Abstract
Introduction: Periodontitis is a set of inflammatory disorders characterized by periodontal attachment loss and alveolar bone resorption. Because of deficiency in periodontitis mechanical therapy, this study was aimed to explore the molecular influence of the erbiumdoped: yttrium aluminum garnet (Er:YAG) laser and cyclosporin A (CsA) on human gingival fibroblasts (HGFs) for improvement in periodontal diseases therapy.
Methods: We focused on articles that studied the proteome profiles of HGFs after treatment with laser irradiation and application of CsA. The topological features of differentially expressed proteins were analyzed using Cytoscape Version 3.4.0 followed by module selection from the protein-protein interaction (PPI) network using Cluster ONE plugin. In addition, we performed gene ontology (GO) enrichment analysis for the densely connected region and key proteins in both PPI networks.
Results: Analysis of PPI network of Er:YAG laser irradiation on HGFs lead to introducing YWHAZ, VCP, HNRNPU, YWHAE, UBA52, CLTC, FUS and IGHG1 as key proteins while similar analysis revealed that ACAT1, CTSD, ALDOA, ANXA2, PRDX1, LGALS3, ARHGDI and EEF1A1 are the crucial proteins related to the effect of drug. GO enrichment analysis of hubbottleneck proteins of the 2 networks showed the different significant biological processes and cellular components. The functional enrichments of module of Er:YAG laser network are included as fatty acid transmembrane transport, cytokinesis, regulation of RNA splicing and asymmetric protein localization. There are not any significant clusters in network of HGF treated by CsA.
Conclusion: The results indicate that there are 2 separate biomarker panels for the 2 treatment methods
Keywords: Er:YAG laser, ; cyclosporin A (CsA), Protein-protein interaction (PPI) network analysis, Gene ontology, human fibroblast cell
Introduction
Periodontitis is a set of inflammatory disorders characterized by periodontal attachment loss and alveolar bone resorption.1,2 Beside tooth loss, periodontitis can affect systemic health, as it can increase the risk of diabetes, rheumatoid arthritis, and atherosclerosis.3-5 Mechanical therapy cannot always completely remove the bacteria from the diseased root surfaces and thus may lead to recurrence of inflammatory periodontal diseases.6 Recently, various types of lasers have been applied to remove subgingival biofilms from periodontal pockets.7 Laser, as a developing method, is widely used in dentistry due to its beneficial effects such as coagulation properties, less pain, and rapid healing.8 Erbium-doped: yttrium aluminum garnet (Er:YAG) laser is considered as one of the safest laser devices in periodontal therapy.9 Favorable results of periodontal wound healing following Er:YAG laser treatment have been reported.7,10
Cyclosporin A (CsA) has been used as an immunosuppressive drug to treat autoimmune diseases and to prevent organ transplant rejection.11 Previous studies demonstrated that CsA has a stimulatory effect on human gingival fibroblasts (HGFs).12,13 In contrast, other reports showed that not only CsA has no effect on proliferation but also it has an inhibitory effect in HGFs.14-16 Despite numerous investigations on periodontitis, the molecular mechanism of therapeutic modalities on HGFs still remains largely unknown.17 HGFs are the most abundant cell type in human periodontal tissue which can influence promotion of gingival health damaged by periodontal diseases.18 HGFs can be used as stem cells for following up therapeutic effects and periodontal tissue engineering.19
Recently, usage of PPI network analysis has increased in medical fields for the detection of novel drug targets in various diseases.20,21 Several proteins influencing periodontitis have been identified,22 but there is little knowledge on the periodontal therapeutic field based on protein-protein interaction (PPI). Protein network analysis provides a desirable model that can elevate comprehension of the mechanisms underlying human diseases.20,21,23,24 This model may represent a logical associational panel which can be efficient in the treatment field.25-27 In the present study, we surveyed the protein networks of HGFs treated using Er:YAG laser and CsA separately to reveal the molecular effects of both modalities on HGFs. The information obtained will hopefully lead to significant improvements in periodontal disease treatment.
Methods
Identification of Differentially Expressed Proteins
To identify differentially expressed proteins in HGFs after treatment by CsA and Er:YAG laser irradiation, we used PubMed database as the main source to study the literature using the following keywords: “gingival fibroblasts” “Proteomics,” “drug,” and “laser therapy.” The inclusion criteria were (1) studies which focus on gingival tissue samples of human species and (2) studies that compared drug or laser treated HGF tissues and non-treated HGF tissues. Papers which surveyed serum or saliva proteome and papers which compared HGF tissues with another cell type, such as human periodontal ligament (PDL) were excluded. A total of 57 differentially expressed proteins after Er:YAG laser irradiation (Table 1)28 and 10 differentially expressed proteins after induction of drug (CsA) were obtained (Table 2).11 The data from reference 28 are retrieved via liquid chromatography-tandem mass spectrometry while the data about the effect of CsA are extracted from a gel based proteomics research. Since the first modality is more sensitive than the second, the number of identified proteins related to the laser effect is higher than for the other investigation. UniProt accession numbers (http://www.uniprot.org) were used to search for annotations and to list proteins in order to carry out the network analysis.
