Abstract
With the eradication of poliovirus, the focus has now shifted to environmental surveillance of poliovirus to determine the circulating polioviruses in an area. L20B and RD cell lines are used for isolation of polioviruses. It is imperative to study the efficacy of these cell line in isolating polioviruses from environmental samples. The present study was carried out to determine the sensitivity and specificity of L20B cell line for isolation of polioviruses from environmental samples. L20B and RD cell lines are used for isolation of polioviruses. Molecular characterization was done by using real time RT-PCR. A total of 432 sewage samples from Delhi and Punjab were processed for the isolation of polioviruses during Jan–Dec 2015. 96.76% of the samples were positive in either of the cell lines. Non-polio enteroviruses were obtained in 50 samples on primary isolation. On RT-PCR, 347 (94.29%) samples yielded polioviruses and the rest (21) non-polio enteroviruses or non-enteroviruses. A total of 703 isolates were obtained. 635 isolates were found polioviruses by PCR (90.33%), 20 isolates were found to be NPEV (2.84%) and 48 (6.83%) were found to be NEV. Out of the 20 NPEV isolates, 14 were from RLR (RD-L20B-RD) route and six isolates were from LR (L20B-RD) route. All 48 NEV isolates were from LR route. Thus L20B cell line is more sensitive as compared to RD cell line for isolation of polioviruses however it is not absolutely specific for polioviruses.
Keywords: Poliovirus, Non-polio enterovirus, Non-enterovirus, RT-PCR
Introduction
Poliovirus is a single-stranded positive-sense RNA virus which belongs to the genus Enterovirus of the family Picornaviridae [1, 2]. Individuals infected with poliovirus excrete viruses in stool for periods exceeding several weeks, whether or not they are symptomatic (http://polioeradication.org/wpcontent/uploads/2016/07/WHO_V-B_03.03_eng.pdf) [3, 4]. Poliomyelitis is a vaccine-preventable disease, and live-attenuated poliovirus vaccine has been used successfully for eliminating the poliovirus from many countries. However, vaccine strains can revert to disease-causing strains and cause poliomyelitis (http://polioeradication.org/wpcontent/uploads/2016/07/WHO_V-B_03.03_eng.pdf). As the route of infection is fecal–oral, therefore, there is a need to assess the prevalence of wild and vaccine-derived poliovirus in the sewage during pulse-immunization against polio. Environmental surveillance has been used successfully in monitoring enteric virus circulation and determining the extent or duration of wild-type and vaccine-derived poliovirus circulation in specific populations [5].
Cell culture is instrumental for diagnosis and surveillance of polioviruses since decades. Isolation of poliovirus has been done using different cell lines. Hep-2 (C) and RD (Rhabdomyosarcoma) cells were used for poliovirus culture from stool specimens of cases of acute flaccid paralysis until 1998 (http://apps.who.int/iris/handle/10665/62186). However, as both the cell lines are susceptible to infection by several enteroviruses including poliovirus, elaborate testing was required for serotyping of isolates [6, 7]. L20B cells are mouse L cells transfected with the gene for the human cell-surface receptor CD 155 which is specific for poliovirus [8]. Expression of the receptor at the cell surface renders L20B cells susceptible to infection with polioviruses. As the cells are of murine origin, very few other human enteric viruses produce cytopathic infection in L20B cells [9]. RD cells are derived from human rhabdomyosarcoma tumor tissue and are susceptible to most human enteroviruses [10, 11]. Environmental surveillance for polioviruses is carried out by subjecting the sewage samples to virus isolation. The surveillance method should be sensitive to detect polioviruses from sewage samples as they may also contain other viruses. Reduced sensitivity of the cell line can result in missing of polioviruses which can be dangerous especially in this period approaching eradication. L20B cell line was introduced for isolation of polioviruses in Global Polio Laboratory Network to increase the detection of polioviruses and reduce the workload due to the isolation of non-polio viruses in RD and Hep-2 cell lines. After the widespread use of L20B cell line throughout the network various studies have been carried out to evaluate the sensitivity and specificity of this cell line in stool samples. The literature reveals that specificity varies in different settings. Also, there are limited numbers of studies in the literature to see the utility of L20B cell lines in sewage samples. There can be toxic elements in the sewage which can affect a particular cell line. The present study was carried out to determine the sensitivity and specificity of L20B cell line for isolation of polioviruses.
