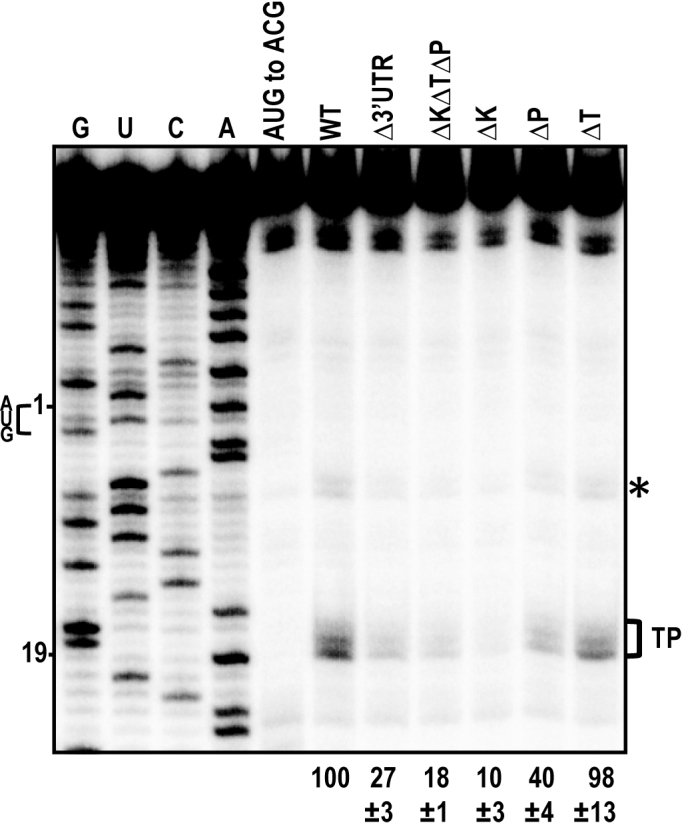
Toeprinting analysis of ribosomes associating with the p33 initiation codon. ΔK, ΔP and ΔT are deletions of the kl-TSS, PTE and TSS, respectively. AUG to ACG has a mutation in the p33 initiation codon. Bracket denotes the p33 AUG toeprint products and relative levels from three independent experiments are given at the bottom. Position of the A in the p33 initiation AUG codon is numbered 1, and numbering from this position is shown at left. G, U, C, A nucleotide ladder lanes. Asterisk denotes an additional toeprint that likely also corresponds with ribosomes occupying the p33 AUG.
The kl-TSS and PTE function by increasing the number of gRNAs available for translation
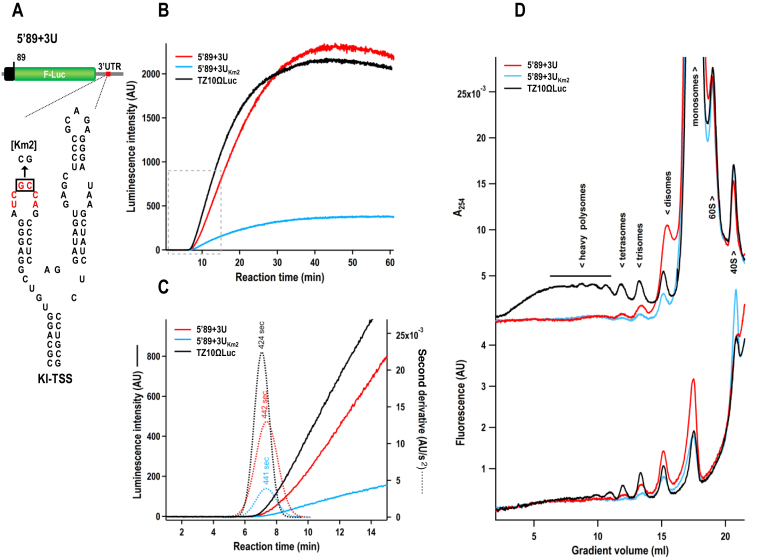
Translation rates can be determined from real-time translation curves by comparing the average time required to complete one round of protein synthesis (full-translation time), which includes scanning, initiation, elongation and termination (58). To determine if the kl-TSS and PTE change the duration of the translation epicycle, two reporter constructs were used: 5′89+3U, containing the PEMV2 5′ terminal 89 nt (including 5H2) followed by the firefly luciferase ORF and the PEMV2 3′UTR; and 5′89+3UKm2, which additionally contains two mutations in the kl-TSS that disrupt the long–distance interaction with 5H2 (Figure 7A). Uncapped transcripts of these reporter constructs, along with TZ10ΩLuc, which was used as an efficient translation control (42), were subjected to continuous in situ measurement of firefly luciferase activity in WGE. Over a 60 min translation period, 5′89+3U translated as efficiently as TZ1010Ωluc, while 5′89+3UKm2, which lacked the long-distance interaction, was translated poorly (Figure 7B). Translation kinetics in the first 15 min showed that all three reporter RNAs had a similar lag time (about 7.5 min) from the start of the reaction to the appearance of active luciferase (Figure 7C, solid lines). After the lag period, TZ10ΩLuc translated efficiently, with a sharp kink in the kinetic curve. 5′89+3U produced a curve similar to TZ10ΩLuc, with a slightly lower slope (Figure 7C). In contrast, 5′89+3UKm2 translated poorly with a much lower curve slope, demonstrating that the absence of the long-distance interaction substantially reduced luciferase synthesis (Figure 7C). This is consistent with translation of these reporter constructs in protoplasts as reported previously (26). The average full-translation time of the reporter RNAs was calculated by secondary-time derivatives of the smooth translation curves with Gaussian fitting (Figure 7C, dotted lines). 5′89+3U and 5′89+3UKm2 had similar full-translation times, 442 and 441 s, respectively, which was 17 and 18 s longer than TZ10ΩLuc (the effect of an extra 69 nt in the reading frame that prolonged the elongation stage). This suggests that the long-distance interaction does not affect the full-translation time, which includes the translation initiation rate.