Table 1. Significantly Changed Expression Proteins of HGF Cells After Irradiation of Low-Level Er:YAG Laser on HGFs .
| Gene Name | Protein Name | Gene Name | Protein Name |
| HRNR | Hornerin | SFN | 14-3-3 protein sigma |
| EIF4A1 | Eukaryotic initiation factor 4A-I | ARG1 | Arginase-1 |
| SERPINB4 | Serpin B4 | CALML5 | Calmodulin-like protein 5 |
| TXN | Thioredoxin | GSTP1 | Glutathione S-transferase P |
| ARHGDIA | Rho GDP-dissociation inhibitor 1 | TGM3 | Protein-glutamine gamma-glutamyltransferase E |
| RACK1 | Guanine nucleotide-binding protein subunit beta-2-like 1 | EEF2 | Elongation factor 2 |
| KRT6C | Keratin, type II cytoskeletal 6C | LDHA | l-Lactate dehydrogenase A chain |
| IGKC | Ig kappa chain C region | CTSD | Cathepsin D |
| EIF1B | Eukaryotic translation initiation factor 1b | DSC1 | Desmocollin-1 |
| LGALS7 | Galectin-7 | RPS3 | 40S ribosomal protein S3 |
| UBA52 | Ubiquitin-60S ribosomal protein L40 | AZGP1 | Zinc-alpha-2-glycoprotein |
| RPL22 | 60S ribosomal protein L22 | EEF1G | Elongation factor 1-gamma |
| RPL17 | 60S ribosomal protein L17 | KRT80 | Keratin, type II cytoskeletal 80 |
| RAB7A | Ras-related protein Rab-7a | PKP1 | Plakophilin-1 |
| YWHAZ | 14-3-3 protein zeta/delta | VCP | Transitional endoplasmic reticulum ATPase |
| TPM3 | Tropomyosin alpha-3 chain | CSTA | Cystatin-A |
| CYR61 | Protein CYR61 | CALML3 | Calmodulin-like protein 3 |
| HNRNPDL | Heterogeneous nuclear ribonucleoprotein D-like | NME2 | Nucleoside diphosphate kinase B |
| DBNL | Drebrin-like protein | ARF1 | ADP-ribosylation factor 1 |
| RPL4 | 60S ribosomal protein L4 | GGCT | Gamma-glutamylcyclotransferase |
| CD44 | CD44 antigen | PRDX6 | Peroxiredoxin-6 |
| HNRNPU | Heterogeneous nuclear ribonucleoprotein U | YWHAE | 14-3-3 protein epsilon |
| FLG2 | Filaggrin-2 | PGAM1 | Phosphoglycerate mutase 1 |
| BLMH | Bleomycin hydrolase | RPS4X | 40S ribosomal protein S4, X isoform |
| FUS | RNA-binding protein FUS | RPL7A | 60S ribosomal protein L7a |
| ATP5A1 | ATP synthase subunit alpha, mitochondrial | IGHG1 | Ig gamma-1 chain C region |
| VCL | Vinculin | SERPINB12 | Serprin B12 |
| CLTC | Clathrin heavy chain 1 | PDIA6 | Protein disulfide-isomerase A6 |
| TUBB4B | Tubulin beta-2C chain |
Table 2. Significantly Changed Expression Proteins of HGF After Treatment of HGFs With Cyclosporin A .
| Gene name | Protein name |
| PGLS | 6-Phosphogluconolactonase |
| GSTO1 | Glutathione S-transferase omega-1 |
| ARHGDIA | Rho GDP-dissociation inhibitor 1 |
| CTSD | Cathepsin D |
| PRDX1 | Peroxiredoxin-1 |
| LGALS3 | Galectin-3 |
| ACAT1 | Acetyl-CoA acetyltransferase, mitochondrial |
| ALDO | A Fructose-bisphosphate aldolase A |
| EEF1A1 | Elongation factor 1-alpha 1 |
| ANXA2 | Annexin A2 |
Construction of the PPI Network and Enrichment Analysis
The network was analyzed based on topological parameters such as node degree and betweenness centrality (BC) using Cytoscape version 3.4.29 High degree values are matched to the hub nodes. The Cluster ONE (a plug-in for Cytoscape) with minimum size 3 and minimum density <0.05 parameters was used to cluster the whole network for identification of densely connected region(s) and further divide the PPI into module(s). To further understand the biological relevance of the hub proteins and proteins in one cluster after treatment by laser or CsA, we performed enrichment analysis using gene ontology (GO) including biological process (BP), molecular function (MF), and cellular component (CC). This analysis was done via ClueGO application of Cytoscape.
