Abstract
PARP1-dependent poly-ADP-ribosylation (PARylation) participates in the repair of many forms of DNA damage. Here, we used atomic force microscopy (AFM) and single molecule fluorescence microscopy to examine the interactions of PARP1 with common DNA repair intermediates. AFM volume analysis indicates that PARP1 binds to DNA at nicks, abasic (AP) sites, and ends as a monomer. Single molecule DNA tightrope assays were used to follow the real-time dynamic behavior of PARP1 in the absence and presence of AP endonuclease (APE1) on AP DNA damage arrays. These experiments revealed that PARP1 conducted damage search mostly through 3D diffusion. Co-localization of APE1 with PARP1 on DNA was found capable of inducing 1D diffusion of otherwise nonmotile PARP1, while excess APE1 also facilitated the dissociation of DNA-bound PARP1. Moreover, auto-PARylation of PARP1 allowed the protein to switch its damage search strategy by causing a 3-fold increase in linear diffusion. Finally, we demonstrated that PARP inhibitor olaparib did not significantly alter the rate of PARP1 dissociation from DNA, but instead resulted in more motility of DNA-bound PARP1 molecules.
INTRODUCTION
Poly(ADP-ribosyl)ation (PARylation) is a unique post-translational modification carried out by a family of poly(ADP-ribose) polymerases (PARPs). During this process, donor NAD molecules are used by PARPs for the synthesis of negatively charged mono- or poly-(ADP-ribose) (PAR) chains, which can be recognized by or covalently attached to target proteins (1). PARP1, the founding member of the PARP family, is a highly abundant nuclear enzyme (∼2 × 105/cell nucleus) whose activity is tightly regulated. PARP1, through PARylation, affects a number of cellular processes including transcription, chromatin remodeling, cell death signaling, and repair of DNA damage (2). In response to certain forms of genomic stress that cause single- or double-strand breaks in DNA, the enzymatic activity of PARP1 is significantly elevated over an extremely low basal level (3–6). Although the primary target of PARP1 is itself (7), many proteins, including DNA processing enzymes, have been shown to either be modified by PARP1 or bind to PAR (8,9). PAR is rapidly degraded and released from PARP1 by the action of poly(ADP-ribose) glycohydrolase (PARG) and ADP-ribosyl-acceptor hydrolases (ARHs) (10).
PARP1 can be activated by transient DNA single-strand break intermediates that occur during base excision repair (BER) (11–14). BER processes a wide range of DNA lesions that are produced by alkylation or oxidation of DNA bases (15,16). The pathway is initiated by DNA glycosylases that specifically recognize and remove modified base lesions, generating apurinic/apyrimidinic (AP) sites. AP endonuclease 1 (APE1) then acts on these AP sites to produce an incision, which results in a 5′-deoxyribose phosphate (5′-dRP) and a 3′-OH. While PARP1 recognizes and binds to AP sites, it is not strongly activated until the lesion is cleaved by APE1 (17,18). Studies indicate that auto-PARylated PARP1 recruits downstream BER components, such as DNA polymerase β, XRCC1, and DNA ligase III to the DNA damage site (19,20). It is widely believed that highly auto-modified PARP1 accumulates sufficient PAR, such that the net negative charge helps PARP1 dissociate from DNA, enabling subsequent repair and ligation steps to restore DNA (21). However, this notion has recently been challenged by work from Luger and coworkers who showed that auto-PARylation of PARP1 only increased the dissociation of PARP1 from chromatin, but not free DNA ends (22). The discrepancy in these reports suggests that there is a significant gap in our knowledge of the dynamics of unmodified and auto-modified PARP1 on DNA.
Also under debate is the stoichiometry of PARP1 binding to DNA nicks, ends, and abasic sites. There are six independent structural domains of the PARP1 protein. The N-terminus contains three zinc-finger (ZnF) domains. ZnF1 and ZnF2 are required for DNA binding (23). While not involved in DNA binding, ZnF3 is essential for activation (24,25). The central auto-modification domain contains a BRCA1 C-terminus (BRCT) fold, and is a major auto-PARylation site. The tryptophan–glycine–arginine (WGR) domain serves as a regulatory domain. The C-terminal catalytic region contains the helical subdomain (HD) and the ADP-ribose transferase (ART) domain. Activation of PARP1 on DNA strand breaks has been described by two mutually exclusive models based on crystal structures and biochemical studies (26,27). Ali and coworkers determined the crystal structure of the intact ZnF1–ZnF2 DNA binding domain of PARP1 bound to a DNA end with a single-nucleotide 5′ overhang, lending support to the model in which ZnF1 and ZnF2 cooperate to recognize DNA ends in a dimeric form (5,28). An X-ray crystal structure of PARP1 from Pascal's laboratory suggests that PARP1 binds to double-strand DNA breaks as a monomer and is activated through domain–domain interactions; where ZnF3 and WGR domains make DNA and protein contacts that induce a structural distortion in the catalytic domain and lead to PARP1 activation (26,29,30). Recently, through a combination of solution NMR and X-ray crystallography on domains of PARP1 including ZnF1 and ZnF2, Eustermann et al. provided further structural evidence and modeled PARP1 binding to single-strand DNA breaks as a monomer (31).
As a ‘DNA damage sensor, ’ PARP1 facilitates DNA repair under low to moderate levels of genomic stress, but can trigger cell death mechanisms upon excessive damage (32), due to depletion of NAD and subsequent alteration in cellular bioenergetics (33). Based on its role in maintaining genome stability, PARP1 has emerged as a promising drug target for cancer treatment (34–36), with most PARP inhibitors designed to compete with NAD at the active site. In addition to catalytic inhibition, some PARP inhibitors may also induce an allosteric conformational change, trapping the protein on DNA (37,38). Cells deficient in the human breast cancer tumor suppressors protein, BRCA1 and BRCA2, were shown to be highly sensitive to PARP inhibition (39–41). Recently, PARP1 inhibitor olaparib has been approved for treatment of ovarian cancer with BRCA mutations (42).
To help resolve some of the key unanswered questions regarding the stoichiometry and the dynamics of PARP1 on DNA during BER, we turned to single molecule approaches using atomic force microscopy (AFM) and oblique angle fluorescence microscopy. In the present study, we sought to first characterize the individual behavior of PARP1 and APE1 on DNA damage-containing DNA, before exploring how the two proteins could collaborate in damage search. Finally, we examined the effects of auto-modification, as well as binding of olaparib, on the interactions between PARP1 and DNA. Our AFM analysis indicates that full-length PARP1 can specifically bind to AP sites, DNA nicks, and ends in a monomeric form. Oblique angle fluorescence microscopy combined with the DNA tightrope platform allows us to observe individual quantum-dot (Qdot) labeled PARP1 and APE1 interacting with DNA arrays containing AP sites at high temporal and spatial resolutions. We found that auto-PARylation of PARP1 switched its interaction with DNA from a three-dimensional (3D) diffusive search in solution to a one-dimensional (1D) anomalous subdiffusion along DNA, increasing its lifetime on DNA. We also provide evidence that PARP1 and wildtype (WT) or catalytically dead APE1 (mAPE1, K87E/E96Q/D210N) can co-localize at AP sites, and that excess WT or catalytically dead APE1 helps to drive PARP1 dissociation from AP-containing DNA. Highly auto-PARylated PARP1 appears to be less stable on DNA and goes through a motile intermediate prior to dissociation. Interestingly, we found that PARP inhibitor olaparib causes PARP1 to undergo constrained motion on DNA, possibly through a conformational change, but does not alter its rate of dissociation. These studies provide new molecular insights into PARP1’s complex behavior on DNA.
MATERIALS AND METHODS
Materials
37mer tetrahydrofuran (THF)-containing oligonucleotides (AP37), with and without the 3′-biotin modification, were purchased from Integrated DNA Technologies (IDT). Recombinant His-tagged APE1 and mAPE1 triple mutant were expressed in Escherichia coli and purified as previously described (43,44) (see Supplementary Information). His-MBP-TEV-tagged PARP1 was expressed in and purified from Sf9 insect cells (see Supplementary Information).