Materials and methods
Sewage sample collection
Sewage samples from 7 sites of Delhi (on a weekly basis) and 4 sites of Punjab (on a fortnightly basis) were collected by Grab method as described in WHO guidelines of Environmental surveillance 2015 (http://polioeradication.org/wpcontent/uploads/2016/07/WHO_V-B_03.03_eng.pdf). Briefly, the sample was collected by lowering the clean bucket in sewage and was filtered through muslin cloth by placing a funnel in the sterile one litre bottle. The bottle was transferred to the laboratory within 2–4 h under cold chain container maintained at 4 °C. The details of sites and number of samples collected from each site are described in Table 1.
Table 1.
Summary of samples collected from different locations of Delhi and Punjab
| Sr. No. | Name of location | No. of samples collected |
|---|---|---|
| 1. | Red Cross Hospital, Delhi | 8 |
| 2. | Wazirpur J J Colony, Delhi | 52 |
| 3. | Bhalaswa Lake, Delhi | 52 |
| 4. | Swarn Cinema, Delhi | 52 |
| 5. | Batla House, Delhi | 52 |
| 6. | SoniyaVihar, Delhi | 52 |
| 7. | Nangloi, Delhi | 52 |
| 8. | Civil Hospital Rajpura, Patiala, Punjab | 35 |
| 9. | Fatehpur Disposal Amritsar, Punjab | 26 |
| 10. | Zirakpur Mohali, Punjab | 25 |
| 11. | Adowal Losara Drain, Maler Kotla, Punjab | 26 |
| Total | 432 | |
Sewage sample processing
Samples were processed by PEG (polyethylene glycol) precipitation method. Briefly, before processing the sample pH of the sample was adjusted to 7.2 by adding 1 N NaOH or 1 N HCl. The sample was then centrifuged at 5000 rpm for 30 min. Supernatant 1 is collected, and sediment containing bottles were rinsed with phosphate buffer saline, and final sediment was collected in a 50 ml conical centrifuge tube. The resultant solution was diluted with five times of its volume of 3% beef extract and half of its volume of chloroform (containing 0.0001% of Dithizone). This was again centrifuged at 5000 rpm for 30 min, and the supernatant was added to supernatant 1. Washing with the addition of beef extract and chloroform was repeated once more, and the supernatant was again added to supernatant 1. To supernatant 1, PEG (MW 8000) and NaCl were added to a final concentration of 80 g/l and 17.5 g/l respectively. The mixture was mixed on a magnetic stirrer for 30 min and allowed to stand at 4 °C for 16 h. Next day, the supernatant was centrifuged at 5000 rpm for 30 min. The supernatant from this was discarded, and the precipitate was washed with PBS. Final sediment was collected in a 50 ml conical centrifuge tube and was re-suspended in five times of its volume of 3% beef extract and half of its volume of chloroform (containing 0.0001% Dithizone). The sample was then centrifuged at 5000 rpm for 30 min. The upper aqueous phase was collected in a 15 ml centrifuge tube and was labeled as supernatant 2. To the sediment was re-suspended in half of its volume of 3% beef extract and centrifuged at 5000 rpm for 30 min. Supernatant from this was added to the supernatant 2 and sediment was discarded. 2% of fetal bovine serum and 1% of antibiotics was added to the final volume of the supernatant 2. Final supernatant 2 is called the sewage extract and was used for virus isolation.
Virus isolation
Virus isolation was performed as per the standard WHO protocol outlined in the flowchart (Fig. 1).
Fig. 1.
Flowchart of virus isolation and identification from sewage samples
Two flasks (25 cm2) of L20B and one flask of RD cell line were used for virus isolation. Adsorption method was used for the inoculation of sewage extract, and 0.4 ml of inoculum is used for each flask. One flask of each cell line was kept as a negative control without adding sewage extract in it. The flasks were incubated at 37 °C. The cell lines were observed under a microscope for development of cytopathic effect for 5 days. If the cytopathic effect was seen in any of cell line, the inoculum was used to cross passage to opposite cell line. If cytopathic effect appeared in L20B cell line at any stage, an inoculum was enriched in RD cell line and was subjected to real-time RT-PCR (version 4.0) for the identification of poliovirus strain. If no cytopathic effect was observed in the first 5 days, re-passage was done in the same cell line and observed for next 5 days. Samples with no CPE were considered as negative. The isolate was considered as NPEV in cases where the RD cell line was positive and L20B negative even after re-passage. Figure 2 shows CPE of poliovirus inoculation observed in RD and L20B cell lines.
Fig. 2.
