Summary
Small ~10 kb microhomology-mediated tandem duplications (“Group 1 TDs”) are abundant in BRCA1-linked but not BRCA2-linked breast cancer genomes. Here, we define the mechanism underlying this “rearrangement signature”. We show that BRCA1, but not BRCA2, suppresses TDs at a Tus/Ter site-specific chromosomal replication fork barrier in primary mammalian cells. BRCA1 has no equivalent role at chromosomal double strand breaks, indicating specificity for the stalled fork response. Tandem duplications in BRCA1 mutant cells arise by a “replication restart-bypass” mechanism terminated by end joining or by microhomology-mediated template switching, the latter forming complex TD breakpoints. We show that solitary DNA ends form directly at Tus/Ter, implicating misrepair of these lesions in TD formation. We find that BRCA1 inactivation is strongly associated with Group 1 TDs in ovarian cancer. The Group 1 TD phenotype may be a general signature of BRCA1-deficient cancer.
Replication fork stalling at abnormal DNA structure or following collision with transcription complexes is a source of genomic instability in cancer and in developmental disorders1–5. Homologous recombination (HR) at stalled or collapsed forks can either suppress or promote genomic instability6,7. To study repair at stalled mammalian replication forks, we previously adapted the Escherichia coli Tus/Ter replication fork barrier (RFB)8,9 to trigger locus-specific fork stalling and HR on a mammalian chromosome10. We uncovered functions for BRCA1, BRCA2 and Rad51 in suppressing aberrant replicative HR responses at stalled forks. In wild type cells, conservative “short tract” gene conversion (STGC) is the major HR product at Tus/Ter. In cells lacking BRCA1 and Rad51, ~85% of all Tus/Ter-induced HR events resolve by aberrant “long tract” gene conversion (LTGC)10—a replicative response to fork stalling potentially analogous to “break-induced replication” (BIR) in yeast11–13. BRCA1, BRCA2, Rad51 and the Fanconi anemia (FA) genes have additional non-HR functions at stalled forks, where they protect DNA from degradation by the MRE11 nuclease14. BRCA1, together with its heterodimeric partner BARD1, has also been implicated in removal of the CMG replicative helicase from the stalled fork15. BRCA1, BARD1 and the BRCA1-interacting protein CtIP have BRCA2-independent functions in DNA end processing16–18. BRCA1/BARD1 interacts with Rad51 directly and also indirectly via PALB2/BRCA219,20. Thus, BRCA1 performs several functions at the stalled fork and in DSB repair, only some of which are shared with BRCA2.
Recently, a novel “rearrangement signature” specifically associated with BRCA1 loss was identified in the human breast cancer genome—the presence of abundant small (~10 kb) tandem duplications (TDs) with microhomologous breakpoints21,22. This chromotype, which differs from the larger (>100 kb) TDs noted previously in the cancer genome23,24, was termed “rearrangement signature 3″ or “Group 1 TD phenotype (TDP)”22,25. We will use the latter term here. Group 1 TDs are strongly associated with loss of BRCA1 but not with BRCA2 loss, and are enriched at loci that disrupt tumor suppressor genes, suggesting that Group 1 TDs promote tumorigenesis in BRCA1-linked breast cancer21,22. However, the mechanisms that connect BRCA1 loss with TD formation remain undefined. Similarly, it is unclear whether suppression of Group 1 TDs is an intrinsic BRCA1 function that operates in primary cells. In this study, we address these questions by analyzing ~2–6 kb microhomology-mediated TDs that arise at a Tus/Ter site-specific chromosomal RFB in primary mouse embryonic stem (ES) cells.
BRCA1 suppresses Tus/Ter-induced TDs
We previously described a ROSA26-targeted 6xTer-HR reporter for simultaneous measurement of STGC and LTGC in mammalian cells, in response to a Tus/Ter-mediated RFB or a chromosomal double strand break (DSB) induced by the rare-cutting I-SceI homing endonuclease10,26. In the reporter shown in Fig. 1a, STGC converts the cell to GFP+RFP–, while LTGC converts it to GFP+RFP+, by replicative duplication of an RFP expression cassette. In response to a Tus/Ter block (following transient Tus expression), we observed extremely low levels of a novel GFP–RFP+ repair product (Fig. 1b). We compared Tus/Ter-induced GFP–RFP+ products in cells expressing wild-type BRCA1 (BRCA1fl/exon11) vs. isogenic cells that express hypomorphic BRCA1 alleles lacking the in-frame exon 11 (BRCA1Δ/exon11). Loss of wtBRCA1 reduced Tus/Ter-induced STGC and increased Tus/Ter-induced LTGC as previously described (Fig. 1b and Extended Data Fig. 1)10. BRCA1Δ/exon11 cells revealed a ~10-fold increase in Tus/Ter-induced GFP–RFP+ products in comparison to BRCA1fl/exon11 cells. GFP–RFP+ products were further increased by siRNA-mediated depletion of the residual BRCA1exon11 hypomorphic gene product and were not suppressed by Rad51 depletion (Fig. 1b and Extended Data Fig. 1). In parallel, I-SceI induced low levels of GFP–RFP+ products that were only marginally increased by loss of BRCA1 (Extended Data Fig. 1). Thus, BRCA1 suppresses a novel Rad51-independent GFP–RFP+ outcome primarily during the stalled fork response.
Figure 1. BRCA1 suppresses Tus/Ter-induced GFP–RFP+ repair products.
a, 6xTer-HR reporter and HR products of Tus-Ter-induced fork stalling. Grey boxes: mutant GFP. Green box: wtGFP. Open circles A and B: 5′ and 3′ artificial RFP exons. 5′Tr-GFP: 5′-truncated GFP. Orange triangle: 6xTer array. Blue line: I-SceI restriction site. STGC/LTGC: short/long tract gene conversion outcomes. LTGC generates wtRFP through RNA splicing (red filled circles). b, Representative primary FACS data for BRCA1fl/exon11 and BRCA1Δ/exon11 6xTer-HR reporter cells co-transfected with wtTus and siLUC or siBRCA1. FACS plots produced from pooled data of duplicate samples from three independent experiments. Numbers represent percentages. See Extended Data Fig. 1 for additional primary data, quantitation and BRCA1 mRNA depletion. Red arrowhead: GFP–RFP+ repair products in BRCA1Δ/exon11 cells depleted of BRCA1.
To determine the rearrangement underlying the GFP–RFP+ outcome, we analyzed 6xTer-HR reporter structure in Tus/Ter-induced GFP–RFP+ clones (Fig. 2). We used fluorescence-activated cell sorting (FACS) to isolate Tus/Ter-induced GFP–RFP+ clones from BRCA1Δ/exon11 6xTer-HR reporter cells, in parallel with Tus/Ter-induced GFP+RFP– (STGC) and GFP+RFP+ (LTGC) controls, and analyzed genomic (g)DNA by Southern blotting. STGC and LTGC products revealed the expected rearrangements (Fig. 2a and 2b)10,26. In contrast, each GFP–RFP+ rearrangement had a unique structure and fell into one of two classes in BglII-restricted gDNA. Class 1 rearrangements contained a single GFP-hybridizing band of ≤10 kb. Class 2 rearrangements contained one invariant band of ~6.6 kb that co-migrated with the BglII-digested parental reporter and one smaller fragment of variable size (Fig. 2b). PCR amplification and sequencing of the rearrangement breakpoints revealed that all Tus/Ter-induced GFP–RFP+ clones contained microhomology (MH)-mediated or non-homologous tandem duplications of the RFP cassette (hereafter termed “TDs”), with predominant use of 1–2 bp MH at the TD breakpoint (Fig. 2c and Extended Data Fig. 2). Class 2 rearrangements reflect inclusion of a BglII site within the TD; in other respects the two classes are similar. A detailed analysis of TD breakpoints is presented below.
Figure 2. Tus/Ter-induced GFP–RFP+ repair products are MH-mediated tandem duplications.
a, STGC, LTGC and GFP–RFP+ products. Elements as in Fig. 1a. Red half-arrows: primers for breakpoint PCR. B: BglII site. Southern blotting with GFP probe fragment sizes indicated. Grey hatched box: breakpoint of GFP–RFP+ product. b, Analysis of GFP–RFP+ repair products. Upper panels: Southern blots of Tus/Ter-induced STGC, LTGC and GFP–RFP+ products in BRCA1Δ/exon11 6xTer-HR reporter cells. MW: molecular weight marker lane. Red asterisk: example of Class 1 GFP–RFP+ repair product. Blue asterisk: example of Class 2 product. See Extended Data Fig. 2 for sequence analysis of these two clones. Lower panels: breakpoint PCR products. c, GFP–RFP+ products are MH-mediated tandem duplications (TDs). Cartoons show typical Class 1 and Class 2 TDs. Elements as in Fig. 1a and Fig. 2a. Green line: TD breakpoint. For gel source data, see Supplementary Figure 1.
Specificity of TD suppression by BRCA1
To determine whether TD suppression at Tus/Ter-stalled forks is specific to BRCA1, we studied the contribution of additional stalled fork metabolism/repair proteins to Tus/Ter-induced repair. We compared, in parallel, the impact of siRNA-mediated depletion of candidates on Tus/Ter-induced vs. I-SceI-induced repair in BRCA1fl/exon11 cells vs. BRCA1Δ/exon11 cells, using siRNA against Luciferase as control. In BRCA1fl/exon11 cells (i.e., expressing wtBRCA1), we identified BRCA1, BARD1 and CtIP as major suppressors of Tus/Ter-induced TDs (Extended Data Fig. 3a). CtIP acts largely independently of BRCA1 as a TD suppressor, as it does in certain other repair functions26,27. In contrast, BRCA2, Rad51, FANCA, FANCD2 or SLX4/FANCP suppressed TDs modestly or not at all, despite evidence that these proteins support Tus/Ter-induced HR, as expected from previous studies (Extended Data Fig. 3b) 10,28. I-SceI-induced GFP–RFP+ products were not regulated by the above-noted proteins (Extended Data Fig. 3a). Depletion of the FANCM translocase29 or the Bloom’s syndrome helicase (BLM)30 did not induce TDs in BRCA1fl/exon11 cells but unexpectedly increased Tus/Ter-induced TDs ~15-fold in BRCA1Δ/exon11 cells (Extended Data Figs. 3a and 3c). FANCM and BLM can each disassemble late recombination intermediates but are also implicated in stalled fork metabolism31. Loss of FANCM or BLM affected Tus/Ter-induced TDs quantitatively but not qualitatively (Extended Data Fig 4a; see also TD breakpoint analysis, below). Co-depletion of FANCM and BLM in BRCA1Δ/exon11 cells produced additive effects on TD formation (Extended Data Fig. 4b), suggesting that the two proteins act independently to suppress Tus/Ter-induced TDs.