Atomic force microscopy
Protein samples (100 nM PARP1) or protein–DNA binding reactions (150 nM DNA and 300 nM PARP1) were diluted and deposited on freshly cleaved mica surfaces and dried in a stream of nitrogen gas. 1 μm × 1 μm images were collected on a MultiMode V microscope (Bruker Corp.) in tapping mode and analyzed for particle volume in Image SXM. TIFF files were exported for bend angle and binding position analysis in ImageJ (NIH). Further details are provided in Supplementary Information.
Single molecule DNA tightrope assay
Data collection and analysis based on the single molecule DNA tightrope platform were carried out as previously described (45–47). Briefly, poly-l-lysine coated silica beads were deposited on the PEG-treated coverslip in a custom flow cell. Lesion-containing DNA substrates were suspended across the beads via hydrodynamic flow. Purified proteins were conjugated to secondary antibody-coated Qdots (Invitrogen) via the appropriate anti-His primary antibody. Images were collected with the appropriate emission filter applied and exported as TIFF stacks for kymograph extraction and single particle tracking in ImageJ (NIH). Mean squared displacement (MSD) of particle trajectories were then calculated and fitted to the general 1D diffusion model  via a custom-written script in MATLAB (Mathworks). See Supplementary Information for details.
via a custom-written script in MATLAB (Mathworks). See Supplementary Information for details.
RESULTS
PARP1 specifically binds DNA lesions as a monomer
Conflicting models exist for the binding stoichiometry of PARP1 at DNA strand breaks, suggesting either a monomer or dimer (26,27). Here, we use AFM to provide a direct measurement of the oligomeric state as PARP1 binds to nicks, ends, and abasic sites in DNA. AFM provides volume measurements of individual molecules, which can be directly related to the molecular weight (MW) of the proteins (48), such that we are able to study the oligomeric states of free PARP1 or PARP1 binding to different types of DNA substrates. Free PARP1 deposited on mica yields a volume measurement consistent with the protein being a monomer in solution (Supplementary Figure S1). Furthermore, most PARP1 molecules remain monomeric when bound to DNA ends or specifically at a nick site within a 514 bp duplex DNA fragment (Supplementary Figure S2C and D). These results are consistent with the PARP1-DNA crystal structure from the Pascal laboratory (26).
We then imaged specific binding of PARP1 to duplex DNA containing an abasic site (Figure 1A). A synthetic stable abasic site analog tetrahydrofuran (THF) was used throughout this study, and is simply termed the AP site for the rest of this work. In this AFM study, the AP site was placed at 30% of the total contour length from one end of a 538 bp DNA duplex (AP538). Experiments involving PARP1 binding to AP538 revealed that PARP1 has similar affinity for DNA ends and internal sites (Figure 1B). The position distribution of internally bound PARP1 along AP538, measured in distance between bound PARP1 and the closest DNA end as a percentage of total DNA contour length (538 bp), is shown in Figure 1C. The Gaussian-fitted internal binding position is centered at  , consistent with the known position of the AP site (30%). Volume analyses of PARP1 particles bound to AP538 at both AP sites and DNA ends also yielded distributions consistent with the expected molecular weight of monomeric proteins (Figure 1D and E). In addition, AFM data showed that PARP1 prefers end binding (68.2%) to internal binding (31.8%) on undamaged 514 bp DNA (N = 145, Supplementary Figure S2G), and that positions of internal binding events exhibit a broader distribution (
, consistent with the known position of the AP site (30%). Volume analyses of PARP1 particles bound to AP538 at both AP sites and DNA ends also yielded distributions consistent with the expected molecular weight of monomeric proteins (Figure 1D and E). In addition, AFM data showed that PARP1 prefers end binding (68.2%) to internal binding (31.8%) on undamaged 514 bp DNA (N = 145, Supplementary Figure S2G), and that positions of internal binding events exhibit a broader distribution ( , N = 46, Supplementary Figure S2H) compared to that of specific binding to nicks on the same DNA sequence (
, N = 46, Supplementary Figure S2H) compared to that of specific binding to nicks on the same DNA sequence ( , N = 96, Supplementary Figure S2B).
, N = 96, Supplementary Figure S2B).
Figure 1.
PARP1 binds to abasic sites and DNA ends as a monomer. (A) 3D view of AFM image of PARP1 binding to 538 bp DNA fragments with abasic (AP) sites at 30% contour length from one end (AP538). Red arrows indicate DNA-bound PARP1, cyan arrows indicate free PARP1, and white arrows indicate free BSA. Scan area 1 μm × 1 μm. (B) Bar graph of end versus internal binding events of PARP1 on AP538 (N = 273). (C) Histogram of internal binding positions of PARP1 along AP538 (N = 136). Solid curve represents a Gaussian fitting to the distribution data, centered at  . (D) and (E) Histograms of volume distributions for PARP1 bound at DNA ends (N = 126) and AP sites (N = 113), respectively. Solid curves represent Gaussian fittings to data. Black arrows indicate the positions of expected dimer volume peaks (451 nm3). For end binding (D), the fitted curve is centered at
. (D) and (E) Histograms of volume distributions for PARP1 bound at DNA ends (N = 126) and AP sites (N = 113), respectively. Solid curves represent Gaussian fittings to data. Black arrows indicate the positions of expected dimer volume peaks (451 nm3). For end binding (D), the fitted curve is centered at  , corresponding to a molecular weight of
, corresponding to a molecular weight of  . For specific binding at AP sites (between 20% and 40%) (E), the fitted curve is centered at
. For specific binding at AP sites (between 20% and 40%) (E), the fitted curve is centered at  corresponding to a molecular weight of
corresponding to a molecular weight of  . The predicted molecular weight of monomeric His-MBP-TEV-PARP1 is
. The predicted molecular weight of monomeric His-MBP-TEV-PARP1 is  .
.
To further characterize PARP1 binding to AP sites embedded in long DNA substrates, we employed the DNA tightrope assay in combination with oblique angle fluorescence microscopy, as previously described (45–47,49,50). DNA damage arrays (>40 kb) were created by end-to-end tandem ligation of 2030 bp linearized plasmids (47,51) containing a site-specific AP lesion. The resulting DNA tightropes contained one AP site every 2030 bp and were suspended across poly-l-lysine-coated 5 μm silica beads using hydrodynamic flow, resulting in ∼90% contour length of B-DNA (Figure 2A) (50). His-MBP-TEV-tagged PARP1 protein was labeled by a Qdot-antibody sandwich approach (Figure 2B, see Supplementary Information) (52). In DNA tightrope assays, 605 nm Qdot-labeled PARP1 molecules were observed binding to long DNA arrays containing AP sites (Figure 2C, Supplementary Movie S1). To confirm the specific binding of PARP1 to AP sites, we used a co-localization assay, in which a deoxy-thymine moiety 16 bp downstream from the AP site was modified with biotin (AP-BiodT) and conjugated to streptavidin (SA)-coated 655 nm Qdots. Figure 2D and Supplementary Movie S2 reveal three labeled AP-BiodT sites (red), and two 605 nm IgG Qdot-labeled PARP1 particles (green), one of which was transiently bound. The kymograph of merged channels shows co-localization of red and green signals (yellow), indicating specific binding of PARP1 at the AP site (Figure 2D). In this co-localization assay (N = 208), we found one-third of all DNA-bound PARP1 particles co-localized with labeled AP-BiodT sites, indicating damage-specific binding events (Figure 2E). The pair-wise distances between Qdot-labeled AP-BiodT sites (Figure 2F, top) and binding events of PARP1 on AP and AP-BiodT DNA were also measured (Figure 2F, middle and bottom, respectively). These data suggest PARP1 displays both specific and non-specific binding to AP DNA damage arrays.
Figure 2.