Cytopathic effect of poliovirus on RD and L20B cell lines
Real-time RT-PCR
Poliovirus identification was done by real-time RT-PCR. The kit used was provided by CDC and identification was done as per the protocol provided in the kit. The primers used were for Pan EV, Pan PV, Duplex WPV1, AFR WPV3 and SOAS WPV3.
Results and discussion
Sample processing and virus isolation
The culture of poliovirus from clinical or environmental samples is the gold-standard method for virological surveillance in the worldwide initiative to eradicate wild poliovirus (http://apps.who.int/iris/bitstream/10665/62186/1/WHO_EPI_CDS_POLIO_90.1.pdf). As per WHO protocol for isolation of polioviruses from environmental samples, two cell lines are used RD and L20B. RD cells are derived from human rhabdomyosarcoma tumor tissue and are susceptible to most enteroviruses excluding the coxsackie B viruses [11]. L20B cell line carries the CD 155 receptor for poliovirus and is susceptible to poliovirus but non-permissive to most other human enteric viruses [9]. Each sample is inoculated into two flasks of L20B and one flask of RD cell line as per the protocol. As all RD positive samples are passaged into L20B cell line which is specific for polioviruses, we are likely to pick only polioviruses. L20B cell line being specific for polioviruses is expected to permit growth of only polioviruses. However, it has been found in recent studies that enteroviruses other than poliovirus and non-enteroviruses also grow in L20B cell line [6, 9, 12, 13]. The current study was carried out to study the specificity of L20B cell line for polioviruses in environmental sewage samples. Studies have been carried out to determine the sensitivity and specificity of cell lines in AFP surveillance; however, there are a limited number of studies on environmental surveillance in this respect. There are many factors like chemical and organic contents of the sewage sample which may affect the cell lines and culture results.
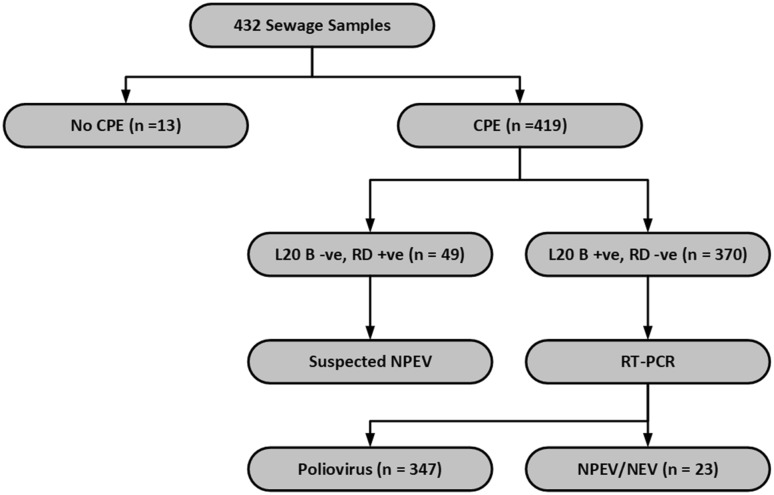
The study was conducted on environmental sewage samples collected from Delhi and Punjab in the year 2015. A total of 432 sewage specimens were processed during this period. Cytopathic effect was seen in either of the cell lines in 419 samples yielding a positivity of 96.99%. Thus 13 samples were negative (3.01%, 13/432). 10 of the 13 negative samples were from Patiala, Punjab (Fig. 3; Table 2).
Fig. 3.
Flowchart of details of results from sewage samples
Table 2.