Figure 7.
Kl-TSS and PTE increase the number of gRNAs available for translation, but have no effect on full-translation time and polysome distribution. (A) Luciferase construct 5′89+3U that contains the PEMV2 5′ terminal 89 nt and 3′UTR flanking firefly luciferase ORF. Secondary structure of kl-TSS and mutant Km2, which contains two mutations in the loop involved in the kissing-loop interaction. (B) Kinetic curves of real-time translation of firefly luciferase in WGE over a 60 min translation period. Uncapped RNAs tested were 5′89+3U, 5′89+3UKm2 and TZ10ΩLuc. (C) Second derivatives of the translation curves from the first 15 min. Solid lines represent the original kinetic curves of luciferase activity. Dashed lines represent the second derivatives of the original kinetic curves. The numbers shown above the dashed lines indicate the average full-translation time corresponding to the Gaussian peaks. (D) Polysome distribution on uncapped, fluorescein-labeled 5′89+3U, 5′89+3UKm2 and TZ10ΩLuc luciferase reporter constructs. RNAs (50 nM) were incubated in WGE for 25 min and then subjected to sedimentation analysis. Ultraviolet absorbance profiles (upper curves) reflect mainly the distribution of ribosomes. Fluorescence profiles (lower curves) represent allocation of the labeled reporter RNAs along gradients.
If the full-translation time is constant, the average number of translating ribosomes on a template depends on the initiation rate, i.e. on the frequency of ribosome recruitment by the RNA template, while the amplitudes of polysome peaks reflect the number of templates involved in translation (42). To determine whether the presence of the kl-TSS and PTE increases the number of templates translated and/or the number of ribosomes per template, fluorescein-labeled reporter RNAs were subjected to polysome sedimentation after 25 min in WGE and the distribution of ribosomes and reporter RNAs in sucrose gradient were determined by measuring ultraviolet absorbance (A254) (Figure 7D, upper panel) or fluorescence intensity (Figure 7D, lower panel), respectively. TZ10ΩLuc was associated with both light (<5 ribosomes per RNA) and heavy (>4 ribosomes) polysomes. A similar profile was observed for capped GloLuc transcripts, with the polysome distribution slightly shifted to the light region (Supplementary Figure S2). In contrast, 5′89+3U and 5′89+3UKm2 were mainly associated with monosomes and light polysomes, and both had similar profiles of absorbance peaks. Fluorescein-labeled 5′89+3UKm2 was also associated with a peak corresponding to 40S subunits, suggesting that in the absence of the long-distance interaction, 40S subunits were likely sequestered by the 3′PTE–eIF4E–eIF4G complex (Figure 7D, lower panel). With the exception of this 40S peak, all peak intensities for 5′89+3U were significantly higher than for 5′89+3UKm2, indicating that, while similar number of ribosomes were occupying both templates, more 5′89+3U templates were being translated. Combined with the toeprinting results (Figure 5) and full-translation times (Figure 7C), our results suggest that the kl-TSS and PTE cooperatively enhance levels of p33 synthesized in WGE by increasing the number of gRNAs available for translation, and not by affecting the translation initiation rate.
DISCUSSION
Unlike animal viruses, plant viruses require that infected cells survive the infection so that progeny virus can exit into adjacent cells through plasmodesmata. For this reason, plant RNA viruses lacking a 5′ cap do not encode products that interfere with cap-recognition, or significantly decrease the abundance of cellular mRNAs, but rather they must directly compete with ongoing cap-dependent translation. Possibly for this reason, many plant viruses have evolved 3′CITEs to enhance translation of the gRNA(s) and any 3′ co-terminal sgRNAs by directly attracting ribosomes or translation initiation factors, followed by their delivery to the 5′ end via long-distance RNA–RNA interactions (5,30). PEMV2 is unusual in possessing three 3′CITEs (17,26,27), including a PTE that is incapable of interacting with the 5′ end of the gRNA, a unique adjacent kl-TSS that provides the long-distance interaction and binds to ribosomes (26,34) and a 3′TSS that binds to 60S ribosomal subunits. The relationship between these 3′CITEs and the reason for their presence and placement within PEMV2 were not known.