PARP1 specifically binds to abasic site arrays embedded in long DNA substrates. (A) Schematic of the DNA tightrope assay. Long DNA substrates with AP sites at defined positions are suspended between 5 μm poly-L-lysine coated silica beads. These lesion sites can be labeled by a streptavidin (SA) Qdot (red dots) through an adjacent biotin or recognized by labeled DNA repair proteins (green dots). Co-localization of protein and lesion is shown as a yellow glow around closely positioned red and green dots. (B) Labeling of His-MBP-TEV-PARP1 by antibody sandwich (see Supplementary Information). Domains of PARP1 are colored as follows: Zinc Fingers 1, 2 and 3 in green, light gray and blue, respectively; BRCT in black; WGR in red; HD in wheat; and ART in dark yellow. (C) Still video frame and kymograph of PARP1 protein array on long DNA substrates containing defined AP sites. Positions of the silica beads are outlined by white circles. Asterisks mark the dissociating particle. Horizontal and vertical scale bars represent 5 s and 2 kp, respectively. (D) Still video frame and kymographs of Qdot605-mHisAB-PARP1 (red) binding to SA-Qdot655 labeled AP-BiodT sites (green). Color-coded red, green, and yellow arrows highlight these binding events where appropriate. Horizontal and vertical scale bars represent 50 s and 2 kb, respectively. (E) Co-localization (yellow) Venn diagrams of Qdot-labeled AP-BiodT sites (red) with Qdot-labeled PARP1 (green) on AP-BiodT DNA tightropes in the dual-color assay (N = 208). (F) Distributions of pair-wise distances between labeled AP-BiodT sites in DNA (white, N = 231, top), labeled PARP1 on AP DNA (orange, N = 26, middle), and labeled PARP1 on AP-BiodT DNA (blue, N = 28, bottom). Solid black curve represents Gaussian fitting to the distance distribution for labeled AP-BiodT sites.
Among all the Qdot-labeled PARP1 particles on AP DNA (N = 129), including both specific and non-specific binding events, 89.1% of them were non-motile, whereas only 10.1% underwent 1D diffusion on DNA (Figure 3A, ‘M’ and B and C). In addition, 34.9% of the 129 DNA-bound PARP1 molecules dissociated during a five-minute observation window (Figure 3A, ‘D’). Finally, 20% of those 1D diffusive PARP1 particles (∼2% of total) also dissociated during the five-minute observation window (Figure 3A, ‘M&D’). Diffusion behavior of each motile PARP1 molecule (N = 12) was further analyzed by characterization of its diffusion coefficient (D) and anomalous diffusion exponent (α factor, see Supplementary Information, Figure 3D, Supplementary Table S1). The mean diffusion coefficient of PARP1 is  and its mean α factor is
and its mean α factor is  .
.
Figure 3.
Dynamic behavior of PARP1 on DNA containing abasic sites. (A) Bar graph showing PARP1 behavior on DNA containing AP sites (N = 129, M: Motile, D: Dissociated, M & D: Motile and Dissociated). Data from three independent experiments are shown as weighted mean ± SEM. Motile population (M) includes protein particles of all three types of movement on DNA (random, constrained, and paused). (B) Kymographs showing four different types of motion of PARP1: (I) random, (II) constrained, (III) paused, and (IV) non-motile. Horizontal and vertical scale bars represent 5 seconds and 2 kbp, respectively. (C) Bar graph showing types of motion of PARP1 on AP DNA. Data from three independent experiments are shown as weighted mean ± SEM. Note that the y-axis breaks between 10% and 80%. (D) Plot of PARP1 anomalous diffusion exponent (α factor) versus diffusion coefficient (log10D).
APE1 facilitates dissociation of PARP1 from abasic DNA
Several single molecule studies have suggested that some types of DNA binding proteins that make multiple contacts with DNA can undergo facilitated dissociation from DNA driven by another DNA binding protein in excess (53,54). Since PARP1 is believed to utilize multiple DNA binding motifs (30), we tested the hypothesis that excess unlabeled APE1 may increase the rate of PARP1 dissociation from AP sites. We found that a 10-fold excess of APE1 (10 nM) led to a 1.8-fold increase in the fraction of PARP1 (1 nM) that dissociated during the five-minute observation window, from 35.1% to 63.8% (P < 0.005; Supplementary Figure S3B). To further test whether the increased dissociation was a result of APE1 incisions at AP sites, we used an excess of mAPE1 (10 nM) and found the presence of this catalytically dead mutant resulted in a similar increase of about 1.7 fold in the fraction of dissociating PARP1 (P < 0.05; Supplementary Figure S3B). These data thus indicate that APE1 binding to abasic sites and not catalysis is sufficient to help facilitate dissociation of PARP1 bound to abasic sites.
PARP1 and APE1 co-localize at abasic sites, facilitating PARP1 movement
Since both APE1 and PARP1 are capable of binding to abasic sites individually, we sought to further investigate their potential interactions at APE1-processed AP sites. To this end, we used a two-color antibody sandwich conjugation strategy in which PARP1 was uniquely labeled with 605 nm goat anti-mouse IgG Qdot, and APE1 with 705 nm goat anti-rabbit IgG Qdot. We began by characterizing behavior of Qdot-labeled wildtype and mutant APE1 individually on AP DNA, where both proteins exhibited more motility compared to PARP1 (Supplementary Data and Supplementary Figure S4). Control experiments indicated that there is no exchange of Qdots between APE1 and PARP1. Since data acquisition occurs over several hours, we wanted to ensure that all AP sites had been incised by APE1 prior to the mixing experiment. APE1 (1 nM) was incubated with DNA tightropes containing AP sites for one hour to create 5′-dRP nick sites before a mixture of PARP1 and APE1 (1 nM each) was introduced into the flow cell. We found that co-localization of APE1 and PARP1 accounted for 6% of all particles on APE1-processed AP DNA (Figure 4A, left). Behavior of PARP1 on APE1-processed AP DNA was similar to that of PARP1 in the absence of APE1 on untreated AP DNA: motile and dissociated fractions were 14.6% and 37.1%, respectively (compare dark green bars in Figure 4B to Figure 3A). Addition of NAD to the reaction had no significant effect on the motile or dissociated fractions (Figure 4B, green). Furthermore, while the overall co-localization pattern of PARP1 and APE1 did not noticeably change in the presence of NAD (Figure 4A, middle), some of the co-localized particles were motile. One such co-localization event is shown in Figure 4D and Supplementary Movie S3, in which one of the two APE1 molecules (red) remained stationary throughout, while the other stationary particle became motile upon co-localization of PARP1 (green). The two particles traveled together for about 80 seconds, covering a distance of ∼8 kbp. To test if PARP1 and APE1 can co-localize at AP sites that are not cleaved by APE1, we used mAPE1 (K87E/E96Q/D210N) in combination with PARP1 on AP DNA. Under this condition, we also observed co-localization for 6% of all molecules (Figure 4A, right and 4E), similar to behavior of WT APE1 and PARP1 on processed AP DNA. Taken together, these data suggest that: (i) PARP1 infrequently interacts with APE1 at AP sites and 5′-dRP nicks; (ii) their co-localization is independent of the presence of NAD and (iii) such interaction with APE1 allows 1D diffusion of PARP1 on DNA.
Figure 4.
Interaction of PARP1 and APE1 on DNA containing APE1 processed abasic sites. (A) Co-localization (yellow) Venn diagrams of PARP1 (green) with APE1 (red) on APE1 processed AP DNA in the absence (left) and presence (middle) of NAD, and PARP1 (green) with mAPE1 (red) on unprocessed AP DNA in the absence of NAD (right). (B) Bar graph showing PARP1 behavior in the presence of APE1 (dark green, N = 89), APE1 and NAD (green, N = 47), and mAPE1 (light green, N = 84). Motile population (M) includes protein particles of all three types of movement on DNA (random, constrained, and paused). (C) Bar graph showing behavior of APE1 (red, N = 44) and mAPE1 (light pink, N = 63) in the presence of PARP1 (M: Motile, D: Dissociated). Motile population (M) includes protein particles of all three types of movement on DNA (random, constrained, and paused). (D) Kymographs of co-localized PARP1 (green) and APE1 (red) in the presence of NAD. (E) Kymographs of co-localized PARP1 (green) and mAPE1 (red). Horizontal and vertical scale bars represent 50 s and 2 kb, respectively.
