Abstract
Pheromone receptors (PRs) are essential in moths to detect sex pheromones for mate finding. However, it remains unknown from which ancestral proteins these specialized receptors arose. The oldest lineages of moths, so-called non-ditrysian moths, use short-chain pheromone components, secondary alcohols, or ketones, so called Type 0 pheromones that are similar to many common plant volatiles. It is, therefore, possible that receptors for these ancestral pheromones evolved from receptors detecting plant volatiles. Hence, we identified the odorant receptors (ORs) from a non-ditrysian moth, Eriocrania semipurpurella (Eriocraniidae, Lepidoptera), and performed functional characterization of ORs using HEK293 cells. We report the first receptors that respond to Type 0 pheromone compounds; EsemOR3 displayed highest sensitivity toward (2S, 6Z)-6-nonen-2-ol, whereas EsemOR5 was most sensitive to the behavioral antagonist (Z)-6-nonen-2-one. These receptors also respond to plant volatiles of similar chemical structures, but with lower sensitivity. Phylogenetically, EsemOR3 and EsemOR5 group with a plant volatile-responding receptor from the tortricid moth Epiphyas postvittana (EposOR3), which together reside outside the previously defined lepidopteran PR clade that contains the PRs from more derived lepidopteran families. In addition, one receptor (EsemOR1) that falls at the base of the lepidopteran PR clade, responded specifically to β-caryophyllene and not to any other additional plant or pheromone compounds. Our results suggest that PRs for Type 0 pheromones have evolved from ORs that detect structurally-related plant volatiles. They are unrelated to PRs detecting pheromones in more derived Lepidoptera, which, in turn, also independently may have evolved a novel function from ORs detecting plant volatiles.
Keywords: Eriocrania semipurpurella, odorant receptor, sex pheromone, HEK293 cells, deorphanization
Introduction
Detection of sex pheromones in insects is crucial when it comes to finding a partner for mating. In moths (Lepidoptera), it is generally the females that release a species-specific sex pheromone blend, which is received by and attractive to conspecific males over long distances (Löfstedt and Kozlov 1997; Ando etal. 2004). Moths, like many other insects, have specialized olfactory systems to detect the components of these mating signals (Baker 2008; Hansson and Stensmyr 2011; Zhang etal. 2016). The species-specificity of pheromone emission and detection forms a robust mate recognition system, which limits heterospecific mating events (Linn and Roelofs 1995).
On the basis of their site of production, distinctive chemical structure, and biosynthetic features, lepidopteran sex pheromones are classified into four major groups: Type I pheromones are C10–C18 acetates, alcohols, and aldehydes, which are used by ∼75% of moth species (Ando etal. 2004; Löfstedt etal. 2016). Type II pheromones are C17–C25 unbranched polyunsaturated hydrocarbons or the corresponding epoxide derivatives. This second major group of pheromones comprises ∼15% of all reported moth pheromones (Ando etal. 2004; Löfstedt etal. 2016). Type III pheromones are thought to have a distinct biosynthetic origin compared with Type I and II pheromones, but all contain one or more methyl branches. These compounds include C17–C23 saturated and unsaturated hydrocarbons, as well as functionalized hydrocarbons. This pheromone type only occurs in a few phylogenetic lineages (Löfstedt etal. 2016). Type 0 pheromones are so called because they have been reported in two of the oldest so-called non-ditrysian lineages of Lepidoptera and in the Trichoptera (caddisflies), the sister group to the Lepidoptera, and thus are thought to represent the ancestral type of pheromone (fig. 1). Type 0 pheromones are short-chain secondary alcohols or ketones that are similar to some general plant volatile compounds (Löfstedt etal. 1994, 2016; Kozlov etal. 1996; Löfstedt and Kozlov 1997).
Fig. 1.
Phylogenetic tree of the major lepidopteran lineages and the sister order Trichoptera with the proposed evolution of the different sex pheromone types mapped onto it using differently colored branches. Only taxa with reported sex pheromones or sex attractants are included in the tree. Adapted from Löfstedt etal. (2016).
Pheromones and other odorants are detected by odorant receptors (ORs) expressed in olfactory sensory neurons (OSNs) located within the olfactory sensilla mainly on the insect antennae. These receptors are seven-transmembrane proteins that form heteromeric complexes of unknown stoichiometry with a highly conserved olfactory coreceptor (Orco) (Vosshall and Hansson 2011). The receptor complexes function as ligand-gated ion channels (Neuhaus etal. 2004; Sato etal. 2008), although some evidence suggests that these complexes also utilize metabotropic signaling (Wicher etal. 2008; Carraher etal. 2015). In general, pheromone compounds in moths and other insects are detected by specialized receptors (Nakagawa etal. 2005; Zhang and Löfstedt 2013; Dweck etal. 2015; Andersson etal. 2016) that generally do not respond to plant volatiles.
New sex pheromone signals have evolved throughout the Lepidoptera during >200 million years since this taxon diverged from the Trichoptera (Löfstedt etal. 2016). The variation of sex pheromone signals has been studied and described extensively in many moth species (Symonds and Elgar 2008; Allison and Cardé 2016). To match the variation in the emitted signal, the responding males have evolved a parallel detection system, however, to date little is known about the evolution of pheromone receptors (PRs) and their specificity. Minor changes in the ratio of pheromone components may be detected using existing PRs. In addition, positive selection can stabilize mutations in OR loci to gain a novel function (Gardiner etal. 2008; Leary etal. 2012; Zhang and Löfstedt 2013; Andersson etal. 2015), and even a single mutation in the sequence of a receptor can modify its specificity (Leary etal. 2012; Steinwender etal. 2015). Each major type of pheromone (Type 0–III), however, represents a distinct chemical class that involves different biosynthetic pathways (Löfstedt etal. 2016), and a transition from one type to another would represent a major evolutionary transition. We sought to find out whether major changes in the pheromone signal recruited new OR lineages or just modifications of the selectivity of existing receptors.
PRs for Type I pheromones have been characterized from an increasing number of moth species (Zhang and Löfstedt 2015), all belonging to the clade Ditrysia that contains over 98% of extant lepidopterans. Together these receptors form a separate phylogenetic clade (the “PR” clade) among lepidopteran ORs (Wanner etal. 2007; Engsontia etal. 2014; Koenig etal. 2015; Zhang and Löfstedt 2015), each containing sequence motifs (Bengtsson etal. 2012; Zhang and Löfstedt 2013) that are well-conserved within the clade. A receptor responding to a typical Type II pheromone component in the winter moth, Operophtera brumata, resides within the PR clade containing receptors for Type I pheromones (Zhang etal. 2016). Type 0 pheromones are considered ancestral to the more derived Type I and Type II pheromones (Löfstedt etal. 2016). Characterization of receptors for Type 0 pheromones could thus provide insight as to whether the PRs of more derived ditrysian moths share an evolutionary origin with those detecting ancestral Type 0 pheromones in older non-ditrysian moth lineages, or whether receptors for different pheromone types have evolved independently. Indeed, since Type 0 pheromones are similar to plant volatiles, the receptors for Type 0 pheromones may have evolved from general plant volatile-detecting ORs through structural mutations in existing receptors or following gene duplication events.
The leaf miner moth, Eriocrania semipurpurella, belongs to the family Eriocraniidae, which is a basal nondistrysian lineage within the Lepidoptera that uses Type 0 sex pheromones (fig. 1). The presence of E. semipurpurella has been reported in birch forests in North America, Europe, and Japan (Bylund and Tenow 1994; Imada etal. 2011) where the females lay eggs in flower buds and the larvae feed on the leaves of the trees (Bylund and Tenow 1994). The fifth abdominal segment of female E. semipurpurella contains a pair of exocrine glands which produce the sex pheromone components (2S, 6Z)-6-nonen-2-ol and (2R, 6Z)-6-nonen-2-ol. These components are attractive to males, but not to females (Larsson etal. 2002). The female pheromone glands also contain the presumed precursors, (Z)-6-nonen-2-one and nonan-2-one, both of which antagonize male sex pheromone attraction (Kozlov etal. 1996). In contrast, Larsson etal. (2002) showed that a low concentration of nonan-2-one increases pheromone attraction, while a similar concentration dependent effect has not been demonstrated for (Z)-6-nonen-2-one. This compound is also an antagonist for the closely related and sympatric species E. sangii, which produces the same compounds as E. semipurpurella, but in different ratios (Kozlov etal. 1996). One of the major pheromone components of E. semipurpurella, (2 R, 6Z)-6-nonen-2-ol, is found in trace amounts in the pheromone gland of E. sangii and acts as an antagonist in this species when added in >5% to the S enantiomer that is the major pheromone component in E. sangii (Kozlov etal. 1996). Electrophysiological recordings previously identified five OSN types in male E. semipurpurella that respond to pheromone components of E. semipurpurella and the two other sympatric species E. cicatricella and E. sparrmannella (Larsson etal. 2002). The pheromones of E. cicatricella and E. sparmennella comprise seven carbon alcohols and ketones, and are thus largely different from the pheromone of E. semipurpurella (Zhu etal. 1995).