As a further test of the relative contributions of BRCA1 and BRCA2 to TD suppression, we depleted BRCA1, BARD1, BRCA2 or Rad51 in combination with FANCM or BLM in BRCA1fl/exon11 6xTer-HR reporter cells. Consistent with the above findings, co-depletion of BRCA1, BARD1 or CtIP with FANCM or BLM induced Tus/Ter-induced TDs, whereas co-depletion of BRCA2 or Rad51 with FANCM or BLM had minimal impact on TDs (Extended Data Figs. 5a and 5b). We made similar observations in BRCA2 mutant (BRCA2lex1/lex2) ES cells32 (Extended Data Fig. 5c). Thus, even when BRCA2 is biallelically mutated, BRCA1 remains the dominant TD suppressor. The Tus/Ter system recapitulates the specific association of BRCA1 loss with small TDs originally noted in the breast cancer genome21,22. We therefore propose that Group 1 TDs in BRCA1 mutant breast cancer are products of aberrant stalled fork repair.
Mechanism of TD formation
Three different mechanisms could mediate TD formation at stalled forks. The first invokes breakage of both sister chromatids and their fusion by end joining (“breakage-fusion”; Fig. 3a). The “partner” sister chromatid (the sister that does not acquire a TD) would be broken and rearranged during this process. A second model invokes TD initiation by MH-mediated synapsis of a free DNA end generated at the stalled/collapsed rightward fork of Fig. 3b, priming TD formation by “microhomology-mediated break-induced replication” (MMBIR)33,34. A third mechanism entails aberrant “replication restart” of the stalled/collapsed leftward fork of Fig. 3c. By analogy with previously described Rad51-independent replication restart mechanisms35–38, processing of the collapsed leftward fork primes extension of the stalled leading strand by a migrating bubble mechanism resembling BIR13 (Fig. 3c). The approaching conventional rightward fork bypasses the restarted leftward nascent strand and re-copies the TD tract before stalling at Tus/Ter (“replication restart-bypass”; Fig. 3c). By this model, the “upstream” site of the TD breakpoint (defined in Extended Data Fig. 2a) marks the site of displacement of the leftward nascent strand and the “Ter-proximal” site (Extended Data Fig. 2a) is derived from a free DNA end formed at the Tus/Ter-stalled rightward fork. Note that fork breakage pictured in Figs. 3b and 3c is not a requirement of these models, since a free DNA end could alternatively be generated at Tus/Ter by fork regression39. Indeed, high frequency rearrangements observed at a site-specific RFB in Schizosaccharomyces pombe are not accompanied by evidence of fork breakage37.
Figure 3. Candidate mechanisms of Tus/Ter-induced TDs.
a, “Breakage-fusion” model. GFP elements not shown. Black lines: parental DNA. Blue lines: nascent strands of conventional replication. Half arrows: nascent strand 3′ ends. Scissors: Sites of fork breakage. Pink dashed arrow: fusion of broken sisters by end joining. b, “MMBIR” model. Red half arrow: repair synthesis during bubble migration. Other symbols as in a. c, “Replication restart-bypass” model. The leftward fork undergoes aberrant “replication restart”—for example, engaging a bubble migration mechanism, as shown. The rightward fork bypasses the leftward nascent strand and stalls at Tus/Ter. End joining (pink dashed arrow) completes the TD. Symbols as in a and b. d, Summary of predictions made by the three TD models.
As summarized in Fig. 3d, the “breakage-fusion”, “MMBIR” and “replication restart-bypass” models predict different fates of the partner sister chromatid during TD formation and/or different dependencies on end joining. To retrieve the partner sister chromatid, we induced mitotic non-disjunction during the cell cycle in which the TD formed, by treating Tus-transfected FANCM-depleted BRCA1Δ/exon11 cells with 30μM cytochalasin B for 24 hours immediately prior to FACS-cloning of GFP–RFP+ cells (Fig. 4a and Materials and Methods). Southern analysis of 60 independent GFP–RFP+ aneuploid clones revealed no off-size GFP-hybridizing bands other than the TD itself in any clone, providing no support for the breakage-fusion model (Fig. 4a and Supplementary Figure 1). In contrast, 11/60 GFP–RFP+ clones contained two copies of the 6xTer-HR reporter: one that had undergone a TD and one that retained the parental structure (Fig. 4b). In 8 of these clones, re-cloning failed to separate the two reporter copies, confirming that the TD and unrearranged reporter were present in the same cell (Extended Data Figs. 6a and 6b). We obtained direct TD breakpoint sequence for 6 of these 8 clones. One TD breakpoint was blunt, one entailed insertion of one nucleotide and four revealed MH. The spontaneous non-disjunction rate for this cell line is ~1/1,000. The fact that 8/60 CB-induced “non-disjunction” TD clones revealed an unaltered partner sister chromatid indicates that TDs form at Tus/Ter primarily via a replicative mechanism, not by breakage-fusion. Interestingly, segmental TDs in Saccharomyces cerevisiae and E. coli are also mediated by replicative mechanisms34,40.
Figure 4. A replicative mechanism involving C-NHEJ mediates Tus/Ter-induced TDs.
a, Aneuploidy induced by 30 μM cytochalasin B (CB) during TD induction. b, Analysis of Tus/Ter-induced TDs in BRCA1Δ/exon11 siFANCM-treated, CB-induced aneuploid clones. Upper panel: Southern blot of gDNA (AseI digest, GFP probe); cartoon indicates fragment sizes. Lanes 1–3, CB-treated TD clones that did not retain a second copy of the reporter; Lane 4, “parental” reporter marks migration of unrearranged reporter; Lanes 5–11, CB-treated TD clones that retained a second copy of the reporter, which co-migrates with parental reporter in all cases. Lower panel: Breakpoint PCR products. c, Tus/Ter-induced TDs in XRCC4fl/fl and XRCC4Δ/Δ ROSA26-targeted 6xTer-HR reporter independent ES cell clones, co-transfected with siRNAs shown. Mean of duplicate samples from nine independent experiments (n=9). Error bars: s.e.m. t-test P values: siFANCM+siBRCA1 vs. any other treatment group within individual clones: <10E-4 (#8 and #39); <0.02 (#11 and #13). siFANCM+siBRCA1 comparisons between any XRCC4fl/fl and any XRCC4Δ/Δ clone: ≤2x10E-4. All other comparisons of same treatment groups between clones: NS. d, Confirmation of genotype of XRCC4fl/fl and XRCC4Δ/Δ clones. Grey boxes: XRCC4 exons. Black triangles: loxP sites. PCR analysis of gDNA using the primer pairs indicated. e, Tus/Ter-induced TDs in XRCC4Δ/Δ #11 transduced with pHIV-EV (empty vector control) or pHIV-mXRCC4 and selected in nourseothricin, co-transfected with siRNAs shown. Mean of duplicates, n=9. Error bars: s.e.m. t-test P values (both pHIV-EV- and pHIV-mXRCC4-transduced cells): siFANCM+siBRCA1 vs. siLUC, siFANCM or siBRCA1: <0.02. siBLM+siBRCA1 vs. siLUC, siBLM or siBRCA1: <0.02. siFANCM+siBRCA1 vs. siBLM+siBRCA1: NS. pHIV-EV vs. pHIV-mXRCC4 siFANCM+siBRCA1: 0.028; siBLM+siBRCA1: 0.047. f, XRCC4 immunoblot in XRCC4fl/fl, XRCC4Δ/Δ cells, and transduced XRCC4Δ/Δ cells as shown. P: parental clone #11. For gel source data, see Supplementary Figure 1. For mRNA quantitation, see Extended Data Figs. 6f and 6g.
To analyze the role of classical non-homologous end joining (C-NHEJ) in TD formation, we targeted a single copy of the 6xTer-HR reporter to the ROSA26 locus of mouse XRCC4fl/fl ES cells, then generated Cre-treated XRCC4fl/fl and XRCC4Δ/Δ derivatives41. The frequency of Tus/Ter-induced TDs in two XRCC4Δ/Δ clones co-depleted of BRCA1/FANCM or BRCA1/BLM was ~30% of that observed in two XRCC4fl/fl clones (Figs. 4c and 4d). We confirmed that BRCA1 and BARD1 are the dominant TD suppressors in XRCC4fl/fl cells (Extended Data Figs. 6c and 6d). Stable lentivirus-mediated expression of wtXRCC4 restored TD frequencies in XRCC4Δ/Δ clones to wild type levels (Figs. 4e and 4f). The involvement of C-NHEJ in TD formation at Tus/Ter suggests that replication restart-bypass, not MMBIR, is the principal mechanism (Fig 3d). Residual Tus/Ter-induced TDs in XRCC4Δ/Δ cells might entail XRCC4-independent alternative end joining. However, we cannot formally exclude contributions by breakage-fusion or MMBIR to a proportion of Tus/Ter-induced TDs.