Results
PPI Network Construction, Key Nodes Identification and Gene Ontology Analysis
The protein codes were searched in Uniprot database (http://www.uniprot.org), the accession numbers of the selected proteins were used for PPI network construction. Topological centralities (degree and BC) were evaluated to distinguish the biological value of proteins. Degree introduced as the number of edges linked to a node and nodes with high degree represented as hub which may have important biological functions.BC indicates the importance of the node based on the number of shortest paths through each node.30 The final network was visualized based on these parameters wherein we mapped the node degree to the size and BC to color in the network view (Figure 1). The number of 20 top nodes of the networks was selected as the critical nodes (Hub proteins) based on degree value (see Tables 3 and 4).
Figure 1.
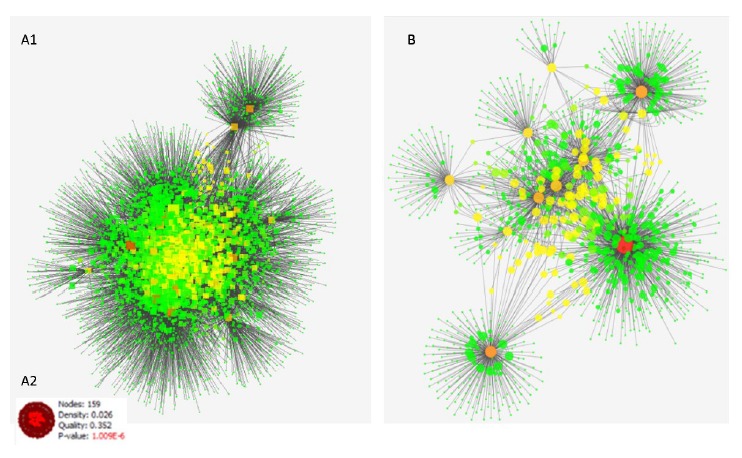
Schema of PPI Networks. (A1) PPI network for significantly changed expression proteins after Er:YAG irradiation in HGFs which consists of 4796 nodes and 15968 edges. (A2) The most significant enrichment module (P = 1.009E-6, node=159 and density =0.02) was extracted from PPI network of Er:YAG laser therapy via Cluster ONE. (B) PPI network for significantly changed expression proteins for HGFs treated by CsA which consists of 1220 nodes and 2247 edges. The node size and color changes indicate the value of proteins based on centrality parameters. As the circles get bigger, their value of degree centrality increases and the nodes which colors change from green to red, their BC value increases.
Table 3. The 20 Top Nodes of Er:YAG Laser Treated Network of HGFs Based on Degree Values .
| ID | Degree | BC value |
| *YWHAZ | 1491 | 0.2123 |
| *VCP | 1160 | 0.1539 |
| *HNRNPU | 1037 | 0.1370 |
| *YWHAE | 869 | 0.0894 |
| *UBA52 | 678 | 0.1290 |
| *CLTC | 644 | 0.0879 |
| *FUS | 604 | 0.0917 |
| RPS3 | 553 | 0.0383 |
| SFN | 543 | 0.0489 |
| RPS4X | 487 | 0.0160 |
| EEF1G | 451 | 0.04550 |
| RACK1 | 412 | 0.0410 |
| RPL22 | 397 | 0.0179 |
| RPL4 | 390 | 0.0179 |
| RPL7A | 369 | 0.0106 |
| *IGHG1 | 356 | 0.0826 |
| EIF4A1 | 351 | 0.0314 |
| RPL17 | 342 | 0.0119 |
| RAB7A | 313 | 0.0440 |
| TPM3 | 310 | 0.0344 |
Note: The data are sorted according to degree score from biggest to smallest. Corresponding BC values are also shown. The asterisked nodes are introduced as hub-bottleneck genes (BC cut off above 0.05).
Table 4. The Top 20 nodes of PPI Network of HGFs Treated by CsA Based on Degree Values .
| ID | Degree | BC Value |
| *EEF1A1 | 713 | 0.5419 |
| *ARHGDIA | 300 | 0.2434 |
| *LGALS3 | 275 | 0.2811 |
| *PRDX1 | 252 | 0.1421 |
| *ANXA2 | 251 | 0.1655 |
| *ALDOA | 201 | 0.1850 |
| *CTSD | 107 | 0.1087 |
| *ACAT1 | 91 | 0.0868 |
| GSTO1 | 48 | 0.0323 |
| PGLS | 29 | 0.0283 |
| VCAM1 | 28 | 0.0162 |
| RAC1 | 23 | 0.0069 |
| MLH1 | 18 | 0.0006 |
| RHOA | 18 | 0.0000 |
| CDC42 | 15 | 0.0025 |
| MDM2 | 14 | 0.0000 |
| EGFR | 13 | 0.0243 |
| STK4 | 13 | 0.0000 |
| FN1 | 12 | 0.0384 |
| ALDOC | 12 | 0.0000 |
Note: The data are sorted according to degree score from biggest to smallest. Corresponding BC values are shown. The asterisked nodes are introduced as hub-bottleneck genes (BC cut off above 0.05).
