Abstract
Botulinum neurotoxins (BoNTs) are causative agents of the life-threatening disease botulism. They are naturally produced by species of the bacteria Clostridium botulinum as stable and non-covalent complexes, in which the BoNT molecule is assembled with several auxiliary non-toxic proteins. Some BoNT serotypes, represented by the well-studied BoNT serotype A (BoNT/A), are produced by Clostridium strains that carry the ha gene cluster, which encodes four neurotoxin-associated proteins (NTNHA, HA17, HA33, and HA70) that play an important role to deliver and protect BoNTs in the gastrointestinal tract during oral intoxication. In contrast, BoNT/E- and BoNT/F-producing strains carry a distinct gene cluster that encodes five proteins (NTNHA, P47, OrfX1, OrfX2, and OrfX3, termed the orfX cluster). The structures and functions of these proteins remain largely unknown. Here, we report the crystal structure of P47 resolved at 2.8 Å resolution. Surprisingly, P47 displays a structural topology that is similar to bactericidal/permeability38 increasing (BPI) like proteins, which were previously identified only in eukaryotes. The similarity of a hydrophobic cleft of P47 with the phospholipid40 binding groove of BPI suggests that P47 might be involved in lipid association to exert its function. Consistently, P47 associates and induces aggregation of asolectin-containing liposomes in a protein- and lipid-concentration dependent manner. These findings laid the foundation for future structural and functional studies of the potential roles of P47 and OrfX proteins in facilitating oral intoxication of BoNTs.
Graphical abstract

1. Introduction
Botulinum neurotoxins (BoNTs) are among the most poisonous toxins, causing the life-threatening disease botulism. They specifically target the presynaptic neurons at neuromuscular junctions, where they degrade SNAREs (soluble N-ethylmaleimide-sensitive factor attachment protein receptors) to block neurotransmission and cause muscle paralysis. BoNTs are classified by Centers for Disease Control and Prevention (CDC) as tier I select agents due to their potential use as bioweapons. Nevertheless, BoNT/A is one of the top selling drugs that is widely used for the treatment of several neuromuscular diseases and as popular cosmetics.
There are seven major serotypes of BoNTs (designated A to G), among which BoNT/A, B, E and F are detected in human patients (Rossetto et al., 2014). Naturally occurring BoNT is synthesized together with several nontoxic neurotoxin-associated proteins (NAPs) in the form of a progenitor toxin complex (PTC). All BoNTs carry non-toxic-non-haemagglutinin (NTNHA) that forms a 1:1 heterodimer with BoNT, the minimally functional PTC (M-PTC) (Gu et al., 2012; Sagane et al., 2012; Gu & Jin, 2013; Matsui et al., 2014; Eswaramoorthy et al., 2016). NTNHA is crucial to protect the toxin in the acidic and protease-rich gastrointestinal (GI) environment during oral intoxication. In addition, BoNT serotypes A1, B, C, D, G, which are encoded in the ha toxin gene cluster, are produced alongside with hemaglutinin-17, 33 and 70 (HA17, 33, 70), which further assemble with the M-PTC to form the large PTC (L-PTC) (Amatsu et al., 2013; Benefield et al., 2013; Lee et al., 2013; Lee et al., 2014; Yao et al., 2014). For BoNT/A, the HA proteins assemble into a dodecameric ~470 kDa complex that facilitates absorption of BoNT/A in the intestine by recognizing cell-surface glycans on the intestinal epithelium and disrupting the integrity of the E-cadherin-mediated epithelial cell junction (Sugawara et la., 2010; Lee et al., 2014; Lee et al., 2015; Lam & Jin, 2015). Intriguingly, bacteria that express BoNT/E, F, and A2-A4 do not encode HA proteins (Jacobson et al., 2008; Popoff & Connan, 2014; Williamson et al., 2016). Instead they carry a distinct gene cluster, known as the orfX toxin gene cluster that encodes NTNHA as well as four other proteins (OrfX1, 2, 3, and P47). The structural and functional roles of these proteins remain poorly understood.
P47 is a hypothetical protein with a molecular weight of ~47 kDa, whose gene is positioned upstream of ntnha in the orfX gene cluster in BoNT/E1-E12 producing C. botulinum (Li et al., 1998; Hill & Smith, 2013). P47 is the only gene co-transcribed with ntnha and bont/E and is selectively expressed at the transition phase together with bont/E (Dineen et al., 2004; Kubota et al., 1998), suggesting they may be involved in BoNT/E function. C. botulinum transformed with p47 antisense mRNA did not affect BoNT/E synthesis, indicating it unlikely serves as a regulator of the operon (Couesnon et al., 2006). P47 was detected in the crude extract of BoNT/A2-producing C. botulinum, suggesting it might be secreted, although this is yet to be reported for BoNT/E or F (Lin et al., 2010; Hines et al., 2005).
Further upstream of p47 are three hypothetical proteins known as OrfX1, 2, and 3 encoded in a separate operon of C. botulinum. It was suggested that these proteins encoded in the orfX cluster may directly or indirectly interact with M-PTC/E (Kukreja & Singh 2007), but the physiological relevance of such interactions and the biological functions of OrfXs and P47 remain mysterious. Interestingly, amino acid sequence analyses of OrfX2 and OrfX3 showed that they have internal segments that share similarities to P47, suggesting that they may carry a P47-like protein fold (Lin et al., 2010). Therefore, we decided to focus our research on P47 first, which will provide new insights into the function of the whole orfX gene cluster.