Auto-modified PARP1 on APE1 processed abasic DNA shows anomalous subdiffusion and increased constrained motion
It has been suggested that auto-PARylation increases the dissociation of PARP1 from DNA (21). Since it was not possible to quantify the amount of modified PARP1 in the DNA tightrope assay flow cell after the addition of NAD and APE1 in the presence of AP DNA, instead, we prepared auto-PARylated PARP1 prior to introducing it in the flow cell. This auto-modification was achieved by incubating two-fold excess of PARP1 (100 nM) with 50 nM 37 bp DNA duplex and 100 μM NAD at room temperature for two hours. PAR synthesis induced by dsDNA ends was confirmed by mouse anti-PAR antibody (Figure 5A, lane 4), while no PAR signal could be detected when NAD was omitted (Figure 5A, lane 3). The interactions of auto-PARylated/auto-modified PARP1 (AM-PARP1) with DNA was investigated by both AFM (Supplementary Figure S5) and DNA tightrope assay (Figure 5).
Figure 5.
Auto-PARylation of PARP1 increases its motility on APE1 processed abasic DNA. (A) Western blot of the in vitro auto-PARylation assay using 37 bp duplex DNA as a substrate for activation of PARP1. (B) Bar graph showing behavior of PARP1 (cyan, N = 118), auto-modified PARP1 from in vitro auto-PARylation reactions (AM-PARP1, purple, N = 126), highly modified PARP1 (HM-PARP1, orange, N = 27), and PARP1 in the presence of olaparib (PARP1 + olaparib, gray, N = 112). Data from three independent experiments are shown as weighted mean ± SEM (M: Motile, D: Dissociated, M & D: Motile and Dissociated). Motile population (M) includes protein particles of all three types of movement on DNA (random, constrained, and paused). Student's t-test was used to test for statistical significance in the difference between behavior of PARP1 (control, denoted by #) and that of AM-PARP1, HM-PARP1, or PARP1 + olaparib (** P < 0.005). (C) Plot of anomalous diffusion exponent (α factor) versus diffusion coefficient (log10D) for PARP1 (cyan, N = 8), AM-PARP1 (purple, N = 40), and HM-PARP1 (orange, N = 11). (D) Bar graph showing motion types of PARP1, AM-PARP1, HM-PARP1, and PARP1 in the presence of olaparib. (E) Plot of anomalous diffusion exponent (α factor) versus diffusion coefficient (log10D) for PARP1 on AP DNA in the presence (red, N = 25) and absence (blue, N = 12) of olaparib. Data in the absence of olaparib are re-plotted from Figure 3D. Still movie frame (top) and kymograph (bottom) of AM-PARP1 on APE1 processed AP DNA (F) and PARP1 on AP DNA in the presence of olaparib (G). Corresponding particles in the movie frame and kymograph are as labeled.
Examination of AFM volumes of PARP1 molecules on a 514 bp duplex with a nick 36% from one end (N514), indicated that a distinct second population corresponding to larger protein volumes emerged only after auto-PARylation (Supplementary Figure S5A and S5B). Fitting volume histogram of AM-PARP1 and comparing to that of unmodified proteins, we quantitatively demonstrate that at least 68% of all PARP1 molecules are PARylated under the experimental conditions used (see Supplementary Information). Surprisingly, we still observed internal binding of AM-PARP1 to N514 DNA fragments (Supplementary Figure S5C and S5D). Compared to unmodified PARP1, internal binding events of AM-PARP1 decreased from 45% to 25% of the total (Supplementary Figure S5C and S5F). Also, the distribution of internal binding positions of AM-PARP1 on N514 ( , Supplementary Figure S5E) is broader than that of PARP1 (Supplementary Figure S2B,
, Supplementary Figure S5E) is broader than that of PARP1 (Supplementary Figure S2B,  , P < 0.05). Both the overall binding behavior and the position distribution of internally bound AM-PARP1 on N514 are similar to that of non-specific binding of unmodified PARP1 on undamaged 514 bp fragments (Supplementary Figure S2). These data suggest that AM-PARP1 may be more motile at nick sites on DNA and exhibit less overall specificity.
, P < 0.05). Both the overall binding behavior and the position distribution of internally bound AM-PARP1 on N514 are similar to that of non-specific binding of unmodified PARP1 on undamaged 514 bp fragments (Supplementary Figure S2). These data suggest that AM-PARP1 may be more motile at nick sites on DNA and exhibit less overall specificity.
To directly test this hypothesis, we imaged AM-PARP1 on APE1-processed AP DNA using the tightrope assay (Figure 5F, and Supplementary Movie S4). Compared to unmodified PARP1, the motile fraction of AM-PARP1 molecules increased ∼3-fold, while 8-fold fewer molecules dissociated during the five-minute observation window (Figure 5B, purple). Furthermore, a large fraction of these motile AM-PARP1 molecules displayed constrained motion with an average diffusion coefficient of  and an anomalous diffusion exponent α factor of
and an anomalous diffusion exponent α factor of  (Figure 5C, purple, and 5D). One important control in these experiments was whether the 37 bp duplex used to activate PARP1 that is present in the flow cell affects PARP1 interaction with DNA. We found that the presence of the 37 bp duplex increased PARP1 dissociation (Supplementary Figure S6), and thus is not contributing to the large increase in the bound population of PARP1 after PARylation.
(Figure 5C, purple, and 5D). One important control in these experiments was whether the 37 bp duplex used to activate PARP1 that is present in the flow cell affects PARP1 interaction with DNA. We found that the presence of the 37 bp duplex increased PARP1 dissociation (Supplementary Figure S6), and thus is not contributing to the large increase in the bound population of PARP1 after PARylation.
To further confirm that this increased motile fraction is due to auto-PARylation, we directly labeled PARylated PARP1 with anti-PAR antibody, which preferentially binds long chains of poly-ADP-ribose. These highly modified PARP1 (HM-PARP1) molecules, selectively labeled through anti-PAR antibody, represent a subset of the AM-PARP1 population, specifically those with higher levels of PARylation. In contrast, AM-PARP1 is visualized by Qdot conjugation through anti-His antibody that recognizes the 6X-His tag present on all PARP1 molecules regardless of their state of modification. Compared to AM-PARP1, HM-PARP1 showed relatively poor (∼83% less) binding to APE1-processed DNA and an increase in the motile fraction, although with a slightly lower average diffusion coefficient of  and α factor of
and α factor of  (Figure 5C orange and Supplementary Table S2). Extensive PARylation of PARP1 also resulted in a 6-fold increase in dissociation as compared to AM-PARP1, and most of these particles were moving along the DNA prior to dissociation (Figure 5B, orange). Taken together these data suggest that relatively lower levels of auto-PARylation help mobilize PARP1 to perform constrained motion around a DNA lesion, which may reduce steric interference for the next repair protein in the cascade. Through PAR binding, auto-PARylated PARP1 may help facilitate the recruitment of other repair proteins to the site. Only after high levels of PARylation of PARP1 does the accumulation of a large net negative charge help release HM-PARP1 from DNA.
(Figure 5C orange and Supplementary Table S2). Extensive PARylation of PARP1 also resulted in a 6-fold increase in dissociation as compared to AM-PARP1, and most of these particles were moving along the DNA prior to dissociation (Figure 5B, orange). Taken together these data suggest that relatively lower levels of auto-PARylation help mobilize PARP1 to perform constrained motion around a DNA lesion, which may reduce steric interference for the next repair protein in the cascade. Through PAR binding, auto-PARylated PARP1 may help facilitate the recruitment of other repair proteins to the site. Only after high levels of PARylation of PARP1 does the accumulation of a large net negative charge help release HM-PARP1 from DNA.