Here we have used transcriptomic analysis, phylogenetic approaches, and an invitro functional assay to identify ORs from E. semipurpurella responding to Type 0 pheromone components and plant volatiles. Our results suggest that sex PRs in Lepidoptera have evolved their ability to detect sex pheromones from ORs that detect plant volatiles.
Results
Transcriptomic Analysis
Sixty-five million reads from male antennal RNA were assembled into 68,151 unigenes with a mean length of 818 bp and an N50-value of 1,761 bp. In total, 37 OR genes, including an Orco orthologue, were identified from the male antennal E. semipurpurella transcriptome. Among the 37 assembled OR transcripts, 24 were full-length. Two OR transcripts lacked the start codon despite encoding predicted proteins of >400 amino acids, three transcripts encoded partial OR sequences ranging from 300 to 400 amino acids, and the remaining eight transcripts encoded shorter OR fragments of <200 amino acids. The EsemORs were labeled EsemOR1-37 and EsemOrco according to the unified nomenclature system (Vosshall and Hansson 2011), with those that have been functionally assayed given the labels EsemOR1, 3-6, and the remaining EsemORs numbered EsemOR7-37 in the order they were identified from the transcriptome. To avoid confusion, no EsemOR was given the label EsemOR2, because this number has previously been used for moth Orco proteins. The raw sequence reads have been deposited in the SRA database at NCBI under the Bioproject accession number SRR5328787, and the sequences of Orco and the five functionally assayed receptors have been deposited in GenBank (accession numbers KY750554–59). The transcriptome assembly was deposited in the TSA database at DDBJ/EMBL/GenBank under the accession GFQP00000000. The version described in this paper is the first version, GFQP01000000. On the basis of RNAseq counts, five E. semipurpurella OR genes (EsemOR3-5, 12, and EsemOrco) are expressed at high levels in male antennae (FPKM > 50). Eleven OR genes had FPKM values between 10 and 50, and the remaining OR genes had relatively lower levels of expression (≤10) (supplementary table S1, Supplementary Material online). The FPKM value for one OR (EsemOR4) was even higher than that of EsemOrco in the antennae.
Phylogenetic Analysis
Multiple sequence alignment (fig. 2) and subsequent phylogenetic analysis of predicted EsemOR proteins, together with ORs from Bombyx mori, Epiphyas postvittana, Plutella xylostella, Manduca sexta, and Spodoptera littoralis revealed two EsemORs, EsemOR1, and EsemOR6, that are related to known lepidopteran PRs, being located at the base of the PR clade with high support (i.e., FastTree support values > 97%; fig. 3). These EsemORs also contain sequence motifs that are similar to those conserved among PRs in more derived moths (fig. 2). The remaining EsemORs either fell individually or in small E. semipurpurella-specific clades across the tree (fig. 3), often being basal to subfamilies containing ORs from more derived moth species. With the exception of the conserved Orco, and perhaps EsemOR25 grouping together with SlitOR50, no simple one-to-one orthologous relationships between EsemORs and the ORs of the other species were evident. Interestingly, three other EsemORs (EsemOR3, 4, and 5) did not group phylogenetically with members of the lepidopteran PR clade, despite them containing sequence similarities within the conserved PR motifs (fig. 2).
Fig. 2.
Aligned C-terminal region of odorant receptors (ORs) from the “PR clade” of Bombyx mori (Bmor), Epiphyas postvittana (Epos), Manduca sexta (Msex), Plutella xylostella (Pxyl), Spodoptera littoralis (Slit), and five PR candidates from Eriocrania semipurpurella (Esem). Colors indicate identical amino acids by amino acid type and conserved motifs are highlighted with black rectangles.
Fig. 3.
(A) Maximum-likelihood phylogram based on protein sequences of odorant receptors (ORs) from Eriocrania semipurpurella (blue), Bombyx mori (orange), Epiphyas postvittana (green), Manduca sexta (black), Plutella xylostella (purple), and Spodoptera littoralis (red). The “pheromone receptor” clade is marked in yellow, the odorant receptors included in the recently extended “pheromone receptor” clade (Koenig etal. 2015) in cyan, and the EsemORs that responded to Type 0 pheromones in purple. Numbers on edges are local support values (0–1) calculated using a Shimodaira–Hasegawa test implemented within FastTree, and are only shown if > 0.70 and only on major branches. Sources of sequences contained in the tree are listed in supplementary table S2, Supplementary Material online. (B) Ligands that have been demonstrated to activate ORs in functional assays, compiled from the present study and previous studies (Nakagawa etal. 2005; Anderson etal. 2009; Tanaka etal. 2009; Sun etal. 2013; Corcoran etal. 2014; de Fouchier etal. 2017; Wicher etal. 2017). Letters beside compounds correspond to superscript notes next to ORs in (A).
To unravel whether ORs for Type 0 pheromones are ancestral to receptors in the PR clade of more derived moths, or if they are evolutionary related to ORs detecting structurally similar plant volatiles, five EsemORs (EsemOR1, 3, 4, 5, and 6) were chosen for functional characterization. This choice was based on several criteria, of which two or more had to be fulfilled for an OR to be selected (see Materials and Methods for additional details): 1) the presence of similarities to conserved PR motifs (Zhang and Löfstedt 2015) (fig. 2); 2) their close relationship with the conserved moth PR clade (EsemOR1 and 6; fig. 3); 3) their relatively high predicted expression levels in male antennae (supplementary table S1, Supplementary Material online); and 4) their relatively close relationship with SlitOR3 and EposOR3 (EsemOR3-5; fig. 3), previously identified ORs that are capable of detecting plant-volatiles (Jordan etal. 2009; Corcoran etal. 2014; de Fouchier etal. 2017).
OR Cloning, Sequence Verification, and Expression in HEK293 Cells
Full-length open reading frames (ORFs) of EsemOrco and EsemOR1, 3, 4, 5, and 6 were successfully cloned from cDNA and their sequences verified by comparison with those obtained from the transcriptome assembly. While some synonymous single nucleotide polymorphisms were found, all amino acid sequences were identical between those from the cDNAs and the transcriptome assembly. HEK293 cell lines expressing EsemOrco in combination with each of EsemOR1, 3, 4, 5, 6 were generated and receptor expression was verified by western blot. Proteins of the corresponding molecular weight of EsemOrco and EsemORs were detected from cell lysates prepared from induced cells for each cell line and not from non-induced cells, indicating that all proteins of interest were expressed in cells, and under proper regulation by the TREx repressor system (supplementary fig. S1, Supplementary Material online).
Functional Testing of EsemOR-Expressing HEK Cells
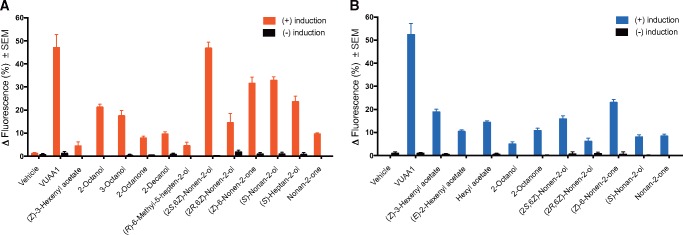
Cell lines expressing EsemOrco in combination with each of the five candidate PRs were tested for responsiveness to a panel of 85 compounds comprised of various plant volatiles, and Type 0 and Type I pheromone compounds, as well as to a vehicle (negative) control and the insect Orco agonist VUAA1. In all of the five cell lines tested, VUAA1 elicited responses in induced cells and not in non-induced cells, whereas no responses to the vehicle control were seen in any cells, whether induced or non-induced. Of the five candidate PRs tested, EsemOR1, EsemOR3, and EsemOR5 responded to compounds in the test panel when screened at the concentration of 30 µM (figs. 4 and 5). Induced cells expressing EsemOR3 responded most strongly to the pheromone component (2S, 6Z)-6-nonen-2-ol, but less so to other Type 0 pheromone compounds, including (S)-nonan-2-ol, (S)-heptan-2-ol, and (2R, 6Z)-6-nonen-2-ol, as well as the behavioral antagonists (Z)-6-nonen-2-one and nonan-2-one. Furthermore, EsemOR3-expressing cells also responded to a lesser extent to several common plant volatile compounds, including 2-octanol, 3-octanol, 2-octanone, 2-decanol, (Z)-3-hexenyl acetate, and (R)-6-methyl-5-hepten-2-ol (fig. 4A).