GO categories were analyzed to identify the function of hubs. The BP, CC and MF enrichment results of these key proteins are shown in Table 5.
Table 5. The Gene Ontology Enrichment Analysis Of Hub- Bottleneck Proteins of Both Therapeutic Modalities (up: Er:YAG Laser and down: cyclosporine A) via ClueGo Application of Cytoscape .
| Gene ID | |
|
YWHAZ
VCP HNRNPU YWHAE UBA52 CLTC FUS IGHG1 |
BP:
1) protein insertion into membrane 2) error-free translation synthesis CC: Not found MF: Not found |
| Gene ID | |
|
EEF1A1
ARHGDIA LGALS3 PRDX1 ANXA2 ALDOA CTSD ACAT1 |
BP: Negative regulation of endocytosis CC: Immunological synapse MF: Not found |
Densely Connected Region in PPI Network and Gene Ontology
Through Cluster ONE Analysis, there was one significant module (P <1 × 10-3) with the parameters of minimum size 3 and minimum density < 0.05. The most significant enrichment module (P =1.009E-6, node = 159 and density = 0.02) was extracted from PPI network of Er:YAG laser irradiation (Figure A1 and Figure A2). GO functional enrichment analysis of the densely connected region (one module) with the threshold of P<0.05 for laser therapy is shown in Table 6. We found that the gene functions were mainly enriched in intracellular signaling cascades such as, fatty acid transmembrane transport, cytokinesis, regulation of RNA splicing, and asymmetric protein localization. No cluster(s) was found for PPI network of CsA therapy.
Table 6. The functional Gene Ontology Enrichment Analysis of Obtained Module Through Cluster ONE .
| Therapeutic Modalities | Obtained Module by Cluster ONE |
| Laser (Er:YAG) | Fatty acid trans membrane transport |
| Cytokinesis | |
| Regulation of RNA splicing | |
| Asymmetric protein localization | |
| Cyclosporine A | Not found |
Discussion
Very little is known about the molecular analysis of the periodontal pocket tissue.31 Periodontitis is caused by bacterial infection and in addition to tooth loss, it can promote diabetic and cardiovascular diseases.3,5 New drug targets and biomarkers that facilitate treatment are essentially needed. The establishment of a network for treatment of periodontal disease would highly be helpful to understand the molecular mechanism and desirable periodontitis therapy. For further understanding of the molecular effect of Er:YAG laser irradiation and CsA therapy on HGFs, we analyzed the PPI network of both modalities separately. We gained 57 and 10 differentially expressed proteins after induction of Er:YAG laser and CsA on HGFs, respectively. PPI network as a powerful tool for analysis and ranking of involved proteins32 in the 2 therapeutic methods led to the construction of 2 scale free networks (see Figure 1). PPI network related to Er:YAG irradiation in HGFs consisted of 4796 nodes and 15 968 edges (about 3 links per node) while the related network to treated HGFs by cyclosporin contained 1220 nodes and 2247 edges (about 2 links per node). As it is shown in the Tbles 2 and 3, topological analyses of the 2 networks revealed 8 hub-bottleneck proteins for each of the networks. YWHAZ, VCP, HNRNPU, YWHAE, UBA52, CLTC, FUS and IGHG1 are hub-bottleneck proteins in the related network to the fibroblast cells treated by laser. In the drug treated cells, EEF1A1, ARHGDI, LGALS3, PRDX1, ANXA2, ALDOA, CTSD and ACAT1 are the crucial proteins. The findings indicate that the key proteins related to the laser treated network are more interacted in the network compared to the CsA network. The high BC values for crucial proteins of CsA therapy demonstrates network relative to the key nodes of the other network (see Tables 3 and 4), and shows that in drug network more shortest paths pass through the nodes. Comparison of the 2 panels shows that the key proteins of the 2 networks are completely different. Similar to these findings, GO enrichment analysis of hub-bottleneck proteins of networks and also functional GO enrichment analysis of obtained module indicate 2 different conditions. It seems that representation of a brief description about biological roles of the top key proteins in the 2 applied methods can provide a useful perspective of the involved mechanisms of treatment procedures.