Olaparib increases constrained motion of PARP1
Olaparib, a competitive inhibitor of PARP1 that occupies the NAD binding pocket at the catalytic site, has been proposed to trap PARP1 on DNA (37). We conducted single molecule experiments to further investigate whether olaparib increases binding of PARP1 to DNA by trapping it at the damage site. Consistent with previous reports, bulk studies using electrophoretic mobility shift assays (EMSAs) showed that addition of NAD caused PARP1 dissociation from a 16 bp DNA duplex, which is completely inhibited by the presence of olaparib (Supplementary Figure S7A). Surprisingly, addition of PARP1 and olaparib (10 μM) into the flow cell with AP-containing DNA tightropes led to an increase in the motile fraction of PARP1, and a similar fraction of dissociated particles as compared to uninhibited PARP1 (Figure 5B). It is interesting to note that dissociating molecules also showed a slight increase in motion prior to dissociation (Figure 5B and 5G and Supplementary Movie S5). Most of the motile PARP1 particles performed constrained motion, as evidenced by limited movements of ∼500–1000 bp around fixed points, as well as α factors significantly lower than 1 (Figure 5E). Compared to free PARP1, we found that addition of olaparib increased both constrained (from 6.9% to 18.8%) and random motion (from 2.3% to 8.0%, Figure 5D and E).
DISCUSSION
Using single molecule approaches, we investigated the dynamic properties of PARP1 and APE1 during the repair of abasic sites and identified several novel features of the interactions involving these two proteins and DNA. First, AFM studies and single molecule fluorescence imaging on the DNA tightrope platform revealed that PARP1 binds DNA as a monomer with significant specificity towards AP sites. Most PARP1 molecules (∼90%) bound to AP-containing DNA tightropes were non-motile, and about half of these molecules dissociated during a five minute observation period. Second, APE1 and PARP1 were found to co-localize on AP DNA, and this interaction facilitated PARP1 movement. Third, auto-PARylation of PARP1 switched its search mode from 3D diffusion to anomalous 1D subdiffusion, increasing constrained motion by about 2-fold over unmodified PARP1. Low levels of PARylation resulted in an 8-fold decrease in dissociation compared to either unmodified or highly-modified PARP1. AFM studies of auto-PARylated PARP1 (68% modified) demonstrated that the specificity of the protein towards AP sites decreased about 2-fold and a significantly higher percentage of PARP1 molecules bound to ends of DNA duplexes. Finally, addition of olaparib, a competitive inhibitor of PARP1, led to a 2.5-fold increase in the observed fraction of motile particles with no significant change in dissociation, as compared to untreated PARP1.
PARP1 binds various DNA lesions as a monomer
While several studies have suggested that PARP1 binds to DNA as a monomer (26,29,31), others have proposed that PARP1 forms dimers at DNA ends and works in trans to activate its catalytic activity (5,28). Our AFM results unequivocally show that PARP1 binds to nicks, AP sites, and DNA ends as a monomer. Single molecule DNA tightrope experiments using PARP1 molecules labeled with Qdots of two different emission wavelengths also did not show any co-localization of the two colors, which would otherwise indicate dimeric binding events.
PARP1 and APE1 interactions on DNA
Since both PARP1 and APE1 have affinity for abasic sites, we reasoned that the two proteins may have the capacity to interact directly at such sites of damage. Data presented here suggest that co-localization of PARP1 with APE1, while infrequent, does occur. Due to the sizes of the Qdots used for protein visualization, it is possible that steric hindrance between the Qdots contributed to the decreased level of PARP1 and APE1 co-localization observed. Control experiments with tandem-ligated undamaged plasmids as a DNA tightrope substrate showed little background binding of either PARP1 or APE1 on DNA, indicating stable binding and co-localization events observed likely occurred only at specific sites of damage. PARP1, which is normally non-motile, can also undergo 1D diffusion in the presence of APE1. Furthermore, our observation that a 10-fold excess of APE1 led to an increase in PARP1 dissociation from DNA is consistent with the concept of ‘facilitated dissociation’. Put forth by Marko et al., facilitated diffusion describes the enhancement of dissociation of proteins that have multiple contacts with DNA, in the presence of other DNA binding domains (54). Thus, we envision that, as portions of PARP1 begin to micro-dissociate, APE1 is able to bind to DNA, blocking the rebinding of a critical domain of PARP1 to DNA and allowing macro-dissociation of the entire PARP1 molecule. It is interesting to note that the addition of 37 bp fragments into the flow cell of the DNA tightrope assay also led to an increase in the number of particles that dissociated. Again, this finding could be interpreted by evoking the same mechanism: micro-dissociation of one of PARP1’s multiple DNA binding domains from the DNA tightrope allows it to bind to a free 37 bp fragment and thus occludes rebinding of the domain to the tightrope. The presence of either excess DNA-binding proteins or a localized high concentration of DNA, as found in the nucleus, would increase the rate of dissociation of PARP1. For repair proteins that are believed to undergo a 3D diffusive process in search for their targets, facilitated dissociation has important implications for increasing the rate at which these proteins can locate and bind to target sites in DNA in vivo. By increasing the off-rate, a repair enzyme could sample a larger fraction of DNA in a shorter time period.
Distinct DNA damage search strategies for APE1 and PARP1
Data presented in this study suggest that PARP1 binds specifically to abasic sites, in agreement with our previous bulk biochemical studies (18). Considering that both APE1 and PARP1 are highly abundant within the cell, it is interesting to compare and contrast how these two proteins search for and bind to AP sites within genomic DNA. Both catalytically active and inactive APE1 were found to be long-lived and slide on AP-containing DNA at  on average, yielding a mean residence time of
on average, yielding a mean residence time of  at each base. Given APE1’s rapid kcat of at least ∼850 s−1 (55), catalysis at AP sites should in theory manifest as a series of pauses punctuating the diffusive kymograph. However, at a temporal resolution of ∼10 fps, such feature could not be discerned from our kymographs. Interestingly, diffusion coefficients of both WT and mutant APE1 were measured to be consistently higher than the theoretical upper limit for diffusion along DNA with rotation (56), which implies that APE1 may slide along the DNA linearly and not track its helical groove. Thus, we cannot conclusively distinguish between particles undergoing linear sliding and those diffusing with helical tracking. Future experiments looking at the salt dependence of APE1 linear diffusion will help reconcile a hopping versus sliding model.
at each base. Given APE1’s rapid kcat of at least ∼850 s−1 (55), catalysis at AP sites should in theory manifest as a series of pauses punctuating the diffusive kymograph. However, at a temporal resolution of ∼10 fps, such feature could not be discerned from our kymographs. Interestingly, diffusion coefficients of both WT and mutant APE1 were measured to be consistently higher than the theoretical upper limit for diffusion along DNA with rotation (56), which implies that APE1 may slide along the DNA linearly and not track its helical groove. Thus, we cannot conclusively distinguish between particles undergoing linear sliding and those diffusing with helical tracking. Future experiments looking at the salt dependence of APE1 linear diffusion will help reconcile a hopping versus sliding model.