The 3′TSS was previously found to be important for translation of reporter constructs in the absence of the kl-TSS and PTE when relocated near the termination codon and when assisted by a nearby sequence that, like the kl-TSS, can interact with 5′ hairpin 5H2 (27). Three hairpin/two pseudoknot structures similar to those that make up the PEMV2 3′TSS are located in identical positions in the 3′UTRs of umbraviruses Tobacco bushy top virus, Carrot mottle virus and Opium poppy mosaic virus; viruses that share little sequence/structure similarity with each other or with PEMV2 beyond this region (27,59). The 3′TSS, however, did not contribute to translation of PEMV2 gRNA in WGE (Figure 1). This suggests either that the 3′TSS does not enhance translation of the gRNA or that this 3′CITE is not functional in WGE. The structurally similar and identically located (relative to the 3′ end) TCV 3′TSS also did not enhance translation of reporter constructs (or the TCV gRNA; unpublished data) in WGE, but strongly enhanced translation of these constructs in protoplasts (28). Several other 3′CITEs are also non-functional in WGE (20,60,61), which may reflect differences in translation conditions between WGE and protoplasts. Interestingly, the PEMV2 3′TSS is important for translation of the PEMV2 sgRNA in WGE and sgRNA-encoded p26 is critical for PEMV2 viability in protoplasts (Gao, F., and Simon, A.E. (2017) Differential use of 3’ CITEs by the subgenomic RNA of Pea enation mosaic virus 2. Virology, in press). Since the 3′TSS enhances translation of the sgRNA in WGE, one possibility is that different 3′CITEs contribute to translation of the PEMV2 gRNA and sgRNA.
Unlike all other PTEs studied, the PEMV2 PTE does not participate in any discernible long-distance interaction; rather it relies on the kl-TSS’s kissing-loop interaction to deliver bound eIF4F to the gRNA 5′ end (26). When the sequence participating in the long-distance interaction was transferred from the kl-TSS to the PTE to allow the PTE to engage in the long-distance interaction similar to other known PTEs, the resultant PTE* was fully functional in vitro as a 3′CITE in WGE and the kl-TSS no longer contributed to translation enhancement (Figure 4D). This result indicates that both canonical PTE activities (eIF4E binding and long-distance kissing-loop interaction) are permissible in the PEMV2 PTE. The negligible effect of the kl-TSS (in the absence of its long-distance interaction) on translation of the gRNA in WGE was apparent both in the presence and absence of abundant levels of ribosomes (Supplementary Figure S1). However, mutation of the kl-TSS that eliminates the kissing-loop interaction (Km1) or complete deletion of the kl-TSS (ΔK) reduced virus accumulation in protoplasts to moderate (27% of WT) or undetectable levels, respectively, even in the presence of the compensating PTE* mutation (Figure 4E). This suggests that the kl-TSS ribosome-binding activity, or an additional property, contributes to virus fitness in vivo. Since the kl-TSS 5′ hairpin loop also engages in a long-distance interaction in the PEMV sgRNA, which is important for translation of critical sgRNA-encoded p26 (Gao, F., and Simon, A.E. (2017) Differential use of 3’ CITEs by the subgenomic RNA of Pea enation mosaic virus 2. Virology, in press), one possibility is that PTE* may not be as efficient as the kl-TSS in mediating the long-distance interaction for the sgRNA, leading to reduced p26 expression.
Exchanging the locations of the PTE and kl-TSS inhibited in vitro translation without significantly altering the structure of these elements. The position of the kl-TSS relative to PTE can either affect its ability to engage in the long-distance interaction, possibly due to steric hindrance of other nearby elements, or is important for proper location and orientation of PTE-bound eIF4F at the RNA 5′ end in the course of initiation. Our results indicate that the kl-TSS long-distance interaction may be compromised when the element is located downstream of the PTE, as kl-TSS 5′ loop residues that should be interacting with 5H2 sequence become more susceptible to NMIA, reflecting more single-stranded character in the ensemble RNAs.
eIF4G is the least abundant initiation factor in WGE and cannot be isolated independent of eIF4E (K. Browning, personal communication). For plant virus gRNAs whose translation depends on eIF4F-binding 3′CITEs, out competing host mRNAs for eIF4F is important since eIF4F is a rate-limiting factor for mRNA translation initiation (48). Some viruses generate nc sgRNAs that sequester eIF4G, thus regulating eIF4G-dependent translation (12,14,62). Other plant viruses, like PEMV2, combine eIF4G sequestration and translation enhancement activities on the same template utilizing the specific mechanism of eIF4G delivery from the 3′ end to the 5′ end of the gRNA. Thus, when the long-distance interaction is abrogated, translation of PEMV2 gRNA itself was significantly reduced in the presence of a functional 3′ PTE (Figure 2). We previously reported that mutations restricting the long-distance interaction between kl-TSS and 5H2 in luciferase reporter construct 5′89+3UKm2 led to poor translation in protoplasts (26). Although others (17) did not observe translation inhibition in either WGE or protoplasts using a luciferase reporter construct with the PEMV2 3′UTR and no discernable long-distance interaction, this could have resulted from the use of 6-fold fewer RNAs in the translation mixture, which may not have sequestered all eIF4F.