Fig. 4.
Responses of HEK293 cells transfected with EsemOrco and EsemOR3 (A) or EsemOrco and EsemOR5 (B) to vehicle control (0.5% DMSO), the Orco agonist VUAA1 (50 μM), and various pheromone compounds and plant volatiles (30 μM). Plotted values are the mean response of three biological replicates (±SEM) from induced cells (red bars, EsemOR3; blue bars, EsemOR5) and non-induced cells (black bars).
Fig. 5.
Response profile of HEK293 cells transfected with EsemOrco and EsemOR1 (A) to vehicle control (0.5% DMSO), the Orco agonist VUAA1 (50 μM), and selected pheromone compounds and plant volatiles (30 μM). Dose–response of HEK293 cells transfected with EsemOrco and EsemOR1 (B) to β-caryophyllene. Plotted values are the mean response of three biological replicates (±SEM) from induced cells (green bars & curve) and non-induced cells (black bars & curve).
Cells expressing EsemOR5 responded most strongly to the pheromone compound (Z)-6-nonen-2-one followed by (2S, 6Z)-6-nonen-2-ol. Weaker responses were elicited by (2R, 6Z)-6-nonen-2-ol, nonan-2-one and (S)-nonan-2-ol (fig. 4B). Similar to EsemOR3, cells expressing EsemOR5 responded to several common green leaf volatiles, including (Z)-3-hexenyl acetate, (E)-2-hexenyl acetate, hexyl acetate, 2-octanone, and 2-octanol, but the responses to the plant volatiles were weaker than those elicited by the pheromone compounds (fig. 4B). Cells expressing EsemOR1, which sits in a position basal to the PR clade, responded only to the plant compound β-caryophyllene (fig. 5A). No responses were observed to any of the compounds when tested on non-induced cells. Cells expressing EsemOR4 or EsemOR6 did not respond to any of the 85 compounds tested.
Both EsemOR3 and EsemOR5-expressing cells responded in a dose-dependent manner to agonists identified in screening experiments (fig. 6A and B). The PR EsemOR3 was the most sensitive to (2S, 6Z)-6-nonen-2-ol with an EC50 of 0.56 µM (95% CI: 0.31–1.02 µM). Apart from the second most active Type 0 pheromone compound (S)-nonen-2-ol, the EC50 of (2S, 6Z)-6-nonen-2-ol for EsemOR3 was significantly lower than for any of the other compounds tested (i.e., the 95% CI of the EC50 values did not overlap) (fig. 6C). The EC50 of (Z)-6-nonen-2-one (EC50: 0.43 µM, 95% CI: 0.25–0.75 µM) for EsemOR5 was significantly lower than for any of its other active ligands (fig. 6C). The dose–response assays also show that response specificity of both EsemOR3 and 5 is concentration-dependent, with fewer ligands being active at lower (compared with higher) stimulus concentrations (fig. 6). The β-caryophyllene response of cells expressing EsemOR1 was dose-dependent with an EC50 of 2.43 µM (95% CI: 1.75–3.40 µM) (fig. 5B).
Fig. 6.
Dose–responses of HEK293 cells transfected with EsemOrco and EsemOR3 (A) or EsemOrco and EsemOR5 (B) to various pheromone compounds and plant volatiles. Data represent the mean response (±SEM) of induced cells from three biological replicates. (C) EC50 with 95% confidence intervals (CI) for each compound. Note the different scales on the y-axes in (A) and (B).
Discussion
We have identified 37 ORs (including the EsemOR coreceptor, Orco) from the male antennal transcriptome of E. semipurpurella. This is fewer than previously identified from the genomes or transcriptomes of more derived species of Lepidoptera. For instance, 70 ORs have been identified in E. postvittana (Corcoran etal. 2015), 66 ORs in B. mori (International Silkworm Genome Consortium 2008), 64 ORs in Danaus plexippus (Zhan etal. 2011), 79 ORs in P. xylostella (Engsontia etal. 2014), and 70 ORs in Heliconius melpomene (The Heliconious Genome Consortium 2012). Because we could only analyze the male antennal transcriptome (females could not be collected in sufficient numbers in the field), the actual number of EsemORs encoded by the genome or expressed in both sexes and other life-stages together is likely to exceed 37. However, being a moth from a basal lineage, it is also possible that E. semipurpurella has fewer OR genes than more derived species. Indeed, extant basal insect taxa express low numbers of ORs (Missbach etal. 2014), whereas different OR lineages appear to have expanded in higher insects as a result of gene duplication (Nei etal. 2008; Missbach etal. 2014; Andersson etal. 2015; Benton 2015). Several studies have found correlations between the number of ORs expressed by adult insects and the number of glomeruli in their antennal lobes (Vosshall etal. 2000; Vosshall and Stocker 2007; Mitchell etal. 2017), so future morphological studies on the E. semipurpurella antennal lobe may help to resolve whether there may be more ORs expressed in adults of this species.
In the absence of receptors grouping within the PR clade, and without a comparable transcriptome from females allowing comparison of OR gene expression levels between the sexes, we had to rely on a different approach to select candidate PRs for functional characterization. Hence, we took a phylogenetic approach to identify the receptors that were more closely related to the lepidopteran PR clade. In addition, since receptors for sex pheromone compounds in moths and other insects with female-produced pheromones typically have male-biased expression (Mitsuno etal. 2008; Widmayer etal. 2009; Grosse-Wilde etal. 2011; Bengtsson etal. 2012; Liu etal. 2013; Andersson etal. 2014, 2016), we also targeted the ORs that were highly expressed in the male transcriptome, especially those that also showed similarity over the conserved sequence motifs common to the PRs of more derived moths, which we reasoned might indicate a role in pheromone detection. Despite the fact that none of the EsemORs grouped within the PR clade, the five selected PR candidates all showed different degrees of similarity to the motifs conserved within the PR clade. Our approach to select PR candidates turned out to be rather successful given that three of five tested receptors were functionally characterized and with two of these detecting pheromone components.
We were successful in attaining functional data for EsemOR1, 3, and 5. Our functional and phylogenetic analyses of EsemOR3 and EsemOR5 demonstrate that these receptors are most sensitive to Type 0 pheromones and support the hypothesis that receptors detecting Type 0 pheromones in E. semipurpurella are evolutionarily related to ORs detecting structurally similar plant volatiles. These PRs are not closely related to the receptors for Type I and II compounds within the lepidopteran PR clade (Wanner etal. 2007; Engsontia etal. 2014; Koenig etal. 2015; Zhang and Löfstedt 2015). In addition, EsemOR1, which in our phylogeny is basal to the PR clade of more derived moths, does not respond to any of the tested Type 0 or Type I pheromone compounds. Instead, this receptor only responded to the plant sesquiterpene, β-caryophyllene, despite the fact that 85 compounds of diverse chemical structure and ecological origin were included in our odor panel. β-caryophyllene is one of the most abundant components of the headspace of the birch tree, Betula pendula (Zhang etal. 1999), the host plant of E. semipurpurella. Unfortunately, EsemOR6, which groups closely with EsemOR1 at the base of the PR clade, did not respond to any of the compounds tested. However, other relatively closely related ORs, such as SlitOR4, SlitOR35, and SlitOR36, do respond to plant volatiles (fig. 3; de Fouchier etal. 2017). In addition, CpomOR3 from codling moth (Cydia pomonella) has a phylogenetic position within the PR clade and responds to pear ester, a common volatile emitted by plants (Bengtsson etal. 2014; Cattaneo etal. 2016). Taken together, these data and EsemOR1’s phylogenetic position basal to the PR clade and specificity for β-caryophyllene are consistent with a scenario where the receptors within the PR clade have evolved their role in detecting sex pheromone components from ORs that detect (longer-chained) plant volatiles. This hypothesis should be tested through further deorphanization of additional ORs of moth species in basal lepidopteran lineages. In addition, it is possible that EsemOR1 is more sensitive to other compounds not included in the present study, perhaps Type I pheromone components from other moth species.