In stark contrast to the rapid linear diffusion of APE1 on DNA containing AP sites, interactions between PARP1 and the same DNA substrate featured very little motion of the protein in addition to appreciable dissociation from DNA. Such behavior indicates that PARP1 likely interrogates DNA mainly through a 3D search mechanism. In addition, the small motile fraction of PARP1 molecules diffused on DNA with a low energy barrier to free diffusion  . Previous work using bulk biochemical approaches suggested that auto-PARylation of PARP1 causes increased dissociation from DNA (9). Surprisingly we found that auto-PARylated PARP1, while being less prone to dissociation from DNA, also switches its search mechanism to facilitated diffusion and exhibits increased diffusion along the DNA. This sliding behavior, constrained to 500–1000 bp in each direction, has an anomalous diffusion exponent consistently less than 1. In the presence of APE1, on APE1-processed AP-containing DNA, unmodified PARP1 diffused with a steeper energy barrier to free diffusion
. Previous work using bulk biochemical approaches suggested that auto-PARylation of PARP1 causes increased dissociation from DNA (9). Surprisingly we found that auto-PARylated PARP1, while being less prone to dissociation from DNA, also switches its search mechanism to facilitated diffusion and exhibits increased diffusion along the DNA. This sliding behavior, constrained to 500–1000 bp in each direction, has an anomalous diffusion exponent consistently less than 1. In the presence of APE1, on APE1-processed AP-containing DNA, unmodified PARP1 diffused with a steeper energy barrier to free diffusion  , similar to that experienced by AM-PARP1
, similar to that experienced by AM-PARP1  . HM-PARP1 was subject to an even more rugged energy landscape
. HM-PARP1 was subject to an even more rugged energy landscape  . This decrease in diffusion coefficients may be due to a combination of the presence of nicked sites in DNA after APE1 processing, conformational changes upon PARylation, and increased hydrodynamic drag due to a larger hydrodynamic radius in the presence of long PAR chains.
. This decrease in diffusion coefficients may be due to a combination of the presence of nicked sites in DNA after APE1 processing, conformational changes upon PARylation, and increased hydrodynamic drag due to a larger hydrodynamic radius in the presence of long PAR chains.
Collapse of search dimensionality from 3D to 1D has long been recognized as an important strategy to accelerate the search process (57). Such a change in diffusive behavior has also been observed for the bacterial UvrA and UvrAB nucleotide excision repair complexes, the former displaying a 3D search, and the latter sliding on DNA (45). Furthermore, the increase in anomalous subdiffusion on DNA, which we term constrained motion, has been recently observed for the Rad4-Rad23 nucleotide excision repair damage sensor (47). Thus, constrained motion may be a general feature of damage searching for DNA repair enzymes (58). Multiple theoretical models have been extensively studied to model and describe experimentally observed anomalous subdiffusion in membranes or systems with molecular crowding effects (59). For proteins that interact with and diffuse one-dimensionally on DNA, subdiffusive behavior has also been predicted to emerge due to position-correlated and Gaussian-distributed random potential energy landscapes (60,61).
Working Model of PARP1 binding to AP sites
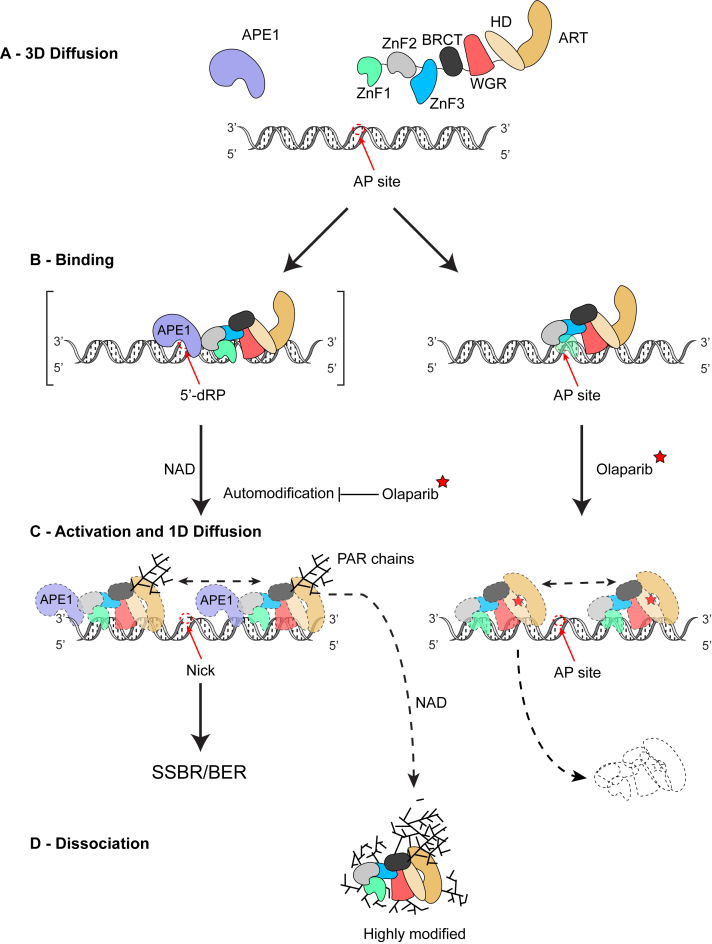
We hereby propose the following working model (Figure 6) for the action of PARP1 at AP sites and nicks in DNA, consistent with all the data presented in this study. Both PARP1 and APE1 search for damage (AP) sites in DNA via 3D diffusion (Figure 6A). PARP1 can bind to the AP site with APE1 and stimulate its activity (62), which in turn produces a 5′-dRP (Figure 6B, left). Direct interaction between PARP1 and APE1 also provides motility to the otherwise non-motile PARP1. In the presence of NAD, PARP1 bound at the 5′-dRP site is activated and undergoes auto-PARylation (Figure 6C, left), which can be blocked by olaparib (red star). PAR chains generated through auto-PARylation decrease the lesion specificity of PARP1 and allow the protein to undergo 1D anomalous subdiffusion on DNA around the lesion (Figure 6C, left), facilitating the recruitment of and hand-off to other repair proteins that might be otherwise sterically blocked to process the 3′-OH and deoxyribose sugar moiety. Finally, highly modified PARP1 undergoes constrained and random motion and is prone to dissociation while sliding along DNA (Figure 6D). Alternatively, PARP1 can directly bind to an AP site in the absence of APE1 (Figure 6B, right). Binding of olaparib in the enzyme's catalytic pocket inhibits auto-modification of PARP1 and likely induces a conformational change of the protein, such that PARP1 can undergo constrained motion around the AP site as well as dissociate from DNA (Figure 6C, right). Since olaparib blocks the active site of PARP1 and therefore any auto-PARylation, PARP1 becomes invisible to other SSBR/BER proteins that require PAR, such as DNA Ligase IIIα (63) and XRCC1 (64).
Figure 6.
Working model for PARP1 and APE1 dynamics at repair intermediates Model for recognition of AP sites by PARP1 (same color scheme as in Figure 2B) and APE1 (purple). See main text for details.
In summary, results from our single molecule experiments answer the question of PARP1 stoichiometry at sites of DNA damage, suggest cooperation between APE1 and PARP1 during BER, and provide evidence that auto-PARylated PARP1, through constrained motion, may serve as a mobile damage sensor for the recruitment of other BER proteins.
Supplementary Material
ACKNOWLEDGEMENTS
The authors wish to thank Emily Beckwitt for careful reading of the manuscript and Dr Elise Fouquerel for helpful discussions.
Footnotes
Present addresses:
Natalie R. Gassman, University of South Alabama Mitchell Cancer Institute, Mobile, AL 36604, USA.
Bret D. Freudenthal, Department of Biochemistry and Molecular Biology, The University of Kansas Medical Center, Kansas City, KS 66160, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [5R01ES019566 to B.V.H., 2P30CA047904 to University of Pittsburgh Cancer Institute, and 1ZIAES050158 and 1ZIAES050159 to S.H.W.]. Funding for open access charge: NIEHS [5R01ES019566 to B.V.H.].
Conflict of interest statement. None declared.