Our results, combined with a molar ratio of eIF4F to 40S ribosomal subunit of 0.03:1 (48) suggest that, in the absence of functional 3′CITEs and/or the long-distance interaction, eIF4G is still needed for translation of p33 through direct interaction with the 5′ end. Human eIF4G1 lacking the eIF4E-binding domain is important for translation of capped and uncapped mRNAs, as it binds to 5′ proximal unstructured sequences via its non-specific RNA-binding domain and recruits the 43S complex to initiate translation (63–65). Wheat eIF4G also has a non-specific RNA-binding domain (17,66) and thus may also have the ability to initiate translation independently of eIF4E by binding to 5′ unstructured sequences. Addition of the PTE sequence in trans at 10-fold molar excess inhibited translation of the WT gRNA by 34% and ΔKΔPΔT by nearly 5-fold. Addition of the PTE (in 100-fold excess) in trans was previously found to inhibit translation of a heterologous template by ∼4-fold (18). The difference in relative inhibition rates between WT PEMV2 gRNA and ΔKΔPΔT is likely the result of a tighter binding of eIF4F to the 3′ PTE than to the uncapped 5′ end, allowing WT gRNA to better compete with the trans-supplied PTE for binding to eIF4F compared with ΔKΔPΔT, which lacks a 3′ PTE. Moreover, PEMV2 gRNA was able to outcompete a capped, polyadenylated RNA for translation (Figure 3), suggesting that the combination of a 3′ PTE and long-distance bridge to the 5′ end gives the viral RNAs a competitive advantage over canonical-capped mRNAs for translation during infection. Translation enhancement and competition with cellular mRNAs through the binding of eIF4G to the 3′UTR is not an exceptional feature of plant viruses. Rotavirus mRNAs contain a special UGACC motif in their 3′UTRs that is recognized by the viral NSP3 protein, which, in turn, interacts with the N-terminus of eIF4G to stimulate viral mRNA translation (67,68). In addition, NSP3 displaces PABP from its complex with eIF4G, and thus inhibits host protein synthesis (67).
Compared with the PEMV2-based 5′89+3U luciferase reporter, mRNA molecules with 5′ caps (GloLuc) or the TMV Ω leader (TZ10ΩLuc) recruited ribosomes more frequently (Figure 7), suggesting at first glance that GloLuc and TZ10ΩLuc might have higher translation initiation rates. The TMV Ω sequence is located upstream of the initiation site, and enhances translation by recruiting eIF4F and heat shock protein 101 directly to the 5′UTR (50,69). In contrast, efficient delivery of eIF4F to the 5′ end of PEMV2 gRNA requires the PTE and regeneration of the long-distance kissing-loop interaction, which is disrupted each time a ribosome translates through the beginning of the p33 ORF. Before the next ribosome can be efficiently recruited, the long-distance interaction would have to be reformed, as proposed previously (70). Such a delay should result in a lower initiation rate and in the ‘dilution’ of translating ribosomes, spreading them more or less evenly along the coding sequence. The reason why a virus like PEMV2 deliberately reduces the ribosome density along the RNA could be to support ribosomal frameshifting necessary for RdRp synthesis. This programmed frameshifting is promoted by three hairpins conserved among umbraviruses (36). Being repeatedly melted by passing ribosomes, these structural elements have to restore their folding before the next translating ribosome arrives. The modulation of ribosome density could help to maintain the RNA structure for a sufficient period of time to provoke occasional frameshifting events. Such spreading of the ribosomes could be a common mechanism preserving important structural peculiarities that are needed for readthrough or frameshifting (depending on the virus) and thus provide for the translation of particular proteins. Nevertheless, despite fewer ribosomes engaging in translation of 5′89+3U compared with capped GloLuc (Figure 7), PEMV2 outcompeted the capped mRNA when co-translated in WGE (Figure 3), suggesting that PEMV2 is more effective than a 5′ cap in attracting translation components.
Interestingly, regardless of the predetermined infrequency of ribosome recruitment, the kl-TSS/PTE-promoted initiation event is an efficient process. This follows from the absence of significant differences between the full-translation times of 5′89+3U, 5′89+3UKm2 and TZ10ΩLuc. The duration of 3′CITE-mediated 40S delivery is similar to that of spontaneous cap-independent initiation and the recognition of a specific 5′ end enhancer of another plant virus. This makes it quite different from IRES-mediated initiation that was shown to be rather time consuming, at least in some cases (58).