14-3-3 protein zeta/delta (YWHAZ) is the most significant hub with highest BC value in PPI network of laser irradiation. 14-3-3 proteins are involved in a wide range of biological processes, including protein transport, transcription, cell cycle,33 and TNF production during inflammation response.34,35 14-3-3 protein sigma and zeta/delta were under-expressed in pathological tissue of periodontal patients.30 In other words, it has been suggested that 14-3-3 proteins may cause wound healing and tissue repair.36 This finding suggests that the gingival fibroblast repair can be facilitated by laser irradiation through overexpression of 14-3-3 proteins. Transitional endoplasmic reticulum ATPase (VCP) segregates protein molecules from large cellular structures and their reassembly after mitosis.37 p97/VCP, a member of the AAA-ATPase super family, has been associated with some wide variety pathways such as cell cycle, suppression of apoptosis, DNA-damage response and Golgi reassembly.38-40 VCP protein expression change via Er:YAG laser in gingival fibroblast is reported.28 In addition to the role of this type of ATPase in cell cycle, laser may influence wound healing through mitochondrial respiration which is followed by ATP synthesis induction.41 Heterogeneous nuclear ribonucleoprotein U (HNRNPU) is a RNA binding protein which influences pre-mRNA processing and other aspects of mRNA metabolism.37 Its role in cell proliferation process is recognized.38 Aleksic et al39 and Pourzarandian et al40 have reported that Er:YAG laser irradiation induced an elevation in the proliferative activity of HGFs. These reports are compatible with our results that are pointed to the roles of key proteins in cell cycle regulation and proliferation induction. The most significant connectivity in PPI network of CsA therapy network belongs to elongation factor 1-alpha 1 which is an isoform of elongation factor 1. It plays a role in translation and is responsible for the enzymatic delivery of amino acyl tRNA to the ribosome.33,34,41 It participates in several processes required for proliferation, mitotic apparatus formation, and signal transduction.34-36 In a previous study, it was reported that elongation factor 1-alpha 1 is differentially expressed in PDL cell in presence of enamel matrix protein in culture medium. Increase was demonstrated in RNA synthesis, which causes periodontal regeneration and wound healing.42 Rho GDP-dissociation inhibitor 1, as an important protein in PPI network of CsA therapy, plays a significant role in the regulation of signaling through Rho GTPases. Activity of this protein is important in a variety of cellular processes and expression of this gene may be altered in tumors.43 This protein has been reported as a common differentially expressed protein after laser irradiation28 and also CsA therapy on HGFs.11 However it was not included as a hub-bottleneck node in the network of laser treated fibroblasts. Galectin-3 is another crucial protein in PPI network of CsA therapy which has a variety of biological functions, including growth regulator, apoptosis inhibitor and interacting with Bcl-2.44 One key protein in PPI network of treated-HGFs by CsA was peroxiredoxin-1 which is reported to protect cells from apoptosis through the inhibition of c-Myc, NF-k B, c-Abl, and TNF-α.45 This protein may play the role of an antioxidant in promoting proliferation in the CsA-treated HGF by reducing cytosolic ROS levels.11
Through functional enrichment analysis of the hub-bottleneck proteins (see Table 5), we identified that protein insertion into membrane and error-free translation synthesis in the laser related network and regulation of endocytosis for drug network are the significant biological processes. Signaling in cells is regulated by endocytosis at multiple levels, such as control cell fate and proliferation.46 Fatty acid transmembrane transport has the most significant score in GO enrichment analysis of proteins in densely connected region in PPI network after Er:YAG laser irradiation (see Table 6). The evidences suggest that saturated fatty acids induce inflammation by imitating the actions of LPS.47 Fatty acids regulate the inflammatory and metabolic genes.48 Some studies showed the down-regulation of inflammatory proteins after Er:YAG laser irradiation.28 This phenomenon suggests that the therapeutic effect of low-level Er:YAG laser can reduce the inflammation of damaged periodontal sites. Further detailed studies are still necessary to validate this hypothesis. Another significant pathway was cytokinesis that is the physical process of cell division and the production of 2 daughter cells which can emphasize the role of Er:YAG laser on cell cycle regulation. In another study, the results of the proteomic investigation on HGFs treated by CsA11 proved the proliferative effect of this drug. We performed quantitative validation of the causal proteins in both PPI networks followed by functional enrichment analysis. The literature validations of hub genes have also confirmed their roles in the cell cycle and proliferation.11,28 It is thus suggested that a proliferation response induced by both mentioned therapeutic methods is oriented to regeneration of gingival tissues and wound healing despite the difference in the elements of the 2 related hub-bottleneck profiles.
Conclusion
Building the PPI network and mining pathways could be regarded as an effective way to explore the biological concepts behind the therapeutic methods. In the present study, the key proteins and their pathways in 2 therapeutic modalities on HGFs, including application of Er:YAG laser irradiation and CsA therapy, were investigated. The results correspond to the induction of proliferation in HGFs as a common effect of both mentioned therapeutic methods, which can be useful in the treatment of periodontal diseases. Introducing 2 different biomarker panels for the 2 methods indicates the possibility of multiple therapeutic procedures for treatment of patients. So investigation about efficiency and side effects of these procedures is an important requirement for selecting the best therapeutic method. It can be concluded that combination of the 2 methods may be an effective procedure to apply in the field. However more investigations are needed about the synergic effects of the 2 methods.