No responses were recorded from EsemORs 4 and 6, despite verification of their presence as proteins in cell lines and the use of a wide range of test compounds. Lack of responses in some ORs is common when expressed in heterologous systems (Hallem and Carlson 2006; Carey etal. 2010; Wang etal. 2010; Andersson etal. 2016; de Fouchier etal. 2017). Several factors could explain the lack of responses for the EsemORs, including that 1) they are expressed in HEK293 cells but in insufficient quantities, 2) they might not incorporate properly in the cell membrane, and 3) they are tuned to other ecologically relevant compounds that were not tested. The lack of response in EsemOR4 was particularly surprising given its high estimated expression in the male antennae, which is common for receptors detecting sex pheromone components (Zhang and Löfstedt 2015). In fact, the FPKM value for EsemOR4 was higher than that for Orco, which is surprising given that Orco is expressed in all OR-expressing neurons. The assembled transcript contains no traces of other genes whose expression could bias its FPKM value. Assuming that the FPKM values for EsemOR4 and Orco are accurately predicted, a possible explanation for this finding is that the FPKM values (estimated at the mRNA level), do not accurately predict actual protein levels. For some genes, mRNA expression and protein expression are not well correlated (Lu etal. 2007; Li etal. 2014; Mayfield etal. 2016). Thus, it is possible that translational regulation is different between Orco and OR4 with the translation of OR4-mRNAs being inefficient or downregulated. Also, the mRNA expression of EsemOR4 and Orco should be verified using quantitative real-time PCR.
With the presence of motifs similar to those found in PRs of higher moths, it was somewhat unexpected that the five EsemORs all grouped outside the PR clade. However, this is likely explained by the phylogenetic analysis considering sequence variation equally across the length of the protein sequence, which seemingly has outweighed the sequence similarity in these few and relatively short motif regions. Whether or not the residues in these motifs have anything to do with pheromone detection or receptor specificity could be tested in future studies using site-directed mutagenesis and functional testing.
Our phylogenetic analysis indicates that the receptors for Type 0 pheromones in E. semipurpurella are evolutionary related to ORs that respond to plant volatiles. In addition, the secondary responses of these ORs to structurally similar plant volatiles, and their grouping within the same clade as the plant odor-responding EposOR3 and SlitOR3 (Jordan etal. 2009; Corcoran etal. 2014; de Fouchier etal. 2017), suggest that mutations altering the specificity of pre-existing or recently duplicated ORs can result in novel functions, and that these PRs might have been recruited from ORs detecting plant volatiles (Löfstedt and Kozlov 1997). Specifically, both EsemOR3 and EsemOR5 respond to seven and nine carbon alcohols and ketones and, with lower sensitivity, to some common green leaf volatiles (e.g., (Z)-3-hexenyl acetate, (E)-2-hexenyl acetate and hexyl acetate), which are structurally similar to the short-chain pheromone compounds of E. semipurpurella (Visser 1986; Löfstedt and Kozlov 1997). On the basis of previous studies showing that one or a few amino acid changes can alter OR specificity significantly (Gardiner etal. 2008; Leary etal. 2012; Zhang and Löfstedt 2013; Steinwender etal. 2015), it is possible that few sequence changes within the OR could underlie an altered response from plant volatiles to structurally similar pheromones. Such changes may not be regarded as a major evolutionary transition. However, a potential major “evolutionary challenge” could have been to change the interpretation of the resulting neuronal signal from one context (“plant”) to a different one (“sex”) for pheromone communication to evolve following mutations in ORs. For example, the OR specificity change might have to be accompanied by modified OR expression, such as novel expression in an OSN already projecting to pre-existing “sex centers” in the central nervous system. Alternatively, if the OR expression pattern remains unchanged, rewiring within the central olfactory centers might have been required to modify the interpretation of the signal. Given that the developmental circuit patterning programs in insects are relatively hard-wired, possibly constraining their response to selection (Imai etal. 2010; Ramdya and Benton 2010; Cande etal. 2013; Andersson etal. 2015), neuronal rewiring to change the interpretation of the signal could be regarded as a major evolutionary transition, similar to the changes occurring at the production side that resulted in a transition from Type 0 to Type I pheromone production in Lepidoptera. On the other hand, the evolutionary transition would have been facilitated if host odor detection by males already was strongly associated with attraction, mate finding, or sex. In other words, males might have been “pre-adapted” to the female pheromonal signal if they aggregated on host plants to find mates prior to the mutations that resulted in specificity changes of their ORs.
We also aimed to link the responses of the functionally characterized EsemORs to previously recorded OSN responses invivo. Previous single-sensillum recordings (SSR) of E. semipurpurella antennae identified five OSN types that respond to pheromone components of E. semipurpurella, E. sangii, E. cicatricella, and E. sparrmannella (Larsson etal. 2002). In the present invitro study, we tested a panel of 85 compounds, including the nine Type 0 pheromone compounds tested in the SSR study. Although the response profiles of EsemOR3 and EsemOR5 did not perfectly match any of the previously described OSN types, there was some correspondence, but with both ORs appearing more broadly tuned than their putative associated OSNs. Both EsemOR3 and OSN type 2 are most sensitive to (2S, 6Z)-6-nonen-2-ol, and the corresponding R-enantiomer is clearly less active (Larsson etal. 2002). Both enantiomers of this compound are crucial for sex pheromone attraction, and the R-enantiomer is detected by another specific OSN type (Larsson etal. 2002). Considering the nine compounds shared between the two studies, EsemOR3, but not OSN type 2, responded to some extent also to (Z)-6-nonen-2-one, (S)-heptan-2-ol, and more weakly to nonan-2-one, suggesting a broader tuning of the OR than the OSN. For EsemOR5, the best match is OSN type 4, with the OR and OSN both being most sensitive to the antagonist (Z)-6-nonen-2-one and less sensitive to nonan-2-one (Larsson etal. 2002). Again, additional compounds shared between the present and previous study activated the OR (to a lesser extent than did (Z)-6-nonen-2-one) but not the OSN (e.g., (2 S, 6Z)-6-nonen-2-ol, and (2 R, 6Z)-6-nonen-2-ol). A possible explanation for the apparent broader response recorded from the receptors invitro compared with the OSNs might be due to the lack of sensory neuron membrane proteins (SNMPs), odorant degrading enzymes (ODEs), and odorant binding proteins (OBPs) in the HEK293 cell system, which could theoretically affect pheromone-PR interactions. It has previously been reported that responses obtained from ORs in heterologous systems do not always match perfectly with those recorded from the native antennal environment. For instance, in S. littoralis, an OSN was found that only responded to (Z, E)-9, 12-tetradecadienyl acetate, however when SlitORs were expressed and tested in the Drosophila empty neuron system the receptor that responded to (Z, E)-9, 12-tetradecadienyl acetate also responded to (Z)-9-dodecenyl acetate (de Fouchier etal. 2015). Likewise, in Agrotis segetum, SSR led to the identification of an OSN that responded solely to (Z)-7-dodecenyl acetate (Löfstedt etal. 1982), however when AsegORs were expressed and tested in Xenopus oocytes the receptors that responded to this compound also responded to several of the other compounds tested (Zhang and Löfstedt 2013). An alternative explanation for the discrepancy between the present OR responses and previous OSN responses for E. semipurpurella might be that OSN type 2 and 4 do not express EsemOR3 and 5, respectively. If so, this would mean that the OSNs expressing these two ORs are still not identified, and that pheromone detection might partly occur via relatively broadly tuned OSNs and thus is more “combinatorial” than in more derived moths where most PRs are relatively specific (Andersson etal. 2015; Zhang and Löfstedt 2015). Additional SSR recordings and further odorant screening of PRs for Type I pheromones using expanded odor panels would be informative in this regard.
In conclusion, Type 0 pheromone compounds and their biosynthetic origins are distinct from those of Type I (and Type II) pheromones, and our results suggest that the receptors that detect these different pheromone types have evolved independently from different ancestral proteins. Furthermore, our results support the hypothesis that receptors for Type 0 pheromones have been recruited from ORs detecting plant volatiles. With the plant odor-specific response of EsemOR1 and its position being basal to the PR clade, a similar scenario might be true for the PRs within the “PR clade” of more derived moths that detect Type I and II pheromones. However, additional studies of ORs from more basal nondistrysian lineages within the Lepidoptera are needed to further test this hypothesis.