The built-in limitation of ribosome recruitment frequency discussed above and the observation that disrupting the kissing-loop interaction did not significantly impact the overall shape of polysome distribution for 5′89+3UKm2 compared with 5′89+3U (Figure 7C) suggest that translation enhancement by the kl-TSS and PTE in WGE is not caused by increasing the translation initiation rate. Since the duration of translation elongation/termination is constant during translation assays (42), the translation enhancement observed could be achieved by increasing the number of viral RNAs translated. This hypothesis is supported by toeprinting, which showed that ΔP or ΔK mutations significantly reduced the number of ribosomes occupying the ensemble RNA p33 initiation codon (Figure 6) and by polysome sedimentation assays showing that peak intensities for 5′89+3U were significantly higher than for 5′89+3UKm2 (Figure 7C). Moreover, although 5′89+3U had less ribosome density along the RNA than TZ10ΩLuc (Figure 7C), it had a similar translation efficiency (Figure 7B), suggesting that more 5′89+3U templates are translated compared with TZ10ΩLuc. This is consistent with the fluorescence profiling data (Figure 7C, lower panel) and demonstrates that 3′CITEs enhance translation using a different mechanism from the TMV Ω translation enhancer. The non-specific interaction of eIF4G with an RNA’s 5′ end is not as efficient as its precise interaction with IRES or 3′CITEs (16,17,71). Thus, the presence of a functional translation enhancer like the PEMV2 PTE increases the opportunity for RNA association with eIF4F, allowing simultaneous translation of more viral RNAs. The absence of full-translation time prolongation, the moderate value of PTE–eIF4E interaction Kd (∼50 nM) (17,18) and limited inhibitory effect of PTE-containing fragments on the translation of WT gRNA (Figure 2D) indicate that binding of eIF4F to the PTE is neither rate limiting nor the event that unambiguously selects RNAs for translation. The reestablishment of the repeatedly broken long-distance kissing-loop interaction is responsible for the stable tempered initiation rate, but once being activated (the 3′–5′ interaction is formed) each viral RNA molecule can undergo a rapid translation initiation event with the assistance of pre-bound eIF4F or through binding one from a free pool. This model may be relevant to other 3′CITEs that enhance translation by binding to eIF4G or eIF4F, and delivering these factors to the 5′ end via a long-distance interaction that must be reformed each time a ribosome passes through, as it would ensure translation of more gRNAs released from virions once cells are infected.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Feng Gao for helpful discussions. We are very grateful to Karen Browning forArabidopsis translation factors.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Science Foundation (NSF) [MCB-1411836]; National Institutes of Health [R21AI117882–01 to A.E.S.]; Program ‘Molecular and Cell Biology’ of the Presidium of the Russian Academy of Sciences (to O.M.A.). Funding for open access charge: NSF Grant MCB-1411836 (to A.E.S).
Conflict of interest statement. None declared.
REFERENCES
- 1. Aitken C.E., Lorsch J.R.. A mechanistic overview of translation initiation in eukaryotes. Nat. Struct. Mol. Biol. 2012; 19:568–576. [DOI] [PubMed] [Google Scholar]
- 2. Shatsky I.N., Dmitriev S.E., Andreev D.E., Terenin I.M.. Transcriptome-wide studies uncover the diversity of modes of mRNA recruitment to eukaryotic ribosomes. Crit. Rev. Biochem. Mol. Biol. 2014; 49:164–177. [DOI] [PubMed] [Google Scholar]
- 3. Terenin I.M., Andreev D.E., Dmitriev S.E., Shatsky I.N.. A novel mechanism of eukaryotic translation initiation that is neither m(7)G-cap-, nor IRES-dependent. Nucleic Acids Res. 2013; 41:1807–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hellen C.U.T., Sarnow P.. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev. 2001; 15:1593–1612. [DOI] [PubMed] [Google Scholar]
- 5. Simon A.E., Miller W.A.. 3′ Cap-independent translation enhancers of plant viruses. Ann. Rev. Microbiol. 2013; 67:21–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shatsky I.N., Dmitriev S.E., Terenin I.M., Andreev D.E.. Cap- and IRES-independent scanning mechanism of translation initiation as an alternative to the concept of cellular IRESs. Mol. Cells. 2010; 30:285–293. [DOI] [PubMed] [Google Scholar]
- 7. Guo L., Allen E., Miller W.A.. Structure and function of a cap-independent translation element that functions in either the 3′ or the 5′ untranslated region. RNA. 2000; 6:1808–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shen R.Z., Miller W.A.. The 3′ untranslated region of tobacco necrosis virus RNA contains a barley yellow dwarf virus-like cap-independent translation element. J. Virol. 2004; 78:4655–4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jan E., Mohr I., Walsh D.. A cap-to-tail guide to mRNA translation strategies in virus-infected cells. Annu. Rev. Virol. 2016; 3:283–307. [DOI] [PubMed] [Google Scholar]