Summary of virus identification for 432 samples
| Site name | Total samples | Negative | NPEV | Sabin-like (%)a | VDPV | NPEV by PCR | NEV by PCR | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P1 | P2 | P3 | P1 | P2 | P3 | |||||||
| 1. | Red Cross Hospital, Delhi | 8 | 1 | 0 | 4 (12.90) | 14 (45.16) | 13 (41.94) | 0 | 0 | 0 | 0 | 0 |
| 2. | Wazirpur J J Colony, Delhi | 52 | 0 | 24 | 3 (4.23) | 38 (53.52) | 30 (42.25) | 0 | 0 | 0 | 4 | 9 |
| 3. | Bhalaswa Lake, Delhi | 52 | 2 | 31 | 2 (3.64) | 32 (58.18) | 21 (38.18) | 0 | 0 | 0 | 3 | 4 |
| 4. | Swarn Cinema, Delhi | 52 | 0 | 9 | 10 (9.26) | 58(53.70) | 40 (37.04) | 0 | 0 | 0 | 5 | 14 |
| 5. | Batla House, Delhi | 52 | 0 | 6 | 14 (9.53) | 73 (49.66) | 60 (40.81) | 0 | 1 | 0 | 1 | 2 |
| 6. | SoniyaVihar, Delhi | 52 | 0 | 12 | 17 (11.72) | 72 (49.66) | 56 (38.62) | 0 | 0 | 0 | 2 | 10 |
| 7. | Nangloi, Delhi | 52 | 0 | 13 | 6 (4.28) | 72 (51.43) | 62 (44.29) | 0 | 1 | 0 | 2 | 2 |
| 8. | Civil Hospital Rajpura, Patiala, Punjab | 35 | 10 | 13 | 0 (0) | 12 (48) | 13 (52) | 0 | 3 | 0 | 2 | 2 |
| 9. | Fatehpur Disposal Amritsar, Punjab | 26 | 0 | 9 | 4 (8.70) | 26 (56.52) | 16 (34.78) | 0 | 1 | 0 | 1 | 3 |
| 10. | Zirakpur Mohali, Punjab | 25 | 0 | 7 | 6 (10.91) | 23 (41.82) | 26 (47.27) | 0 | 0 | 0 | 0 | 2 |
| 11. | Adowal Losara Drain, Maler Kotla, Punjab | 26 | 0 | 2 | 6 (7.89) | 38 (50) | 32 (42.11) | 0 | 1 | 0 | 0 | 0 |
aThe percentage is the number of type-specific positives with respect to the total number of positive samples
The positivity in sewage samples was expected to be higher as compared to AFP stool surveillance as sewage is a multi-source sample and it contains enteroviruses. In the current study, the positivity was 96.99% which is comparable to other studies in the literature. In a 2 year study on environmental surveillance of sewage specimens in Egypt, Hovi et al. [14] have reported an overall positivity of 97% (284/293) with polioviruses being isolated in 84% of the samples. There are several factors affecting isolation of viruses in sewage sample such as time of collection, the chemical composition of the sewage, environmental temperature, pH, transport to the laboratory, etc. Sewage is a heterogeneous material rich in factors which can be toxic for cells. Toxicity in cell culture can be variable; excessive toxicity causes widespread cell death while limited toxicity may adversely affect virus attachment to cells. Detergents, as demonstrated by Richards [15], may damage viral capsid proteins, without causing damage to specific genome [16].
Cell line sensitivity
Out of the 419 positive samples, 49 samples showed CPE in RD cell line but were negative in L20B cell line both on primary isolation and passage from RD cell line. Thus they were most likely to be non-polio enteroviruses. These were not processed further (Fig. 2; Table 2).
The remaining 370 samples demonstrated CPE in the L20B cell line also resulting in L20B positivity of 85.65% (370/432). Out of these 370 samples, 347 (93.78%) samples yielded polioviruses, and the rest (23) were NPEV/NEV. Thus out of 432, 16.67% samples probably contained NPEV/NEV (49 by primary isolation and 23 by RT-PCR from L20B positive isolates). The current algorithm aims at isolating poliovirus from a sewage sample. As a result, even if a sewage sample contains a mixture of NPEV and poliovirus, only poliovirus would be isolated as L20B cell line is selective for polioviruses. In the current study, on primary isolation 49 samples probably contained NPEV (RD +ve, L20B −ve) and 23 samples which were L20B positive, were NPEV/NEV on RT-PCR. Thus a total of 72 samples of the 432 (16.67%) probably contained NPEV/NEV. In addition to this, there may be samples which had a mixture of poliovirus and NPEV which were reported as poliovirus positive as the algorithm will pick up only PV from the mixture (Table 2).
Out of 370 L20B positive samples obtained after primary isolation, 77 samples were poliovirus positive only in L20B cell line and the rest 293 samples were positive in both cell lines. Thus L20B cell line could detect additional 20.81% (77/370) isolates as compared to RD cell line. Out of 77 samples, 2 samples were vaccine-derived polioviruses (VDPV) as identified by RT-PCR. The positivity of RD cell line for polioviruses amongst L20B positive samples was 79.18% (293/370). L20B cell line could isolate poliovirus on primary isolation in 77 (20.81%) samples which were negative in RD. On the other hand, RD cell line could isolate poliovirus on primary isolation in 11 (2.97%) which were negative in L20B. Thus in our study L20B cell line was more sensitive for detection of polioviruses as compared to RD cell line. Many studies have demonstrated the selective power of L20B cell line for polioviruses. Hovi et al. [11] in their study on sewage specimens have reported L20B cell line to be much more selective for the isolation of poliovirus as compared to RD cell line. As sewage usually contains abundant non-polio enteroviruses, a selective cell line such as L20B is necessary to avoid masking of small amounts of polioviruses in the samples (Table 2).