Materials and Methods
Insect Material
Adult male insects were collected using live traps baited with female pheromone compounds (2S, 6Z)-6-nonen-2-ol and (2R, 6Z)-6-nonen-2-ol (50 + 50 µg on a rubber septum) in a birch forest during spring 2014, 15 km east of Lund, Sweden (55°38′51.0″N 13°41′28.1″E, 39.53 m alt.).
RNA Preparation, Sequencing, and Unigene Annotation
Total RNA was extracted and purified from pools of 100 pairs of male antennae using Trizol reagent (Thermo Fisher Scientific, Carlsbad, CA) following the manufacturer‘s instructions. RNA yield and quality were verified, and DNase treatment was performed prior to sequencing. The resulting RNA sample was used for library construction and 100 bp paired-end RNA-seq was performed at the Beijing Genomics Institute (BGI, Hong Kong Co., Ltd.) on an Illumina HiSeq 2000 platform (Illumina, San Diego, CA), yielding 6 Gb of clean data. Adaptor sequences were trimmed, and low quality reads filtered out. Sequenced reads were then de novo assembled using the short reads assembling program Trinity (version 20121005, Grabherr etal. 2011) and clustered by TGICL (Pertea etal. 2003). Initial functional annotation of unigenes was performed by searching all unigene sequences against a pooled database of non-redundant (nr) proteins at NCBI (National Center for Biotechnology Information) (e-value cut-off at <1e-5), as well as KEGG (Kyoto Encyclopedia of Genes and Genomes), COG (Orthologous Groups of proteins) and SwissProt annotation with e-value cut-off at <1e-5. The expression levels of unigenes were estimated using the FPKM method (fragments per kb transcript per million mapped reads; FPKM = 106 C/NL/103, where C is the number of mappable reads on a transcript, L is the length of a transcript (kb), and N is the total number of mappable reads in millions).
Identification of EsemORs and Candidate EsemPRs
Initially, putative OR genes were identified from the above-mentioned annotations, ORFs identified, and the receptor sequence verified by BlastP searches against the nr database at NCBI, transmembrane predictions (TMHMM2), and Pfam searches. To ensure that no OR sequences had been missed from this annotation, we also performed exhaustive local blast searches against the E. semipupurella unigene database, using previously identified moth OR and PR sequences as queries. The amino acid sequences and predicted start and stop codons of identified EsemORs were then verified in multiple sequence alignments, including OR sequences from additional moth species. Finally, all identified EsemORs were also used as queries in additional local blast searches against the E. semipurpurella transcriptome. For unigenes encoding putative ORs sharing > 99% amino acid identity, only one “copy” (the one with the longest ORF) was included. Similarly, two short fragments (<75 amino acid) that did not overlap in multiple sequence alignments were discarded from further analyses because unigene status could not be confirmed.
Phylogenetic Analysis and Selection of EsemORs for Functional Assays
The amino acid sequences of ORs from E. semipurpurella (Eriocraniidae), B. mori (Bombycidae; International Silkworm Genome Consortium 2008), E. postvittana (Tortricidae; Corcoran etal. 2015), P. xylostella (Plutellidae; Engsontia etal. 2014), M. sexta (Sphingidae; Koenig etal. 2015), and S. littoralis (Noctuidae; de Fouchier etal. 2017; unpublished S. littoralis sequences SlitOR47-60 were kindly provided by Dr William Walker) were aligned using MAFFT (built-in plugin, Geneious R7: Biomatters, http://www.geneious.com; last accessed June 7, 2017). Misaligned sequences for which the alignment could not be corrected manually were removed to improve the quality of the analysis. The alignment was then used to build a maximum likelihood phylogram using FastTree 2.1.5 (Price etal. 2010), implemented in the Geneious R7 software (Biomatters, http://www.geneious.com). The phylogenetic tree was rooted with the subfamily of conserved olfactory coreceptors, Orco. The tree was rendered and color coded using FigTree V 1.4.2 (http://tree.bio.ed.ac.uk/software/figtree/; last accessed July 12, 2017). The sources for reference protein sequences used in the tree are presented in supplementary table S2, Supplementary Material online.
To test the hypothesis that the receptors for Type 0 pheromones might be ancestral to the receptors in the PR clade of more derived Lepidoptera, EsemOR1, and EsemOR6 that fell out as basal to the PR clade (see also Results) were chosen for functional analyses. These receptors also contain motifs that are similar (fig. 2), but not identical, to the motifs characteristic of PRs in more derived moths (Zhang and Löfstedt 2015). Three additional EsemORs (EsemOR3, 4, and 5) were included in the functional assays to test the alternative hypothesis that receptors for Type 0 pheromones are related to, or have evolved from, receptors detecting structurally similar plant volatiles. These three particular receptors were chosen based on their relatively high predicted expression levels in the male antennae (as expected for sex PRs; see supplementary table S1, Supplementary Material online for FPKM values of all unigenes encoding EsemORs), and because they form a clade that is sister to ORs previously shown to respond to plant volatiles (i.e., EposOR3 and SlitOR3) (Jordan etal. 2009; Corcoran etal. 2014; de Fouchier etal. 2017). These EsemORs also included motifs resembling those of PRs in more derived moths, despite these receptors falling well outside the defined PR clade. Some ORs with relatively high FPKM values (such as OR12, 23, 26, and 32) were not included in the functional assays because these ORs do not contain similarity to the PR motifs and are not phylogenetically related to the PR clade of more derived moths.
First Strand cDNA Synthesis and Gene Cloning
First-strand cDNA was synthesized from 1 μg total RNA using the ThermoScript RT-PCR system (Thermo Fisher Scientific) following the manufacturer‘s protocol, except that both oligo-dT primers and random hexamers were used in the reaction mixture. Prior to cDNA synthesis, the RNA was treated with DNAse (Thermo Fisher Scientific) and RT-PCRs were carried out in parallel with and without reverse transcriptase to confirm the elimination of genomic DNA. Full-length gene-specific primers were designed for EsemOrco and the EsemORs of interest based on the sequences from the transcriptome. The full-length genes were amplified using Platinum Pfu Polymerase (Thermo Fisher Scientific), and adenosine residues were added to the ends of PCR products using GoTaq Green Master mix (Thermo Fisher Scientific), analyzed on 0.7% TAE agarose gels, and purified using the Wizard SV Gel and PCR Clean-Up System (Promega, Madison, WI), following the manufacturer‘s instructions. The purified DNA fragments were ligated into pTZ57R/T (Thermo Fisher Scientific) overnight at 4 °C and transformed into TOP10 competent cells (Thermo Fisher Scientific). Successful transformation of colonies was confirmed by colony PCR using gene-specific primers. Positive colonies were grown in LB broth overnight with ampicillin, and plasmids were extracted using GeneJET plasmid miniprep kit (Thermo Fisher Scientific). Plasmids were then Sanger sequenced using a capillary 3130xL Genetic Analyser (Thermo Fisher Scientific) at the Department of Biology sequencing facility (Lund University, Lund, Sweden). Obtained sequences were aligned to transcriptomic sequences using MAFFT alignment in Geneious R7.1.5 (Biomatters, http://www.geneious.com; Kearse etal. 2012) software and plasmids containing target EsemOR ORFs with the correct sequence were used for further cloning. In a second round of PCR, a new set of primers was used to add a NotI restriction site, a Kozak sequence and an N-terminal epitope tag (c-Myc for EsemOrco, V5 for EsemORs) to the 5′ end, as well as 3′ ApaI restriction sites to each EsemOR using Platinum Pfu Polymerase (Thermo Fisher Scientific). PCR products were then analyzed by gel electrophoresis, DNA of expected band length purified, and modified EsemOrco and EsemOR DNA was then double digested using NotI and ApaI restriction enzymes (NEB, Ipswich, MA), purified and ligated into the expression vectors pcDNA4/TO (EsemOrco) and pcDNA5/TO (EsemORs), respectively (all Thermo Fisher Scientific). Transformation of pcDNA4/5 ligations, transformation, colony PCR, minipreps, and sequencing were performed as described above. Large quantities of purified plasmids were obtained using the PureLinkTM HiPure Plasmid Filter Midiprep Kit (Thermo Fisher Scientific).