- 10. Bercovich-Kinori A., Tai J., Gelbart I.A., Shitrit A., Ben-Moshe S., Drori Y., Itzkovitz S., Mandelboim M., Stern-Ginossar N.. A systematic view on influenza induced host shutoff. Elife. 2016; 5:e18311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Abernathy E., Glaunsinger B.. Emerging roles for RNA degradation in viral replication and antiviral defense. Virology. 2015; 479:600–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Miller W.A., Shen R.Z., Staplin W., Kanodia P.. Noncoding RNAs of plant viruses and viroids: sponges of host translation and RNA interference machinery. Mol. Plant Microbe Interact. 2016; 29:156–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shen R.Z., Miller W.A.. Subgenomic RNA as a riboregulator: negative regulation of RNA replication by Barley yellow dwarf virus subgenomic RNA 2. Virology. 2004; 327:196–205. [DOI] [PubMed] [Google Scholar]
- 14. Iwakawa H.-o., Mizumoto H., Nagano H., Imoto Y., Takigawa K., Sarawaneeyaruk S., Kaido M., Mise K., Okuno T.. A viral noncoding RNA generated by cis-element-mediated protection against 5′- 3′ RNA decay represses both cap-independent and cap-dependent translation. J. Virol. 2008; 82:10162–10174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hashem Y., des Georges A., Dhote V., Langlois R., Liao H.Y., Grassucci R.A., Pestova T.V., Hellen C.U.T., Frank J.. Hepatitis-C-virus-like internal ribosome entry sites displace eIF3 to gain access to the 40S subunit. Nature. 2013; 503:539–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Treder K., Kneller E.L.P., Allen E.M., Wang Z.H., Browning K.S., Miller W.A.. The 3′ cap-independent translation element of Barley yellow dwarf virus binds eIF4F via the eIF4G subunit to initiate translation. RNA. 2008; 14:134–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang Z., Treder K., Miller W.A.. Structure of a viral cap-independent translation element that functions via high affinity binding to the eIF4E subunit of eIF4F. J. Biol. Chem. 2009; 284:14189–14202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang Z., Parisien M., Scheets K., Miller W.A.. The cap-binding translation initiation factor, elF4E, binds a pseudoknot in a viral cap-independent translation element. Structure. 2011; 19:868–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gazo B.M., Murphy P., Gatchel J.R., Browning K.S.. A novel interaction of cap-binding protein complexes eukaryotic initiation factor (eIF) 4F and eIF(iso)4F with a region in the 3′-untranslated region of satellite tobacco necrosis virus. J. Biol. Chem. 2004; 279:13584–13592. [DOI] [PubMed] [Google Scholar]
- 20. Nicholson B.L., Wu B., Chevtchenko I., White K.A.. Tombusvirus recruitment of host translational machinery via the 3′ UTR. RNA. 2010; 16:1402–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nicholson B.L., Zaslaver O., Mayberry L.K., Browning K.S., White K.A.. Tombusvirus Y-shaped translational enhancer forms a complex with eIF4F and can be functionally replaced by heterologous translational enhancers. J. Virol. 2013; 87:1872–1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Browning K.S. The plant translational apparatus. Plant Mol. Biol. 1996; 32:107–144. [DOI] [PubMed] [Google Scholar]
- 23. Martin F., Barends S., Jaeger S., Schaeffer L., Prongidi-Fix L., Eriani G.. Cap-assisted internal initiation of translation of histone H4. Mol. Cell. 2011; 41:197–209. [DOI] [PubMed] [Google Scholar]
- 24. Wells S.E., Hillner P.E., Vale R.D., Sachs A.B.. Circularization of mRNA by eukaryotic translation initiation factors. Mol. Cell. 1998; 2:135–140. [DOI] [PubMed] [Google Scholar]
- 25. Kahvejian A., Svitkin Y.V., Sukarieh R., M’Boutchou M.N., Sonenberg N.. Mammalian poly(A)-binding protein is a eukaryotic translation initiation factor, which acts via multiple mechanisms. Genes Dev. 2005; 19:104–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gao F., Kasprzak W., Stupina V.A., Shapiro B.A., Simon A.E.. A ribosome-binding, 3′ translational enhancer has a T-shaped structure and engages in a long-distance RNA-RNA interaction. J. Virol. 2012; 86:9828–9842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gao F., Kasprzak W.K., Szarko C., Shapiro B.A., Simon A.E.. The 3′ untranslated region of Pea enation mosaic virus contains two T-shaped, ribosome-binding, cap-independent translation enhancers. J. Virol. 2014; 89:11696–11712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Stupina V.A., Meskauskas A., McCormack J.C., Yingling Y.G., Shapiro B.A., Dinman J.D., Simon A.E.. The 3′ proximal translational enhancer of Turnip crinkle virus binds to 60S ribosomal subunits. RNA. 2008; 14:2379–2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Stupina V.A., Yuan X., Meskauskas A., Dinman J.D., Simon A.E.. Ribosome binding to a 5′ translational enhancer is altered in the presence of the 3′ untranslated region in cap-independent translation of turnip crinkle virus. J. Virol. 