Ethical Considerations
The proposal of study was approved by the Ethics Committee, deputy of research, Shahid Beheshti University of Medical Sciences (#IR.SBMU.Retech.REC.1395.387)
Conflict to Interests
Authors declare that they have no Conflict to interests.
Acknowledgments
The present research is supported by Shahid Beheshti University of Medical Sciences, Tehran, Iran.
Please cite this article as follows: Abbaszadeh HA, Peyvandi AA, Sadeghi Y, et al. Er:YAG laser and cyclosporin A effect on cell cycle regulation of human gingival fibroblast cells. J Lasers Med Sci. 2017;8(3):143-149. doi:10.15171/jlms.2017.26.
References
- 1.Apsey DJ, Kaciroti N, Loesche WJ. The diagnosis of periodontal disease in private practice. J Periodontol. 2006;77(9):1572–1581. doi: 10.1902/jop.2006.050449. [DOI] [PubMed] [Google Scholar]
- 2.Flemmig TF. Periodontitis. Ann Periodontol. 1999;4(1):32–37. doi: 10.1902/annals.1999.4.1.32. [DOI] [PubMed] [Google Scholar]
- 3.Ozden FO, Ozgönenel O, Ozden B, Aydogdu A. Diagnosis of periodontal diseases using different classification algorithms: a preliminary study. Niger J Clin Pract. 2015;18(3):416–421. doi: 10.4103/1119-3077.151785. [DOI] [PubMed] [Google Scholar]
- 4.Genco RJ, Van Dyke TE. Prevention: reducing the risk of CVD in patients with periodontitis. Nat Rev Cardiol. 2010;7(9):479–480. doi: 10.1038/nrcardio.2010.120. [DOI] [PubMed] [Google Scholar]
- 5.Ahmed U, Tanwir F. Association of periodontal pathogenesis and cardiovascular diseases: a literature review. Oral Health Prev Dent. 2015;13(1):21–27. doi: 10.3290/j.ohpd.a32823. [DOI] [PubMed] [Google Scholar]
- 6.Oda S, Nitta H, Setoguchi T, Izumi Y, Ishikawa I. Current concepts and advances in manual and power‐driven instrumentation. Periodontol 2000. 2004;36(1):45–58. doi: 10.1111/j.1600-0757.2004.03674.x. [DOI] [PubMed] [Google Scholar]
- 7.Krohn-Dale I, Boe OE, Enersen M, Leknes KN. Er:YAG laser in the treatment of periodontal sites with recurring chronic inflammation: a 12-month randomized, controlled clinical trial. J Clin Periodontol. 2012;39(8):745–52. doi: 10.1111/j.1600-051X.2012.01912.x. [DOI] [PubMed] [Google Scholar]
- 8.Strauss RA, Fallon SD. Lasers in contemporary oral and maxillofacial surgery. Dent Clin North Am. 2004;48(4):861–888. doi: 10.1016/j.cden.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 9.Passanezi E, Damante CA, DeRezende ML, Greghi SL. Lasers in periodontal therapy. Periodontol 2000. 2015;67(1):268–291. doi: 10.1111/prd.12067. [DOI] [PubMed] [Google Scholar]
- 10.Crespi R, Capparè P, Toscanelli I, Gherlone E, Romanos GE. Effects of Er: YAG laser compared to ultrasonic scaler in periodontal treatment: a 2-year follow-up split-mouth clinical study. J Periodontol. 2007;78(7):1195–1200. doi: 10.1902/jop.2007.060460. [DOI] [PubMed] [Google Scholar]
- 11.Jung JY, Kang GC, Jeong YJ, Kim SH, Kwak YG, Kim WJ. Proteomic analysis in cyclosporin A-induced overgrowth of human gingival fibroblasts. Biol Pharm Bull. 2009;32(8):1480–5. doi: 10.1248/bpb.32.1480. [DOI] [PubMed] [Google Scholar]
- 12.Sobral LM, Aseredo F, Agostini M. et al. Molecular events associated with ciclosporin A-induced gingival overgrowth are attenuated by Smad7 overexpression in fibroblasts. J Periodontal Res. 2012;47(2):149–158. doi: 10.1111/j.1600-0765.2011.01412.x. [DOI] [PubMed] [Google Scholar]
- 13.Ponnaiyan D, Jegadeesan V. Cyclosporine A: novel concepts in its role in drug-induced gingival overgrowth. Dent Res J (Isfahan) 2015;12(6):499–506. doi: 10.4103/1735-3327.170546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Williamson MS, Miller EK, Plemons J, Rees T, Iacopino AM. Cyclosporine A upregulates interleukin-6 gene expression in human gingiva: possible mechanism for gingival overgrowth. J Periodontol. 1994;65(10):895–903. doi: 10.1902/jop.1994.65.10.895. [DOI] [PubMed] [Google Scholar]