2. Materials and Methods
2.1 Cloning, expression and purification
Genomic DNA of Clostridium botulinum E1 strain Beluga (Accession code: ACSC01000002) was isolated. P47 gene (locus_tag CLO_2650) was amplified by PCR using the corresponding primers and cloned into the pQE (Qiagen) expression vector with an N-terminal His6 tag fused to a thrombin cleavage site and a C-terminal Strep tag, yielding pH6tP47-ES. The recombinant H6tP47-ES (Protein ID ZP_04823541.1) was expressed in the E. coli strain BL21-star (DE3) (Calsbad, CA, USA). Bacteria were grown at 37 °C in LB (Luria-Bertani) medium containing 100 µg /ml ampicillin. The temperature was decreased to 18 °C when OD600 reached 0.4 – 0.6. Expression was induced with 0.5 mM isopropyl-β-D-thiogalactopyranoside (IPTG) for 16 hrs. The cells were harvested by centrifugation and stored at - 20 °C until use. The selenomethionine (Se-Met) labeled H6tP47-ES was expressed as previously described (Van Duyne et al., 1993). Bacteria were grown in M9 minimal media supplemented with amino acids (lysine, threonine, phenylalanine, leucine, isoleucine, and valine) and Se-Met.
For purification of native and Se-Met labeled pH6tP47-ES, proteins were bound to a Ni-NTA (nitrilotriacetic acid, Qiagen) affinity column in a buffer containing 50 mM Tris, pH 8.0, 400 mM NaCl, 10 mM imidazole, and 1 mM TCEP, and subsequently eluted in the same buffer containing 300 mM imidazole. The eluted fractions were pooled and dialyzed for 16 hrs at 4 °C against a buffer containing 20 mM Tris, pH 8.8, 20 mM NaCl, and 1 mM TCEP and then further purified by MonoQ ion-exchange chromatography (GE Healthcare) in a buffer containing 20 mM Tris, pH 8.8 and eluted with a NaCl gradient. The peak fractions were then subjected to Superdex 200 size- exclusion chromatography (SEC) in a buffer containing 20 mM Hepes, pH 6.8, 150 mM NaCl, 1 mM TCEP and subsequently concentrated to ~7 mg/ml using Amicon Ultra centrifugal filters (Millipore) and stored at −80 °C until use.
2.2 Crystallization and X-ray diffraction data collection
Initial crystallization screens for P47 with the intact His- and Strep-tag were carried out using a Gryphon crystallization robot (Art Robbins Instrument, Sunnyvale, CA, USA) and commercial high-throughput crystallization screen kits from Hampton Research or Qiagen. After extensive manual optimization, the best P47 crystals were grown by hanging-drop vapor diffusion at 18 °C, in which the protein (2.5 mg/ml) was mixed in 1:1 (v/v) ratio with a reservoir solution containing 12 % Polyethylene glycol (PEG) 6000 (Hampton Research), 0.1 M Hepes, pH 7.5. Micro-seeding was employed to obtain single crystals.
The P47 crystals were cryoprotected in the original mother liquor supplemented with 15 % (v/v) ethylene glycol and flash-frozen in liquid nitrogen. The crystals belong to space group P41212, with unit cell dimensions of a = b = 74.08 Å, c = 308.18 Å; α = β = γ = 90°. The best native data set was collected at 2.8 Å. The X-ray diffraction data were collected at 100 K at beam line 24-ID-E, Advanced Photon Source (APS), using detector ADSC Q315. All data sets were processed and scaled using XDS (Kabsch 2010) and CCP4i (Potterton et al., 2003). Data collection statistics are summarized in Table 1.
Table 1.
Data collection and refinement statistics.
| Se-Met P47 | P47 | |
|---|---|---|
| Data collection | ||
| Wavelength (Å) | 0.97918 | 0.97918 |
| Resolution (Å) | 51.34–2.88 (3.04–2.88) | 43.32–2.80 (2.9–2.8) |
| Space group | P 41212 | P 41212 |
| Cell dimensions | ||
| a, b, c (Å) | 74.1, 74.1, 308 | 74.1, 74.1, 308.2 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 |
| Completeness (%) | 100.0 (100.0) | 99.08 (100.0) |
| Redundancy | 6.8 (6.9) | 5.5 (5.6) |
| Rmerge (%) | 15.9 (143.3) | 10.2 (166.3) |
| Mean I/σ(I) | 14.4 (2.4) | 12.48 (1.6) |
| Autosol FOM | 0.281 | |
| Refinement | ||
| Resolution (Å) | 43.32–2.80 | |
| No. reflections | 21, 917 | |
| Rwork/Rfree (%) | 23.10/27.48 | |
| No. atoms | ||
| Protein | 3, 250 | |
| Ligand/ion | 0 | |
| Water | 0 | |
| B-factors (Å2) | ||
| Protein | 105.4 | |
| Ligand/ion | - | |
| Water | - | |
| r.m.s. deviations | ||
| Bond lengths (Å) | 0.009 | |
| Bond angles (°) | 1.246 |
Values in parentheses are for the highest resolution shell.
2.3 Structure determination
Using the Se-Met derivative crystals, we solved the structure of P47 to 2.8 Å by SAD (single-wavelength anomalous dispersion) using PHENIX (Adams et al., 2010). Except for the first Met, 8 out of 9 Se atoms were found that yielded the SAD-phased map with a figure of merit of 0.281. The manual model building and refinements were performed in COOT (Emsley & Cowtan, 2004) and PHENIX (Adams et al., 2010) in an iterative manner. The refinement progress was monitored with the free R value using a 5% randomly selected test set (Brunger 1992). P47 structure was refined to 2.8 Å with Rwork/Rfree = 0.239 / 0.275. The structure was validated through the MolProbity web server (Chen et al., 2010) and showed excellent stereochemistry. Structural refinement statistics are listed in Table 1. The coordinate and diffraction data have been deposited in the Protein Data Bank under accession code 5WIX. All structure figures were prepared with PyMol (http://www.pymol.org).