TD breakpoint analysis
To better understand the mechanisms underlying TD formation, we analyzed in detail the sequence of Tus/Ter-induced TDs from BRCA1Δ/exon11 cells depleted of FANCM, BRCA1 or BLM. TD spans varied from ~2 kb to ~6 kb, which represent the technical boundaries of TD detection using this reporter (Extended Data Fig. 7a). Tus/Ter-induced TD breakpoints revealed a modest MH bias (Extended Data Fig. 7b). 14/237 (5.9%) breakpoints were homeologous, containing 1–2 bp internal mismatches within longer MH tracts of 4–10 bp, with no consistent strand preference of mismatch correction (Extended Data Fig. 7c). Notably, 6/231 (2.6%) TDs contained complex breakpoints (Extended Data Fig. 7d), suggestive of MH-mediated template switching 42. Template switching is associated with TD formation in E. coli, BIR in S. cerevisiae and alternative end joining in mammalian cells40,43–47. It has been invoked to explain complex breakpoints associated with replication stress in the cancer genome33,48. Our findings provide direct evidence of MH-mediated template switching at stalled mammalian replication forks.
Solitary DNA ends form at Tus/Ter
The “Ter-proximal” site of the TD represents the product of rightward fork stalling at Tus/Ter (Fig. 3c). Indeed, Ter-proximal sites were clustered near the first Ter elements encountered by the rightward fork, a minority being distributed upstream (Extended Data Fig. 7e). In contrast, “upstream” TD sites were more widely distributed (Extended Data Fig. 7f). To determine whether Ter-proximal TD sites correspond to detectable DNA lesions at Tus/Ter, we used high throughput genome-wide translocation sequencing (HTGTS)49,50 to map translocation-competent DNA ends at Tus/Ter. As “bait” for HTGTS, we induced a Cas9/CRISPR-mediated DSB ~30 kb from the 6xTer array at ROSA26 (Fig. 5). (The “translocations” studied here are, strictly, intrachromosomal rearrangements.) Control I-SceI-induced two-ended DSBs should produce equal representation of (+) and (–) DNA ends in HTGTS mapping (Fig. 5a). In contrast, rightward forks arriving at Tus/Ter (Fig. 5b) might generate predominantly (+) DNA ends, while leftward forks (not shown) would generate (–) DNA ends. This polarity is expected whether the DNA end is generated directly by breakage at the branch-point of the stalled fork (Fig. 5b) or indirectly via fork regression (Fig. 5c). Notably, if either sister chromatid were broken anywhere other than at the branch-point of the stalled fork, this would generate a conventional two-ended DSB with equal representation of (+) and (–) DNA ends.
Figure 5. Solitary DNA ends form at Tus/Ter-stalled forks.
CRISPR/Cas9 induces “bait” DSB ~30 kb from 6xTer array + I-SceI site at ROSA26. a, I-SceI-induced two-ended DSB produces balanced (+) and (–) ends in HTGTS. Half-arrow: HTGTS sequencing primer. Weighted black line: duplex DNA. b, Focused rightward fork breakage produces (+) orientation DNA ends in HTGTS. c, Alternatively, solitary (+) DNA end forms via regression of rightward fork. Thus, stalled rightward forks generate (+) ends, irrespective of mechanism. Stalled leftward forks (not shown) generate (–) ends. d, HTGTS breakpoints in FANCM-depleted BRCA1Δ/exon11 cells harboring a single ROSA26-targeted 6xTer-I-SceI-GFP cassette. Grey area/orange triangles: 6xTer-array. I-SceI-induced DSBs produce expected symmetrical pattern in HTGTS. Tus/Ter-induces asymmetrical pattern with (+) ends ≫ (–) ends, indicating presence of solitary DNA ends. Note virtual absence of signal in EV controls. Maps represent pooled data from two (I-SceI), three (Tus), or two (EV) independent replicates. e, Tus/Ter-induced HTGTS in BRCA1fl/exon11 or BRCA1Δ/exon11 cells receiving siRNAs shown. For all BRCA1Δ/exon11 groups and for BRCA1fl/exon11 cells depleted of FANCM, data pooled from three independent replicates. All other BRCA1fl/exon11 groups, data pooled from two independent replicates.
As expected, FANCM-depleted BRCA1Δ/exon11 cells co-transected with control I-SceI and the CRISPR/Cas9 “bait” vectors revealed symmetrical HTGTS distributions of (+) and (–) DNA ends that mapped to the I-SceI site adjacent to the Ter array (Fig. 5d)49. In contrast, translocations into Tus/Ter in FANCM-depleted BRCA1Δ/exon11 cells were highly asymmetric. We noted a ~7-fold excess of (+) ends over (–) ends (Fig. 5d), indicating that solitary DNA ends predominate at Tus/Ter-stalled forks. Tus/Ter HTGTS breakpoints were tightly focused on the Ter array and were MH biased in comparison to I-SceI HTGTS breakpoints, revealing a 1–2 bp MH preference reminiscent of Tus/Ter-induced TD breakpoints (Extended Data Fig. 8a; compare with Extended Data Fig. 7b). Further, translocations at Tus/Ter were more abundant into the Ter sites first encountered by the approaching replication fork (Fig. 5d). In all treatment groups, including cells containing wtBRCA1, the distributions of Tus/Ter HTGTS breakpoints were similar (Fig. 5e and Extended Data Fig. 8b). However, a quantitative impact of BRCA1 on the formation of DNA ends at Tus/Ter is not excluded. In all treatment groups, the distribution of Ter-proximal TD sites (products of rightward fork stalling) was significantly shifted in comparison to the distribution of Tus/Ter HTGTS (+) ends (also products of rightward fork stalling; Extended Data Fig. 8b). Taken together, these findings suggest that the Ter-proximal site of the TD breakpoint arises from a solitary DNA end generated at the Tus/Ter RFB, which is further processed before being misrepaired in BRCA1 mutants to form a TD.
TD phenotype in BRCA1 mutant cancer
Our data suggest that “TD suppression” at stalled replication forks is an intrinsic function of BRCA1. If so, the TD phenotype might be a general feature of BRCA1 loss in cancer. To test this idea, we analyzed TDs occurring in 92 cancers from the Australian Ovarian Cancer Study (URL: http://www.aocstudy.org/), for which whole genome sequence, BRCA1 promoter methylation status and transcriptome data are available. We noted a strong association between loss of BRCA1 by mutation or promoter methylation and Group 1 TDP (Extended Data Fig. 9). Re-analysis of the Sanger Institute dataset22 using our TD algorithm confirmed that TDP Group 1 is strongly associated with BRCA1 loss but not with BRCA2 loss. Indeed, in the Sanger dataset filtered to include only TNBC (which included almost all the Group 1 TDP breast cancers), BRCA2 inactivation was negatively associated with Group 1 TDP (Extended Data Fig. 9b). In the AOCS and Sanger_TNBC datasets, we observed no association between Group 1 TDP and either mutation or aberrant expression of FANCM or BLM (Extended Data Fig. 10). Whether these genes function as TD suppressors in human tumorigenesis therefore remains to be determined.
Conclusion
In work described here, we demonstrate that BRCA1 but not BRCA2 is a major suppressor of TDs at a Tus/Ter RFB in primary mammalian cells. These findings recapitulate the Group 1 TD phenotype of BRCA1 mutant breast cancers21,22,25. We therefore propose that Group 1 TDs in BRCA1-linked cancer arise by defective processing of stalled replication forks. Extending these observations across tumor types, we observed a strong Group 1 TDP in ovarian cancers lacking BRCA1. The Group 1 TDP may therefore serve as a useful biomarker of BRCA1 loss in other cancer types51. Furthermore, our findings suggest that inactivation of BARD1 or RBBP8 (encoding CtIP) may also be associated with Group 1 TDP cancers.
Our analysis suggests that Tus/Ter-induced TDs in BRCA1 mutant cells arise by an aberrant “replication restart-bypass” mechanism terminated by end joining. Certain key elements of this mechanism are conserved in yeast7,13,37,38. However, it remains to be determined precisely how BRCA1 and BARD1, of which there are no yeast orthologs, suppress these aberrant stalled fork responses. BRCA1/BARD1 has BRCA2-independent roles in DNA end processing and in CMG helicase unloading at the stalled fork15–18. Thus, several distinct BRCA1-mediated functions might suppress TD formation at stalled forks. A notable aspect of this study is the finding that solitary DNA ends predominate at Tus/Ter-stalled forks. We detected these lesions in both TD-prone and control cells, suggesting that the production of solitary DNA ends is a generalized, perhaps physiological, response to fork stalling39. The element of TD formation that is specific to BRCA1 loss therefore appears to be the “licensing” of an aberrant replication restart mechanism at stalled forks.
Methods
Molecular biology and siRNAs
The vectors for Ter HR reporters described were constructed by conventional cloning methods using a previously described 6xTer-HR and RFP-SCR reporters10,26. pHIV-NAT-CD52 vectors were derived from pHIV-Zsgreen, a gift from Bryan Welm and Zena Werb (Addgene plasmid # 18121) 52. Ter-containing plasmids were amplified in JJC33 (Tus–) strains of E. coli. siRNA SMARTpools were purchased from Dharmacon. All plasmids used for transfection were prepared by endotoxin-free maxiprep (QIAGEN Sciences, Maryland, MD).