- 15.Mitic K, Popovska M, Pandilova M, Jovanovic R, Spasovski G. The role of inflammation and apoptosis in cyclosporine A – induced gingival overgrowth. Bosn J Basic Med Sci. 2013;13(1):14–20. doi: 10.17305/bjbms.2013.2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bartold P. Cyclosporine and gingival overgrowth. J Oral Pathol Med. 1987;16(9):463–468. doi: 10.1111/j.1600-0714.1987.tb00718.x. [DOI] [PubMed] [Google Scholar]
- 17.Song L, Yao J, He Z, Xu B. Genes related to inflammation and bone loss process in periodontitis suggested by bioinformatics methods. BMC Oral Health. 2015;15:105. doi: 10.1186/s12903-015-0086-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ara T, Kurata K, Hirai K. et al. Human gingival fibroblasts are critical in sustaining inflammation in periodontal disease. J Period Res. 2009;44(1):21–27. doi: 10.1111/j.1600-0765.2007.01041.x. [DOI] [PubMed] [Google Scholar]
- 19.Lee I-k, Lee M-j, Jang H-S. The interrelationship between human gingival fibroblast differentiation and cultivating time. Tissue Eng Regen Med. 2013;10(2):60–64. [Google Scholar]
- 20.Tavirani MR. Meningioma protein-protein interaction network. Arch Iran Med. 2014;17(4):262. [PubMed] [Google Scholar]
- 21.Safaei A, Tavirani MR, Oskouei AA, Azodi MZ, Mohebbi SR, Nikzamir AR. Protein-protein interaction network analysis of cirrhosis liver disease. Gastroenterol Hepatol Bed Bench. 2016;9(2):114. [PMC free article] [PubMed] [Google Scholar]
- 22.Lackey B, Nunes QM, Higham SM, Fernig DG, Valappil SP. In silico analyses of heparin binding proteins expression in human periodontal tissues. BMC Res Notes. 2016;9:53. doi: 10.1186/s13104-016-1857-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zamanian-Azodi M, Rezaei-Tavirani M, Rahmati-Rad S, Hasanzadeh H, Tavirani MR, Seyyedi SS. Protein-protein interaction network could reveal the relationship between the breast and colon cancer. Gastroenterol Hepatol Bed Bench. 2015;8(3):215. [PMC free article] [PubMed] [Google Scholar]
- 24.Safari-Alighiarloo N, Taghizadeh M, Rezaei-Tavirani M, Goliaei B, Peyvandi AA. Protein-protein interaction networks (PPI) and complex diseases. Gastroenterol Hepatol Bed Bench. 2014;7(1):17–31. [PMC free article] [PubMed] [Google Scholar]
- 25.Bakail M, Ochsenbein F. Targeting protein–protein interactions, a wide open field for drug design. C R Chim. 2016;19(1):19–27. doi: 10.1016/j.crci.2015.12.004. [DOI] [Google Scholar]
- 26.Zamanian–Azodi M, Rezaei–Tavirani M, Hasanzadeh H. et al. Introducing biomarker panel in esophageal, gastric, and colon cancers; a proteomic approach. Gastroenterol Hepatol Bed Bench. 2015;8(1):6–18. [PMC free article] [PubMed] [Google Scholar]
- 27.Ballatore C, Brunden KR, Trojanowski JQ, Lee VM, Smith AB 3rd, Huryn DM. Modulation of protein-protein interactions as a therapeutic strategy for the treatment of neurodegenerative tauopathies. Curr Top Med Chem. 2011;11(3):317–30. doi: 10.2174/156802611794072605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ogita M, Tsuchida S, Aoki A. et al. Increased cell proliferation and differential protein expression induced by low-level Er: YAG laser irradiation in human gingival fibroblasts: proteomic analysis. Lasers Med Sci. 2015;30(7):1855–1866. doi: 10.1007/s10103-014-1691-4. [DOI] [PubMed] [Google Scholar]
- 29.Shannon P, Markiel A, Ozier O. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Proulx SR, Promislow DE, Phillips PC. Network thinking in ecology and evolution. Trends Ecol Evol. 2005;20(6):345–353. doi: 10.1016/j.tree.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 31.Bertoldi C, Bellei E, Pellacani C. et al. Non‐bacterial protein expression in periodontal pockets by proteome analysis. J Clini Periodontol. 2013;40(6):573–582. doi: 10.1111/jcpe.12050. [DOI] [PubMed] [Google Scholar]