Generation of EsemOrco & EsemOR-Expressing HEK Cells
Human Embryonic Kidney (HEK293) cells expressing EsemOrco in combination with different EsemORs were produced and cultured as previously described (Corcoran etal. 2014). Briefly, EsemOrco-expressing pcDNA4/TO was linearized using FspI (New England Biolabs), resolved on a 0.7% TAE agarose gel, and purified using the Wizard SV Gel and PCR clean-up systems (Promega). Five micrograms of linearized plasmid and 15 µL of Lipofectamine 2000 transfection reagent (Thermo Fisher Scientific) were each diluted separately into 500 µL of Optimem medium (Thermo Fisher Scientific) and incubated for 10 min at room temperature. The two solutions were then combined and incubated for 60 min at RT. The mixture was then added to the culture flask containing ∼70% confluent isogenic tetracycline repressor-expressing (TREx) cells (allowing controlled expression of exogenous OR genes; Corcoran etal. 2014) and incubated overnight at 37 °C with 5% CO2. A heterogenic TREx/HEK293 cell line expressing EsemOrco (i.e., TEO) was obtained after culturing the transfected cells for approximately four weeks with vector-specific antibiotics. This TEO cell line was then used in five separate transfections with pcDNA5/TO/EsemORs (EsemOR1, 3, 4, 5, and 6) following the same transfection conditions as above (Corcoran etal. 2014), except that different restriction enzymes were used to linearize plasmids (BstZ17I for OR1, FspI for OR5, and PciI for OR3, 4, and 6; all from New England Biolabs). All newly transformed cell lines were passaged three times then frozen and stored at -80 °C until future use.
Ligands
In total, 85 pheromone and plant volatile compounds were obtained from different sources (supplementary table S3, Supplementary Material online). The test odor panel included 53 plant compounds, 13 Type 0 pheromones, and 18 Type I pheromones and comprised of 1) Type 0 pheromone compounds of E. semipurpurella and closely related species, 2) plant compounds structurally similar to Type 0 pheromone compounds, 3) major headspace compounds of the birch tree, and 4) commonly occurring Type I pheromone compounds with various chain-lengths, functional groups, and degrees of unsaturation. Stock solutions were prepared by diluting each compound to 100 mM in DMSO. In screening experiments, compounds were diluted from DMSO stocks into assay buffer and tested at 30 μM, and in dose–response experiments compounds were tested from 150 μM downwards with 1/2 dilutions. The Orco agonist VUAA1 (Jones etal. 2011) was used as a positive assay control and tested at 50 μM. In all cases, the final concentration of DMSO in functional assays was 0.5%, and assay buffer containing 0.5% DMSO (vehicle) was used as a negative control.
Fluorescent Calcium Assays and Data Analysis
Five cell lines expressing EsemORs were tested for responses to sex pheromone and plant volatile compounds in the previously described fluorescent calcium assay (Corcoran etal. 2014). Briefly, cells were plated into each well of a poly-d-lysine-coated 96-well plate, induced to express the EsemOrco and EsemORs (or left non-induced, as a negative control). Before the fluorescence assay, the plates were rinsed with assay buffer (DPBS, containing Ca2+, Mg2+, and 1 mM probenecid, pH 7.1) and incubated with loading buffer (assay buffer containing 1 µM Fluo4-AM and 0.2% pluronic acid) for 30 min at RT. Cells were then rinsed twice with assay buffer, loaded with 100 µL of assay buffer and allowed to incubate for 30 min at RT prior to functional testing. Upon addition of pheromones and other ligands, ligand-induced receptor activation was measured for 1 min using a FLUOstar Omega plate reader (BMG Labtech, Ortenberg, Germany). The mean response of three induced or non-induced wells of cells was calculated for each compound or dose of compound, per experiment (technical replicates). Naïve cells were plated, induced (or non-induced), and tested for responsiveness to compounds as described above in three independent experiments (biological replicates). The mean response (±SEM) of the three biological replicates was analyzed and graphed in GraphPad Prism 6 (GraphPad Software Inc., La Jolla, CA). In addition, half maximal effective concentrations (EC50) with 95% CI from dose–response assays were estimated using the nonlinear curve fit regression function in GraphPad Prism 6. Responses of EsemORs to the various active ligands were regarded as significantly different if the 95% CI of the EC50 values did not overlap.
Western Blot
Western blots were used to verify the regulated expression of transfected EsemOrco and ORs in HEK293 cells using the methods previously described (Corcoran etal. 2014; Andersson etal. 2016). Briefly, total protein was purified from induced and non-induced cells for each cell line by resuspending cell pellets in 200 µl of lysis buffer containing 1× PBS, 1% DDM detergent (Glycon Biochemicals GmbH), and 1× protease inhibitor cocktail (Roche, Basel, Switzerland), and incubated at 4 °C for 1.5 h, with occasional inversion to mix contents. The samples were then centrifuged at 21,000 × g for 30 min at 4 °C and the supernatants were transferred to new tubes. Using the BioRad DC Protein Assay kit, total protein content was quantified according to the manufacturer’s instructions. Twenty micrograms of total protein from induced and non-induced cells for each cell line was mixed with 5× loading solution (50 mM Tris pH 6.8, 50% glycerol, 2% SDS, 5% 2-mercaptoethanol, and 0.125% bromophenol blue) and incubated at 37 °C for 30 min. Samples were then loaded onto a 4–15% Criterion TGX Precast gel (BioRad) and run for 35 min, 200 V, after which they were transferred to a PVDF membrane using a BioRad Trans Blot Turbo. Membranes were then blocked with 5% nonfat milk powder in TBST buffer (50 mM Tris–Cl, 150 mM NaCl, 0.05% Tween 20, pH 7.5) for 1 h at RT, rinsed with TBST buffer and incubated with a primary antibody (rabbit antimyc antibody for myc-tagged EsemOrco; rabbit antiV5 antibody for EsemORs [Cell Signaling]) for 1 h at RT. Membranes were then rinsed again and incubated with an HRP-conjugated secondary antibody (Cell Signaling). Epitope-containing bands were developed using the Pierce ECL western blotting substrate and imaged with a BioRad ChemiDoc imaging system.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Author Contributions
J.K.Y., J.A.C, M.N.A., O.A., R.D.N., and C.L. conceived and designed the study. J.K.Y and O.A. collected the biological material. J.K.Y. and M.N.A performed transcriptomic data analysis and constructed the phylogenetic tree in figure 3. J.K.Y and J.A.C. performed molecular work, cell line generation, and culturing. J.K.Y performed functional assays. J.K.Y. drafted the manuscript with contributions from M.N.A., J.A.C., O.A., R.D.N., and C.L. All authors read and approved the final version of the manuscript.
Supplementary Material
Acknowledgments
The authors would like to thank Erling Jirle for assistance in collecting biological material, Dan-Dan Zhang for helping with the sample extraction, and Colm Carraher for assistance with the western blot analyses. We would like to thank Wittko Francke for generous gifts of compounds and valuable discussions. Also, we would like to thank Niklas Wahlberg for providing the phylogenetic tree with mapped pheromone types. We are thankful to William Walker for kindly providing sequences of SlitOR47 to 60 for the phylogenetic analysis. This work was supported by the Swedish Royal Physiographic Society in Lund (to J.K.Y.), The Swedish Foundation for International Cooperation in Research and Higher Education (grant number IB2013-5256 and IG2013-5483), and the Swedish Research Council VR (grant number VR-621-2013-4355 to C.L.). M.N.A. acknowledges FORMAS for funding (grant number 217-2014-689).