Mouse cell lines and cell culture
Mouse embryonic stem (ES) cells were authenticated as described in the text and were periodically tested for mycoplasma contamination. Only mycoplasma-free cells were used in experiments described here. Cells were thawed on MEF feeders and maintained in ES medium on gelatinized plates. 10 μg of Ter/HR reporter ROSA26 targeting plasmids per 1 x 107 cells were linearized using Kpn I and introduced by electroporation. ES cells were seeded onto 6-cm dishes containing puromycin-resistant feeders and plates supplemented with 4 μg/ml puromycin 24 hours. Individual colonies were picked 7–10 days later. ROSA26 targeted lines were screened for by PCR. Reporter cassette integration and overall structure was verified for targeted lines by Southern blotting. Multiple BRCA1-deficient or XRCC4-deficient ES clones were generated by transient adenovirus-mediated Cre expression. ROSA26 genotyping primers: ROSA26-sense-(CAT CAA GGA AAC CCT GGA CTA CTG); Ter-HR reporter antisense-(CCT CGG CTA GGT AGG GGA TC). BRCA1 exon11 status was determined by PCR: BRCA1 5′-sense-(CTG GGT AGT TTG TAA GCA TCC); BRCA1 exon11- antisense-(CAA TAA ACT GCT GGT CTC AGG C); BRCA1 exon11-sense-(GGA AAT GGC AAC TTG CCT AG); BRCA1 3′-antisense-(CTG CGA GCA GTC TTC AGA AAG). XRCC4 status was determined by PCR: XRCC4 5′-sense-(TTC AGC TAA CCA GCA TCA ATA G); floxed allele, XRCC4 3′-antisense-(GCA CCT TTG CCT ACT AAG CCA TCT CAC); Exon 3-deleted allele, XRCC4 3′-antisense-(TAA GCT ATT ACT CCT GCA TGG AGC ATT ATC ACC)41. BRCA2 Exons 26 and 27 status was determined by PCR: BRCA2 Intron 25 5′-sense-(TTC AGC TAA CCA GCA TCA ATA G); BRCA2 Exon 27 3′-antisense-(CGT TCT CTC CAC TCC AAG ACT TTG C); BRCA2 PGK promoter 3′-antisense-(TCC ATT TGT CAC GTC CTG CAC GAC G)32. Exon3-deleted, XRCC4-deficient mES cells were transduced with lentivirus expressing a single mRNA encoding nuorseothricin acetyl transferase and human CD52 (the CAMPATH antigen), with or without wild type mouse XRCC4: pHIV-NAT-hCD52-EV (empty vector control) or pHIV-NAT-hCD52-mXRCC4. Stable cultures were selected and maintained in 100 μg/mL nourseothricin (Jenna Bioscience cat#AB-102L).
Recombination assays
1.6 x 105 cells were co-transfected in suspension with 0.35 μg empty vector, pcDNA3β-myc NLS-Tus10, or pcDNA3β-myc NLS-I-SceI53, and 20 pmol ONTargetPlus-smartpool using Lipofectamine 2000 (Invitrogen). GFP+RFP–, GFP+RFP+ and GFP–RFP+ frequencies were scored 72 hours after transfection by flow cytometry using a Becton Dickinson 5 Laser LSRII in duplicate. For each duplicate sample, 3–6 x 105 total events were scored. Repair frequencies presented are corrected for background events and for transfection efficiency (50–85%). Transfection efficiency was measured by parallel transfection with 0.05 μg wild type GFP expression vector, 0.30 μg control vector and 20 pmol siRNA. Data presented represents the arithmetic mean and error bars represent the standard error of the mean (s.e.m.) of between five (n=5) and eleven (n=11) independent experiments (n values given in figure legends).
Statistical methods
Figure legends specify the sample number in terms of the number of replicates within each individual experiment (typically two) and the number of independent experiments (n) that were performed to generate the data presented. For repair frequency statistical analysis, the arithmetic mean of samples collected for independent experiments was calculated and data points for each independent experiment were used to calculate the mean and standard error of the mean (s.e.m.), calculated as standard deviation/√n, in which n indicates the number of independent experiments. Differences between sample pairs repair frequencies were analyzed by Student’s two-tailed unpaired t-test, assuming unequal variance using GraphPad Prism v6.0d software. P-values are indicated in each figure legend. Additional statistical analyses are as described in figure legends.
RT-qPCR analysis
RNA from transfected cells was extracted using QIAGEN RNeasy Mini Kit (QIAGEN Sciences, Maryland, MD) 48 hours after transfection. First-strand cDNA analysis was performed on an ABI 7300 Real time PCR System using Power SYBR Green RNA-to CTTM 1-Step Kit (Applied Biosystems, Foster City, CA). SYBR green RT-qPCR assays of GAPDH and siRNA-targeted gene was performed. We used the NIH NCBI Nucleotide utility to generate gene-specific primer sequences for mouse BRCA1, BRCA2, RAD51, BARD1, CTIP, SLX4, FANCA, FANCD2, and GAPDH. Primers for RT-PCR: BRCA1-sense-(ATG AGC TGG AGA GGA TGC TG); BRCA1-antisense-(CTG GGC AGT TGC TGT CTT CT); BRCA2-sense-( TCT GCC ACT GTG AAA AAT GC); BRCA2-antisense-(TCA AGC TGG GCT GAA GAT T; SLX4-sense-(GTG GGA CGA CTG GAA TGA GG); SLX4-antisense-(GCA CCT TTT GGT GTC TCT GG); CTIP-sense-(AGG AGA AGG AGG GGA CGC); CTIP-antisense-(TGA AAT ACC TCG GCG GGT G); FANCA-sense- (GGC AGC CCT GTA CAA CTG AT); FANCA-antisense- (GCC AGC AGC TCT GTC ATG TT); FANCD2-sense- (CAG ATT CGC AGC AGG TTC AC); FANCD2-antisense- (ACA CAC ATG CAG AAC AGG AT); GAPDH-sense-(CGT CCC GTA GAC AAA ATG GT); GAPDH-antisense-(TCG TTG ATG GCA ACA ATC TC). We used the Roche ProbeFinder utility based on Primer 3 software (Whitehead Institute, MIT) to generate gene-specific primer sequences for mouse FANCM and BLM: FANCM-sense-(GTC GTT ATC CTC GCT GAA GG); FANCM-antisense-(TTT GTT GGA CTG ACT CTG ATT ATA TGT); BLM-sense-(CGC GAC GTA AGC CTG AGT); BLM-antisense-(TGG CTG AGT GTC GCT GTA GT). mRNA was measured in triplicates. Target gene expression level was normalized to GAPDH and expressed as a fold difference from siLUCIFERASE treated sample from the same experiment (x=−2ΔΔCt, with ΔΔCt= [Ct target-CtGapdh]-[CtsiLUCIFERASE-CtsiGAPDH]). Error-bars represent the standard deviation of the ΔCt value (SDEV = √[SDEVTARGET2 + SDEVGAPDH2]).
Western blotting
Cells were lysed using RIPA buffer (50mM Tris-HCl, pH 8.0, 250 mM NaCl, 0.1% sodium dodecyl sulfate, 1% NP-40 containing the protease inhibitors, PMSF, and Roche complete protease inhibitor tablet) and resolved by 10 % bis-Tris SDS-PAGE (Invitrogen). Protein expression was analyzed by immunoblotting using the following antibodies; beta-tubulin (Abcam ab6046, 1:4,000), hRad51 (aliquot B32, 1:500), mXRCC4 (Abcam ab97351, 1:3,000).
Southern Blotting
Southern blotting of BglII or AseI digested genomic DNA was performed using a GFP cDNA probe by methods described previously10,26,53. For all experiments, mouse ES cell clones harboring a single, intact copy of the reporter integrated at the ROSA26 locus on chromosome 6 were used. Genomic DNA was extracted from ES cells grown to confluence on gelatinized 6-well plates (~5–10 x 106 cells) using a Puregene DNA Isolation Kit (QIAGEN Sciences, Maryland, MD).
Individual Repair Clone Capture and Molecular Analysis
Individual GFP+ RFP–, GFP+RFP+, or GFP–RFP+ cells were FACS captured 72 hrs post transfection using a FACSAria II SORP running FACSDiva software v6.1.3. To capture aneuploid “non-disjunction” clones, individual GFP+RFP+, or GFP–RFP+ cells were FACS sorted 48 hrs post transfection. Cytochalasin B induced mitotic arrest and nondisjunction, 24 hours post transfection cells were incubated for 22hrs and FACS sorted for 2hr in 30 μM dihydrocytochalasin B (Sigma Aldrich D1641). Isolated colonies from single cells were picked from 6 cm dishes containing feeder MEFs and individual repair clones expanded onto 24-well plates also containing feeder MEFs. Genomic DNA was extracted from ES clones subsequently expanded and grown to confluence on gelatinized 6-well plates (~5–10 x 106 cells) using a Puregene DNA Isolation Kit (QIAGEN Sciences, Maryland, MD). LTGC and TD breakpoint junction PCR was performed using Taq DNA Polymerase (QIAGEN Sciences, Maryland, MD) according to manufacturer’s instructions using primers unique to HR cassette synthetic RFP exons: RFP-exonA-sense-(ATG TAC GGC TCC AAG GCC TAC GTG AAG CAC); RFP-exonB-antisense-(TCG TAC TGT TCC ACG ATG GTG TAG TCC TCG). Unpurified PCR product sequencing was performed by Eton Bioscience (Cambridge, MA) using nested primers: sense-(TGC ACG CTT CAA AAG CGC ACG); antisense-(CAA GTT AAC AAC AAC AAT TGC ATT C). TD breakpoint sequence analysis and alignment was performed manually. Exact duplicate clones of individual TDs from within one experiment were removed prior to subsequent analysis. No exact duplicates of individual TDs were identified between different experiments.