2011; 85:4638–4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nicholson B.L., White K.A.. Functional long-range RNA-RNA interactions in positive-strand RNA viruses. Nat. Rev. Microbiol. 2014; 12:493–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rakotondrafara A.M., Polacek C., Harris E., Miller W.A.. Oscillating kissing stem-loop interactions mediate 5′ scanning-dependent translation by a viral 3′-cap-independent translation element. RNA. 2006; 12:1893–1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Das Sharma S., Kraft J.J., Miller W.A., Goss D.J.. Recruitment of the 40S ribosomal subunit to the 3′-untranslated region (UTR) of a viral mRNA, via the eIF4 complex, facilitates cap-independent translation. J. Biol. Chem. 2015; 290:11268–11281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Miras M., Sempere R.N., Kraft J.J., Miller W.A., Aranda M.A., Truniger V.. Interfamilial recombination between viruses led to acquisition of a novel translation-enhancing RNA element that allows resistance breaking. New Phytol. 2014; 202:233–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gao F., Gulay S.P., Kasprzak W., Dinman J.D., Shapiro B.A., Simon A.E.. The kissing-loop T-shaped structure translational enhancer of pea enation mosaic virus can bind simultaneously to ribosomes and a 5′ proximal hairpin. J. Virol. 2013; 87:11987–12002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Demler S.A., Rucker D.G., de Zoeten G.A.. The chimeric nature of the genome of pea enation mosaic virus: the independent replication of RNA 2. J. Gen. Virol. 1993; 74:1–14. [DOI] [PubMed] [Google Scholar]
- 36. Gao F., Simon A.E.. Multiple cis-acting elements modulate programmed-1 ribosomal frameshifting in pea enation mosaic virus. Nucleic Acids Res. 2016; 44:878–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ryabov E.V., Robinson D.J., Taliansky M.. Umbravirus-encoded proteins both stabilize heterologous viral RNA and mediate its systemic movement in some plant species. Virology. 2001; 288:391–400. [DOI] [PubMed] [Google Scholar]
- 38. Simon A.E. 3′UTRs of carmoviruses. Virus Res. 2015; 206:27–36. [DOI] [PubMed] [Google Scholar]
- 39. Chattopadhyay M., Shi K., Yuan X., Simon A.E.. Long-distance kissing loop interactions between a 3′ proximal Y-shaped structure and apical loops of 5′ hairpins enhance translation of saguaro cactus virus. Virology. 2011; 417:113–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Liu H., Naismith J.H.. An efficient one-step site-directed deletion, insertion, single and multiple-site plasmid mutagenesis protocol. BMC Biotech. 2008; 8:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Patrick R.M., Mayberry L.K., Choy G., Woodard L.E., Liu J.S., White A., Mullen R.A., Tanavin T.M., Latz C.A., Browning K.S.. Two Arabidopsis loci encode novel eukaryotic initiation factor 4E isoforms that are functionally distinct from the conserved plant eukaryotic initiation factor 4E. Plant Phys. 2014; 164:1820–1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Alekhina O.M., Vassilenko K.S., Spirin A.S.. Translation of non-capped mRNAs in a eukaryotic cell-free system: acceleration of initiation rate in the course of polysome formation. Nucleic Acids Res. 2007; 35:6547–6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. McCormack J.C., Simon A.E.. Cur. Prot. Microbiol. 2005; 16: [Google Scholar]
- 44. Du Z.Y., Chen A.Z., Chen W.H., Liao Q.S., Zhang H.M., Bao Y.M., Roossinck M.J., Carr J.P.. Nuclear-cytoplasmic partitioning of cucumber mosaic virus protein 2b determines the balance between its roles as a virulence determinant and an RNA-silencing suppressor. J. Virol. 2014; 88:5228–5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kuhlmann M.M., Chattopadhyay M., Stupina V.A., Gao F., Simon A.E.. An RNA element that facilitates programmed ribosomal readthrough in turnip crinkle virus adopts multiple conformations. J. Virol. 2016; 90:8575–8591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Das R., Laederach A., Pearlman S.M., Herschlag D., Altman R.B.. SAFA: semi-automated footprinting analysis software for high-throughput quantification of nucleic acid footprinting experiments. RNA. 2005; 11:344–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Laederach A., Das R., Vicens Q., Pearlman S.M., Brenowitz M., Herschlag D., Altman R.B.. Semiautomated and rapid quantification of nucleic acid footprinting and structure mapping experiments. Nat. Protoc. 2008; 3:1395–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Browning K.S., Humphreys J., Hobbs W., Smith G.B., Ravel J.M.. Determination of the amounts of the protein-synthesis initiation and elongation-factors in wheat-germ. J. Biol. Chem. 1990; 265:17967–17973. [PubMed] [Google Scholar]
- 49. Shirokov V.A., Kommer A., Kolb V.A., Spirin A.S.. Grandi G. Methods in Molecular Biology. 2007; 375:2nd edn, Totowa: Humana Press Inc; 19–55. [DOI] [PubMed] [Google Scholar]
- 50. Gallie D.R. The 5′-leader of tobacco mosaic virus promotes translation through enhanced recruitment of eIF4F. Nucleic Acids Res. 2002; 30:3401–3411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Gallie D.R. The CAP and poly(A) tail function synergistically to regulate messenger-RNA translational efficiency. Genes Dev. 1991; 5:2108–2116. [DOI] [PubMed] [Google Scholar]