2.4 Protein melting assay
The thermal stability of P47 was measured using a fluorescence-based thermal shift assay on a StepOne real-time PCR machine (Life Technologies). The protein (~2 µM) was mixed with the fluorescent dye SYPRO Orange (Sigma-Aldrich) immediately before the experiment. The samples were heated from 25 °C to 90 °C over 45 mins. The midpoint of t he protein-melting curve (Tm) was determined using the analysis software provided by the instrument manufacturer. Data obtained from three independent experiments were averaged to generate the bar graph.
2.5 Preparation of liposomes
Liposomes were prepared by extrusion method using Avanti Mini Extruder according to manufacturer’s protocol. Briefly, lipids containing 80 % asolectin (Sigma-Aldrich) and 20 % cholesterol (Avanti Polar Lipid) were dissolved in chloroform and mixed. The lipid mixture was dried under nitrogen gas and placed under vacuum for 2 hrs. The dried lipids were rehydrated and were subjected to five rounds of freezing and thawing cycles. Liposomes were prepared by extruding through 200 nm filter membrane.
2.6 Turbidity measurements
Optical density measurements were performed at 25 °C on P47-lipid mixtures. P47 at different concentration (0–200 nM) was mixed with 0.2 mM liposome in a buffer containing 150 mM NaCl, 10 mM Hepes, pH 7. Optical density of the solution was continuously monitored at 450 nm (Spectramax M2e, Molecular Devices) for 10 mins. The measured intensities were corrected from the absorbance of the buffer. The experiment was duplicated.
2.7 Co-sedimentation experiment
The protein-liposome co-sedimentation experiment was performed by mixing 400 nM of P47 with varying concentrations of liposomes diluted in the buffer (150 mM NaCl, 10 mM Hepes, pH 7) for 10 mins at room temperature. The mixture was subjected to centrifugation at ~21, 000 g for 10 mins. The supernatant was removed and the pellet was re-suspended in the same buffer for SDS-PAGE analysis. The experiment was triplicated.
3. Result
3.1 Recombinant production and characterization of P47
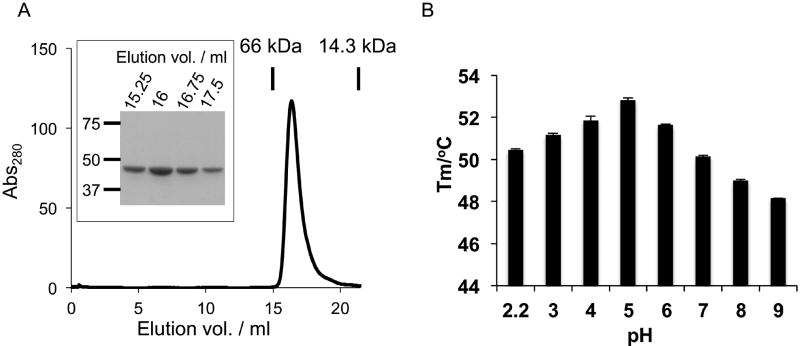
P47 fused with an N-terminal His6-tag and a C-terminal Strep tag was overexpressed in Escherichia coli and was purified subsequently by Ni-NTA affinity chromatography, anion exchange chromatography, and size-exclusion chromatography (SEC). The purified P47 is highly homogeneous and exists as a monomer as judged by SDS-PAGE and SEC analyses (Figure 1A). We next examined the stability of P47 using a thermal shift assay, where the thermal denaturation of the protein was examined by the fluorescence dye Sypro orange. Sypro orange is an environmental sensitive dye that binds to exposed hydrophobic surfaces on a partially unfolded protein and therefore is useful for measuring the protein stability (Niesen et al., 2007). We found that P47 is stable over a wide range of pH between 2.2 and 9 (Figure 1B). Interestingly, P47 displays the highest melting temperature (Tm) of 52.8 °C at pH 5. These data demonstrate that the recombinant P47 is pure, stable, and amenable for structural studies.
Fig. 1. Biochemical characterization of P47.
(A) SEC analysis of the recombinant P47. P47 is homogeneously monomeric with purity higher than 95 % judged by SDS-PAGE (Boxed). (B) Thermo stability of P47 as a function of pH. The data are presented as mean ± S.D., n = 3.
3.2 Crystal structure of P47
We next carried out structural studies of P47. After performing extensive crystallization screening and optimization, single crystals of P47 were obtained under a condition containing 12 % PEG 6000 and 0.1 M Hepes, pH 7.5. The best crystal diffracted to 2.8 Å. Since there was no known protein homologous to P47 based on the amino acid sequence analyses, we solved the phase problem by single-wavelength anomalous dispersion (SAD), using selenomethionine (SeMet) derived P47 crystals. Eight Se atoms were identified in the SAD-phased map with a figure of merit of 0.28. One protein molecule was identified in the asymmetric unit. The structure of P47 is well resolved except for solvent-exposed residues 114-119, which have no visible electron densities likely due to their high structural flexibility. The final model is refined to Rwork and Rfree of 23.9% and 27.5% with good geometry (Table 1).