LAM-HTGTS Sample Preparation and Analysis
24x 1.6 x 105 cells mouse ES reporter cells containing a single copy of the 1xGFP Ter/HR reporter cassette targeted to the ROSA26 locus were co-transfected in suspension with 0.35 μg empty vector, pcDNA3β-myc NLS-Tus, or pcDNA3β-myc NLS-I-SceI, 10 pmol ONTargetPlus-smartpool and 0.15 μg pX330 CRISPR/Cas9 expression plasmid targeting bait sequence ~30 kb distant to the ROSA26 locus. CRISPR/Cas9 sgRNA sequence-(GGC AGG AGT AAC TTG CTT CC*T GG), 30 kb distance to ROSA26. Underlined nucleotides identify the positions of the Cas9-induced “bait” DSB. “*” indicates the boundary between the protospacer and PAM sequences. For all conditions, 48 hours after transfection, gDNA was isolated using a Puregene DNA Isolation Kit (QIAGEN Sciences, Maryland, MD) with the following modifications: 2x volume of Protein Precipitation Buffer was added to each sample; samples were gently inverted and never vortexed; protein was removed by consecutive incubations on ice for 30 min followed by 30 min centrifugation, 2,000xg, 4˚C; genomic DNA was rehydrated in 125ul TE and allowed to dissolve overnight at room temperature; samples were incubated with 13 μg/ml RNase A overnight at 55˚C. 35–50 μg gDNA for each sample was diluted in a final volume of 100 μl TE. LAM-HTGTS libraries were prepared and analyzed as outlined in50. Primers used: LAM-PCR, CRISPR/Cas9-(Biotin-GGC GTC ACC ACA TAG TAG GC); on-bead ligation, bridge adapter-sense-(GCG ACT ATA GGG CAC GCG TGG NNNNNN-NH2) bridge-adapter-antisense (5-Phos CCA CGC GTG CCC TAT AGT CGC-NH2); nested-PCR, I5-nested-(ACA CTC TTT CCC TAC ACG ACG CTC TTC CGA TCT-5nt BARCODE-NESTEDPRIMER), CRISPR/Cas9 nested primer-(CAT GGC GGA AAG TAG ATA CC), I7-blue-( CTC GGC ATT CCT GCT GAA CCG CTC TTCCGA TCT GAC TAT AGG GCA CGC GTG G); tagged-PCR, P5-I5-(AAT GAT ACG GCG ACC ACC GAG ATC TAC ACT CTT TCC CTA CAC GAC GCT CTT CCG ATC T), P7-I7-(CAA GCA GAA GAC GGC ATA CGA GAT CGG TCT CGG CAT TCC TGC TGA ACC GCT CTT C).
Code Availability Statement
The code used to analyze the HTGTS data was published in Hu et al. 201650.
Data Availability Statement
The datasets generated during and/or analysed during the current study (e.g., the recombination/repair assays analysed throughout the paper, with quantitation by FACS) are available from the corresponding author on reasonable request. Figure source data is available in Supplementary Figure 1. The HTGTS datasets (10 datasets, corresponding to a total of 25 independent HTGTS experiments) are deposited in the GEO repository (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi), accession number GSE103624.
Extended Data
Extended Data Figure 1. BRCA1 suppresses Rad51-independent Tus/Ter-induced GFP–RFP+ repair outcomes.
a, Repair frequencies in BRCA1fl/exon11 and BRCA1Δ/exon11 6xTer-HR reporter cells transfected with Tus or I-SceI and with either control Luciferase siRNA (siLUC) or BRCA1 SMARTpool (siBRCA1). Columns represent mean of duplicate samples from ten independent experiments (i.e., n=10). Error bars: s.e.m. Tus-induced HR, BRCA1fl/exon11 cells, t-test siBRCA1 vs. siLUC: All measurements p<0.01; BRCA1Δ/exon11 cells, siBRCA1 vs. siLUC: Total HR: p=0.0470; STGC: p=0.0003; LTGC: not significant (NS); LTGC/Total HR: p<0.0001; GFP–RFP+: p=0.0010. I-SceI-induced HR, BRCA1fl/exon11 cells, t-test siBRCA1 vs. siLUC: All measurements P<0.05; BRCA1Δ/exon11 cells, t-test siBRCA1 vs. siLUC: All measurements p<0.02. b, Representative primary FACS data for BRCA1fl/exon11 and BRCA1Δ/exon11 6xTer-HR reporter cells transfected with empty vector (EV), Tus or I-SceI and with siLUC or siBRCA1. Tus-transfected samples reproduced from Fig. 1b. FACS plots produced from pooled data of duplicate samples from three independent experiments. Numbers represent percentages. c, RT-qPCR analysis of BRCA1 mRNA in siRNA-treated cells. Data normalized to GAPDH and expressed as fold difference from siLUC sample from the same experiment (x=−2ΔΔCt, with ΔΔCt= [Ct target-CtGapdh]-[CtsiLUC-CtsiGAPDH]). Error-bars: standard deviation of ΔCt value (SDEV = √[SDEVTARGET2 + SDEVGAPDH2]). d, Frequencies of GFP–RFP+ events in BRCA1fl/exon11 and BRCA1Δ/exon11 6xTer-HR reporter cells transfected with Tus or I-SceI and with either siLUC, siBRCA1, or RAD51 SMARTpool (siRAD51). Columns represent mean of duplicate samples, n=5. Error bars: s.e.m. Tus-induced GFP–RFP+, BRCA1fl/exon11 cells, t-test: All comparisons p<0.05. Tus-induced GFP–RFP+, BRCA1Δ/exon11 cells, t-test: All comparisons p<0.03. Abundance of Rad51 protein in siRNA-treated BRCA1fl/exon11 and BRCA1Δ/exon11 6xTer-HR reporter ES cells. For gel source data, see Supplementary Fig. 1.
Extended Data Figure 2. Examples of breakpoint sequence analysis of Tus/Ter-induced GFP–RFP+ products: Class 1 and Class 2 rearrangements are microhomology-mediated tandem duplications (TDs).
a, Structure of the GFP–RFP+ Class 1 rearrangement marked with red asterisk in Fig. 2. Cartoon elements as in Figs. 1 and 2; orange triangle represents 6xTer array. Right cartoon: Schematic of TD breakpoint. Grey number: site of Ter-proximal breakpoint relative to Ter array. In this TD clone, this breakpoint is located 333 bp upstream of the first nucleotide of the first Ter site encountered by the rightward replication fork (i.e., position −333). Black number: number of base pairs of MH at the breakpoint (in this clone, MH=2). Grey arrows identify the orientation of the segments of the TD, relative to the reporter. Upper text box: direct sequence of TD breakpoint. Green bold text: fragments of GFP open reading frame (ORF). Red bold letters: 2 bp MH breakpoint. Black text: other reporter sequences. Lower text box: overlay of TD breakpoint ends (green bold for GFP sequences + red bold for 2 bp MH breakpoint) on full-length wild type GFP (grey). b, Structure of the GFP–RFP+ Class 2 rearrangement marked with blue asterisk in Fig. 2. Blue letter “B”: BglII site retained within TD breakpoint. Right cartoon: Schematic of TD breakpoint, elements as in panel a. In this TD clone, the Ter-proximal TD breakpoint is located 8 bp downstream of the first nucleotide of the first Ter site encountered by the rightward replication fork (i.e., position +8). Text box: direct sequence of TD breakpoint. Green bold text: fragments of GFP ORF. Orange highlighting: 8 bp fragment of first Ter element retained within TD breakpoint. Red bold letter: 1 bp MH breakpoint. Blue highlighting: BglII site retained within TD. Black text: other reporter sequences.
Extended Data Figure 3. Specificity of BRCA1 loss on Tus/Ter-induced TDs.
a, Tus/Ter-induced and I-SceI-induced TD (GFP–RFP+) products in BRCA1fl/exon11 or BRCA1Δ/exon11 6xTer-HR cells depleted of repair proteins indicated. Induction of repair products was calculated relative to siLUC controls (which therefore score as 1). Data represents mean of between eight and ten independent experiments, each experimental data point collected in duplicate (replicates: BRCA1, n=10; BARD1, n=9; CtIP, n=9; BLM, n=8; FANCM, n=9; BRCA2, n=8; FANCA, n=9; FANCD2, n=10; RAD51, n=9; SLX4, n=9). Error bars: s.e.m. b, Tus-induced and I-SceI-induced STGC (GFP+RFP–) products in BRCA1fl/exon11 or BRCA1Δ/exon11 6xTer-HR cells depleted of repair proteins indicated. Replicates and error bars as in panel a. c, Representative primary FACS data for BRCA1Δ/exon11 6xTer-HR reporter cells co-transfected with empty vector (EV), Tus or I-SceI expression vectors (as shown) and siRNAs as shown. FACS plots pooled from duplicate samples of four independent experiments. Numbers represent percentages. d, RT qPCR analysis of BLM, FANCM, BRCA2, FANCA, SLX4, CTIP, BARD1 mRNA. Data normalized to GAPDH and expressed as a fold difference from siLUC treated sample from the same experiment (x=−2ΔΔCt, with ΔΔCt= [Ct target-CtGapdh]-[CtsiLUC-CtsiGAPDH]). Error-bars represent the standard deviation of the ΔCt value (SDEV = √[SDEVTARGET2 + SDEVGAPDH2]).
Extended Data Figure 4. Tus/Ter-induced TDs in FANCM- or BLM-depleted BRCA1Δ/exon11 6xTer-HR reporter cells.
a, Southern blot analysis of Tus/Ter-induced LTGC and GFP–RFP+ TD products in FANCM or BLM-depleted BRCA1Δ/exon11 6xTer-HR reporter cells (BglII digest, GFP probe). MW: molecular weight lane. TD Breakpoints were identified by PCR product sequencing. b, Repair frequencies in BRCA1fl/exon11 and BRCA1Δ/exon11 6xTer-HR reporter cells transfected with siLUC, siFANCM, siBLM or siFANCM+siBLM in combination. Columns represent mean of duplicate samples, n=7. Error bars: s.e.m. Tus/Ter-induced Total HR, BRCA1fl/exon11 cells, t-test: siFANCM vs. siLUC and siBLM vs. all others p<0.0001; BRCA1Δ/exon11 cells, t-test: siBLM or siFANCM+siBLM vs. siLUC: p<0.005. Tus/Ter-induced STGC, BRCA1fl/exon11 cells, t-test: siFANCM vs. siLUC and siBLM vs. all others p<0.0010; BRCA1Δ/exon11 cells, t-test: siFANCM+siBLM vs. siLUC: p=0.01. Tus/Ter-induced LTGC, BRCA1fl/exon11 cells, t-test: siFANCM or siBLM vs. siLUC: p<0.0001; siFANCM+siBLM vs. all others p<0.005; BRCA1Δ/exon11 cells, t-test: siFANCM or siBLM vs. siLUC: p<0.01; siFANCM+siBLM vs. all others p<0.03. Tus/Ter-induced Ratio LTGC:Total HR, BRCA1fl/exon11 cells, t-test: all siFANCM samples vs. those with no siFANCM: p<0.001; BRCA1Δ/exon11 cells, t-test: all samples vs. siLUC: p<0.002; siFANCM vs. siFANCM+siBLM: p=0.0420; siBLM vs. siFANCM+siBLM: p=0.0294. Tus/Ter-induced TD, BRCA1Δ/exon11 cells, t-test: siFANCM or siBLM vs. siLUC: p<0.002; siFANCM vs. siBLM: NS; siFANCM+siBLM vs. all others: p<0.0001. I-SceI-induced Total HR, BRCA1fl/exon11 cells, t-test: siFANCM vs. siBLM: p=0.0265. I-SceI-induced STGC, BRCA1fl/exon11 cells, t-test: siFANCM vs. siLUC or siBLM: p<0.05; siBLM vs. siFANCM+siBLM: p=0.0445. I-SceI-induced LTGC: NS. I-SceI-induced Ratio LTGC:Total HR, BRCA1fl/exon11 cells, t-test: all samples vs. siLUC: p<0.03; siFANCM vs. siFANCM+siBLM: p=0.0305; BRCA1Δ/exon11 cells, t-test: all samples vs. siLUC: p<0.05; siFANCM vs. siBLM: p=0.0245. I-SceI-induced TD, BRCA1Δ/exon11 cells, t-test: all samples vs. siLUC: p<0.02. For gel source data, see Supplementary Fig. 1.