- 52. Gallie D.R., Walbot V.. RNA pseudoknot domain of tobacco mosaic virus can functionally substitute for a poly(A) tail in plant and animal cells. Genes Dev. 1990; 4:1149–1157. [DOI] [PubMed] [Google Scholar]
- 53. Jan E., Sarnow P.. Factorless ribosome assembly on the internal ribosome entry site of cricket paralysis virus. J. Mol. Biol. 2002; 324:889–902. [DOI] [PubMed] [Google Scholar]
- 54. Murray J., Savva C.G., Shin B.-S., Dever T.E., Ramakrishnan V., Fernandez I.S.. Structural characterization of ribosome recruitment and translocation by type IV IRES. Elife. 2016; 5:e13567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wilson J.E., Powell M.J., Hoover S.E., Sarnow P.. Naturally occurring dicistronic cricket paralysis virus RNA is regulated by two internal ribosome entry sites. Mol. Cell. Biol. 2000; 20:4990–4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Battiste J.L., Pestova T.V., Hellen C.U.T., Wagner G.. The eIF1A solution structure reveals a large RNA-binding surface important for scanning function. Mol. Cell. 2000; 5:109–119. [DOI] [PubMed] [Google Scholar]
- 57. Pisareva V.P., Pisarev A.V., Komar A.A., Hellen C.U.T., Pestova T.V.. Translation initiation on mammalian mRNAs with structured 5′ UTRs requires DExH-Box protein DHX29. Cell. 2008; 135:1237–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Vassilenko K.S., Alekhina O.M., Dmitriev S.E., Shatsky I.N., Spirin A.S.. Unidirectional constant rate motion of the ribosomal scanning particle during eukaryotic translation initiation. Nucleic Acids Res. 2011; 39:5555–5567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. McCormack J.C., Yuan X., Yingling Y.G., Kasprzak W., Zamora R.E., Shapiro B.A., Simon A.E.. Structural domains within the 3′ untranslated region of Turnip crinkle virus. J. Virol. 2008; 82:8706–8720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wu B.D., White K.A.. A primary determinant of cap-independent translation is located in the 3′-proximal region of the tomato bushy stunt virus genome. J. Virol. 1999; 73:8982–8988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kraft J.J., Treder K., Peterson M.S., Miller W.A.. Cation-dependent folding of 3′ cap-independent translation elements facilitates interaction of a 17-nucleotide conserved sequence with eIF4G. Nucleic Acids Res. 2013; 41:3398–3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Shen R., Rakotondrafara A.M., Miller W.A.. Trans regulation of cap-independent translation by a viral subgenomic RNA. J. Virol. 2006; 80:10045–10054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Ali I.K., McKendrick L., Morley S.J., Jackson R.J.. Truncated initiation factor eIF4G lacking an eIF4E binding site can support capped mRNA translation. EMBO J. 2001; 20:4233–4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Prevot D., Decimo D., Herbreteau C.H., Roux F., Garin J., Darlix J.L., Ohlmann T.. Characterization of a novel RNA-binding region of eIF4GI critical for ribosomal scanning. EMBO J. 2003; 22:1909–1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Terenin I.M., Dmitriev S.E., Andreev D.E., Royall E., Belsham G.J., Roberts L.O., Shatsky I.N.. A cross-kingdom internal ribosome entry site reveals a simplified mode of internal ribosome entry. Mol. Cell. Biol. 2005; 25:7879–7888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Gallie D.R. Cap-independent translation conferred by the 5′ leader of tobacco etch virus is eukaryotic initiation factor 4G dependent. J. Virol. 2001; 75:12141–12152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Piron M., Vende P., Cohen J., Poncet D.. Rotavirus RNA-binding protein NSP3 interacts with eIF4GI and evicts the poly(A) binding protein from eIF4F. EMBO J. 1998; 17:5811–5821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Vende P., Piron M., Castagne N., Poncet D.. Efficient translation of rotavirus mRNA requires simultaneous interaction of NSP3 with the eukaryotic translation initiation factor eIF4G and the mRNA 3′ end. J. Virol. 2000; 74:7064–7071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Wells D.R., Tanguay R.L., Le H., Gallie D.R.. HSP101 functions as a specific translational regulatory protein whose activity is regulated by nutrient status. Genes Dev. 1998; 12:3236–3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Fabian M.R., White K.A.. Analysis of a 3′-translation enhancer in a tombusvirus: a dynamic model for RNA-RNA interactions of mRNA termini. RNA. 2006; 12:1304–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Lomakin I.B., Hellen C.U.T., Pestova T.V.. Physical association of eukaryotic initiation factor 4G (eIF4G) with eIF4A strongly enhances binding of elF4G to the internal ribosomal entry site of encephalomyocarditis virus and is required for internal initiation of translation. Mol. Cell. Biol. 2000; 20:6019–6029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.