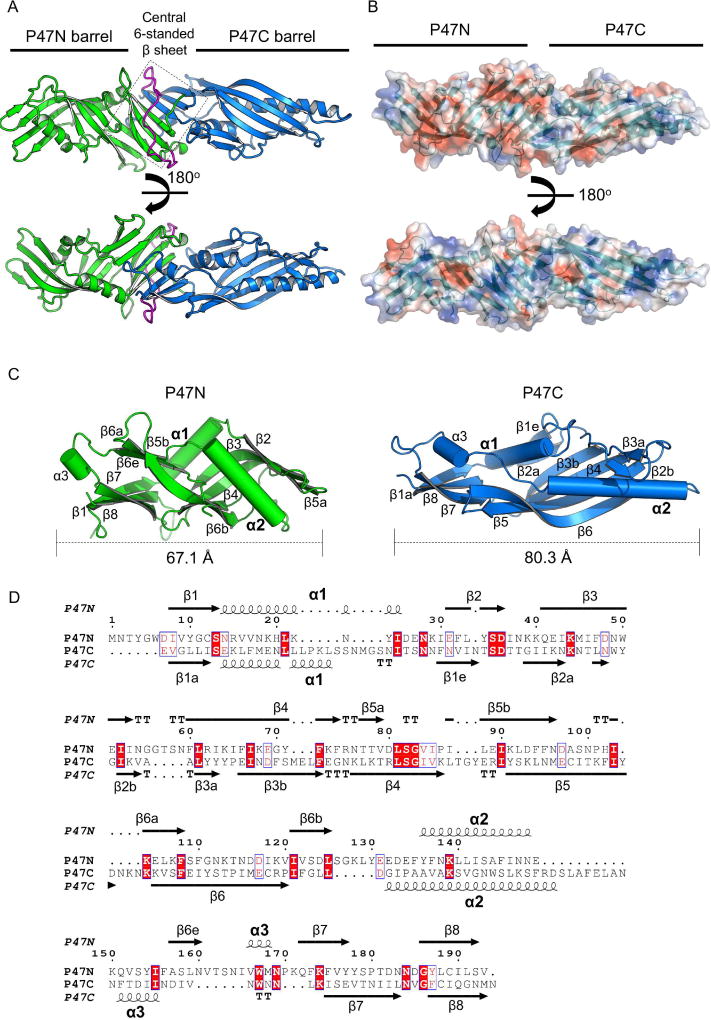
The P47 structure adopts an elongated tubular shape with a dimension of ~122.3 Å × 40.5 Å × 36.7 Å (Figure 2A). P47 can be divided into two major domains: an N-terminal domain (P47N) covering residues M1–V193 and a C- terminal domain (P47C) covering residues E216–N416, which are connected by a loop (V194–S215). These two domains share a similar topology despite their low sequence identity of ~11.7 % (Figure 2C, D). P47N and P47C each has an incomplete β-barrel composed of five twisted anti-parallel β-strands (β2-β6), which surrounds two amphipathic α-helices (α1-α2) (Figure 2C). The two incomplete β-barrels are connected through a central β-sheet platform that includes three anti-parallel β-strands from each domain.
Fig. 2. Crystal structure of P47.
(A) P47 has two major domains (green and blue) that are connected by a flexible loop (magenta). Three anti-parallel strands from each domain interact to form a six-stranded β-sheet in the middle of P47 (highlighted in a box). (B) Molecular surface of P47 is shown and colored according to the electrostatic potential (±3kT/e, red: negative; blue: positive) that was calculated using APBS (Baker et al., 2001). P47N is more electronegative than P47C. (C) Structural comparison of P47N and P47C. The secondary structures are labeled to highlight the similar topology of these two domains. The two β-strands that are unique to P47N and P47C are labeled as β6e and β1e, respectively. (D) Sequence alignment of P47N and P47C performed by PROMALS3D (Pei et al., 2008) and displayed using ESPript 3.0 (Robert & Gouet, 2014). Their secondary structures are placed on the top and the bottom, respectively. Identical residues are white-colored and red-boxed. Conserved residues are red-colored and blue-framed.
Despite the similarity in overall organization of the secondary structures, P47N and P47C are poorly superimposable with a root-mean-square- deviation (rmsd) of ~5.3 Å over 132 Cα atom pairs (DaliLite v.3). Most noticeable, P47N is more compact than P47C (~67 and 80 Å in length, respectively), and the helices α1 and α2 are “squeezed” into an almost perpendicular orientation in P47N. Moreover, the five-stranded β-barrel platform of P47N is highly twisted and bulged compared to that in P47C. We also notice that the two domains show significantly different isoelectric points (pI ~5.8 and 7.9 for P47N and P47C, respectively), as well as different electrostatic surface properties, where P47N has a more negative charged surface (Figure 2B).