Extended Data Figure 5. BRCA2 is not a major suppressor of Tus/Ter-induced TDs.
a, GFP–RFP+ products in BRCA1fl/exon11 6xTer-HR cells transfected with siFANCM or siBLM alone or together with siBRCA1, siBARD1, siBRCA2 or siRAD51. Columns represent mean of triplicate samples, n=5. Error bars: s.e.m. Tus-induced TDs, t-test: siFANCM+siBRCA1 or siBARD1 vs. all other samples: p<0.01. siBLM+siBRCA1 or siBARD1 vs. all other samples: p<0.03. I-SceI-induced TDs, t-test: all comparisons not significant (NS). b, GFP–RFP+ products in BRCA1fl/exon11 6xTer-HR cells following depletion of CtIP. Columns represent mean of duplicate samples, n=11. Error bars: s.e.m. Tus-induced TD t-test: all samples vs. siLUC: p<0.01; siFANCM+siCtIP vs. siCtIP or siFANCM: p<0.001; siFANCM+siBRCA1 vs. all other siFANCM samples: p<0.0001; siBLM+siCtIP vs. siBLM: p<0.0001; siBLM+siBRCA1 vs. all other siBLM samples: p<0.0001. I-SceI-induced TD t-test: all samples vs. siLUC: p<0.05; siFANCM+siCtIP vs. siCtIP p=0.0311; siFANCM+siBRCA1 vs. all other siFANCM samples: p<0.01; siFANCM+siCtIP vs. siFANCM NS; siBLM+siBRCA1 vs. all other siBLM samples: p<0.01; siBLM+siCtIP vs. siBLM p=NS. c, GFP–RFP+ products in two independently derived BRCA2lex1/lex2 single-copy 6xTer-HR reporter clones transfected with siRNAs as shown. Columns represent mean of duplicate samples, n=8. Error bars: s.e.m. Clone #3 Tus-induced TD t-test: siFANCM+siBRCA1 vs. all other samples: p<0.01; siLUC vs. siFANCM+siBRCA2: p=0.0131; siFANCM vs. siFANCM+siBRCA2: NS. Clone #56 Tus-induced TD t-test: siFANCM+siBRCA1 vs. all other samples: p<0.003; siFANCM vs. siFANCM+siBRCA2: NS. Clone #3 and clone #56 I-SceI-induced TD: NS. d, RT qPCR analysis of siRNA-treated BRCA2lex1/lex2 6xTer-HR cells. Data normalized to GAPDH and expressed as a fold difference from siLUC sample (x=−2ΔΔCt, with ΔΔCt= [Ct target-CtGapdh]-[CtsiLUC-CtsiGAPDH]). Error-bars: standard deviation of the ΔCt value (SDEV = √[SDEVTARGET2 + SDEVGAPDH2]). e, BRCA2 gene structure in BRCA2lex1/lex2 reporter cells. Grey boxes: BRCA2 exons. PCR primers a, b, and c indicated by arrows. neo: neomycin resistance gene. HPRT: hypoxanthine-guanine phosphoribosyl-transferase gene. *Partial Exon26 deletion. For gel source data, see Supplementary Fig. 1.
Extended Data Figure 6. Tus/Ter-induced TDs arise by a replicative mechanism involving canonical end-joining.
a, Southern blot analysis of aneuploid TD clones (AseI digest of gDNA, full length GFP probe). Same data as Fig. 4b. Parental Ter-HR reporter (“P”) marks size of unaltered reporter. b, Southern blot analysis of 19 reclones of aneuploid TD clones (AseI digest of gDNA, full length GFP probe) that contained a second reporter copy. M: molecular weight; R: original aneuploid clone; lanes 3–20, nineteen independent re-clones. For parental and TD structure, see Fig. 4b. c, Tus/Ter-induced TDs in FANCM-depleted XRCC4fl/fl (#8) and XRCC4Δ/Δ (#11) cells co-transfected with siRNAs shown. Mean of duplicates, n=5. Error bars: s.e.m. t-test P values apply to #8 and #11 data unless otherwise stated. siFANCM+siBRCA1 or siFANCM+siBARD1 vs. all other samples: <0.02, except for clone #11 siFANCM+siBRCA1 vs. siFANCM+siRAD51: NS; siFANCM+siBRCA1 vs. siFANCM+siBARD1: NS; siFANCM+siBRCA2 or siFANCM+siRAD51 vs: siLUC or siFANCM: NS. d, Tus/Ter-induced TDs in BLM-depleted XRCC4fl/fl (#8) and XRCC4Δ/Δ (#11) cells co-transfected with siRNAs shown. Mean of duplicates, n=5. Error bars: s.e.m. t-test P values apply to both #8 and #11 data unless otherwise stated. siBLM+siBRCA1 or siBLM+siBARD1 vs. all other samples in clone #8: p<0.05. In clone #11 siBLM+siBRCA1 or siBLM+siBARD1 vs. siBLM+siRAD51 or siBLM+siBRCA2: NS; siBLM+siBRCA1 vs. siBLM+siBARD1: NS. siBLM+siBRCA2 or siBLM+siRAD51 vs: siLUC or siBLM: NS. e, Rad51 western blot in siRNA-treated #8 and #11 cells. f, RT qPCR analysis of FANCM, BRCA1, BARD1, BLM, and BRCA2 mRNA in siRNA-treated #8 and #11 cells. Data normalized to GAPDH and expressed as fold difference from siLUC sample (x=−2ΔΔCt, with ΔΔCt= [Ct target-CtGapdh]-[CtsiLUC-CtsiGAPDH]). Error-bars: standard deviation of ΔCt value (SDEV = √[SDEVTARGET2 + SDEVGAPDH2]). g, RT qPCR analysis of BRCA1, FANCM and BLM mRNA in siRNA-treated XRCC4Δ/Δ (#11) cells lentivirally transduced with pHIV-EV or pHIV-mXRCC4 (“X4”). See f for normalization and error-bar detail. For gel source data, see Supplementary Fig. 1.
Extended Data Figure 7. Breakpoint analysis of Tus/Ter-induced TDs.
a, Span of TDs in BRCA1Δ/exon11 6xTer-HR reporter siFANCM (121 independent TDs), siBRCA1 (44 independent TDs), or siBLM (66 independent TDs) treatment groups. b, Microhomology (MH) usage at breakpoint of Tus/Ter-induced TDs for BRCA1Δ/exon11 cells depleted of FANCM, BRCA1 or BLM. Numbers in panel count total number of breakpoints with MH≤5, excluding untemplated insertions. Grey line: expected MH usage by chance alone. c, Strand preference of mismatch correction in 14 homeologous breakpoints (i.e., MH with internal mismatches) of Tus/Ter-induced TDs from BRCA1Δ/exon11 cells transfected with siRNAs shown. “C/T” indicates C-T mismatch. TD site (i.e., Ter-proximal or upstream) that underwent mismatch correction is noted. d, Template switches associated with six TD breakpoints. Cartoon format as in Extended Data Fig. 2a. Light grey arrows identify orientation of TD segments relative to the parental reporter. Grey numbers: position of Ter-proximal site relative to first Ter site encountered by rightward fork. Black numbers: Breakpoint MH use (bp). Template switch insertions as shown. e, Distribution of Ter-proximal sites of TD breakpoints in BRCA1Δ/exon11 cells for each treatment group, relative to first Ter site encountered by rightward fork. 10bp binned data. Grey area/orange triangles: 6xTer-array. Bottom panel, distribution of Ter-proximal TD sites in BRCA1Δ/exon11 6xTer-HR reporter cells transfected with siFANCM, siBRCA1, or siBLM. The source data is identical to that used for histograms in upper panels, but has been re-presented as “survival” curves, scoring the probability that a Ter-proximal TD site will be positioned to the right of the nucleotide in question. Hence, all groups at nucleotide position –800 are at 100% and all reach 0% by position +300. Mantel-Cox log-rank statistical test between all pairs: not significant. f, Distribution of “Upstream” sites of TD breakpoints in BRCA1Δ/exon11 cells for each treatment group, relative to splice acceptor adjacent to RFP exon B. 100bp binned data.