REFERENCES
- 1. Ame J.C., Spenlehauer C., de Murcia G.. The PARP superfamily. Bioessays. 2004; 26:882–893. [DOI] [PubMed] [Google Scholar]
- 2. Ko H.L., Ren E.C.. Functional aspects of PARP1 in DNA repair and transcription. Biomolecules. 2012; 2:524–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Benjamin R.C., Gill D.M.. Poly(ADP-ribose) synthesis in vitro programmed by damaged DNA. A comparison of DNA molecules containing different types of strand breaks. J. Biol. Chem. 1980; 255:10502–10508. [PubMed] [Google Scholar]
- 4. Dantzer F., Ame J.C., Schreiber V., Nakamura J., Menissier-de Murcia J., de Murcia G.. Poly(ADP-ribose) polymerase-1 activation during DNA damage and repair. Methods Enzymol. 2006; 409:493–510. [DOI] [PubMed] [Google Scholar]
- 5. Pion E., Ullmann G.M., Ame J.C., Gerard D., de Murcia G., Bombarda E.. DNA-induced dimerization of poly(ADP-ribose) polymerase-1 triggers its activation. Biochemistry. 2005; 44:14670–14681. [DOI] [PubMed] [Google Scholar]
- 6. Liu C., Vyas A., Kassab M.A., Singh A.K., Yu X.. The role of poly ADP-ribosylation in the first wave of DNA damage response. Nucleic Acids Res. 2017; 45:8129–8141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gibson B.A., Kraus W.L.. New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nat. Rev. Mol. Cell. Biol. 2012; 13:411–424. [DOI] [PubMed] [Google Scholar]
- 8. Daniels C.M., Ong S.E., Leung A.K.. The promise of proteomics for the study of ADP-ribosylation. Mol. Cell. 2015; 58:911–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. D’Amours D., Desnoyers S., D'Silva I., Poirier G.G.. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem. J. 1999; 342:249–268. [PMC free article] [PubMed] [Google Scholar]
- 10. Davidovic L., Vodenicharov M., Affar E.B., Poirier G.G.. Importance of poly(ADP-ribose) glycohydrolase in the control of poly(ADP-ribose) metabolism. Exp. Cell Res. 2001; 268:7–13. [DOI] [PubMed] [Google Scholar]
- 11. Woodhouse B.C., Dianov G.L.. Poly ADP-ribose polymerase-1: an international molecule of mystery. DNA Repair (Amst.). 2008; 7:1077–1086. [DOI] [PubMed] [Google Scholar]
- 12. Dantzer F., Schreiber V., Niedergang C., Trucco C., Flatter E., De La Rubia G., Oliver J., Rolli V., Menissier-de Murcia J., de Murcia G.. Involvement of poly(ADP-ribose) polymerase in base excision repair. Biochimie. 1999; 81:69–75. [DOI] [PubMed] [Google Scholar]
- 13. Lavrik O.I., Prasad R., Sobol R.W., Horton J.K., Ackerman E.J., Wilson S.H.. Photoaffinity labeling of mouse fibroblast enzymes by a base excision repair intermediate. Evidence for the role of poly(ADP-ribose) polymerase-1 in DNA repair. J. Biol. Chem. 2001; 276:25541–25548. [DOI] [PubMed] [Google Scholar]
- 14. Ziegler M., Oei S.L.. A cellular survival switch: poly(ADP-ribosyl)ation stimulates DNA repair and silences transcription. Bioessays. 2001; 23:543–548. [DOI] [PubMed] [Google Scholar]
- 15. Wallace S.S. Base excision repair: a critical player in many games. DNA Repair (Amst.). 2014; 19:14–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sander M., Wilson S.H.. eLS. 2001; John Wiley & Sons, Ltd. [Google Scholar]
- 17. Prasad R., Horton J.K., Chastain P.D. 2nd, Gassman N.R., Freudenthal B.D., Hou E.W., Wilson S.H.. Suicidal cross-linking of PARP-1 to AP site intermediates in cells undergoing base excision repair. Nucleic Acids Res. 2014; 42:6337–6351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Khodyreva S.N., Prasad R., Ilina E.S., Sukhanova M.V., Kutuzov M.M., Liu Y., Hou E.W., Wilson S.H., Lavrik O.I.. Apurinic/apyrimidinic (AP) site recognition by the 5′-dRP/AP lyase in poly(ADP-ribose) polymerase-1 (PARP-1). Proc. Natl. Acad. Sci. U.S.A. 2010; 107:22090–22095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mortusewicz O., Ame J.C., Schreiber V., Leonhardt H.. Feedback-regulated poly(ADP-ribosyl)ation by PARP-1 is required for rapid response to DNA damage in living cells. Nucleic Acids Res. 2007; 35:7665–7675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Okano S., Lan L., Tomkinson A.E., Yasui A.. Translocation of XRCC1 and DNA ligase IIIalpha from centrosomes to chromosomes in response to DNA damage in mitotic human cells. Nucleic Acids Res. 2005; 33:422–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Satoh M.S., Lindahl T.. Role of poly(ADP-ribose) formation in DNA repair. Nature. 1992; 356:356–358. [DOI] [PubMed] [Google Scholar]
- 22. Muthurajan U.M., Hepler M.R., Hieb A.R., Clark N.J., Kramer M., Yao T., Luger K.. Automodification switches PARP-1 function from chromatin architectural protein to histone chaperone. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:12752–12757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Langelier M.F., Planck J.L., Roy S., Pascal J.M.. Crystal structures of poly(ADP-ribose) polymerase-1 (PARP-1) zinc fingers bound to DNA: structural and functional insights into DNA-dependent PARP-1 activity. J. Biol. Chem. 2011; 286:10690–10701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Langelier M.F., Ruhl D.D., Planck J.L., Kraus W.L., Pascal J.M.. The Zn3 domain of human poly(ADP-ribose) polymerase-1 (PARP-1) functions in both DNA-dependent poly(ADP-ribose) synthesis activity and chromatin compaction. J. Biol. Chem. 2010; 285:18877–18887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Langelier M.F., Servent K.M., Rogers E.E., Pascal J.M.. A third zinc-binding domain of human poly(ADP-ribose) polymerase-1 coordinates DNA-dependent enzyme activation. J. Biol. Chem. 2008; 283:4105–4114. [DOI] [PubMed] [Google Scholar]
- 26. Langelier M.F., Planck J.L., Roy S., Pascal J.M.. Structural basis for DNA damage-dependent poly(ADP-ribosyl)ation by human PARP-1. Science. 2012; 336:728–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ali A.A., Timinszky G., Arribas-Bosacoma R., Kozlowski M., Hassa P.O., Hassler M., Ladurner A.G., Pearl L.H., Oliver A.W.. The zinc-finger domains of PARP1 cooperate to recognize DNA strand breaks. Nat. Struct. Mol. Biol. 2012; 19:685–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mendoza-Alvarez H., Alvarez-Gonzalez R.. Poly(ADP-ribose) polymerase is a catalytic dimer and the automodification reaction is intermolecular. J. Biol. Chem. 1993; 268:22575–22580. [PubMed] [Google Scholar]
- 29. Eustermann S., Videler H., Yang J.C., Cole P.T., Gruszka D., Veprintsev D., Neuhaus D.. The DNA-binding domain of human PARP-1 interacts with DNA single-strand breaks as a monomer through its second zinc finger. J. Mol. Biol. 2011; 407:149–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Langelier M.F., Pascal J.M.. PARP-1 mechanism for coupling DNA damage detection to poly(ADP-ribose) synthesis. Curr. Opin. Struct. Biol. 2013; 23:134–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Eustermann S., Wu W.F., Langelier M.F., Yang J.C., Easton L.E., Riccio A.A., Pascal J.M., Neuhaus D.. Structural basis of detection and signaling of DNA single-strand breaks by human PARP-1. Mol. Cell. 2015; 60:742–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. De Vos M., Schreiber V., Dantzer F.. The diverse roles and clinical relevance of PARPs in DNA damage repair: current state of the art. Biochem. Pharmacol. 2012; 84:137–146. [DOI] [PubMed] [Google Scholar]
- 33. Fouquerel E., Sobol R.W.. ARTD1 (PARP1) activation and NAD(+) in DNA repair and cell death. DNA Repair (Amst.). 2014; 23:27–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Peralta-Leal A., Rodriguez M.I., Oliver F.J.. Poly(ADP-ribose)polymerase-1 (PARP-1) in carcinogenesis: potential role of PARP inhibitors in cancer treatment. Clin. Transl. Oncol. 2008; 10:318–323. [DOI] [PubMed] [Google Scholar]