21 samples were positive by RD route and negative by L20B route on primary isolation. However, when the passage was done from RD to L20B cell line, CPE was seen. On RT-PCR, out of these 21 samples, 11 samples were polioviruses, and the rest were NPEV by PCR.
Out of 293 samples which showed CPE in both cell lines, in 118 samples CPE in L20B cell line appeared late as compared to RD cell line. First preference is given to L20B cell line for RT-PCR as per WHO protocol, but due to late CPE and timeliness, these 118 samples were tested by RT-PCR for both cell lines. Out of these 118 samples, 9 samples (7.63%) were found to be polioviruses in RD cell line, but they were NEV/NPEV in L20B cell line by RT-PCR. Of the remaining 109 samples, 104 samples were positive for polioviruses by both cell lines, 3 samples yielded polioviruses in L20B cell line, but NEV/NPEV in RD cell line and 2 samples were NEV by L20B route and negative by RD route. We observed that in general, the CPE appears early and also progresses fast in RD cell line as compared to L20B cell line. In 118 samples the CPE appeared late (after 5 days) in L20B cell line as compared to RD. Out of these 104 samples yielded polioviruses in both cell lines. Thus the samples giving late positivity in L20B cell lines cannot be ignored.
In the current study a total of 68 L20B positive isolates (9.67%) were either NPEV (20/703, 2.85%) or NEV (48/703, 6.83%) by PCR. Out of the 20 NPEV isolates, 14 were obtained after the passage from RD to L20B cells, indicating that a high multiplicity of infection is required. Only six of the NPEV produced CPE primarily in L20B cells which may be due to a high titer of virus in these samples. Similar findings have also been reported by other studies in literature [10]. On the other hand, all the 48 (6.83%) NEV grew primarily in the L20B cell line. This demonstrates that L20B cell line helps in primary isolation of non-enteroviruses whereas RD cell line does not. This finding has also been supported by another study in which reoviruses were found growing in L20B cell line [17].
RT-PCR sensitivity
A total of 703 isolates were obtained from 370 sewage samples. These were subjected to real-time RT-PCR. 635 isolates were found polioviruses by PCR (90.33%). 20 isolates were found to be NPEV by PCR (2.84%), and 48 (6.83%) were found to be NEV by PCR. Out of the 20 NPEV isolates, 14 were from RLR (RD- L20B- RD) route and six isolates were from LR (L20B-RD) route. All 48 NEV isolates were from LR route. Out of 703 isolates, some of isolates contained mixture of more than one polio type. Therefore a total of 974 individual virus types were obtained from 703 isolates (Table 3).
Table 3.
Summary of virus identification of 974 cell line isolates obtained
| Total number of cell line isolates | 974 |
| P1 Sabin-like | 72 |
| P2 Sabin-like | 458 |
| P3 Sabin-like | 369 |
| P1 VDPV | 0 |
| P2 VDPV | 7 |
| P3 VDPV | 0 |
| NPEV by PCR | 20 |
| NEV by PCR | 48 |
Site-wise distribution of poliovirus isolates
The type wise details of polio isolates obtained from different sewage sites in Delhi and Punjab is depicted in Fig. 4. Type 2 poliovirus was predominant type in Batla House, Soniya Vihar and Nangloi regions of Delhi and Type 3 poliovirus was predominant type in Batla House, Nangloi, and Soniya Vihar regions of Delhi.
Fig. 4.
Graphical representation of virus load in different locations
To conclude the current algorithm for isolation of polioviruses from sewage samples appears to be satisfactory for isolating polioviruses as over 90% of the isolates were polioviruses, with a CPE positivity of 96.99%. The L20B cell line is more sensitive as compared to RD cell line for isolation of polioviruses however it is not specific for polioviruses. L20B cell line appears to facilitate primary isolation of non-enteroviruses in some samples. Further studies on non-enteroviruses growing in L20B cell line need to be carried out.
Acknowledgements
Guidance and support of Polio Laboratory Experts at CDC Atlanta, ERC Global Specialized Laboratory Mumbai, Polio Laboratory Network WHO, HOD Microbiology Division, NCDC and Director, NCDC is gratefully acknowledged.