3.3 P47 shares a structural-fold with BPI-like protein family
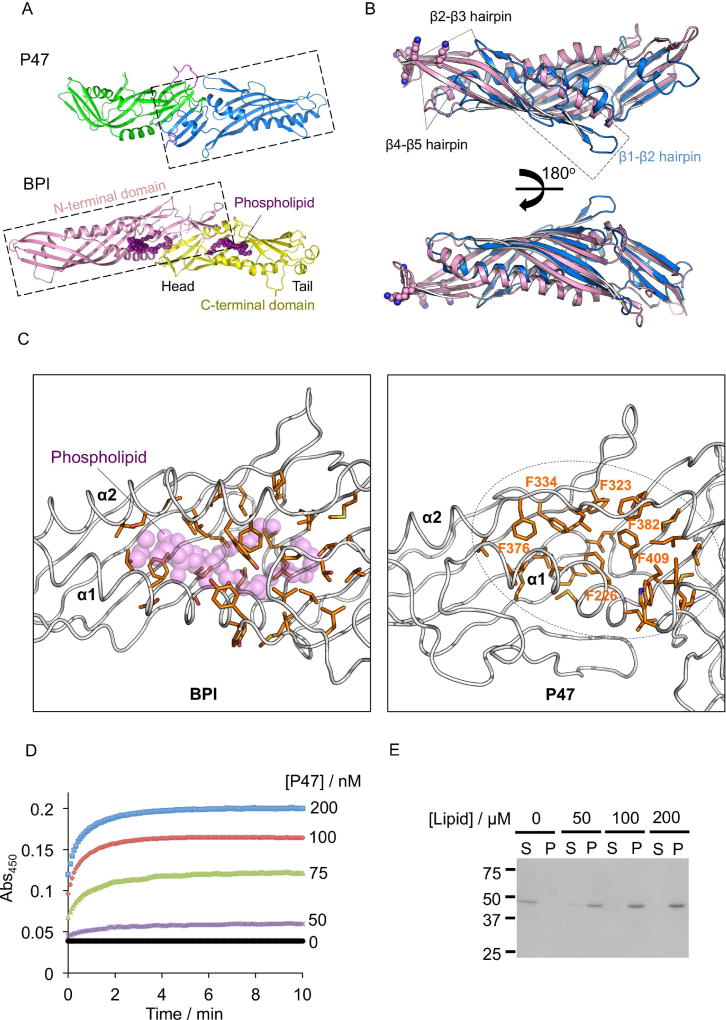
The lack of sequence and structural similarity between P47 and the HA proteins of BoNT/A implicated that P47 may have a distinct function that is currently unknown (Lee et al., 2013; Lam & Jin, 2015). To look for the potential function of P47, we performed pairwise structural comparison between P47 and other proteins with known crystal structures deposited in the Protein Data Bank (PDB) using DaliLite v.3 (Holm & Laakso 2016) and FATCAT (Li et al., 2006). Interestingly, both results showed that P47 is distantly related to the members of BPI-like protein family of the Tulip (tubular lipid-binding) domain superfamily (Kopec et a., 2011; Alva & Lupas, 2016). The closest structural homologs to P47 are BPI (Dali Z score: 11.8), LBP (lipopolysaccharide binding protein) (Z score: 11.5), and CETP (Cholesteryl ester transfer protein) (Z score: 11.2) (Figure 3A), but P47 has very low primary sequence identity to these proteins (~10 %).
Fig. 3. Structural comparison of P47 with BPI reveals a potential lipid-binding pocket.
(A) The P47 domains are colored the same as in Fig. 2. The N- and C- terminal domains of BPI (PDB code: 1BP1) are colored pink and yellow respectively. The phospholipid is represented as purple spheres. (B) Structural alignment of P47C with the N-terminal domain of BPI (DaliLite v.3). β1-β2 hairpin of P47, β2-β3 and β4- β5 hairpins of BPI and P47 are marked. Positively charged residues on BPI that are crucial for LPS binding are shown as spheres. (C) Hydrophobic residues in BPI that interact with the bound phospholipid are shown as sticks (left panel). The corresponding region in P47 is shown in the right panel, where six phenylalanine residues occupy the potential lipid-binding pocket. (D) P47 induces aggregation of asolectin-liposome. P47 at indicated concentration was added to 0.2 mM liposomes, and the changes of solution turbidity were measured. (E) Protein-liposome aggregates were analyzed by SDS-PAGE. 400 nM of P47 was incubated with liposomes at indicated concentration and the aggregates were isolated by centrifugation. The supernatant (S) and the pellet fractions (P) were separated and analyzed by SDS-PAGE.
BPI-like proteins are lipid-binding proteins, although they have distinct functions. Among them, BPI and LBP are lipopolysaccharide (LPS)-binding proteins that are involved in innate immunity defense against gram-negative bacteria; CETP functions in the transport of lipids between lipoprotein particles in the bloodstream (Weiss 2003). These proteins are mostly studied in mammals and their homologs have only been reported in eukaryotes (Krasity et al., 2011).
Structurally, BPI-like proteins are composed of two domains in a head-to-head orientation and the two domains have a similar Tulip domain topology (Figure 3A) (Beamer et al., 1997, 2008; Eckert et al., 2013). Superposition of P47 with BPI revealed that P47C shares a clear structural homology with the N- terminal domain of BPI with rmsd ~3.2 Å over 123 Cα pairs (PDB code: 1BP1, Beamer et al., 1997) (Figure 3B), while the more compact P47N did not align well with BPI. Notably, the β-barrels of P47C and N-terminal domain of BPI are largely superimposable except for two distinct regions. First, BPI has more extended β2-β3 and β4-β5 hairpins compared with P47C (Figure 3B). These two hairpins contain conserved positively charged residues at the tip that are important for LPS-binding of BPI and LBP (Beamer et al., 1998). Therefore P47 does not seem to have a LPS-binding site. On the other hand, P47C has an extra β1e-β2 hairpin that packs against the core β-barrel that is not found in BPI.
Interestingly, the crystal structures of all three BPI-like proteins (BPI, LBP, and CETP) contain lipid molecules bound at hydrophobic clefts, which are situated between the N-terminal and C-terminal barrels and the central β-sheet with α1 and α2 forming the entrance of the pocket (Beamer et al., 1997) (Figure 3A & C). Detailed structural analysis showed that the corresponding region in P47C is predominantly occupied by several hydrophobic residues including six phenylalanine (Figure 3C). We suspect that rearrangement of the α1 and α2 in P47 would permit the exposure of the hydrophobic cleft for potential ligand binding. The structural similarities between P47 and BPI indicate that P47 might have a function related to lipid association, which is the direction for further experimental work.