References
- Allison JD, Cardé RT.. 2016. Pheromone communication in moths: evolution, behavior and application. Berkeley: University of California Press. [Google Scholar]
- Anderson AR, Wanner KW, Trowell SC, Warr CG, Jaquin-Joly E, Zagatti P, Robertson H, Newcomb RD.. 2009. Molecular basis of female-specific odorant responses in Bombyx mori. Insect Biochem Mol Biol. 39:189–197. [DOI] [PubMed] [Google Scholar]
- Andersson MN, Videvall E, Walden KKO, Harris MO, Robertson HM, Löfstedt C.. 2014. Sex- and tissue-specific profiles of chemosensory gene expression in a herbivorous gall-inducing fly (Diptera: Cecidomyiidae). BMC Genomics. 15:501.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson MN, Löfstedt C, Newcomb RD.. 2015. Insect olfaction and the evolution of receptor tuning. Front Ecol Evol. 3:53. [Google Scholar]
- Andersson MN, Corcoran JA, Zhang D-D, Hillbur Y, Newcomb RD, Löfstedt C.. 2016. A sex pheromone receptor in the Hessian fly Mayetiola destructor (Diptera, Cecidomyiidae). Front Cell Neurosci. 10:212.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ando T, Inomate SI, Yamamoto M.. 2004. Lepidopteran sex pheromones In: Schulz S, editor. The chemistry of pheromones and other semiochemicals. Berlin, Heidelberg, New York: Springer; p. 51–96. [Google Scholar]
- Baker TC. 2008. Balanced olfactory antagonism as a concept for understanding evolutionary shifts in moth sex pheromone blends. J Chem Ecol. 34:971–981. [DOI] [PubMed] [Google Scholar]
- Bengtsson JM, Trona F, Montagne N, Anfora G, Ignell R, Witzgall P, Jacquin-Joly E.. 2012. Putative chemosensory receptors of the codling moth, Cydia pomonella, identified by antennal transcriptome analysis. PLoS ONE. 7:e31620.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengtsson JM, Gonzalez F, Cattaneo AM, Montagné N, Walker WB, Bengtsson M, Anfora G, Ignell R, Jacquin-Joly E, Witzgall P.. 2014. A predicted sex pheromone receptor of codling moth Cydia pomonella detects the plant volatile pear ester. Front Ecol Evol. 2:33. [Google Scholar]
- Benton R. 2015. Multigene family evolution: perspectives from insect chemoreceptors. Trends Ecol Evol. 30:590–600. [DOI] [PubMed] [Google Scholar]
- Bylund H, Tenow O.. 1994. Long-term dynamics of leaf-miners, Eriocrania spp., on mountain birch: alternate year fluctuations and interaction with Epirrita autumnata. Ecol Entomol. 19:310–318. [Google Scholar]
- Cande J, Prud'homme B, Gompel N.. 2013. Smells like evolution: the role of chemoreceptor evolution in behavioral change. Curr Opin Neurobiol. 23:152–158. [DOI] [PubMed] [Google Scholar]
- Carey AF, Wang G, Su CY, Zwiebel LJ, Carlson JR.. 2010. Odorant reception in the malaria mosquito Anopheles gambiae. Nature 464:66–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carraher C, Dalziel J, Jordan MD, Christie DL, Newcomb RD, Kralicek AV.. 2015. Towards an understanding of the structural basis for insect olfaction by odorant receptors. Insect Biochem Mol Biol. 66:31–41. [DOI] [PubMed] [Google Scholar]
- Cattaneo AM, Gonzalez F, Bengtsson JM, Corey EA, Jacquin-Joly E, Montagné N, Salvagnin U, Walker WB, Witzgall P, Anfora G, et al. 2017. Candidate pheromone receptors of codling moth Cydia pomonella respond to pheromones and kairomones. Sci Rep. 7:41105.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corcoran JA, Jordan MD, Carraher C, Newcomb RD.. 2014. A novel method to study insect olfactory receptor function using HEK293 cells. Insect Biochem Mol Biol. 54:22–32. [DOI] [PubMed] [Google Scholar]
- Corcoran JA, Jordan MD, Thrimawithana AH, Crowhurst RN, Newcomb RD.. 2015. The peripheral olfactory repertoire of the lightbrown apple moth, Epiphyas postvittana. PLoS ONE. 10:e0128596.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Fouchier A, Sun X, Monsempes C, Mirabeau O, Jacquin-Joly E, Montagné N.. 2015. Evolution of two receptors detecting the same pheromone compound in crop pest moths of the genus Spodoptera. Front Ecol Evol. 3:472. [Google Scholar]
- de Fouchier A, Walker WB, Montagné N, Steiner C, Binyameen M, Schlyter F, Chertemps T, Maria A, François M-C, Monsempes C, et al. 2017. Functional evolution of Lepidoptera olfactory receptors revealed by deorphanization of a moth repertoire. Nat Commun. 8:15709.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dweck HKM, Ebrahim SAM, Thoma M, Mohamed AAM, Keesey IW, Trona F, Lavista-Llanos S, Svatoš A, Sachse S, Knaden M, et al. 2015. Pheromones mediating copulation and attraction in Drosophila. Proc Natl Acad Sci U S A. 112:E2829–E2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engsontia P, Sangket U, Chotigeat W, Satasook C.. 2014. Molecular evolution of the odorant and gustatory receptor genes in lepidopteran insects: implications for their adaptation and speciation. J Mol Evol. 79:21–39. [DOI] [PubMed] [Google Scholar]
- Gardiner A, Barker D, Butlin RK, Jordan WC, Ritchie MG.. 2008. Drosophila chemoreceptor gene evolution: selection, specialization and genome size. Mol Ecol. 17:1648–1657. [DOI] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, et al. 2011. Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat Biotechnol. 29:644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosse-Wilde E, Kuebler LS, Bucks S, Vogel H, Wicher D, Hansson BS.. 2011. Antennal transcriptome of Manduca sexta. Proc Natl Acad Sci U S A. 108:7449–7454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallem EA, Carlson JR.. 2006. Coding of odors by a receptor repertoire. Cell 125:143–160. [DOI] [PubMed] [Google Scholar]
- Hansson BS, Stensmyr MC.. 2011. Evolution of insect olfaction. Neuron 72:698–711. [DOI] [PubMed] [Google Scholar]
- Imada Y, Kawakita A, Kato M.. 2011. Allopatric distribution and diversification without niche shift in a bryophyte-feeding basal moth lineage (Lepidoptera: Micropterigidae). Proc R Soc B. 278:3026–3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Silkworm Genome Consortium. 2008. The genome of a lepidopteran model insect, the silkworm Bombyx mori. Insect Biochem Mol Biol. 38:1036–1045. [DOI] [PubMed] [Google Scholar]
- Imai T, Sakano H, Vosshall LB.. 2010. Topographic mapping—the olfactory system. Cold Spring Harb Perspect Biol. 2:a001776.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PL, Pask GM, Rinker DC, Zwiebel LJ.. 2011. Functional agonism of insect odorant receptor ion channels. Proc Natl Acad Sci U S A. 108:8821–8825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan MD, Anderson A, Begum D, Carraher C, Authier A, Marshall SDG, Kiely A, Gatehouse LN, Greenwood DR, Christie DL, et al. 2009. Odorant Receptors from the light brown apple moth (Epiphyas postvittana) recognize important volatile compounds produced by plants. Chem Senses. 34:383–394. [DOI] [PubMed] [Google Scholar]
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenig C, Hirsh A, Bucks S, Klinner C, Vogel H, Shukla A, Mansfield JH, Morton B, Hansson BS, Grosse-Wilde E.. 2015. A reference gene set for chemosensory receptor genes of Manduca sexta. Insect Biochem Mol Biol. 66:51–63. [DOI] [PubMed] [Google Scholar]
- Kozlov MV, Zhu J, Philipp P, Francke W, Zvereva E, Hansson B, Löfstedt C.. 1996. Pheromone specificity in Eriocrania semipurpurella (Stephens) and E. sangii (Wood) (Lepidoptera: Eriocraniidae) based on chirality of semiochemicals. J Chem Ecol. 22:431–454. [DOI] [PubMed] [Google Scholar]
- Larsson MC, Hallberg E, Kozlov MV, Francke W, Hansson BS, Löfstedt C.. 2002. Specialized olfactory receptor neurons mediating intra-and interspecific chemical communication in leafminer moths Eriocrania spp. (Lepidoptera: Eriocraniidae). J Exp Biol. 205:989–998. [DOI] [PubMed] [Google Scholar]
- Leary GP, Allen JE, Bunger PL, Luginbill JB, Linn CE Jr, Macallister IE, Kavanaugh MP, Wanner KW.. 2012. Single mutation to a sex pheromone receptor provides adaptive specificity between closely related moth species. Proc Natl Acad Sci U S A. 109:14081–14086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JJ, Bickel PJ, Biggin MD.. 2014. System wide analyses have underestimated protein abundances and the importance of transcription in mammals. PeerJ 2:e270.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linn CE, Roelofs WL.. 1995. Pheromone communication in moths and its role in the speciation process In: Lambert DM, Spencer HG, editors. Speciation and the recognition concept: theory and application. Baltimore and London: John Hopkins University Press; p. 263–300. [Google Scholar]