References
- 1.Buenz EJ, Howe CL. Picornaviruses and cell death. Trends Microbiol. 2006;14(1):28–36. doi: 10.1016/j.tim.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 2.Melnick JL, Dalldorf G, Enders JF, Gelfand HM, Hammon WM, Huebner RJ, et al. A. Classification of human enteroviruses. Virology. 1962;16(4):501–504. doi: 10.1016/0042-6822(62)90233-7. [DOI] [Google Scholar]
- 3.Nakamura T, Hamasaki M, Yoshitomi H, Ishibashi T, Yoshiyama C, Maeda E, et al. Environmental surveillance of poliovirus in sewage water around the introduction period for inactivated polio vaccine in Japan. Appl Environ Microbiol. 2015;81(5):1859–1864. doi: 10.1128/AEM.03575-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.http://www.virology.ws/2004/08/18/poliovirus/. Accessed 11 Aug 2017.
- 5.Deshpande JM, Shetty SJ, Siddiqui ZA. Environmental surveillance system to track wild poliovirus transmission. Appl Environ Microbiol. 2003;69(5):2919–2927. doi: 10.1128/AEM.69.5.2919-2927.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nadkarni SS, Deshpande JM. Recombinant murine L20B cell line supports multiplication of group A coxsackieviruses. J Med Virol. 2003;70(1):81–85. doi: 10.1002/jmv.10360. [DOI] [PubMed] [Google Scholar]
- 7.Schmidt NJ, Ho HH, Lennette EH. Propagation and isolation of group A coxsackieviruses in RD cells. J Clin Microbiol. 1975;2(3):183–185. doi: 10.1128/jcm.2.3.183-185.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mendelsohn CL, Wimmer E, Racaniello VR. Cellular receptor for poliovirus: molecular cloning, nucleotide sequence, and expression of a new member of the immunoglobulin superfamily. Cell. 1989;56(5):855–865. doi: 10.1016/0092-8674(89)90690-9. [DOI] [PubMed] [Google Scholar]
- 9.Pipkin PA, Wood DJ, Racaniello VR, Minor PD. Characterisation of L cells expressing the human poliovirus receptor for the specific detection of polioviruses in vitro. J Virol Methods. 1993;41(3):333–340. doi: 10.1016/0166-0934(93)90022-J. [DOI] [PubMed] [Google Scholar]
- 10.Bell EJ, Cosgrove BP. Routine enterovirus diagnosis in a human rhabdomyosarcoma cell line. Bull World Health Organ. 1980;58(3):423. [PMC free article] [PubMed] [Google Scholar]
- 11.Hovi T, Stenvik M, Partanen H, Kangas A. Poliovirus surveillance by examining sewage specimens. Quantitative recovery of virus after introduction into sewerage at remote upstream location. Epidemiol Infect. 2001;127(01):101–106. doi: 10.1017/S0950268801005787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hovi T, Stenvik M. Selective isolation of poliovirus in recombinant murine cell line expressing the human poliovirus receptor gene. J Clin Microbiol. 1994;32(5):1366–1368. doi: 10.1128/jcm.32.5.1366-1368.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wood DJ, Hull B. L20B cells simplify culture of polioviruses from clinical samples. J Med Virol. 1999;58(2):188–192. doi: 10.1002/(SICI)1096-9071(199906)58:2<188::AID-JMV15>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 14.Hovi T, Blomqvist S, Nasr E, Burns CC, Sarjakoski T, Ahmed N, Savolainen C, Roivainen M, Stenvik M, Laine P, Barakat I. Environmental surveillance of wild poliovirus circulation in Egypt—balancing between detection sensitivity and workload. J Virol Methods. 2005;126(1):127–134. doi: 10.1016/j.jviromet.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 15.Richards GP. Limitations of molecular biological techniques for assessing the virological safety of foods. J Food Prot. 1999;62(6):691–697. doi: 10.4315/0362-028X-62.6.691. [DOI] [PubMed] [Google Scholar]
- 16.Wieczorek M, Ciąćka A, Witek A, Kuryk Ł, Żuk-Wasek A. Environmental surveillance of non-polio enteroviruses in Poland, 2011. Food Environ Virol. 2015;7(3):224–231. doi: 10.1007/s12560-015-9195-3. [DOI] [PubMed] [Google Scholar]
- 17.Grabow WOK, Botma KL, De Villiers JC, Clay CG, Erasmus B. Assessment of cell culture and polymerase chain reaction procedures for the detection of polioviruses in wastewater. Bull World Health Organ. 1999;77(12):973–980. [PMC free article] [PubMed] [Google Scholar]