3.4 P47 associates with asolectin-containing liposome
Inspired by our structural finding, we would like to experimentally explore the potential P47-lipid interaction. As a proof-of-concept, we examined the association of P47 with liposomes composed of asolectin and cholesterol. Asolectin contains a mixture of different phospholipids, therefore is a commonly used model to study protein-lipid interaction especially when the lipid-binding specificity of the target protein is unknown. Notably, mixing P47 with liposomes at neutral pH caused an immediate change in turbidity of the solution due to liposome aggregation. We monitored the time course of solution turbidity change by measuring absorbance at wavelength 450 nm and found that the degree of turbidity increased with protein concentration, suggesting the vesicle aggregation was induced by P47 (Figure 3D). We also pelleted the liposome aggregates by centrifugation and analyzed them by SDS-PAGE (Figure 3E). P47 was clearly co-sedimented with liposomes in a lipid-concentration dependent manner, further supporting the P47-lipid interaction.
4. Discussion
Here, we report the first crystal structure of P47, encoded in the orfX toxin gene cluster of BoNT/E producing C. botulinum. Surprisingly, we found that P47 shares a structural fold that is similar to BPI-like proteins, and P47 may also have a similar function in lipid binding. To the best of our knowledge, this is the first example showing that the BPI-like proteins are not restricted to eukaryotes. Based on the annotation by Pfam protein families database (Bateman et al., 2002), P47 is a representative member of the Clostridium_P47 protein family (Accession code: pfam06597). The related protein members could be found in Pseudomonas putida, Nitrobacter winogradskyi and Streptomyces scabiei, further suggesting that the BPI-like fold exists across various microbes.
BPI and LBP are important in host defense by binding LPS of gram-negative bacteria. However, structural analysis showed that P47 does not have a homologous LPS-binding site. We speculate that there may be two possible functional roles of P47 in BoNT intoxication that are worthy of further study. First, P47 might modulate the membrane integrity of the C. botulinum cell membrane to facilitate the secretion of BoNT and NTNHA, as these proteins are co-expressed at the transition phase. Second, P47 may directly or indirectly interact with the host cells to facilitate intestinal absorption of the toxin complexes during BoNT oral intoxication, similar to the function of the HA proteins. Interestingly, amino acid sequence analysis showed that part of OrfX2 and OrfX3 may adopt a P47-like fold. Since Tulip domain could mediate protein-protein interactions to form higher order complexes (AhYoung et al., 2015), it is possible that P47 and OrfX2/OrfX3 that carry a P47-like fold could interact with each other. It also raised another intriguing question if P47 and OrfX proteins may interact with BoNT (Kalb et al., 2017), which should be of great interest for future research.
Supplementary Material
Highlights.
The first crystal structure of a hypothetical protein P47 in the orfX toxin gene cluster from human pathogenic Clostridium botulinum E1 strain Beluga is solved.
P47 shares a conserved structural topology with bactericidal/permeability-increasing (BPI) like protein.
P47 is the first protein found in the bacteria kingdom that has a Tulip domain fold.
P47 might bind phospholipid to exert its function
Acknowledgments
This work was partly supported by National Institute of Allergy and Infectious Diseases (NIAID) grants R01AI091823 (R.J.), R01AI125704 (R.J.), and R21AI123920 (R.J.); and by the Swiss Federal Office for Civil Protection BABS #353005630 (A.R.). NE-CAT at the Advanced Photon Source (APS) is supported by a grant from the National Institute of General Medical Sciences (P41 GM103403). The Pilatus 6M detector on 24-ID-C beam line is funded by a NIH-ORIP HEI grant (S10 RR029205). Use of the APS, an Office of Science User Facility operated for the U.S. Department of Energy (DOE) Office of Science by Argonne National Laboratory, was supported by the U.S. DOE under Contract No. DE-AC02-06CH11357. Some crystal screens were performed at Stanford Synchrotron Radiation Lightsource, SLAC National Accelerator Laboratory, which is supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract No. DE- AC02-76SF00515. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research, and by the National Institutes of Health, National Institute of General Medical Sciences (including P41GM103393).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Ethical Statement
No animal was used in this study.