- 35. Peralta-Leal A., Rodriguez-Vargas J.M., Aguilar-Quesada R., Rodriguez M.I., Linares J.L., de Almodovar M.R., Oliver F.J.. PARP inhibitors: new partners in the therapy of cancer and inflammatory diseases. Free Radic. Biol. Med. 2009; 47:13–26. [DOI] [PubMed] [Google Scholar]
- 36. Curtin N. PARP inhibitors for anticancer therapy. Biochem. Soc. Trans. 2014; 42:82–88. [DOI] [PubMed] [Google Scholar]
- 37. Murai J., Huang S.Y., Das B.B., Renaud A., Zhang Y., Doroshow J.H., Ji J., Takeda S., Pommier Y.. Trapping of PARP1 and PARP2 by Clinical PARP Inhibitors. Cancer Res. 2012; 72:5588–5599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Murai J., Huang S.Y., Renaud A., Zhang Y., Ji J., Takeda S., Morris J., Teicher B., Doroshow J.H., Pommier Y.. Stereospecific PARP trapping by BMN 673 and comparison with olaparib and rucaparib. Mo.l Cancer Ther. 2014; 13:433–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Farmer H., McCabe N., Lord C.J., Tutt A.N., Johnson D.A., Richardson T.B., Santarosa M., Dillon K.J., Hickson I., Knights C. et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005; 434:917–921. [DOI] [PubMed] [Google Scholar]
- 40. Bryant H.E., Schultz N., Thomas H.D., Parker K.M., Flower D., Lopez E., Kyle S., Meuth M., Curtin N.J., Helleday T.. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005; 434:913–917. [DOI] [PubMed] [Google Scholar]
- 41. Horton J.K., Stefanick D.F., Prasad R., Gassman N.R., Kedar P.S., Wilson S.H.. Base excision repair defects invoke hypersensitivity to PARP inhibition. Mol. Cancer Res. 2014; 12:1128–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kim G., Ison G., McKee A.E., Zhang H., Tang S., Gwise T., Sridhara R., Lee E., Tzou A., Philip R. et al. FDA approval summary: olaparib monotherapy in patients with deleterious germline BRCA-mutated advanced ovarian cancer treated with three or more lines of chemotherapy. Clin. Cancer Res. 2015; 21:4257–4261. [DOI] [PubMed] [Google Scholar]
- 43. Strauss P.R., Beard W.A., Patterson T.A., Wilson S.H.. Substrate binding by human apurinic/apyrimidinic endonuclease indicates a Briggs-Haldane mechanism. J. Biol. Chem. 1997; 272:1302–1307. [DOI] [PubMed] [Google Scholar]
- 44. Freudenthal B.D., Beard W.A., Cuneo M.J., Dyrkheeva N.S., Wilson S.H.. Capturing snapshots of APE1 processing DNA damage. Nat. Struct. Mol. Biol. 2015; 22:924–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kad N.M., Wang H., Kennedy G.G., Warshaw D.M., Van Houten B.. Collaborative dynamic DNA scanning by nucleotide excision repair proteins investigated by single- molecule imaging of quantum-dot-labeled proteins. Mol. Cell. 2010; 37:702–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ghodke H., Wang H., Hsieh C.L., Woldemeskel S., Watkins S.C., Rapic-Otrin V., Van Houten B.. Single-molecule analysis reveals human UV-damaged DNA-binding protein (UV-DDB) dimerizes on DNA via multiple kinetic intermediates. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:E1862–1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kong M., Liu L., Chen X., Driscoll K.I., Mao P., Bohm S., Kad N.M., Watkins S.C., Bernstein K.A., Wyrick J.J. et al. Single-Molecule Imaging Reveals that Rad4 Employs a Dynamic DNA Damage Recognition Process. Mol Cell. 2016; 64:376–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ratcliff G.C., Erie D.A.. A novel single-molecule study to determine protein–protein association constants. J. Am. Chem. Soc. 2001; 123:5632–5635. [DOI] [PubMed] [Google Scholar]
- 49. Lin J., Countryman P., Buncher N., Kaur P., E L., Zhang Y., Gibson G., You C., Watkins S.C., Piehler J. et al. TRF1 and TRF2 use different mechanisms to find telomeric DNA but share a novel mechanism to search for protein partners at telomeres. Nucleic Acids Res. 2014; 42:2493–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hughes C.D., Wang H., Ghodke H., Simons M., Towheed A., Peng Y., Van Houten B., Kad N.M.. Real-time single-molecule imaging reveals a direct interaction between UvrC and UvrB on DNA tightropes. Nucleic Acids Res. 2013; 41:4901–4912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Geng H., Du C., Chen S., Salerno V., Manfredi C., Hsieh P.. In vitro studies of DNA mismatch repair proteins. Anal. Biochem. 2011; 413:179–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wang H., Tessmer I., Croteau D.L., Erie D.A., Van Houten B.. Functional characterization and atomic force microscopy of a DNA repair protein conjugated to a quantum dot. Nano Lett. 2008; 8:1631–1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Gibb B., Ye L.F., Gergoudis S.C., Kwon Y., Niu H., Sung P., Greene E.C.. Concentration-dependent exchange of replication protein A on single-stranded DNA revealed by single-molecule imaging. PLoS One. 2014; 9:e87922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Graham J.S., Johnson R.C., Marko J.F.. Concentration-dependent exchange accelerates turnover of proteins bound to double-stranded DNA. Nucleic Acids Res. 2011; 39:2249–2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Maher R.L., Bloom L.B.. Pre-steady-state kinetic characterization of the AP endonuclease activity of human AP endonuclease 1. J. Biol. Chem. 2007; 282:30577–30585. [DOI] [PubMed] [Google Scholar]
- 56. Blainey P.C., Luo G., Kou S.C., Mangel W.F., Verdine G.L., Bagchi B., Xie X.S.. Nonspecifically bound proteins spin while diffusing along DNA. Nat. Struct. Mol. Biol. 2009; 16:1224–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Adam G., Delbrück M.. Reduction of dimensionality in biological diffusion processes. Struct. Chem. Mol. Biol. 1968; 198–215. [Google Scholar]
- 58. Kong M., Van Houten B.. Rad4 recognition-at-a-distance: Physical basis of conformation-specific anomalous diffusion of DNA repair proteins. Prog. Biophys. Mol. Biol. 2017; 127:93–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Metzler R., Jeon J.H., Cherstvy A.G., Barkai E.. Anomalous diffusion models and their properties: non-stationarity, non-ergodicity, and ageing at the centenary of single particle tracking. Phys. Chem. Chem. Phys. 2014; 16:24128–24164. [DOI] [PubMed] [Google Scholar]
- 60. Slutsky M., Kardar M., Mirny L.A.. Diffusion in correlated random potentials, with applications to DNA. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2004; 69:061903. [DOI] [PubMed] [Google Scholar]
- 61. Goychuk I., Kharchenko V.O.. Anomalous features of diffusion in corrugated potentials with spatial correlations: faster than normal, and other surprises. Phys. Rev. Lett. 2014; 113:100601. [DOI] [PubMed] [Google Scholar]
- 62. Prasad R., Dyrkheeva N., Williams J., Wilson S.H.. Mammalian Base Excision Repair: Functional Partnership between PARP-1 and APE1 in AP-Site Repair. PLoS One. 2015; 10:e0124269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Leppard J.B., Dong Z., Mackey Z.B., Tomkinson A.E.. Physical and functional interaction between DNA ligase IIIalpha and poly(ADP-Ribose) polymerase 1 in DNA single-strand break repair. Mol. Cell. Biol. 2003; 23:5919–5927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. El-Khamisy S.F., Masutani M., Suzuki H., Caldecott K.W.. A requirement for PARP-1 for the assembly or stability of XRCC1 nuclear foci at sites of oxidative DNA damage. Nucleic Acids Res. 2003; 31:5526–5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.