Extended Data Figure 8. Analysis of TD and HTGTS breakpoints.
a, MH usage in HTGTS (+) end breakpoints for Tus/Ter-induced translocations from BRCA1Δ/exon11 cells treated with siLUC (655), siFANCM (612), siBRCA1 (548) or siBLM (633) or BRCA1fl/exon11 cells treated with siLUC control (636) siFANCM (658), siBRCA1 (403) or siBLM (405) or I-SceI-induced HTGTS breakpoints for BRCA1Δ/exon11 cells treated with siFANCM (all: 954; +: 506; –: 403). Breakpoints with insertions or with MH use>6 were not included in this analysis. Note that HTGTS breakpoints at Tus/Ter are MH skewed in comparison to HTGTS breakpoints at I-SceI. b, Comparison of distributions of Ter-proximal TD sites and HTGTS (+) end breakpoint distribution for BRCA1Δ/exon11 6xTer cells treated with siFANCM (679), siBRCA1 (630), or siBLM (724). Mantel-Cox log-rank test for TD vs. HTGTS: siFANCM, p<0.0001; siBRCA1, p<0.0001; siBLM, p<0.0001. Gehan-Breslow-Wilcoxon log-rank statistical test: siFANCM TD vs. HTGTS, p<0.0001; siBRCA1 TD vs. HTGTS, p<0.0001; siBLM TD vs. HTGTS, p<0.0001. Right panel: distribution of Tus-induced HTGTS (+) end breakpoint distributions relative to the Ter array in BRCA1Δ/exon11 6xTer cells transfected with siLUC (786). Mantel-Cox log-rank test for HTGTS: siLUC vs. siFANCM, p=0.0171; siLUC vs. siBRCA1, p=0.0003; siLUC vs. siBLM, p<0.0001; siFANCM vs. siBRCA1, p=0.1528; siFANCM vs. siBLM, p=0.0017; siBLM vs. siBRCA1, p=0.1213. Gehan-Breslow-Wilcoxon log-rank test for HTGTS: siLUC vs. siFANCM, p=0.3108; siLUC vs. siBRCA1, p=0.0009; siLUC vs. siBLM, p<0.0001; siFANCM vs. siBRCA1, p=0.0166; siFANCM vs. siBLM, p<0.0001; siBLM vs. siBRCA1, p=0.0751. 6xTer array: grey-shaded region. Orange triangles: individual Ter sites within the 6xTer array. Nucleotide position 0 represents first nucleotide of first Ter site encountered by the rightward fork. For all BRCA1Δ/exon11 treatment groups and BRCA1fl/exon11 cells depleted of FANCM, each sample group represents pooled data from three independent biological replicates. For all other BRCA1fl/exon11 treatment groups, data shown is from two pooled biological replicates.
Extended Data Figure 9. BRCA1 loss in ovarian and breast carcinomas is associated with wide-spread tandem duplications of ~10 kb (Group 1 TDs).
a, Analysis of 92 human ovarian carcinoma genomes (available through the Australian Ovarian Cancer Study, AOCS – URL: http://www.aocstudy.org) and 560 breast carcinoma (BC) genomes (available through the Wellcome Trust Sanger Institute – URL: http://cancer.sanger.ac.uk/cosmic), including 163 triple negative breast cancer (TNBC) genomes. For each dataset, samples are sorted on the x-axis based on increasing number of somatic TDs. y-axis: log10 of TD span (in kb) within each cancer genome, with median marked with circle. Samples featuring a TDP group 1 profile are indicated in orange. Abrogation of BRCA1 and BRCA2 (by germ line mutation, somatic mutation or promoter methylation), and of CDK12 (by somatic mutation) is noted according to key. b, Upper panel: exact numbers of samples analyzed for each dataset and each genetic/genomic subgroup indicated in boxes, with digits color-coded according to key in a. Orange boxes: Group 1 TDP. White boxes: not Group 1 TDP. The numbers comprise only samples for which the relevant genetic annotation is available. Bar charts show percentages of cancer samples with abrogation of BRCA1 (red) or BRCA2 (blue) among the two cancer subsets with or without a TDP group 1 profile; P values calculated by Fisher’s exact test. c, Percentages of cancer samples with (orange) or without (grey) a TDP group 1 profile among the entire datasets and the subsets of samples showing abrogation of BRCA1 (B1m) or BRCA2 (B2m); P values calculated by probability mass function.
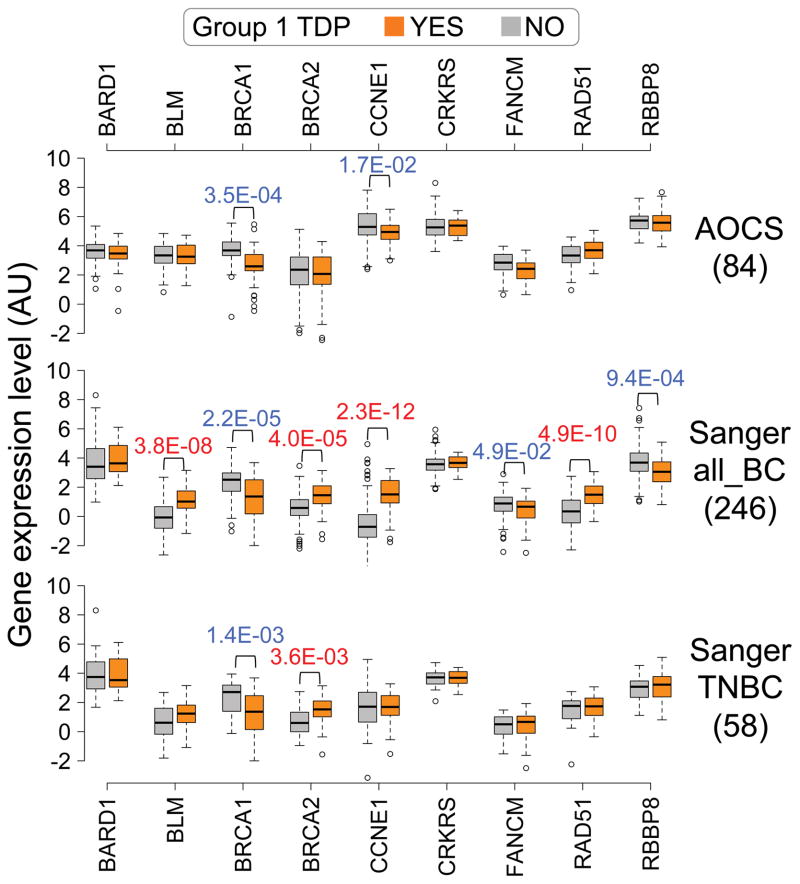
Extended Data Figure 10. Down-regulation of BRCA1 expression is the most prominent and consistent transcriptional feature of ovarian and breast carcinomas associated with TDP group 1 profile.
Box-plots comparing expression levels between cancer samples with (orange) or without (grey) a TDP group 1 profile, relative to nine DNA replication/DNA repair genes whose role as potential contributors to the wide-spread TD formation in cancer has been investigated or suggested. Numbers under each dataset represent number of cancers for which expression data is available. P values calculated by Student’s t-test.
Supplementary Material
Acknowledgments
We thank Drs. Jim Haber, Lorraine Symington and Serena Nik-Zainal for discussions. This work was supported by NCI/DFCI SPORE in Breast Cancer Developmental Research Project Award DF/HCC 5 P50 CA 168504-03 (to NAW), ACS postdoctoral research fellowship PF-12-248-01-DMC (to NAW), R01 ES022054 and R01 CA188032-01 (to EPH), NCI grant P30CA034196 and Andrea Branch and David Elliman Cancer Study Fund (to ETL), grants R01CA095175, R01CA217991, CDMRP OC160440 and HeritX funding (to RS), a BIDMC-JAX pilot grant and CDMRP grant BC160172 (to RS and ETL). FWA is an investigator of the Howard Hughes Medical Institute.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author contributions
NAW and RS developed the overall experimental plan. NAW performed or participated in all experiments with the exception of cancer genome analysis. NAW and RS planned and designed all the experiments, with additional contributions as follows. HTGTS experiments: Plan and design: RLF and FWA; execution: RLF and NAW. Cancer genome analysis: Plan and design: FM and ETL; execution: FM. Analysis of BRCA2 mutant cells: Plan and design: EPH; execution: NAW. Optimization of FACS analysis and FACS sorting protocols: VC; execution: NAW and VC. Construction and characterization of pHIV lentiviral vectors for expression of XRCC4: NAW, EED, AP and RS. NAW and RS wrote the manuscript. Individual figure panels were generated by NAW, RLF, FM and RS.
The authors declare no competing financial interests.