References
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D. Biol. Crystallogr. 2010;66:213–21. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AhYoung AP, Jiang J, Zhang J, Khoi Dang X, Loo JA, Zhou ZH, Egea PF. Conserved SMP domains of the ERMES complex bind phospholipids and mediate tether assembly. Proc. Natl. Acad. Sci. 2015;112:E3179–E3188. doi: 10.1073/pnas.1422363112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alva V, Lupas AN. The TULIP superfamily of eukaryotic lipid-binding proteins as a mediator of lipid sensing and transport. Biochim. Biophys. Acta - Mol. Cell Biol. Lipids. 2016;1861:913–923. doi: 10.1016/j.bbalip.2016.01.016. [DOI] [PubMed] [Google Scholar]
- Amatsu S, Sugawara Y, Matsumura T, Kitadokoro K, Fujinaga Y. Crystal Structure of Clostridium botulinum Whole Hemagglutinin Reveals a Huge Triskelion-shaped Molecular Complex. J. Biol. Chem. 2013;288:35617–35625. doi: 10.1074/jbc.M113.521179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. U. S. A. 2001;98:10037–41. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beamer LJ, Carroll SF, Eisenberg D. Crystal structure of human BPI and two bound phospholipids at 2.4 angstrom resolution. Science. 1997;276:1861–4. doi: 10.1126/science.276.5320.1861. [DOI] [PubMed] [Google Scholar]
- Beamer LJ, Carroll SF, Eisenberg D. The BPI/LBP family of proteins: A structural analysis of conserved regions. Protein Sci. 2008;7:906–914. doi: 10.1002/pro.5560070408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benefield DA, Dessain SK, Shine N, Ohi MD, Lacy DB. Molecular assembly of botulinum neurotoxin progenitor complexes. Proc. Natl. Acad. Sci. 2013;110:5630–5635. doi: 10.1073/pnas.1222139110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brünger AT. Free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature. 1992;355:472–5. doi: 10.1038/355472a0. [DOI] [PubMed] [Google Scholar]
- Chen VB, Arendall WB, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couesnon A, Raffestin S, Popoff MR. Expression of botulinum neurotoxins A and E, and associated non-toxin genes, during the transition phase and stability at high temperature: analysis by quantitative reverse transcription-PCR. Microbiology. 2006;152:759–70. doi: 10.1099/mic.0.28561-0. [DOI] [PubMed] [Google Scholar]
- Dineen S, Bradshaw M, Karasek CE, Johnson EA. Nucleotide sequence and transcriptional analysis of the type A2 neurotoxin gene cluster in Clostridium botulinum. FEMS Microbiol. Lett. 2004;235:9–16. doi: 10.1016/j.femsle.2004.04.002. [DOI] [PubMed] [Google Scholar]
- Eckert JK, Kim YJ, Kim JI, Gürtler K, Oh D-Y, Sur S, Lundvall L, Hamann L, van der Ploeg A, Pickkers P, Giamarellos-Bourboulis E, Kubarenko AV, Weber AN, Kabesch M, Kumpf O, An H-J, Lee J-O, Schumann RR. The crystal structure of lipopolysaccharide binding protein reveals the location of a frequent mutation that impairs innate immunity. Immunity. 2013;39:647–60. doi: 10.1016/j.immuni.2013.09.005. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. Sect. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Eswaramoorthy S, Sun J, Li H, Singh BR, Swaminathan S. Molecular Assembly of Clostridium botulinum progenitor M complex of type E. Sci. Rep. 2016;5:17795. doi: 10.1038/srep17795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu S, Rumpel S, Zhou J, Strotmeier J, Bigalke H, Perry K, Shoemaker CB, Rummel A, Jin R. Botulinum Neurotoxin Is Shielded by NTNHA in an Interlocked Complex. Science (80-.) 2012;335:977–981. doi: 10.1126/science.1214270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu S, Jin R. Assembly and Function of the Botulinum Neurotoxin Progenitor Complex. Current Topics in Microbiology and Immunology. 2013:21–44. doi: 10.1007/978-3-642-33570-9_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill KK, Smith TJ. Genetic Diversity Within Clostridium botulinum Serotypes, Botulinum Neurotoxin Gene Clusters and Toxin Subtypes. Current Topics in Microbiology and Immunology. 2012:1–20. doi: 10.1007/978-3-642-33570-9_1. [DOI] [PubMed] [Google Scholar]
- Hines HB, Lebeda F, Hale M, Brueggemann EE. Characterization of Botulinum Progenitor Toxins by Mass Spectrometry. Appl. Environ. Microbiol. 2005;71:4478–4486. doi: 10.1128/AEM.71.8.4478-4486.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L, Laakso LM. Dali server update. Nucleic Acids Res. 2016;44:W351–W355. doi: 10.1093/nar/gkw357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson MJ, Lin G, Raphael B, Andreadis J, Johnson EA. Analysis of neurotoxin cluster genes in Clostridium botulinum strains producing botulinum neurotoxin serotype A subtypes. Appl. Environ. Microbiol. 2008;74:2778–86. doi: 10.1128/AEM.02828-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. XDS. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalb S, Baudys J, Smith T, Smith L, Barr J. Characterization of Hemagglutinin Negative Botulinum Progenitor Toxins. Toxins (Basel) 2017;9:193. doi: 10.3390/toxins9060193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopec KO, Alva V, Lupas AN. Bioinformatics of the TULIP domain superfamily: Figure 1. Biochem. Soc. Trans. 2011;39:1033–1038. doi: 10.1042/BST0391033. [DOI] [PubMed] [Google Scholar]
- Krasity BC, Troll JV, Weiss JP, McFall-Ngai MJ. LBP/BPI proteins and their relatives: conservation over evolution and roles in mutualism. Biochem. Soc. Trans. 2011;39:1039–1044. doi: 10.1042/BST0391039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kukreja RV, Singh BR. Comparative Role of Neurotoxin-Associated Proteins in the Structural Stability and Endopeptidase Activity of Botulinum Neurotoxin Complex Types A and E †. Biochemistry. 2007;46:14316–14324. doi: 10.1021/bi701564f. [DOI] [PubMed] [Google Scholar]