- Liu C, Liu Y, Walker WB, Dong S, Wang G.. 2013. Identification and functional characterization of sex pheromone receptors in beet armyworm Spodoptera exigua (Hübner). Insect Biochem Mol Biol. 43:747–754. [DOI] [PubMed] [Google Scholar]
- Lu P, Vogel C, Wang R, Yao X, Marcotte EM.. 2007. Absolute protein expression profiling estimates the relative contributions of transcriptional and translational regulation. Nat Biotechnol. 25:117–124. [DOI] [PubMed] [Google Scholar]
- Löfstedt C, Van Der Pers JNC, Löfqvist J, Lanne BS, Appelgren M, Bergström G, Thelin B.. 1982. Sex pheromone components of the turnip moth, Agrotis segetum, chemical identification, electrophysiological evaluation, and behavioural activity. J Chem Ecol. 8:1305–1322. [DOI] [PubMed] [Google Scholar]
- Löfstedt C, Hansson BS, Petersson E, Valeur P, Richards A.. 1994. Pheromonal secretions from glands on the 5th abdominal sternite of hydropsychid and rhyacophilid caddisflies (Trichoptera). J Chem Ecol. 20:153–169. [DOI] [PubMed] [Google Scholar]
- Löfstedt C, Kozlov M.. 1997. A phylogenetic analysis of pheromone communication in primitive moths In: Cardé RT, Minks AK, editors. Insect pheromone research-new directions. New York: Chapman & Hall; p. 473–489. [Google Scholar]
- Löfstedt C, Wahlberg N, Millar JM.. 2016. Evolutionary patterns of pheromone diversity in lepidoptera. In: Allison JD, Cardé RT, editors. Pheromone communication in moths: evolution, behavior and application. Berkeley: University of California Press; p. 43–78. [Google Scholar]
- Mayfield AB, Wang YB, Chen CS, Chen SH, Lin CY.. 2016. Dual-compartmental transcriptomic + proteomic analysis of a marine endosymbiosis exposed to environmental change. Mol Ecol. 25:5944–5958. [DOI] [PubMed] [Google Scholar]
- Missbach C, Dweck HKM, Vogel H, Vilcinskas A, Stensmyr MC, Hansson BS, Grosse-Wilde E.. 2014. Evolution of insect olfactory receptors. eLife 3:e02115.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell RF, Hall LP, Reagel PF, McKenna DD, Baker TC, Hildebrand JG.. 2017. Odorant receptors and antennal lobe morphology offer a new approach to understanding olfaction in the Asian longhorned beetle. J Comp Physiol A. 203:99–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuno H, Sakurai T, Murai M, Yasuda T, Kugimiya S, Ozawa R, Toyohara H, Takabayashi J, Miyoshi H, Nishioka T.. 2008. Identification of receptors of main sex-pheromone components of three lepidopteran species. Eur J Neurosci. 28:893–902. [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Sakurai T, Nishioka T, Touhara K.. 2005. Insect sex-pheromone signals mediated by specific combinations of olfactory receptors. Science 307:1638–1642. [DOI] [PubMed] [Google Scholar]
- Nei M, Niimura Y, Nozawa M.. 2008. The evolution of animal chemosensory receptor gene repertoires: roles of chance and necessity. Nat Rev Genet. 9:951–963. [DOI] [PubMed] [Google Scholar]
- Neuhaus EM, Gisselmann G, Zhang W, Dooley R, Störtkuhl K, Hatt H.. 2005. Odorant receptor heterodimerization in the olfactory system of Drosophila melanogaster. Nat Neurosci. 8:15–17. [DOI] [PubMed] [Google Scholar]
- Pertea G, Huang X, Liang F, Antonescu V, Sultana R, Karamycheva S, Lee Y, White J, Cheung F, Parvizi B, et al. 2003. TIGR gene indices clustering tools (TGICL): a software system for fast clustering of large EST datasets. Bioinformatics 19:651–652. [DOI] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP.. 2010. FastTree 2 – approximately maximum-likelihood trees for large alignments. PLoS ONE. 5:e9490.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramdya P, Benton R.. 2010. Evolving olfactory systems on the fly. Trends Genet. 26:307–316. [DOI] [PubMed] [Google Scholar]
- Sato K, Pellegrino M, Nakagawa T, Nakagawa T, Vosshall LB, Touhara K.. 2008. Insect olfactory receptors are heteromeric ligand-gated ion channels. Nature 452:1002–1009. [DOI] [PubMed] [Google Scholar]
- Steinwender B, Thrimawithana AH, Crowhurst RN, Newcomb RD.. 2015. Pheromone receptor evolution in the cryptic leafroller species, Ctenopseustis obliquana and C. herana. J Mol Evol. 801:42–56. [DOI] [PubMed] [Google Scholar]
- Symonds MRE, Elgar MA.. 2008. The evolution of pheromone diversity. Trends Ecol Evol. 23:220–228. [DOI] [PubMed] [Google Scholar]
- Sun M, Liu Y, Walker WB, Liu C, Lin K, Gu S, Zhang Y, Zhou J, Wang G.. 2013. Identification and characterization of pheromone receptors and interplay between receptors and pheromone binding proteins in the diamondback moth, Plutella xyllostella. PLoS ONE. 8:e62098.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Uda Y, Ono Y, Nakagawa T, Suwa M, Yamaoka R, Touhara K.. 2009. Highly selective tuning of a silkworm olfactory receptor to a key mulberry leaf volatile. Curr Biol. 19:881–890. [DOI] [PubMed] [Google Scholar]
- The Heliconius Genome Consortium. 2012. Butterfly genome reveals promiscuous exchange of mimicry adaptations among species. Nature. 487:94–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visser JH. 1986. Host odor perception in phytophagous insects. Annu Rev Entomol. 31:121–144. [Google Scholar]
- Vosshall LB, Wong AM, Axel R.. 2000. An olfactory sensory map in the fly brain. Cell 102:147–159. [DOI] [PubMed] [Google Scholar]
- Vosshall LB, Stocker RF.. 2007. Molecular architecture of smell and taste in Drosophila. Annu Rev Neurosci. 30:505–533. [DOI] [PubMed] [Google Scholar]
- Vosshall LB, Hansson BS.. 2011. A unified nomenclature system for the insect olfactory coreceptor. Chem Senses. 36: 497–498. [DOI] [PubMed] [Google Scholar]
- Wang G, Carey AF, Carlson JR, Zwiebel LJ.. 2010. Molecular basis of odor coding in the malaria vector mosquito Anopheles gambiae. Proc Natl Acad Sci U S A. 107:4418–4423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanner KW, Anderson AR, Trowell SC, Theilmann DA, Robertson HM, Newcomb RD.. 2007. Female-biased expression of odourant receptor genes in the adult antennae of the silkworm, Bombyx mori. Insect Mol Biol. 16:107–119. [DOI] [PubMed] [Google Scholar]
- Wicher D, Schäfer R, Bauernfeind R, Stensmyr MC, Heller R, Heinemann SH, Hansson BS.. 2008. Drosophila odorant receptors are both ligand-gated and cyclic-nucleotide-activated cation channels. Nature 452:1007–1011. [DOI] [PubMed] [Google Scholar]
- Wicher D, Morinaga S, Halty-deLeon L, Funk N, Hansson B, Touhara 2, Stengl M.. 2017. Identification and characterization of the bombykal receptor in the hawkmoth Manduca sexta. J Exp Biol. 220:1781–1786. [DOI] [PubMed] [Google Scholar]
- Widmayer P, Heifetz Y, Breer H.. 2009. Expression of a pheromone receptor in ovipositor sensilla of the female moth (Heliothis virescens). Insect Mol Biol. 18:541–547. [DOI] [PubMed] [Google Scholar]
- Zhan S, Merlin C, Boore JL, Reppert SM.. 2011. The monarch butterfly genome yields insights into long-distance migration. Cell 147:1171–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang QH, Birgersson G, Zhu JW, Löfstedt C, Löfqvist J, Schlyter F.. 1999. Leaf volatiles from nonhost deciduous trees: variation by tree species, season, and temperature and electrophysiological activity in Ips typographus. J Chem Ecol. 25:1923–1943. [Google Scholar]
- Zhang D-D, Löfstedt C.. 2013. Functional evolution of a multigene family: orthologous and paralogous pheromone receptor genes in the turnip moth, Agrotis segetum. PLoS ONE. 8:e77345.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D-D, Löfstedt C.. 2015. Moth pheromone receptors: gene sequences, function and evolution. Front Ecol Evol. 3:105. [Google Scholar]
- Zhang D-D, Wang HL, Schultze A, Froß H, Francke W, Krieger J, Löfstedt C.. 2016. Receptor for detection of a Type II sex pheromone in the winter moth Operophtera brumata. Sci Rep. 6:18576.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JW, Kozlov MV, Philipp P, Francke W, Löfstedt C.. 1995. Identification of a novel moth sex pheromone in Eriocrania cicatricella (Zett.) (Lepidoptera: Eriocraniidae) and its phylogenetic implications. J Chem Ecol. 21:29–43. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.