References
- 1.Zeman MK, Cimprich KA. Causes and consequences of replication stress. Nat Cell Biol. 2014;16:2–9. doi: 10.1038/ncb2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Duxin JP, Walter JC. What is the DNA repair defect underlying Fanconi anemia? Current opinion in cell biology. 2015;37:49–60. doi: 10.1016/j.ceb.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.D’Andrea AD. Susceptibility pathways in Fanconi’s anemia and breast cancer. The New England journal of medicine. 2010;362:1909–1919. doi: 10.1056/NEJMra0809889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. S1097-2765(10)00747-1 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garcia-Muse T, Aguilera A. Transcription-replication conflicts: how they occur and how they are resolved. Nat Rev Mol Cell Biol. 2016 doi: 10.1038/nrm.2016.88. [DOI] [PubMed] [Google Scholar]
- 6.Carr AM, Lambert S. Replication Stress-Induced Genome Instability: The Dark Side of Replication Maintenance by Homologous Recombination. J Mol Biol. 2013 doi: 10.1016/j.jmb.2013.04.023. S0022-2836(13)00271–4 [pii] [DOI] [PubMed] [Google Scholar]
- 7.Mayle R, et al. Mus81 and converging forks limit the mutagenicity of replication fork breakage. Science. 2015;349:742–747. doi: 10.1126/science.aaa8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mulcair MD, et al. A molecular mousetrap determines polarity of termination of DNA replication in E. coli. Cell. 2006;125:1309–1319. doi: 10.1016/j.cell.2006.04.040. [DOI] [PubMed] [Google Scholar]
- 9.Larsen NB, Sass E, Suski C, Mankouri HW, Hickson ID. The Escherichia coli Tus-Ter replication fork barrier causes site-specific DNA replication perturbation in yeast. Nat Commun. 2014;5:3574. doi: 10.1038/ncomms4574. [DOI] [PubMed] [Google Scholar]
- 10.Willis NA, et al. BRCA1 controls homologous recombination at Tus/Ter-stalled mammalian replication forks. Nature. 2014;510:556–559. doi: 10.1038/nature13295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Llorente B, Smith CE, Symington LS. Break-induced replication: what is it and what is it for? Cell Cycle. 2008;7:859–864. doi: 10.4161/cc.7.7.5613. [DOI] [PubMed] [Google Scholar]
- 12.Anand RP, Lovett ST, Haber JE. Break-induced DNA replication. Cold Spring Harb Perspect Biol. 2013;5 doi: 10.1101/cshperspect.a010397a010397. [pii] cshperspect.a010397 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saini N, et al. Migrating bubble during break-induced replication drives conservative DNA synthesis. Nature. 2013;502:389–392. doi: 10.1038/nature12584. nature12584 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schlacher K, Wu H, Jasin M. A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell. 2012;22:106–116. doi: 10.1016/j.ccr.2012.05.015. S1535-6108(12)00214-0 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Long DT, Joukov V, Budzowska M, Walter JC. BRCA1 promotes unloading of the CMG helicase from a stalled DNA replication fork. Mol Cell. 2014;56:174–185. doi: 10.1016/j.molcel.2014.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stark JM, Pierce AJ, Oh J, Pastink A, Jasin M. Genetic steps of mammalian homologous repair with distinct mutagenic consequences. Mol Cell Biol. 2004;24:9305–9316. doi: 10.1128/MCB.24.21.9305-9316.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bunting SF, et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell. 2010;141:243–254. doi: 10.1016/j.cell.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pathania S, et al. BRCA1 is required for postreplication repair after UV-induced DNA damage. Mol Cell. 2011;44:235–251. doi: 10.1016/j.molcel.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sy SM, Huen MS, Chen J. PALB2 is an integral component of the BRCA complex required for homologous recombination repair. Proc Natl Acad Sci U S A. 2009;106:7155–7160. doi: 10.1073/pnas.0811159106. 0811159106 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao W, et al. Promotion of RAD51-mediated homologous DNA 1 pairing by the tumor suppressor complex BRCA1-BARD1. Nature. 2017 doi: 10.1038/nature24060. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Menghi F, et al. The tandem duplicator phenotype as a distinct genomic configuration in cancer. Proc Natl Acad Sci U S A. 2016;113:E2373–2382. doi: 10.1073/pnas.1520010113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nik-Zainal S, et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature. 2016;534:47–54. doi: 10.1038/nature17676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Popova T, et al. Ovarian Cancers Harboring Inactivating Mutations in CDK12 Display a Distinct Genomic Instability Pattern Characterized by Large Tandem Duplications. Cancer research. 2016;76:1882–1891. doi: 10.1158/0008-5472.CAN-15-2128. [DOI] [PubMed] [Google Scholar]
- 24.Watkins J, Tutt A, Grigoriadis A. Tandem duplications contribute to not one but two distinct phenotypes. Proc Natl Acad Sci U S A. 2016;113:E5257–5258. doi: 10.1073/pnas.1610228113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Menghi F, Liu ET. Reply to Watkins et al.: Whole-genome sequencing-based identification of diverse tandem duplicator phenotypes in human cancers. Proc Natl Acad Sci U S A. 2016;113:E5259–5260. doi: 10.1073/pnas.1610624113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chandramouly G, et al. BRCA1 and CtIP suppress long-tract gene conversion between sister chromatids. Nat Commun. 2013;4:2404. doi: 10.1038/ncomms3404. ncomms3404 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Polato F, et al. CtIP-mediated resection is essential for viability and can operate independently of BRCA1. The Journal of experimental medicine. 2014;211:1027–1036. doi: 10.1084/jem.20131939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakanishi K, et al. Homology-directed Fanconi anemia pathway cross-link repair is dependent on DNA replication. Nature structural & molecular biology. 2011;18:500–503. doi: 10.1038/nsmb.2029. nsmb.2029 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xue X, Sung P, Zhao X. Functions and regulation of the multitasking FANCM family of DNA motor proteins. Genes Dev. 2015;29:1777–1788. doi: 10.1101/gad.266593.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Larsen NB, Hickson ID. RecQ Helicases: Conserved Guardians of Genomic Integrity. Advances in experimental medicine and biology. 2013;767:161–184. doi: 10.1007/978-1-4614-5037-5_8. [DOI] [PubMed] [Google Scholar]
- 31.Deans AJ, West SC. DNA interstrand crosslink repair and cancer. Nature reviews. 2011;11:467–480. doi: 10.1038/nrc3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morimatsu M, Donoho G, Hasty P. Cells deleted for Brca2 COOH terminus exhibit hypersensitivity to gamma-radiation and premature senescence. Cancer research. 1998;58:3441–3447. [PubMed] [Google Scholar]
- 33.Hastings PJ, Ira G, Lupski JR. A microhomology-mediated break-induced replication model for the origin of human copy number variation. PLoS genetics. 2009;5:e1000327. doi: 10.1371/journal.pgen.1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Payen C, Koszul R, Dujon B, Fischer G. Segmental duplications arise from Pol32-dependent repair of broken forks through two alternative replication-based mechanisms. PLoS genetics. 2008;4:e1000175. doi: 10.1371/journal.pgen.1000175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bhowmick R, Minocherhomji S, Hickson ID. RAD52 Facilitates Mitotic DNA Synthesis Following Replication Stress. Mol Cell. 2016;64:1117–1126. doi: 10.1016/j.molcel.2016.10.037. [DOI] [PubMed] [Google Scholar]
- 36.Doe CL, Osman F, Dixon J, Whitby MC. DNA repair by a Rad22-Mus81-dependent pathway that is independent of Rhp51. Nucleic Acids Res. 2004;32:5570–5581. doi: 10.1093/nar/gkh853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lambert S, et al. Homologous recombination restarts blocked replication forks at the expense of genome rearrangements by template exchange. Mol Cell. 2010;39:346–359. doi: 10.1016/j.molcel.2010.07.015. [DOI] [PubMed] [Google Scholar]
- 38.Nguyen MO, Jalan M, Morrow CA, Osman F, Whitby MC. Recombination occurs within minutes of replication blockage by RTS1 producing restarted forks that are prone to collapse. eLife. 2015;4:e04539. doi: 10.7554/eLife.04539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Neelsen KJ, Lopes M. Replication fork reversal in eukaryotes: from dead end to dynamic response. Nat Rev Mol Cell Biol. 2015;16:207–220. doi: 10.1038/nrm3935. [DOI] [PubMed] [Google Scholar]
- 40.Slack A, Thornton PC, Magner DB, Rosenberg SM, Hastings PJ. On the mechanism of gene amplification induced under stress in Escherichia coli. PLoS genetics. 2006;2:e48. doi: 10.1371/journal.pgen.0020048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yan CT, et al. XRCC4 suppresses medulloblastomas with recurrent translocations in p53-deficient mice. Proc Natl Acad Sci U S A. 2006;103:7378–7383. doi: 10.1073/pnas.0601938103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee JA, Carvalho CM, Lupski JR. A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell. 2007;131:1235–1247. doi: 10.1016/j.cell.2007.11.037. [DOI] [PubMed] [Google Scholar]
- 43.Smith CE, Llorente B, Symington LS. Template switching during break-induced replication. Nature. 2007;447:102–105. doi: 10.1038/nature05723. [DOI] [PubMed] [Google Scholar]
- 44.Anand RP, et al. Chromosome rearrangements via template switching between diverged repeated sequences. Genes Dev. 2014;28:2394–2406. doi: 10.1101/gad.250258.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sakofsky CJ, et al. Translesion Polymerases Drive Microhomology-Mediated Break-Induced Replication Leading to Complex Chromosomal Rearrangements. Mol Cell. 2015;60:860–872. doi: 10.1016/j.molcel.2015.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Simsek D, Jasin M. Alternative end-joining is suppressed by the canonical NHEJ component Xrcc4-ligase IV during chromosomal translocation formation. Nature structural & molecular biology. 2010;17:410–416. doi: 10.1038/nsmb.1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hartlerode AJ, Willis NA, Rajendran A, Manis JP, Scully R. Complex Breakpoints and Template Switching Associated with Non-canonical Termination of Homologous Recombination in Mammalian Cells. PLoS genetics. 2016;12:e1006410. doi: 10.1371/journal.pgen.1006410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Arlt MF, Ozdemir AC, Birkeland SR, Wilson TE, Glover TW. Hydroxyurea induces de novo copy number variants in human cells. Proc Natl Acad Sci U S A. 2011;108:17360–17365. doi: 10.1073/pnas.1109272108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Frock RL, et al. Genome-wide detection of DNA double-stranded breaks induced by engineered nucleases. Nature biotechnology. 2015;33:179–186. doi: 10.1038/nbt.3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hu J, et al. Detecting DNA double-stranded breaks in mammalian genomes by linear amplification-mediated high-throughput genome-wide translocation sequencing. Nat Protoc. 2016;11:853–871. doi: 10.1038/nprot.2016.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Davies H, et al. HRDetect is a predictor of BRCA1 and BRCA2 deficiency based on mutational signatures. Nat Med. 2017;23:517–525. doi: 10.1038/nm.4292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Welm BE, Dijkgraaf GJ, Bledau AS, Welm AL, Werb Z. Lentiviral transduction of mammary stem cells for analysis of gene function during development and cancer. Cell stem cell. 2008;2:90–102. doi: 10.1016/j.stem.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Puget N, Knowlton M, Scully R. Molecular analysis of sister chromatid recombination in mammalian cells. DNA Repair (Amst) 2005;4:149–161. doi: 10.1016/j.dnarep.2004.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analysed during the current study (e.g., the recombination/repair assays analysed throughout the paper, with quantitation by FACS) are available from the corresponding author on reasonable request. Figure source data is available in Supplementary Figure 1. The HTGTS datasets (10 datasets, corresponding to a total of 25 independent HTGTS experiments) are deposited in the GEO repository (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi), accession number GSE103624.