- Lam K-H, Jin R. Architecture of the botulinum neurotoxin complex: a molecular machine for protection and delivery. Curr. Opin. Struct. Biol. 2015;31:89–95. doi: 10.1016/j.sbi.2015.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, Zhong X, Gu S, Kruel AM, Dorner MB, Perry K, Rummel A, Dong M, Jin R. Molecular basis for disruption of E-cadherin adhesion by botulinum neurotoxin A complex. Science (80-.) 2014;344:1405–1410. doi: 10.1126/science.1253823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, Gu S, Jin L, Le TTN, Cheng LW, Strotmeier J, Kruel AM, Yao G, Perry K, Rummel A, Jin R. Structure of a Bimodular Botulinum Neurotoxin Complex Provides Insights into Its Oral Toxicity. PLoS Pathog. 2013;9:e1003690. doi: 10.1371/journal.ppat.1003690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, Lam K-H, Kruel AM, Perry K, Rummel A, Jin R. High-resolution crystal structure of HA33 of botulinum neurotoxin type B progenitor toxin complex. Biochem. Biophys. Res. Commun. 2014;446:568–573. doi: 10.1016/j.bbrc.2014.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, Lam K-H, Kruel A-M, Mahrhold S, Perry K, Cheng LW, Rummel A, Jin R. Inhibiting oral intoxication of botulinum neurotoxin A complex by carbohydrate receptor mimics. Toxicon. 2015;107:43–49. doi: 10.1016/j.toxicon.2015.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Qian X, Sarkar HK, Singh BR. Molecular characterization of type E Clostridium botulinum and comparison to other types of Clostridium botulinum. Biochim. Biophys. Acta. 1998;1395:21–7. [PubMed] [Google Scholar]
- Li Z, Ye Y, Godzik A. Flexible Structural Neighborhood--a database of protein structural similarities and alignments. Nucleic Acids Res. 2006;34:D277–D280. doi: 10.1093/nar/gkj124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin G, Tepp WH, Pier CL, Jacobson MJ, Johnson EA. Expression of the Clostridium botulinum A2 neurotoxin gene cluster proteins and characterization of the A2 complex. Appl. Environ. Microbiol. 2010;76:40–7. doi: 10.1128/AEM.01882-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui T, Gu S, Lam K, Carter LG, Rummel A, Mathews II, Jin R. Structural Basis of the pH-Dependent Assembly of a Botulinum Neurotoxin Complex. J. Mol. Biol. 2014;426:3773–3782. doi: 10.1016/j.jmb.2014.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niesen FH, Berglund H, Vedadi M. The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat. Protoc. 2007;2:2212–21. doi: 10.1038/nprot.2007.321. [DOI] [PubMed] [Google Scholar]
- Pei J, Kim B-H, Grishin NV. PROMALS3D: a tool for multiple protein sequence and structure alignments. Nucleic Acids Res. 2008;36:2295–2300. doi: 10.1093/nar/gkn072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirazzini M, Rossetto O, Eleopra R, Montecucco C. Botulinum Neurotoxins: Biology, Pharmacology, and Toxicology. Pharmacol. Rev. 2017;69:200–235. doi: 10.1124/pr.116.012658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popoff MR, Connan C. Molecular Aspects of Botulinum Neurotoxin. Springer; New York, New York, NY: 2014. Absorption and Transport of Botulinum Neurotoxins; pp. 35–68. [Google Scholar]
- Potterton E, Briggs P, Turkenburg M, Dodson E. A graphical user interface to the CCP4 program suite. Acta Crystallogr D. Biol. Crystallogr. 2003;59:1131–7. doi: 10.1107/s0907444903008126. [DOI] [PubMed] [Google Scholar]
- Robert X, Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014;42:W320–4. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossetto O, Pirazzini M, Montecucco C. Botulinum neurotoxins: genetic, structural and mechanistic insights. Nat. Rev. Microbiol. 2014;12:535–549. doi: 10.1038/nrmicro3295. [DOI] [PubMed] [Google Scholar]
- Sagane Y, Miyashita S-I, Miyata K, Matsumoto T, Inui K, Hayashi S, Suzuki T, Hasegawa K, Yajima S, Yamano A, Niwa K, Watanabe T. Small-angle X-ray scattering reveals structural dynamics of the botulinum neurotoxin associating protein, nontoxic nonhemagglutinin. Biochem. Biophys. Res. Commun. 2012;425:256–260. doi: 10.1016/j.bbrc.2012.07.077. [DOI] [PubMed] [Google Scholar]
- Sugawara Y, Matsumura T, Takegahara Y, Jin Y, Tsukasaki Y, Takeichi M, Fujinaga Y. Botulinum hemagglutinin disrupts the intercellular epithelial barrier by directly binding E-cadherin. J. Cell Biol. 2010;189:691–700. doi: 10.1083/jcb.200910119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Duyne GD, Standaert RF, Karplus PA, Schreiber SL, Clardy J. Atomic Structures of the Human Immunophilin FKBP-12 Complexes with FK506 and Rapamycin. J. Mol. Biol. 1993;229:105–124. doi: 10.1006/jmbi.1993.1012. [DOI] [PubMed] [Google Scholar]
- Weiss J. Bactericidal/permeability-increasing protein (BPI) and lipopolysaccharide-binding protein (LBP): structure, function and regulation in host defence against Gram-negative bacteria. Biochem. Soc. Trans. 2003;31:785–90. doi: 10.1042/bst0310785. [DOI] [PubMed] [Google Scholar]
- Williamson CHD, Sahl JW, Smith TJ, Xie G, Foley BT, Smith LA, Fernández RA, Lindström M, Korkeala H, Keim P, Foster J, Hill K. Comparative genomic analyses reveal broad diversity in botulinum-toxin-producing Clostridia. BMC Genomics. 2016;17:180. doi: 10.1186/s12864-016-2502-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao G, Lee K, Gu S, Lam K-H, Jin R. Botulinum Neurotoxin A Complex Recognizes Host Carbohydrates through Its Hemagglutinin Component. Toxins (Basel) 2014;6:624–635. doi: 10.3390/toxins6020624. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.