- 32.Xia J, Benner MJ, Hancock RE. NetworkAnalyst--integrative approaches for protein-protein interaction network analysis and visual exploration. Nucleic Acids Res. 2014;42(Web Server issue):W167–W74. doi: 10.1093/nar/gku443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang J, Fuller PJ, Morgan J. et al. Use of phage display to identify novel mineralocorticoid receptor-interacting proteins. Mol Endocrinol. 2014;28(9):1571–1584. doi: 10.1210/me.2014-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hamrita B, Nasr HB, Hammann P. et al. An elongation factor-like protein (EF-Tu) elicits a humoral response in infiltrating ductal breast carcinomas: an immunoproteomics investigation. Clin Biochem. 2011;44(13):1097–1104. doi: 10.1016/j.clinbiochem.2011.06.005. [DOI] [PubMed] [Google Scholar]
- 35.Morrissey C, Schwefel D, Ennis-Adeniran V, Taylor IA, Crow YJ, Webb M. The eukaryotic elongation factor eEF1A1 interacts with SAMHD1. Biochem J. 2015;466(1):69–76. doi: 10.1042/BJ20140203. [DOI] [PubMed] [Google Scholar]
- 36.Liu H, Ding J, Chen F. et al. Increased expression of elongation factor-1α is significantly correlated with poor prognosis of human prostate cancer. Scand Urol Nephrol. 2010;44(5):277–283. doi: 10.3109/00365599.2010.492787. [DOI] [PubMed] [Google Scholar]
- 37.Ye J, Beetz N, O’Keeffe S. et al. hnRNP U protein is required for normal pre-mRNA splicing and postnatal heart development and function. Proc Natl Acad Sci. 2015;112(23):E3020–E3029. doi: 10.1073/pnas.1508461112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bao K, Belibasakis GN, Selevsek N, Grossmann J, Bostanci N. Proteomic profiling of host-biofilm interactions in an oral infection model resembling the periodontal pocket. Sci Rep. 2015;5:15999. doi: 10.1038/srep15999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aleksic V, Aoki A, Iwasaki K. et al. Low-level Er: YAG laser irradiation enhances osteoblast proliferation through activation of MAPK/ERK. Lasers Med Sci. 2010;25(4):559–569. doi: 10.1007/s10103-010-0761-5. [DOI] [PubMed] [Google Scholar]
- 40.Pourzarandian A, Watanabe H, Ruwanpura SM, Aoki A, Ishikawa I. Effect of low-level Er: YAG laser irradiation on cultured human gingival fibroblasts. J Periodontol. 2005;76(2):187–193. doi: 10.1902/jop.2005.76.2.187. [DOI] [PubMed] [Google Scholar]
- 41.Becker M, Kuhse J, Kirsch J. Effects of two elongation factor 1A isoforms on the formation of gephyrin clusters at inhibitory synapses in hippocampal neurons. Histochem Cell Biol. 2013;140(6):603–609. doi: 10.1007/s00418-013-1122-9. [DOI] [PubMed] [Google Scholar]
- 42.Brett P, Parkar M, Olsen I, Tonetti M. Expression profiling of periodontal ligament cells stimulated with enamel matrix proteins in vitro: a model for tissue regeneration. J Dent Res. 2002;81(11):776–783. doi: 10.1177/0810776. [DOI] [PubMed] [Google Scholar]
- 43.Boulter E, Garcia-Mata R, Guilluy C. et al. Regulation of Rho GTPase crosstalk, degradation and activity by RhoGDI1. Nat Cell Biol. 2010;12(5):477–483. doi: 10.1038/ncb2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Califice S, Castronovo V, Van Den Brûle F. Galectin-3 and cancer (Review) Int J Oncol. 2004;25(4):983–992. [PubMed] [Google Scholar]
- 45.Zhang P, Liu B, Seo MS, Rhee SG, Obeid LM. Thioredoxin peroxidase is a novel inhibitor of apoptosis with a mechanism distinct from that of Bcl-2. J Biol Chem. 1997;272(49):30615–30618. doi: 10.1074/jbc.272.49.30615. [DOI] [PubMed] [Google Scholar]
- 46.Barbieri E, Di Fiore PP, Sigismund S. Endocytic control of signaling at the plasma membrane. Curr Opin Cell Biol. 2016;39:21–27. doi: 10.1016/j.ceb.2016.01.012. [DOI] [PubMed] [Google Scholar]
- 47.Fritsche KL. The science of fatty acids and inflammation. Adv Nut. 2015;6(3):293S–301S. doi: 10.3945/an.114.006940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Masi LN, Rodrigues AC, Curi R. Fatty acids regulation of inflammatory and metabolic genes. Curr Opin Clin Nut Metab Care. 2013;16(4):418–424. doi: 10.1097/MCO.0b013e32836236df. [DOI] [PubMed] [Google Scholar]


